Abstract
Langerhans cell histiocytosis (LCH) is a rare neoplasm predominantly affecting children. It occupies a hybrid position between cancers and inflammatory diseases, which makes it an attractive model for studying cancer development. To explore the molecular mechanisms underlying the pathophysiology of LCH and its characteristic clinical heterogeneity, we investigated the transcriptomic and epigenomic diversity in primary LCH lesions. Using single-cell RNA sequencing, we identified multiple recurrent types of LCH cells within these biopsies, including putative LCH progenitor cells and several subsets of differentiated LCH cells. We confirmed the presence of proliferative LCH cells in all analyzed biopsies using immunohistochemistry, and we defined an epigenomic and gene regulatory basis of the different LCH cell subsets by chromatin accessibility profiling. In summary, our single-cell analysis of LCH uncovered an unexpected degree of cellular, transcriptomic, and epigenomic heterogeneity among LCH cells, indicative of complex developmental hierarchies in LCH lesions.
Keywords: Langerhans cell histiocytosis, single-cell sequencing, tumour heterogeneity, epigenome mapping
Introduction
Langerhans cell histiocytosis (LCH) is a rare hematopoietic neoplasm driven by gain-of-function mutations in the mitogen-activated protein kinase (MAPK) pathway1,2. The name of the disease originates from the characteristic expression of CD207, also known as langerin, which is conventionally associated with Langerhans cells – a type of resident epidermal immune cell3. However, the relationship between LCH cells and Langerhans cells remains controversial, and LCH has also been linked to dendritic cells and other myeloid cell types4–6. LCH is phenotypically characterized by an accumulation of CD1A and CD207 double-positive cells in various tissues (Fig.1a). These lesions can develop in almost any organ, but are most common in skin and bone1.
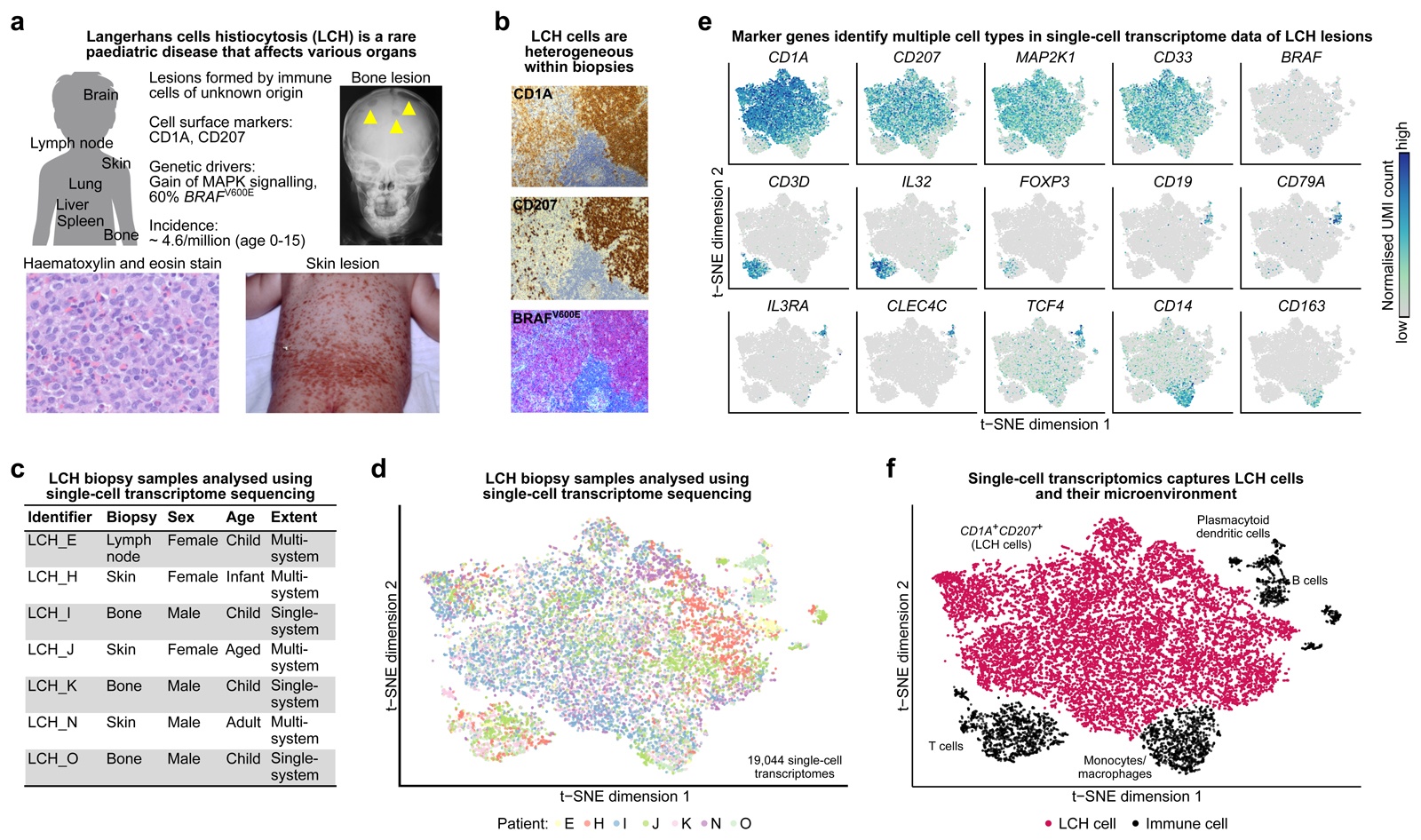
Figure 1. Single-cell transcriptome analysis captures cellular and molecular diversity of LCH lesions.
A) Clinical presentation and incidence2 of Langerhans cell histiocytosis (LCH). Shown are an x-ray image of LCH bone lesions in the skull (top right), a photography of extensive LCH skin lesions (bottom right), and a hematoxylin and eosin staining of an LCH biopsy (bottom left).
B) Cellular heterogeneity in an LCH biopsy (lymph node), as revealed by immunostaining for CD1A, CD207, and an antibody that specifically detects mutated BRAFV600E protein.
C) Overview the of patient biopsy samples analyzed by single-cell RNA-sequencing in this study.
D) Low-dimensional projection (t-SNE plot) of the combined single-cell RNA-seq dataset across all analyzed biopsies, comprising a total of 19,044 single-cell transcriptome profiles.
E) Molecular heterogeneity in LCH illustrated by low-dimensional projection (as in panel D) of all single-cell transcriptome profiles overlaid with the expression levels of selected marker genes (dark blue color indicates high expression levels).
F) Cellular heterogeneity in LCH illustrated by low-dimensional projection (as in panel D), annotated with cell types inferred from marker gene expression.
LCH severity is clinically assessed by the extent of the disease (involvement of one or more organs, i.e. single-system vs. multisystem disease) and the involvement of known risk organs (liver, spleen, hematopoietic system). Treatment decisions are currently based solely on clinical presentation, as no reliable molecular markers for risk stratification have been identified yet. LCH is most commonly driven by the BRAFV600E mutation6–8, but the pathophysiology of LCH is different from cancers with oncogenic BRAFV600E mutations, such as melanoma, non-small-cell lung cancer, and colorectal cancer. LCH cells carry few genetic alterations beyond BRAFV600E or another oncogenic driver in the MAPK pathway (MAP2K1, ARAF), and they show no progressive accumulation of recurrent somatic mutations9. Instead, high levels of inflammatory infiltrate suggest that cell-extrinsic, immunological stimuli could be key contributors to the formation of LCH lesions10–12. LCH is lethal in up to 20% of patients with risk-organ-positive, multisystem disease treated according to current standard of care. In contrast, the disease often resolves spontaneously following unknown mechanisms in patients with single lesions1,12.
Results
Langerhans Cell Histiocytosis Lesions Are Composed of Distinct Cell Populations
LCH cells within the same lesion show wide variability in their CD1A and CD207 levels based on IHC (Fig. 1B) and flow cytometry (Supplementary Fig. S1). To characterize the cellular and molecular landscape of LCH lesions, we performed droplet-based single-cell transcriptome sequencing of seven LCH lesions. The biopsies were obtained from patients with multisystem disease (n = 4) and single-system disease (n = 3), and they were collected from different sites: lymph node (n = 1), skin (n = 3), and bone (n = 3), thus representing the wide clinical spectrum of the disease (Fig. 1C; Supplementary Table S1). Because the percentage of LCH cells within lesions can be low1, two samples were enriched for cell populations with specific expression levels of the canonical LCH surface markers CD1A and CD207 (Supplementary Fig. S1). A total of 19,044 single-cell transcriptomes passed quality control and collectively gave rise to a detailed cellular and molecular portrait of LCH lesions, spanning multiple patients and diverse clinical presentations (Fig. 1D, Supplementary Fig. S2A and S2B). In the single-cell transcriptome dataset, we identified several distinct clusters of immune cells, which we labelled based on known cell-type-specific marker genes (Fig. 1E and F; Supplementary Fig. S2C). As expected, we observed T cells (some expressing FOXP3, indicating that these could be regulatory T cells as described before in LCH13), B cells, monocytes/macrophages, and dendritic cells, in addition to LCH cells.
Focusing initially on LCH-specific transcriptional profiles that were shared across cells and across patients, we compared CD1A and CD207 marker-positive LCH cells with four immune-cell populations identified in all biopsies (Supplementary Fig. 2D). The LCH cells showed high expression of multiple genes previously reported as specifically expressed in LCH cells14,15, including the MMP9 gene, several genes relevant for antigen presentation (for instance, CD1E), and genes encoding members of the HLA complex (Fig. 2A,b). We combined the results of the individual comparisons into an LCH gene signature derived from our single-cell transcriptome data, which distinguished LCH cells from immune cells across patients (Fig. 2B; Supplementary Fig. S3A and S3B). We validated this gene signature on published LCH bulk transcriptome datasets8,14,15 and were able to confirm its robustness for a wider range of LCH samples (Supplementary Fig. S3C and S3D) and in comparison to other histiocytoses, namely Erdheim-Chester disease and juvenile xanthogranuloma (Supplementary Fig. S3E).
