Abstract
Vascular cambium, a lateral plant meristem is a central producer of woody biomass. Although a few transcription factors (TFs) have been shown to regulate cambial activity1, the phenotypes of the corresponding loss-of-function mutants are relatively modest, highlighting our limited understanding of the underlying transcriptional regulation. Here, we utilize cambium cell-specific transcript profiling followed by a combination of TF network and genetic analyses to identify 62 novel TF genotypes displaying an array of cambial phenotypes. This approach culminated in virtual loss of cambial activity when both WUSCHEL-RELATED HOMEOBOX 4 (WOX4) and KNOTTED-like from Arabidopsis thaliana 1 (KNAT1; also known as BREVIPEDICELLUS (BP) were mutated, thereby unlocking the genetic redundancy in the regulation of cambium development. We also identified TFs with dual functions in cambial cell proliferation and xylem differentiation, including WOX4, SHORT VEGETATIVE PHASE (SVP) and PETAL LOSS (PTL). Using the TF network information, we combined overexpression of the cambial activator WOX4 and removal of the putative inhibitor PTL to engineer Arabidopsis for enhanced radial growth. This line also showed ectopic cambial activity, thus further highlighting the central roles of WOX4 and PTL in cambium development.
Vascular cambium is a secondary meristem which originates from the primary meristems, the shoot apical meristem (SAM) and the root apical meristem (RAM)2. In roots, cambium is derived from xylem associated procambium and xylem pole pericycle (XPP) cells3. Radial growth involves various processes from cambial cell proliferation (i.e. cambial activity) to the maturation of xylem and phloem. The herbaceous plant Arabidopsis thaliana serves as an excellent model to study cambium development because activation and subsequent radial growth can be accurately traced in time3,4 (Supplementary Fig. 1). It is known that TFs play crucial roles in regulating the maintenance of the SAM and RAM; for example, a single mutation in TF encoding genes, such as WUSCHEL(WUS)5 or SHOOT MERISTEMLESS (STM)6 in the SAM or SHORT-ROOT (SHR)7–10 or SCARECROW (SCR)7,11 in the RAM, can result in the termination of apical growth. Several TFs have been found to be required for cambial meristematic activity, including WOX412–14, WOX1415, AINTEGUMENTA (ANT)16, ETHYLENE RESPONSE FACTOR 01817 and ERF10917 as well as a set of AUXIN RESPONSE FACTORS (ARFs)3,18. However, only relatively modest proliferation phenotypes were seen in mutants of these genes, even in mutants like wox4 wox1415, erf018 erf10917 or arf combinations3,18. This indicates a high degree of functional redundancy in the transcriptional regulation of cambial activity.
To identify novel cambium TFs, we performed a genome-wide transcript profiling in Arabidopsis roots using Fluorescence-activated cell sorting (FACS) in combination with microarray technology19 (Fig. 1a), based on procambium and cambium expressing ARABIDOPSIS RESPONSE REGULATOR 15 (ARR15)::GFP (Fig. 1a; Supplementary Fig. 2a). Subsequent meta-analysis including assessment of Differentially Expressed Genes (DEG)19 and gene regulatory Modules20,21 predicted 41 TF-encoding genes that were expressed in the cambium (Supplementary Note; Fig. 1a; Supplementary Fig. 2b; Supplementary Tables 1, 2). To experimentally validate the radial expression patterns of the candidate TFs, we examined their transcriptional GUS fusions in early stage (1-2 week) and transcript localization with RNA in situ hybridisation in late stage (5-week). This resulted in a list of 32 genes with diverse expression patterns in cambium (Supplementary Table 2), which we considered as “Cambium TFs” in subsequent analyses (Supplementary Note; Fig.1b; Supplementary Fig. 3).
Fig. 1. Identification and validation of cambium transcription factors (TFs) during radial growth in Arabidopsis roots.
a, Schematic workflow for the discovery of “cambium TFs”. Cells marked by pARR15::GFP were isolated from roots at three stages of cambium development and used for genome-wide transcript profiling. Candidate cambium TFs were selected based on the “meta-analysis”: an initial 25 TFs were identified based on a combination of two bioinformatic methods, Differentially Expressed Genes (DEG) and Module analysis (shown as Venn diagram), followed by an analysis which added homologues of the initial TFs and cytokinin-induced TFs to obtain a candidate list of 41 TFs (Supplementary Note; Supplementary Table 2). After experimental validation (shown in b), the list of 32 “cambium TFs” were obtained. b, Radial expression patterns of a subset of cambium TFs. Cross sections of the transcriptional GUS reporter line in 1-2 week old roots (left) and RNA in situ hybridisation in 5-week old roots (right) were displayed are shown for each gene. The primary xylem axis is marked in red on the GUS sections. Black arrow-heads indicate the in situ signals. In pLBD4::nYFP-GUS, nYFP-GUS refers to Nuclear Localization Sequence tagged eYFP-GUS. All GUS staining was repeated at least three times with similar results. For in situ hybridisation, at least 10 Col (wild type) plants were examined for each gene with similar patterns. Scale bars, 100µm.
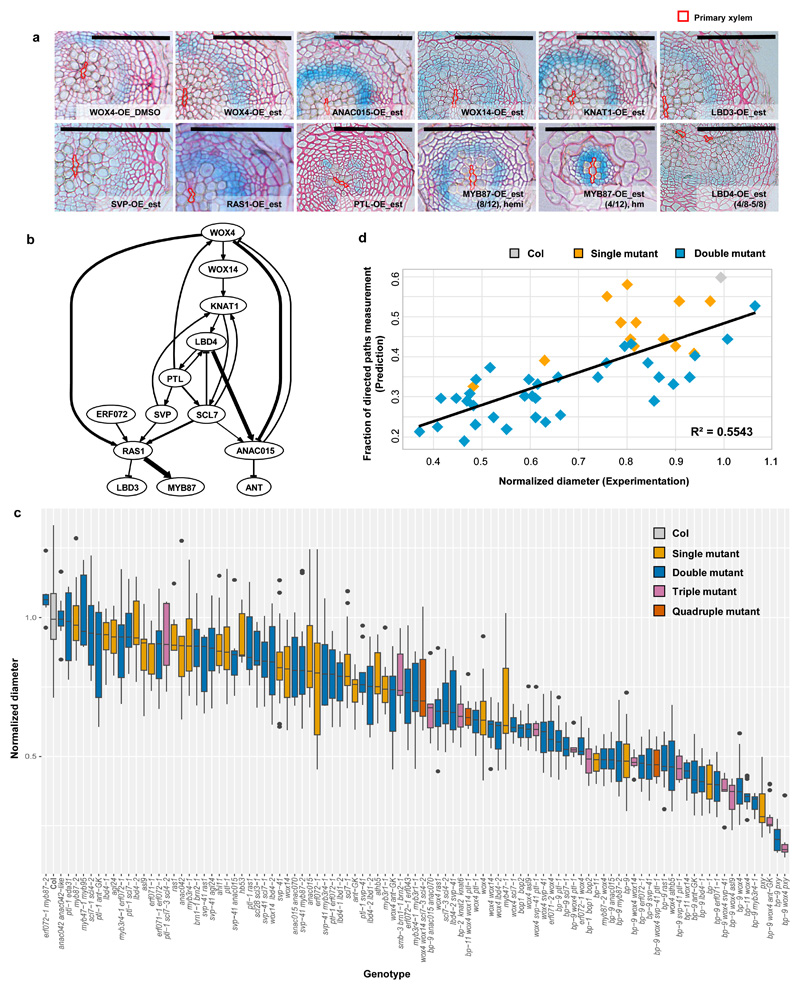
Next, we investigated the effect of individual candidate TFs on cambial activity by generating estradiol inducible overexpression constructs under the 35S promoter22 in the line expressing a cambial cell proliferation marker line pCYCD3;1::GUS (Ler, referred to as CGL hereafter)16. Phenotypic characterization of each overexpression line after a 7-day induction suggested that a few TFs were sufficient to control cambial activity. Based on changes in the pCYCD3;1::GUS patterns or in radial growth, we found that WOX4, WOX14, NAC DOMAIN CONTAINING PROTEIN 15 (ANAC015), KNAT1 and LOB DOMAIN-CONTAINING PROTEIN 3 (LBD3) promote whereas SVP, RESPONSE TO ABA AND SALT 1 (RAS1), PTL and MYB87 inhibit cambial activity. Furthermore, we observed that LBD4 seems to affect vascular patterning (Supplementary Note; Fig. 2a; Supplementary Fig. 4).
