Short abstract
Underlying genetic influences may affect perinatal pain, depression, or both. We investigated the role of 59 single-nucleotide polymorphisms on 20 quantitative traits measured in perinatal women. Moreover, 183 pregnant women (28–37 weeks’ gestation) were prospectively genotyped for single-nucleotide polymorphisms with known prior associations with either pain or depression in nonpregnant populations. Prenatal saliva samples were collected. Phenotypic data were gathered during prenatal, labor and delivery, and postpartum (six weeks and three months) periods, capturing labor pain, Edinburgh Postnatal Depression Score, and Brief Pain Inventories. Following quality control, genotypes were used as predictors and phenotypes as dependent variables in multiple linear regression analyses to detect associations. Three statistical models were tested: additive allele effects, deviation from dominant allele effects, and the joint test of both. rs4633 (a synonymous single-nucleotide polymorphism in COMT) associated with “pain right now” scores at six weeks postpartum. Single-nucleotide polymorphisms rs1135349 (a single-nucleotide polymorphism within a small noncoding RNA that has many prior associations for depression) and rs7548151 (intronic in ASTN1) were associated with the maximum pain unpleasantness score experienced during labor (a measure of the emotional valence of labor pain), controlling for the Holm–Bonferroni family-wise error rate. Sensory dimensions of labor pain (i.e., pain intensity) and postpartum depression scores were not associated with genotyped single-nucleotide polymorphisms. Identifying genomic components of these perinatal complex disorders may produce insights into relevant pathways or novel treatment options.
Keywords: Acute pain, childbirth pain, pain, psychology
Introduction
Labor pain has been associated with postpartum depression,1 but the biological basis of this relationship is unclear. Like other complex human traits, pain is multifactorial, mediated by environmental and genetic factors.2 Pain heritability has been estimated by twin studies (heritability = 16%–50%).3,4 Within DNA studies, patterns of genetic association for pain are apparent. Variants in chaperonin containing TCP1 subunit 5 (CCT5) and family with sequence similarity 173 member B (FAM173B) were associated with chronic widespread pain in a genome-wide association study (GWAS) performed on 7099 European subjects.5 A GWAS performed on 11,891 European women found association of dysmenorrhea in a locus proximal to nerve growth factor (NGF).6
Genetic studies can potentially shed light on common factors between perinatal pain and depression. For example, catechol-O-methyltransferases (COMT) plays a role in degrading catecholamines, via methylation transfer, including neurotransmitters (dopamine, epinephrine, or norepinephrine) and has prior association with depression. Gene variants in COMT have been associated with opioid efficacy in pain studies and has prior association with pain.7,8
Comorbid chronic pain and depression is extremely common in the population (estimated at 30%–60%).9 This comorbidity is strongly associated with females by approximately two-fold.10 The public health burden is considerable, as the costs to treat depression or pain can be up to $560 billion.11 Like pain, depression is a common, multifactorial disorder with a considerable element of heredity, evidenced by prior twin and family studies.12 A case/control meta-analysis identified 44 loci associated with major depression in approximately one half-million research subjects using GWAS.13
A distinct group of depressive symptoms may arise in pregnant women before, during, and after labor and delivery. Past literature supports a potential genetic component. A linkage study for postpartum pain used microsatellite typing and found evidence of linkage in two genomic loci on chromosomes 1q and 9p.14 One single-nucleotide polymorphism (SNP) from chromosome 1q was subsequently replicated in an independent sample.15 In addition, several candidate gene studies identified SNPs associated with perinatal depression, including COMT.16 Candidate gene studies focusing on labor pain have been undertaken with associative findings in ADRB2 (β2-andrenergic receptor), OPRM1 (µ-opioid receptor), and GCH1 (guanosine triphosphate cyclohydrolase).17
Pain has also been linked to postpartum depression.1,18–21 Furthermore, prior studies report the use of epidural analgesia with decreased postpartum depression; however, lack of association has also been reported.22–25 Although the literature correlates chronic pain and depression with common genetic variants, we are unaware of any data that associates labor pain and postpartum depression with a common genetic explanation. Perinatal pain and postpartum depression possess biological and physiological characteristics that are distinct from chronic pain and depression outside of pregnancy and the postpartum period. Identifying common genetic components for pain and depression is important to inform the potential development of novel or innovative therapeutic targets in the perinatal period and to provide insights into risk stratification strategies for perinatal depression and pain management.
In this study, we explored genetic factors associated with pain and depression in perinatal women, to provide preliminary information identifying loci to target in larger genetic studies. We genotyped DNA variants previously associated with either pain or depression in nonpregnant populations, in a cohort of perinatal women who were prospectively phenotyped for prenatal, labor, and postpartum pain and depression. We hypothesized that SNPs associated with either pain or depression in nonpregnant populations are associated with clinical measures of pain or depression in perinatal women.
Methods
A prospective observational approach was chosen. Written informed consent was obtained from all research participants and this study was approved by the University of Pittsburgh Institutional Review Board. A convenience sampling of perinatal women aged 18 years or older were recruited at the third trimester prenatal clinic visit (28–40 weeks estimated gestational age), and prospectively followed throughout labor and delivery, six weeks postpartum, and three months postpartum. Participants were included if they were nulliparous, single gestation, vertex fetal presentation, English literate (due to the use of English language-validated surveys), and presented for spontaneous or induced labor and delivery at term gestation (estimated gestational age ≥37 weeks). Exclusion criteria included chronic pain, current opioid maintenance therapy, preeclampsia or emergency cesarean delivery, body mass index of ≥40 kg/m2 (given data suggesting a relationship between obesity and depression26), and known fetal disease.
Self-reported demographic data included gravidity, parity, race, ethnicity, prenatal history of anxiety or depression, mental illness other than anxiety or depression, and marital status. Obstetric data included mode of delivery, perinatal lacerations, number of supplemental epidural dosing requirements during labor and delivery, and breastfeeding. Duration of labor was measured from labor and delivery data collected from the medical record (see below). Participants completed surveys at baseline (at the time of recruitment), six weeks postpartum, and three months postpartum.
Upon enrollment, depression was measured by the Edinburgh Postnatal Depression Scale (EPDS). Pain assessments were assessed with the pain catastrophizing (PCS) and pain inventory (Brief Pain Inventory, BPI). These instruments are validated and reliable for the constructs they are designed to measure.27–30 For labor and delivery pain data collection, an electronic “pain diary” application was programmed and available via personal electronic device (Google Nexus 7, Android version 4.3 “Jelly Bean,” Mountain View, CA). After admission into the labor and delivery unit, baseline assessment of pain intensity (sensory dimension) and pain unpleasantness (affective dimension) were established, each using a 100 mm horizontal visual analog scale for patient-reported ratings (e.g., “Over the last hour, how intense has your pain been?” and “Over the last hour, how unpleasant has your pain been?”). Zero mm indicated “no intensity (or unpleasantness) at all” and 100 mm indicated “the most intense (or unpleasant) pain I can imagine.” Pain ratings were assessed in this fashion every hour, and the application produced an audible reminder every hour for patients to capture these data. Pain unpleasantness is a measure of the emotional valence of pain31; this affective dimension of pain is distinct from the sensory aspect of pain and is differentially impacted by psychological factors.
Postpartum assessments occurred during hospitalization at zero to three days after childbirth (clinically important time points for acute pain and breastfeeding), at six weeks (this time period corresponds to persistent acute pain and is a common time point for postpartum depression screening), and at three months (this time period corresponds to diagnosis of chronic pain).1,21,32,33 These surveys were sent by e-mail for assessment of pain (BPI) and breastfeeding (yes/no) at each time point. In-hospital postpartum pain and opioid requirement (in milligram morphine equivalents, MME) data were collected. Postpartum pain variables were calculated as described below.
Withdrawal criteria included incomplete baseline or follow-up survey responses, incomplete labor pain diaries for any reason, lack of electronic data transfer from pain diaries, and/or request and receipt of epidural labor analgesia prior to reporting first/baseline pain score in labor pain diary.
