Abstract
Collapsin response mediator proteins (CRMPs) are intracellular proteins that mediate signals for several extracellular molecules, such as Semaphorin3A and neurotrophins. The phosphorylation of CRMP1 and CRMP2 by Cdk5 at Ser522 is involved in axonal guidance and spine development. Here, we found that the Ser522-phosphorylated CRMP1 and/or CRMP2 are enriched in the dendrites of cultured cortical neurons and P7 cortical section. To determine the physiological role of CRMPs in dendritic development, we generated CRMP2 knock-in mutant mice (crmp2ki/ki) in which the Ser residue at 522 was replaced with Ala. Strikingly, the cortical basal dendrites of double mutant crmp2ki/ki and crmp1−/− mice exhibited severe abnormal dendritic patterning, which we defined as “curling phenotype.” These findings demonstrate that the function of CRMP1 and CRMP2 synergistically control dendritic projection, and the phosphorylation of CRMP2 at Ser522 is essential for proper dendritic field organization in vivo.
Introduction
Cortical pyramidal neurons are the most highly polarized cells composed of two structurally and functionally distinct parts, the axon and dendrites (Craig and Banker, 1994). The dendrites are further classified into apical and basal dendrites, based on their origins and covered fields. Cortical apical dendrites are transformed from the leading process of migrating neurons and elongated toward the pial surface (Hatanaka and Murakami, 2002). Basal dendrites are emerged from the cell bodies and are extended into deeper layer of the cortical plate. Both apical and basal dendrites develop inherent stereotaxic morphology and complexity in their respective fields. In addition to axonal guidance, the dendritic field organization is required for proper neuronal circuits (Parrish et al., 2007). However, the mechanism governing the dendritic guidance and patterning is largely unknown.
Collapsin response mediator proteins (CRMP1-5) are the intracellular phospho-proteins that mediate signals for numerous extracellular cues such as Semaphorins, Ephrins, neurotrophins, and Reelin. CRMPs are involved in cell migration (Yamashita et al., 2006), axonal guidance (Goshima et al., 1995; Arimura et al., 2005; Uchida et al., 2005; Yoshimura et al., 2005), dendritic spine development (Yamashita et al., 2007) and synaptic plasticity (Yamashita et al., 2011) through its phosphorylation. In Semaphorin3A (Sema3A) signaling, CRMP2 is phosphorylated by Cdk5 at Ser522. This phosphorylation is necessary for the subsequent phosphorylation by glycogen synthase kinase-3β (GSK-3β) at Ser518, Thr514, and Thr509 (Brown et al., 2004; Cole et al., 2004; Uchida et al., 2005; Yoshimura et al., 2005). In cdk5−/− mice brain, the phosphorylation of CRMP1 and CRMP2 at Ser522 is largely suppressed (Uchida et al., 2005). Cortex-specific conditional cdk5−/− mice exhibit disorganized dendrites as well as inverted cortical layer formation (Ohshima et al., 2007). These findings suggest that Cdk5-CRMP signaling may be involved in the cortical dendritic development.
To elucidate in vivo significance of the Ser522 phosphorylation of CRMP2, we here generated CRMP2 S522A knock-in (crmp2ki/ki) mice. Strikingly, the layer V cortical neurons in the double mutant crmp2ki/ki and crmp1−/− mice showed severe irregular basal dendritic projection. This finding indicates that CRMP1 and CRMP2 are both necessary for, and the phosphorylation of CRMP2 at Ser522 is involved in proper dendritic field organization.
Materials and Methods
Generation of crmp2ki/ki mice.
A targeting vector construct was designed to insert a neomycin resistance cassette flanked by two loxP sites into the crmp2 gene. A point mutation was introduced to exon 13. Codon TCC coding for Ser at 522 was replaced with GCC coding for Ala. The diphtheria toxin A (DT-A) gene was used for negative selection against random integration. The vector was electroporated into the mouse embryonic stem (ES) cell line, E14. Selected clones possessing mutated allele were injected into blastocysts from C57BL/6J to generate chimeric mice. Heterozygous mice were obtained by crossing chimeric mice with C57BL/6J females. The heterozygous mice appeared normal and were bred for homozygous. To remove the neomycin- resistant gene, we mated the heterozygous mice with EIIaCre transgenic mice (Williams-Simons and Westphal, 1999), and then backcross with C57BL/6J. For genotyping, we used following primers, P1: 5′-GAGCGTGGTGAACATCGCGT-3′, P2: 5′-GTGGTGCCTGCTGCTTGGCA-3′, and P3: 5′-CTAGACTGGACGTAAACTCCTCTTC-3′. For DNA sequencing of mutant mice, we amplified genomic fragment by PCR using following primers, P4: 5′-GGCCGTCGACCCACACATCATAGTCCTGTCTCGG-3′, and P5: 5′-GTGTAGGGACTTGGGTGAGAGAAT-3′. The DNA sequence was analyzed using primer P6: 5′-CACATCATAGTCCTGTCTCGGCTA-3′. To confirm whether the loss of phosphorylation of CRMP2 at Ser522 is observed in the crmp2ki/ki mice, immunoblot analysis was performed as previously described (Yamashita et al., 2006).The primary antibodies were as follows: anti-CRMP2 mouse monoclonal (C4G, IBL), anti-pCRMP1/2(S522) rabbit polyclonal (Uchida et al., 2005), anti-CRMP1 hamster monoclonal (2E7G) (Yamashita et al., 2006). Anti-pCRMP1(T509) and anti-pCRMP2(T509) rabbit polyclonal antisera were raised by injection of synthetic phosphopeptides; VYEVPApTPKHAAPC and VCEVSVpTPKTVTPC (amino acids 503–515 plus Cys for conjugation), respectively. The antisera were purified with protein A beads.
