Abstract
Methamphetamine (MA) use is a major public health problem in the United States, especially among people living with HIV (PLWH). Many MA-induced neurotoxic effects are mediated by inflammation and gut microbiota may play a role in this process, yet the effects of MA on the microbiome have not been adequately explored. Therefore, we performed 16S rRNA gene sequencing on rectal swab samples from 381 men who have sex with men, 48% of whom were PLWH and 41% of whom used MA. We compared microbiome composition between MA users and non-users while testing for potential interactions with HIV and controlling for numerous confounders using inverse probability of treatment weighting. We found that MA use explained significant variation in overall composition (R2 = 0.005, p = 0.008) and was associated with elevated Finegoldia, Parvimonas, Peptoniphilus, and Porphyromonas and reduced Butyricicoccus and Faecalibacterium, among others. Genera including Actinomyces and Streptobacillus interacted with HIV status, such that they were increased in HIV+ MA users. Finegoldia and Peptoniphilus increased with increasing frequency of MA use, among others. In summary, MA use was associated with a microbial imbalance favoring pro-inflammatory bacteria, including some with neuroactive potential and others that have previously been associated with poor HIV outcomes.
Subject terms: Inflammation, Epidemiology, Preclinical research
Introduction
The 2017 National Survey on Drug Use and Health estimated that nearly one million people in the United States were current users of methamphetamine (MA) or had a MA use disorder in the past year1. MA use is a major public health concern with myriad negative health consequences ranging from anxiety and confusion to psychosis and violent behavior; chronic abuse may even result in severe and lasting structural changes in the brain affecting emotional regulation and cognition2. MA use is much more prevalent among people living with HIV, with rates of recent use 30 times higher and rates of dependence 33 times higher than the general population (0.3% vs. 9% recent use and 0.4% vs. 13% dependence)1,3,4. MA increases susceptibility to HIV infection by altering immune activity5,6 inhibiting neurocognitive processes involved in judgement and decision-making7 and increasing the frequency of risky sex acts4,8. MA use among people living with HIV is associated with reduced likelihood of achieving viral suppression9, faster disease progression10, and increased risk of transmission to others11.
Many MA-induced neurotoxic effects are mediated by inflammation5,12, and the microbiome, which is involved in inducing and regulating the immune system13, may play a role in this process. Exposure to MA impacts both innate and adaptive immunity, increasing production of inflammatory cytokines, inhibiting T-cell proliferation, altering gene expression of immune cells, modifying cytokine signaling pathways, and increasing blood-brain barrier permeability5,14,15. MA damages gut wall integrity and increases intestinal permeability leading to the translocation of microbiota into the body15. This process disrupts symbiotic interactions between the host immune system and microbiota, inducing an immune response that may cyclically exacerbate intestinal permeability and further inflammation16. Microbial translocation has been cited as one of the key drivers of chronic inflammation described in many other diseases, including HIV, and may play a similar role in MA-induced inflammation and toxicity. Furthermore, mediated by inflammation, disruption of gut bacterial communities (termed “dysbiosis”) may be a mechanistic link between methamphetamine use and HIV transmission and disease progression.
Dysbiosis has been described in individuals with substance use disorders17, chronic prescription opioid18 and cocaine users19, as well as people living with HIV20 and those practicing anal intercourse21. We recently showed that MA use was associated with microbiome changes in a small sample of HIV-infected MSM22. However, no large studies into the effects of MA on the microbiome have been completed, and no studies have examined the potential role of HIV in MA-induced dysbiosis. In order to address this gap, we studied the effects of MA on the gastrointestinal microbiome in a cohort of young men who have sex with men (MSM), approximately half of whom were HIV-infected, and all of whom were engaging in anal intercourse. We hypothesized that MA use would be associated with increased relative abundance of pro-inflammatory and pathogenic bacterial taxa as well as alterations to those with neurologic effects. We also hypothesized that MA use and HIV would interact to increase the severity of dysbiosis.
Materials and Methods
Study population
Specimens and data for this study were drawn from a larger cohort, the NIDA-funded Minority Men who have Sex with Men Cohort at UCLA Linking Infections Noting Effects (MASCULINE, or mSTUDY). Subject selection procedures have been previously described23. Briefly, participants were all men born males, aged 18–45, with approximately one-half of the sample purposefully included due to current substance use (the other half non-substance users) and one-half of the sample purposefully included due to HIV-infection (the other half being HIV-negative). The mSTUDY was approved by the UCLA South General Institutional Review Board (IRB), and the current study was approved by the UCLA Medical IRB 1. All participants provided written informed consent prior to participation and all study procedures were done in accordance with ethical principles for human subjects research.
Specimen collection and DNA preparation
As previously described23, samples included in this study were rectal swabs (FLOQSwabs, Copan Diagnostics, Murrieta, CA). The majority (76%) were collected via anoscopy under direct mucosal visualization and without preparatory enema at approximately 8 cm from the anal verge. Due to a protocol change, others (24%) were participant self-collected at approximately 4–5 cm from the anal verge. Collection method was taken into account in the analysis (Table 1; Supplemental content). Swabs were immediately frozen neat at −80 °C until processing in bulk. For DNA processing the samples were transferred to Lysing Matrix E tubes (MP Biomedicals, Burlingame, CA) containing RLT lysis buffer (Qiagen, Hilden, Germany) and bead-beated on a TissueLyser (Qiagen). DNA was then extracted using the AllPrep DNA/RNA/Protein kit (Qiagen) per manufacturer’s protocol.
Table 1.
Participant characteristics, N = 381 men who have sex with men in Los Angeles, CA.