Pain and analgesia variables
Labor pain diary data were used to calculate the following pain and analgesia variables per individual patient: first labor and delivery pain score (mm); postepidural analgesia average pain score (mm); labor pain intensity max score (mm); labor pain unpleasantness max score (mm); labor pain intensity burden (area under curve, AUC); and labor pain unpleasantness burden (AUC). Pain AUC has been used to assess aspects of pain burden in multiple studies in obstetric, perioperative, and hospitalized patient populations.34,35 For postpartum pain and analgesia assessments, the following data were collected and calculated. In-hospital opioid requirements were calculated as MME. Postpartum pain management strategies were unchanged from standard clinical care during the study period and were specifically comprised of acetaminophen 650–1000 mg oral every 6 h (vaginal and cesarean deliveries), ibuprofen 800 mg oral every 6 h (vaginal and cesarean deliveries), and oxycodone 5–10 mg orally every 4–6 h as needed for severe breakthrough pain (cesarean deliveries). Pain during the postpartum hospital stay was reported by nursing assessments in the context of routine clinical care and was assessed by an 11-point (0–10) numeric rating scale, where 0 is no pain and 10 is the worst pain imaginable. Pain scores in the postpartum period are expected to be measured at least twice per shift (every 4 h) by nursing staff. For variability in frequency of pain assessment sampling, time-weighted assessments of pain were calculated, as previously described by our group (percent change in pain and time-weighted percent change in pain).1
Labor epidural analgesia was protocolized for this study as follows. Epidural analgesia was initiated at time of patient request. Epidural space was localized by loss-of-resistance to saline and catheters were inserted at a 5 cm depth in the epidural space. Activation was by lidocaine 1.5% with 1:200,000 epinephrine (3 mL “test dose” followed by additional 2 mL), bupivacaine 0.0825% with fentanyl 2 mcg/mL (8 mL), and fentanyl 100 mcg. Continuous patient-controlled epidural analgesia was bupivacaine 0.0825% infusion with fentanyl 2 mcg/mL (8 mL/h), demand 8 mL, and lockout of 24 mL/h. Supplemental epidural dosing protocol included bupivacaine 0.125% with 10–15 mL in 5 mL increments at first request, lidocaine 1% 10 mL in 5 mL increments with adjustment to basal infusion rate to 11 mL/h at second request, and clinician judgment at third request. For women choosing not to use epidural labor analgesia, no other medications were used for pain relief during labor.
Candidate SNP selection
Given sample availability, this cohort was not sufficiently powered to perform GWAS for complex traits such as pain and depression; rather, the goal of this preliminary study is to generate hypotheses and inform future replication studies in larger cohorts. Given the recent wealth of knowledge obtained from pain and depression GWAS that used hundreds of thousands of subjects, we sought to replicate findings in our cohort of perinatal women, a population that has not had extensive genetic mechanistic evaluations for both labor pain and depression. In this study, we focused on genotyping SNPs that were previously associated with either pain or depression. Recently, a GWAS meta-analysis conducted on ∼480,000 subjects revealed 44 risk loci significantly associated with major depression.13 In addition, we considered SNPs that were previously associated with major depression in other studies.13,36,37 Furthermore, prior studies associated several loci with pain perception or pain sensitivity, including COMT, sodium voltage-gated channel alpha subunit 9 (SCN9A), and GTP cyclohydrolase 1 (GCH1).38,39 Based upon these prior associations demonstrated in the published literature, a total of 59 candidate SNPs associated with pain and/or depression were selected for testing and are completely listed in Supplemental Table 1.
DNA extraction and genotyping
Genomic DNA was extracted from saliva of n = 183 perinatal women (collected at 28–37 weeks estimated gestational age; Oragene Saliva Kit DNA Genotek, Inc., Ottawa, Canada) and quantified with PICOgreen dsDNA quantitation kit (ThermoFisher, Inc., Waltham, MA) using manufacturer’s instruction. Subjects from the Centre d’Etude du Polymorphisme Humain (CEPH) were included as positive control DNAs. CEPH is a collection of European ancestry individuals and family members residing in Utah, USA, that were genotyped as part of the 1000 Genomes Project. Their SNP genotypes are freely available to the public (www.internationalgenome.org), and we used three subjects to check for genotype concordance (NA12778, NA12340, and NA12751; Coriell Institute for Medical Research).40 Oligo pools were designed using Agena’s Assay Design Suite (https://agenacx.com). The oligo pool design was able to accommodate n = 59 SNPs into two oligo pools: pool 1 contained 30 SNPs and pool 2 contained 29 SNPs (see Supplemental Table 1 for oligo sequences). SNPs were genotyped by Sequenom iPLEX MALDI-TOF mass spectrometric assays using manufacturer’s protocols (Agena Bioscience, San Diego, CA). Genotyping was conducted by the Genomics Research Core at the University of Pittsburgh.
Genotyping quality control
SNP genotypes were converted into PLINK formatted files, and analysis was performed in PLINK v1.90b4.5 64-bit (25 July 2017).41 SNPs were removed that had >5% failure rate (n = 11), failed Hardy Weinberg expectations (P < 0.001; n = 4), or were discordant for CEPH genotyping (n = 1). Individuals were removed if >5% of markers failed genotyping across both oligo pools (n = 11). Three individuals were chosen at random as duplicates per plate; concordance was 100%. Following quality control measures, there remained n = 173 subjects and 43 markers; the SNP genotyping rate was >99%.
Statistical methods
Linear regression was used to test the null hypothesis that the regression coefficient was not equal to zero across the three genotypes for each clinical phenotype, using race, ethnicity (due to known genetic heterogeneity across racial and ethnic groups), age, and a prior history of anxiety or depression as covariates. Variables were assessed for multicollinearity. PLINK software was used to assess additive gene effects (ADD), deviation from dominance effects (DOMDEV), and the joint effect of both ADD and DOMDEV (GENO-2DF). Each clinical measure was standardized using the “–standard-beta” command, which set the mean of each clinical variable to zero and unit variance. To reduce type I errors, multiple comparisons were addressed using Holm–Bonferroni method (H–B).42 Tests were considered significant when the H–B family-wise error rate was less than 0.05. Furthermore, for any SNP reaching significance from the regression analysis, the mean differences were assessed using a one-way analysis of variance (ANOVA) in GraphPad Prism version 7. ANOVA was performed on the clinical variables among the three genotype groups (alpha level of significance = 0.05), and then a post hoc Tukey’s test was used to assess the mean differences of each genotype group by comparing the mean of each genotype group to the means of the other two genotype groups. Significance for Tukey’s testing was assessed for each comparison using family-wise significance level P < 0.05.
Missing data
We evaluated data for missingness. Where data were missing for labor pain, we evaluated the nature of missingness and compared demographic characteristics between participants who completed the study and participants who had missing data. Where patterns of missingness could be established or where uncertainty existed in type of missing data, a conservative assumption was made that the data was missing not at random.
Results
Cohort characteristics and quantitative traits
Of the 183 subjects genotyped, 173 perinatal women passed genotyping quality control filters and were included in the final analyses. Table 1 represents demographic characteristics of the study cohort. All women had not previously experienced childbirth (parity = 0). Of these, 65.3% (n = 113) planned to receive labor epidural analgesia; the remaining 30.6% (n = 53) planned not to use labor epidural analgesia. In this cohort, 62.4% had no history of depression or anxiety in the prenatal period, and 87.3% had no history of other mental illness. There was no significant relationship between history of anxiety or depression and postpartum pain intensity maximum score (estimate 4.27, 95% confidence interval −9.26 to 17.8, P = 0.53). Results for quantitative traits obtained before, during, and after labor and delivery are reported in Tables 1 and 2.
Table 1.