Mutant mice.
The crmp1−/− mice were generated as described previously (Charrier et al., 2006). YFP-H mice (Feng et al., 2000) were purchased from Jackson laboratory. EIIaCre mice were kindly provided by Dr. Heiner Westphal (NIH). Genotypes of the offspring of these mutant lines were assessed using PCR. All mice were housed in the standard mouse facility and fed autoclaved diet and water. All procedures were performed according to the guidelines outlined in the Institutional Animal Care and Use Committee of Yokohama City University School of Medicine and RIKEN Brain Science Institute.
Cell Culture and Transfection.
Primary culture of cortical neurons was prepared from E16.5 ICR and mutant mice as described previously (Yamashita et al., 2007). In cultured neurons from mutant mice, the neurons were transfected with EGFP using Lipofectamine 2000 (Invitrogen) at 7 DIV, and fixed for immunocytochemistry at 14 DIV.
Immunostaining and histology.
As for immunohistochemistry, the C57BL6 mouse was perfused intracardially with 4% (v/v) PFA in 0.1 m phosphate buffer, pH7.4, at P7. The brain tissue was fixed with 4% PFA for 24 h at 4°C. Sections were cut with a vibratome (50 μm). Immunostaining was performed using anti-pCRMP1/2(S522) and anti-MAP2 antibodies. TO-PRO-3 iodide (Invitrogen) was used for nuclei staining. Immunocytochemistry was performed by a standard protocol as described previously (Yamashita et al., 2007). The primary antibodies used for staining the cultured neurons were as follows: anti-GFP rabbit polyclonal (MBL), anti-Tau-1 mouse monoclonal (Millipore), anti-MAP2 mouse monoclonal (Sigma), anti-pCRMP1/2(S522) and anti-CRMP2 (C4G).
As for in vivo morphological analysis, the brain tissues of mutant mice were fixed at P17–P18 and sections were then cut with a vibratome (100 μm). They were analyzed using confocal microscopy (Zeiss LSM510), and the z-axis series of images were superimposed to one image using Zeiss software.
Image analysis.
To quantify the overlap between pCRMP1/2S522 and total CRMP2 (C4G)-, MAP2,- or Tau-1- immunoreactive signals, we calculate colocalization coefficients using WCIF ImageJ software. To evaluate whether cultured cortical neurons have curling dendrite(s), we measured the radius of curvature (R) of the most winding dendrite of each EGFP-transfected neurons using the ThreePointCircularROI plug-in (developed by Dr. G. Landini, University of Birmingham, Birmingham, UK) from ImageJ software (see Fig. 2E). The neurons with R < 60 from the winding dendrite(s) were defined as curling phenotype. To perform quantitative evaluation of the irregular dendritic pattering in vivo, the cell body was placed in the center of a square (200 μm). Each square was further divided into 16 (4 by 4) smaller squares of 50 μm (see Fig. 3C). We then plotted the tips of basal dendrites and quantified how the basal dendrites were distributed in these squares. Sholl analysis was performed as described previously (Sholl, 1953).
Figure 2.