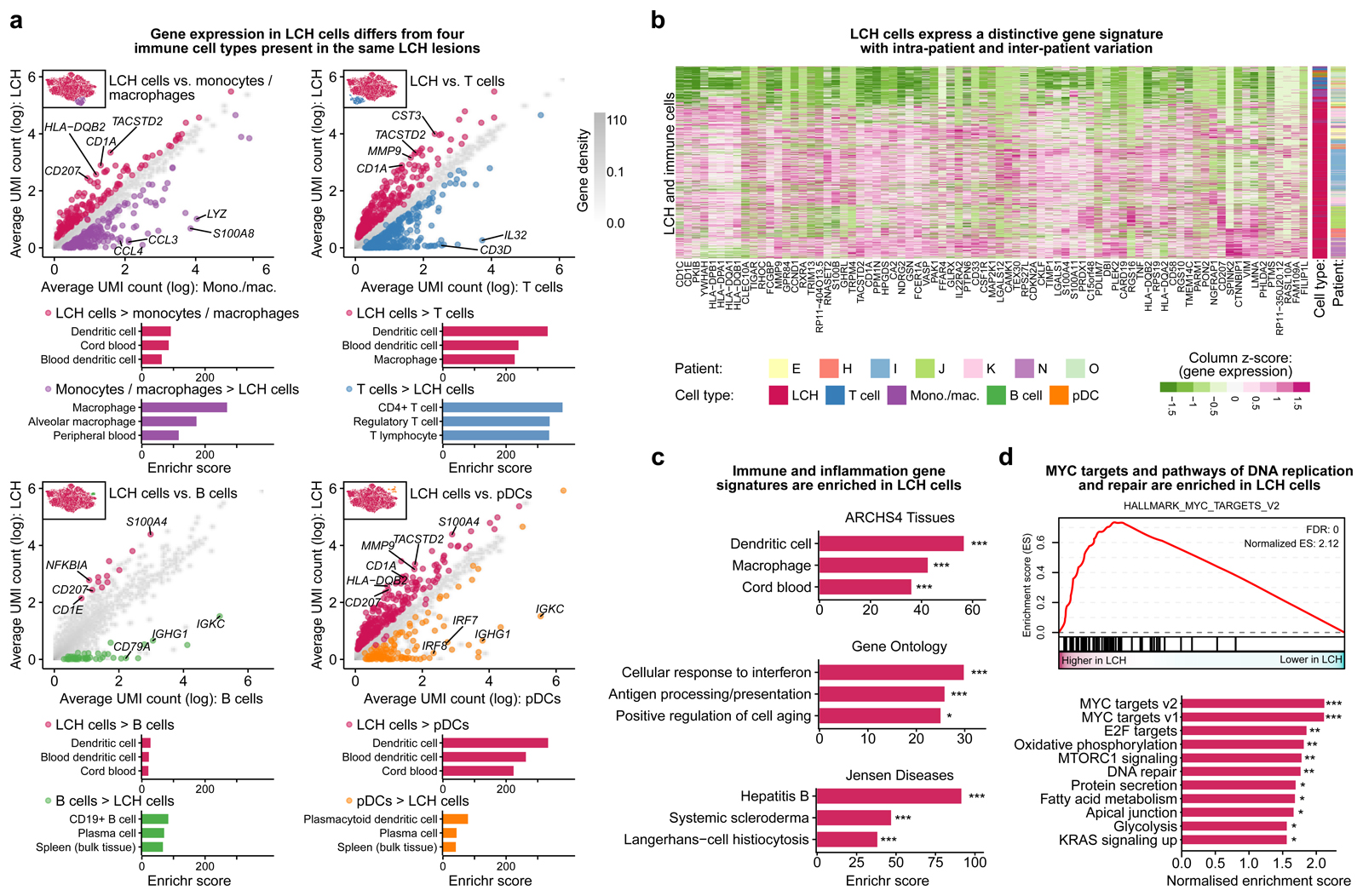
Figure 2. Characteristic gene expression patterns distinguish LCH cells from other immune cells present in LCH lesions.
A) Scatterplots comparing gene expression (mean normalized UMI counts plotted on logarithmic scale) between LCH cells and non-LCH immune cell populations. Differentially expressed genes specific for LCH cells are colored in red, whereas genes specific for the immune cell populations are colored in violet (monocyte/macrophages), blue (T cells), green (B cells), and orange (plasmacytoid dendritic cells). The top three enriched categories from the ARCHS4 tissue database based on Enrichr are shown below.
B) Heatmap showing the expression of the genes in the LCH gene signature (genes overexpressed in LCH cells compared to at least three of the four immune cell populations in panel A) in the LCH and immune cells analyzed (rows). Cell type and patient are indicated by the colored bars on the right.
C) Enrichment analysis with Enrichr for the LCH signature genes using three databases (top to bottom: ARCHS4 Tissues, Gene Ontology, or Jensen Diseases), ordered by the Enrichr combined score58. FDR-adjusted p-value: *, p < 0.1; ***, p < 0.001. Further details are provided in Supplementary Table S3.
D) Gene set enrichment analysis (GSEA) for LCH cell gene expression compared to immune cells using MSigDB hallmark signatures, including a GSEA plot for the most significant gene set (MYC_TARGETS_V2, top) and all significant enrichments visualized in the bar plot (bottom). FDR-adjusted p-value: *, p < 0.1; **, p < 0.05; ***, p < 0.001
Our LCH gene signature was enriched for genes associated with macrophages as well as dendritic cells (Fig. 2C), supporting that LCH cells share relevant properties with both cell types. We also observed an enrichment of genes involved in IFN signaling and antigen presentation, reflecting the inflammatory nature of LCH lesions. Moreover, MYC-associated genes were characteristically enriched in the LCH cells, and the same was true for pathways linked to cell cycle and DNA repair (Fig. 2D). In aggregate, our analysis identified an LCH gene signature that is consistent with the neoplastic (cell cycle, DNA replication, MYC) as well as the inflammatory (immunity, inflammation, macrophages, dendritic cells) phenotype of the disease, it confirmed and extended previous reports describing gene expression profiles of bulk LCH samples, and it established a catalog of potential targets for future research on disease mechanisms and molecularly guided therapies in LCH.
LCH Lesions Harbor a Developmental Hierarchy of LCH Progenitors and Their Progeny
We next exploited the single-cell resolution of our dataset to investigate cellular and molecular heterogeneity among LCH cells. To that end, we clustered the single-cell transcriptomes of all biopsies and identified 14 discernible subsets (Fig. 3A). Each of these subsets was represented in all samples, despite differences in sample characteristics such as disease extent and site of the biopsied lesion (Fig. 3B; Supplementary Fig. S4A). Given that LCH is generally considered a clonal disease16,17, we hypothesized that the observed heterogeneity with its recurrent subsets of LCH cells may arise from developmental processes such as cellular differentiation, de-differentiation, or transdifferentiation. This would be in line with the clinical observation of a dynamic nature of LCH lesions, which often develop and resolve spontaneously1,12.
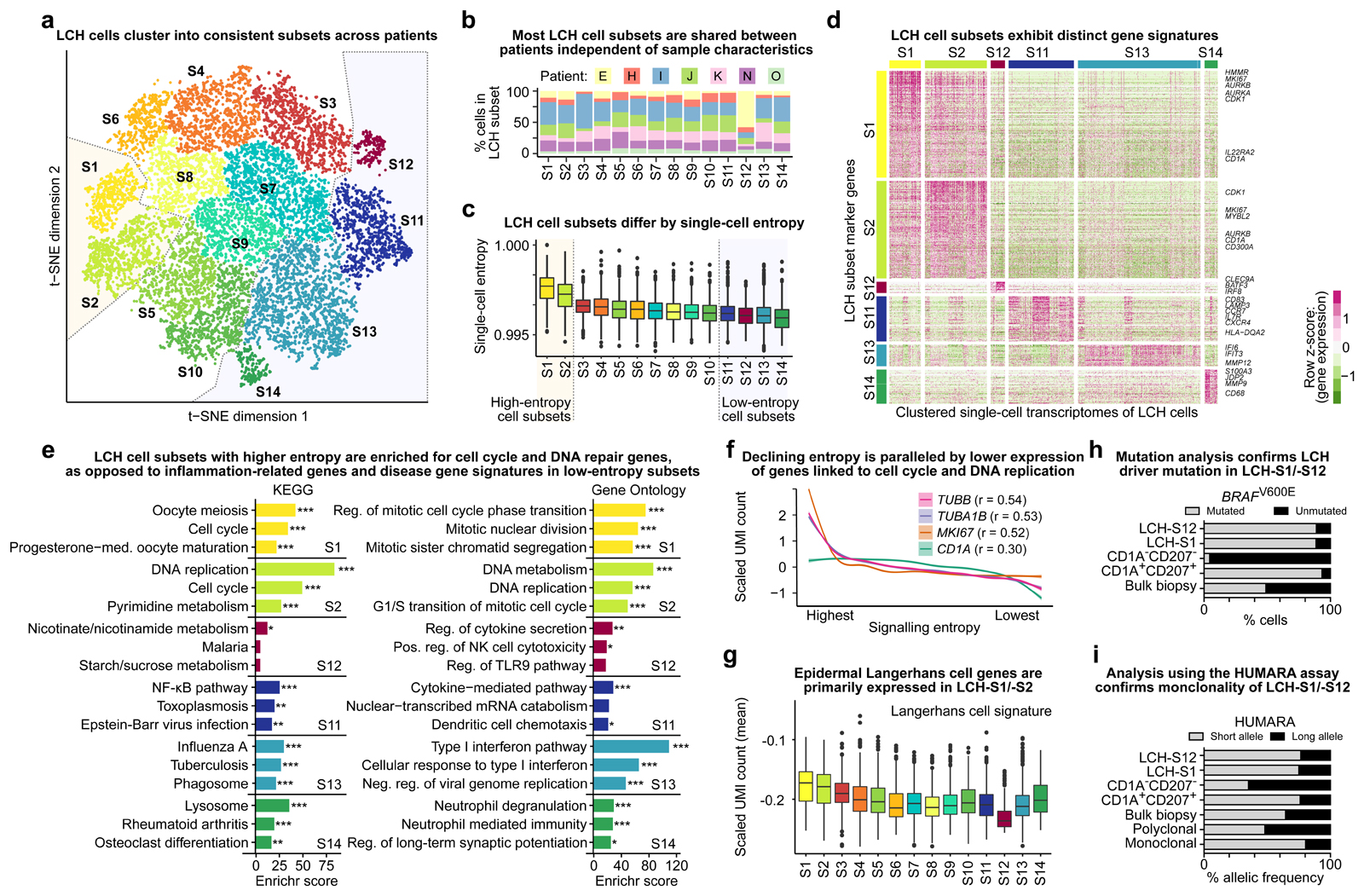
Figure 3. LCH lesions comprise proliferating LCH progenitors and multiple differentiated LCH cell subsets.
A) Low-dimensional projection (t-SNE plot) of LCH cell transcriptomes, excluding non-LCH immune cells. LCH cells were clustered into 14 subsets. Dashed lines highlight the putative progenitor cell subsets (LCH-C1, LCH-C2) and the most differentiated cell subsets (LCH-C11 to LCH-C14) based on the analysis of transcriptome entropy (panel C).
B) Percentage of cells assigned to each of the 14 LCH cell subset (from panel A) for each analyzed patient biopsy sample.
C) LCH cell subsets ordered by the median single-cell entropy of the corresponding single-cell transcriptomes. High entropy indicates promiscuity of gene expression, which has been described as a hallmark as undifferentiated cells. Dashed lines highlight the putative progenitor cell subsets (LCH-C1, LCH-C2) and the most differentiated cell subsets (LCH-C11 to LCH-C14) as in panel A.
D) Heatmap showing expression levels of differentially expressed genes between the two LCH subsets with highest entropy and the four subsets with lowest entropy. Selected hallmark genes are highlighted for each subset.
E) Enrichment analysis with Enrichr for the LCH subset signature genes (from panel D) using KEGG pathways and Gene Ontology, ordered by the Enrichr combined score. FDR-adjusted p-value: *, p < 0.1; **, p < 0.05; ***, p < 0.001. Further details are provided in Supplementary Table S3.
F) Smoothed line plots displaying the relation between decreasing entropy (x-axis; ranked) and the expression of selected genes. The cell cycle genes TUBB, TUBA1B, and MKI67 showed the highest Pearson correlation (r) between entropy and expression. A weaker correlation was found also between entropy and the expression of the canonical LCH marker gene CD1A.
G) Expression of genes characteristic of (non-LCH) epidermal Langerhans cells in each LCH cell subset, displaying the mean of the scaled UMI counts of all genes in the LCH subset signature.
H) Assessment of BRAFV600E mutation burden (in percent) based on allele-specific qPCR in sorted LCH-S12 cells, LCH-S1 cells, CD1A/CD207-negative (non-LCH) cells, CD1A/CD207-double-positive LCH cells, and bulk biopsy cells. All data refer to patient sample LCH_E.
I) Assessment of cell clonality using the HUMARA assay in the same sorted cell populations as in panel H, and in known polyclonal and monoclonal cell lines as negative and positive controls, respectively.
