Abstract
Methods for investigating DNA methylation nowadays either require a reference genome and high coverage, or investigate only CG methylation. Moreover, no large-scale analysis can be performed for N6-methyladenosine (6 mA) at an affordable price. Here we describe the methylation content sensitive enzyme double-digest restriction-site-associated DNA (ddRAD) technique (MCSeEd), a reduced-representation, reference-free, cost-effective approach for characterizing whole genome methylation patterns across different methylation contexts (e.g., CG, CHG, CHH, 6 mA). MCSeEd can also detect genetic variations among hundreds of samples. MCSeEd is based on parallel restrictions carried out by combinations of methylation insensitive and sensitive endonucleases, followed by next-generation sequencing. Moreover, we present a robust bioinformatic pipeline (available at https://bitbucket.org/capemaster/mcseed/src/master/) for differential methylation analysis combined with single nucleotide polymorphism calling without or with a reference genome.
Subject terms: DNA methylation, Epigenomics
Introduction
DNA methylation is one of the fastest mechanisms that organisms use to rapidly adapt to new conditions1–3. Indeed, the methylation of cytosine residues in genomic DNA has a pivotal role in regulation of genome expression4–7, particularly for the cytosines in promoter sequences of specific genes. Generally, methylation is correlated with silencing of genes and transposable elements, while demethylation is correlated with active transcription4, although the reverse has also been documented8. Moreover, methylation patterns along a gene can have specific effects on the gene expression: body-methylated genes tend to be constitutively expressed, whereas promoter-methylated genes are preferentially expressed in a tissue-specific manner5.
Cytosine methylation is conventionally classified in terms of CG, CHG, and CHH sequence contexts (where H is A, C, or T), which are subjected to the actions of different DNA methyltransferases9–12. Methylation on adenine (N6-methyladenosine; 6 mA)7,13 has been recently found in Chlamydomonas and in several multicellular eukaryotes, including flowering plants7,14–18. While in prokaryotes and ancient eukaryotes, 6 mA serves as major marker to discriminate invasive foreign DNA19, its role in eukaryotes has recently been associated with transcriptional activation in response to stress7,14, and with transgenerational chromatin regulation20.
There are many technologies available to obtain genome-wide information on differential DNA methylation. Some of these provide qualitative information on the methylation state, while others are based on the chemical conversion of unmethylated cytosine to thymine. This chemical conversion defines the level of unconverted cytosines, which provides a measure of the level of DNA methylation21–23.
Whole-genome bisulfite sequencing (WGBS) is a technique that can assess virtually every cytosine methylation state in the genome, although this requires high coverage (at least 5–10–fold)21,24,25, which makes it expensive in species with a large genome and/or in experiments with many samples26. Reduced representative bisulfite sequencing (RRBS)27 has been proposed as an alternative to WGBS for large genomes. RRBS introduces a DNA digestion step with a methylation-insensitive enzyme that is followed by size filtration of the restricted fragments, and chemical conversion. This technique investigates only a fraction of the genome, but the increased sequencing coverage of the represented fraction provides greater confidence in such methylation measurements. Although RRBS has been shown to be extremely powerful, there remain technical and financial bottlenecks that challenge the feasibility of these approaches, especially in species with a large genome and lacking a reference genome27,28.
Further reductions in sequencing efforts can be achieved through adoption of methylation-sensitive endonucleases, as in methylation-sensitive restriction-enzyme digestion and sequencing (MRE-seq)29, or the EpiRADseq30 variation of Double digest RADseq (ddRADseq31). These techniques involve DNA digestion with a methylation-sensitive enzyme followed by size selection and sequencing. However, these methylation-sensitive enzymes target non-methylated sites, and thus they act more specifically on sites with low level of methylation, which are the most difficult to detect with conventional techniques29. Such read counts for each locus do not provide absolute measures of cytosine methylation, although they can be useful to infer methylation differences between samples at specific sites. A limitation of MRE-seq is that it is specific for CG methylation. Importantly, none of these methods address analysis of the methylation status of adenines, which is currently being studied using either costly or complex approaches, such as PacBio sequencing mass spectrometry, immunoprecipitation, and 6mA-RE-seq7,14,32.
To overcome some of these limitations, we present the methylation content sensitive enzyme double-digest restriction-site-associated DNA (ddRAD) technique (MCSeEd), a very simple, highly scalable, cost-effective extension of the original ddRAD protocol that allows the detection of methylation changes for the CG, CHG, CHH, and 6 mA contexts.
This MCSeEd technique was tested on leaves of a commercial maize hybrid grown under normal irrigation (well watered; WW) and under drought stress (DS), and collected 60 days after sowing (DAS). The relative methylation changes estimated by MCSeEd for differentially methylated positions (DMPs) and differentially methylated regions (DMRs) clearly discriminated between WW and DS samples with both genome-dependent and genome-independent approaches. The DMRs identified by MCSeEd showed gene enrichments related to the experimental system under investigation. Shifts in single-nucleotide polymorphism (SNP) allele frequencies were also identified, and were related to the specific methylated/unmethylated alleles.
Results
MCSeEd efficiently identifies methylation variations induced by drought stress in maize leaves
Differentially methylated positions
The MCSeEd technique was used to monitor DNA methylation changes induced by drought stress in maize leaves. To this end, we constructed next-generation sequencing (NGS) libraries from genomic DNA purified from the leaves of the WW and DS maize plants. A total of 24 libraries were produced by double restriction–ligations, each using MseI in combination with one of the four methylation-sensitive enzymes AciI, PstI, EcoT22I, and DpnII, for the CG, CHG, and CHH and 6 mA contexts, respectively14,29,33–36 (Supplementary Table S1) as outlined in Fig. 1 and in Supplementary Table S2.
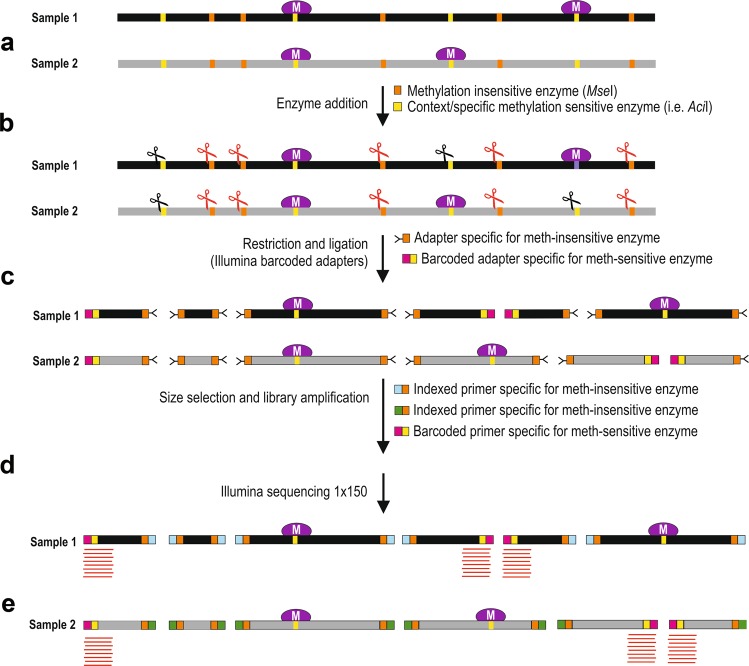
Figure 1.