| MA- negative n = 225 mean (sd), median n (%) | MA-positive n = 156 | P d | SMDe (pre, post IPTW) | |
|---|---|---|---|---|
| Age | 30.17 (6.85), 29 | 32.58 (6.75), 33 | <0.001 | 0.35, 0.16 |
| HIV+ | 80 (35.6) | 102 (65.4) | <0.001 | N/A |
| Race/ethnicity | 0.6 | 0.1, 0.07 | ||
| Black-Non Hispanic | 93 (41.3) | 57 (36.5) | ||
| Hispanic | 107 (47.6) | 79 (50.6) | ||
| Other-Non Hispanic | 25 (11.1) | 20 (12.8) | ||
| Homeless in past 6 months | 52 (23.1) | 77 (49.4) | <0.001 | 0.57, 0.28 |
| Had RAI in last 7 days | 102 (45.3) | 65 (41.7) | 0.5 | 0.07, 0.03 |
| Number of RAI acts in past month | 2.09 (4.94), 0 | 2.88 (5.33), 1 | <0.001 | 0.15, 0.03 |
| Number of anal sex partners in past 6 months | 6.17 (7.60), 3 | 8.79 (9.28), 5 | <0.001 | 0.31, 0.18 |
| Positive for STIa | 19 (8.4) | 28 (17.9) | 0.006 | 0.28, 0.18 |
| Marijuana use in past 6 months | 0.1 | 0.23, 0.21 | ||
| Daily/Weekly | 69 (30.7) | 60 (38.5) | ||
| Monthly/less | 52 (23.1) | 41 (26.3) | ||
| Never | 104 (46.2) | 55 (35.3) | ||
| Cocaine use in past 6 months | 40 (17.8) | 60 (38.5) | <0.001 | 0.47, 0.24 |
| Tobacco smoker | 73 (32.4) | 95 (60.9) | <0.001 | 0.6, 0.38 |
| Binge drinking in past 6 monthsb | 138 (61.3) | 91 (58.3) | 0.6 | 0.06, 0.04 |
| Antibiotic use in past month | 15 (6.7) | 16 (10.3) | 0.2 | 0.13, 0.07 |
| Sample collection strategy | 0.5 | 0.07, 0.01 | ||
| Anoscopy | 169 (75.1) | 122 (78.2) | ||
| Self-collected | 56 (24.9) | 34 (21.8) | ||
| Type of ART | <0.001 | 0.56, 0.28 | ||
| INSTI + NRTI | 30 (13.3) | 39 (25.0) | ||
| NNRTI + NRTI | 25 (11.1) | 23 (14.7) | ||
| NRTI + PI | 15 (6.7) | 15 (9.6) | ||
| Other | 4 (1.8) | 12 (7.7) | ||
| HIV+ and missing ART data | 6 (2.7) | 14 (9.0) | ||
| HIV− pre-exposure prophylaxis (PrEP) user | 30 (13.3) | 7 (4.5) | ||
| HIV−, no PrEP | 115 (51.1) | 47 (30.1) | ||
| Among HIV+ participants only | ||||
| HIV RNA log10 copies/mL (median, IQR) c | 1.03 (0.7) | 1.03 (1.7) | N/A | |
| CD4 cells/mm3 (median, IQR)c | 590.5 (267) | 635 (424.3) | N/A | |
| CD4 cells/mm3 <200 | 5 (2.2) | 9 (5.8) | 0.18, 0.15 |
MA = Methamphetamine; SMD = Standardized mean difference; RAI = Receptive anal intercourse; STI = Sexually transmitted infection; ART = Antiretroviral therapy; INSTI = Integrase strand transfer inhibitor; NRTI = Nucleoside reverse transcriptase inhibitor; NNRTI = Non-nucleoside reverse transcriptase inhibitor; PI = Protease inhibitor.
aSexually transmitted infections include rectal gonorrhea, rectal chlamydia as well as primary/secondary syphilis.
bBinge drinking defined as 6 or more drinks on one occasion.
cHIV status, HIV RNA, and continuous CD4 cell count were not included in the inverse probability of treatment weight model, all other variables in the table were included. HIV status was taken into account in the analyses by stratifying on it (if there was evidence for an interaction between MA and HIV) or conditioning on it (if there was no evidence for an interaction).
dp values are from Wilcoxon tests or Chi-square tests.
eSMD is a measure of imbalance across groups; higher SMDs indicate greater imbalance. Average SMD before weighting = 0.28, after weighting = 0.14.
16S rRNA gene sequencing and data processing
Microbiome profiling was performed by sequencing of the V4 region of the 16S rRNA gene as previously described23–25. Briefly, the V4 region was amplified in triplicate reactions using Golay-barcoded primers 515F/806R. PCR products were then pooled and sequenced on the Illumina MiSeq platform using 2 × 150 bp v2 chemistry. The sequences were demultiplexed with Golay error correction using QIIME v1.9.126, and Divisive Amplicon Denoising Algorithm (DADA2) version 1.8 was used for error correction, exact sequence inference, read merging, and chimera removal27. The resultant amplicon sequence variant (ASV) table comprised 19,955,039 total merged read pairs (mean per sample = 52,375; range 10,906 to 124,889). Taxonomic assignment was performed using RDP trainset 16 (10.5281/zenodo.810827). Rarefaction was performed at a depth of 10,906 reads for alpha diversity analyses. To normalize all other analyses, estimates of relative library sizes (“size factors”) were obtained by calculating geometric means of pairwise read count ratios28.
Measurement of MA use
MA use was measured using an adapted version of the NIDA-modified ASSIST29. Participants were asked how often they used MA in the previous six months; response choices were “Daily”, “Weekly”, “Monthly”, “Less often”, “Once”, and “Never.” For most analyses we categorized participants as MA users if they indicated any use in the past six months, and non-users if they responded “Never.” For the dose-response analysis, we combined “Monthly”, “Less often”, and “Once” into “Monthly or less often,” given that infrequent exposures are likely to have similar effects on the microbiome. In addition, participants were screened for MA use via urinalysis [Fastect® II (Branan Medical Corporation); iScreen® Dip Card (Alere)]. We did not use the urinalysis results as our primary exposure variable because the detection window for MA is 48–72 hours and no exposure quantification (and thus dose-response analysis) could be done. We instead compared the self-report and urinalysis results as a sensitivity analysis (Supplementary Figs S4, S5 and S6). HIV status was ascertained upon entry into the cohort by medical record review (for known HIV-infected participants) or the OraQuick Advance® HIV 1/2 (OraSure Technologies, Bethlehem, PA).
Behavioral and clinical covariates
Analyses controlled for a large set of behavioral and clinical covariates including age, race/ethnicity, homelessness in past six months, number of receptive anal intercourse (RAI) acts in past month, number of anal sex partners in past six months, an indicator for RAI in the past seven days, an indicator for a positive STI test (including PCR tests for rectal gonorrhea and chlamydia and serology for primary or secondary syphilis), self-reported use of marijuana and cocaine, tobacco smoking, and binge drinking (defined as 6+ drinks on more than one occasion) in the past six months, and use of antibiotics in the past month. We also controlled for type of antiretroviral therapy (including use of pre-exposure prophylaxis if HIV−) and an indicator for CD4 cell count <200 (Table 1). Measures and assays have been previously described23.