Cohort demographics and obstetric, labor and delivery, and postpartum characteristics of the total cohort.
| Variable | n = 173 |
|---|---|
| Prenatal assessment | |
| Body mass index (kg/m2) | 31.9 (6.0) |
| Education (years) | 15.3 (2.0) |
| Parity | |
| 0 | 173 (100) |
| Race | |
| American Indian | 1 (0.58) |
| Asian | 12 (6.94) |
| African American | 34 (19.65) |
| European/Caucasian | 117 (67.63) |
| Other | 1 (0.58) |
| Not reported | 8 (4.62) |
| Ethnicity | |
| Hispanic or Latino | 4 (2.31) |
| Not Hispanic or Latino | 161 (93.06) |
| Not reported | 8 (4.62) |
| Gravidity | |
| 1 | 119 (68.79) |
| 2 | 16 (9.25) |
| 3 | 6 (3.47) |
| 4 | 3 (1.73) |
| Not reported | 29 (16.76) |
| Marital status | |
| Single | 43 (24.85) |
| Married | 117 (67.63) |
| Divorced | 3 (1.73) |
| Not reported | 10 (5.78) |
| Prenatal history of anxiety or depression | |
| Yes | 57 (32.95) |
| No | 108 (62.43) |
| Not reported | 8 (4.62) |
| History of mental illness other than anxiety or depression | |
| Yes | 12 (6.94) |
| No | 151 (87.28) |
| Not reported | 10 (5.78) |
| Plan to use labor epidural analgesia | |
| Yes | 113(65.32) |
| No | 53 (30.64) |
| Not reported | 7 (4.05) |
| Childbirth/labor assessment | |
| Labor characteristics | |
| Estimated gestational age (weeks) | 39.3 (1.3) |
| Duration of labor (hours) | 16.0 (8.3) |
| Mode of delivery | |
| Normal spontaneous vaginal delivery | 94 (54.34) |
| Assisted vaginal—vacuum | 3 (1.73) |
| Cesarean—nonreassuring fetal tracing | 7 (4.05) |
| Cesarean—arrest of dilation/descent | 12 (6.93) |
| Cesarean—other | 9 (5.20) |
| Not reported | 48 (27.75) |
| Perineal lacerations | |
| None | 41 (23.70) |
| First degree | 19 (10.98) |
| Second degree | 60 (34.68) |
| Third degree | 3 (1.73) |
| Fourth degree | 1 (0.58) |
| Not reported | 49 (28.32) |
| Number of supplemental labor epidural analgesia doses | |
| 0 | 84 (48.55) |
| 1 | 13 (7.51) |
| 2 | 5 (2.89) |
| 3 | 1 (0.58) |
| 4 | 1 (0.58) |
| 5 | 0(0) |
| 6 | 1 (0.58) |
| Not reported | 68 (39.30) |
| Postpartum assessment | |
| Breastfeeding, postpartum days 1–2 | |
| Yes | 75 (43.35) |
| No | 5 (2.89) |
| Not reported | 93 (53.76) |
| Breastfeeding, six weeks postpartum | |
| Yes | 67 (38.73) |
| No | 9 (5.61) |
| Not reported | 97 (56.07) |
| Breastfeeding, three months postpartum | |
| Yes | 56 (32.37) |
| No | 17 (9.83) |
| Not reported | 100 (57.80) |
Note: Data are reported as frequency (%) or mean (standard deviation).
Table 2.
Perinatal pain and depression phenotypes reported across prenatal, labor, and postpartum time points.
| Pain and depression traits (units) | n | Mean (SD) | 95% CI |
|---|---|---|---|
| Depression | |||
| EPDS (baseline score) | 167 | 5 (4) | 4.4–5.6 |
| EPDS (six weeks postpartum score) | 82 | 4.1 (3.8) | 3.3–5.0 |
| EPDS (three months postpartum score) | 74 | 4 (3.7) | 3.1–4.8 |
| Labor pain | |||
| Initial labor pain score (mm) | 88 | 6.4 (6) | 5.1–7.6 |
| Labor pain intensity max (mm) | 88 | 77.8 (20.1) | 73.5–82.1 |
| Labor pain intensity burden (AUC) | 88 | 429 (333.3) | 358.4–499.7 |
| Labor pain unpleasantness max (mm) | 88 | 79.7 (19.5) | 75.6–83.8 |
| Labor pain unpleasantness burden (AUC) | 88 | 443.1 (349.8) | 369.0–517.2 |
| Supplemental labor epidural analgesia doses (number) | 106 | 2.8 (4.2) | 2.0–3.6 |
| Postpartum pain, zero to three days after delivery | |||
| Postpartum opioid requirements (MME)a | 106 | 30 (50.5) | 20.3–39.7 |
| Postpartum percent change in pain (%)a | 118 | −11 (47.1) | −19.6 to −2.4 |
| Time-weighted postpartum percent change in pain (%)a | 118 | 3.9 (1.5) | 3.6–4.2 |
| Postpartum pain, six weeks after delivery | |||
| Pain at worst in last 24 h (score) | 81 | 4.9 (2.7) | 4.3–5.5 |
| Pain at least in last 24 h (score) | 81 | 5.4 (2.8) | 4.8–6.0 |
| Pain on average (score) | 81 | 2.9 (2.3) | 2.4–3.4 |
| Pain right now (score) | 80 | 2.9 (2.3) | 2.4–3.4 |
| Postpartum pain, three months after delivery | |||
| Pain at worst in last 24 h (score) | 71 | 1.1 (2) | 0.6–1.5 |
| Pain at least in last 24 h (score) | 71 | 1.1 (2) | 0.6–1.6 |
| Pain on average (score) | 71 | 0.7 (1.6) | 0.3–1.1 |
| Pain right now (score) | 71 | 0.5 (1.5) | 0.1–0.8 |
EPDS: Edinburgh Postnatal Depression Scale; AUC: area under curve; MME: milligram morphine equivalents; SD: standard deviation; CI: confidence interval.
aMeasurements included in-hospital data only.
Multiple linear regression testing
We fit three regression models for each quantitative trait per SNP: (i) additive genetic effects (ADD), (ii) deviation from dominance (DOMDEV), and (iii) a two-degree-of-freedom joint test of ADD and DOMDEV (GENO-2DF). In the ADD model, increased quantitative phenotype scores (the dependent variables) as a function of the number of minor alleles counted is observed when the regression coefficient, β, is positive. The DOMDEV model tests whether a significant proportion of risk is attributable to the individuals who have a heterozygous genotype, that is, a “heterozygote advantage.” The data are coded AA = 0, Aa = 1, and aa = 0, where a negative β (slope) in the DOMDEV model indicate the recessive allele is associated with increased quantitative trait scores. The GENO-2DF is a t test accounting for the slopes of both the ADD and DOMDEV together, where the two groups tested are homozygotes or heterozygotes versus homozygotes of the opposite allele. There were 69 SNP/phenotype pairs that were unable to be tested due to detection of multicollinearity.
SNP rs4633 is associated with “pain right now” score at 6 weeks postpartum (BPI #6)
Using the genotypes for rs4633 as predictors and “pain right now” score at 6 weeks postpartum as the dependent variable, we detected an association using linear regression analysis, considering race, ethnicity, age, and a history of anxiety or depression as covariates, for additive genetic effects (ADD; β = −0.33, 95% confidence interval (CI) = −0.54 to −0.13, P = 0.002), for deviation from dominance genetic effects (DOMDEV; β = −0.23, 95% CI = −0.43 to −0.02, P = 0.03), and for the joint effects (GENO_2DF; unadjusted P = 0.008, H–B family-wise error rate = 0.029; Table 3). The slopes (β) of the regression lines were negative for both the ADD and the DOMDEV models, suggesting the major allele is associated with higher “pain right now” score at six weeks postpartum, and individuals with heterozygous genotypes have lower “pain right now” score at six weeks postpartum than individuals with either homozygous genotypes.
Table 3.