Schematic representation of MCSeEd. Two samples (e.g., control, stressed samples) (a) are subjected to double digestion (b) with a methylation-insensitive (MseI) and a methylation-sensitive enzyme (AciI, PstI, EcoT22, DpnII; see Supplementary Table S1) and ligated (c) with specific adapters: a Y adapter for the MseI site and a barcoded adapter specific for the methylation-sensitive enzyme (c). After size selection, the fragments are pooled and amplified with adapter-specific primers (MseI primers carry library-specific indices) and sequenced using the 1 × 150 Illumina chemistry (d). Demultiplexed reads (e) are analyzed with the design-appropriate MCSeEd pipeline.
A mean of 7.8 million 150-bp-long reads were obtained from each library (Supplementary Table S3). Of these, 98% passed quality control and were aligned to the B73 reference genome. To avoid bias due to paralogous sequences that can align at multiple genomic sites, only the reads that mapped at unique genomic positions were retained. Thus, a total of 89,935,677 reads were mapped uniquely on the reference genome (48.49% of the total reads, with a minimum of 30.13% for DpnII, and a maximum of 78.70% for PstI). We named these reads the MCSeEd loci (Supplementary Table S3). They identified 992,320 loci containing cytosines (705,341 in symmetric, and 286,889 in asymmetric contexts) and 1,629,894 loci containing adenines (Supplementary Table S4).
The mapping location of each MCSeEd locus was investigated to determine whether it fell into an extended gene body (EGB) that included the region within 2.5 kb upstream of the transcription start site (TSS), the transcribed region (i.e., the gene body), and the region within 2.5 kb downstream of the transcription termination site (TTS). Of note, in all, 92.9% (AciI), 78.0% (PstI), 82.3% (EcoT22I), and 98.1% (DpnII) of the identified MCSeEd loci fell into EGBs. Specifically, a mean of 6.28 sites per EGB was recorded, with a minimum of 2.96 for PstI, and a maximum of 12.58 for DpnII (Supplementary Table S5).
After normalization of the MCSeEd loci, the sites covered by a total number of reads < 4 or showing excessive read-count variation among the replicates (standard deviation > 8%) were discarded (Supplementary Table S4). The remaining sites were used to estimate the total of 62,489 DMPs, out of the 992,230 investigated cytosines, with significantly altered methylation levels between the WW and DS samples (false discovery rate, ≤0.05). Of these, 44,176 belonged to symmetric, and 18,313 to asymmetric contexts. With similar filtering, out of 1.6 million on 6 mA, 118,269 DMPs were detected (Supplementary Tables S6, S7).
Principal component analysis was used to cluster the samples based on the methylation levels of the DMPs (Supplementary Fig. S1). The first latent component (PC1) accounted for 53%, 84%, 44%, and 47% of the total variance, for the CG, CHG, CHH, and 6 mA contexts, respectively, and clearly discriminated between WW and DS, which indicated that the drought stress leads to genome-wide methylation changes in maize leaves.
Accordingly, complete linkage clustering of the methylation levels at DMPs clearly separated the DS from the WW samples (Supplementary Fig. S2). Altogether, these data indicate that the MCSeEd pipeline can infer the effects of drought stress for each methylated context. Considering all of the methylation changes as being induced by water deficiency in the drought-stressed replicates, we observed 1.6-fold (CG) to 3.4-fold (CHH) more methylation increases than decreases as responses to this stress, whereas for CHG and 6 mA, the proportion of methylation changes in each direction was equivalent (Supplementary Fig. S3a).
Differentially methylated regions
Genomic regions with co-regulated methylation changes upon drought-stress were identified by an adjacent window approach that targeted adjacent DMPs with concordant methylation changes (at least 2). After validation by logistic regression, the identified genomic regions were investigated as DMRs. In total, 5,726 DMRs were identified for the CG (347), CHG (836), CHH (205), and mA (4,338) contexts (Supplementary Tables S6, S8). The DMR median length was similar for CG (485 bp), CHH (484 bp), and 6 mA (506 bp), and a little lower for CHG (325 bp) (Supplementary Table S9).
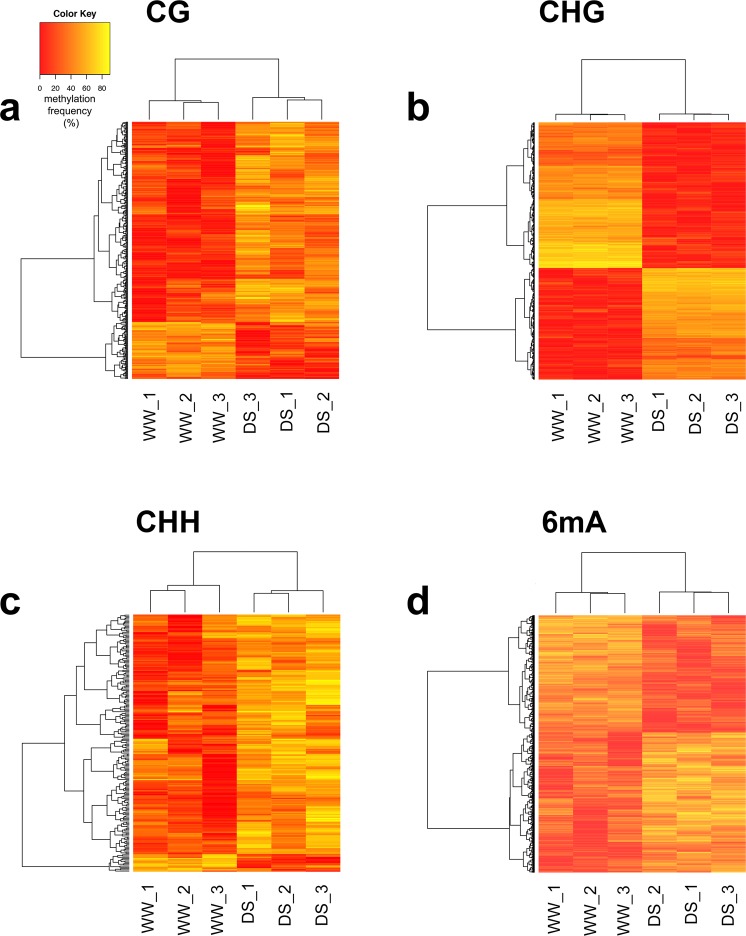
The estimated relative methylation levels of the DMPs belonging to each DMR were hierarchically clustered, and as expected, clustered according to treatment, as WW or DS (Fig. 2). In particular, for the CG and CHH sites, the number of DMRs with higher methylation levels in the DS samples (relative to the WW samples) was higher than the number of DMRs that showed a lower level in the DS samples (Fig. 2). In contrast, for the CHG and 6 mA contexts, the number of DMRs with higher methylation levels in the DS samples (relative to the WW samples) was equivalent to the number of DMRs with a lower level in the DS samples.
Figure 2.
Relative methylation frequencies of differentially methylated regions identified from the comparison between the well watered (WW) and drought stressed (DS) samples. Relative methylation frequencies of the differentially methylated positions contained in each differentially methylated region were averaged and used in complete linkage clustering analysis of samples derived from WW and DS based on 347 (a; CG), 836 (b; CHG), 205 (c; CHH) and 4338 (d; 6 mA) differentially methylated regions.