Statistical analyses
We compared clinical and behavioral characteristics between MA users and non-users using descriptive statistics, Wilcoxon or Chi-square tests, and standardized mean differences (see Table 1; Supplemental content). All analyses of microbiome outcomes were adjusted for clinical and behavioral confounders using inverse probability of treatment weighting (IPTW). IPTW is a technique in which the study sample is re-weighted to achieve balance between exposure groups (here, MA users vs. non-users) on important covariates so that their confounding effects are substantially reduced30. Covariates included in the IPTW model are listed in Table 1, and further information about the IPTW calculation and adjustment process is available in the supplemental content as well as our previous publication23. Weights were estimated using generalized boosted models (R package ‘twang’31) and robust standard errors for IPTW-adjusted analyses were obtained using the sandwich estimator (R package ‘sandwich’32). All analyses in this study proceeded by first testing for interactions between MA use and HIV status using multiplicative interaction terms. An alpha level of 0.1 was used as a cutoff for significance of interaction tests; if significant, comparisons of MA users vs. non-users are presented stratified by HIV status (retaining HIV status and the interaction term in the model). If no significant interaction was detected, comparisons of MA users vs. non-users were completed controlling for HIV status (retaining HIV status as a covariate but dropping the interaction term).
The R package ‘phyloseq’33 was used to calculate distance matrices, alpha diversity metrics, and for ordination. Permutational multivariate ANOVA (PERMANOVA; R package ‘vegan’34) was used to test for differences in overall microbial composition. Cluster analysis was performed using the partitioning around mediods method with the optimal number of clusters chosen according to the Calinski-Harabasz statistic (R packages ‘cluster’35 and ‘fpc’36). Logistic regression was used to assess the relationship between MA use and cluster membership. Linear regression was used to test for differences in alpha diversity between MA users and non-users. Zero-inflated negative binomial (ZINB) models were used to test for differences in individual bacterial genera between groups (R package ‘pscl’37). We employed a previously described model selection strategy23 to choose the optimal ZINB model for each genus. A pre-filtering step excluded genera appearing in less than 10% of samples as well as those with less than 100 total reads across all samples, resulting in 78 genera included in ZINB analyses. Dose-response analysis was completed by regressing bacterial counts on frequency of MA use using orthogonal polynomial coding of linear and quadratic curves. As sensitivity analyses, all analyses were repeated redefining MA use according to urine drug screen results (except for dose-response, owing to the qualitative nature of urine drug screening). Finally, we examined short-term interactions between MA use and the microbiome using a correlation analysis among individuals testing positive for MA on urine drug screen. Correlations were calculated with the sparCC method38 with bootstrapped standard errors (R package ‘SpiecEasi’39).
PERMANOVA, alpha diversity, and dose-response analyses utilized a threshold of p < 0.05 to determine statistical significance. In order to account for the large amount of genera tested, p values obtained in ZINB analyses and sparCC correlations were corrected using Benjamini & Hochberg’s False Discovery Rate (FDR) method40. FDR-adjusted p values are labelled as q values, and q < 0.1 was used as a threshold to determine statistical significance. Accordingly, we display 90% false coverage rate (FCR)-adjusted confidence intervals41 to accompany these analyses. All statistical analyses were performed using R v.3.5.142 and graphics were generated with ggplot243.
Results
Participant characteristics
This study included 381 participants, 156 MA users (41%) and 225 non-users (59%). All participants were MSM, the mean age was 31, and most were Hispanic (49%) or non-Hispanic Black (39%). One hundred eighty-two participants were HIV+ (48%); sixty-five percent of MA users were HIV+ as compared to 36% of non-users. MA users were also older than non-users, were more likely to have experienced homelessness, had RAI more frequently, had more anal sex partners, were more likely to have recently used cocaine, and were more likely to be tobacco smokers (Table 1).
Effects of MA use on overall microbial composition and diversity
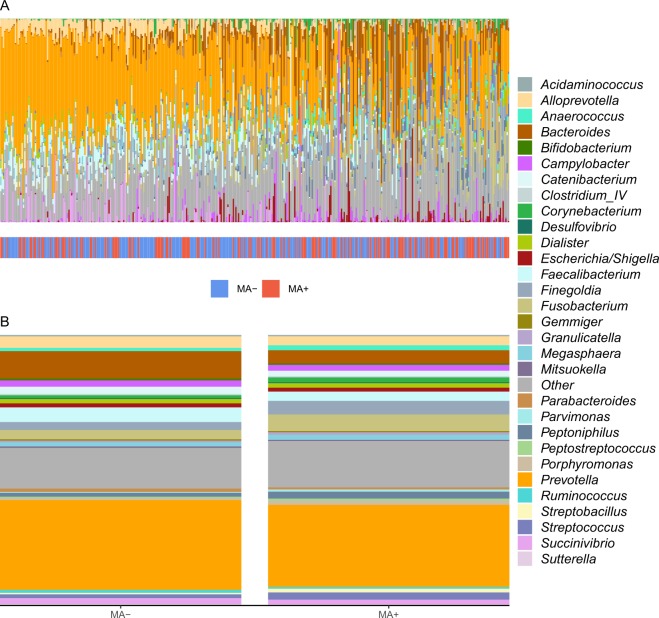
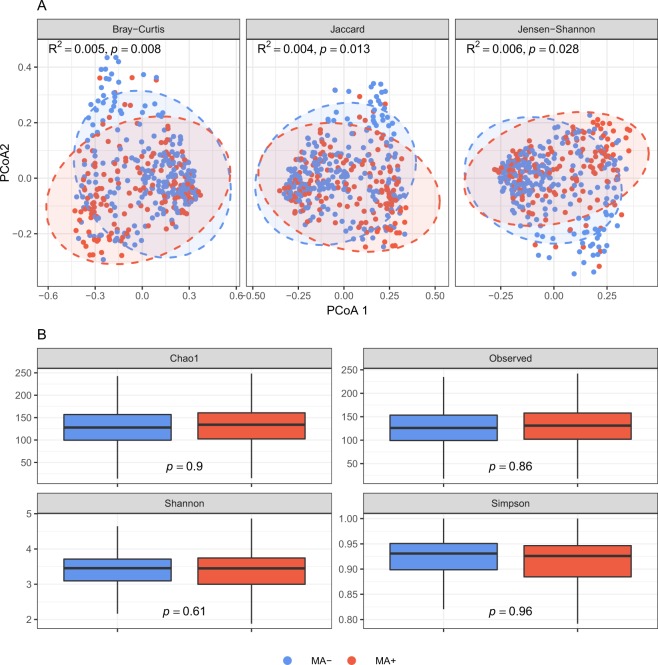
PERMANOVA analyses with Bray-Curtis, Jaccard, and Jensen-Shannon distances did not reveal significant evidence supporting an interaction between MA and HIV on overall microbial composition (all p > 0.1; interaction p values supplied in Supplementary Table S1). Therefore, we described and compared overall composition between MA users and non-users while controlling for HIV status. Qualitative examination of descriptive barplots suggested increased Finegoldia, Fusobacterium, Peptoniphilus, Porphyromonas, Streptobacillus, and Streptococcus and decreased Bacteroides, Faecalibacterium, and Succinivibrio in MA users compared to non-users (Fig. 1A,B). Ordination of Bray-Curtis, Jaccard, and Jensen-Shannon distances by principal coordinates analyses revealed clustering by MA status (Fig. 2A). These findings were supported by PERMANOVA analyses showing that MA explained significant variation in overall microbial composition (Bray-Curtis R2 = 0.005, p = 0.008; additional results in Fig. 2A). Additionally, a clustering analysis using partitioning around medoids of Bray-Curtis distances revealed two distinct clusters in the microbiome data, which were significantly related to MA use. Adjusted for HIV status and our behavioral and clinical covariate set, the likelihood of classification into the second cluster was 2.24 times higher for MA users compared to non-users (OR = 2.24, p = 0.011, pseudo-R2 = 0.008).