Fig. 2. A transcriptional network based on inducible overexpression analysis largely predicts the severity of cambial loss-of-function mutants.
a, Inducible overexpression of certain cambium transcription factors (TFs) affected cambial activity. Cross sections of GUS stained followed by Ruthenium red contrast stained roots of control (DMSO) and 5µM estradiol (est) treated plants from overexpression (OE) lines. Each gene was induced for 7 days in 7-day old seedlings. All lines were generated in a cambial cell proliferation marker line pCYCD3,1::GUS (Ler) background. The GUS signal intensity represents the change in the expression level of CYCD3;1. Control sections from all OE lines showed similar GUS patterns, and only the DMSO section from WOX4-OE is shown. Phenotypes were characterized in T2 or T3 segregating lines, and similar results were found in at least two independent lines (See Methods for details). Segregation of the phenotype was observed in MYB87-OE. “hemi” refers to hemizygous plants; “hm” refers to homozygous plants. The primary xylem axis is marked in red. Scale bars, 100µm. b, The transcriptional regulatory network was constructed based on quantitative Reverse Transcription PCR (qRT-PCR) analysis in inducible overexpression lines of thirteen selected transcription factors (Supplementary Table 3a and b). Positive and negative regulation are shown as arrow-headed line and bar-headed lines, respectively. The width of the line indicates the strength of the regulation. c, An array of vascular phenotype (vascular diameter) was displayed among wild-type (Col) and all mutants characterized at 17-days under long day conditions. Data were collected from a number of independent experiments, and phenotype data for each mutant were normalized to the Col within the same analysis. Box-and-whisker plots are centred at the median, which splits first and third quartiles. Whiskers show range and black dots represent outliers. For each genotype, the median of vascular diameter, the frequency of significance (examined by a two-tailed Student’s t-test in each experiment) observed in multiple experiments and the exact total number of independent roots (N) were listed in Supplementary Table 6a. d, The correlation between the phenotype severities predicted by the network (indicated as “Fraction of directed paths measurement”) (Supplementary Table 3c) and the observed phenotype (Normalized vascular diameter, Supplementary Table 6a) for Col and related single and double mutants. Linear regression model was adopted and R-squared (R2) was used to measure the correlation. The genotypes used in the analysis was listed in Supplementary Dataset 1 and the exact total number of independent roots (N) for each genotype was listed in Supplementary Table 6a.
We next selected a subset of 13 cambium TFs which displayed prominent overexpression phenotypes (Fig. 2a) or otherwise represented one of the TF gene families expressed in the cambium (Supplementary Note; Supplementary Table 2) and used these to construct a cambium transcriptional regulatory network. A 24-hour induction of these 13 genes, and the reciprocal Quantitative Reverse Transcription PCR (qRT-PCR) analysis produced a square matrix of their transcriptional interdependence (Supplementary Table 3a, b), which served as the input for the network construction (Methods). A pared-back version of the network was constructed to highlight connections that were more likely to correspond to direct interactions (Methods; Supplementary Note).We took this to be our final network (Fig. 2b). Multiple levels of hierarchical interactions among target TFs were revealed in the network, and several major nodes were identified, including WOX4, KNAT1, LBD4 and PTL.
We then tested the cambium network experimentally. To this end, we carried out network perturbation analysis (Supplementary Note) to test whether the network could predict the severity of the cambial phenotypes of single and double mutant combinations of the 13 genes (Supplementary Table 3c). Cambial phenotype, that is, vascular “diameter” representing radial growth and “cell count” illustrating cambial cell proliferation (Supplementary Note) correlated well with the prediction (Fig. 2d; Supplementary Fig. 5a), indicating that our network specifically represents cambium regulation. The network analysis guided us to investigate additional combinatorial mutants involving homologous TFs and higher order mutants of the cambium TFs, resulting in roughly 100 genotypes with an array of vascular phenotypes, including the mutants characterized above (Supplementary Note; Fig. 2c; Supplementary Fig. 5). This included 62 novel TF genotypes with cambial phenotypes (Supplementary Note; Supplementary Table 6a).
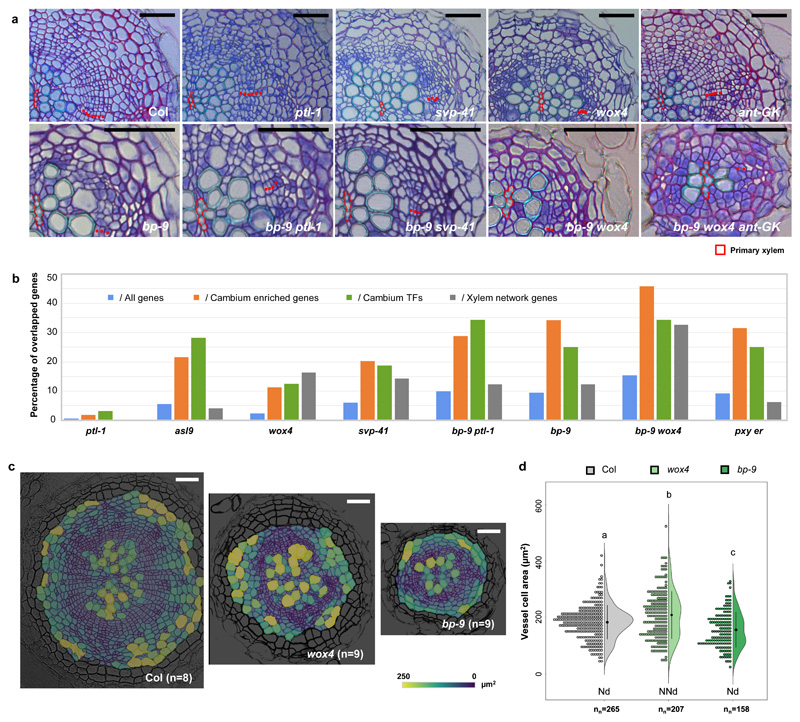
Of the analysed single mutants, the KNAT1 mutant (referred to as bp hereafter) exhibited the greatest reduction in cambial activity (Fig. 3a; Supplementary Table 6a,b). Several other mutants, including wox4, svp and ant, displayed defects in cambial cell proliferation (Fig. 3a; Supplementary Table 6a), while svp also showed increased vessel density (Supplementary Note; Supplementary Fig. 5c; Supplementary Table 6a). Even though radial growth is reduced in these mutants, meristematic activity is maintained, even in bp alleles (Fig. 3a), suggesting functional redundancy within the transcriptional regulation of cambial activity. Next we analysed in detail double or triple mutants of closely homologous genes to examine the redundancy within each TF family. Anatomical characterization revealed functional redundancy within the GRAS, ERF, LBD, Trihelix and MYB families, as certain vascular phenotypes were strengthened by a second mutation (Supplementary Note; Supplementary Fig. 6a,b; Supplementary Table 6a,b); however, antagonistic or epistatic interactions were found in KNOX or MADS family, respectively (Supplementary Fig. 6c; Supplementary Table 6a,b).