Differentially methylated genes
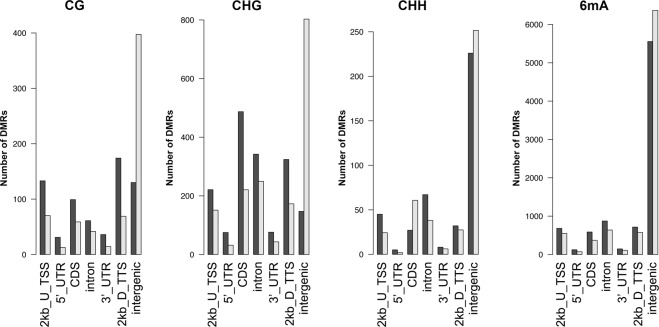
To analyze how water stress impacts the methylation patterns typical of genic regions, we analyzed the DMP and DMR distributions in relation to the coding and regulatory genomic sequences. In particular, we compared the distribution of DMPs and DMRs in transcribed genic regions extended by 2 kb at both ends (EGBs) (Supplementary Fig. S4, Fig. 3) and found that DMRs mapped preferentially to EGBs (Fig. 3). In particular 243, 731, and 307 EGBs were overlapped at least once by 1,388 DMRs in the 2 kb windows upstream of TSS, within the gene body, or in the 2 kb windows downstream of TTS, respectively. The genes belonging to these EGBs were defined as differentially methylated genes (DMGs, Supplementary Table S10) and were investigated as DMGs upon drought stress. Moreover, 418, 929, and 448 EGBs were overlapped at least once by 4,338 DMRs in the 2 kb windows upstream of TSS, within the gene body, or in the 2 kb windows downstream of TTS, respectively.
Figure 3.
Enrichment analysis of DMRs in different genomic regions. Enrichment analysis was performed using the binomial distribution of all of the MCSeEd loci as expected and the differentially methylated regions (CG, CHG, CHH, 6 mA contexts; note that scales for each context differ), as the observed datasets. U, upstream; D, downstream. Light gray = expected number of DMRs, Dark gray = observed number of DMRs
Panther enrichment analysis using all of the DMGs identified in all of the contexts identified the gene ontology (GO) terms, which were mainly related to regulation of transcription, biosynthetic and metabolic processes, responses to stimuli, oxidoreductase activity, and binding of nucleic acids (Supplementary Table S11). Several DMGs identified from DMRs located within 2 kb upstream of the TSS have orthologs in other species (i.e., Arabidopsis) including DRE-binding protein 3 (Zm00001d021207), WRKY 36 (Zm00001d039532), ethylene-responsive transcription factor WIN1 (Zm00001d046501), Myb family transcription factor PHL6 (Zm00001d015226), ISWI chromatin-remodeling complex ATPase CHR11 (Zm00001d040831), with role related to response to water deprivation, defense responses, drought stress tolerance, dehydration stress memory, and response to drought stress, respectively (Supplementary Table S10).
Validation of differentially methylated positions inferred by MCSeEd
The MCSeEd technique was validated using a quantitative (q)MRE14,37–40. In this technique, if the cytosine/adenine of the restriction site within a PCR-amplification target is methylated, the enzyme cannot cut the DNA, and the relative amplicon is produced; conversely, when the cytosine is not methylated, the DNA is digested by the enzyme and the amplicon cannot occur.
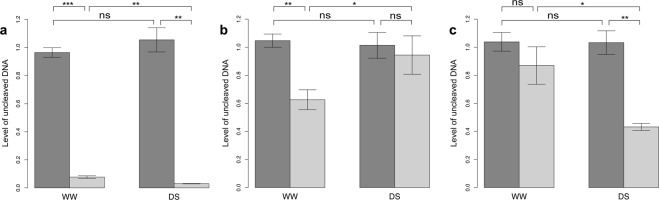
For this approach, 10 randomly chosen DMPs were used. Nine of these 10 DMPs showed methylation differences comparable to those obtained after methylKit analysis (Supplementary Table S12, Fig. 4), while the remaining one showed no significant differences between the WW and DS samples.
Figure 4.
The MCSeEd validation by quantitative methylation-sensitive PCR (a–c), for the well-watered (WW) and drought stressed (DS) samples. Differentially Methylated Positions with different behavior, DMP_B (a), DMP_F (b), DMP_J (c) (details in Supplementary Table S12) were tested. Dark shading, mock (no digestion, equivalent to a fully methylated DNA site); light shading, digested DNA. Error bars indicate standard deviation. *p < 0.05; **p < 0.01; ***p < 0.001; ns, p > 0.05 (unpaired two-tailed Student’s t-tests).
Reference-free strategy
Since one of our goals was to develop a high-throughput technique that can also be applied to species without a reference genome, the MCSeEd bioinformatic pipeline was tested for mapping of filtered reads to a pseudo-reference genome autogenerated by a pipeline, hereafter indicated as the genome independent strategy.
Under the DS conditions, a total of 2,258,361 pseudo-genome contigs were generated for mCs (686,548 contigs, 30.4%) and 6 mA (1,571,813, 69.6%) libraries (Supplementary Table S4). After MCSeEd loci normalization, the sites covered by a total number of reads either <4 or showing an excessive read count variation among the replicates (standard deviation >8%) were discarded (Supplementary Table S4), and the remaining sites were used to estimate the total of 30,092 DMPs, out of 686,548 investigated cytosines, with significantly altered methylation levels between the WW and DS samples. Of these, 27,203 belonged to symmetric and 10,889 to asymmetric contexts. Moreover, out of 1.6 million on 6 mA, 143,389 DMPs were detected (Supplementary Tables S6, S7).
In the genome-independent approach, both principal component analysis and heatmaps (Supplementary Figs S5, S6) clearly discriminated between the WW and DS samples. The CG and CHH contexts showed higher methylation in response to stress (2.89-fold, 5.09-fold, respectively), while for CHG and 6 mA, the differences there were effectively no differences (0.92, 0.88, respectively; Supplementary Fig. S3b).
Variant calling and shift in allelic frequency
Allele-specific methylation responsive (ASMR) sites to drought stress were identified based on the allelic origins of the reads. Using SNP information from PstI libraries as an example, the relative allelic contributions to the total read counts were inferred by counting the number of reads that originated from either SNP allele (Supplementary Table S13). This highlighted 10,861 heterozygous SNP loci. Among these, 287 showed significant shifts in their relative allelic contributions between the WW and DS samples (P < 0.05), and were defined as ASMR sites (Fig. 5).
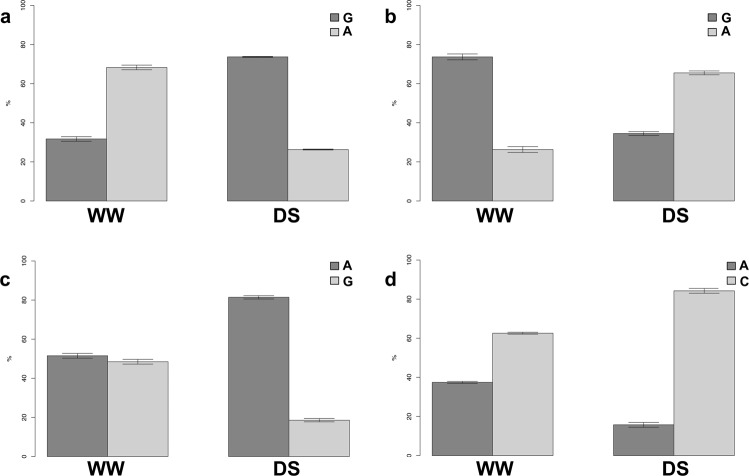
Figure 5.