Multiple regression of phenotypes was performed on perinatal women, adjusted for race, ethnicity, age, and history of anxiety or depression, using coefficients from terms on additive gene effects (ADD), deviation from dominance (DOMDEV), and the joint effect of ADD and DOMDEV (GENO-2DF).
| Coefficient |
GENO_2DF |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clinical outcome | Chr: base (hg19) | SNP | Allele | Symbol | Transcript | global genomeAD MAF (n subjects) | Test | β | SE | 95% CI | T | P | T | P | H–B |
| Pain right now (six weeks postpartum) | chr22: 19,950,485 | rs4633 | C/T | COMT | ENST 00000361682 | 0.4626 (140,690) | ADD | −0.33 | 0.11 | −0.54 to −0.13 | −3.18 | 0.002 | 14.35 | 0.0008 | 0.029 |
| DOMDEV | −0.23 | 0.10 | −0.43 to −0.02 | −2.19 | 0.03 | ||||||||||
| Labor pain unpleasantness max score (labor) | chr5: 164,523,472 | rs11135349 | A/C | Intergenic | ENST 00000519570 | 0.614 (15,683) | ADD | −0.43 | 0.11 | −0.65 to −0.21 | −3.83 | 0.0003 | 16.69 | 0.0002 | 0.009 |
| DOMDEV | 0.20 | 0.10 | −0.004 to 0.40 | 1.93 | 0.058 | ||||||||||
| chr1: 177,026,733 | rs7548151 | G/A | ASTN1 | ENST 00000361833 | 0.1345 (15,684) | ADD | −0.60 | 0.15 | −0.89 to −0.31 | −4.1 | 0.0001 | 18.8 | 8.3 × 10−5 | 0.003 | |
| DOMDEV | 0.59 | 0.15 | 0.29 to 0.88 | 3.9 | 0.0002 | ||||||||||
Note: These findings support that alleles increase risk for the pain phenotypes, specifically for additive gene effects (both “pain right now at six weeks postpartum” and “labor pain unpleasantness maximum score” phenotypes) and for DOMDEV effects (“pain right now at six weeks postpartum” phenotype). The relationships between rs4633 and “pain right now at six weeks postpartum” and rs11135349, rs7548151, and labor pain unpleasantness maximum score were significant for the joint test that accounted for both additive and dominant-deviance models, after considering multiple comparisons using Holm–Bonferroni family-wise error rate (H–B), where tests with H–B rate < 0.05 were considered significant. H–B: Holm–Bonferroni family-wise error rate; CI: confidence interval; Chr: chromosome; SE: standard error; T: student t test; β: regression coefficient; ADD: additive gene effect model; DOMDEV: deviation from dominance effect model; GENO_2DF: joint test of the coefficients for ADD and DOMDEV models; SNP: single-nucleotide polymorphism; MAF: minor allele frequency.
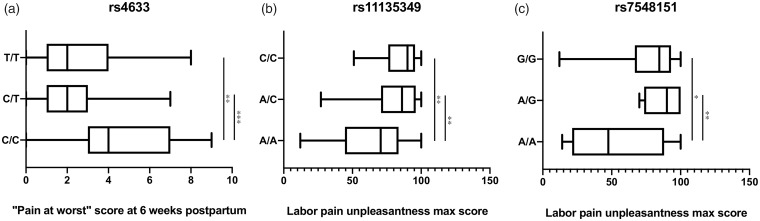
Furthermore, a one-way ANOVA was performed on “pain right now” score at six weeks postpartum for each of the three genotype groups and yielded a significant result (F(2,76)=9.058, P = 0.0003; Figure 1(a)). A post hoc Tukey’s test for “pain right now” score at six weeks postpartum showed group differences between the genotype groups (C/C vs. T/C, adjusted P = 0.0002 and C/C vs. T/T, adjusted P = 0.0077; Figure 1(a)). The T/C versus T/T group comparison did not have significantly different means (adjusted P = 0.676; Figure 1(a)). Indeed, as indicated by the regression analysis, the heterozygous individual group had the lowest mean value of “pain right now” score at six weeks postpartum (mean = 2.189, standard deviation (SD) = 1.838) compared to either homozygous group (C/C group mean = 4.632, SD = 2.543 and T/T group mean = 2.652, SD = 1.968).
Figure 1.
Box plot of clinical variables, BPI short 6pp (a) and pain unpleasantness max (b and c), by genotype of rs4633, rs11135349, and rs7548151, respectively, in perinatal women. Tukey’s multiple comparisons post hoc test was performed on the clinical variables between each genotype group per SNP. *P ≤ 0.05, **0.005< P <0.01, and ***P < 0.0005.
SNPs rs11135349 and rs7548151 are associated with labor pain unpleasantness maximum score
Using the genotypes for rs11135349 or rs7548151 as predictors and “labor pain unpleasantness maximum score” as the dependent variable, we detected an association using linear regression analysis, considering race, ethnicity, age, and history of anxiety or depression as covariates. SNP rs11135349 was significant for ADD (β = −0.43, 95% CI = −0.65 to −0.21, P = 0.003) and for GENO_2DF (t = 16.69, unadjusted P = 0.0002, H–B family-wise error rate = 0.009). The DOMDEV genetic effects model approached significance (P = 0.062; Table 3).
SNP rs7548151 was significant for ADD (β = −0.60, 95% CI = −0.89 to −0.31, P = 0.001), DOMDEV (β = 0.59, 95% CI = 0.29 to 0.88, P = 0.0002), and GENO_2DF (t = 18.8, P = 8.3 × 10−5, H–B family-wise error rate = 0.003; Table 3).
The slopes (β) of the regression lines for the ADD models for rs11135349 or rs7548151 were negative, indicating the minor allele for each SNP was associated with an increased score for labor pain unpleasantness maximum.
For group differences, one-way ANOVA was performed on the labor pain unpleasantness maximum score values for each of the three genotype groups and yielded a significant finding for rs11135349 and rs7548151 (F(2,84) = 7.673, P = 0.0009 and F(2,84)=7.182, P = 0.028, respectively; Figure 1(b) and (c)). The post hoc Tukey’s test for labor pain unpleasantness maximum score showed group differences between the genotype groups for rs11135349 and rs7548151 (A/A vs. A/C, adjusted P = 0.0021 and A/A vs. C/C, adjusted P = 0.0013; and A/A vs. A/G, adjusted P = 0.005 and A/A vs. G/G, adjusted P = 0.02; Figure 1(b) and (c)). The A/C versus C/C or A/G versus GG group comparisons for rs11135349 and rs7548151, respectfully, did not have significantly different mean differences (adjusted P = 0.813; Figure 1(b)).
Pertinent negative findings
We failed to reject the null hypothesis for the remaining quantitative traits across the genotyped SNPs in this cohort. Specifically, we note labor pain intensity traits (i.e., sensory components of pain), as well as the EPDS scores (at baseline, six weeks postpartum, and three months postpartum) were not significant following multiple comparisons correction for any of the genotyped SNPs. The full list of linear regression test results is available upon request.
Sensitivity analyses for missing data
There were missing data for “pain unpleasantness maximum score” (50%) and for “pain right now at six weeks” (54%). These variables are related to each other in that if a participant could not complete the labor pain diaries for any reason, she was withdrawn from the study, and subsequently “pain right now” assessments at six weeks were also missing. For the participants who did not compete the labor pain diaries (n = 61), whereby the “pain unpleasantness maximum score” was calculated, these data were conservatively judged to be missing not at random as they were logged as missing due to the following reasons: participants progressing through labor quickly (not enough time to complete the diary or incomplete entries; n = 42), pain diary data transfer problems (network issues; n = 7), requirement for cesarean delivery without labor for reasons of breech, development of preeclampsia, or other obstetric indications (n = 10), or receiving epidural analgesia prior to completing the diary (n = 2).
Results of the comparisons of demographic characteristics between missing and nonmissing data are shown in Supplemental Table 2. Groups of missingness were different for duration of labor (hours) and statistically, but not clinically, significantly different for level of education. Given the above considerations and these findings, we therefore assumed that the missing data for pain unpleasantness maximum score were missing not at random (MNAR). Given MNAR, coupled with the frequency of missing data exceeding 50%, it was judged that imputation was not appropriate, and results were interpreted within the context of the limitations presented with this information.
Discussion
In this study, we identified genetic factors contributing to depression and pain in perinatal women. Both pain and depression are complex, multifactorial disorders encompassing a wide range of symptoms and mechanisms: cognitive, emotional, and physical. While our cohort is a small, exploratory cohort, we were able to identify loci with modest associations. We identified three SNPs that were associated with two pain phenotypes: (1) rs4633 (a variant in the coding region of COMT) associated with “pain right now” at six weeks postpartum; (2) rs11135349 (a variant in an intergenic region previously associated with major depressive disorder) was associated with labor pain unpleasantness max score (a measure of the emotional valence or affective component of pain); and (3) rs7548151, an intronic SNP that maps to astroactin-1 (ASTN1), was previously associated with depression in a British study.37
Together, these data suggest that genetic variants are a common factor between depression and perinatal pain, and they provide a biological basis for continued research to explain the observed associations between perinatal pain and depression.