Figure 1.
Rectal microbial composition of study participants, N = 381. (A) Columns represent the relative composition of each subject’s microbiome at the genus level. Methamphetamine (MA) use by the subjects is indicated by a colored line below their composition. Subjects are ordered by the first principal coordinate of a Bray-Curtis pairwise distance matrix. Genera representing less than 1% of the composition on average across samples were combined into “Other.” (B) Average microbial composition within each MA use group. Bacterial genera representing less than 1% of the overall relative composition or present in less than 10% of the samples were grouped into “Other.”
Figure 2.
Associations between methamphetamine (MA) use and overall microbial composition and diversity. (A) Ordination of the samples using principal coordinates analysis. PCoA = Principal coordinate axis. Ellipses are 95% confidence regions for each group assuming points follow a multivariate t distribution. R2 and p values are from PERMANOVA analyses of distance metrics. (B) Boxplots of diversity metrics. Boxes represent the inverse probability of treatment weight-adjusted lower, median, and upper quartiles of the data and whiskers are 1.5*interquartile range. p values are from IPTW-adjusted linear regression analyses comparing diversity metrics between MA users and non-users.
No significant interactions between MA and HIV were detected in observed diversity or Chao1, Shannon, or Simpson indices (all p > 0.1; Supplementary Table S1), and no differences in diversity were detected between MA users and non-users in any metric (Fig. 2B). Despite lack of evidence for an interaction between HIV and MA use, we display descriptive, ordination and alpha diversity plots stratified by HIV status in Supplementary Figs S1 and S2.
Effects of MA use on specific genera
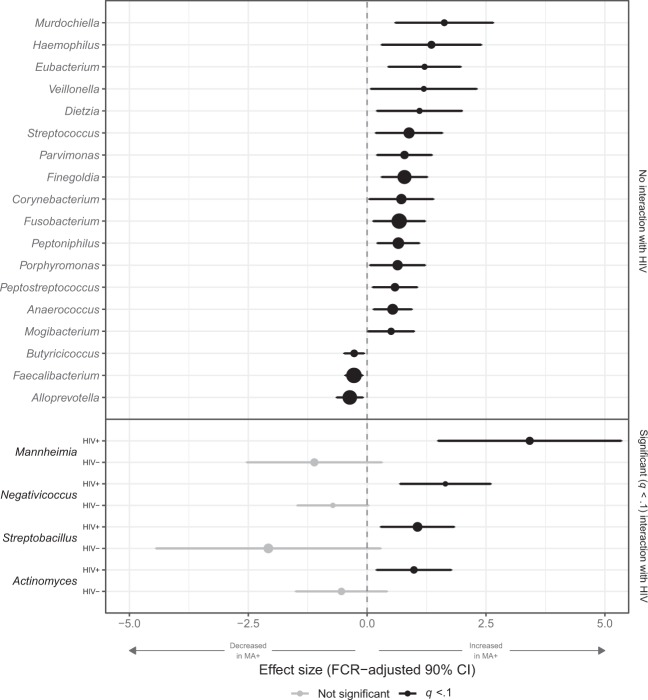
Using ZINB models with IPTW adjustment, we found differences between MA users and non-users in multiple genera. For some, there was no evidence for an interaction between MA and HIV: Regardless of HIV status, MA users had higher levels of Finegoldia, Fusobacterium, Parvimonas, Peptoniphilus, Peptostreptococcus, and Porphyromonas, and lower levels of Butyricicoccus and Faecalibacterium, among others (all q < 0.1; Fig. 3; estimates and q values supplied in Supplementary Table S2). For four genera, a significant (q < 0.1; interaction q values supplied in Supplementary Table S2) interaction between HIV and MA was detected. Actinomyces, Mannheimia, Negativicoccus, and Streptobacillus were increased in HIV+ MA users compared to HIV+ non-users, but no difference was found in the absence of HIV. No genera were significant only in the HIV− stratum.
Figure 3.
Comparisons of individual genera between methamphetamine (MA) users and non-users. Forest plots of results of zero-inflated negative binomial models comparing genus-level bacterial counts between methamphetamine (MA) users and non-users. Inverse probability of treatment-weighted effect sizes (log normalized count ratios) and false coverage rate (FCR)-adjusted 90% confidence intervals are plotted, with statistical significance (q < 0.1) indicated in black. Genera with no effect are not shown. Dots are sized proportionally to overall mean abundance across samples, i.e., genera with larger dots are, on average, more abundant.
Dose-response analysis of bacterial counts on increasing frequency of MA use
Of the 156 MA users in the study, 40 were daily users, 35 used weekly, and 81 used monthly or less often. Counts of Anaerococcus, Corynebacterium, Dietzia, Finegoldia, Mannheimia and Peptoniphilus increased linearly with increasing frequency of MA use (q < 0.1 for test of linear trend). No significant quadratic dose-response curves were noted in any genera (Fig. S3).
Sensitivity analysis using urine toxicology screening to define MA use
Our findings, which were based on participant self-report of MA use, were consistent when we re-defined MA use using urine drug screening results. All participants completed urine drug screening, and fourteen percent of study participants (n = 52) tested positive for MA including 3 individuals who self-reported no MA use (49/52 who tested positive also self-reported using MA). One hundred seven self-reported using MA, but tested negative, likely because their last use was outside the drug screen window of detection. HIV+ participants were more likely to have a positive MA urine drug screen (21% among HIV+ vs. 7% among HIV−, p < 0.001).