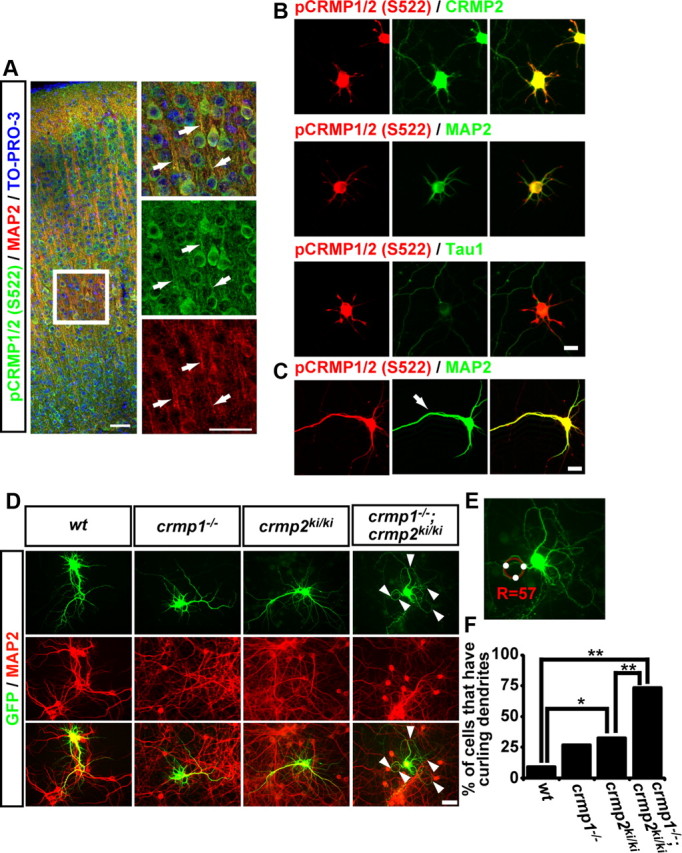
Subcellular localization of pCRMP1/2 S522 and dendritic abnormality in cultured cortical neurons from crmp1−/−;crmp2ki/ki. A, Vibratome slice (50 μm thick) of P7 mouse brain was coimmunostained with anti-pCRMP1/2(S522) antibody and anti-MAP2 antibody. The nuclei were stained with TO-PRO-3. The arrows show colocalization of phosphorylated CRMP1/2 and MAP2 in dendritic processes. Scale bar, 50 μm. B, Cultured cortical neurons from wt mice at DIV8 were coimmunostained with anti-pCRMP1/2(S522) antibody and anti-CRMP2 (C4G), anti-MAP2, or anti-Tau-1 antibodies. Scale bar, 20 μm. C, Cultured hippocampal neurons from E16.5 wt mice at DIV8 were coimmunostained with anti-pCRMP1/2(S522) antibody and anti-MAP2 antibody. The apical dendrite-like process was defined by anti-MAP2 staining (arrow). Scale bar, 20 μm. D, Typical morphology of cortical neurons from each genotype at DIV14. EGFP labeled neurons were double-stained with anti-GFP antibody and anti-MAP2 antibody. The arrowheads show abnormal dendrites. Scale bar, 50 μm. E, Measurement of the radius of curvature (R) of the most winding dendrite. F, Quantitative data of cultured neurons in the percentage of the neurons that have curling dendrites from each genotype. *p < 0.08, **p < 0.001 by χ2 distribution. n = 40–101 neurons.
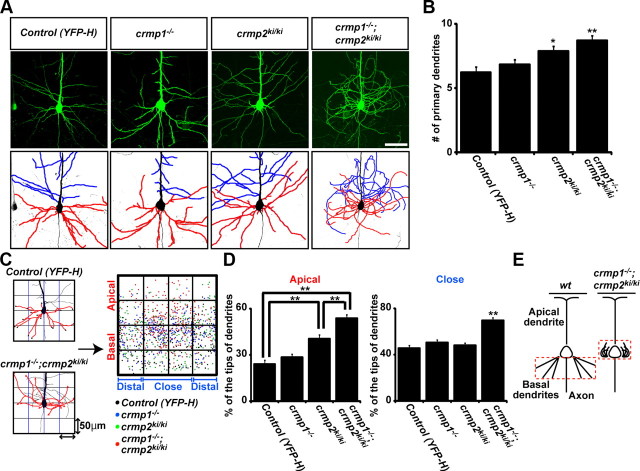
Figure 3.
Double mutant crmp1−/−;crmp2ki/ki mice show defective basal dendritic field organization. A, YFP-labeled neurons (top) and dendritic tracings from these cells (bottom). Tracings depict basal dendrites (red) and collaterals of apical dendrite (blue). The double mutant mice show disorientation of basal dendrites and collaterals from apical trunk. Scale bar, 50 μm. B, The quantitative analysis of the number of primary dendrites of layer V cortical neurons from each mutant and control mice. C, Quantitative analysis method for the basal dendritic phenotype. The confocal images were divided into four boxes of 50 μm each from the center of the cell body. The number of basal dendrite tips in each box was counted. The percentage of basal dendrite tips per box was calculated in apical to basal and close to distal boxes. D, The ratio of dendritic tips in apical or closer part from center of cell body. Definitions of apical and basal, or close and distal are shown in C. E, Summary of the dendritic phenotype of double mutant mice. Data represent the mean values ± SEM for n = 24–30 from 3 individuals. *p < 0.05, **p < 0.001 by one-way ANOVA.
Statistical analysis.
Data were analyzed by one-way ANOVA or χ2 distribution. The p < 0.05 was used as a criterion for statistical significance (one-way ANOVA). In the case of χ2 distribution, the p < 0.008 was statistical significance according to Bonferroni correction.