Examples of the differential changes in the methylation states due to drought stress, between the alleles at the single SNP loci. PstI libraries were screened for SNP polymorphisms at each of four random loci: (a) Chr1_254513757; (b) Chr1_34649135; (c) Chr8_150564756; and (d) Chr8_169547803. WW, well watered; DS, drought stressed.
A total of 128 of the 287 identified ASMR sites also showed significant net changes in total methylation status (i.e., the DMP–ASMR loci). The remaining 159 ASMR loci were not related to DMPs. This might have been because the methylation changes between WW and DS for one allele was compensated for by a similar change in the opposite direction for the other allele, therein not altering the total relative methylation levels between WW and DS. Interestingly, the ASMR sites were preferentially located (223 out of 287) in EGBs (P < 0.05): 128 within genes, 52 within 2 kb upstream of the TSS, and 43 within 2 kb downstream of the TTS (Supplementary Table S14).
Discussion
We have developed a reduced-representation, reference-free approach for characterizing whole genome methylation patterns across different methylation contexts. While WGBS is the gold standard, it is cost prohibitive at most experimental scales, and requires a reference genome28.
Alternative approaches aimed at reducing the sequencing demands are based on genomic digestion with either methylation-sensitive or methylation-insensitive endonucleases, before direct NGS (e.g., EpiRADseq30, MRE-seq29), or bisulfite treatments and NGS (e.g., RRBS27, methylation-sensitive restriction enzyme bisulfite sequencing [MREBS]41). However, both RRBS and MREBS still require a reference genome and a certain level of coverage, at least for estimation of the methylation status of the cytosines. Although EpiRADseq and MRE-seq do not require a reference genome, these consider only the CG sites, and the methylation status is deduced by counting the unmethylated cytosines within the recognition site. The scenario for 6 mA is even worse to date, with the level of 6 mA determined by immunoprecipitation or liquid chromatography coupled with tandem mass spectrometry7,32. The only sequencing technique that can simultaneously infer the cytosine-5 methylation and 6 mA is PacBio single-molecule real-time sequencing, but this is prohibitively expensive for most applications7,32.
To address these limitations, we developed a new method: MCSeEd. Similar to other reduced-representation methods, MCSeEd is based upon parallel restrictions carried out by combinations of a methylation-insensitive endonuclease (MseI) and one of four methylation-sensitive endonucleases, directed to one of CG, CHG, CHH, and 6 mA. This is completed using NGS.
With MCSeEd, the read counts can be readily used to estimate the differential methylation between two samples, which is often of primary interest41. Indeed, the cytosine libraries here described were equally represented, and they covered a mean of 2.6 million sites, about 7% of which were differentially methylated between the samples. This MCSeEd pipeline was tested without and with the support of a reference genome and in both cases, this resulted in identification of DMPs at very high rates. The genome-dependent approach resulted in a similar number of DMPs with respect to the genome-independent approach (180,758 vs 181,481).
As validation, we used qMRE to confirm the changes in the DNA methylation recorded in the WW versus DS comparison. To confirm our DS experiment data, we randomly chose 10 DMPs and applied the qMRE method. Of note, this system was able to confirm the methylation levels within the samples and the differential methylation between the WW and DS samples in nine out of the 10 cases.
Methylation of adenines is still a poorly investigated field. For instance, Fu et al.14 reported that the consensus motifs for 6 mA are only partially conserved in different eukaryotic organisms, and enzymes might have evolved to catalyze 6 mA modifications in the evolutionary process. They reported that GATC appears to be the most ancient 6 mA motif, which exists in both lower eukaryotes and bacteria, but is lost in higher eukaryotes14. Instead, GAGG is present in both plantae and animalia, and AGAA might be specific to animals15,42. Liang et al.7 demonstrated that Arabidopsis contains two specific methylated motifs, ANYGA and ACCT, which have not been found in other organisms to date. On the other hand, the presence of adenine methylation at GATC sequences was shown in rice43, tobacco44, and Arabidopsis45. In particular, both Dhar et al.43 and Ashapkin et al.45 used DpnI, which digests the DNA only if adenine is methylated in the sequence GATC, to demonstrate the extensive digestion of rice and Arabidopsis DNAs. Our results confirm these findings, and demonstrate that methylation at the GATC site is not only present in maize, but also that the mechanisms of methylation/demethylation are still active, as the levels of differentially methylated positions due to stress are very high (118,269 and 143,389 DMPs in the genome-dependent and genome-independent approaches).
CG methylation is prevalent in the transcribed regions of many constitutively expressed plant genes (i.e., gene body methylation), and it shows characteristic patterns within genic regions46,47. In maize, CG methylation has been shown for exons more than introns, which suggests a defensive function versus transposon insertion in the coding sequence, while allowing insertions into introns and other noncoding regions48. Moreover, both in maize and Arabidopsis, the CHG and CHH methylation contexts are significantly enriched at intron-exon junctions48. Our data show that all cytosine-5 methylation contexts are enriched within 2 kb upstream of the TSS; moreover, while CG and CHG are highly enriched in exons, CHH is enriched in introns.
For 6 mA, Liang et al.7 reported that these sites in Arabidopsis genes are mainly located in exons, while those in transposable element genes show a local reduction at the TSS, followed by an immediate increase, as also seen in Chlamydomonas genes14. This is in contrast to what was seen in C. elegans, where 6 mA are distributed equally in genomic regions, including introns, exons, and TSS regions, and in Mus musculus and Xenopus laevis, where 6 mA are primarily excluded from coding regions. Our data show an enrichment of 6 mA in EGBs, and lower levels of 6 mA in intergenic regions. This is particular true for DMRs, but less evident for DMPs. It is worth nothing that the 6 mA located in EGBs appears to respond to water restriction more than the 6 mA located in intergenic regions.
The efficiency of MCSeEd for detection of changes in DNA methylation between two sets of contrasting samples was evident in both principal component analysis and linkage clustering, which clearly discriminated the WW samples from the DS samples, for both DMPs and DMRs.
Boyko et al.49 reported an increase in global genome methylation in Arabidopsis plants exposed to stress, including salt, UVC, cold, heat and flood stresses. In particular, the progenies of these stress-treated plants showed increased global methylation, even in the absence of the stress, but these transgenerational effects did not persist in successive generations in the absence of stress. Moreover, induction of transient DNA methylation was related to drought stress in pea50 and in drought susceptible rice genotypes51. By using MCSeEd on these maize samples with different water status (i.e., WW vs. DS), we found that DS appears to induce methylation rather than demethylation, particularly in the CHH and CG contexts. In total, we identified 1,240 DMGs, most of which were related to regulation of transcription, biosynthetic and metabolic processes, and response to stimuli, processes that suggest that changes in DNA methylation are correlated with stress responses to water deprivation.
Genomic DNA cytosine/adenine methylation polymorphism studies can be tested for their ability to reveal “epigenetic heterosis” effects for tolerance to drought and other stress factors, and as a tool to identify novel stress-responsive genes. Indeed, methylation changes can be allele specific or genome specific (in case of polyploids). Therefore, inferred ASMR sites might be very important for this purpose. In our study, many ASMR sites for drought stress were identified, as a genome-wide situation that already existed in the control plants (i.e., WW). It is known than genomes can show selective allelic imbalance, as seen for humans52 and plants53–55.
