Abstract
Background
Mitogen-activated protein kinase (MAPK) cascade is a conserved and universal signal transduction module in organisms. Although it has been well characterized in many plants, no systematic analysis has been conducted in barley.
Results
Here, we identified 20 MAPKs, 6 MAPKKs and 156 MAPKKKs in barley through a genome-wide search against the updated reference genome. Then, phylogenetic relationship, gene structure and conserved protein motifs organization of them were systematically analyzed and results supported the predictions. Gene duplication analysis revealed that segmental and tandem duplication events contributed to the expansion of barley MAPK cascade genes and the duplicated gene pairs were found to undergone strong purifying selection. Expression profiles of them were further investigated in different organs and under diverse abiotic stresses using the available 173 RNA-seq datasets, and then the tissue-specific and stress-responsive candidates were found. Finally, co-expression regulatory network of MAPK cascade genes was constructed by WGCNA tool, resulting in a complicated network composed of a total of 72 branches containing 46 HvMAPK cascade genes and 46 miRNAs.
Conclusion
This study provides the targets for further functional study and also contribute to better understand the MAPK cascade regulatory network in barley and beyond.
Keywords: Barley, Gene family, MAPK cascade, Regulatory network
Background
To coordinate the biotic and abiotic stresses during growth and development, plants have evolved to form the complex mechanisms to perceive and transmit environmental stimuli by inducing or repressing a series of genes to express [1]. The Mitogen-activated protein kinase (MAPK) cascades are characterized as evolutionarily conserved and fundamentally universal signaling transduction pathways, playing the vital roles as diverse receptors/sensors from the extracellular environment to intracellular transcriptional and metabolic centers in eukaryotes [2]. The canonical MAPK cascade is composed of three specific kinases, namely MAPK, MAPK kinase (MAPKK) as well as MAPKK kinase (MAPKKK), which was activated sequentially by phosphorylation at certain activation sites [3, 4]. In general, MAPKs are phosphorylated at their conserved threonine and tyrosine residues in the activation loop (T-loop) by MAPK kinase, and in turn, MAPKK are activated by MAPKKKs when development or environmental signals incurred as their serine and serine/threonine residues located in the S/T activation site are phosphorylated [1, 2].
In plants, extensive studies have revealed that the MAPK cascades widely involved in regulating many biological processes, including cell division, plant development, growth and hormonal response as well as in response to diverse biotic and abiotic stresses, such as drought, salt, heat and pathogen infection [5–7]. In light of their importance, a large number of MAPK genes have been functionally identified in several plants, including Arabidopsis [8], rice [9–11], Brachypdoium [12, 13] and maize [14, 15]. At the same time, a series of plant MAPK signaling cascades have also been well constructed and studied. The AtMEKK1-MKK4/5-MPK3/6 cascades is the first identified MAPK signaling module in plants, which was involved in plant innate immunity of flg22 signal transmission [16, 17]. The complete MAPK signaling cascade of ANP3-MKK6-MPK4 and YDA-MKK4/5-MPK3/6 is determined to control the stomatal development and patterning in Arabidopsis [18]. MEKK1-MKK1/2-MPK4 module was found to play the important role in the defenses against abiotic stresses and contributed to the freezing tolerance in Arabidopsis [19–21]. The ABA(abscisic acid)-activated MEKK17/18-MKK3-MPK1/2/7/14 module displayed stress signaling to ABA and regulated the expression of a series of ABA-dependent genes [22]. In tobacco, the NPK1-MEK1-Ntf6 cascade was identified to confer the resistance to tobacco mosaic virus via mediating the resistant protein N [23]. Additionally, the NPK1-NQK1/NtMEK1-NRK1 module is found to be a positive regulator of tobacco cytokinesis during meiosis as well as mitosis [24]. Barley (Hordeum vulgare L.) is one of the earliest domesticated and also one of the most important staple crops, which holds the significance for agriculture drawn and human civilization [25, 26]. Furthermore, barley is also well-studied in terms of cytology, genetics and genomics and thus qualifies as the model for Triticeae research [27]. The survey of MAPK family in barley has also been conducted and a total of 16 HvMAPKs were identified based on the full-length cDNA, EST(expressed sequence tag) and genomic survey database [28]. However, the incomplete data used by Krenek et al. [28] might cause the incomplete prediction and identification of MAPKK and MAPKKK family is not performed in barley up to now. The recently published reference-quality barley genome [26] makes it possible to conduct a comprehensive identification of its MAPK cascade gene families at whole genome scale and then construct the MAPK signal transduction pathway.
In this study, we systematically identified the MAPK, MAPKK and MAPKKK gene family based on a genome-wide search against barley reference genome. Then, the gene structures, chromosomal locations, gene duplication events and evolutionary dynamics were investigated. Furthermore, the expression patterns at diverse development stages and under different abiotic stresses were also analyzed. Finally, we constructed the regulatory networks of MAPK-MAPKK-MAPKKK signal pathway based on the co-expression patterns from a total of 173 RNA-seq datasets. This study reported the genomic organization, expression and phylogenetic relationships of the MAPK, MAPKK and MAPKKK gene families in barley, which could provide the candidates for further functional analysis and also contribute to illuminate the MAPK signal cascade-mediated pathway of barley and beyond.