Fig. 3. Among the studied TFs, WOX4 and KNAT1 emerged as major regulators of radial growth.
a, Toluidine blue O-stained cross sections of roots of 17-day old wild-type (Col) and various mutants displaying radial growth defects. Red dots indicate cambium cells. The primary xylem axis is marked in red. For each genotype, the exact number of independent experiments and the number of roots in each experiment was listed in Supplementary Table 6a. Scale bars, 50µm. b, The overlapping genes between Differentiated Expressed Genes (DEGs) identified by RNA-Seq profiling and all Arabidopsis genes (TAIR10), Cambium enriched genes (Supplemental Table 1c), Cambium transcription factors (TFs) (Supplementary Table 2) and xylem network genes (Supplemental Table 8b) were obtained by Venn Diagram. The percentage of overlapping genes against all gene lists was calculated and compared in each mutant (Supplemental Table 8a). c, Characterization of radial growth phenotypes in 17-day old Col, wox4 and bp-9 using LithoGraphX. A cell-size heatmap is overlaid on its original greyscale microscopic cross section. The colour scale represents the size of vascular cells. The experiment was repeated twice. n, independent roots. Scale bars, 100µm. d, A half-violin plot showing the cell size distribution of differentiated vessels characterized by LithoGraphX. The black dot with lines represents the mean value with confidence interval. Normality was assessed with the Shapiro-Wilk test. Nd, normally distributed; NNd, non-normally distributed; “nn” indicated the number of all cells analysed. Different letters indicate a significant difference (P<0.05) based on a one-way ANOVA (the exact P value for each comparison can be found in Supplementary Dataset 2a).
Furthermore, we characterized a number of combinatorial mutants involving TFs from different families. Inspection of the vascular phenotypes demonstrated genetic interactions between these TF families (Supplementary Note; Fig. 3a; Supplementary Figs. 7-9; Supplementary Table 6). In particular, wox4, wox14, myb3r4 and erf072 enhanced the cambial activity defect of bp (Fig. 3a; Supplementary Fig. 7b). Unexpectedly, ptl-1 partially suppressed bp phenotypes, indicating that it might function as an inhibitor of cambial activity (Fig. 3a; Supplementary Figs. 7b, 8d). We also found that ptl-1 svp-41 had an outstandingly high total vessel number (Supplementary Fig. 5b) with slightly reduced cambial activity (Supplementary Fig. 5a; Supplementary Table 6a), indicating specific, inhibitory roles for PTL and SVP on vessel production in addition to cambial activity regulation. Altogether, these data indicate that a number of cambium TFs regulate cambium development in a redundant manner.
Among all the double mutants, bp wox4 (bp-9 wox4 and bp-11 wox4) consistently demonstrated the most severe defects in cambial cell proliferation (Figs. 2c, 3a; Supplementary Table 6a). We therefore explored the progression of the bp-9 wox4 phenotype in detail (Supplementary Fig. 8). Cambium initiation seemed normal in bp-9 wox4; however, during the transition stage we observed the differentiation defect (i.e. xylem adjacent to phloem) at a higher frequency (6/10) than in either bp-9 (2/10) or wox4 (1/10). At later stage, vessel differentiation along the xylem axis was delayed in bp-9 wox4, probably due to reduced cambial activity from XPP cells3. Importantly, gaps in the vasculature were discovered in 7-week old bp-9 wox4 plants (Supplementary Fig. 8d), suggesting that cambial activity had been abolished in certain circumferential positions. Therefore, our data support the roles of KNAT1 and WOX4 as two master regulators of cambial activity.
Despite the strong phenotype of bp-9 wox4, the double mutant displayed residual vascular cambial activity, indicating further redundancy among cambium TFs. Analyses of various higher order mutants showed that only ant could slightly enhance the defects in cambial activity in bp-9 wox4 (Figs. 2c, 3a; Supplementary Table 6a), whereas ptl-1 suppressed the defects (Supplementary Fig. 9a). When the mutant of PHLOEM INTERCALATED WITH XYLEM (PXY; also known as TDIF RECEPTOR (TDR)), the receptor-like kinase acting upstream of WOX413,15, was introduced into bp-9 wox4, cambial activity was virtually eliminated (Supplementary Fig. 9b). This extreme phenotype indicates the synergistic effect between the KNAT1-mediated regulatory pathway and the PXY-WOX4 pathway and their fundamental roles on cambial activity. Collectively, our systematic phenotypic characterization of combinatorial mutants demonstrated functional redundancy among various TF families in regulating cambial activity and revealed some of the key players and interactions constituting this redundancy.
To inspect the potential link between the radial growth phenotypes and changes in gene expression, we used RNA-Seq to examine the whole-genome transcriptome in 12-day old wild-type and selected mutants. Since the two receptor kinases PXY and ER are known to be important regulators of radial growth15,23, pxy er15 was included as an additional control of reduced cambial activity (Supplementary Fig. 10). Our analysis showed that the vascular diameter correlated positively with the network prediction and negatively with the number of DEGs (Supplementary Fig. 10c, d). The latter observation suggests that plants with a more severe vascular phenotype had more genes with altered expression. We found that all of the mutants had a much higher proportion of perturbed genes in the cambium enriched genes and our list of 32 cambium TFs than in either the whole transcriptome or in xylem network genes24 (Fig. 3b; Supplementary Table 8), highlighting the link between the TFs identified in this study and cambium development. The fact that the xylem network was largely interfered in wox4 reflects the roles of WOX4 in xylem differentiation. We therefore investigated the xylem phenotypes of wox4 and bp-9, the two single mutants exhibiting the strongest cambial phenotypes (Supplementary Fig. 10), using LithoGraphX25,26, a previously published tool for vascular phenotype quantification (Figs. 3c, d; Supplementary Fig. 11). This analysis showed that the distribution of vessel cell size in wox4 differed from Col and bp-9, and the mean vessel cell size was larger in wox4 than in Col (Fig. 3c, d). Taken together the cell dimension and transcriptomic data, in addition to its well-known role in regulating cambial cell proliferation, WOX4 may also have a role in xylem vessel expansion. Given that the cells defining the stem cell organizer also possess xylem identity3, the dual roles of WOX4 suggest that it may regulate stem cell or its organizer function.
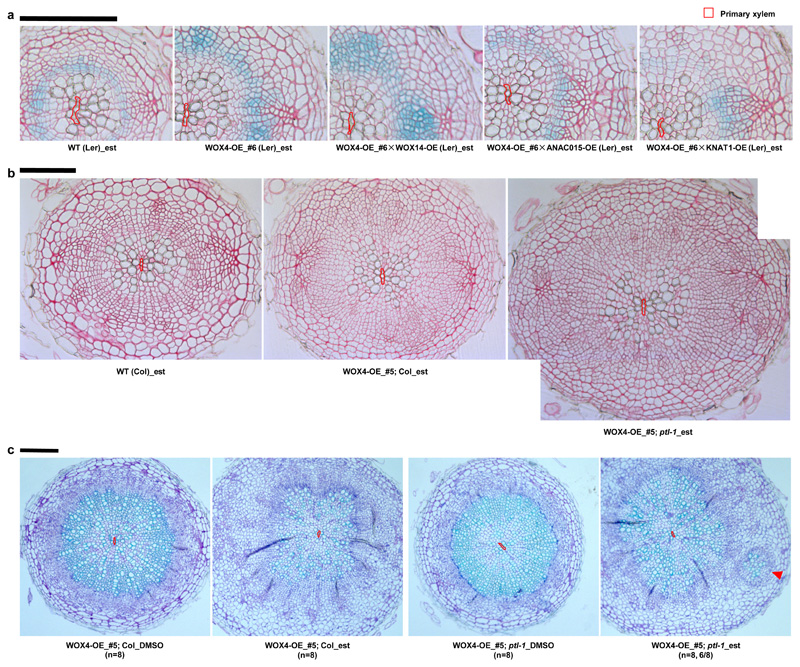
Previous work has shown that it is possible to influence cambial activity by overexpressing TFs27,28. Therefore, we asked whether more prominent radial growth could be achieved by combining multiple positive factors and removing negative factors. First, we made double overexpression (OE) lines by crossing WOX4-OE with other OE lines. The results showed that cambial activity was significantly increased in WOX4-OE (#6) and all double overexpressing lines compared with WT (CGL) (Fig. 4a; Supplementary Fig. 12). Although the total cell number did not increase further in the double-overexpressors, each overexpressor exerted diverse effects combined with WOX4-OE (Fig.4a; Supplementary Fig. 12). In combination with WOX4-OE (#6), WOX14-OE increased both the number and percentage of cambium cells while ANAC015-OE and KNAT1-OE increased the numbers and percentages of differentiated vessels which are the main source of biomass. Intriguingly, in the double-overexpressors including KNAT1-OE and ANAC015-OE, the cambium domain was narrower and there was less ectopic cell proliferation in the phloem and periderm region, resulting in a more organized pattern, as in WT (Fig. 4a; Supplementary Fig. 12). These data suggest that WOX14 acts jointly with WOX4 to promote cambial cell proliferation while ANAC015 and KNAT1 might accelerate xylem differentiation in the WOX4-OE background to maintain the balance between cambial cell proliferation and differentiation.