Pursuing the hypothesis that a developmental hierarchy exists within LCH lesions, we exploited previous findings that transcriptional promiscuity is a hallmark of undifferentiated cells, including stem cell and progenitor cell populations18–21. We quantified this property by calculating the “single-cell entropy” for each single-cell transcriptome, which provides a measure of lack of order (interpreted here as a lack of specialization), in analogy to the term’s use in physics (thermodynamics). We found that the 14 LCH-cell subsets differed in their levels of entropy, forming a gradient of LCH cells with progressively lower entropy, corresponding to more restricted transcriptomes and more differentiated cell states (Fig. 3C). We therefore labelled the 14 LCH cell subsets by decreasing entropy, starting from LCH-S1 and LCH-S2 as the least differentiated LCH subsets and ending with LCH-S11 to LCH-S14 as the most differentiated subsets. We also identified multiple intermediate states, indicative of a continuous developmental process unfolding within LCH lesions. For further analysis, we focused on the extreme points of this developmental hierarchy, which exhibited markedly distinct gene expression (Fig. 3A and D; Supplementary Fig. S4B).
Comparing the highest-entropy, least differentiated cell subsets (LCH-S1 and LCH-S2) with the four lowest-entropy subsets (LCH-S11 to LCH-S14), we identified characteristic transcriptional profiles associated with each LCH cell subset (Fig. 3D; Supplementary Table S2). In both LCH-S1 and LCH-S2, we detected high expression of CD1A and of genes associated with cell proliferation, including MKI67 (which encodes the canonical proliferation marker Ki-67) and the aurora kinases AURKA and AURKB – consistent with the interpretation that these two subsets constitute proliferative, progenitor-like LCH cells. Pathway enrichment analyses corroborated their proliferative nature with specific enrichment for DNA replication and cell-cycle-regulated genes (Fi. 3E).
In contrast, the lowest-entropy and putatively more differentiated LCH cell subsets LCH-S11 to LCH-S14 were characterized by high expression of immune genes involved in cellular processes such as cytokine signaling, chemotaxis, and IFN signaling. Specifically, LCH-S11 cells expressed markers of mature dendritic cells such as CD83 and LAMP3, as well as two receptors involved in Langerhans cell migration, CXCR4 and CCR73,22; LCH-S12 cells expressed CLEC9A, BATF3, and IRF8, which are characteristic of certain dendritic cell populations23; LCH-S13 cells expressed genes associated with interferon response; and LCH-S14 cells were characterized by expression of MMP9, CD68, CD63, and the peptidase ANPEP (CD13), as well as pathway enrichment for osteoclast differentiation and rheumatoid arthritis, suggesting that this LCH cell subset may be involved in osteolysis and tissue destruction (Fig. 3D and 3E).
We applied our LCH subset gene signatures to published LCH bulk transcriptome datasets8,14,15 (Supplementary Fig. S4C–S4E) and observed patient-to-patient heterogeneity in the relative strength of these signatures, supporting that LCH lesions can differ in their composition of LCH cell subsets. Moreover, this analysis showed that many of the genes in the LCH-S11 signature were also highly expressed in epidermal Langerhans cells, and genes in the LCH-S12 signature were highly expressed in dendritic cells, while the signatures of these subsets were only weakly detectable in the other histiocytoses analyzed (Supplemental Fig. 4E).
Across all LCH cells (independent of their subset), we observed a gradual loss of expression for cell proliferation genes such as TUBB, TUBA1B, and MKI67 as entropy levels decreased and cells became more differentiated (Fig. 3F). This trend was further associated with a decrease of CD1A expression in the lowest-entropy cells (Fig. 3F) and with a reduction in the expression of genes associated with epidermal Langerhans cells, which was most prominent in the LCH-S12 subset (Fig. 3G). Notably, the LCH-S11 subset displayed reduced expression of the overall Langerhans cell signature despite high expression of individual Langerhans cell genes in the LCH-S11 gene signature. Taken together, these observations lend further support to a model where LCH progenitor cells with high cell proliferation and high levels of CD1A marker expression give rise, through a gradual process, to differentiated cell subsets that are less proliferative and carry gene expression profiles reminiscent of differentiated immune cells, including that of dendritic cells (most pronounced in the LCH-S12 subset).
To confirm that the analyzed cell subsets indeed constitute bona fide LCH cells, we performed two complementary validations, assaying BRAFV600E mutation status as well as cell clonality for representative LCH subsets. We prospectively enriched cells from the LCH-S1 and LCH-S12 subsets, as well as CD1A+CD207+ LCH cells and CD1A-CD207- non-LCH cells, for the patient sample with the highest percentage of LCH-S12 cells (Supplementary Fig. S4F and S4G). We then quantified the BRAFV600E mutation rate in each sorted cell population using allele-specific droplet digital PCR24. Reassuringly, both LCH subsets as well as the bulk LCH cell population displayed a BRAFV600E mutation rate in the range of 85% to 90% (Fig. 3H). We further assessed clonality for the same cell populations using the HUMARA assay16,17, which evaluates X chromosome inactivation status in female-derived samples (such as the tested LCH lesion). Indeed, we found that both LCH subsets as well as the bulk LCH cell population showed substantial skewing similar to the positive (monoclonal) control, while non-LCH cells were more similar to the negative (polyclonal) control (Fig. 3I). These results demonstrate that the LCH-S1 and LCH-S12 cell subsets constitute bona fide LCH cells of clonal origin that carry the BRAFV600E driver mutation.
We next tested whether the results obtained on the merged dataset comprising all seven LCH patients were replicated in the individual LCH lesions (Supplementary Fig. S5A-S5C). Indeed, cells corresponding to the progenitor-like LCH-S1 subset consistently exhibited high levels of entropy in all seven lesion-specific single-cell transcriptome datasets, while cells corresponding to the more differentiated cell subsets (LCH-S11 to LCH-S14) had substantially lower levels of entropy in each analyzed LCH patient sample.
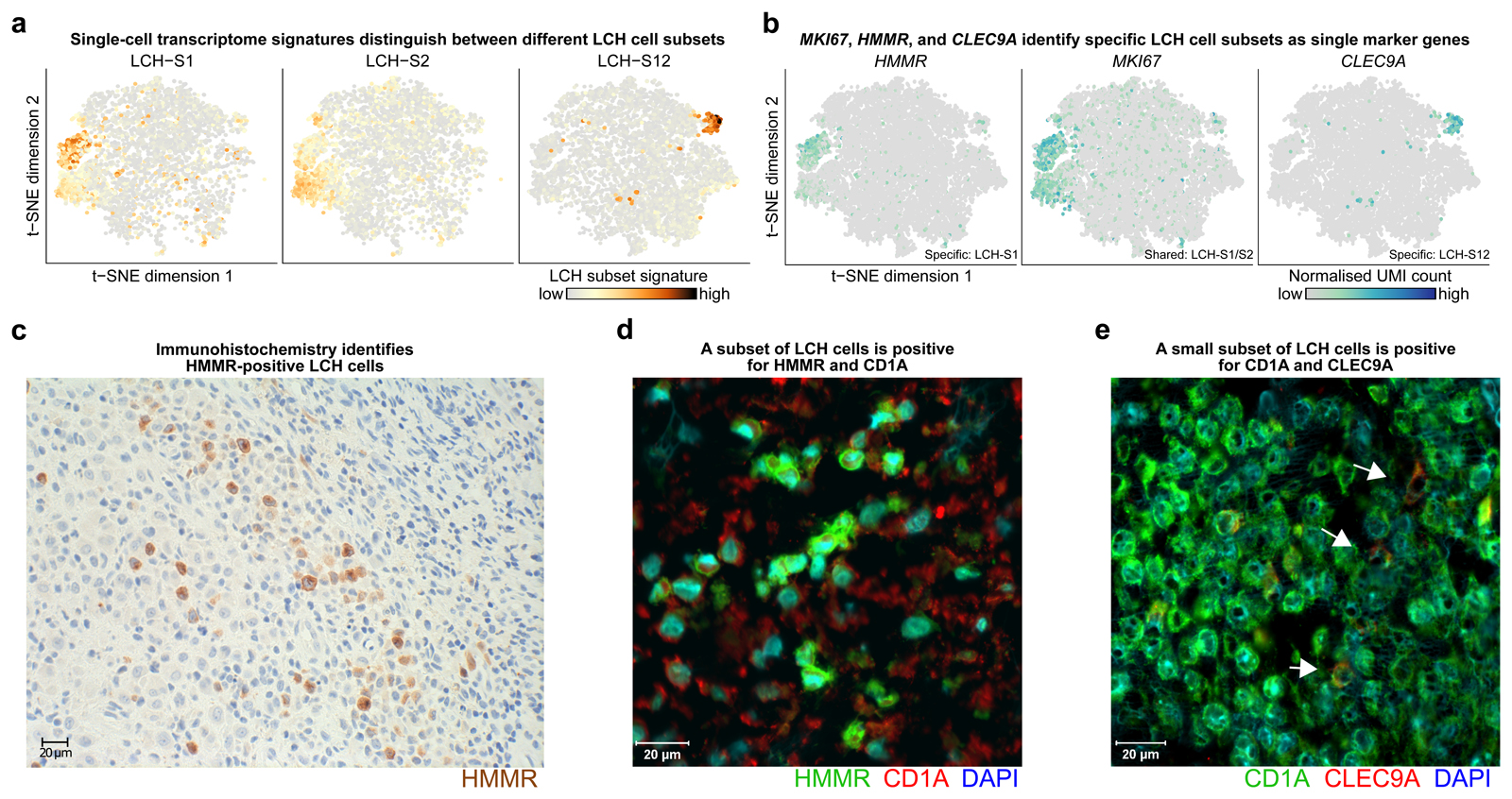
Finally, to visualize the different LCH-cell subsets in tissue slices of LCH lesions (with potential future relevance for clinical diagnostics), we searched our transcriptome dataset for subset-specific markers that can be analyzed using IHC or immunofluorescence, and we found promising candidates (Fig. 4A and B). Ki-67 (MKI67) and HMMR, in combination with the LCH-specific markers CD1A and CD207, were identified as candidate markers for the LCH-S1 and LCH-S2 subsets; and CLEC9A, in combination with CD1A and CD207, was identified as a candidate marker for the LCH-S12 cell subset (Fig. 4C-E; Supplementary Fig. S6A-S6G).
Figure 4. The cell surface markers HMMR and CLEC9A identify specific LCH cell subsets.
A) Low-dimensional projection (t-SNE plot) of LCH cell transcriptomes (same layout as in Fig. 3A) overlaid with the intensity of LCH-subset-specific gene expression signatures (calculated as the mean scaled UMI count for all genes in the signature). Dark red color indicates high expression levels.
B) Low-dimensional projection (t-SNE plot) of the LCH cell transcriptomes (as in panel A) overlaid with the expression of selected LCH-subset-specific marker genes (dark blue color indicates high expression levels). Expression of HMMR is specific to LCH-S1 cells, MKI67 expression overlaps strongly with both the LCH-S1 and LCH-S2 cell subsets, and CLEC9A is detected almost exclusively in LCH-S12 cells.
C) Immunohistochemistry image of HMMR (brown) staining of an LCH biopsy.
D) Immunofluorescence image of HMMR (green), CD1A (red) and DAPI (blue) staining of an LCH biopsy.
E) Immunofluorescence image of CD1A (green), CLEC9A (red) and DAPI (blue) staining of an LCH biopsy.
In aggregate, our results support a developmental hierarchy within LCH lesions that is seeded by progenitor-like LCH-S1 / LCH-S2 cells and gives rise to more differentiated LCH cell subsets, which we interpret as maturing LCH cells (LCH-S11 and LCH-S12), based on transcriptome features resembling epidermal Langerhans cell activation and dendritic cell maturation25,26, and as aggressive LCH cells (LCH-S13 and LCH-S14), based on the expression of genes linked to destructive inflammatory behaviour15,27,28.