Results and discussion
Genome-wide identification of MAPK cascade genes in barley
Availability of the reference-quality barley genome [26] made it possible for the first time to systematically identify all the MAPK cascade genes in this model crop species. Using the methods as described below, a total of 20 HvMAPK, 6 HvMAPKKs and 156 HvMAPKKKs were obtained, respectively (Table 1). The conserved domain analysis showed that all of them have the serine/threonine-protein kinase-like domain (PFAM accession No. PF00069) (Additional file 7: Table S1). We further validated the identified genes using the public ESTs to provide the expression support. Results showed that majority (19 out of 20 HvMAPKs, 5 out of 6 HvMAPKKs and 103 out of 156 HvMAPKKKs) of the predicted genes had the existing EST hit supports (Table 1). Given the limit of available ESTs, the non-supported HvMAPK cascade gene might not be detected under specific conditions or low levels of expression that can’t be investigated experimentally. Compared to previous study that only 16 HvMAPKs were identified by Krenek et al [28], this study found 20 HvMAPKs, which covered the 16 previous predicted ones, suggesting the whole genome-search could provide more comprehensive prediction of barley MAPK family.
Table 1.
List of MAPK cascade genes identified in barley
| No. | MAPK names | Family | Sub_Family | Ensemble barley Gene_ID | Chromosome Location | Amino acid size | EST | PI | Mw (kD) | Subcellular location | GRAGY | Ortholog |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HvMAPK1 | MAPK | – | HORVU1Hr1G049500.1 | chr1H | 378 | 3 | 5.75 | 42,892.79 | Cytoplasmic | − 0.335 | AtMPK4 |
| 2 | HvMAPK2 | MAPK | – | HORVU1Hr1G088510.1 | chr1H | 560 | 15 | 9.33 | 63,157.25 | Cytoplasmic | −0.489 | – |
| 3 | HvMAPK3 | MAPK | – | HORVU1Hr1G090940.17 | chr1H | 621 | 11 | 8.38 | 69,944.49 | Nuclear | −0.584 | – |
| 4 | HvMAPK4 | MAPK | – | HORVU1Hr1G091890.1 | chr1H | 700 | 21 | 9.55 | 77,086.97 | Nuclear | −0.475 | – |
| 5 | HvMAPK5 | MAPK | – | HORVU3Hr1G056200.1 | chr3H | 615 | 23 | 9.04 | 69,867.66 | Nuclear | −0.539 | AtMPK20 |
| 6 | HvMAPK6 | MAPK | – | HORVU3Hr1G057660.34 | chr3H | 400 | 12 | 9.33 | 44,831.54 | Mitochondrial | −0.336 | – |
| 7 | HvMAPK7 | MAPK | – | HORVU3Hr1G060390.3 | chr3H | 585 | 19 | 9.32 | 66,919.8 | Nuclear | −0.546 | – |
| 8 | HvMAPK8 | MAPK | – | HORVU4Hr1G049430.1 | chr4H | 370 | 6 | 6.67 | 42,327.22 | Nuclear | −0.18 | AtMPK1 |
| 9 | HvMAPK9 | MAPK | – | HORVU4Hr1G057200.4 | chr4H | 370 | 10 | 5.46 | 42,811.12 | Cytoplasmic | −0.299 | AtMPK3 |
| 10 | HvMAPK10 | MAPK | – | HORVU5Hr1G078060.3 | chr5H | 172 | – | 5.02 | 19,465.59 | Cytoplasmic | −0.028 | – |
| 11 | HvMAPK11 | MAPK | – | HORVU5Hr1G120960.1 | chr5H | 441 | 5 | 5.93 | 50,322.66 | Cytoplasmic | −0.336 | – |
| 12 | HvMAPK12 | MAPK | – | HORVU6Hr1G017820.5 | chr6H | 213 | 1 | 7.88 | 24,578.45 | Cytoplasmic | −0.076 | – |
| 13 | HvMAPK13 | MAPK | – | HORVU6Hr1G021480.1 | chr6H | 386 | 6 | 7.46 | 43,983.12 | Nuclear | −0.222 | – |