Fig. 4. Activation of positive transcriptional regulators and removal of negative regulator synergistically stimulate cambial activity.
a, Cross sections of GUS stained followed by Ruthenium red contrast stained roots from wild-type (cambial cell proliferation marker line pCYCD3,1::GUS (Ler), marked as WT (Ler)), WOX4 single overexpression (OE) line (#6) and three double OE lines. All lines were treated with 5µM estradiol (est) to induce the expression of WOX4 and co-expression of WOX4 and WOX14, ANAC015 or KNAT1 respectively for 7 days in 7-day old seedlings. Phenotypes checked in F1 lines from two independent crosses produced similar results. b, Cross sections of Ruthenium red stained roots of wild-type (Col, marked as WT (Col)), WOX4-OE in the Col background (WOX4-OE_#5; Col) and in the ptl-1 background (WOX4-OE_#5; ptl-1). All lines were treated with 5µM est to induce the expression of WOX4 for 7 days in 10-day old seedlings. Phenotypes analysed in F1 lines from two independent crosses produced similar results. The exact number of independent roots (n) for each line in a and b can be found in Supplementary Table 5b. c, Ectopic cambium islands were observed at a high frequency (6/8) in the roots of WOX4-OE_#5; ptl-1 upon long-term induction but not in WOX4-OE_#5; Col. The expression of WOX4 was induced for 4-week upon 5µM est treatment. DMSO was used as control. Sections were taken from the main roots 5mm below hypocotyl-root junction. The experiment was repeated twice. The primary xylem axis is marked in red. Scale bars, 100µm.
We also combined WOX4-OE and ptl-1 to test whether cambial activity could be further enhanced when a putative negative factor was removed. The comparison between WOX4-OE (#5) in Col (WOX4-OE_#5;Col) and in ptl-1 (WOX4-OE_#5;ptl-1) demonstrated that cambial activity was significantly enhanced by ptl mutation (Fig. 4b). The total cell number in WOX4-OE_#5;ptl-1 was almost double than in WT (Col) (Supplementary Fig. 13a). Surprisingly, longer-term (4-week) induction of WOX4-OE;ptl-1 led to the formation of ectopic cambium islands comprising cambium-like cells, vessels and sieve elements in most analysed plants (Fig. 4c; Supplementary Fig. 13e), indicating that in the absence of PTL, enables WOX4 to be sufficient to induce vascular cambium identity. Given that PTL is expressed more broadly towards the phloem domain than WOX4 (Fig. 1b), our data suggest that PTL prevents WOX4 activity in the phloem side.
Our genetic and gene expression data demonstrate that cambium development is orchestrated through a sophisticated transcriptional network involving a large number of TFs. Simultaneous removal of multiple factors resulted in a partial abolishment in cambial activity, indicating that several pathways mediated by these TFs function redundantly during cambium development. Future studies will uncover whether these TFs interact with the regulators defining the stem cell organiser of the vascular cambium3 at the cellular level. By combining overexpressors with loss-of-function mutants, we were able to generate transgenic lines with enhanced cambial activity. Since these lines also slightly inhibited apical growth (Supplementary Figs. 12d and 13d), a future challenge is to optimize the genetic engineering of these regulators (for example by using tissue-specific promoters29–31) to increase biomass, particularly in woody species.
Methods
Fluorescence-activated cell sorting (FACS) and microarray expression data
The genome-wide transcript profiles in Arabidopsis roots have been generated by integration of microarray datasets in the cambium generated in this study (Supplementary Table 1b) and other individual cell types reported previously 19,32–34. To generate the cambium expression data, about 200 roots of 8-day old seedlings were dissected into three segments (about 1cm per segment) and the basal (ARRI, cambium), middle (ARRII, developing cambium) and apical (ARRIII, procambium) regions were separately collected (Supplementary Fig. 2a). All visible lateral roots were removed with razor blade. Protoplasts from these root segments were isolated and sorted through FACS. RNA transcripts collected from GFP expressing protoplasts were profiled on the Affymetrix ATH1 GeneChips. Parallel profiling was done upon treatment of synthetic cytokinin 6-Benzylaminopurine (BAP) for 48 hours, marked as BAP_ARRI, BAP_ARR II and BAP_ARR III (Supplementary Table 1a). The obtained CEL files (GEO accession number GSE125244) were combined with CEL files for other 17 non-overlapping cell types and normalized using mixed model ANOVA35. Genes that are enriched in any of the six ARR15 data compared with non ARR15 data with more than 1.2 fold and False Discovery Rate (FDR) adjusted P value (Padj) less than 0.01 were defined as differentially expressed genes (DEGs). 543 DEGs were identified and marked as cambium enriched genes (Supplementary Table 1c).
Gene regulatory module identification
23 non-overlapping cell type specific profiling data were normalized at the probe level using the GC content based robust multi-array algorithm (GCRMA)36. The log ratios between each cell type and other non-overlapping cell types using the LIMMA test, which is based on linear models and empirical Bayes methods37, were calculated. 12,828 genes, which are expressed in at least one cell type with more than 2-fold difference with FDR less than 0.01 were identified and further processed for module identification. To identify regulatory modules, we applied Genomica (http://genomica.weizmann.ac.il), an evolved version of the GeneXPress38 that clusters the genes and samples simultaneously and build the best regulation programs by learning regression tree using the expression profiles of TFs. This algorithm assumes that TFs regulate gene expression in a combinatorial manner, which likely well reflects TF regulation in higher organisms. 150 modules with predicted gene members (Supplementary Table 1d) were identified, among which 16 showed gene expression patterns highly enriched in the developing cambium and cambium (ARRI/II and/or BAP_ARRI/II). 174 TFs were also identified from the 16 gene modules (Supplementary Table 1e). To find functional enrichment in the identified modules, we extracted significant GO terms in genes belonging to each module using BiNGO Gene Ontology tool, a Cytoscape plugin39 (http://www.psb.ugent.be/cbd/papers/BiNGO/). The significant GO terms related to biological processes were retrieved by applying hypergeometric distribution and Padj less than 0.05 and various GO terms were identified in the cambium enriched gene modules (Supplementary Table 1f)
Plant materials and cloning
All mutants and transcriptional reporter lines analysed in this study were in Arabidopsis Columbia (Col) ecotype. Unless otherwise stated, inducible overexpression lines were transformed in pCYCD3;1::GUS (Landsberg, referred to as CGL hereafter) 16 background. Seeds published previously were listed in Supplementary table 4b. Mutant alleles purchased from seed stock centre (NASC) or requested were genotyped using primers listed in Supplementary Table 4a.