Aberrant Regulatory Programs Orchestrate the Developmental Hierarchy in LCH Lesions
To explore the regulatory basis of the observed developmental hierarchy, we prospectively enriched selected LCH subsets from the same sample for which we confirmed BRAFV600E mutation rates and cell clonality (Fig. 3H and I). We performed chromatin profiling by assay for transposase-accessible chromatin using sequencing (ATAC-seq)29 on enriched cells of the progenitor-like cell subset LCH-S1 and of two differentiated cell subsets, LCH-S11 and LCH-S12 (Supplementary Fig. S7A and S7B). Transcriptional regulators and signaling pathways identified in this analysis may help understand the developmental processes in LCH lesions and potentially identify candidate targets for future molecularly guided therapies of LCH.
We integrated the ATAC-seq data with our matched single-cell RNA-sequencing (RNA-seq) data, to connect chromatin-accessible regulatory regions to the genes that they may regulate. To link regulatory regions to their most likely target genes, we considered not only proximity in the DNA sequence but also proximity in the chromatin 3D structure according to HiC data for hematopoetic cells30. We then plotted the average chromatin accessibility of gene-linked regulatory regions against the expression of the respective gene, focusing on differences between the LCH-S1 and LCH-S11, and between the LCH-S1 and LCH-S12 subsets (Fig. 5A). LCH-S12 cells displayed increased gene expression, and a concordant gain in chromatin accessibility, for the key myeloid regulatory gene IRF8, whereas LCH-S11 cells displayed increased expression and chromatin accessibility for the NF-κB genes REL and RELB as well as the immune regulatory genes JUNB and ETV3.
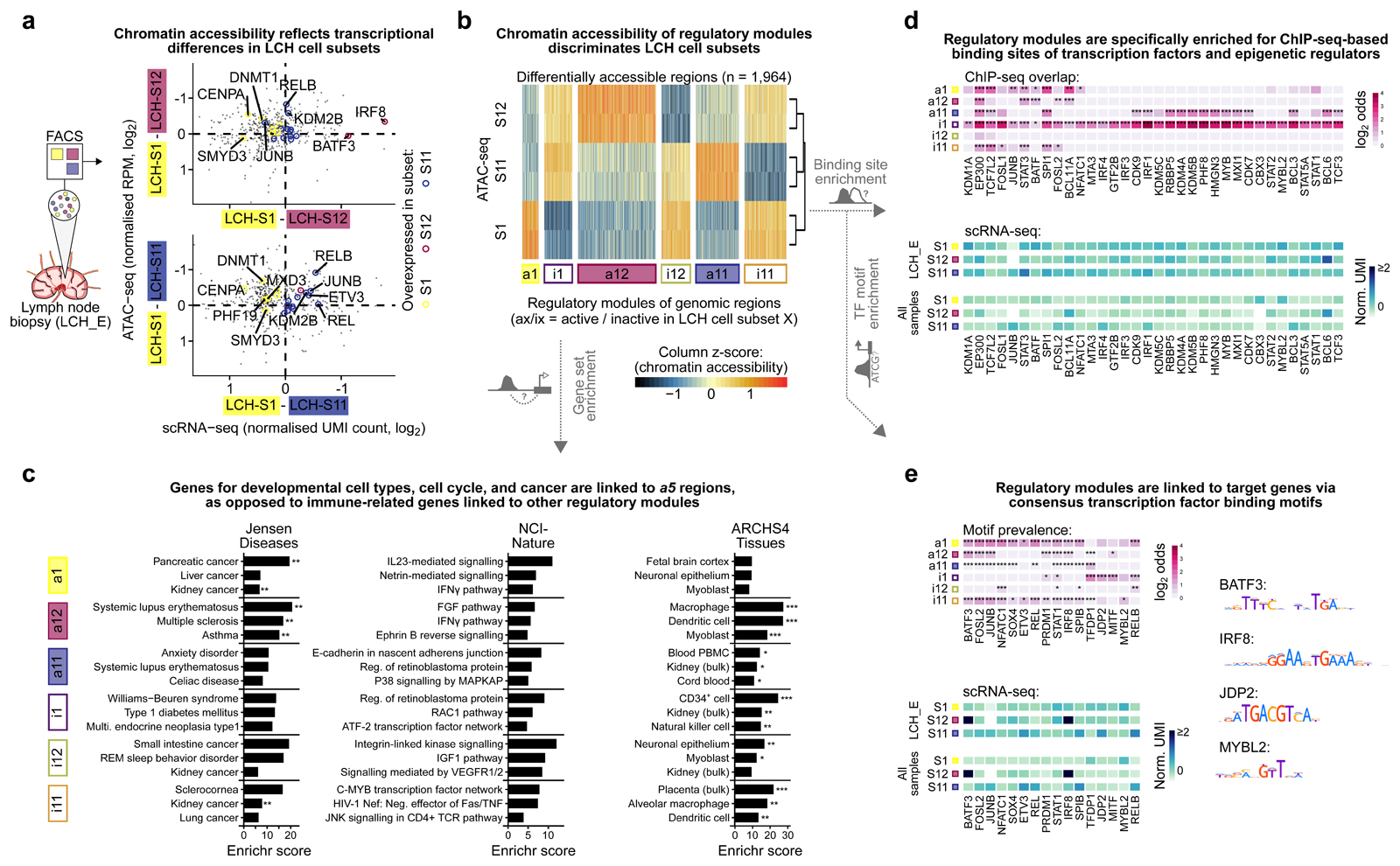
Figure 5. Characteristic patterns of chromatin accessibility distinguish between LCH cell subsets.
A) Scatterplots contrasting differential gene expression with differential chromatin accessibility between progenitor-like LCH-S1 cells and the more differentiated LCH subsets LCH-S12 (top) and LCH-S11 (bottom). The x-axis denotes differences in gene expression (mean normalized UMI count based on scRNA-seq) and the y-axis displays the mean difference in chromatin accessibility over all regulatory regions linked to the respective gene (mean of two replicates, mean normalized reads per million [RPM]). Colored points denote LCH-subset-specific marker genes (from Fig. 3D).
B) Heatmap showing ATAC-seq signal intensity for 1,964 differentially accessible regions identified in pair-wise comparisons of one LCH cell subset against the two other subsets. Regions are grouped into six modules based on differential chromatin accessibility. ATAC-seq signal intensity scores are scaled by column (ATAC-seq peaks) for better visualization of differences.
C) Enrichment analysis with Enrichr for genes linked to each of the six modules of LCH-subset-specific chromatin accessible regions, showing the top-3 most enriched terms ordered by the Enrichr combined score58. FDR-adjusted p-value: *, p < 0.1; **, p < 0.05. Further details are provided in Supplementary Table S3.
D) Heatmap of transcription factor binding enrichment for LCH-subset-specific modules, based on LOLA31 analysis using a large collection of ChIP-seq peak profiles. Top: LOLA enrichments colored by the log2 odds ratio of enriched overlap between the region modules and ChIP-seq peaks for the corresponding transcription factor, compared to the background of all regulatory regions in the ATAC-seq dataset. Bottom: Mean expression (normalized UMI count) of the gene encoding each transcription factor in different LCH subsets (showing gene expression in LCH_E, which matches the ATAC-seq data, as well as the mean expression across all patient samples). Further details are provided in Supplementary Table S3.
E) Enrichment of transcription factor binding motifs for LCH-subset-specific modules, based on HOCOMOCO32 motif occurrences identified using FIMO59. Top: Motif enrichments colored by the log2 odds ratio of enriched overlap between the module regions and DNA motif hits for the corresponding transcription factor, compared to the background of all regulatory regions identified in the ATAC-seq dataset. Bottom: Expression levels of the genes encoding the corresponding transcription factors (as in panel D). Right: Consensus DNA motif for selected transcription factors. FDR-adjusted p-value, Fisher’s exact test: *, p < 0.1; **, p < 0.05, ***, p < 0.005.
For a more systematic analysis of the regulatory dynamics in LCH lesions, we identified all genomic regions that displayed differential accessibility between the LCH-S1, LCH-S11, and/or LCH-S12 subsets (n = 1,964; Fig. 5B; Supplementary Table S2). We refer to the set of regulatory regions that showed increased chromatin accessibility (activation) in LCH subset LCH-S1 relative to LCH-S11 or LCH-S12 as a1 (for “active”) and to those that were characterized by decreased accessibility (inactivation) as i1 (for “inactive”). We analogously defined region modules a11, i11, a12, and i12 as those specifically active or inactive in the corresponding LCH subsets. We characterized these regulatory modules by three complementary lines of bioinformatic analysis.
First, enrichment analysis of genes associated with the regulatory regions identified gene sets implicated in cancer (LCH-S1) and in immune diseases (LCH-S12), consistent with the dual nature of LCH as a neoplasm with a strong inflammatory component (Fig. 5C; Supplementary Table S3). We also identified genes involved in various immune-related pathways, including interleukin and interferon signaling. Regions that were differentially accessible in the more differentiated LCH-S11 or LCH-S12 cell subsets (region modules a11 and a12) were frequently associated with genes expressed in macrophages, dendritic cell, and other mononuclear blood cells, consistent with the transcriptional signatures we discovered in the gene expression analysis.
Second, we sought to identify the key regulators of each module using enrichment analysis for transcription factor (TF) binding sites. To that end, we performed genomic region enrichment analysis using the LOLA software31 for a large collection of TF binding sites experimentally determined by chromatin immunoprecipitation sequencing (ChIP-seq; Supplementary Table S3). Of all TFs identified as significantly enriched in at least one module, 34 TFs were either among the genes previously identified as differentially expressed in one of the LCH subsets or directly linked to such a gene (Fig. 5D; Supplementary Table S3). Notably, all of these TFs were enriched in regulatory regions with inaccessible chromatin in LCH-S1 cells (module i1), while some were accessible in LCH-S11 (module a11), underlining the contrasting nature of these two LCH subsets. Several TFs were enriched in multiple region modules indicating broad relevance for LCH cells, which included the AP-1 factors JUNB and FOSL2 as well as the STAT and IRF families of TFs. We further observed enrichment of SPI1 (also known as PU.1) binding sites in all modules except those linked to LCH-S12 (both accessible [a12] and inaccessible [i12]), indicative of a central role of this hematopoietic master regulator in LCH. Interestingly, binding sites of the histone lysine demethylase KDM1A (also known as LSD1) were significantly enriched in module i1 (inaccessible in LCH-S1 progenitors) but not in other modules, suggesting a role of this repressive histone modifier in the development of the more differentiated LCH subsets.
Third, we performed enrichment analysis for a comprehensive catalogue of DNA sequence binding motifs of TFs obtained from the HOCOMOCO database32 (Fig. 5E). Motif enrichment analysis is complementary to LOLA analysis (which uses cell-type-specific ChIP-seq data) as it focuses on DNA binding propensities that are intrinsic to the TF and largely independent of cell type. Regulatory regions specifically active in LCH-S1 (module a1) were strongly enriched for the MYBL2 motif, which has a known regulatory role in cancer stem cells33. In contrast, the a12 module (active in LCH-S12 cells) was enriched for motifs of the hematopoietic TFs IRF8 and BATF3. These factors are critical for myeloid and dendritic cell differentiation and function34–36, possibly explaining the more mature myeloid identity of LCH-S12 cells. Moreover, we observed enrichment for the JDP2 motif in the i1 module. This suggests that an inactivation of AP-1 may regulate differentiation, in line with upregulation of JDP2 transcription in more differentiated subsets (Fig. 3D).