In biomarker analyses, MA use was still a significant driver of variation in overall microbial composition (Bray-Curtis R2 = 0.008, p = 0.008), and no differences in alpha diversity were noted between MA users and non-users. Many of the same genera were elevated in MA users, e.g., Finegoldia, Fusobacterium, Peptoniphilus, Peptostreptococcus, and Porphyromonas, and elevations in Streptobacillus in the HIV+ stratum were noted in both analyses. Depletion in Faecalibacterium was consistent across analyses; the biomarker analysis also identified depletions in Clostridium cluster XI and Lactobacillus in MA users. Results from this sensitivity analysis are presented in Supplementary Figs S4–S6.
Finally, to examine short-term interactions between MA use and the microbiome, we performed a genera correlation analysis among individuals testing positive for recent MA use. Large, significant positive correlations were noted between Parvimonas and Campylobacter as well as Peptoniphilus and Campylobacter, among others (sparCC correlation >0.6 and q < 0.1). Moderate-to-large negative correlations were noted between Alloprevotella and Butyricicoccus as well as Parabacteroides and Campylobacter, among others (sparCC correlation < −0.4 and q < 0.1). Full results are presented in Supplemental Table S3 and Fig. S7.
Discussion
This study of 381 MSM who were either HIV-infected or at high risk for HIV acquisition found that MA use significantly impacted gut microbial composition after controlling for multiple clinical and behavioral confounders. Measures of overall composition were altered by MA use, but measures of diversity were not, and the associations between MA and overall composition and diversity did not depend on participants’ HIV status. Several genera were increased in MA users regardless of HIV status, many of them considered pro-inflammatory and pathogenic, while others were increased among HIV+ participants only. We found that the abundance of some pro-inflammatory taxa increased and commensals decreased with increasing frequency of MA use. Finally, we were able to replicate our findings using a biomarker confirming recent MA use (urine drug screen). MA effect sizes were slightly larger in the biomarker analysis, likely because the window of detection for MA is short, making frequent users more likely to test positive. Our analyses utilized a novel method of confounder control, IPTW, to account for several factors that have previously been associated with dysbiosis (e.g. RAI21, cocaine use19, and alcohol use44), making our findings more likely to be truly attributable to MA use.
Although little research has been done on the effects of MA on the microbiome, our results are mostly consistent with previously published literature. A study of 37 HIV+ individuals from the same cohort as the current data22 (none of the individuals in this current sample were included in the previous study) reported a MA effect size (PERMANOVA R2) of 0.1, larger than the effect we found. As in our study, there were no significant differences in diversity between MA users and non-users. Enrichment in Porphyromonas in MA users was consistent across studies. As a well-known modifier of inflammatory cytokines and a potential cause of intestinal permeability45, Porphyromonas may play a role in MA-associated inflammation and deserves further investigation. Another study in which MA was administered to rats reported an overall effect of MA that is consistent with our findings (R2 of 0.008)46. This study also reported higher alpha diversity in the MA-conditioned group, which was not replicated in our study, and taxonomic differences that do not overlap with our findings, likely because of differences between the mouse model and a human cohort. Finally, a study comparing individuals with substance use disorders (SUDs) to healthy controls found a large effect of SUD (R2 of 0.067), higher observed diversity among individuals with SUDs, and differences in specific genera that do not match our findings17. However, participants with MA use disorder only accounted for 30% of the SUD group, and the study did not control for large differences in lifestyle and clinical confounders between individuals with SUDs and healthy controls.
MA use is associated with increases in production and alterations in gene expression of many pro-inflammatory cytokines6,47, which may contribute to neurological deficits, anxiety, and impaired memory48. Many of the bacterial genera that were elevated in MA users, such as Porphyromonas45, Veillonella49, and Fusobacterium50 have been correlated with increases in pro-inflammatory cytokines. MA also exacerbates systemic inflammation by damaging gastrointestinal barrier integrity and inducing permeability, allowing the translocation of microbes and microbial products into the body. Our study identified depletions in the butyrate-producing genera Faecalibacterium and Butyricicoccus in MA users, which have been inversely correlated with biomarkers of microbial translocation51,52. A study of patients with alcohol use disorder showed that those with higher levels of gut permeability had lower levels of Bifidobacterium and Faecalibacterium species and exhibited more symptoms of alcohol dependence and cravings53.
Emerging preclinical research has demonstrated a complex interplay between the microbiome and the central nervous system54, leading to inquiries about the role of dysbiosis in addiction pathology. Gut bacteria produce neuroactive substances, including serotonin, epinephrine and dopamine55, which may access the brain’s reward centers via gut-innervating vagal neurons56. Streptococcus, which was elevated in MA users in our study, can produce serotonin and Lactobacillus, which was depleted in MA users (in our biomarker analysis), can produce gamma-aminobutyric acid (GABA)55. Laboratory experiments of other drugs of abuse have suggested a connection between dysbiosis and addiction pathology. For example, increased sensitivity to cocaine reward through alterations in dopaminergic pathways has been observed in mice with experimentally disrupted microbiome57. Another study showed that manipulation of the microbiome resulted in several characteristics of opioid dependence in mice, such as reduced opioid analgesic potency and impaired reward behavior58. In addition, there is some preclinical evidence that “repairing” the microbiome might alleviate addiction pathology, e.g., Lactobacillus species restored chemically-depressed dopamine levels in the prefrontal cortex when administered to rats as a probiotic59. However, these studies have not involved MA, and the preclinical connection between dysbosis and MA dependence is speculative.
In humans, potential mechanisms linking dysbiosis to the pathology of MA dependence remain largely unexplored. There is limited clinical evidence linking dysbiosis with symptoms associated with MA dependence, such altered stress response and increased depression. Butyrate-producing Faecalibacterium, which was decreased in MA users in our study, has been associated with reduced depression and higher quality of life60. Common symptoms of MA withdrawal, including depression, anxiety, and fatigue, have been correlated with imbalances in gut microbiota61, and probiotics have been used to successfully reduce anxiety and depression in clinical trials62. It is plausible that targeting dysbiosis may ease these symptoms among individuals undergoing treatment for MA use disorders.