| 14 | HvMAPK14 | MAPK | – | HORVU6Hr1G068270.1 | chr6H | 462 | 2 | 9.73 | 51,836.95 | Mitochondrial | − 0.423 | – |
| 15 | HvMAPK15 | MAPK | – | HORVU7Hr1G008690.19 | chr7H | 484 | 9 | 9.44 | 55,037.08 | Nuclear | −0.491 | – |
| 16 | HvMAPK16 | MAPK | – | HORVU7Hr1G023760.3 | chr7H | 280 | 7 | 6.33 | 31,878.74 | Cytoplasmic | −0.116 | – |
| 17 | HvMAPK17 | MAPK | – | HORVU7Hr1G082510.1 | chr7H | 276 | 9 | 8.61 | 31,502.72 | Cytoplasmic | 0.015 | – |
| 18 | HvMAPK18 | MAPK | – | HORVU7Hr1G095810.7 | chr7H | 579 | 21 | 6.85 | 65,275.1 | Nuclear | −0.49 | AtMPK9 |
| 19 | HvMAPK19 | MAPK | – | HORVU7Hr1G097740.1 | chr7H | 370 | 7 | 7.17 | 42,188.13 | Nuclear | −0.165 | – |
| 20 | HvMAPK20 | MAPK | – | HORVU0Hr1G016660.4 | chrUn | 462 | 2 | 9.73 | 51,836.95 | Mitochondrial | −0.423 | – |
| 21 | HvMAPKK1 | MAPKK | – | HORVU1Hr1G086310.1 | chr1H | 331 | – | 9.11 | 35,272.53 | Mitochondrial | −0.153 | – |
| 22 | HvMAPKK2 | MAPKK | – | HORVU5Hr1G067100.3 | chr5H | 233 | 1 | 8.85 | 26,401.41 | Nuclear | −0.201 | – |
| 23 | HvMAPKK3 | MAPKK | – | HORVU5Hr1G125270.1 | chr5H | 375 | 2 | 5.92 | 42,093.32 | Cytoplasmic | −0.214 | – |
| 24 | HvMAPKK4 | MAPKK | – | HORVU5Hr1G125290.3 | chr5H | 524 | 4 | 5.62 | 58,532.46 | Cytoplasmic | −0.249 | AtMKK3 |
| 25 | HvMAPKK5 | MAPKK | – | HORVU7Hr1G031720.3 | chr7H | 266 | 1 | 8.26 | 29,177.39 | Mitochondrial | −0.165 | AtMKK4 |
| 26 | HvMAPKK6 | MAPKK | – | HORVU0Hr1G038850.2 | chrUn | 295 | 1 | – | – | Nuclear | −0.131 | – |
| 27 | HvMEKK1 | MAPKKK | MEKK | HORVU1Hr1G071060.1 | chr1H | 339 | 4 | 5.41 | 38,432.96 | Cytoplasmic | −0.343 | – |
| 28 | HvMEKK2 | MAPKKK | MEKK | HORVU1Hr1G078710.3 | chr1H | 542 | 3 | 5.43 | 57,335.41 | Chloroplast | −0.153 | AtMAPKKK17 |
| 29 | HvMEKK3 | MAPKKK | MEKK | HORVU1Hr1G078720.3 | chr1H | 444 | 1 | 4.97 | 46,771.58 | Chloroplast | −0.037 | – |
| 30 | HvMEKK4 | MAPKKK | MEKK | HORVU1Hr1G078760.1 | chr1H | 271 | 1 | 9.51 | 28,242.39 | Chloroplast | −0.05 | – |
| 31 | HvMEKK5 | MAPKKK | MEKK | HORVU1Hr1G078790.1 | chr1H | 414 | 1 | 4.67 | 43,185.31 | Chloroplast | −0.093 | – |
| 32 | HvMEKK6 | MAPKKK | MEKK | HORVU1Hr1G078860.6 | chr1H | 402 | 1 | 4.22 | 42,770.5 | Cytoplasmic | −0.15 | – |
| 33 | HvMEKK7 | MAPKKK | MEKK | HORVU2Hr1G039070.1 | chr2H | 586 | 3 | 6.69 | 65,825.69 | Cytoplasmic | −0.37 | – |
| 34 | HvMEKK8 | MAPKKK | MEKK | HORVU2Hr1G110900.9 | chr2H | 1332 | 6 | 6.05 | 147,411.07 | Nuclear | −0.318 | AtMAPKKK6 |
| 35 | HvMEKK9 | MAPKKK | MEKK | HORVU2Hr1G047960.2 | chr2H | 693 | 3 | 7 | 76,096.19 | Nuclear | −0.671 | – |
| 36 | HvMEKK10 | MAPKKK | MEKK | HORVU3Hr1G065620.1 | chr3H | 516 | 2 | 5.14 | 54,444.88 | Chloroplast | −0.187 | AtMAPKKK16 |
| 37 | HvMEKK11 | MAPKKK | MEKK | HORVU3Hr1G065630.1 | chr3H | 470 | 4 | 5.11 | 49,969.2 | Cytoplasmic | −0.182 | – |
| 38 | HvMEKK12 | MAPKKK | MEKK | HORVU3Hr1G065640.1 | chr3H | 490 | 1 | 5.21 | 52,802.78 | Chloroplast | −0.286 | – |