Regulatory Networks Identify Subset-Specific Regulatory Hubs in LCH Lesions
Finally, we integrated our data into gene regulatory network models of LCH, in order to combine and visualize the regulatory programs that we identified in the different LCH subsets. We assigned the identified TFs (selected based on genomic region enrichment and motif analysis) to the regions that they are predicted to bind (based on ChIP-seq data and sequence motifs), and then linked regulatory regions to their putative target genes (using sequence proximity and HiC data), which gave rise to a network model that incorporates data from all LCH subsets. Next, we derived network models specific to each LCH subset by parameterizing this global network with the measured chromatin accessibility and gene expression levels in each LCH subset using the matched ATAC-seq and single-cell RNA-seq data for the most comprehensively characterized biopsy LCH_E (Fig. 6A).
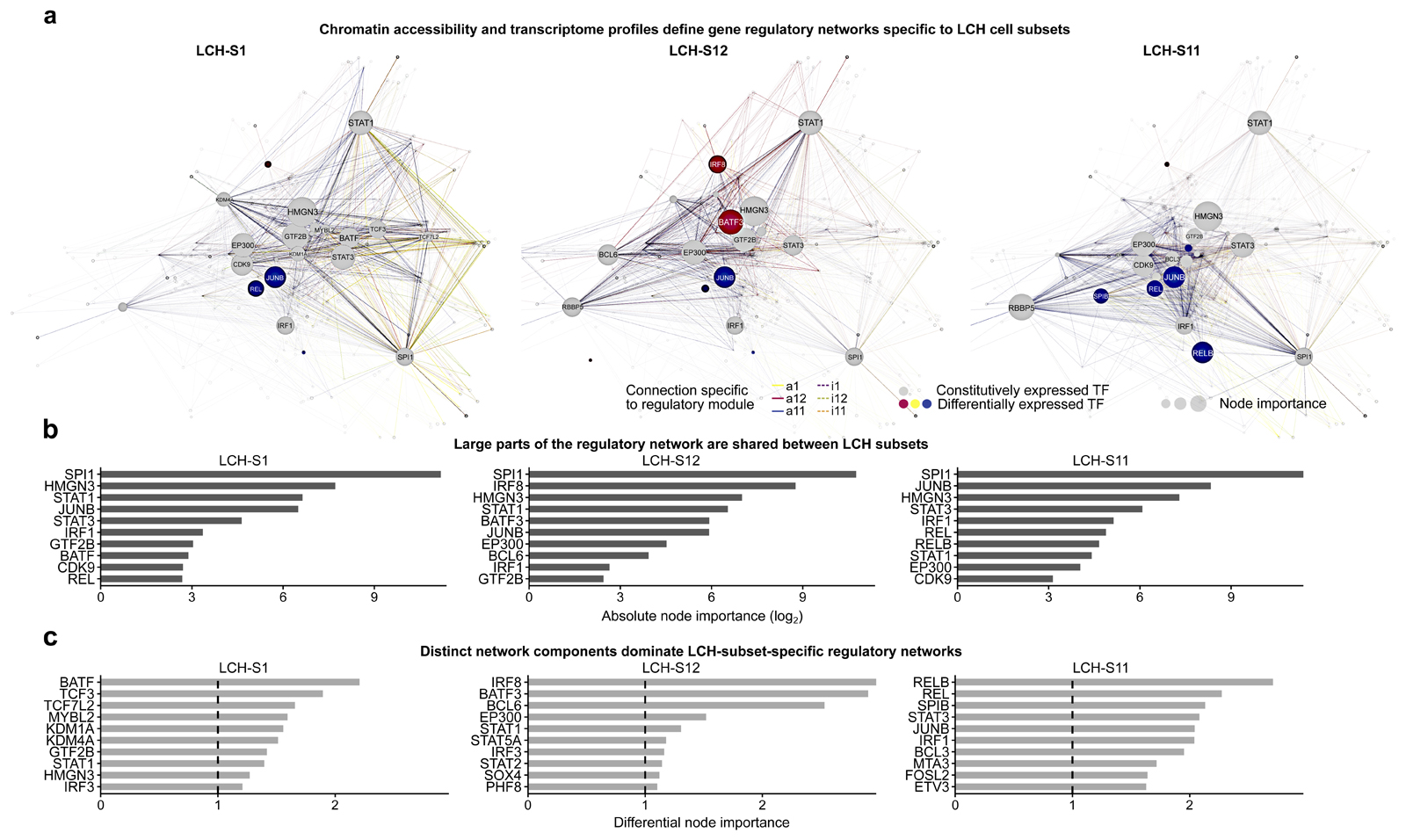
Figure 6. Characteristic gene regulatory networks underlie the observed LCH developmental hierarchy.
A) Gene regulatory networks inferred for three LCH cell subsets (LCH-S1, LCH-S12, LCH-S11), based on single-cell transcriptome and ATAC-seq data, and the key regulators identified by the enrichment analysis in Fig. 5D and E. Nodes in the network correspond to the enriched transcription factors as well as their putative target genes (based on sequence proximity and chromatin 3D structure). Node size is proportional to both gene expression level and node out-degree (i.e., number of outgoing connections from the transcription factor) in the respective LCH subset. Edge colors indicate the module of the corresponding peak, and edge visibility is proportional to chromatin accessibility. The network layout was automatically generated using the igraph package. A browser-based version for interactive data exploration is available in Supplementary File S1 and on the Supplementary Website: http://LCH-hierarchy.computational-epigenetics.org).
B) Bar plots showing the top-10 transcription factors ranked by node importance (calculated as a combination of gene expression level and node out-degree) in the networks in panel A.
C) Bar plots showing the top-10 transcription factors ranked by differential node importance (node importance in one network relative to the mean across all three networks) in the networks in panel A.
This network-based analysis revealed gene regulatory hubs shared across all LCH subsets, such as the hematopoietic TF SPI1 (PU.1; Fig. 6B), but also subset-specific differences (Fig. 6C). For example, the TFs BATF, TCF3, TCF7L2, and MYBL2 were highly prominent in the regulatory network of LCH-S1. MYBL2 is known to be involved in cancer initiation and cell proliferation, often associated with poor survival33,37,38. Lysine demethylases (KDMs) including KDM1A and KDM4A were also most active in LCH-S1 cells, consistent with the progenitor-like state of LCH-S1 and suggesting that developmental TFs may be retained in a poised state39–42. The regulatory network of LCH-S12 was instead dominated by the TFs IRF8 (an interferon-dependent mediator of macrophage and dendritic cell function and differentiation34,36,43), BATF3, and BCL6, which were all closely connected to STAT1. Moreover, many connections of SPI1 to its target genes in other LCH subsets were altered in LCH-S12 cells, possibly contributing to the specific expression of dendritic cell genes in these cells. For the LCH-S11 subset, the regulatory network is centered on NF-κB (REL and RELB), STAT3, and AP-1 (JUNB and FOSL2), which are present in all networks but most pronounced in LCH-S11 cells. Finally, we observed differential node importance for the histone acetyltransferase and transcriptional coactivator EP300 in both LCH-S11 and LCH-S12 cells, implicating this epigenetic modifier in the specification of more differentiated LCH cell subsets.
In summary, we used chromatin accessibility profiling in prospectively purified LCH cell subsets in combination with single-cell RNA-seq data and integrative bioinformatic analysis, in order to reconstruct the gene regulatory networks that may control LCH-subset-specific gene expression. Gene expression of the discussed regulators was largely consistent across all analyzed lesions (Supplementary Fig. S7C-S7E), suggesting that the prototype gene regulatory networks put forward here indeed capture the epigenomic program underlying the developmental hierarchy in LCH lesions.
Discussion
LCH lesions are thought to arise from hematopoietic progenitor cells that carry an activating mutation in the MAPK signaling pathway2,44. Despite their clonality and dependence on a single oncogenic driver (most frequently the activating BRAFV600E mutation), LCH is pathophysiologically and clinically very different from hematopoietic cancers and from BRAFV600E-driven solid tumors. In this study, we systematically investigated the cellular and molecular heterogeneity in primary LCH lesions, using single-cell transcriptome sequencing in combination with epigenome profiling of prospectively enriched LCH cell subsets. Our dataset comprises 19,044 single-cell transcriptomes across seven LCH biopsies, representing the wide clinical spectrum of LCH. Integrative bioinformatic analysis of this rich resource uncovered an unexpected degree of heterogeneity among LCH cells, as well as evidence of a shared developmental hierarchy that underlies all analyzed lesions.
Most notably, we identified two subsets of LCH cells with hallmarks of progenitor cells (LCH-S1 and LCH-S2), including high single-cell entropy (indicative of an undifferentiated cell state) and higher cell proliferation. Based on our data, we put forward a conceptual model in which these progenitor-like LCH cells seed a hierarchy of LCH cells that develop into more differentiated cell states (Fig. 7). The four subsets of LCH cells with lowest transcriptome entropy (indicative of a differentiated cell state) expressed genes reminiscent of different immune cell types, suggesting that hematopoietic differentiation programs remain active in LCH cells and contribute to the cellular and molecular hierarchy observed in LCH lesions. Specifically, LCH-S12 cells expressed genes associated with dendritic cells, including CLEC9A and BATF323; LCH-S11 cells carried features of maturing epidermal Langerhans cells, such as expression of CCR7 and CXCR422; and LCH-S13 as well as LCH-S14 cells were characterized by high expression of pro-inflammatory genes, metalloproteinases (MMP9, MMP12), and aminopeptidases (ANPEP), indicating that these LCH cell subsets may contribute to tissue destruction as observed in LCH45. Finally, we inferred preliminary gene regulatory network models for three of the LCH cell subsets (LCH-S1, LCH-S12, LCH-C11), which implicated key regulators of immunity (including JAK-STAT, AP-1, and NF-κB signaling) and development (including several epigenetic modifiers) in the regulation of the developmental hierarchy in LCH cells. While the gene regulatory networks presented here are based on the analysis of one biopsy by ATAC-seq and on external data (ChIP-seq, DNA motifs) from other biological systems, we have observed consistent transcriptional activity of key regulators in the other examined biopsies, supporting relevance to our conceptual understanding of LCH development.
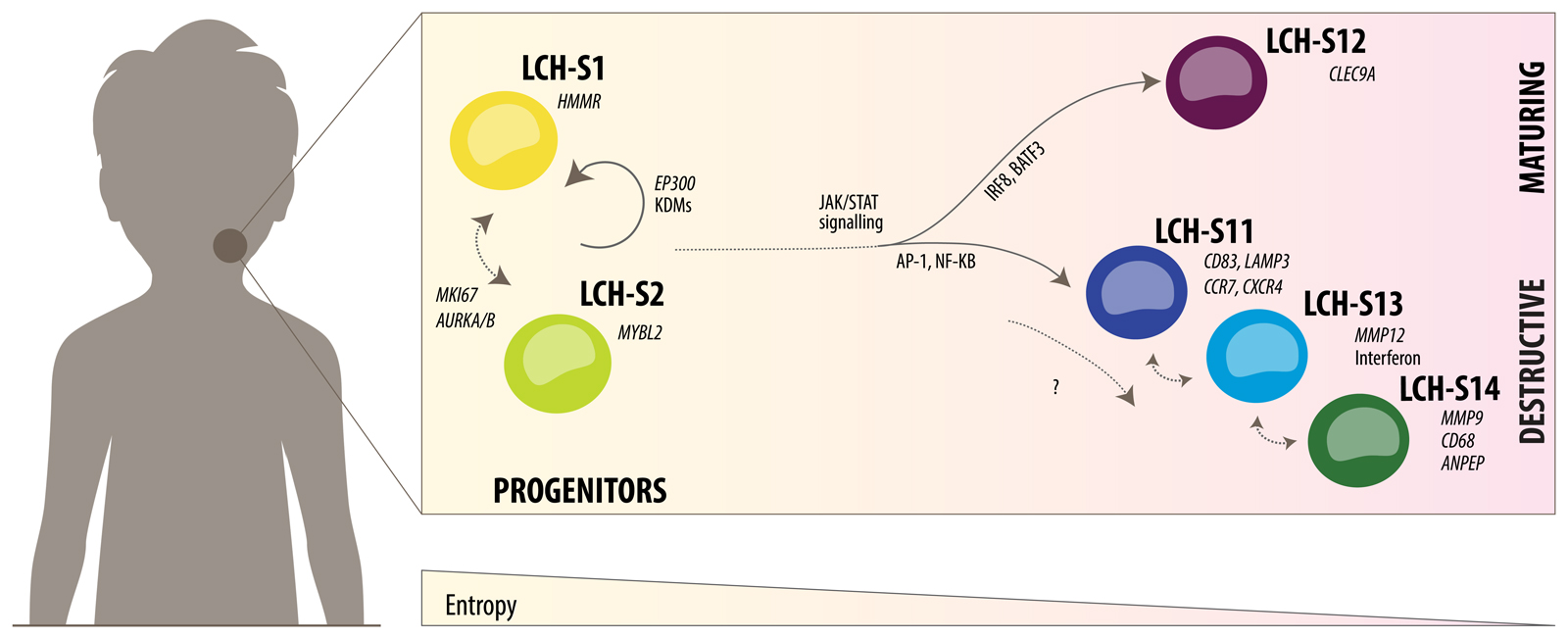
Figure 7. Speculative model of the LCH developmental hierarchy and underlying regulatory mechanisms.