Results
Generation of crmp2ki/ki mice
Since Ser522 residue of CRMP2 is the major phosphorylation site for Cdk5 in vivo, we generated CRMP2 knock-in (crmp2ki/ki) mice in which the serine residue is replaced with alanine (Fig. 1A–C). In this mutant mice brain lysates, not only Ser522 phosphorylation but also Thr509 phosphorylation of CRMP2 was completely suppressed (Fig. 1D). This is consistent with our in vitro observation that CRMP2 phosphorylation at Ser522 is required for subsequent CRMP2 phosphorylation at Thr509 by GSK-3β (Uchida et al., 2005). Because the major phenotype of cdk5−/− mice is inverted cortical layer formation (Ohshima et al., 1996), we first examined the cortical layer formation of crmp2ki/ki mice. We also intercrossed crmp2ki/ki mice with crmp1−/− mice (Charrier et al., 2006) to eliminate functional redundancy of Ser522 signaling from CRMP1. The gross cytoarchitecture of cortical layer of both crmp2ki/ki and crmp1−/−;crmp2ki/ki mice was essentially normal (data not shown).
Figure 1.
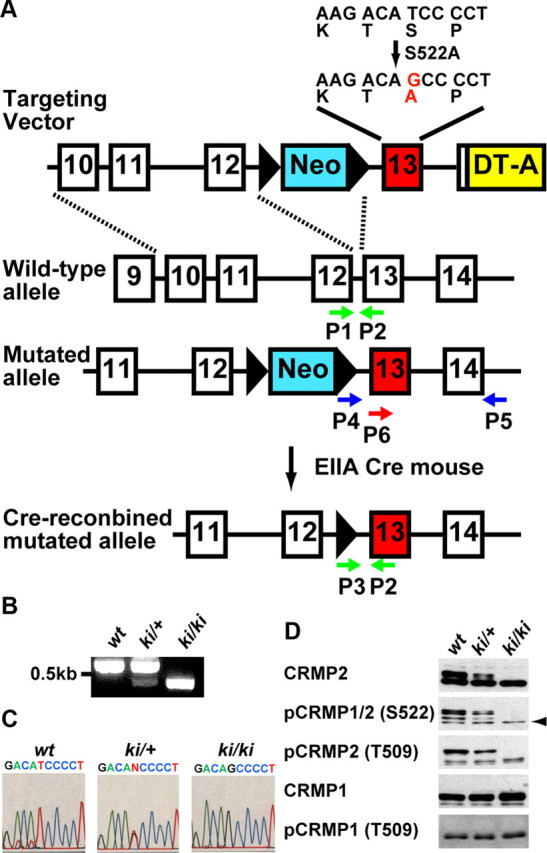
Generation of CRMP2S522A knock-in mice. A, Strategy for the generation of the CRMP2–S522A knock-in mutation in mouse ES cells. PCR primers used in the genotypic analysis are shown by arrows (P1–P6). B, Genotyping analysis for mutated allele by PCR. PCR primers were used in A (P1–P3). C, DNA sequencing. The genomic fragment of around exon13 was amplified by PCR using primers P4 and P5. The DNA sequence was analyzed using primer P6. D, Immunoblot analysis of CRMP1/2 protein and phosphorylation. Immunoblot analysis of P0 wt, Heterozygous (ki/+), and Homozygous (ki/ki) mouse brain lysates with anti-CRMP2 antibody (C4G), anti-pCRMP1/2(S522), anti-pCRMP2(T509), anti-CRMP1, and anti-pCRMP1(T509) antibodies. Arrow indicates pCRMP1 S522.
Phosphorylated CRMP1 and/or CRMP2 are localized in the dendrites of cortical and hippocampal neurons
To elucidate the physiological role of CRMP2 phosphorylation in neuronal development, we performed coimmunostaining of mouse P7 brains with anti-pCRMP1/2(S522) and anti-MAP2 antibodies. The localization of pCRMP1/2(S522) was well overlapped with MAP2, a marker of dendrites (Fig. 2A). We also performed coimmunostaining with anti-pCRMP1/2(S522) antibody and anti-CRMP2 (C4G), anti-MAP2, or anti-Tau-1 antibodies in cultured cortical neurons. The colocalization coefficients with anti-pCRMP1/2 were 0.671 ± 0.008 (anti-CRMP2), 0.807 ± 0.009 (anti-MAP2), and 0.463 ± 0.013 (anti-Tau-1) (n = 60 neurons), indicating that pCRMP1/2 at Ser522 were mainly localized at the MAP2-positive neurites (Fig. 2B). We also examined the localization of pCRMP1/2(S522) in cultured hippocampal neurons possessing apical dendrite-like processes. Again, pCRMP1/2(S522) was mainly localized in MAP2-positive neurites (colocalization coefficients was 0.860 ± 0.005, n = 60 neurons), and was evenly localized in the apical dendrite-like process (Nakamura et al., 2009) as well as other dendritic processes (Fig. 2C). These results suggest that the phosphorylation of CRMP1 and/or CRMP2 at Ser522 may play some roles during dendritic development.