The drought stress influenced the methylation status of cytosines and adenines, which created a level of imbalanced heterozygosis between the stress (DS) and control (WW) conditions. This imbalance might impact on the level of gene expression, as methylation can have a general role in the regulation of gene expression and contribute to the hybrid vigor phenomenon56–59. MCSeEd can thus mine SNPs associated with selectively methylated sites, to highlight allelic imbalance. This has implications for breeding programs, wherein MCSeEd can provide information about which of two genomes in a hybrid has been methylated, and through the targeting of candidate ASMR sites, allow a “MAS by methylation” approach.
Here we tested MCSeEd on maize, as an important crop that has a large genome and is thus very rich in transposable elements. Also, in its cultivated form, maize is a hybrid, and hence a very challenging species to work with. We have demonstrated that even in such a complex scenario, use of MCSeEd identified differentially methylated regions both between stress and control conditions in a hybrid genotype, and between developing organs in an inbred line (B73). Benchmarking experiments indicate that MCSeEd can be reliably used for differential methylation analyses with consistent results.
Materials and Methods
Plant material
For the drought-stress study, plants of a commercial maize hybrid variety were subjected to normal irrigation (well-watered; WW) and drought stress (DS), as reported by Bocchini et al.60. At 60 DAS, portions of the leaves were collected, bulked into three biological replicas of five plants each (WW1, WW2, WW3, DS1, DS2, DS3), and then stored at −80 °C, until further processing.
For MCSeEd validation, B73 seeds were germinated at 25 °C in the dark on wet paper towels in glass Pyrex dishes. At 5 days after germination, the shoots at the coleoptile stage and the roots were excised and stored at −80 °C, until further processing.
DNA purification, library construction and sequencing
Genomic DNA was purified from each sample using DNeasy Plant Mini kits (Qiagen GmbH, old (Qubit; Life Technologies, Grand Island, NY, USA). The library set-up protocol was performed according to Peterson et al.31 with some modifications, as described below. Four specific enzyme combinations were chosen (as one of four methylation-sensitive enzymes, each combined with methylation-insensitive MseI) to infer the CG (AciI/MseI), CHG (PstI/MseI), CHH (EcoT22I/MseI), and 6 mA (DpnII/MseI) methylation contexts, respectively (Supplementary Table S1). To define the efficacy of the enzyme combinations, we developed a program that scanned the genome in silico and calculated the size distribution of the restriction fragments (Supplementary Fig. S7). Briefly, the user was asked to insert the name of a desired restriction enzyme combination, and a virtual digestion was performed. Fragments with the optimal length range can then be selected and counted.
For each library, 150 ng DNA were double-digested with one of these four enzyme combinations. In the same reaction, a sample-specific barcoded adapter was ligated to the methylation-sensitive restriction end, while a common Y adapter was ligated to the sticky end left by MseI (Supplementary Table S15) as follow (e.g. for AciI enzyme): 150 ng of genomic DNA was added with 5U of AciI restriction enzyme (New England BioLabs), 5 U of MseI restriction enzyme (New England BioLabs), 2 μM of unique barcoded adapter, 2 μM of unique common Y adapter, 1U of T4 DNA ligase (ThermoFisher), 0.2 mM ATP and 1x RL buffer (5x CutSmart Buffer, New England Biolabs, 25 mM DTT, Invitrogen) for a final volume of 50 μL. The libraries were then pooled, as reported in the experimental design (Supplementary Table S2), purified using magnetic beads (Agencourt AMPure XP; Beckman Coulter, MA, USA), size selected by gel electrophoresis, and purified using QIAquick Gel Extraction kits (Qiagen) for fragments in the range of 250 bp to 600 bp. Size-selected libraries were quantified using a fluorometer (Qubit; Life Technologies), and a normalized DNA amount (15 ng) was amplified with a primer that introduced an Illumina index (at the Y common adapter site) for demultiplexing. Following PCR with uniquely indexed primers, multiple samples were pooled. PCR-enrichment was performed as described by Peterson et al.31. Specifically, 15 ng of pooled DNA was amplified using 1 U of Phusion® High-Fidelity DNA polymerase (New England BioLabs) in a final volume of 50 μL containing 1X Phusion HF Buffer, 0.2 μM PCR1_MCSeEd oligo (Supplementary Table S15), 0.2 μM PCR_index oligo (Supplementary Table S15), 0.2 μM dNTPs. Amplified libraries were purified with magnetic beads (AMPure; Beckman Coulter, Brea, CA, USA), and then quantified (Qubit and Bioanalyzer 2100: Agilent Technologies, Santa Cruz. CA, USA). The grouped libraries were pooled in an equimolar fashion, and the final library was Illumina-sequenced using 150-bp single-end chemistry.
Genome-dependent workflow
Raw reads from the Illumina sequencing of the CG, CHG, CHH and adenine methylation libraries were demultiplexed using the process_radtags tool (STACKS v.2.3b package)61. This identifies and assigns reads to each individual on the basis of 7-bp custom barcode sequences (removed after analysis). After processing the raw reads, the MCSeEd pipeline was run following either genome-dependent or genome-independent procedures, as detailed below. The MCSeEd pipeline consisted of a bash wrapper using different algorithms or Perl scripts. Sequences from each library were mapped to the reference maize genome (AGPv4; https://www.maizegdb.org) with the bwa mem algorithm using the default settings62. Bam sorted and indexed files of uniquely mapped reads were produced with Samtools63.
Loci-counting approach
To create a count matrix where the columns are the sample libraries and the rows represent the locations in the genome hit by the sequencing, we created a merged bam file that acted as a guide for creating an “experiment-wise annotation”. Here, all of the genomic positions sequenced were stored, whereby all of the uniquely mapped reads were recorded for each genomic location covered in the experiment. Briefly, for each alignment in the bam file, genome coordinates and CIGAR63 fields were processed to produce meaningful intervals with a Perl script. Then, redundant coordinates were collapsed and sorted, and overlapping intervals were merged with the bedtools suite, maintaining strandedness64. This information was converted to a formal GFF file, and then used as the input in featureCounts65, along with the bam files previously described, to count the occurrences on the experiment-wise annotation.
The count matrix consisted of one row per locus and one column per sample, and it was then filtered and processed by two in-house–built Perl scripts. The following operations were performed: (1) the libraries were normalized and balanced in a reads per million fashion; (2) loci with a coverage of at least 10 reads were retained; (3) relative methylation level per locus were estimated; and (4) the filtered data were parsed for use by the methylKit R package66. In particular, the relative methylation levels at each site (point 3) were calculated following a rescaling procedure that was based on the maximum number of observed read counts. In practice, the sample showing the highest number of reads was assumed to be the not-methylated reference for the site, or to have 0% methylcytosine and 100% cytosine, and all of the remaining samples were rescaled proportionally (Supplementary Table S16). As the reference was common to all samples, the methylation level estimates can be used to infer relative methylation changes between the samples. DMPs were therefore identified as sites that showed significant differences in the methylation levels between the treatments, using logistic regression as implemented in methylKit. The DMPs were called following the methylKit manual best practices.