Schematic representation of the developmental hierarchy in LCH lesions based on the combined analysis of single-cell transcriptomes (Fig. 2 and 3) and epigenome data from prospectively sorted LCH cell subsets (Fig. 5 and 6).
Our proposed model (Fig. 7) provides a framework for understanding the developmental and regulatory dynamics in LCH. This model will need to be refined, validated, and improved by subsequent research, taking into account three important points. First, the precise number of distinct LCH cell subsets remains somewhat arbitrary, given that cellular differentiation is a gradual process, and any separation into a number of distinct cell populations is necessarily a simplification. While our bioinformatic analysis identified 14 LCH cell subsets in a data-driven manner, for many investigations it will make sense to combine related cell subsets, as we have done for the two progenitor-like subsets (LCH-S1 and LCH-S2). Second, it remains to be studied to what degree the cells that proceed along the developmental hierarchy in LCH lesions undergo irreversible cell fate decisions and to what degree they retain the plasticity to change from one LCH subset to another. For example, it seems possible that the three closely related cell subsets LC-S11, LCH-S13, and LCH-S14 constitute different manifestations (e.g., depending on the cellular microenvironment) of one developmentally defined LCH cell subset (Fig. 7, center right). Third, our study focused on (peripheral) LCH lesions, while it is entirely possible that the developmental hierarchy in LCH lesions is seeded by an unseen LCH cell precursor that is located in different anatomical location. For example, a plausible model of multi-system LCH may include an LCH cell precursor in the bone marrow, which has acquired the clonal driver mutation and seeds LCH lesions at distal sites that support a pro-inflammatory microenvironment. To address the many questions posed by our preliminary model, it will be necessary to develop new in vitro (e.g., patient-derived organoids) and in vivo (e.g., xenograft experiments46) experimental systems for LCH research, and to closely align these systems with our observations in primary patient samples.
Our model has several potential implications for the future clinical management of LCH (and possibly other histiocytic diseases). First, we speculate that inter-individual differences in the composition of LCH cell subsets reflect differences in the aggressiveness of the disease and may correlate with relevant clinical characteristics including prognosis. There are currently no established molecular markers for risk stratification in LCH, which constitute an unmet clinical need given the range of treatment options (from watch-and-wait to intensive chemotherapy). While single-cell transcriptome sequencing may not be practical in a routine clinical setting, we were able to observe differences in LCH subset composition in bulk RNA-seq (Supplementary Fig. S4C-S4E), immunohistochemistry, and immunofluorescence imaging (Fig.4; Supplementary Fig. S6). After validation in large patient cohorts, such markers may help inform patient-specific treatment decisions in LCH. Second, our data implicate several immune-regulatory signaling pathways in the regulation of the developmental hierarchy, including the potentially destructive LCH-S13 and LCH-S14 cell subsets. This observation may explain the success of anti-inflammatory drugs such as indomethacin in the treatment of LCH, suggesting that such treatment might not only alleviate symptoms but indeed interfere with key pathogenic processes. This underlines the importance of the current randomization between chemotherapy (mercaptopurine and methotrexate) versus an anti-inflammatory drug (indomethacin) in the ongoing international study for the treatment of LCH (LCH-IV; ClinicalTrials.gov Identifier: NCT02205762). Third, a thorough dissection of developmental hierarchies in other histiocytic disorders such as Erdheim-Chester Disease may refine our understanding of the similarities and differences between these diseases and potentially give rise to more precise molecular diagnoses of diseases of the histiocytic spectrum.
In summary, our study charts a detailed molecular map of LCH lesions, uncovering a cellular hierarchy that shows evidence of developmental, immunological, and oncogenic regulatory mechanisms. Our results reinforce the view that LCH shares important aspects with cancers as well as immune diseases, which makes it a particularly interesting model for biomedical research. Moreover, this study demonstrates the power of combining single-cell sequencing and epigenome profiling for dissecting complex developmental hierarchies and their regulatory underpinnings, thereby providing a rational basis for the development of personalized therapies.
Methods
Patient Cohort and Sample Collection
LCH samples were obtained with informed consent from patients undergoing routine diagnostic biopsies. Diagnosis was confirmed by central pathology review. Cell suspensions were prepared by dissociating collagenase IV (Worthington Biochemical) and dispase II (Sigma-Aldrich) treated tissue using a 70 μm cell dissociation sieve (Sigma-Aldrich). Cells were then pelleted, re-suspended in CellGro medium (CellGenix), and immunostained for fluorescence-activated cell sorting (FACS) using the antibodies specified below. Sorting was performed on a BD FACS Aria 1 as described previously15. All protocols for obtaining and studying patient material were approved by the responsible institutional review board and the ethics committee of the Medical University of Vienna. Sampling was done according to the regulations of the Declaration of Helsinki after written informed consent.
IHC
IHC stainings were done on formalin-fixed, paraffin-embedded tissue sections with antibodies against CD1A (NCL-L-CD1A-235, Leica Biosystems), CD207 (CMC39221020, Cell Marque), and the mutation-specific BRAFV600E antibody (06965814001, Spring Bioscience). Staining was performed on an automated Leica BOND III immunohistologic stainer. Images were captured with a Leica DM6B using LEICA LAS X software.
Immunofluorescence
OCT-embedded (Tissue-Plus O.C.T. compound, Scigen Scientific), fresh-frozen tissue was cut into 7 μm sections and mounted on microscope slides (Dako). After 20 minutes of air-drying, the sections were fixed in ice-cold acetone (Sigma-Aldrich) for 10 minutes, rehydrated in phosphate-buffered saline (PBS) (Gibco Life Technologies) and incubated with PBS/2% goat serum (Dako) for 30 minutes at room temperature. This was followed by an overnight incubation at 4°C with either CD168 FITC (bs-4736R-FITC, Bioss) or CLEC9a PE (30-101-294, Miltenyi Biotec) diluted in PBS/2% bovine serum albumin (BSA, Sigma-Aldrich). After washing with PBS, the second step antibodies (i.e. goat anti-rabbit IgG Alexa Fluor488 and goat anti-mouse A546, respectively) were applied for one hour at room temperature, washed with PBS, blocked, and then stained with CD1a PE or CD1a FITC for 1h at room temperature. After washing with PBS, a second round of secondary antibody staining was performed (goat anti-mouse A546 or goat anti-FITC A488, respectively) for 1h at room temperature. The slides were then counterstained with 4,6-diamidino-2-phenylindole dihydrochloride (DAPI, Roche Diagnostics), washed again, and mounted in aqueous mounting medium (PermaFluor, Thermo Scientific). Negative controls were obtained by substituting isotype-matched IgG for the respective primary antibodies and by applying the second step antibodies and the anti-FITC antibodies alone. Image acquisition was performed on a Zeiss LSM510 equipped with four lasers and a Zeiss Axiovert 200M using a Zeiss Plan-Neofluar 40x objective with oil immersion. Images were exported from Zen software (Zeiss).
Flow Cytometry and Cell Sorting
The following antibodies were used for FACS purification of primary LCH samples: CD45-PerCP (345809, BD Biosciences), CD1A-FITC (555806, BD Biosciences), CD207 PE (IM3577, Beckman Coulter), CD14 Alexa Fluor 700 (A7-212-T100, Exbio). To selected antibodies for the analysis of LCH subsets, we cross-checked marker gene lists for commercially available, high-quality antibodies. The following antibodies were used for FACS purification of LCH subsets: CD207 APC-Vio770 (130-112-371, Miltenyi Biotec), CD1A PE-Vio 770 (130-112-872, Miltenyi Biotec), CD45 eFluor 506 (69-0459-42, eBioscience), CD168 FITC (bs-4736R-FITC, Bioss), CD208 PE (130-104-392, Miltenyi Biotec), CD298 PerCP-Vio770 (130-101-294, Miltenyi Biotec), CD300A Alexa 647 (566342, BD Biosciences), and CD370 VioBlue (130-097-406, Miltenyi Biotec). LIVE/DEAD stain (Invitrogen) was used for live/dead staining according to manufacturer’s description. Cell sorting was performed on a FACSAria instrument (BD Biosciences). The FACSDiva software (BD Biosciences) was used for data analysis.
HUMARA Assay
HUMARA assays were performed as previously described47. Briefly, DNA isolated from unsorted LCH biopsy cells and LCH-cell subsets was digested with HpaII (NEB) or mock digested. Amplification of the HUMARA STR region was performed using Phusion HS II Polymerase (ThermoFisher) and the following primers: 5‘-6FAM-TCCAGAATCTGTTCCAGAGCGTGC-3‘, 5’-GCTGTGAAGGTTGCTGTTCCTCAT-3‘. PCR products were analysed and quantified using an Applied Biosystems 3730XL DNA Analyzer. The ratio of the active to the inactive X chromosome was calculated using the area under the peak. To account for the amplification bias of the smaller allele, a corrected ratio was calculated by dividing the ratio of the digested sample (allele 1 / allele 2) by the ratio of the mock digested sample (allele 1 / allele 2). Ratios close to 1 indicate a polyclonal cell population and ratios above 3 are considered evidence of clonality48. DNA isolated from a single-cell-derived cell line and from whole blood of a healthy donor was used as monoclonal control (ratio 4.1) or polyclonal control (ratio 0.9), respectively.
Allele-Specific Droplet Digital PCR
Droplet digital PCR for BRAFV600E was performed as previously described24, using the Mutation Assay BRAF p.V600E c.1799T-A, Human (dHsaMDV2010027, Bio-Rad Laboratories) and the QX200 Droplet Digital PCR System (Bio-Rad Laboratories).