SNP rs4633 is a common (gnomeAD43 global minor allele frequency (MAF) = 0.4626), missense variant in the coding region of COMT (EC2.1.1.6) on chromosome 22q11, resulting in increased translation rate.44 COMT functions as a methyltransferase that mediates methyl group transfer from S-adenosylmethionine to catecholamines, including neurotransmitters like dopamine,45 which can result in catecholamine transmitter degradation.46 A total of 11 COMT transcripts have been described in GTEx47 with transcripts ENST00000467943.1 and ENST00000406520.3 having nearly ubiquitous and high levels of expression across the 53 human body tissues analyzed.
In our study, the mean pain score (millimeters) value of “pain right now” at six weeks postpartum for individuals with C/C genotype (reference allele) was 4.6 mm (standard error of the mean (SEM) = 0.58), with T/C genotype (variant) was 2.2 mm (SEM = 0.30), with T/T genotype (variant) was 2.7 mm (SEM = 0.41). In other words, mutant alleles were associated with less pain than wild-type alleles. We know that rs4633 mutations result in CAC-CAT codon change, which has implications for molecular binding motifs (proteins, RNA, etc.). Altogether, this information points to required research to test what binding partners exist for rs4633 that could impact pain.
To our knowledge, this is the first study to find such an association between rs4633 and pain at six weeks postpartum. Our data suggest that individuals with C/C genotypes for rs4633 had higher mean pain scores when assessed at six weeks postpartum. A prior study found that rs4633 was associated with “longer” durations of latent labor (approximately 5 h)48; however, the association was for the opposite alleles than our study. A potential reason for observing opposite alleles may be due to differences in minor allele frequencies between study cohorts: our study was comprised of individuals from North America (United States), while Terkawi’s cohort was from Saudi Arabia. In that study, the duration of latent labor was dependent on the time of admission to the hospital. Presentation and admission to the hospital for childbirth may, in fact, have been driven by the degree of pain experienced by women during the early latent phase of labor. Therefore, it is possible that the findings of Terkawi et al. are not necessarily a function of latent labor duration, but rather a function of severity of pain in latent labor, that may then dictate a woman’s presentation to the hospital for delivery. If it was in fact the pain (and not necessarily the duration of the latent phase of labor) that correlated with rs4633, then those findings are consistent with our finding of rs4633 associations with pain.
We found that two SNPs (rs11135349 and rs7548151) are associated with maximum labor pain unpleasantness score (a measurement of the emotional valence of pain). SNP rs11135349 maps to an intergenic region on chromosome 5q34 and was selected for genotyping in this cohort due to its prior association with major depressive disorder in the largest GWAS meta-analysis of major depression to date.13 SNP rs11135349 is a common SNP (gnomeAD global MAF = 0.6155) across population groups, where it maps to a noncoding/antisense RNA locus, as supported by GTEx RNA-seq Gene Expression (ENSG00000241956.5) and Transcript Expression (ENST00000519267, ENST00000519570, ENST00000522646, and ENST00000522303) analyses performed on 570 participant donors (Figure 2).47 These noncoding RNA transcripts, identified by GTEx, were present in CNS tissues.47 When considering a 200 kb flanking region around rs11135349, there are a striking number of past GWAS that find significant association with depression,36,37,49,50 neuroticism,51,52 subjective well-being,49 and neuropathic pain in post total joint replacement surgery for osteoarthritis (Figure 2).53 Given these considerations, this locus may be an important region that dictates associations between both phenotypes of pain and depression, not only in perinatal women but also in the broader population. Genetic studies examining pain phenotypes in larger cohorts are justified to establish this relationship.
Figure 2.
The 4.3 mega-base region of Chromosome 5 that flanks rs11135349 (filled star) (chr5:162614336–166985335). This area is absent of any known, documented UCSC gene coding loci (considered an intergenic region). Documented UCSC genes are mapped in the first panel “UCSC Genes.” The second panel displays SNPs, within a 200 kb flanking region, that had prior association for depression (filled circle and filled star),13,36,37,49,50 neuroticism (filled square),51,52 subjective well-being (filled rectangle),49 and pain (filled diamond).53 GTEx RNA-sequencing analysis describes gene and transcript expression from this region, directly overlying the present SNP, rs11135349 (third and fourth panels, respectively), indicating rs11135349 maps to noncoding RNA species. In the present study, rs11135349 was associated with pain phenotype (“pain unpleasantness maximum score” measured during labor). In prior work,13 rs11135349 was associated with depression. Image modified from the UCSC Genome Browser (assembly release date: Human Feb 2009 (GRCh37/hg19); http://genome.ucsc.edu/).61
Although this region is intergenic, it may be functional for noncoding RNAs, supported by the above studies, for pain and depression. Other disorders resulting from variation in noncoding RNAs have been documented, such as cancer, Alzheimer disease, Prader–Willi syndrome, Parkinson disease, and deafness.54 However, the function of these noncoding RNA transcripts on 5q34 is presently unknown, and validation is warranted. The present study identifies a relationship between this noncoding RNA SNP (rs11135349) and labor pain unpleasantness; our own data have similarly found that labor pain unpleasantness is significantly associated with depression scores at six weeks postpartum, in women planning/receiving labor epidural analgesia (R2 = 0.41; P = 0.001).55 Together, these findings suggest that this noncoding RNA species may contribute to disease etiology not only for depression phenotypes but also for labor pain.
The other SNP associated with labor pan unpleasantness (the emotional valence of pain) was rs7548151. Both SNPs have been associated with depression phenotypes in GWAS studies.37 SNP rs7548151 maps to astrotactin-1 (ASTN1), a neuronal adhesion molecule that functions in the migration of postmitotic neuroblasts.56 While these SNPs do not map to an exonic region, they may be in linkage disequilibrium with and tagging truly causal variants. These findings provide further support for a central nervous system component that may underlie a relationship between depression and pain.
Notably, we did not find associations between SNPs that were previously associated with major depression, with the depression scores (EPDS) measures obtained at any time point (prenatal, six weeks postpartum, or three months postpartum). This failure to detect an association does not necessarily rule out a common genetic explanation for both perinatal pain and depression, but rather, may be due to (1) the small sample size of the current study and the small effect size of each SNP on phenotypic risk or (2) fundamental biological/mechanistic differences between perinatal depression and depression outside of pregnancy and the postpartum period. Prior association studies on major depression need sample sizes that contain hundreds of thousands of subjects.13 Alternatively, perinatal depression may be unique and distinct from major depression. To our knowledge, there have been no GWAS findings for perinatal depression reported in The GWAS Catalog (accessed 12 August 2018).57 This may be due to the temporal nature of depression onset, weaker biological evidence for SNP associations, or perhaps an etiological model that is epigenetic in nature.58
Some shortcomings of this study should be noted. The genotyping failure of some SNPs in COMT prevented us from performing haplotype analyses. Prior studies find associations of COMT haplotypes with metrics of pain59 and pain experienced by female individuals diagnosed with major depressive disorder.60 Furthermore, the use of self-reported ancestry cannot rule out type 1 error. Moreover, SNP MAFs were not compared in this study due to population stratification (i.e., MAF differences between groups may be due to group differences in ancestry and not due to a positive association of a SNP to a trait). Furthermore, introducing subgroups would have dramatically decreased the power to detect associations, and therefore we did not pursue these analyses for this study. Future studies incorporating an increased number of perinatal women and an increased number of SNP markers, including those missed in COMT, are justified and will add power to this study. Finally, there was missing data for labor pain. The missing data potentially may have biased our sample toward an underestimation of relationship due to omission of patients who had rapid labors (potentially short but intense periods of labor pain with subsequent omission of pain diary data), or who may have otherwise had immeasurably different characteristics with respect to risk for both pain and depression.
Conclusions
In conclusion, we have identified genetic loci that are associated with perinatal pain and which require replication in a larger cohort of obstetric patients. These identified loci have known associations with pain and depression in nonpregnant individuals, and they map to genetic regions that may have functional consequences, including to mechanisms underlying the relationships between perinatal pain and depression. Continued research to identify the genomic components of these complex disorders may produce insights into relevant pathways or novel treatment options in the obstetric population.