Dendritic abnormality in cultured cortical neurons from crmp1−/−;crmp2ki/ki
To investigate whether phosphorylation of CRMP1 and/or CRMP2 at Ser522 is involved in proper dendritic development, we examined the dendritic morphology of cultured cortical neurons from wt, crmp1−/−, crmp2ki/ki, and crmp1−/−;crmp2ki/ki mice. While the basic properties, number of primary dendrite and number of branching point did not change in each genotype (data not shown), we noticed that the neurons from crmp1−/−;crmp2ki/ki mice exhibited irregular curling dendritic elongation (Fig. 2D). Since this dendritic phenotype is quite unique, we define this phenotype as “curling phenotype.” To estimate the percentage of neurons that exhibit curling phenotype, we defined the neurons with R < 60 from the winding dendrite(s) as curling phenotype (Fig. 2E). The percentage of curling phenotype was slightly but significantly increased in crmp2ki/ki than wt neurons. Furthermore, this was significantly increased in the double mutant crmp1−/−;crmp2ki/ki compared with wt as well as crmp2ki/ki neurons (Fig. 2F). This unique phenotype in double mutant crmp1−/−;crmp2ki/ki mice suggests that the Ser522 phosphorylation on CRMP2 or both CRMP1 and CRMP2 phosphorylation are important for proper dendritic growth of cortical neurons.
The crmp1−/−;crmp2ki/ki mice show defects in the basal dendritic field of cortical pyramidal neurons
Next, we investigated the in vivo dendritic morphology of cortical neuron in the series of mutant mice. For this purpose, we intercrossed these mutant mice with YFP-H transgenic mice (Feng et al., 2000) to visualize layer V cortical pyramidal neurons. The basal dendrites of Layer V pyramidal neurons in crmp1−/−;crmp2ki/ki;YFP-H were wandering around the neuronal soma (Fig. 3A). To analyze this phenotype in detail, we traced both basal dendrites (red) and branches from apical dendrite (blue) of layer V cortical neurons of each genotype. In control mice, basal dendrites grew to the basal side and collaterals from apical dendrites elongated horizontally. However, in the double mutant crmp1−/−;crmp2ki/ki;YFP-H mice, the basal dendrites grew to the apical side with curling. The collaterals from apical trunks also showed disorientation and winding (Fig. 3A). Cortical neurons in crmp2ki/ki;YFP-H mice showed an increase in the number of primary dendrites (Fig. 3B). In crmp1−/−;crmp2ki/ki;YFP-H mice, an even greater number of primary dendrites was observed, compared with that of YFP-H mice (Fig. 3B). The Sholl analysis indicated that in control as well as crmp1−/−;YFP-H and crmp2ki/ki;YFP-H mice, the highest number of dendrite crossings is found at 60–80 μm distance from the cell soma. This was shifted to ∼40 μm in crmp1−/−;crmp2ki/ki;YFP-H mice which may be due to the curling phenotype (data not shown). To further perform quantitative evaluation of the curling phenotype, we plotted the tips of basal dendrites and quantified how the basal dendrites were distributed around the cell body (Fig. 3C). In control mice, the basal dendritic field of layer V cortical neurons was organized in the basal side of the cell body and spread evenly. In contrast, in crmp2ki/ki;YFP-H mice, the basal dendritic field was slightly shifted to the apical side of the cell body. Additionally, in double mutant crmp1−/−;crmp2ki/ki;YFP-H mice, the basal dendritic field was further shifted to the apical part of the cell body and to the close part of cell body (Fig. 3D,E). These results demonstrate that the phosphorylation of CRMP2 by Cdk5 or both CRMP1 and CRMP2 phosphorylation are essential for proper dendrite orientation and guidance in vivo.
Discussion
In this study, to elucidate the physiological role of phosphorylation of CRMP2 at Ser522, we newly generated CRMP2 knock-in mutant mice in which the Ser residue at 522 was replaced with Ala (Fig. 1). We here demonstrate, for the first time, phosphorylation of CRMP2 at S522 plays an essential role in regulating dendritic branch trajectories in the cerebral cortical neurons. Furthermore, by analyzing the phenotypes of crmp1−/−;crmp2ki/ki double mutants, we showed that both CRMP1 and CRMP2 are required for proper dendritic patterning (Fig. 3). Since this is quite a unique dendritic phenotype compared with others that have been reported, we define this phenotype as curling phenotype.