| 39 | HvMEKK13 | MAPKKK | MEKK | HORVU3Hr1G087600.1 | chr3H | 533 | 4 | 6.65 | 59,672.15 | Cytoplasmic | −0.433 | – |
| 40 | HvMEKK14 | MAPKKK | MEKK | HORVU3Hr1G109290.2 | chr3H | 100 | 1 | 5.36 | 11,266.28 | Chloroplast | 0.002 | – |
| 41 | HvMEKK15 | MAPKKK | MEKK | HORVU4Hr1G004540.4 | chr4H | 327 | – | 5.75 | 36,885.2 | Cytoplasmic | −0.318 | – |
| 42 | HvMEKK16 | MAPKKK | MEKK | HORVU4Hr1G056120.2 | chr4H | 482 | 2 | 5.94 | 50,614.24 | Extracellular | −0.113 | AtMAPKKK14 |
| 43 | HvMEKK17 | MAPKKK | MEKK | HORVU4Hr1G088910.12 | chr4H | 741 | 3 | 6.37 | 81,763.31 | Nuclear | −0.491 | – |
| 44 | HvMEKK18 | MAPKKK | MEKK | HORVU5Hr1G059030.1 | chr5H | 583 | – | 6.22 | 63,951.39 | Chloroplast | −0.2 | – |
| 45 | HvMEKK19 | MAPKKK | MEKK | HORVU5Hr1G059840.4 | chr5H | 617 | – | – | – | Nuclear | −0.566 | AtMAPKKK1 |
| 46 | HvMEKK20 | MAPKKK | MEKK | HORVU5Hr1G094350.1 | chr5H | 1105 | 4 | 6.28 | 123,492.11 | Nuclear | −0.357 | – |
| 47 | HvMEKK21 | MAPKKK | MEKK | HORVU5Hr1G095970.2 | chr5H | 837 | 1 | 5.33 | 90,903.5 | Nuclear | −0.507 | – |
| 48 | HvMEKK22 | MAPKKK | MEKK | HORVU5Hr1G110900.3 | chr5H | 536 | 11 | 6.31 | 60,056.37 | Cytoplasmic | −0.475 | – |
| 49 | HvMEKK23 | MAPKKK | MEKK | HORVU6Hr1G002500.1 | chr6H | 409 | – | 5.24 | 43,108.97 | Chloroplast | −0.385 | – |
| 50 | HvMEKK24 | MAPKKK | MEKK | HORVU6Hr1G029780.1 | chr6H | 536 | 3 | 7.65 | 60,327.14 | Cytoplasmic | −0.53 | – |
| 51 | HvMEKK25 | MAPKKK | MEKK | HORVU6Hr1G084460.23 | chr6H | 419 | – | 6.12 | 47,039.57 | Nuclear | −0.374 | – |
| 52 | HvMEKK26 | MAPKKK | MEKK | HORVU7Hr1G047720.7 | chr7H | 703 | 5 | 6.22 | 78,253.71 | Nuclear | −0.585 | – |
| 53 | HvMEKK27 | MAPKKK | MEKK | HORVU0Hr1G030360.3 | chrUn | 429 | 1 | 4.68 | 45,136.42 | Chloroplast | −0.06 | – |
| 54 | HvMEKK28 | MAPKKK | MEKK | HORVU0Hr1G030380.1 | chrUn | 354 | – | – | – | Chloroplast | −0.084 | – |
| 55 | HvRaf-like1 | MAPKKK | Raf-like | HORVU1Hr1G000090.3 | chr1H | 661 | 2 | 9.28 | 70,543.87 | Nuclear | −0.43 | – |
| 56 | HvRaf-like2 | MAPKKK | Raf-like | HORVU1Hr1G005720.7 | chr1H | 763 | – | 6.29 | 85,499.18 | PlasmaMembrane | −0.281 | – |
| 57 | HvRaf-like3 | MAPKKK | Raf-like | HORVU1Hr1G015770.2 | chr1H | 398 | 2 | 6.81 | 44,301.65 | Cytoplasmic | −0.217 | – |
| 58 | HvRaf-like4 | MAPKKK | Raf-like | HORVU1Hr1G035440.2 | chr1H | 958 | 16 | 5.74 | 106,350.79 | Nuclear | −0.475 | – |
| 59 | HvRaf-like5 | MAPKKK | Raf-like | HORVU1Hr1G065310.1 | chr1H | 445 | – | 5.58 | 49,026.03 | Chloroplast | −0.216 | – |
| 60 | HvRaf-like6 | MAPKKK | Raf-like | HORVU1Hr1G066190.1 | chr1H | 857 | 3 | 6.02 | 92,569.39 | PlasmaMembrane | −0.052 | – |
| 61 | HvRaf-like7 | MAPKKK | Raf-like | HORVU1Hr1G074310.4 | chr1H | 695 | 2 | 6.57 | 75,011.62 | PlasmaMembrane | −0.084 | – |
| 62 | HvRaf-like8 | MAPKKK | Raf-like | HORVU1Hr1G075670.4 | chr1H | 1047 | 8 | 7.77 | 110,771.06 | PlasmaMembrane | 0.06 | – |
| 63 | HvRaf-like9 | MAPKKK | Raf-like | HORVU1Hr1G076110.4 | chr1H | 602 | 2 | 9.37 | 66,839.21 | Nuclear | −0.492 | – |