The mapping of the DMPs in the same scaffold and as closer than a given threshold provided their clustering together to identify the DMRs, based on the following procedure. Briefly, the first step was to maximize the number of DMRs in a set of adjacent windows, to identify the best window length for each context. We therefore tested a range of windows, from 100 bp to 2,000 bp. To do so, each potential window (i.e., 100 bp) was screened for DMPs that were significantly differentially methylated (false discovery rate, ≤0.05). The 5′-end of the window was therefore registered to start at the DMP position. Additional DMPs that were mapped within the re-positioned window (i.e., 100 bp) were included in the cluster, provided that the following conditions were met: (i) the direction of the methylation change agreed with the preceding DMP included in the cluster; and (ii) the DMPs to be included were called with a given significance threshold (false discovery rate, ≤0.05). After the additional DMPs were included in the cluster, the window start was registered to the position of the most 3′ of the DMPs included, and the procedure was repeated as described. If no additional DMPs were identified based on the described condition, the scanning procedure was restarted until a DMP was identified. These clusters of DMPs that were composed of a number of DMPs that exceeded a given threshold were analyzed using logistic regression, to identify and define the DMRs.
Once the data for each window length was produced, the operator chose the best length, i.e., the one that maximized the number of DMRs per window (Supplementary Table S9). At this point, the script was re-stared for each context using the adjacent window of the chosen length.
Reference-free workflow
For the genome-independent part of the pipeline, we relied on the robust and simple approach of Schield et al.30, with some modifications. Briefly, as no reference genome was available, the raw reads were collapsed using Rainbow 2.0.4 (https://sourceforge.net/projects/bio-rainbow)67 and CDHit (https://github.com/weizhongli/cdhit)68, to create a pseudo-reference genome that consists of a multi-fasta file that contained the read contigs: this serves as a guide for the mapping algorithm. After mapping the reads to the pseudo-reference using the bwa mem algorithm with its default settings62, a result matrix was created for each sample using Samtools, which counts how many sequences per contig are mapped. This matrix was then used following the loci counting approach described above.
Variant calling and shift in allelic frequency
For both genome-dependent and genome-independent approaches, MCSeEd can perform a variant calling procedure using the Stacks suite61,69. Briefly, after the creation of a population file and a catalog of mutations, a VCF file with frequencies and reference/alternative alleles was created, which was ready to be transformed into any population-genetics exchange file (i.e., PED/MAP).
To highlight unbalanced allelic frequencies in heterozygous loci among the WW and DS samples, and putatively due to differential methylation, the reference (REF) and alternative (ALT) allele counts were extracted from the vcf files and normalized across the samples. Only sites covered by at least 25 reads were considered. Then, at each site, the frequency of the reference allele was computed for each allele across the three replicates, as in Equation (1):
| 1 |
This was then used to calculate the mean frequency at each locus, for each enzymatic context. The shifts in the allele contribution indices between the WW and DS samples were calculated based on Student’s t-tests, followed by Benjamini-Hochberg correction for multiple tests (P < 0.05).
MCSeEd validation
As described by Hashimoto et al.40, for each MCSeEd sequence to be validated, the DNA was digested using the methylation-sensitive enzyme for the corresponding methylcytosine context. The reaction mixture of 25 μL contained 100 ng DNA, 0.5 U of either AciI, PstI, or EcoT22I in the specific buffer defined for each enzyme. For the non-enzyme control (mock), distilled water was added instead of the enzyme. All of the samples were then incubated at 37 °C for 4 h, follow by heat inactivation at 65 °C/80 °C for 20 min.
Real-time PCR for the methylation status was performed (Mx3000P QPCR system; Stratagene, La Jolla, CA, USA) with the SYBR Green JumpStart Taq ReadyMix for Quantitative PCR (Sigma Aldrich). Using the Primer3 software70, specific primers were designed for each randomly chosen DMP to be validated. The sequence information of the primers that bracketed the enzyme site of each DMP are reported in Supplementary Table S15.
The PCR fragments were analyzed using a dissociation protocol, to ensure that each amplicon was a single product. The amplicons were also sequenced to verify the specificities of the targets. The amplification efficiency was calculated from the raw data using the LingRegPCR software71.
All of the qMRE were performed in a final volume of 25 µL that contained 20 ng DNA template (previously digested/mock), 0.2 µM of each primer, and 12.5 µL 2 × PCR Master Mix, according to the manufacturer instructions. The following thermal cycling profile was used: 95 °C for 10 min, followed by 40 cycles of 95 °C for 10 s, 57 °C for 15 s, and 72 °C for 15 s. Following the cycling, the melting curve was determined in the range of 57 °C to 95 °C, with the temperature increment of 0.01 °C/s. Each reaction was run in triplicate (as technical replicates).
The raw Ct data from the real-time PCR were exported to a data file and analyzed using the GeneEx Pro software72. During the pre-processing phase, the data were corrected for PCR efficiency, with the means of the three biological samples calculated. The selected reference gene, GAPDH (GenBank accession no. X15596.1), was subsequently used to normalize the Ct values73–75, and the quantities were calculated relative to the maximum Ct value. As our interest was in fold-changes in the amplification between the mock and treated samples and the WW and DS samples, we ultimately converted the quantities to a logarithmic scale using log base 2 conversion, which also allowed the normal distribution of the values to be tested76.
Statistical analysis
Statistical analyses were performed in R version 3.3.2 (www.r-project.org) using the ‘stats’, ‘factoextra’, and ‘gplots’ packages. The ‘stats’ package was used to estimate correlations and binomial and logistic regression, and ‘factorextra’ was used to perform the principal component analysis. Complete linkage clustering was carried out using the ‘heatmap.2’ function of the ‘gplots’ package, in combination with the ‘hclust’ and ‘dist’ functions, and with ‘ward.D2’ as the clustering method. The MethylKit R package was used to estimate the methylation changes between the WW and DS samples.
Supplementary information
Acknowledgements
The authors acknowledge David Haak (Virginia Tech, VA, USA), Jose Gutierrez-Marcos (University of Warwick, UK), Cristian Forestan (University of Padova, PD, Italy), Daniele Rosellini and all the other people at University of Perugia (Italy) for the critical revision of the manuscript.
Author contributions
G.M. and E.A. devised the study, and funded the experiments. G.M., C.C., E.P., and E.A. developed the technique. G.M., A.P. and S.C., developed the bioinformatic pipeline. G.M. and C.C. performed the library preparations. G.M., A.P., S.C. and A.A. performed the bioinformatic analysis. Investigation. G.M., A.P. and E.A. designed and interpreted all the experimental data. G.M., A.P. and E.A. drafted the manuscript. All authors reviewed and edited the manuscript. All authors read and approved the final manuscript.
Data availability
All sequencing data that support the findings of this study can be found under the bioproject PRJNA533220. The MCSeEd suite and ancillary scripts are available online at https://bitbucket.org/capemaster/mcseed/src/master/.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available this paper at 10.1038/s41598-019-51423-2.