We also found a number of genera that were impacted by MA which have previously been shown to play a role in HIV acquisition, disease progression, and pathogenesis. Increased abundances of Finegoldia and Peptoniphilus in the penile microbiome have been associated with elevated risk for HIV seroconversion in men63, and Parvimonas has been shown to increase genital tract inflammation64 and the risk of HIV acquisition in women65. The implications of enrichment of these bacteria in the rectal microbiome have not been explored; however, it is likely that inflammation exacerbated by dysbiosis underlies the increase in seroconversion risk, which would be highly relevant to at-risk MSM. We also found that MA use increased Fusobacterium, which has been correlated with decreased CD4+ T-cell count and increased T-cell activation in HIV+ individuals as well as reduced T-cell recovery following ART initiation66. HIV and MA interacted to multiplicatively increase Actinomyces, which may play a role in reactivating HIV in latently infected cells67. Parvimonas and Peptostreptococcus are oral pathogens that have been implicated in periodontal infections among HIV+ individuals68,69, and their damaging effects may be heightened in MA users due to MA’s proclivity to reduce saliva production. Finally, increased abundance of Veillonella has been linked with HIV-associated pulmonary diseases70. Our study, showing that MA impacted the relative abundance of each of these genera, may highlight mechanisms underlying the relationship between MA use, HIV acquisition and transmission risk, and HIV disease progression which warrant further investigation.
Our results should be interpreted considering the following limitations. Primarily, no diet data is collected for this cohort. We controlled for race/ethnicity and homelessness, which may impact diet and thus mitigate the effects of this limitation; however, we were unable to fully account for the effects of diet in our analyses. Using IPTW, our study accounts for a plethora of other clinical and behavioral confounders which may have masked true findings or generated spurious associations in previous studies. However, IPTW cannot achieve perfect balance between exposure groups in real-world research applications, and thus we cannot rule out residual confounding even by variables included in our analyses. Our study was also conducted in a cohort comprised entirely of MSM, all of whom were practicing anal intercourse, which increases internal validity by eliminating the effects of some important confounders (e.g. gender, sexual behavior). However, this may limit the generalizability of our findings to other groups, such as women who use MA. Finally, because the ability of 16 S gene sequencing to identify bacterial species is limited, we conducted our analyses at the genus level. We caution that differences in genera do not necessarily correspond to differences in functionally important species.
MA use remains a significant public health challenge, especially among people living with HIV. Our study found that MA use was associated with an imbalance in gut microbial composition favoring pro-inflammatory, potentially pathogenic bacteria, including some with neuroactive potential. There is currently no accepted pharmaceutical treatment for MA use disorder and limited evidence for the effectiveness of cognitive-behavioral therapy; further research into changes in the microbiome associated with MA use may inform therapeutic approaches for individuals with MA use disorder. Moreover, increases in multiple taxa that have been previously associated with poor HIV outcomes or HIV transmission and acquisition are particularly concerning in our study population of MSM who were either HIV-infected or at high risk for infection. Additional investigation into the mechanisms underlying these associations may improve HIV prognosis and prevent future infections among this vulnerable group.
Supplementary information
Acknowledgements
This work was supported by the National Institute on Drug Abuse (1R36 DA046310 and 2U01 DA036267) and the UCLA Center for HIV Identification, Prevention, and Treatment Services (CHIPTS; National Institute of Mental Health P30 MH58107). Additional support provided by the UCLA AIDS Institute and UCLA CFAR Microbiome and Mucosal Immunology Core (P30 AI028697). J.A.F. was supported in part by National Institute of Allergy and Infectious Diseases (NIAID) (K08 AI124979). Overall support for the International Maternal Pediatric Adolescent AIDS Clinical Trials Group (IMPAACT) was provided by the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH) under Award Numbers UM1AI068632 (IMPAACT LOC), UM1AI068616 (IMPAACT SDMC) and UM1AI106716 (IMPAACT LC), with co-funding from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) and the National Institute of Mental Health (NIMH). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author Contributions
R.R.C., J.F., N.H.T., F.L., M.J., R. Brookmeyer, G.M.A. and P.M.G. were responsible for the study concept and design. S.S., R. Bolan, and P.M.G. were instrumental in data and sample acquisition and J.F., N.H.T., C.W. and G.M.A. performed the 16S gene sequencing. F.L. and D.L. performed the bioinformatic and genomic analyses, R.R.C. and R. Brookmeyer performed the statistical analyses, and all authors assisted with interpretation of the results. R.R.C. drafted the manuscript and J.F., N.H.T., F.L., M.J., R. Brookmeyer, S.S., G.M.A. and P.M.G. provided critical revision of the manuscript for important intellectual content. All authors critically reviewed content and approved the final version for publication.
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request. All sequencing data has been deposited into BioProject with the accession number PRJNA422134.
Competing Interests
Dr. Steve Shoptaw declares that he has received supplies for clinical trials from Pfeizer and Medicinova.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Grace M. Aldrovandi and Pamina M. Gorbach jointly supervised this work.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-51142-8.
References
- 1.Substance Abuse and Mental Health Services Administration. Key Substance Use and Mental Health Indicators in the United States: Results from the 2017 National Survey on Drug Use and Health, https://www.samhsa.gov/data/sites/default/files/cbhsq-reports/NSDUHFFR2017/NSDUHFFR2017.pdf (2018).
- 2.National Institute on Drug Abuse. Research Report Series: Methamphetamine, https://d14rmgtrwzf5a.cloudfront.net/sites/default/files/methrrs.pdf (2013).