For new transgenic plants, most constructs were generated using Gateway cloning system as described earlier22. A two-step PCR amplification protocol was used for all entry clone construction as previously reported3. The promoter sequence or coding sequence with stop codes of a candidate gene was amplified from genomic DNA or cDNA from Col roots. For procambium and cambium marker that used for the FACS experiment, the promoter sequence of ARR15 was cloned into pDONR-221 vector and then recombined into Gateway destination vector pMDC204 40 (https://www.botinst.uzh.ch/en/research/development/grossnik/vectors/Markd GatewayVectors.html). All gene specific primers and adaptor sequences for cloning were listed in Supplementary Table 4c. For WOX4 overexpression line construction, genomic fragment containing both exons and introns was amplified from Col DNA. For other genes, only coding sequences were amplified from Col cDNA. For most transcriptional GUS reporter cloning, the promoter sequence was cloned into pDONR-221 entry vector and recombined with pBGWFS7-GFP/GUS (basta selection) or pHGWFS7-GFP/GUS (hygromycin selection) in a single site LR-reaction. For all the overexpression lines and some GUS reporter lines (marked with asterisks in Supplementary Table 4c), MultiSite Gateway technology (Invitrogen) was used. Entry clones of 35S::XVE22 or candidate promoter (1st box), gene of interest (GOI) or reporter (2nd box) and terminator (3rd box) were combined with destination vector (pBm43GW or pHm43GW) in a MultiSite LR-reaction. For pLBD4::NYG cloning, a 4-kb promoter fragment was amplified using primers pLBD4_4kb_XhoI_FW and pLBD4_BamHI_REW and the obtained PCR product was cloned into a destination vector harbouring Nuclear Localization Sequence (NLS) tagged EYFP-GUS, pBI-NYG), using the restriction sites. All generated vectors in this study were listed in Supplementary Table 4c.
Knock-out mutants for LBD3/ASL9 were generated using CRISPR-CAS9 system. pJY-35SpCas9-RG1 binary construct was used to target LBD3/ASL9 via agrobacterium-mediated transformation, in which 35S promoter and AtU6 promoter drive Cas9 and sgRNA, respectively. Three 20-mer targeting regions were selected for CRISPR guide sequences, based on the prediction results from CRISPR RGEN Tools (www.rgenome.net): RG1 (5’-AGACAAAAGGGTCACAGACA-3’); RG2 (5’-TCGCCGGAGAAGTTTACAGC-3’); and RG3 (5’-GGCTCTTTGGCCGGAAAATA -3’). Eight hygromycin-resistant transgenic T1 plants harbouring the CRISPR cassette either with sgRNA-RG1, RG2, or RG3 were grown for T2 germline transmission of potentially edited alleles. Sanger sequencing analyses on targeted regions from T2 progenies revealed that targeted RG1 successfully generated homozygous alleles with an insertion of a single nucleotide “A”, thereby leading to a premature stop codons at the 19th codon. This asl9 mutant was used in this study.
Plant growth and sampling
For surface-sterilization of seeds, two protocols were used in this study. Seeds were surface-sterilized first with 70% ethanol, then in 20% chlorine followed by washing in H2O three times before plating, alternatively seeds were rinsed with 100% ethanol and dried on sterilized filter paper before plating. ½ MS medium with 1% sucrose, 0,5g/L MES pH 5.7-5.8 and 1% Difco agar (BD™) was used in all experiments. Seeds were stratified for two days at 4°C under dark conditions. Seedlings were grown vertically in the growth cabinets (Sanyo) at 22°C under long day conditions (16-hour light and 8-hour dark). The age of the plants was counted from the next day after putting them into the growth cabinets for all experiments. Randomization of plates was carried out every 2-3 days.
For GUS reporter line generation, the corresponding construct was transformed into Col plants and at least two lines with relatively strong signals were analysed for each construct, the similar GUS staining patterns were shown in seedlings at 1-2 week (Fig. 1b, Supplementary Fig. 3).
To generate inducible overexpression lines, all 35S::XVE>>GOI (GOI-OE was used as short name) constructs were transformed into a cambium marker line CGL16. T1 plants were screened based on the changed expression of the cambium marker, pCYCD3;1::GUS16, upon 5µM 17-β-estradiol (est, Sigma) induction. Subsequently, 6-12 independent T1 seedlings for each inducible lines were selected for further investigation. To investigate the vascular phenotypes of each inducible cambial TF, at least two independent T2 or T3 lines with Mendelian segregation were carefully analysed and the most representative images were selected for the Fig. 2a and Supplementary Fig. 4a. Homozygous lines obtained from the same progeny present in Fig. 2a were used for Quantitative Reverse Transcription PCR (qRT-PCR) (Supplementary Table 3a, b) except for WOX4-OE, another line (# 6) was used for qRT-PCR. To produce double overexpression lines, homozygous line of WOX4-OE, # 6 was crossed with homozygous lines of ANAC015-OE, WOX14-OE and KNAT1-OE and obtained F1 seeds were used for further examination. To inspect the effect of WOX4 in ptl, WOX4-OE construct was transformed in ptl-1 (Col) allele and the obtained homozygous lines (#5) were crossed back to both ptl-1 (referring as WOX4-OE_#5; ptl-1) and Col (referring as WOX4-OE_#5; Col) and the F1 plants were used for further study.
For T2 or T3 segregating OE lines (CGL background), transformants were selected from antibiotic plates after 3-4 days and then transferred to a new ½ MS plate for recovery until they were 7-day old. Seedlings with similar growth conditions were then transferred to ½ MS plates containing either 5µM est or an equal volume of DMSO and plants were sampled after 7-day induction. For homozygous or F1 lines, after confirmation the antibiotics resistance on a separate plate, seeds were directly plated on ½ MS before transferring to DMSO or est plates. For most overexpression lines, similar phenotypes were observed in all transformants on estradiol plates. For MYB87-OE, clear segregation of phenotypes was observed after estradiol induction (Fig. 2a) and homozygous plants displayed smaller shoots (Supplementary Fig. 4f), which could be confirmed in the T3 homozygous line.
Genotyping for mutants
Genomic DNA was purified from Arabidopsis leaves with either a Whatman FTA classic card method (Sigma-aldrich /Merck) or a precipitation method with Precellys 24 tissue homogenizer (Bertin Technologies) in a buffer containing 3 M Tris-HCl (pH 8.0), 0.5 M EDTA and 3 M NaCl.
For T-DNA insertion lines, genotyping PCR using genomic DNA isolated as described above was carried out to amplify either mutant band or wild-type band using the genotyping primers listed in Supplementary Table 4a. For ptl-1 and svp-41, dCAPs primers were used. For asl9, a PCR product flanking the insertion position was amplified and sequenced to identify the mutant allele. All related primers can be found in Supplementary Table 4a. For mutants containing ant-GK allele, genotyping was done from the segregation population to identify homozygous plants for further phenotype characterization in each experiment.
Histological analysis and GUS staining
Root samples were collected and vacuum-infiltrated using a fix solution (1% glutaraldehyde, 4% formaldehyde, 0.05M sodium phosphate). After keeping in the fix solution for at least overnight and subsequent ethanol dehydration, the samples were infiltrated and then embedded with Leica basic resin using home-made chambers. 5-µm thin cross sections were cut with a Leica microtome at fixed position in the root (5mm below the hypocotyl-root junction) unless otherwise mentioned and stained with Toluidine blue O or Ruthenium red before imaging. GUS staining and subsequent analysis was performed as previously described3 with modifications. The collected root samples were vacuum-infiltration with GUS solution for 30min to 1 hour and then incubated at 37°C until the desired level of GUS staining was observed (2 to 16 hours). Fixed and resin (Leica) embedded samples were cut on the microtome into 7-µm sections.