Summary Statement
SNPs in COMT (rs4633) and in an intergenic region associated with depression (rs1135349) are linked to pain experienced at six weeks postpartum and to the emotional valence of labor pain, respectively.
Supplemental Material
Supplemental material, MPX882139 Supplemental Material1 for Genetic associations of perinatal pain and depression by Lora McClain, Lia Farrell, Kelsea LaSorda, Lisa A. Pan, David Peters and Grace Lim in Molecular Pain
Supplemental Material
Supplemental material, MPX882139 Supplemental Material2 for Genetic associations of perinatal pain and depression by Lora McClain, Lia Farrell, Kelsea LaSorda, Lisa A. Pan, David Peters and Grace Lim in Molecular Pain
Acknowledgments
The authors sincerely thank the research participants for their involvement in this study. This project used the University of Pittsburgh HSCRF Genomics Research Core to genotype DNA using the iPLEX genotyping platform, and the authors also thank Deborah Hollingshead and Erica Fong for their assistance.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by the Department of Anesthesiology & Perioperative Medicine, University of Pittsburgh School of Medicine. The project was supported by a grant from the University of Pittsburgh Physicians Academic Foundation to Dr. Lim. Dr. Lim is supported by an award from the NIH/ORWH Building Interdisciplinary Research Careers in Women’s Health (BIRCWH), NIH K12HD043441, and by the NIH Ruth Kirschstein National Service Award, NIH T32MG075770. Funding was also provided by donations administered via the Children’s Hospital of Pittsburgh Foundation to Drs. Pan and Peters.
Supplemental Material
Supplemental material for this article is available online.
References
- 1.Lim G, Farrell LM, Facco FL, Gold MS, Wasan AD. Labor analgesia as a predictor for reduced postpartum depression scores: a retrospective observational study. Anesth Analg 2018; 126: 1598–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mogil JS. The genetic mediation of individual differences in sensitivity to pain and its inhibition. Proc Natl Acad Sci U S A 1999; 96: 7744–7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trost Z, Strachan E, Sullivan M, Vervoort T, Avery AR, Afari N. Heritability of pain catastrophizing and associations with experimental pain outcomes: a twin study. Pain 2015; 156: 514–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nielsen CS, Knudsen GP, Steingrimsdottir OA. Twin studies of pain. Clin Genet 2012; 82: 331–340. [DOI] [PubMed] [Google Scholar]
- 5.Peters MJ, Broer L, Willemen HLDM, Eiriksdottir G, Hocking LJ, Holliday KL, Horan MA, Meulenbelt I, Neogi T, Popham M, Schmidt CO, Soni A, Valdes AM, Amin N, Dennison EM, Eijkelkamp N, Harris TB, Hart DJ, Hofman A, Huygen FJPM, Jameson KA, Jones GT, Launer LJ, Kerkhof HJM, de Kruijf M, McBeth J, Kloppenburg M, Ollier WE, Oostra B, Payton A, Rivadeneira F, Smith BH, Smith AV, Stolk L, Teumer A, Thomson W, Uitterlinden AG, Wang K, van Wingerden SH, Arden NK, Cooper C, Felson D, Gudnason V, Macfarlane GJ, Pendleton N, Slagboom PE, Spector TD, Völzke H, Kavelaars A, van Duijn CM, Williams FMK, van Meurs JBJ. Genome-wide association study meta-analysis of chronic widespread pain: evidence for involvement of the 5p15.2 region. Ann Rheum Dis 2013; 72: 427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones AV, Hockley JRF, Hyde C, Gorman D, Sredic-Rhodes A, Bilsland J, McMurray G, Furlotte NA, Hu Y, Hinds DA, Cox PJ, Scollen S. Genome-wide association analysis of pain severity in dysmenorrhea identifies association at chromosome 1p13.2, near the nerve growth factor locus. Pain 2016; 157: 2571–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao P, Ding YY, Wang ZB, Ma JM, Hong T, Pan SN. Effect of gene polymorphism of COMT and OPRM1 on the preoperative pain sensitivity in patients with cancer. Int J Clin Exp Med 2015; 8: 10036–10039. [PMC free article] [PubMed] [Google Scholar]
- 8.Schmahl C, Ludascher P, Greffrath W, Kraus A, Valerius G, Schulze TG, Treutlein J, Rietschel M, Smolka MN, Bohus M. COMT val158met polymorphism and neural pain processing. PLoS One 2012; 7: e23658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnow BA, Hunkeler EM, Blasey CM, Lee J, Constantino MJ, Fireman B, Kraemer HC, Dea R, Robinson R, Hayward C. Comorbid depression, chronic pain, and disability in primary care. Psychosom Med 2006; 68: 262–268. [DOI] [PubMed] [Google Scholar]
- 10.Substance Abuse and Mental Health Services Administration. 2016 National Survey on Drug Use and Health Washington, DC: National Institutes of Health; 2017.
- 11.Gaskin DJ, Richard P. The economic costs of pain in the United States. J Pain 2012; 13: 715–724. [DOI] [PubMed] [Google Scholar]
- 12.Lohoff FW. Overview of the genetics of major depressive disorder. Curr Psychiatry Rep 2010; 12: 539–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, Adams MJ, Agerbo E, Air TM, Andlauer TMF, Bacanu S-A, Bækvad-Hansen M, Beekman AFT, Bigdeli TB, Binder EB, Blackwood DRH, Bryois J, Buttenschøn HN, Bybjerg-Grauholm J, Cai N, Castelao E, Christensen JH, Clarke T-K, Coleman JIR, Colodro-Conde L, Couvy-Duchesne B, Craddock N, Crawford GE, Crowley CA, Dashti HS, Davies G, Deary IJ, Degenhardt F, Derks EM, Direk N, Dolan CV, Dunn EC, Eley TC, Eriksson N, Escott-Price V, Kiadeh FHF, Finucane HK, Forstner AJ, Frank J, Gaspar HA, Gill M, Giusti-Rodríguez P, Goes FS, Gordon SD, Grove J, Hall LS, Hannon E, Hansen CS, Hansen TF, Herms S, Hickie IB, Hoffmann P, Homuth G, Horn C, Hottenga J-J, Hougaard DM, Hu M, Hyde CL, Ising M, Jansen R, Jin F, Jorgenson E, Knowles JA, Kohane IS, Kraft J, Kretzschmar WW, Krogh J, Kutalik Z, Lane JM, Li Y, Li Y, Lind PA, Liu X, Lu L, MacIntyre DJ, MacKinnon DF, Maier RM, Maier W, Marchini J, Mbarek H, McGrath P, McGuffin P, Medland SE, Mehta D, Middeldorp CM, Mihailov E, Milaneschi Y, Milani L, Mill J, Mondimore FM, Montgomery GW, Mostafavi S, Mullins N, Nauck M, Ng B, Nivard MG, Nyholt DR, O’Reilly PF, Oskarsson H, Owen MJ, Painter JN, Pedersen CB, Pedersen MG, Peterson RE, Pettersson E, Peyrot WJ, Pistis G, Posthuma D, Purcell SM, Quiroz JA, Qvist P, Rice JP, Riley BP, Rivera M, Saeed Mirza S, Saxena R, Schoevers R, Schulte EC, Shen L, Shi J, Shyn SI, Sigurdsson E, Sinnamon GBC, Smit JH, Smith DJ, Stefansson H, Steinberg S, Stockmeier CA, Streit F, Strohmaier J, Tansey KE, Teismann H, Teumer A, Thompson W, Thomson PA, Thorgeirsson TE, Tian C, Traylor M, Treutlein J, Trubetskoy V, Uitterlinden AG, Umbricht D, Van der Auwera S, van Hemert AM, Viktorin A, Visscher PM, Wang Y, Webb BT, Weinsheimer SM, Wellmann J, Willemsen G, Witt SH, Wu Y, Xi HS, Yang J, Zhang F, Arolt V, Baune BT, Berger K, Boomsma DI, Cichon S, Dannlowski U, de Geus ECJ, DePaulo JR, Domenici E, Domschke K, Esko T, Grabe HJ, Hamilton SP, Hayward C, Heath AC, Hinds DA, Kendler KS, Kloiber S, Lewis G, Li QS, Lucae S, Madden PFA, Magnusson PK, Martin NG, McIntosh AM, Metspalu A, Mors O, Mortensen PB, Müller-Myhsok B, Nordentoft M, Nöthen MM, O’Donovan MC, Paciga SA, Pedersen NL, Penninx BWJH, Perlis RH, Porteous DJ, Potash JB, Preisig M, Rietschel M, Schaefer C, Schulze TG, Smoller JW, Stefansson K, Tiemeier H, Uher R, Völzke H, Weissman MM, Werge T, Winslow AR, Lewis CM, Levinson DF, Breen G, Børglum AD, Sullivan PF. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet 2018; 50: 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahon PB, Payne JL, MacKinnon DF, Mondimore FM, Goes FS, Schweizer B, Jancic D, Coryell WH, Holmans PA, Shi J, Knowles JA, Scheftner WA, Weissman MM, Levinson DF, DePaulo JR, Zandi PP, Potash JB. Genome-wide linkage and follow-up association study of postpartum mood symptoms. Am J Psychiatry 2009; 166: 1229–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alvim-Soares AM, Miranda DM, Campos SB, Figueira P, Correa H, Romano-Silva MA. HMNC1 gene polymorphism associated with postpartum depression. Rev Bras Psiquiatr 2014; 36: 96–97. [DOI] [PubMed] [Google Scholar]