References
- 1.Causevic A, et al. DNA methylating and demethylating treatments modify phenotype and cell wall differentiation state in sugarbeet cell lines. Plant Physiol. Biochem. 2005;43:681–691. doi: 10.1016/j.plaphy.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 2.Feil R, Fraga MF. Epigenetics and the environment: Emerging patterns and implications. Nat. Rev. Genet. 2012;13:97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- 3.Giuliani C, et al. The epigenetic side of human adaptation: Hypotheses, evidences and theories. Ann. Hum. Biol. 2015;42:1–9. doi: 10.3109/03014460.2014.961960. [DOI] [PubMed] [Google Scholar]
- 4.Paszkowski J, Whitham SA. Gene silencing and DNA methylation processes. Curr. Opin. Plant Biol. 2001;4:123–129. doi: 10.1016/S1369-5266(00)00147-3. [DOI] [PubMed] [Google Scholar]
- 5.Zhang X, et al. Genome-wide High-Resolution Mapping and Functional Analysis of DNA Methylation in Arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Hasbún R, et al. HPCE quantification of 5-methyl-2′-deoxycytidine in genomic DNA: Methodological optimization for chestnut and other woody species. Plant Physiol. Biochem. 2008;46:815–822. doi: 10.1016/j.plaphy.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 7.Liang Z, et al. DNA N 6 -Adenine Methylation in Arabidopsis thaliana. Dev. Cell. 2018;45:406–416.e3. doi: 10.1016/j.devcel.2018.03.012. [DOI] [PubMed] [Google Scholar]
- 8.Lang Z, et al. Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc. Natl. Acad. Sci. 2017;114:E4511–E4519. doi: 10.1073/pnas.1705233114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gent JI, et al. CHH islands: De novo DNA methylation in near-gene chromatin regulation in maize. Genome Res. 2013;23:628–637. doi: 10.1101/gr.146985.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gouil, Q. & Baulcombe, D. C. DNA Methylation Signatures of the Plant Chromomethyltransferases. PLoS Genet. 12 (2016). [DOI] [PMC free article] [PubMed]
- 12.Corem, S. et al. Redistribution of CHH methylation and small interfering RNAs across the genome of tomato ddm1 mutants. Plant Cell30, 1628–1644 (2018). [DOI] [PMC free article] [PubMed]
- 13.Iyer LM, Zhang D, Aravind L. Adenine methylation in eukaryotes: Apprehending the complex evolutionary history and functional potential of an epigenetic modification. BioEssays. 2016;38:27–40. doi: 10.1002/bies.201500104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu Y, et al. N 6 -methyldeoxyadenosine marks active transcription start sites in Chlamydomonas. Cell. 2015;161:879–892. doi: 10.1016/j.cell.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greer EL, et al. DNA methylation on N6-adenine in C. elegans. Cell. 2015;161:868–878. doi: 10.1016/j.cell.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mondo SJ, et al. Widespread adenine N6-methylation of active genes in fungi. Nat. Genet. 2017;49:964–968. doi: 10.1038/ng.3859. [DOI] [PubMed] [Google Scholar]
- 17.Xiao C-L, et al. N6-Methyladenine DNA Modification in the Human Genome. Mol. Cell. 2018;71:306–318.e7. doi: 10.1016/j.molcel.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 18.Zhou C, et al. Identification and analysis of adenine N 6 -methylation sites in the rice genome. Nat. Plants. 2018;4:554–563. doi: 10.1038/s41477-018-0214-x. [DOI] [PubMed] [Google Scholar]
- 19.Luo G-Z, Blanco MA, Greer EL, He C, Shi Y. DNA N6-methyladenine: A new epigenetic mark in eukaryotes? Nat. Rev. Mol. Cell Biol. 2015;16:705–710. doi: 10.1038/nrm4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang Z, Geng Y, Gu X. Adenine Methylation: New Epigenetic Marker of DNA and mRNA. Mol. Plant. 2018;11:1219–1221. doi: 10.1016/j.molp.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 21.Frommer M, et al. A genomic sequencing protocol that yields a positive display of 5- methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacinto FV, Ballestar E, Esteller M. Methyl-DNA immunoprecipitation (MeDIP): Hunting down the DNA methylome. BioTechniques. 2008;44:35–43. doi: 10.2144/000112708. [DOI] [PubMed] [Google Scholar]
- 23.Laird PW. Principles and challenges of genome-wide DNA methylation analysis. Nat. Rev. Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 24.Beck S. Taking the measure of the methylome. Nat. Biotechnol. 2010;28:1026–1028. doi: 10.1038/nbt1010-1026. [DOI] [PubMed] [Google Scholar]
- 25.Libertini E, et al. Information recovery from low coverage whole-genome bisulfite sequencing. Nat. Commun. 2016;7:11306. doi: 10.1038/ncomms11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ziller MJ, Hansen KD, Meissner A, Aryee MJ. Coverage recommendations for methylation analysis by whole-genome bisulfite sequencing. Nat. Methods. 2015;12:230–232. doi: 10.1038/nmeth.3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meissner A, et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yong, W.-S., Hsu, F.-M. & Chen, P.-Y. Profiling genome-wide DNA methylation. Epigenetics Chromatin9 (2016). [DOI] [PMC free article] [PubMed]
- 29.Maunakea AK, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schield DR, et al. Incipient speciation with biased gene flow between two lineages of the Western Diamondback Rattlesnake (Crotalus atrox) Mol. Phylogenet. Evol. 2015;83:213–223. doi: 10.1016/j.ympev.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 31.Peterson, B. K., Weber, J. N., Kay, E. H., Fisher, H. S. & Hoekstra, H. E. Double digest RADseq: An inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE7 (2012). [DOI] [PMC free article] [PubMed]
- 32.Zhang Q, et al. N 6 -Methyladenine DNA Methylation in Japonica and Indica Rice Genomes and Its Association with Gene Expression, Plant Development, and Stress Responses. Mol. Plant. 2018;11:1492–1508. doi: 10.1016/j.molp.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 33.Bhushan A, Mukherjee T, Joshi J, Shankar P, Kalia VC. Insights into the Origin of Clostridium botulinum Strains: Evolution of Distinct Restriction Endonuclease Sites in rrs (16S rRNA gene) Indian J. Microbiol. 2015;55:140–150. doi: 10.1007/s12088-015-0514-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Gurp TP, et al. EpiGBS: Reference-free reduced representation bisulfite sequencing. Nat. Methods. 2016;13:322–324. doi: 10.1038/nmeth.3763. [DOI] [PubMed] [Google Scholar]
- 35.You, W. et al. Atypical DNA methylation of genes encoding cysteine-rich peptides in Arabidopsis thaliana. BMC Plant Biol. 12 (2012). [DOI] [PMC free article] [PubMed]
- 36.Luo, G.-Z. et al. Characterization of eukaryotic DNA N 6 -methyladenine by a highly sensitive restriction enzyme-assisted sequencing. Nat. Commun. 7 (2016). [DOI] [PMC free article] [PubMed]
- 37.Pogribny, I. P., Pogribna, M., Christman, J. K. & James, S. J. Single-site methylation within the p53 promoter region reduces gene expression in a reporter gene construct: Possible in vivo relevance during tumorigenesis. Cancer Res. 60, 588–594 (2000). [PubMed]
- 38.Roach HI, et al. Association between the abnormal expression of matrix-degrading enzymes by human osteoarthritic chondrocytes and demethylation of specific CpG sites in the promoter regions. Arthritis Rheum. 2005;52:3110–3124. doi: 10.1002/art.21300. [DOI] [PubMed] [Google Scholar]
- 39.Moore HR, Meehan RR, Young LE. Methylation-sensitive polymerase chain reaction. Methods Mol. Biol. Clifton NJ. 2006;325:239–249. doi: 10.1385/1-59745-005-7:239. [DOI] [PubMed] [Google Scholar]