| 64 | HvRaf-like10 | MAPKKK | Raf-like | HORVU1Hr1G080600.2 | chr1H | 635 | 2 | 5.72 | 70,783.85 | PlasmaMembrane | −0.142 | – |
| 65 | HvRaf-like11 | MAPKKK | Raf-like | HORVU1Hr1G087050.1 | chr1H | 677 | – | 6.34 | 76,355.28 | Cytoplasmic | −0.341 | – |
| 66 | HvRaf-like12 | MAPKKK | Raf-like | HORVU1Hr1G091230.12 | chr1H | 236 | 2 | 8.46 | 25,840.97 | Cytoplasmic | −0.131 | – |
| 67 | HvRaf-like13 | MAPKKK | Raf-like | HORVU1Hr1G092250.3 | chr1H | 691 | 2 | 7.18 | 75,448.29 | PlasmaMembrane | −0.081 | – |
| 68 | HvRaf-like14 | MAPKKK | Raf-like | HORVU1Hr1G092290.2 | chr1H | 694 | – | 5.77 | 75,163.36 | PlasmaMembrane | −0.096 | – |
| 69 | HvRaf-like15 | MAPKKK | Raf-like | HORVU2Hr1G008140.6 | chr2H | 789 | 10 | 6.45 | 87,315.15 | PlasmaMembrane | −0.199 | – |
| 70 | HvRaf-like16 | MAPKKK | Raf-like | HORVU2Hr1G038790.1 | chr2H | 694 | – | 6.95 | 77,384.09 | PlasmaMembrane | −0.147 | – |
| 71 | HvRaf-like17 | MAPKKK | Raf-like | HORVU2Hr1G044270.3 | chr2H | 754 | 5 | 7.76 | 81,472.98 | PlasmaMembrane | −0.099 | – |
| 72 | HvRaf-like18 | MAPKKK | Raf-like | HORVU2Hr1G044520.4 | chr2H | 668 | 6 | 6.8 | 72,527.68 | PlasmaMembrane | −0.068 | – |
| 73 | HvRaf-like19 | MAPKKK | Raf-like | HORVU2Hr1G044590.1 | chr2H | 679 | 3 | 6.86 | 74,113.13 | PlasmaMembrane | −0.159 | – |
| 74 | HvRaf-like20 | MAPKKK | Raf-like | HORVU2Hr1G044640.4 | chr2H | 254 | 4 | 5.4 | 28,342.25 | Nuclear | −0.213 | – |
| 75 | HvRaf-like21 | MAPKKK | Raf-like | HORVU2Hr1G044650.1 | chr2H | 702 | 16 | 6.15 | 76,575.16 | PlasmaMembrane | −0.114 | – |
| 76 | HvRaf-like22 | MAPKKK | Raf-like | HORVU2Hr1G044870.3 | chr2H | 713 | 2 | 6.75 | 78,483.04 | Extracellular | −0.228 | – |
| 77 | HvRaf-like23 | MAPKKK | Raf-like | HORVU2Hr1G087930.2 | chr2H | 398 | 1 | 5.69 | 44,310.08 | Cytoplasmic | −0.389 | – |
| 78 | HvRaf-like24 | MAPKKK | Raf-like | HORVU2Hr1G099570.11 | chr2H | 765 | 2 | 8.25 | 83,699.77 | Chloroplast | −0.17 | AtRaf-like6 |
| 79 | HvRaf-like25 | MAPKKK | Raf-like | HORVU2Hr1G104030.4 | chr2H | 1114 | – | 6 | 118,546.3 | PlasmaMembrane | 0.09 | – |
| 80 | HvRaf-like26 | MAPKKK | Raf-like | HORVU2Hr1G107250.1 | chr2H | 1005 | 3 | 6.11 | 109,068.87 | Extracellular | 0.018 | – |
| 81 | HvRaf-like27 | MAPKKK | Raf-like | HORVU2Hr1G123850.10 | chr2H | 277 | – | 6.22 | 31,174.32 | Nuclear | −0.075 | – |
| 82 | HvRaf-like28 | MAPKKK | Raf-like | HORVU2Hr1G124370.9 | chr2H | 291 | 2 | 9.03 | 32,082.2 | Mitochondrial | 0.019 | – |
| 83 | HvRaf-like29 | MAPKKK | Raf-like | HORVU2Hr1G124530.35 | chr2H | 348 | – | 7.73 | 39,465.15 | Nuclear | −0.395 | – |
| 84 | HvRaf-like30 | MAPKKK | Raf-like | HORVU2Hr1G125210.1 | chr2H | 666 | 4 | 8.47 | 73,491.31 | PlasmaMembrane | −0.106 | – |
| 85 | HvRaf-like31 | MAPKKK | Raf-like | HORVU3Hr1G000350.4 | chr3H | 367 | – | 9.17 | 40,921.23 | Nuclear | −0.26 | – |
| 86 | HvRaf-like32 | MAPKKK | Raf-like | HORVU3Hr1G000770.2 | chr3H | 686 | 1 | 6.39 | 71,693.86 | Nuclear | −0.374 | – |
| 87 | HvRaf-like33 | MAPKKK | Raf-like | HORVU3Hr1G002820.2 | chr3H | 688 | 2 | 5.78 | 74,705.12 | PlasmaMembrane | −0.066 | – |