It has been reported that extra- and intracellular signaling molecules play an important role in regulating dendritic patterning in the cerebral cortex (Wong and Ghosh, 2002; Jan and Jan, 2010). Because the curling phenotype in our study differs from that observed in sema3A-deficient mice (Polleux et al., 2000; Sasaki et al., 2002), this phenotype cannot simply be explained by the lack of Sema3A signaling. We also sought to examine the role of other signaling molecules such as α and β subunit of soluble guanylyl cyclase (sGC) and Notch, because these molecules play important roles in establishing neural cell polarity, and might be involved in the curling phenotype (Breunig et al., 2007; Shelly et al., 2010). However, the subcellular localizations of sGC was not altered in crmp1−/−;crmp2ki/ki cultured cortical neurons. Furthermore, introduction of wild-type or a dominant-negative mutant of Notch did not induce the curling phenotype (our unpublished observation). These findings strongly suggest that both CRMP1 and CRMP2 are important and unique key players in regulating dendritic patterning. Since the curling phenotype was enhanced in crmp1−/−;crmp2ki/ki mice (Figs. 2, 3), it is likely that both CRMP2S522 phosphorylation and CRMP1 phosphorylation are synergistically involved in regulating dendritic field organization.
There are some inconsistencies between in vivo and in vitro data about number of primary dendrite. The cultured neurons did not show any difference in the number of primary dendrites and number of branching points among each genotype. On the other hand, the number of primary dendrites is increased in the layer V neurons of the double mutant mice in vivo (Fig. 3). The exact reasons for this discrepancy are unknown at present. Recently, it has been well recognized that extracellular environments play important roles in regulating cell fate, proliferation, polarity and other cellular functions in non-neuronal as well as neuronal tissues (Craig and Banker, 1994; Breunig et al., 2007; Parrish et al., 2007; Jan and Jan, 2010). Since extracellular environments of the cortical neurons in vivo probably differ from those of the cultured cortical neurons, it is possible that the lack of the CRMP2 phosphorylation and of CRMP1 may affect dendritic outgrowth and/or formation of the cortical neurons quite differently depending on their extracellular environments in vivo and in vitro. Although secreted Sema3A may regulate dendritic outgrowth or formation in cultured neurons (Polleux et al., 2000; Sasaki et al., 2002; Morita et al., 2006), the effects of Sema3A in conditioned cultured media may be different from those of Sema3A in environment surrounding the cortical neurons in vivo, which may underlie the phenotypic difference in vitro and in vivo. Alternatively, CRMPs are involved in microtubule assembly (Fukata et al., 2002; Uchida et al., 2005); it is also possible that the curling phenotype might be related to basic microtubule polymerization. If this curling phenotype was caused from basic tubulin polymerization defect, and was totally independent from outside signals, curling phenotype can be observed in every dendrite at any developmental time period in cultured neurons from double mutant mice. However, only a minor subset of dendrites showed curling phenotype (Fig. 2). And the morphology of apical trunks are not altered in crmp1−/−;crmp2ki/ki cortex in vivo (Fig. 3). These findings argued against the idea that the curling phenotype was caused from basic tubulin polymerization defect in a cell-autonomous manner.
Double mutant mice showed basal dendrites and apical branches defects, but not apical trunks of cortical neurons in vivo (Fig. 3A). However, the pCRMP1/2(S522) signal was observed in apical dendrites in tissue section and evenly localized in apical dendrite-like processes as well as other dendritic processes in cultured neurons (Fig. 2). These results probably reflect a different developmental mechanism for the control of basal and apical dendritic trajectories. In fact, an apical dendrite extends vertically toward the pial surface, and this defines the path along which the cell will migrate. The leading processes of radially migrating neurons develop into apical dendrites after completing their radial migration (Hatanaka and Murakami, 2002). On the other hand, neurons start to elongate the basal dendrites and apical branches after migration to their final location and these processes exhibited curling phenotype (Fig. 3). These facts suggest that Cdk5-CRMP1/2 signaling regulates dendritic morphology after neurons have migrated to their final location. Supporting this idea, the major phenotype of cdk5−/− mice is lamina formation defect in the cortex (Ohshima et al., 1996, 2007), though the Nissl-stained sections of crmp1−/−;crmp2S522A/S522 mice cortex did not show lamina formation defect like cdk5−/− mice (data not shown). In addition, emerging data clearly demonstrate distinct distribution patterns of receptors and ion channels in apical and basal dendrites (Tran et al., 2009; Shah et al., 2010). Hence, our data further suggest that the different molecular mechanism(s) may exist in apical and basal dendritic formation.
In conclusion, we propose a novel regulatory mechanism of basal dendrite patterning in which the phosphorylation of CRMP1 and/or CRMP2 at Ser522 is involved. Our mutant mice will be useful to get insight into the functional and physiological significance of the proper dendritic field organization.
Footnotes
This work was supported by Grants-in-Aid for Scientific Research on Priority Areas from The Ministry of Education, Culture, Sports, Science and Technology (Y.G.) and Core Research for Evolutional Science and technology (CREST) of Japan Science and Technology Agency (Y.G.). We thank Research Resources Center members in RIKEN BSI for advice and technical support with the generation of knock in mice. We also thank Drs. Heiner Westphal (NIH) for EIIaCre transgenic mice. We acknowledge Dr. Masaharu Ogawa (RIKEN BSI) and Dr. Satoshi Morita (Yokohama City University) for advice on mouse phenotypic analysis and statistic analysis, respectively. We thank Dr. Reina Aoki, Sandy Chen, and Miyuki Ogawara for advice and technical assistance on cell culture and immunocytochemistry.