- 16.Doornbos B, Dijck-Brouwer DA, Kema IP, Tanke MA, van Goor SA, Muskiet FA, Korf J. The development of peripartum depressive symptoms is associated with gene polymorphisms of MAOA, 5-HTT and COMT. Prog Neuro-Psychopharmacol Biol Psychiatry 2009; 33: 1250–1254. [DOI] [PubMed] [Google Scholar]
- 17.Landau R, Smiley R. Pharmacogenetics in obstetric anesthesia. Best Pract Res Clin Anaesthesiol 2017; 31: 23–34. [DOI] [PubMed] [Google Scholar]
- 18.Gaudet C, Wen SW, Walker MC. Chronic perinatal pain as a risk factor for postpartum depression symptoms in Canadian women. Can J Public Health 2013; 104: e375–e387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henderson J. Sixteen per cent of women have depression symptoms in the year after childbirth and are more likely to report physical symptoms, including tiredness and back pain. Evid Based Nurs 2015; 18: 36. [DOI] [PubMed] [Google Scholar]
- 20.Safadi RR, Abushaikha LA, Ahmad MM. Demographic, maternal, and infant health correlates of post-partum depression in Jordan. Nurs Health Sci 2016; 18: 306–313. [DOI] [PubMed] [Google Scholar]
- 21.Eisenach JC, Pan PH, Smiley R, Lavand’homme P, Landau R, Houle TT. Severity of acute pain after childbirth, but not type of delivery, predicts persistent pain and postpartum depression. Pain 2008; 140: 87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu YM, McArthur E, Dixon S, Dirk JS, Welk BK. Association between intrapartum epidural use and maternal postpartum depression presenting for medical care: a population-based, matched cohort study. Int J Obstet Anesth 2018; 35: 10–16. [DOI] [PubMed] [Google Scholar]
- 23.Orbach-Zinger S, Landau R, Harousch AB, Ovad O, Caspi L, Kornilov E, Ioscovich A, Bracco D, Davis A, Fireman S, Hoshen M, Eidelman LA. The relationship between women’s intention to request a labor epidural analgesia, actually delivering with labor epidural analgesia, and postpartum depression at 6 weeks: a prospective observational study. Anesth Analg 2018; 126: 1590–1597. [DOI] [PubMed] [Google Scholar]
- 24.Nahirney M, Metcalfe A, Chaput KH. Administration of epidural labor analgesia is not associated with a decreased risk of postpartum depression in an urban Canadian population of mothers: a secondary analysis of prospective cohort data. Local Region Anesth 2017; 10: 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding T, Wang DX, Qu Y, Chen Q, Zhu SN. Epidural labor analgesia is associated with a decreased risk of postpartum depression: a prospective cohort study. Anesth Analg 2014; 119: 383–392. [DOI] [PubMed] [Google Scholar]
- 26.Jantaratnotai N, Mosikanon K, Lee Y, McIntyre RS. The interface of depression and obesity. Obes Res Clin Pract 2017; 11: 1–10. [DOI] [PubMed] [Google Scholar]
- 27.Cleeland CS, Ryan KM. Pain assessment: global use of the Brief Pain Inventory. Ann Acad Med Singapore 1994; 23: 129–138. [PubMed] [Google Scholar]
- 28.Osman A, Barrios FX, Kopper BA, Hauptmann W, Jones J, O’Neill E. Factor structure, reliability, and validity of the Pain Catastrophizing Scale. J Behav Med 1997; 20: 589–605. [DOI] [PubMed] [Google Scholar]
- 29.Kernot J, Olds T, Lewis LK, Maher C. Test-retest reliability of the English version of the Edinburgh Postnatal Depression Scale. Arch Womens Mental Health 2015; 18: 255–257. [DOI] [PubMed] [Google Scholar]
- 30.Shrestha SD, Pradhan R, Tran TD, Gualano RC, Fisher JR. Reliability and validity of the Edinburgh Postnatal Depression Scale (EPDS) for detecting perinatal common mental disorders (PCMDs) among women in low-and lower-middle-income countries: a systematic review. BMC Pregnancy Childbirth 2016; 16: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price DD, Harkins SW, Baker C. Sensory-affective relationships among different types of clinical and experimental pain. Pain 1987; 28: 297–307. [DOI] [PubMed] [Google Scholar]
- 32.Classification of chronic pain. Descriptions of chronic pain syndromes and definitions of pain terms. Prepared by the International Association for the Study of Pain, Subcommittee on Taxonomy. Pain Suppl 1986; 3: S1–S226. [PubMed] [Google Scholar]
- 33.World Health Organization. Depression and Other Common Mental Disorders. Geneva, Switzerland: World Health Organization; 2017: 24. [Google Scholar]
- 34.Komatsu R, Carvalho B, Flood PD. Recovery after nulliparous birth: a detailed analysis of pain analgesia and recovery of function. Anesthesiology 2017; 127: 684–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kannampallil T, Galanter WL, Falck S, Gaunt MJ, Gibbons RD, McNutt R, Odwazny R, Schiff G, Vaida AJ, Wilkie DJ, Lambert BL. Characterizing the pain score trajectories of hospitalized adult medical and surgical patients: a retrospective cohort study. Pain 2016; 157: 2739–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hyde CL, Nagle MW, Tian C, Chen X, Paciga SA, Wendland JR, Tung JY, Hinds DA, Perlis RH, Winslow AR. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat Genet 2016; 48: 1031–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howard DM, Adams MJ, Shirali M, Clarke T-K, Marioni RE, Davies G, Coleman JRI, Alloza C, Shen X, Barbu MC, Wigmore EM, Gibson J, Hagenaars SP, Lewis CM, Ward J, Smith DJ, Sullivan PF, Haley CS, Breen G, Deary IJ, McIntosh AM. Genome-wide association study of depression phenotypes in UK Biobank identifies variants in excitatory synaptic pathways. Nat Commun 2018; 9: 1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Candiotti KA, Yang Z, Buric D, Arheart K, Zhang Y, Rodriguez Y, Gitlin MC, Carvalho E, Jaraba I, Wang L. Catechol-o-methyltransferase polymorphisms predict opioid consumption in postoperative pain. Anesth Analg 2014; 119: 1194–1200. [DOI] [PubMed] [Google Scholar]
- 39.Diatchenko L, Nackley AG, Slade GD, Bhalang K, Belfer I, Max MB, Goldman D, Maixner W. Catechol-O-methyltransferase gene polymorphisms are associated with multiple pain-evoking stimuli. Pain 2006; 125: 216–224. [DOI] [PubMed] [Google Scholar]
- 40.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature 2015; 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat 1979; 6: 65–70. [Google Scholar]