Single-Cell RNA-seq
Single-cell RNA-seq was performed using the 10x Genomics Chromium Single Cell Controller with the Chromium Single Cell 3’ V2 Kit following the manufacturer’s instructions. After quality control, libraries were sequenced on the Illumina HiSeq 4000 platform in 2x75bp paired-end mode. Supplementary Table S1 includes an overview of sequencing data and performance metrics. Raw sequencing data were processed with the CellRanger v1.3.0 software (10x Genomics) for demultiplexing and alignment to the GRCh38 human reference transcriptome. Processed data were analyzed using the R statistics software and various Bioconductor packages. Specifically, we used Seurat v2.3.449 to load pre-processed results from CellRanger into R and to perform quality control (removing cells with less than 1,000 genes or mitochondrial content greater than 10%). Unless otherwise stated, we used default parameters throughout. We merged all datasets, log-transformed, and normalized UMI counts, regressing out UMI count, mitochondrial content, cell cycle scores, patient ID, biopsy, sex, and age. We then used canonical correlation analysis (CCA; using the RunMultiCCA function) using variable genes in the merged dataset and seven canonical vectors. The CCA-aligned space was used as input for low-dimensional projection using t-SNE with all seven dimensions. Cells were clustered with the FindClusters function (resolution = 0.2). Clusters in which at least 25% of cells were double-positive for CD1A and CD207 (“positive list”; normalized UMI greater 1), and no more than 60% of cells were positive for either CD19, CD3D, CD27, IL32, CD7, NKG7, or CD163 (“negative list”) were defined as LCH cells. These criteria select cells with high expression of canonical LCH markers and excludes cells which are clearly identified as non-LCH immune cell types. We used the same procedure and thresholds to define immune cells using CD1A and CD207 as a negative marker list and as positive markers: CD19 for B cells, CD3D for T cells, CD14 for monocytes/macrophages, and IL3RA for plasmacytoid dendritic cells. To compare gene expression of these immune cell types with that of LCH cells (Fig. 2), we used the FindMarkers function. To avoid possible confounding effects by sex differences (cells of both sexes are present at slightly variable rates between cell types in our dataset), we performed this analysis stratified by sex and used the mean log2 fold change and maximum (worst) p-value for each gene. An FDR-adjusted p-value threshold of q ≤ 0.005 was used to select differentially expressed genes. To identify LCH cell subsets, we repeated the CCA, clustering, and dimensionality reduction after filtering out non-LCH cells and used the FindAllMarkers function to perform differential analysis between LCH cell subsets (Fig. 3), again stratified by sex and using the same parameters. Differentially expressed genes and LCH cell subset markers are listed in Supplementary Table S2. To quantify the single-cell entropy (also known as “signaling entropy”) of each cell18,21,50, we used the CompSRana function in the LandSCENT v0.99.2 package. Signaling entropy has been described as a robust measure of differentiation potential / progress. To be consistent with our other analyses, we modified the code of the LandSCENT package to allow use of our pre-calculated clusters and dimensionality reduction. For the alternative analysis of LCH subsets in each sample separately (Supplementary Fig. S5), we filtered the LCH dataset to contain only cells from one sample at a time and performed principal component analysis, t-SNE dimensionality reduction, and clustering independently (as in the standard Seurat workflow). External microarray and RNA-seq for comparisons (Gene Expression Omnibus: GSE35340, GSE16395, GSE74442) were downloaded using the GEOquery package and quantile-normalised. Finally, to define reference gene expression signatures of immune cells, we used microarray data from our previous research15 (Gene Expression Omnibus: GSE35340, GSE114181). These data were normalised using frma2 (parameters: summarize="robust_weighted_average") and ComBat from the sva package51. The probe set with the highest variance per gene was chosen for further analysis, and we excluded genes with a z-score below 2 in all samples. We performed differential expression analysis with the limma52 package and used the mean rank of p-values and fold changes to determine the top genes associated with each of the four immune cell types (CD1C+ dendritic cells [n = 7], plasmacytoid dendritic cells [n = 3], monocytes [n = 6], and Langerhans cells [n = 3]), limited to genes with an FDR-adjusted p-value less than 0.05 and an absolute log2 fold change greater than 1.
Chromatin-Accessibility Mapping
ATAC-seq was performed as described previously53. Briefly, 20,000 to 50,000 cells were lysed in a buffer containing digitonin and Tn5 transposase enzyme (Illumina). After incubation at 37°C for 30 minutes, tagmented DNA was purified and enriched. After final purification, the libraries were quality checked using a 2100 Bioanalyzer (Agilent) with high-sensitivity DNA chips. DNA concentration was examined using Qubit Fluorometer, and the libraries were sequenced on the Illumina HiSeq 4000 platform in 1x50bp single-read mode. Supplementary Table S1 includes an overview of the sequencing data and performance metrics. Raw sequencing data were trimmed using Skewer v0.1.12654, followed by alignment to the GRCh38 assembly of the human genome with Bowtie 2 v2.2.455 (parameters: --very-sensitive --no-discordant). Only deduplicated, uniquely mapped reads with mapping quality ≥30 were kept for further analysis. To identify accessible genomic regions, we used MACS2 v2.1.056 (parameters: -q 0.1 -g hs). Following initial data processing, all subsequent analyses were performed in R using Bioconductor packages. After merging peaks across all ATAC-seq datasets and removing peaks that overlapped blacklisted regions from ENCODE (https://sites.google.com/site/anshulkundaje/projects/blacklists) or repetitive regions (obtained from the UCSC Genome Browser), we quantified for each input dataset the number of reads overlapping the retained peaks. Raw read counts were loaded into DESeq2 v1.22.257 for normalization and differential analysis (using a minimum absolute log2 fold change of 1 as threshold; n = 2 replicates per LCH subset). We refer to the set of regulatory regions that showed increased chromatin accessibility (activation) in LCH subset LCH-S1 compared to LCH-S11 or LCH-S12 as a1 (for “active”) and to those that were characterized by decreased accessibility (inactivation) in the same comparison as i1 (for “inactive”). Analogously, we defined region modules a11, i11, a12, and i12 as those that are specifically active or inactive in one LCH subset. To associate ATAC-seq peaks with putative target genes, we used chromatin conformation data from 17 blood cell types30, considering each interaction between a promoter and another genomic region observed in these cells as a possible regulatory interaction. Additionally, we assigned each peak also to the closest gene promoter in its vicinity. Annotated regulatory regions from the analysis of ATAC-seq data are listed in Supplementary Table S2.
Enrichment Analysis for Genes and Genomic aegions
To help understand the biological roles of differentially expressed genes and differentially active genomic regions, we employed four types of enrichment analysis (Supplementary Table S3). First, we used Gene Set Enrichment Analysis (GSEA; v3.0) to test genes in LCH cells compared to immune cells (using the Wilcoxon test statistic as a ranking criterion) for an enrichment of annotations from the list of hallmark gene signatures from MSigDB (Fig. 2D). We considered pathways with an FDR-adjusted p-value below 0.05 significant. Second, we used the Enrichr API58 (v1.0) to test differentially expressed genes and genes linked to enhancers of interest for significant enrichment across a broad range of gene sets (Figs. 2A and C, 3E, and 5C). In all plots, we report the Enrichr combined score calculated as log(Old.P.value)×Z.score by Enrichr. This amounts to a product of the significance estimate and the magnitude of enrichment. Third, we used Locus Overlap Analysis31 (LOLA; v1.12.0) to test regulatory regions identified in our ATAC-seq data for significant overlaps with experimentally determined transcription factor binding sites from publicly available ChIP-seq data (Fig. 5D). To this end, we used the codex, encode_tfbs, and cistrome_cistrome databases contained in the LOLA Core database. We considered only terms with an FDR-adjusted p-value below 0.005 and a minimum absolute log2 fold change of 1 significant. We focused on transcription factors that were found in our lists of differentially expressed LCH subset markers or that were closely related to those (by performing a fuzzy string match, e.g. including STAT3 when only STAT1 was in the marker list). Fourth, we searched the DNA sequences underlying sets of ATAC-seq peaks for matches to known DNA binding motifs from the HOCOMOCO database v1132 (Fig. 5E). For this search, we used FIMO (v4.10.2)59 (parameters: --no-qvalue --text --bgfile motif-file), and regions with at least one hit (p < 0.0001) were counted. To test for differential motif enrichment, we compared the number of regions with at least one match to a given motif with the respective frequency of motif hits in the set of all ATAC-seq peaks by using Fisher’s exact test. Motifs with an FDR-adjusted p-value below 0.05 and log2 odds above log2(1.5) were considered significant. We focused on motifs matching factors that were differentially expressed.
Network analysis
Integrating the results of gene-centric and region-centric enrichment analyses as well as the DNA motif search, we inferred a gene regulatory network of LCH subsets. In this network, we defined “regulatory edges” (arrows in the network) as connections pointing from a transcription factor to its targets. The source nodes of these edges were ATAC-seq peaks which either had an overlap with one of the enriched ChIP-seq datasets (LOLA analysis) or a match with an enriched DNA binding motif. All source nodes belonging to the same TF were merged into one node. Targets of the edges were all genes linked to the respective ATAC-seq peak. LCH-subset-specific networks were constructed by using the single-cell RNA-seq and ATAC-seq data for the respective subset. We defined node sizes proportional to expression level and out-degree (“node importance”) as follows: ng = Ng2 × Eg2 where Ng = max(0, xg – xmed) is the difference of expression of a gene (xg) from the median expression (xmed) of all genes (normalized and scaled UMI counts) and is the sum of edge weights (y) over the 80th percentile (y80th) of all weights (normalized ATAC-seq read count). Edges were weighted by accessibility (mean of all data for the respective cell subset). Furthermore, browser-based network visualizations were generated to support interactive exploration of the regulatory networks using the visNetwork package in R (Supplementary File S1, Supplementary Website: http://LCH-hierarchy.computational-epigenetics.org). Node size in these networks is proportional to node importance, which was calculated in the same way as described above.
Supplementary Material
Significance.
This study sketches a molecular portrait of LCH lesions by combining single-cell transcriptomics with epigenome profiling. We uncovered extensive cellular heterogeneity, explained in part by an intrinsic developmental hierarchy of LCH cells. Our findings provide new insights and hypotheses for advancing LCH research and a starting point for personalizing therapy.
Acknowledgments
We would like to thank all patients and their families who gave their permission to include samples and clinical data in this study. We also thank the Biomedical Sequencing Facility at CeMM for assistance with next-generation sequencing, Dieter Prinz and Elke Zipperer for flow cytometry support, and the members of the Bock lab for help and advice. This work was supported by DFG Research Fellowship (HA 7723/1-1; to F. Halbritter), Innovation Fund of the Austrian Academy of Sciences (IF_2015_36; to M. Farlik), Austrian Science Fund (FWF) Special Research Programme grant (FWF SFB F 6102-B21; to M. Farlik. and C. Bock), EMBO Long-Term Fellowship (ALTF 241-2017; to N. Fortenly), New Frontiers Group award of the Austrian Academy of Sciences and European Research Council (ERC) Starting Grant (European Union’s Horizon 2020 Research and Innovation Programme, grant agreement no. 679146; to C. Bock), clinical investigator grant by St. Anna Kinderkrebsforschung and Histiocytosis Association research grant (to C. Hutter).
Financial Support:
F.H.: DFG Research Fellowship (HA 7723/1-1). M.F.: Innovation Fund of the Austrian Academy of Sciences (IF_2015_36). M.F. and C.B.: Austrian Science Fund (FWF) Special Research Programme grant (FWF SFB F 6102-B21). N.F.: EMBO Long-Term Fellowship (ALTF 241-2017). C.B.: New Frontiers Group award of the Austrian Academy of Sciences; European Research Council (ERC) Starting Grant (European Union’s Horizon 2020 Research and Innovation Programme, grant agreement n° 679146). C.H.: Clinical investigator grant by St. Anna Kinderkrebsforschung; Histiocytosis Association research grant.