References
- Arimura N, Ménager C, Kawano Y, Yoshimura T, Kawabata S, Hattori A, Fukata Y, Amano M, Goshima Y, Inagaki M, Morone N, Usukura J, Kaibuchi K. Phosphorylation by Rho kinase regulates CRMP-2 activity in growth cones. Mol Cell Biol. 2005;25:9973–9984. doi: 10.1128/MCB.25.22.9973-9984.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breunig JJ, Silbereis J, Vaccarino FM, Sestan N, Rakic P. Notch regulates cell fate and dendrite morphology of newborn neurons in the postnatal dentate gyrus. Proc Natl Acad Sci U S A. 2007;104:20558–20563. doi: 10.1073/pnas.0710156104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M, Jacobs T, Eickholt B, Ferrari G, Teo M, Monfries C, Qi RZ, Leung T, Lim L, Hall C. Alpha2-chimaerin, cyclin-dependent Kinase 5/p35, and its target collapsin response mediator protein-2 are essential components in semaphorin 3A-induced growth-cone collapse. J Neurosci. 2004;24:8994–9004. doi: 10.1523/JNEUROSCI.3184-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier E, Mosinger B, Meissirel C, Aguera M, Rogemond V, Reibel S, Salin P, Chounlamountri N, Perrot V, Belin MF, Goshima Y, Honnorat J, Thomasset N, Kolattukudy P. Transient alterations in granule cell proliferation, apoptosis and migration in postnatal developing cerebellum of CRMP1(−/−) mice. Genes Cells. 2006;11:1337–1352. doi: 10.1111/j.1365-2443.2006.01024.x. [DOI] [PubMed] [Google Scholar]
- Cole AR, Knebel A, Morrice NA, Robertson LA, Irving AJ, Connolly CN, Sutherland C. GSK-3 phosphorylation of the Alzheimer epitope within collapsin response mediator proteins regulates axon elongation in primary neurons. J Biol Chem. 2004;279:50176–50180. doi: 10.1074/jbc.C400412200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig AM, Banker G. Neuronal polarity. Annu Rev Neurosci. 1994;17:267–310. doi: 10.1146/annurev.ne.17.030194.001411. [DOI] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Fukata Y, Itoh TJ, Kimura T, Ménager C, Nishimura T, Shiromizu T, Watanabe H, Inagaki N, Iwamatsu A, Hotani H, Kaibuchi K. CRMP-2 binds to tubulin heterodimers to promote microtubule assembly. Nat Cell Biol. 2002;4:583–591. doi: 10.1038/ncb825. [DOI] [PubMed] [Google Scholar]
- Goshima Y, Nakamura F, Strittmatter P, Strittmatter SM. Collapsin-induced growth cone collapse mediated by an intracellular protein related to UNC-33. Nature. 1995;376:509–514. doi: 10.1038/376509a0. [DOI] [PubMed] [Google Scholar]
- Hatanaka Y, Murakami F. In vitro analysis of the origin, migratory behavior, and maturation of cortical pyramidal cells. J Comp Neurol. 2002;454:1–14. doi: 10.1002/cne.10421. [DOI] [PubMed] [Google Scholar]
- Jan YN, Jan LY. Branching out: mechanisms of dendritic arborization. Nat Rev Neurosci. 2010;11:316–328. doi: 10.1038/nrn2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita A, Yamashita N, Sasaki Y, Uchida Y, Nakajima O, Nakamura F, Yagi T, Taniguchi M, Usui H, Katoh-Semba R, Takei K, Goshima Y. Regulation of dendritic branching and spine maturation by semaphorin3A-Fyn signaling. J Neurosci. 2006;26:2971–2980. doi: 10.1523/JNEUROSCI.5453-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura F, Ugajin K, Yamashita N, Okada T, Uchida Y, Taniguchi M, Ohshima T, Goshima Y. Increased proximal bifurcation of CA1 pyramidal apical dendrites in sema3A mutant mice. J Comp Neurol. 2009;516:360–375. doi: 10.1002/cne.22125. [DOI] [PubMed] [Google Scholar]
- Ohshima T, Ward JM, Huh CG, Longenecker G, Veeranna, Pant HC, Brady RO, Martin LJ, Kulkarni AB. Targeted disruption of the cyclin-dependent kinase 5 gene results in abnormal corticogenesis, neuronal pathology and perinatal death. Proc Natl Acad Sci U S A. 1996;93:11173–11178. doi: 10.1073/pnas.93.20.11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima T, Hirasawa M, Tabata H, Mutoh T, Adachi T, Suzuki H, Saruta K, Iwasato T, Itohara S, Hashimoto M, Nakajima K, Ogawa M, Kulkarni AB, Mikoshiba K. Cdk5 is required for multipolar-to-bipolar transition during radial neuronal migration and proper dendrite development of pyramidal neurons in the cerebral cortex. Development. 2007;134:2273–2282. doi: 10.1242/dev.02854. [DOI] [PubMed] [Google Scholar]