- 3.Hartzler B, et al. Prevalence and Predictors of Substance Use Disorders Among HIV Care Enrollees in the United States. AIDS Behav. 2017;21:1138–1148. doi: 10.1007/s10461-016-1584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mimiaga MJ, et al. Substance use among HIV-infected patients engaged in primary care in the United States: findings from the Centers for AIDS Research Network of Integrated Clinical Systems cohort. American journal of public health. 2013;103:1457–1467. doi: 10.2105/ajph.2012.301162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salamanca SA, Sorrentino EE, Nosanchuk JD, Martinez LR. Impact of methamphetamine on infection and immunity. Frontiers in neuroscience. 2014;8:445. doi: 10.3389/fnins.2014.00445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fulcher JA, et al. Brief Report: Recent Methamphetamine Use Is Associated With Increased Rectal Mucosal Inflammatory Cytokines, Regardless of HIV-1 Serostatus. Journal of acquired immune deficiency syndromes (1999) 2018;78:119–123. doi: 10.1097/qai.0000000000001643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Droutman Vita, Xue Feng, Barkley-Levenson Emily, Lam Hei Yeung, Bechara Antoine, Smith Benjamin, Lu Zhong-Lin, Xue Gue, Miller Lynn C., Read Stephen J. Neurocognitive decision-making processes of casual methamphetamine users. NeuroImage: Clinical. 2019;21:101643. doi: 10.1016/j.nicl.2018.101643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maxwell S, Shahmanesh M, Gafos M. Chemsex behaviours among men who have sex with men: A systematic review of the literature. The International journal on drug policy. 2018;63:74–89. doi: 10.1016/j.drugpo.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 9.Hood JE, et al. The Changing Burden of HIV Attributable to Methamphetamine Among Men Who Have Sex with Men in King County, Washington. AIDS Patient Care STDS. 2018;32:223–233. doi: 10.1089/apc.2017.0306. [DOI] [PubMed] [Google Scholar]
- 10.Carrico AW, et al. Stimulant use and progression to AIDS or mortality after the initiation of highly active antiretroviral therapy. Journal of acquired immune deficiency syndromes (1999) 2014;67:508–513. doi: 10.1097/qai.0000000000000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott HM, et al. Age, race/ethnicity, and behavioral risk factors associated with per contact risk of HIV infection among men who have sex with men in the United States. Journal of acquired immune deficiency syndromes (1999) 2014;65:115–121. doi: 10.1097/QAI.0b013e3182a98bae. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loftis JM, Janowsky A. Neuroimmune basis of methamphetamine toxicity. International review of neurobiology. 2014;118:165–197. doi: 10.1016/b978-0-12-801284-0.00007-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Potula R, Haldar B, Cenna JM, Sriram U, Fan S. Methamphetamine alters T cell cycle entry and progression: role in immune dysfunction. Cell death discovery. 2018;4:44. doi: 10.1038/s41420-018-0045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prakash MD, et al. Methamphetamine: Effects on the brain, gut and immune system. Pharmacological research. 2017;120:60–67. doi: 10.1016/j.phrs.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 16.Brenchley JM, Douek DC. Microbial translocation across the GI tract. Annual review of immunology. 2012;30:149–173. doi: 10.1146/annurev-immunol-020711-075001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Y, et al. Bacterial Diversity of Intestinal Microbiota in Patients with Substance Use Disorders Revealed by 16S rRNA Gene Deep Sequencing. Scientific reports. 2017;7:3628. doi: 10.1038/s41598-017-03706-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banerjee S, et al. Opioid-induced gut microbial disruption and bile dysregulation leads to gut barrier compromise and sustained systemic inflammation. Mucosal immunology. 2016;9:1418–1428. doi: 10.1038/mi.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Volpe GE, et al. Associations of cocaine use and HIV infection with the intestinal microbiota, microbial translocation, and inflammation. Journal of studies on alcohol and drugs. 2014;75:347–357. doi: 10.15288/jsad.2014.75.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dillon SM, Frank DN, Wilson CC. The gut microbiome and HIV-1 pathogenesis: a two-way street. AIDS (London, England) 2016;30:2737–2751. doi: 10.1097/qad.0000000000001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noguera-Julian M, et al. Gut Microbiota Linked to Sexual Preference and HIV Infection. EBioMedicine. 2016;5:135–146. doi: 10.1016/j.ebiom.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fulcher JA, et al. Effects of Substance Use and Sex Practices on the Intestinal Microbiome During HIV-1 Infection. The Journal of infectious diseases. 2018;218:1560–1570. doi: 10.1093/infdis/jiy349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cook RR, et al. Effects of HIV viremia on the gastrointestinal microbiome of young MSM. AIDS (London, England) 2019;33:793–804. doi: 10.1097/qad.0000000000002132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bender JM, et al. Maternal HIV infection influences the microbiome of HIV-uninfected infants. Science translational medicine. 2016;8:349ra100. doi: 10.1126/scitranslmed.aaf5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pannaraj PS, et al. Shared and Distinct Features of Human Milk and Infant Stool Viromes. Frontiers in microbiology. 2018;9:1162. doi: 10.3389/fmicb.2018.01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Callahan BJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nature methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen L, et al. GMPR: A robust normalization method for zero-inflated count data with application to microbiome sequencing data. PeerJ. 2018;6:e4600. doi: 10.7717/peerj.4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.National Institute on Drug Abuse. NIDA-Modified ASSIST V2.0, https://www.drugabuse.gov/sites/default/files/pdf/nmassist.pdf (2012).
- 30.Robins JM, Hernan MA, Brumback B. Marginal structural models and causal inference in epidemiology. Epidemiology (Cambridge, Mass.) 2000;11:550–560. doi: 10.1097/00001648-200009000-00011. [DOI] [PubMed] [Google Scholar]
- 31.Ridgeway, D., McCaffrey, D. F., Morral, A., Griffin, B. A. & Burgette, L. F. Toolkit for Weighting and Analysis of Nonequivalent Groups v. R package version 1.5, https://CRAN.R-project.org/package=twang (2017).
- 32.Zeileis, A. Econometric Computing with HC and HAC Covariance Matrix Estimators. Journal of statistical software1 (2004).
- 33.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS one. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oksanen, J. et al. vegan: Community Ecology Package v. R package version 2.5-4, https://CRAN.R-project.org/package=vegan (2019).
- 35.Maechler, M., Rousseeuw, P., Struyf, A., Hubert, M. & Hornik, K. cluster: Cluster Analysis Basics and Extensions v. R package version 2.1.0, https://CRAN.R-project.org/package=cluster (2019).
- 36.Hennig, C. fpc: Flexible Procedures for Clustering v. R package version 2.2-3, https://CRAN.R-project.org/package=fpc (2019).
- 37.Zeileis, A., Kleiber, C. & Jackman, S. Regression Models for Count Data in R. Journal of statistical software1 (2008).
- 38.Friedman J, Alm EJ. Inferring Correlation Networks from Genomic Survey Data. PLoS computational biology. 2012;8:e1002687. doi: 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurtz ZD, et al. Sparse and Compositionally Robust Inference of Microbial Ecological Networks. PLoS computational biology. 2015;11:e1004226. doi: 10.1371/journal.pcbi.1004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the royal statistical society. Series B (Methodological), 289–300 (1995).
- 41.Benjamini Y, Yekutieli D. False Discovery Rate–Adjusted Multiple Confidence Intervals for Selected Parameters. Journal of the American Statistical Association. 2005;100:71–81. doi: 10.1198/016214504000001907. [DOI] [Google Scholar]
- 42.R Core Team R: A language and environment for statistical computing (R Foundation for Statistical Computing, Vienna, Austria, 2018), https://www.R-project.org/.
- 43.Wickham, H. ggplot2: Elegant Graphics for Data Analysis. (Springer-Verlag, 2016).