Phenotypic characterization of mutants
A standard procedure was established for mutant phenotype screening as follows. In each experiment, wild-type (Col) and mutants were plated and germinated on ½ MS plates separately as descried above. 5-day old seedlings with similar growth status were transferred to new ½ MS plates. Five or six seedlings were put onto one plate and sampled when they were 17-day old. Randomization of plates was carried out every 2-3 days. Root apical growth was monitored by marking the root tips every 2-3 days until they reached the bottom of the plate. For all analysed mutants, no defect of root apical growth was observed except for smb-3 brn1-1 brn2-1. Initially, 10 to 18 seedlings were used for each genotype in each experiment. More plants were used for ant-GK combination due to the segregation of the population, for which genotyping PCR was done to identify the homozygous individuals for phenotypic analysis.
Root samples were collected at 17-day, plants with overall growth defects un-related to the genotypes were excluded. Samples were then processed following the histology protocol as described above and obtained cross sections were imaged and analysed. For 6 or 7 week old root phenotype examination, seedling was grown in the pot with soil individually at 23°C under long day conditions in the glass house. Randomization of pots was carried out every 2-3 days.
RNA in situ hybridisation
5-week old Arabidopsis roots growing in soil under long day conditions in the growth room were harvested and immediately fixed in FAA (3.7% formaldehyde, 5% acetic acid, 50% ethanol) on ice. The samples were embedded in wax and cut into 8-μm sections. The sections were then processed by dewaxing, rehydration and dehydration, as described in the online protocol (http://www.its.caltech.edu/~plantlab/protocols/insitu.pdf). To generate the probes for mRNA in situ hybridisation, cDNAs were amplified with gene-specific primers (Supplementary Table 4d) and ligated into the pGEM®-T Easy vector (Promega). The plasmids were verified by sequencing and then used as templates for PCR with the primers T7 and SP6. The PCR products were purified and used for in vitro transcription with the DIG RNA Labeling Kit (Roche). Gene-specific probes were applied onto the sections and hybridisation was performed at 55 °C overnight. After hybridisation, the slices were washed with SSC and incubated with anti-digoxigenin-AP antibody (Roche) for 2 hours at room temperature. Following brief washing with SSC, the signals were detected by overnight colour reaction at 28 °C using NBT/BCIP (Roche). Images were taken with the Zeiss AxioImager M2 microscope fitted with a Zeiss Axiocam MRc colour camera and a PlanApochromat 20x/ 0.8 NA objective. In total, 28 antisense probes were generated in this study and used as cross reference to each other and therefore no sense probe was presented.
qRT-PCR
Inducible overexpression lines for 13 TFs were used for qRT-PCR. T3 homozygous lines were used for most of overexpression lines except that T2 segregating lines were used for ANT-OE and ERF072-OE (Supplementary Table 3a). 8-day old plants were transferred to the plates containing either 5μM est or DMSO control to induce the expression of the target gene for 24 hours. Two third higher part of the primary roots from 20 individual plants were harvested from either est or DMSO plates for RNA purification. For each line, experiment was repeated three times. Total RNA from root samples was isolated with RNeasy Plant Mini Kit (QIAGEN) with an on-column DNase I treatment. cDNA was synthesized using the First Strand cDNA Synthesis Kit (Roche Life Science) according to the manufacturer’s instructions. qRT-PCR experiment was performed in 10μl reaction volume using the LightCycler® 480 SYBR Green I Master Mix (Roche Life Science) in a LightCycler® 480 instrument II (Roche Life Science). PCR program was as follows: 95°C for 5 min, 45 cycles (95°C for 10s, 60°C for 10s, 72°C for 10s), melting curve analysis. Each sample was analysed with three technical replicates. Detected expression levels were normalized using the Comparative CT Method (ΔΔCT method)41 using multiple reference genes (ACTIN 2, PP2AA3, TIP41, and UBQ 10). All statistical analyses (unpaired parametric t-test) were performed using GraphPad PRISM v.7 (GraphPad Software). All primers used for qRT-PCR are listed in Supplementary Table 4e. Relative expression level (fold change) and significance values were calculated based on the three independent analyses and put in in the form of a matrix for the subsequent network analysis (See below).
Transcriptional regulatory network construction
The matrix of fold change obtained from qRT-PCR data (Supplementary Table 3a) together with the matrix of P value (Supplementary Table 3b) collected from 24-hour induction in the overexpression lines formed the basis of the network construction. We denoted the fold change matrix as F and the significance matrix of P values as P, then the directed, signed adjacency matrix A of the network was given by aij = 1 if fij > 1 and pij ≤ 0.05, aij = -1 if fij < 1 and pij ≤ 0.05 and aij = 0 if pij > 0.05. For the pared-back network, we removed all directed connections between two nodes if a directed path of length between those two nodes exists, with the same overall significance. Hence a direct inhibitory link from gene A to gene B would be removed if there was an activatory link from gene A to another gene C, and an inhibitory link from gene C to gene B (or an inhibitory link from A to C, and an activatory one from C to B). Links were only removed if they do not cut off nodes entirely, which can happen if a connection is both part of a direct and an indirect path.
Network perturbation analysis
The network perturbation analysis was undertaken by measuring the fraction “f” of directed network paths that exist in a network, out of the maximum of N (N-1) directed paths that could possibly exist in a directed network with N nodes. The value of “f” was defined as “Fraction of directed paths measurement”. We calculate this value of “f” for the network described above (wild-type) as well as for alternative versions of the same network with single or double knockouts, where we implement a knockout by removing all interactions of the knocked out gene(s). The deviation in the value of “f” between a knockout network and the full network in the wild-type is a measurement of the disruption caused by that knockout in terms of the network topology.
The code for network construction and perturbation analysis can be found on Github (https://github.com/Zhangcambium2019/Zhang2019).
Microscope image processing
To check the expression of pARR15::GFP in Arabidopsis root, the 8-day old seedling were imaged with Leica SP5 confocal microscopy. To examine the radial patter of pARR15::GFP, vibratome sectioning was carried out as previously described3 and 200-µm sections taken from three positions in the root of the 8-day old seedling (Supplementary Fig. 2a) were cleared with ClearSee solution and stained with 1µg/ml Calcofluor white (Sigma) to visualize the cell wall as reported3. Confocal images were acquired with the Leica LAS AF Software.
Images of resin embedded histological cross sections were taken with Leica 2500 or Zeiss Axioimager M2 with 10×, 20× or 40× objectives. The quantification of diameter, area, cell count was done using Fiji (1.0)/ImageJ (1.47v) or LithoGrapX (described later) for specific analysis.
Vascular phenotype quantification with LithoGraphX
LithoGraphX 1.2.2 with the Builder 1.2.2.7 (http://www.lithographx.com/) was employed to analyse the specified experiments following the protocol described earlier25 with modification. Images were pre-processed in Fiji (1.0)/ImageJ (1.47v) to meet the requirement of LithoGraphX. The details of parameters used in the segmentation pipeline can be found on Github (https://github.com/Zhangcambium2019/Zhang2019). The raw segmentation and measurement data were included in Supplementary Dataset 3. All quantification was restricted to the vasculature so that periderm was excluded (Fig. 4c). The number of all vascular cells, number of vessels and the area of each cell (cell size) was measured with the program and a heatmap was generated based on the size of each cell (Fig. 4c). The “total vascular area” was calculated by making the sum of cell area of all vascular cells, therefore intercellular spaces were excluded. The number of independent roots for analysis was denoted as “n” and the number of cells was denoted as “nn”.
Plots and statistical analysis
Statistical analyses were carried out in SPSS Statistics 24 and R 3.5.1 (http://www.r-project.org/). Bar plots and scatter plots were generated with Microsoft Excel and all other plots were generated with necessary packages using RStudio (1.1.423). All the box-and-whisker plot were created using standard settings 'geom_boxplot' using ggplot2 (3.1.0) package. The median splits first and third quartiles; whiskers extend about 1.5 interquartile range beyond the box and the black dots represent outliers. The half-violin plot is made by combining raw data with violin plot splitting each category in two location. The black dot with whisker indicates the mean value. The related codes can be found on Github (https://gist.github.com/jbburant/b3bd4961f3f5b03aeb542ed33a8fe062 and https://gist.github.com/dgrtwo/eb7750e74997891d7c20).