- Parrish JZ, Emoto K, Kim MD, Jan YN. Mechanisms that regulate establishment, maintenance, and remodeling of dendritic fields. Annu Rev Neurosci. 2007;30:399–423. doi: 10.1146/annurev.neuro.29.051605.112907. [DOI] [PubMed] [Google Scholar]
- Polleux F, Morrow T, Ghosh A. Semaphorin 3A is a chemoattractant for cortical apical dendrites. Nature. 2000;404:567–573. doi: 10.1038/35007001. [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Cheng C, Uchida Y, Nakajima O, Ohshima T, Yagi T, Taniguchi M, Nakayama T, Kishida R, Kudo Y, Ohno S, Nakamura F, Goshima Y. Fyn and Cdk5 mediate semaphorin-3A signaling, which is involved in regulation of dendrite orientation in cerebral cortex. Neuron. 2002;35:907–920. doi: 10.1016/s0896-6273(02)00857-7. [DOI] [PubMed] [Google Scholar]
- Shah MM, Hammond RS, Hoffman DA. Dendritic ion channel trafficking and plasticity. Trends Neurosci. 2010;33:307–316. doi: 10.1016/j.tins.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelly M, Lim BK, Cancedda L, Heilshorn SC, Gao H, Poo MM. Local and long-range reciprocal regulation of cAMP and cGMP in axon/dendrite formation. Science. 2010;327:547–552. doi: 10.1126/science.1179735. [DOI] [PubMed] [Google Scholar]
- Sholl DA. Dendritic organization in the neurons of the visual and motor cortices of the cat. J Anat. 1953;87:387–406. [PMC free article] [PubMed] [Google Scholar]
- Tran TS, Rubio ME, Clem RL, Johnson D, Case L, Tessier-Lavigne M, Huganir RL, Ginty DD, Kolodkin AL. Secreted semaphorins control spine distribution and morphogenesis in the postnatal CNS. Nature. 2009;462:1065–1069. doi: 10.1038/nature08628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida Y, Ohshima T, Sasaki Y, Suzuki H, Yanai S, Yamashita N, Nakamura F, Takei K, Ihara Y, Mikoshiba K, Kolattukudy P, Honnorat J, Goshima Y. Semaphorin3A signalling is mediated via sequential Cdk5 and GSK3beta phosphorylation of CRMP2: implication of common phosphorylating mechanism underlying axon guidance and Alzheimer's disease. Genes Cells. 2005;10:165–179. doi: 10.1111/j.1365-2443.2005.00827.x. [DOI] [PubMed] [Google Scholar]
- Williams-Simons L, Westphal H. EIIaCre—utility of a general deleter strain. Transgenic Res. 1999;8:53–54. doi: 10.1023/a:1008994831937. [DOI] [PubMed] [Google Scholar]
- Wong RO, Ghosh A. Activity-dependent regulation of dendritic growth and patterning. Nat Rev Neurosci. 2002;3:803–812. doi: 10.1038/nrn941. [DOI] [PubMed] [Google Scholar]
- Yamashita N, Uchida Y, Ohshima T, Hirai S, Nakamura F, Taniguchi M, Mikoshiba K, Honnorat J, Kolattukudy P, Thomasset N, Takei K, Takahashi T, Goshima Y. Collapsin response mediator protein 1 mediates reelin signaling in cortical neuronal migration. J Neurosci. 2006;26:13357–13362. doi: 10.1523/JNEUROSCI.4276-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita N, Morita A, Uchida Y, Nakamura F, Usui H, Ohshima T, Taniguchi M, Honnorat J, Thomasset N, Takei K, Takahashi T, Kolattukudy P, Goshima Y. Regulation of spine development by semaphorin3A through cyclin-dependent kinase 5 phosphorylation of collapsin response mediator protein 1. J Neurosci. 2007;27:12546–12554. doi: 10.1523/JNEUROSCI.3463-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita N, Mosinger B, Roy A, Miyazaki M, Ugajin K, Nakamura F, Sasaki Y, Yamaguchi K, Kolattukudy P, Goshima Y. CRMP5 (collapsin response mediator protein 5) regulates dendritic development and synaptic plasticity in the cerebellar Purkinje cells. J Neurosci. 2011;31:1773–1779. doi: 10.1523/JNEUROSCI.5337-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura T, Kawano Y, Arimura N, Kawabata S, Kikuchi A, Kaibuchi K. GSK-3beta regulates phosphorylation of CRMP-2 and neuronal polarity. Cell. 2005;120:137–149. doi: 10.1016/j.cell.2004.11.012. [DOI] [PubMed] [Google Scholar]