| 88 | HvRaf-like34 | MAPKKK | Raf-like | HORVU3Hr1G003920.7 | chr3H | 401 | 1 | 6.76 | 43,580.58 | Cytoplasmic | −0.224 | – |
| 89 | HvRaf-like35 | MAPKKK | Raf-like | HORVU3Hr1G006640.3 | chr3H | 644 | 5 | 6.25 | 70,581.73 | PlasmaMembrane | −0.001 | – |
| 90 | HvRaf-like36 | MAPKKK | Raf-like | HORVU3Hr1G006790.4 | chr3H | 655 | 4 | 6.43 | 71,454.11 | PlasmaMembrane | 0.062 | – |
| 91 | HvRaf-like37 | MAPKKK | Raf-like | HORVU3Hr1G006800.2 | chr3H | 697 | 6 | 6.97 | 75,564.22 | PlasmaMembrane | −0.021 | – |
| 92 | HvRaf-like38 | MAPKKK | Raf-like | HORVU3Hr1G017420.4 | chr3H | 414 | 3 | 8.84 | 45,791.61 | Chloroplast | −0.077 | – |
| 93 | HvRaf-like39 | MAPKKK | Raf-like | HORVU3Hr1G026870.3 | chr3H | 577 | 2 | 9.59 | 64,845.42 | Mitochondrial | −0.525 | – |
| 94 | HvRaf-like40 | MAPKKK | Raf-like | HORVU3Hr1G057190.5 | chr3H | 527 | – | 6.92 | 56,137.72 | Chloroplast | −0.28 | – |
| 95 | HvRaf-like41 | MAPKKK | Raf-like | HORVU3Hr1G057440.1 | chr3H | 500 | 2 | 8.8 | 55,275.02 | Mitochondrial | −0.416 | AtRaf-like33 |
| 96 | HvRaf-like42 | MAPKKK | Raf-like | HORVU3Hr1G061400.1 | chr3H | 844 | – | 6.03 | 94,593.52 | PlasmaMembrane | −0.285 | – |
| 97 | HvRaf-like43 | MAPKKK | Raf-like | HORVU3Hr1G061410.7 | chr3H | 841 | – | 7.54 | 94,745.11 | PlasmaMembrane | −0.233 | – |
| 98 | HvRaf-like44 | MAPKKK | Raf-like | HORVU3Hr1G061450.4 | chr3H | 781 | – | 6.07 | 87,268.33 | PlasmaMembrane | −0.142 | – |
| 99 | HvRaf-like45 | MAPKKK | Raf-like | HORVU3Hr1G061480.1 | chr3H | 835 | 1 | 6.59 | 91,656.76 | PlasmaMembrane | −0.153 | – |
| 100 | HvRaf-like46 | MAPKKK | Raf-like | HORVU3Hr1G061860.2 | chr3H | 482 | 2 | 6.49 | 54,487.91 | Nuclear | −0.484 | AtRaf-like15 |
| 101 | HvRaf-like47 | MAPKKK | Raf-like | HORVU3Hr1G071240.2 | chr3H | 603 | 2 | 9.1 | 67,617.07 | Nuclear | −0.593 | AtRaf-like36 |
| 102 | HvRaf-like48 | MAPKKK | Raf-like | HORVU3Hr1G077110.18 | chr3H | 813 | 1 | 5.97 | 89,761.84 | PlasmaMembrane | −0.044 | – |
| 103 | HvRaf-like49 | MAPKKK | Raf-like | HORVU3Hr1G077130.1 | chr3H | 831 | 1 | 6.08 | 92,032.95 | PlasmaMembrane | −0.125 | – |
| 104 | HvRaf-like50 | MAPKKK | Raf-like | HORVU3Hr1G093140.3 | chr3H | 622 | 1 | 7.23 | 69,718.6 | Nuclear | −0.504 | – |
| 105 | HvRaf-like51 | MAPKKK | Raf-like | HORVU3Hr1G098910.5 | chr3H | 308 | – | 5.4 | 35,484.92 | Cytoplasmic | −0.369 | – |
| 106 | HvRaf-like52 | MAPKKK | Raf-like | HORVU3Hr1G109370.13 | chr3H | 823 | 12 | 5.84 | 91,309.94 | PlasmaMembrane | −0.195 | – |
| 107 | HvRaf-like53 | MAPKKK | Raf-like | HORVU4Hr1G001850.2 | chr4H | 774 | – | 6.56 | 84,022.73 | Chloroplast | −0.201 | AtRaf-like1 |
| 108 | HvRaf-like54 | MAPKKK | Raf-like | HORVU4Hr1G010030.27 | chr4H | 875 | 4 | 8.49 | 95,121.28 | Chloroplast | −0.234 | – |
| 109 | HvRaf-like55 | MAPKKK | Raf-like | HORVU4Hr1G020000.1 | chr4H | 374 | – | 6.32 | 40,364.96 | Chloroplast | −0.094 | – |
| 110 | HvRaf-like56 | MAPKKK | Raf-like | HORVU4Hr1G026160.7 | chr4H | 844 | – | 5.83 | 95,761.31 | PlasmaMembrane | −0.263 | – |