- 44.Mutlu EA, et al. Colonic microbiome is altered in alcoholism. American journal of physiology. Gastrointestinal and liver physiology. 2012;302:G966–978. doi: 10.1152/ajpgi.00380.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakajima M, et al. Oral Administration of P. gingivalis Induces Dysbiosis of Gut Microbiota and Impaired Barrier Function Leading to Dissemination of Enterobacteria to the Liver. PloS one. 2015;10:e0134234. doi: 10.1371/journal.pone.0134234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ning T, Gong X, Xie L, Ma B. Gut Microbiota Analysis in Rats with Methamphetamine-Induced Conditioned Place Preference. Frontiers in microbiology. 2017;8:1620. doi: 10.3389/fmicb.2017.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li X, et al. Methamphetamine causes neurotoxicity by promoting polarization of macrophages and inflammatory response. Human & experimental toxicology. 2018;37:486–495. doi: 10.1177/0960327117714039. [DOI] [PubMed] [Google Scholar]
- 48.Grigoleit JS, et al. Dose-dependent effects of endotoxin on neurobehavioral functions in humans. PloS one. 2011;6:e28330. doi: 10.1371/journal.pone.0028330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abe K, et al. Dysbiosis of oral microbiota and its association with salivary immunological biomarkers in autoimmune liver disease. PloS one. 2018;13:e0198757. doi: 10.1371/journal.pone.0198757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Proenca MA, et al. Relationship between Fusobacterium nucleatum, inflammatory mediators and microRNAs in colorectal carcinogenesis. World journal of gastroenterology. 2018;24:5351–5365. doi: 10.3748/wjg.v24.i47.5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dillon SM, et al. Low abundance of colonic butyrate-producing bacteria in HIV infection is associated with microbial translocation and immune activation. AIDS (London, England) 2017;31:511–521. doi: 10.1097/qad.0000000000001366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mutlu EA, et al. A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS pathogens. 2014;10:e1003829. doi: 10.1371/journal.ppat.1003829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leclercq S, et al. Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc Natl Acad Sci USA. 2014;111:E4485–4493. doi: 10.1073/pnas.1415174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burokas A, Moloney RD, Dinan TG, Cryan JF. Microbiota regulation of the Mammalian gut-brain axis. Advances in applied microbiology. 2015;91:1–62. doi: 10.1016/bs.aambs.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 55.Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nature reviews. Neuroscience. 2012;13:701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 56.Han W, et al. A Neural Circuit for Gut-Induced Reward. Cell. 2018;175:665–678.e623. doi: 10.1016/j.cell.2018.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kiraly DD, et al. Alterations of the Host Microbiome Affect Behavioral Responses to Cocaine. Scientific reports. 2016;6:35455. doi: 10.1038/srep35455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee K, et al. The gut microbiota mediates reward and sensory responses associated with regimen-selective morphine dependence. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2018;43:2606–2614. doi: 10.1038/s41386-018-0211-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wei Chia-Li, Wang Sabrina, Yen Jui-Ting, Cheng Yun-Fang, Liao Chia-Li, Hsu Chih-Chieh, Wu Chien-Chen, Tsai Ying-Chieh. Antidepressant-like activities of live and heat-killed Lactobacillus paracasei PS23 in chronic corticosterone-treated mice and possible mechanisms. Brain Research. 2019;1711:202–213. doi: 10.1016/j.brainres.2019.01.025. [DOI] [PubMed] [Google Scholar]
- 60.Valles-Colomer Mireia, Falony Gwen, Darzi Youssef, Tigchelaar Ettje F., Wang Jun, Tito Raul Y., Schiweck Carmen, Kurilshikov Alexander, Joossens Marie, Wijmenga Cisca, Claes Stephan, Van Oudenhove Lukas, Zhernakova Alexandra, Vieira-Silva Sara, Raes Jeroen. The neuroactive potential of the human gut microbiota in quality of life and depression. Nature Microbiology. 2019;4(4):623–632. doi: 10.1038/s41564-018-0337-x. [DOI] [PubMed] [Google Scholar]
- 61.Stevens BR, et al. Increased human intestinal barrier permeability plasma biomarkers zonulin and FABP2 correlated with plasma LPS and altered gut microbiome in anxiety or depression. Gut. 2018;67:1555–1557. doi: 10.1136/gutjnl-2017-314759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pirbaglou M, et al. Probiotic supplementation can positively affect anxiety and depressive symptoms: a systematic review of randomized controlled trials. Nutrition research (New York, N.Y.) 2016;36:889–898. doi: 10.1016/j.nutres.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 63.Liu, C. M. et al. Penile Anaerobic Dysbiosis as a Risk Factor for HIV Infection. mBio8, 10.1128/mBio.00996-17 (2017). [DOI] [PMC free article] [PubMed]
- 64.Lennard, K. et al. Microbial Composition Predicts Genital Tract Inflammation and Persistent Bacterial Vaginosis in South African Adolescent Females. Infection and immunity86, 10.1128/iai.00410-17 (2018). [DOI] [PMC free article] [PubMed]
- 65.McClelland RS, et al. Evaluation of the association between the concentrations of key vaginal bacteria and the increased risk of HIV acquisition in African women from five cohorts: a nested case-control study. The Lancet. Infectious diseases. 2018;18:554–564. doi: 10.1016/s1473-3099(18)30058-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee SC, et al. Enrichment of gut-derived Fusobacterium is associated with suboptimal immune recovery in HIV-infected individuals. Scientific reports. 2018;8:14277. doi: 10.1038/s41598-018-32585-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang CB, Emerson KA, Gonzalez OA, Ebersole JL. Oral bacteria induce a differential activation of human immunodeficiency virus-1 promoter in T cells, macrophages and dendritic cells. Oral microbiology and immunology. 2009;24:401–407. doi: 10.1111/j.1399-302X.2009.00533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brito LC, et al. Microbiologic profile of endodontic infections from HIV− and HIV+ patients using multiple-displacement amplification and checkerboard DNA-DNA hybridization. Oral diseases. 2012;18:558–567. doi: 10.1111/j.1601-0825.2012.01908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Paster BJ, et al. Bacterial diversity in necrotizing ulcerative periodontitis in HIV-positive subjects. Annals of periodontology. 2002;7:8–16. doi: 10.1902/annals.2002.7.1.8. [DOI] [PubMed] [Google Scholar]
- 70.Shenoy MK, Lynch SV. Role of the lung microbiome in HIV pathogenesis. Current opinion in HIV and AIDS. 2018;13:45–52. doi: 10.1097/coh.0000000000000427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request. All sequencing data has been deposited into BioProject with the accession number PRJNA422134.