- 43.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won H-H, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016; 536: 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsao D, Shabalina SA, Gauthier J, Dokholyan NV, Diatchenko L. Disruptive mRNA folding increases translational efficiency of catechol-O-methyltransferase variant. Nucleic Acids Res 2011; 39: 6201–6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Napolitano A, Cesura AM, Da Prada M. The role of monoamine oxidase and catechol O-methyltransferase in dopaminergic neurotransmission. J Neural Transm Suppl 1995; 45: 35–45. [PubMed] [Google Scholar]
- 46.Axelrod J, Tomchick R. Enzymatic O-methylation of epinephrine and other catechols. J Biol Chem 1958; 233: 702–705. [PubMed] [Google Scholar]
- 47. The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013; 45: 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Terkawi AS, Jackson WM, Thiet MP, Hansoti S, Tabassum R, Flood P. Oxytocin and catechol-O-methyltransferase receptor genotype predict the length of the first stage of labor. Am J Obstet Gynecol 2012; 207: 184.e1–184.e8. [DOI] [PubMed] [Google Scholar]
- 49.Turley P, Walters RK, Maghzian O, Okbay A, Lee JJ, Fontana MA, Nguyen-Viet TA, Wedow R, Zacher M, Furlotte NA, Magnusson P, Oskarsson S, Johannesson M, Visscher PM, Laibson D, Cesarini D, Neale BM, Benjamin DJ. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat Genet 2018; 50: 229–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X, Luo Z, Gu C, Hall LS, McIntosh AM, Zeng Y, Porteous DJ, Hayward C, Li M, Yao Y-G, Zhang C, Luo X-J. Common variants on 6q16.2, 12q24.31 and 16p13.3 are associated with major depressive disorder. Neuropsychopharmacol 2018; 43: 2146–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Moor MH, van den Berg SM, Verweij KJ, Krueger RF, Luciano M, Arias Vasquez A, Matteson LK, Derringer J, Esko T, Amin N. Meta-analysis of genome-wide association studies for neuroticism, and the polygenic association with major depressive disorder. JAMA Psychiatry 2015; 72: 642–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okbay A, Baselmans BML, De Neve J-E, Turley P, Nivard MG, Fontana MA, Meddens SFW, Linnér RK, Rietveld CA, Derringer J, Gratten J, Lee JJ, Liu JZ, de Vlaming R, Ahluwalia TS, Buchwald J, Cavadino A, Frazier-Wood AC, Furlotte NA, Garfield V, Geisel MH, Gonzalez JR, Haitjema S, Karlsson R, van der Laan SW, Ladwig K-H, Lahti J, van der Lee SJ, Lind PA, Liu T, Matteson L, Mihailov E, Miller MB, Minica CC, Nolte IM, Mook-Kanamori D, van der Most PJ, Oldmeadow C, Qian Y, Raitakari O, Rawal R, Realo A, Rueedi R, Schmidt B, Smith AV, Stergiakouli E, Tanaka T, Taylor K, Thorleifsson G, Wedenoja J, Wellmann J, Westra H-J, Willems SM, Zhao W, Amin N, Bakshi A, Bergmann S, Bjornsdottir G, Boyle PA, Cherney S, Cox SR, Davies G, Davis OSP, Ding J, Direk N, Eibich P, Emeny RT, Fatemifar G, Faul JD, Ferrucci L, Forstner AJ, Gieger C, Gupta R, Harris TB, Harris JM, Holliday EG, Hottenga J-J, De Jager PL, Kaakinen MA, Kajantie E, Karhunen V, Kolcic I, Kumari M, Launer LJ, Franke L, Li-Gao R, Liewald DC, Koini M, Loukola A, Marques-Vidal P, Montgomery GW, Mosing MA, Paternoster L, Pattie A, Petrovic KE, Pulkki-Råback L, Quaye L, Räikkönen K, Rudan I, Scott RJ, Smith JA, Sutin AR, Trzaskowski M, Vinkhuyzen AE, Yu L, Zabaneh D, Attia JR, Bennett DA, Berger K, Bertram L, Boomsma DI, Snieder H, Chang S-C, Cucca F, Deary IJ, van Duijn CM, Eriksson JG, Bültmann U, de Geus EJC, Groenen PJF, Gudnason V, Hansen T, Hartman CA, Haworth CMA, Hayward C, Heath AC, Hinds DA, Hyppönen E, Iacono WG, Järvelin M-R, Jöckel K-H, Kaprio J, Kardia SLR, Keltikangas-Järvinen L, Kraft P, Kubzansky LD, Lehtimäki T, Magnusson PKE, Martin NG, McGue M, Metspalu A, Mills M, de Mutsert R, Oldehinkel AJ, Pasterkamp G, Pedersen NL, Plomin R, Polasek O, Power C, Rich SS, Rosendaal FR, den Ruijter HM, Schlessinger D, Schmidt H, Svento R, Schmidt R, Alizadeh BZ, Sørensen TIA, Spector TD, Starr JM, Stefansson K, Steptoe A, Terracciano A, Thorsteinsdottir U, Thurik AR, Timpson NJ, Tiemeier H, Uitterlinden AG, Vollenweider P, Wagner GG, Weir DR, Yang J, Conley DC, Smith GD, Hofman A, Johannesson M, Laibson DI, Medland SE, Meyer MN, Pickrell JK, Esko T, Krueger RF, Beauchamp JP, Koellinger PD, Benjamin DJ, Bartels M, Cesarini D. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat Genet 2016; 48: 624–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Warner SC, van Meurs JB, Schiphof D, Bierma-Zeinstra SM, Hofman A, Uitterlinden AG, Richardson H, Jenkins W, Doherty M, Valdes AM. Genome-wide association scan of neuropathic pain symptoms post total joint replacement highlights a variant in the protein-kinase C gene. Eur J Hum Genet 2017; 25: 446–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 2011; 12: 861–874. [DOI] [PubMed] [Google Scholar]
- 55.Lim G, Lasorda KR, Farrell LM, Nam S, Wasan AD. Pernatal, labor, and postpartum pain are associated with postpartum depression scores: a prospective observational study. J Womens Health 2018; 27: 1424. [Google Scholar]
- 56.Fink JM, Hirsch BA, Zheng C, Dietz G, Hatten ME, Ross ME. Astrotactin (ASTN), a gene for glial-guided neuronal migration, maps to human chromosome 1q25.2. Genomics 1997; 40: 202–205. [DOI] [PubMed] [Google Scholar]
- 57.MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, Junkins H, McMahon A, Milano A, Morales J, Pendlington ZM, Welter D, Burdett T, Hindorff L, Flicek P, Cunningham F, Parkinson H. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res 2017; 45: D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Osborne L, Clive M, Kimmel M, Gispen F, Guintivano J, Brown T, Cox O, Judy J, Meilman S, Braier A, Beckmann MW, Kornhuber J, Fasching PA, Goes F, Payne JL, Binder EB, Kaminsky Z. Replication of epigenetic postpartum depression biomarkers and variation with hormone levels. Neuropsychopharmacol 2016; 41: 1648–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Diatchenko L, Slade GD, Nackley AG, Bhalang K, Sigurdsson A, Belfer I, Goldman D, Xu K, Shabalina SA, Shagin D, Max MB, Makarov SS, Maixner W. Genetic basis for individual variations in pain perception and the development of a chronic pain condition. Hum Mol Genet 2005; 14: 135–143. [DOI] [PubMed] [Google Scholar]
- 60.Fijal B, Perlis RH, Heinloth AN, Houston JP. The association of single nucleotide polymorphisms in the catechol-O-methyltransferase gene and pain scores in female patients with major depressive disorder. J Pain 2010; 11: 910–915, 915.e1–915.e9. [DOI] [PubMed] [Google Scholar]
- 61.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res 2002; 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, MPX882139 Supplemental Material1 for Genetic associations of perinatal pain and depression by Lora McClain, Lia Farrell, Kelsea LaSorda, Lisa A. Pan, David Peters and Grace Lim in Molecular Pain
Supplemental material, MPX882139 Supplemental Material2 for Genetic associations of perinatal pain and depression by Lora McClain, Lia Farrell, Kelsea LaSorda, Lisa A. Pan, David Peters and Grace Lim in Molecular Pain