| 111 | HvRaf-like57 | MAPKKK | Raf-like | HORVU4Hr1G026170.1 | chr4H | 836 | – | 5.66 | 91,551.28 | PlasmaMembrane | −0.13 | – |
| 112 | HvRaf-like58 | MAPKKK | Raf-like | HORVU4Hr1G026230.1 | chr4H | 842 | – | 8.45 | 92,169.14 | PlasmaMembrane | −0.07 | – |
| 113 | HvRaf-like59 | MAPKKK | Raf-like | HORVU4Hr1G029350.13 | chr4H | 742 | 1 | 7.26 | 82,634.14 | Nuclear | −0.596 | – |
| 114 | HvRaf-like60 | MAPKKK | Raf-like | HORVU4Hr1G069020.1 | chr4H | 392 | – | 6.15 | 43,297.77 | Cytoplasmic | −0.261 | – |
| 115 | HvRaf-like61 | MAPKKK | Raf-like | HORVU4Hr1G069890.1 | chr4H | 190 | – | 4.64 | 21,123.19 | Cytoplasmic | −0.236 | – |
| 116 | HvRaf-like62 | MAPKKK | Raf-like | HORVU4Hr1G070190.1 | chr4H | 396 | – | 5.98 | 43,979.3 | Cytoplasmic | −0.342 | – |
| 117 | HvRaf-like63 | MAPKKK | Raf-like | HORVU4Hr1G073290.3 | chr4H | 1014 | 6 | 6.14 | 110,526.93 | Nuclear | −0.516 | AtRaf-like2 |
| 118 | HvRaf-like64 | MAPKKK | Raf-like | HORVU4Hr1G075550.1 | chr4H | 671 | – | 6.12 | 72,129.41 | PlasmaMembrane | 0.095 | – |
| 119 | HvRaf-like65 | MAPKKK | Raf-like | HORVU4Hr1G079950.13 | chr4H | 346 | 8 | 6.12 | 39,038.93 | Cytoplasmic | −0.13 | – |
| 120 | HvRaf-like66 | MAPKKK | Raf-like | HORVU4Hr1G083590.2 | chr4H | 865 | 1 | 7.81 | 94,015.33 | PlasmaMembrane | −0.082 | – |
| 121 | HvRaf-like67 | MAPKKK | Raf-like | HORVU4Hr1G089460.1 | chr4H | 113 | – | 4.72 | 12,557.55 | Cytoplasmic | 0.185 | – |
| 122 | HvRaf-like68 | MAPKKK | Raf-like | HORVU5Hr1G001800.2 | chr5H | 690 | – | 5.76 | 76,845.9 | PlasmaMembrane | −0.292 | – |
| 123 | HvRaf-like69 | MAPKKK | Raf-like | HORVU5Hr1G001920.1 | chr5H | 1024 | 2 | 6.71 | 106,899.38 | Chloroplast | 0.134 | – |
| 124 | HvRaf-like70 | MAPKKK | Raf-like | HORVU5Hr1G016840.6 | chr5H | 1127 | 1 | 5.55 | 123,895.7 | Nuclear | −0.484 | AtRaf-like16 |
| 125 | HvRaf-like71 | MAPKKK | Raf-like | HORVU5Hr1G022360.3 | chr5H | 758 | 1 | 8.18 | 83,590.48 | Nuclear | −0.649 | AtRaf-like11 |
| 126 | HvRaf-like72 | MAPKKK | Raf-like | HORVU5Hr1G040040.6 | chr5H | 458 | 1 | 5.17 | 50,925.78 | Cytoplasmic | −0.219 | – |
| 127 | HvRaf-like73 | MAPKKK | Raf-like | HORVU5Hr1G061150.2 | chr5H | 438 | – | – | – | Nuclear | −0.473 | – |
| 128 | HvRaf-like74 | MAPKKK | Raf-like | HORVU5Hr1G061460.1 | chr5H | 388 | – | 5.41 | 43,267.93 | Cytoplasmic | −0.459 | – |
| 129 | HvRaf-like75 | MAPKKK | Raf-like | HORVU5Hr1G077430.5 | chr5H | 523 | – | 6.27 | 58,965.62 | Nuclear | −0.209 | – |
| 130 | HvRaf-like76 | MAPKKK | Raf-like | HORVU5Hr1G077450.7 | chr5H | 336 | – | 6.8 | 38,284.21 | Cytoplasmic | −0.221 | – |
| 131 | HvRaf-like77 | MAPKKK | Raf-like | HORVU5Hr1G084880.1 | chr5H | 665 | 3 | 7.79 | 72,493.17 | PlasmaMembrane | −0.066 | – |
| 132 | HvRaf-like78 | MAPKKK | Raf-like | HORVU5Hr1G085020.10 | chr5H | 270 | 1 | 5.39 | 30,562.59 | Cytoplasmic | −0.311 | – |
| 133 | HvRaf-like79 | MAPKKK | Raf-like | HORVU5Hr1G085070.61 | chr5H | 555 | 4 | 5.75 | 60,676.75 | Cytoplasmic | −0.232 | – |