For large scale characterization of mutants, a two-tailed Student’s t-test was carried out using the normalized values to calculate P value. The number of independent roots in each experiment was indicated as “n” (Supplementary Table 6b) while the total number of roots from the pools of all experiments was denoted as “N" (Supplementary Table 6a). The pooled normalized data from all the experiments used to generate the box-and-whisker plot for each genotype. One-way ANOVA analysis was done in SPSS Statistics 24, multiple comparisons were performed using Post-hoc tests Tuckey HSD (for variables with homogenous variance) and Games-Howell (for variables with non-homogenous variance). Homogeneity of variance was defined by Levene's Test. All ANOVA results were included in Supplementary Dataset 2.
RNA-seq profiling and data analysis
For RNA-seq experiment, main root fragments (1 mm to 20 mm below root-hypocotyl junction) of 12-day old plants, including wild-type (Col), ptl-1, asl9, svp-41, wox4, bp-9, bp-9 ptl-1, bp-9 wox4 and pxy er, undergoing radial growth were collected from 30-60 seedlings depending on the genotype. Visible lateral roots were excised with razor blades. The radial growth status was checked and quantified at the middle position (10mm) of the root fragments used for profiling. For each genotype, three independent replicates were collected. Total RNA was isolated with RNeasy Plant Mini Kit (QIAGEN) with an on-column DNase I treatment. RNA concentration was measured with Qubit® 2.0 Fluorometer together with Qubit RNA BR Assay Kit while the RNA purity was assessed with NanoDrop™ 1000 Spectrophotometer (ThermoFisher Scientific). RNA integrity was assess with RNA integrity number equivalent (RINe) using Agilent 2200 TapeStation system following the manufacturer’s instructions. RNA samples giving a RINe>9 were sent for downstream processing. Library preparation with NEB Next® Ultra™ RNA Library Prep Kit and subsequent sequencing with Illumina HiSeq PE150 (pair-end sequencing) platform were performed by Novogene (China, https://en.novogene.com/). Data Quality Control, Data Filtering, Mapping to Reference Genome, Expression Quantification and DEGs identification were carried out using pipelines from Novogene. TopHat2 algorithm was used for mapping sequence to Arabidopsis reference genome (TAIR10). HTSeq software was used to analyse the gene expression levels (read_counts) using the union mode. After normalization, the relative expression (fold change) of one gene was obtained by comparing the average read_counts in each mutant against that in Col with DESeq. Genes with more than 1.5 fold change and Padj less than 0.05 was defined as DEGs (Supplementary Table 7). Venn Diagram was generated using Venny 2.1.0 (http://bioinfogp.cnb.csic.es/tools/venny/index.html). All raw data (FASTQ) can be found in NCBI (BioProject ID: PRJNA523600) and normalized read_counts for all samples can be found in Supplementary Dataset 4.
Supplementary Material
Acknowledgments
We thank O. Smetana for providing the root cross section sketches for making vector images in Fig. 1a; S. Miyashima for providing the pBI-nlsYFP-GUS vector for the cloning of pLBD4::nYFP-GUS; H. Fukuda, J. Murray, D. Smyth, U. Fischer, H. Yu, T. Mizuno, J. Lim, B. Scheres, M. Ito, S. Hepworth, V. Pautot, M. Kater and S. Turner for providing published seeds (Supplementary Table 4a, b); K. Kainulainen and M. Herpola for technical assistance; O. Smetana and L-L. Ye for providing help on finalizing the figures; S. El-Showk for language correction. This work was supported by Finnish Centre of Excellence in Molecular Biology of Primary Producers (Academy of Finland CoE program 2014-2019) decision #271832, the Gatsby Foundation (GAT3395/PR3)), the University of Helsinki (award 799992091) and the European Research Council Advanced Investigator Grant SYMDEV (No. 323052) to Y.H.; Academy of Finland (grants #132376, #266431, #271832), University of Helsinki HiLIFE fellowship to A.P.M.; National Research Foundation of Korea (2018R1A5A1023599 and 2016R1A2B2015258) to J-Y. L.
Footnotes
Code availability
Codes for network construction and LithoGraphX analysis can be found on Github (https://github.com/Zhangcambium2019/Zhang2019). R-codes for Boxplot, Half Violin Plot, Median calculation, Average calculation and P value calculation can also be found on Github.
Data availability
Gene accession numbers are as follows: ARR15, AT1G74890; CYCD3;1, AT4G34160; ANT, AT4G37750; WOX14, AT1G20700; KNAT1 (also known as BP), AT4G08150; PTL, AT5G03680; SVP, AT2G22540; AGL24, AT4G24540; LBD4, AT1G31320; SCL7, AT3G50650; AGL14, AT4G11880; ATHB53, AT5G66700; ATHB5, AT5G65310; WRI3, AT1G16060; AHL11, AT3G61310; MYB47, AT1G18710; MYB95, AT1G74430; MYB87, AT4G37780; STY2, AT4G36260; RAS1, AT1G09950; AtERF019, AT1G22810; AtERF021, AT1G71450; AtERF029, AT4G25490; AtERF032, AT1G63030; AtERF043, AT4G32800; AtERF071, AT2G47520; AtERF072, AT3G16770; WOX4, AT1G46480; WOX13, AT4G35550; STM, AT1G62360; KNAT2, AT1G70510; KNAT6, AT1G23380; BOP1, AT3G57130; BOP2, AT2G41370; EDA31, AT3G10000; LBD3 (also known as ASL9), AT1G16530; LBD1, AT1G07900; SCL4, AT5G66770; SCL3, AT1G50420; SCL28, AT1G63100; ANAC015 (also known as BRN1), AT1G33280; ANAC070 (also known as BRN2), AT4G10350; SMB, AT1G79580; ANAC042 (also known as JUNGBRUNNEN 1), AT2G43000; ANAC042-like, AT3G12910; MYB3R4, AT5G11510; MYB3R1, AT4G32730; PXY (also known as TDR), AT5G61480; ERECTA/ER, AT2G26330. Raw data of Affymetrix ATH1 GeneChip and RNA-seq data can be found in NCBI as described above. Source data related to LithoGraphX analysis (Fig. 3c, d; Supplementary Fig. 11) and ANOVA analysis (Fig. 4d; Supplementary Figs. 8e, 11, 12, 13) can be found in Supplementary Datasets. The data that support the findings of this study are available from the corresponding author upon request.
Author contributions
J.Z., J-Y.L., A.P.M. and Y.H. designed the experiments. J.Z. carried out most of the experiments with the help of other co-authors. K.N. constructed the pARR15::GFP line for FACS. K.N., A.E. and J-Y. L. performed FACS and microarray hybridisation. J-Y. L. and J-G. J. analysed the microarray data from the FACS. J.Z. generated and analysed all transgenic lines with input from A.E. on GUS reporter lines. W.Y. and J.Z. did the RNA in situ analysis. M.K. conducted the qRT-PCR and analysed the data for network construction. S.E.A constructed the network and carried out perturbation analysis. J.Z. and G.E. generated combinatorial mutants. J-Y. Y. and J-H. L. produced the asl9 CRISPR mutant. J.Z., G.E., J.X. and T.D. performed mutant genotyping. J.Z. characterized the mutant and overexpression line phenotypes with the assistance from J.X., G.E. and J.A-S. For phenotypic examination of mutants and overexpression lines, J.Z., G.E. and J.A-S conducted quantification and statistical analyses. J. A-S ran the LithoGraphX analysis with input from G.E., L.R. and P.B.R. J.Z. performed the RNA-seq experiment and analysed the RNA-seq data with S.E.A. G.E. drew all box plot figures. J.Z., J-Y.L., A.P.M. and Y.H. wrote the manuscript with the help of co-authors. All authors read and commented on the manuscript.