Footnotes
Data and code availability
Processed single-cell RNA-seq and ATAC-seq data are openly available via Gene Expression Omnibus (GEO, accession: GSE133706). Raw sequencing reads are available as controlled access via the European Genome-Phenome Archive (EGA, accession number: EGAS00001003822) to safeguard patient privacy. Additional materials including interactive network visualizations and genome browser tracks provided on the Supplementary Website: http://LCH-hierarchy.computational-epigenetics.org.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors’ Contributions
Conception and design: F. Halbritter, M. Farlik, C. Bock, C. Hutter Development of methodology: F. Halbritter, M. Farlik, L.C. Schuster, C. Bock, C. Hutter
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): M. Farlik, R. Schwentner, G. Jug, T. Schnoller, A. Reinprecht, T. Czech, J. Gojo, W. Holter, M. Minkov, W.M. Bauer, I. Simonitsch-Klupp, C. Hutter
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): F. Halbritter, M. Farlik, R. Schwentner, N. Fortelny, T. Schnoller, H. Pisa, I. Simonitsch-Klupp, C. Hutter
Writing, review, and/or revision of the manuscript: F. Halbritter, M. Farlik, N. Fortelny, L.C. Schuster, M. Minkov, W.M. Bauer, I. Simonitsch-Klupp, C. Bock, C. Hutter
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): F. Halbritter, R. Schwentner, G. Jug, T. Schnoller, J. Gojo, M. Minkov, W.M. Bauer, C. Hutter
Study supervision: C. Bock, C. Hutter
References
- 1.Allen CE, Merad M, McClain KL. Langerhans-Cell Histiocytosis. N Engl J Med. 2018;379:856–868. doi: 10.1056/NEJMra1607548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Emile JF, et al. Revised classification of histiocytoses and neoplasms of the macrophage-dendritic cell lineages. Blood. 2016;127:2672–2681. doi: 10.1182/blood-2016-01-690636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doebel T, Voisin B, Nagao K. Langerhans Cells - The Macrophage in Dendritic Cell Clothing. Trends Immunol. 2017;38:817–828. doi: 10.1016/j.it.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 4.Durham BH, et al. Functional evidence for derivation of systemic histiocytic neoplasms from hematopoietic stem/progenitor cells. Blood. 2017;130:176–180. doi: 10.1182/blood-2016-12-757377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mass E, et al. A somatic mutation in erythro-myeloid progenitors causes neurodegenerative disease. Nature. 2017;549:389–393. doi: 10.1038/nature23672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Milne P, et al. Hematopoietic origin of Langerhans cell histiocytosis and Erdheim-Chester disease in adults. Blood. 2017;130:167–175. doi: 10.1182/blood-2016-12-757823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakraborty R, et al. Mutually exclusive recurrent somatic mutations in MAP2K1 and BRAF support a central role for ERK activation in LCH pathogenesis. Blood. 2014;124:3007–3015. doi: 10.1182/blood-2014-05-577825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diamond EL, et al. Diverse and targetable kinase alterations drive histiocytic neoplasms. Cancer Discov. 2016;6:154–165. doi: 10.1158/2159-8290.CD-15-0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allen CE, Parsons DW. Biological and clinical significance of somatic mutations in Langerhans cell histiocytosis and related histiocytic neoplastic disorders. Hematology. 2015;2015:559–564. doi: 10.1182/asheducation-2015.1.559. [DOI] [PubMed] [Google Scholar]
- 10.Braier J. Is Langerhans cell histiocytosis a neoplasia? Pediatr Blood Cancer. 2017;64:e26267. doi: 10.1002/pbc.26267. [DOI] [PubMed] [Google Scholar]
- 11.Degar BA, Rollins BJ. Langerhans cell histiocytosis: malignancy or inflammatory disorder doing a great job of imitating one? Dis Model Mech. 2009;2:436–439. doi: 10.1242/dmm.004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haroche J, et al. Histiocytoses: emerging neoplasia behind inflammation. Lancet Oncol. 2017;18:e113–e125. doi: 10.1016/S1470-2045(17)30031-1. [DOI] [PubMed] [Google Scholar]
- 13.Senechal B, et al. Expansion of regulatory T cells in patients with langerhans cell histiocytosis. PLoS Med. 2007;4:1374–1384. doi: 10.1371/journal.pmed.0040253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allen CE, et al. Cell-Specific Gene Expression in Langerhans Cell Histiocytosis Lesions Reveals a Distinct Profile Compared with Epidermal Langerhans Cells. J Immunol. 2010;184:4557–4567. doi: 10.4049/jimmunol.0902336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hutter C, et al. Notch is active in Langerhans cell histiocytosis and confers pathognomonic features on dendritic cells. Blood. 2012;120:5199–5208. doi: 10.1182/blood-2012-02-410241. [DOI] [PubMed] [Google Scholar]
- 16.Willman CL, et al. Langerhans’-Cell Histiocytosis (Histiocytosis X) -- A Clonal Proliferative Disease. N Engl J Med. 1994;331:154–160. doi: 10.1056/NEJM199407213310303. [DOI] [PubMed] [Google Scholar]
- 17.Yu RC, Chu C, Buluwela L, Chu AC. Clonal proliferation of Langerhans cells in Langerhans cell histiocytosis. Lancet (London, England) 1994;343:767–8. doi: 10.1016/s0140-6736(94)91842-2. [DOI] [PubMed] [Google Scholar]
- 18.Banerji CRS, et al. Cellular network entropy as the energy potential in Waddington’s differentiation landscape. Sci Rep. 2013;3:3039. doi: 10.1038/srep03039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo M, Bao EL, Wagner M, Whitsett JA, Xu Y. SLICE: Determining cell differentiation and lineage based on single cell entropy. Nucleic Acids Res. 2017;45 doi: 10.1093/nar/gkw1278. gkw1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi J, Teschendorff AE, Chen W, Chen L, Li T. Quantifying Waddington’s epigenetic landscape: a comparison of single-cell potency measures. Brief Bioinform. 2018 doi: 10.1093/bib/bby093. [DOI] [PubMed] [Google Scholar]
- 21.Teschendorff AE, Enver T. Single-cell entropy for accurate estimation of differentiation potency from a cell’s transcriptome. Nat Commun. 2017;8 doi: 10.1038/ncomms15599. 15599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Villablanca EJ, Mora JR. A two-step model for Langerhans cell migration to skin-draining LN. Eur J Immunol. 2008;38:2975–2980. doi: 10.1002/eji.200838919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Villani A-C, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356:eaah4573. doi: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwentner R, et al. Longitudinal assessment of peripheral blood BRAFV600E levels in patients with Langerhans cell histiocytosis. Pediatr Res. 2018;85:856–864. doi: 10.1038/s41390-018-0238-y. [DOI] [PubMed] [Google Scholar]
- 25.Hieronymus T, Zenke M, Baek JH, Seré K. The clash of Langerhans cell homeostasis in skin: Should I stay or should I go? Semin Cell Dev Biol. 2015;41:30–38. doi: 10.1016/j.semcdb.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 26.Konradi S, et al. Langerhans cell maturation is accompanied by induction of N-cadherin and the transcriptional regulators of epithelial-mesenchymal transition ZEB1/2. Eur J Immunol. 2014;44:553–560. doi: 10.1002/eji.201343681. [DOI] [PubMed] [Google Scholar]
- 27.Murakami I, et al. IL-17A receptor expression differs between subclasses of Langerhans cell histiocytosis, which might settle the IL-17A controversy. Virchows Arch. 2013;462:219–228. doi: 10.1007/s00428-012-1360-6. [DOI] [PubMed] [Google Scholar]
- 28.Murakami I, et al. Interleukin-1 loop model for pathogenesis of Langerhans cell histiocytosis. Cell Commun Signal. 2015;13:13. doi: 10.1186/s12964-015-0092-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Javierre BM, et al. Lineage-Specific Genome Architecture Links Enhancers and Non-coding Disease Variants to Target Gene Promoters. Cell. 2016;167:1369–1384.e19. doi: 10.1016/j.cell.2016.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheffield NC, Bock C. LOLA: Enrichment analysis for genomic region sets and regulatory elements in R and Bioconductor. Bioinformatics. 2015;32:587–589. doi: 10.1093/bioinformatics/btv612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kulakovskiy IV, et al. HOCOMOCO: Towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic Acids Res. 2018;46:D252–D259. doi: 10.1093/nar/gkx1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Musa J, Aynaud M-M, Mirabeau O, Delattre O, Grünewald TG. MYBL2 (B-Myb): a central regulator of cell proliferation, cell survival and differentiation involved in tumorigenesis. Cell Death Dis. 2017;8:e2895. doi: 10.1038/cddis.2017.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee J, et al. Lineage specification of human dendritic cells is marked by IRF8 expression in hematopoietic stem cells and multipotent progenitors. Nat Immunol. 2017;18:877–888. doi: 10.1038/ni.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy TL, Tussiwand R, Murphy KM. Specificity through cooperation: BATF-IRF interactions control immune-regulatory networks. Nat Rev Immunol. 2013;13:499–509. doi: 10.1038/nri3470. [DOI] [PubMed] [Google Scholar]
- 36.Sichien D, et al. IRF8 Transcription Factor Controls Survival and Function of Terminally Differentiated Conventional and Plasmacytoid Dendritic Cells, Respectively. Immunity. 2016;45:626–640. doi: 10.1016/j.immuni.2016.08.013. [DOI] [PubMed] [Google Scholar]
- 37.Björk JK, et al. Heat-shock factor 2 is a suppressor of prostate cancer invasion. Oncogene. 2016;35:1770–1784. doi: 10.1038/onc.2015.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ren F, et al. MYBL2 is an independent prognostic marker that has tumor-promoting functions in colorectal cancer. Am J Cancer Res. 2015;5:1542–1552. [PMC free article] [PubMed] [Google Scholar]
- 39.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 40.Bernstein BE, et al. A Bivalent Chromatin Structure Marks Key Developmental Genes in Embryonic Stem Cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 41.O’Carroll D, et al. The Polycomb-Group Gene Ezh2 Is Required for Early Mouse Development. Mol Cell Biol. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valk-Lingbeek ME, Bruggeman SWM, Van Lohuizen M. Stem cells and cancer: The polycomb connection. Cell. 2004;118:409–418. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 43.Langlais D, Barreiro LB, Gros P. The macrophage IRF8/IRF1 regulome is required for protection against infections and is associated with chronic inflammation. J Exp Med. 2016;213:585–603. doi: 10.1084/jem.20151764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berres ML, Allen CE, Merad M. Pathological Consequence of Misguided Dendritic Cell Differentiation in Histiocytic Diseases. Adv Immunol. 2013;120:127–161. doi: 10.1016/B978-0-12-417028-5.00005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rust R, et al. Gene expression analysis of dendritic/Langerhans cells and Langerhans cell histiocytosis. J Pathol. 2006;209:474–483. doi: 10.1002/path.2003. [DOI] [PubMed] [Google Scholar]
- 46.Zitvogel L, Pitt JM, Daillère R, Smyth MJ, Kroemer G. Mouse models in oncoimmunology. Nat Rev Cancer. 2016;16:759–773. doi: 10.1038/nrc.2016.91. [DOI] [PubMed] [Google Scholar]
- 47.Swierczek SI, et al. Hematopoiesis is not clonal in healthy elderly women. Blood. 2008;112:3186–3193. doi: 10.1182/blood-2008-03-143925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boudewijns M, van Dongen JJM, Langerak AW. The human androgen receptor X-chromosome inactivation assay for clonality diagnostics of natural killer cell proliferations. J Mol Diagnostics. 2007;9:337–344. doi: 10.2353/jmoldx.2007.060155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teschendorff AE, Morabito SJ, Kessenbrock K, Meyer K. Integrated single-cell potency and expression landscape in mammary epithelium reveals novel bipotent-like cells associated with breast cancer risk. bioRxiv. 2018 doi: 10.1101/496471. 496471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The SVA package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ritchie ME, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Corces MR, et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods. 2017;14:959–962. doi: 10.1038/nmeth.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang H, Lei R, Ding SW, Zhu S. Skewer: A fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics. 2014;15:182. doi: 10.1186/1471-2105-15-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9 doi: 10.1186/gb-2008-9-9-r137. R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:55. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen EY, et al. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grant CE, Bailey TL, Noble WS. FIMO: Scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.