- 40.Hashimoto K, Kokubun S, Itoi E, Roach HI. Improved quantification of DNA methylation using methylation-sensitive restriction enzymes and real-time PCR. Epigenetics. 2007;2:86–91. doi: 10.4161/epi.2.2.4203. [DOI] [PubMed] [Google Scholar]
- 41.Bonora, G. et al. DNA methylation estimation using methylation-sensitive restriction enzyme bisulfite sequencing (MREBS). bioRxiv 217208, 10.1101/217208 (2017). [DOI] [PMC free article] [PubMed]
- 42.Wu TP, et al. DNA methylation on N 6 -adenine in mammalian embryonic stem cells. Nature. 2016;532:329–333. doi: 10.1038/nature17640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dhar MS, Pethe VV, Gupta VS, Ranjekar PK. Predominance and tissue specificity of adenine methylation in rice. Theor. Appl. Genet. 1990;80:402–408. doi: 10.1007/BF00210080. [DOI] [PubMed] [Google Scholar]
- 44.Brodzik R, Hennig J. Adenine methylation at GATC sequences regulates activity of tobacco PR-1 and PR-2 promoters in electroporated protoplasts. Plant Physiol. Biochem. 1998;36:401–406. doi: 10.1016/S0981-9428(98)80082-X. [DOI] [Google Scholar]
- 45.Ashapkin VV, Kutueva LI, Vanyushin BF. The gene for domains rearranged methyltransferase (DRM2) in Arabidopsis thaliana plants is methylated at both cytosine and adenine residues. FEBS Lett. 2002;532:367–372. doi: 10.1016/S0014-5793(02)03711-0. [DOI] [PubMed] [Google Scholar]
- 46.Kim MY, Zilberman D. DNA methylation as a system of plant genomic immunity. Trends Plant Sci. 2014;19:320–326. doi: 10.1016/j.tplants.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 47.Bewick AJ, et al. On the origin and evolutionary consequences of gene body DNA methylation. Proc. Natl. Acad. Sci. USA. 2016;113:9111–9116. doi: 10.1073/pnas.1604666113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Regulski M, et al. The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res. 2013;23:1651–1662. doi: 10.1101/gr.153510.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boyko, A. et al. Transgenerational adaptation of Arabidopsis to stress requires DNA methylation and the function of dicer-like proteins. PLoS ONE5 (2010). [DOI] [PMC free article] [PubMed]
- 50.Labra M, et al. Analysis of cytosine methylation pattern in response to water deficit in pea root tips. Plant Biol. 2002;4:694–699. doi: 10.1055/s-2002-37398. [DOI] [Google Scholar]
- 51.Gayacharan & Joel, A. J. Epigenetic responses to drought stress in rice (Oryza sativa L.). Physiol. Mol. Biol. Plants19, 379–387 (2013). [DOI] [PMC free article] [PubMed]
- 52.Shoemaker R, Deng J, Wang W, Zhang K. Allele-specific methylation is prevalent and is contributed by CpG-SNPs in the human genome. Genome Res. 2010;20:883–889. doi: 10.1101/gr.104695.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peng, X. et al. CCCH-type zinc finger family in maize: Genome-wide identification, classification and expression profiling under abscisic acid and drought treatments. PLoS ONE7 (2012). [DOI] [PMC free article] [PubMed]
- 54.Shen H, et al. OsWRKY30 is activated by MAP kinases to confer drought tolerance in rice. Plant Mol. Biol. 2012;80:241–253. doi: 10.1007/s11103-012-9941-y. [DOI] [PubMed] [Google Scholar]
- 55.Von Korff M, et al. Asymmetric allele-specific expression in relation to developmental variation and drought stress in barley hybrids. Plant J. 2009;59:14–26. doi: 10.1111/j.1365-313X.2009.03848.x. [DOI] [PubMed] [Google Scholar]
- 56.Chodavarapu RK, et al. Transcriptome and methylome interactions in rice hybrids. Proc. Natl. Acad. Sci. USA. 2012;109:12040–12045. doi: 10.1073/pnas.1209297109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen SX, He H, Deng XW. Allele-specific DNA methylation analyses associated with siRNAs in Arabidopsis hybrids. Sci. China Life Sci. 2014;57:519–525. doi: 10.1007/s11427-014-4629-y. [DOI] [PubMed] [Google Scholar]
- 58.Gao, X. et al. Comparison of TALE designer transcription factors and the CRISPR/dCas9 in regulation of gene expression by targeting enhancers. Nucleic Acids Res. 42 (2014). [DOI] [PMC free article] [PubMed]
- 59.Shivaprasad PV, Dunn RM, Santos BACM, Bassett A, Baulcombe DC. Extraordinary transgressive phenotypes of hybrid tomato are influenced by epigenetics and small silencing RNAs. EMBO J. 2012;31:257–266. doi: 10.1038/emboj.2011.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bocchini, M. et al. Soil selenium (Se) biofortification changes the physiological, biochemical and epigenetic responses to water stress in Zea mays L. by inducing a higher drought tolerance. Front. Plant Sci. 9 (2018). [DOI] [PMC free article] [PubMed]
- 61.Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA. Stacks: An analysis tool set for population genomics. Mol. Ecol. 2013;22:3124–3140. doi: 10.1111/mec.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinforma. Oxf. Engl. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liao Y, Smyth GK, Shi W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 66.Akalin A, et al. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13:R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chong Z, Ruan J, Wu C-I. Rainbow: An integrated tool for efficient clustering and assembling RAD-seq reads. Bioinformatics. 2012;28:2732–2737. doi: 10.1093/bioinformatics/bts482. [DOI] [PubMed] [Google Scholar]
- 68.Li W, Godzik A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 69.Catchen JM, Amores A, Hohenlohe P, Cresko W, Postlethwait JH. Stacks: Building and genotyping loci de novo from short-read sequences. G3 Genes Genomes Genet. 2011;1:171–182. doi: 10.1534/g3.111.000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. Clifton NJ. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 71.Ramakers C, Ruijter JM, Lekanne Deprez RH, Moorman AFM. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003;339:62–66. doi: 10.1016/S0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 72.Kubista M, et al. The real-time polymerase chain reaction. Mol. Aspects Med. 2006;27:95–125. doi: 10.1016/j.mam.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 73.Manjunath S, Sachs MM. Molecular characterization and promoter analysis of the maize cytosolic glyceraldehyde 3-phosphate dehydrogenase gene family and its expression during anoxia. Plant Mol. Biol. 1997;33:97–112. doi: 10.1023/A:1005729112038. [DOI] [PubMed] [Google Scholar]
- 74.Qi J, et al. Reference gene selection for real-time quantitative polymerase chain reaction of mRNA transcript levels in Chinese cabbage (Brassica rapa L. ssp. pekinensis) Plant Mol. Biol. Report. 2010;28:597–604. doi: 10.1007/s11105-010-0185-1. [DOI] [Google Scholar]
- 75.Kumar R, et al. Functional screening of cDNA library from a salt tolerant rice genotype Pokkali identifies mannose-1-phosphate guanyl transferase gene (OsMPG1) as a key member of salinity stress response. Plant Mol. Biol. 2012;79:555–568. doi: 10.1007/s11103-012-9928-8. [DOI] [PubMed] [Google Scholar]
- 76.Capomaccio S, et al. Athletic humans and horses: comparative analysis of interleukin-6 (IL-6) and IL-6 receptor (IL-6R) expression in peripheral blood mononuclear cells in trained and untrained subjects at rest. BMC Physiol. 2011;11:3. doi: 10.1186/1472-6793-11-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data that support the findings of this study can be found under the bioproject PRJNA533220. The MCSeEd suite and ancillary scripts are available online at https://bitbucket.org/capemaster/mcseed/src/master/.