Competing Interests statement
The authors declare no competing interests.
References
- 1.Zhang J, Nieminen K, Serra JAA, Helariutta Y. The formation of wood and its control. Current Opinion in Plant Biology. 2014;17:56–63. doi: 10.1016/j.pbi.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Nieminen K, Blomster T, Helariutta Y, Mahonen AP. Vascular Cambium Development. Arabidopsis Book. 2015;13:e0177. doi: 10.1199/tab.0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smetana O, et al. High levels of auxin signalling define the stem-cell organizer of the vascular cambium. Nature. 2019;565:485–489. doi: 10.1038/s41586-018-0837-0. [DOI] [PubMed] [Google Scholar]
- 4.Zhang J, Elo A, Helariutta Y. Arabidopsis as a model for wood formation. Current Opinion in Biotechnology. 2011;22:293–299. doi: 10.1016/j.copbio.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 5.Laux T, Mayer KFX, Berger J, Jurgens G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development. 1996;122:87–96. doi: 10.1242/dev.122.1.87. [DOI] [PubMed] [Google Scholar]
- 6.Clark SE, Jacobsen SE, Levin JZ, Meyerowitz EM. The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development. 1996;122:1567–1575. doi: 10.1242/dev.122.5.1567. [DOI] [PubMed] [Google Scholar]
- 7.Benfey PN, et al. Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development. 1993;119:57–70. doi: 10.1242/dev.119.Supplement.57. [DOI] [PubMed] [Google Scholar]
- 8.Helariutta Y, et al. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell. 2000;101:555–67. doi: 10.1016/s0092-8674(00)80865-x. [DOI] [PubMed] [Google Scholar]
- 9.Lucas M, et al. SHORT-ROOT Regulates Primary, Lateral, and Adventitious Root Development in Arabidopsis. Plant Physiology. 2011;155:384–398. doi: 10.1104/pp.110.165126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheres B, et al. Mutations Affecting the Radial Organization of the Arabidopsis Root Display Specific Defects Throughout the Embryonic Axis. Development. 1995;121:53–62. [Google Scholar]
- 11.DiLaurenzio L, et al. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell. 1996;86:423–433. doi: 10.1016/s0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
- 12.Ji JB, et al. WOX4 Promotes Procambial Development. Plant Physiology. 2010;152:1346–1356. doi: 10.1104/pp.109.149641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirakawa Y, Kondo Y, Fukuda H. TDIF Peptide Signaling Regulates Vascular Stem Cell Proliferation via the WOX4 Homeobox Gene in Arabidopsis. Plant Cell. 2010;22:2618–2629. doi: 10.1105/tpc.110.076083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suer S, Agusti J, Sanchez P, Schwarz M, Greb T. WOX4 Imparts Auxin Responsiveness to Cambium Cells in Arabidopsis. Plant Cell. 2011;23:3247–3259. doi: 10.1105/tpc.111.087874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Etchells JP, Provost CM, Mishra L, Turner SR. WOX4 and WOX14 act downstream of the PXY receptor kinase to regulate plant vascular proliferation independently of any role in vascular organisation. Development. 2013;140:2224–2234. doi: 10.1242/dev.091314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Randall RS, et al. AINTEGUMENTA and the D-type cyclin CYCD3;1 regulate root secondary growth and respond to cytokinins. Biology Open. 2015;4:1229–1236. doi: 10.1242/bio.013128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Etchells JP, Provost CM, Turner SR. Plant Vascular Cell Division Is Maintained by an Interaction between PXY and Ethylene Signalling. Plos Genetics. 2012;8 doi: 10.1371/journal.pgen.1002997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brackmann K, et al. Spatial specificity of auxin responses coordinates wood formation. Nature Communications. 2018;9 doi: 10.1038/s41467-018-03256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Birnbaum K, et al. Cell type-specific expression profiting in plants via cell sorting of protoplasts from fluorescent reporter lines. Nature Methods. 2005;2:615–619. doi: 10.1038/nmeth0805-615. [DOI] [PubMed] [Google Scholar]
- 20.Wagner GP, Pavlicev M, Cheverud JM. The road to modularity. Nat Rev Genet. 2007;8:921–31. doi: 10.1038/nrg2267. [DOI] [PubMed] [Google Scholar]
- 21.Wang XW, Dalkic E, Wu M, Chan C. Gene module level analysis: identification to networks and dynamics. Current Opinion in Biotechnology. 2008;19:482–491. doi: 10.1016/j.copbio.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siligato R, et al. MultiSite Gateway-Compatible Cell Type-Specific Gene-Inducible System for Plants. Plant Physiology. 2016;170:627–641. doi: 10.1104/pp.15.01246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uchida N, Tasaka M. Regulation of plant vascular stem cells by endodermis-derived EPFL-family peptide hormones and phloem-expressed ERECTA-family receptor kinases. Journal of Experimental Botany. 2013;64:5335–5343. doi: 10.1093/jxb/ert196. [DOI] [PubMed] [Google Scholar]
- 24.Taylor-Teeples M, et al. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature. 2015;517:571–U307. doi: 10.1038/nature14099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wunderling A, Ben Targem M, de Reuille PB, Ragni L. Novel tools for quantifying secondary growth. Journal of Experimental Botany. 2017;68:89–95. doi: 10.1093/jxb/erw450. [DOI] [PubMed] [Google Scholar]
- 26.de Reuille PB, R L. Vascular Morphodynamics During Secondary Growth. In: de Lucas M, E J, editors. Xylem Methods in Molecular Biology. Humana Press; New York, NY: 2017. pp. 103–125. [DOI] [PubMed] [Google Scholar]
- 27.Guo Y, Qin GJ, Gu HY, Qu LJ. Dof5.6/HCA2, a Dof Transcription Factor Gene, Regulates Interfascicular Cambium Formation and Vascular Tissue Development in Arabidopsis. Plant Cell. 2009;21:3518–3534. doi: 10.1105/tpc.108.064139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yordanov YS, Regan S, Busov V. Members of the LATERAL ORGAN BOUNDARIES DOMAIN Transcription Factor Family Are Involved in the Regulation of Secondary Growth in Populus. Plant Cell. 2010;22:3662–3677. doi: 10.1105/tpc.110.078634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Etchells JP, Mishra LS, Kumar M, Campbell L, Turner SR. Wood Formation in Trees Is Increased by Manipulating PXY-Regulated Cell Division. Current Biology. 2015;25:1050–1055. doi: 10.1016/j.cub.2015.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J, Serra JAA, Helariutta Y. Wood development: Growth through knowledge. Nature Plants. 2015;1 15060. [Google Scholar]
- 31.Immanen J, et al. Cytokinin and Auxin Display Distinct but Interconnected Distribution and Signaling Profiles to Stimulate Cambial Activity. Current Biology. 2016;26:1990–1997. doi: 10.1016/j.cub.2016.05.053. [DOI] [PubMed] [Google Scholar]
- 32.Brady SM, et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 33.Nawy T, et al. Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell. 2005;17:1908–1925. doi: 10.1105/tpc.105.031724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee JY, et al. Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6055–6060. doi: 10.1073/pnas.0510607103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levesque MP, et al. Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. Plos Biology. 2006;4:1284–1284. doi: 10.1371/journal.pbio.0040143. vol 4, pg 739, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu ZJ, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A model-based background adjustment for oligonucleotide expression arrays. Journal of the American Statistical Association. 2004;99:909–917. [Google Scholar]
- 37.Smyth GK. Bioinformatics and computational biology solutions using R and Bioconductor. Springer; 2005. Limma: linear models for microarray data; pp. 397–420. [Google Scholar]
- 38.Segal E, et al. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nature Genetics. 2003;34:166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 39.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in Biological Networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 40.Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiology. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.