| 134 | HvRaf-like80 | MAPKKK | Raf-like | HORVU5Hr1G089400.1 | chr5H | 355 | 2 | – | – | Cytoplasmic | −0.23 | – |
| 135 | HvRaf-like81 | MAPKKK | Raf-like | HORVU5Hr1G093370.3 | chr5H | 374 | 4 | 8.52 | 41,309.32 | Nuclear | −0.336 | AtRaf-like39 |
| 136 | HvRaf-like82 | MAPKKK | Raf-like | HORVU5Hr1G094510.2 | chr5H | 389 | – | 6.6 | 42,568.27 | Cytoplasmic | −0.42 | – |
| 137 | HvRaf-like83 | MAPKKK | Raf-like | HORVU5Hr1G095120.2 | chr5H | 1113 | 1 | 6.74 | 121,029.31 | PlasmaMembrane | 0.138 | – |
| 138 | HvRaf-like84 | MAPKKK | Raf-like | HORVU5Hr1G097010.3 | chr5H | 740 | 1 | 7.24 | 81,842.19 | Nuclear | −0.414 | – |
| 139 | HvRaf-like85 | MAPKKK | Raf-like | HORVU5Hr1G106710.1 | chr5H | 249 | – | 7.01 | 27,607.12 | Mitochondrial | −0.143 | – |
| 140 | HvRaf-like86 | MAPKKK | Raf-like | HORVU5Hr1G111670.1 | chr5H | 420 | 2 | 8 | 45,614.15 | Nuclear | −0.305 | AtRaf-like31 |
| 141 | HvRaf-like87 | MAPKKK | Raf-like | HORVU5Hr1G119060.5 | chr5H | 918 | 2 | 5.29 | 99,360.82 | Nuclear | −0.441 | – |
| 142 | HvRaf-like88 | MAPKKK | Raf-like | HORVU5Hr1G122950.2 | chr5H | 1065 | 1 | – | – | PlasmaMembrane | 0.077 | – |
| 143 | HvRaf-like89 | MAPKKK | Raf-like | HORVU5Hr1G123540.2 | chr5H | 673 | 2 | 5.96 | 72,900.5 | PlasmaMembrane | 0.069 | – |
| 144 | HvRaf-like90 | MAPKKK | Raf-like | HORVU5Hr1G123550.1 | chr5H | 285 | 4 | 5.13 | 32,333.85 | Cytoplasmic | −0.168 | – |
| 145 | HvRaf-like91 | MAPKKK | Raf-like | HORVU5Hr1G125710.2 | chr5H | 1228 | 3 | 5.37 | 133,759 | Nuclear | −0.55 | AtRaf-like20 |
| 146 | HvRaf-like92 | MAPKKK | Raf-like | HORVU6Hr1G012800.9 | chr6H | 542 | – | 5.83 | 60,241.36 | Cytoplasmic | −0.304 | AtRaf-like30 |
| 147 | HvRaf-like93 | MAPKKK | Raf-like | HORVU6Hr1G025940.2 | chr6H | 798 | – | 6.36 | 89,926.43 | Cytoplasmic | −0.245 | – |
| 148 | HvRaf-like94 | MAPKKK | Raf-like | HORVU6Hr1G039740.15 | chr6H | 133 | 5 | 5.77 | 14,810.17 | Extracellular | −0.048 | – |
| 149 | HvRaf-like95 | MAPKKK | Raf-like | HORVU6Hr1G045360.5 | chr6H | 429 | 8 | 8.19 | 48,710.48 | PlasmaMembrane | −0.155 | – |
| 150 | HvRaf-like96 | MAPKKK | Raf-like | HORVU6Hr1G053310.1 | chr6H | 353 | 1 | 6.68 | 39,662.74 | Cytoplasmic | −0.231 | AtRaf-like34 |
| 151 | HvRaf-like97 | MAPKKK | Raf-like | HORVU6Hr1G069710.4 | chr6H | 422 | 1 | 8.28 | 46,692.96 | Chloroplast | −0.128 | – |
| 152 | HvRaf-like98 | MAPKKK | Raf-like | HORVU6Hr1G070880.1 | chr6H | 820 | – | 6.01 | 92,509.74 | Extracellular | −0.232 | – |
| 153 | HvRaf-like99 | MAPKKK | Raf-like | HORVU6Hr1G078810.22 | chr6H | 646 | 1 | 6.21 | 71,929.9 | Nuclear | −0.497 | – |
| 154 | HvRaf-like100 | MAPKKK | Raf-like | HORVU6Hr1G083270.16 | chr6H | 1097 | 4 | 5.4 | 120,927.14 | Nuclear | −0.633 | AtRaf-like35 |
| 155 | HvRaf-like101 | MAPKKK | Raf-like | HORVU6Hr1G085710.2 | chr6H | 995 | – | 5.81 | 110,316.57 | PlasmaMembrane | −0.005 | – |
| 156 | HvRaf-like102 | MAPKKK | Raf-like | HORVU6Hr1G091540.1 | chr6H | 465 | – | 9.45 | 49,316.59 | Chloroplast | −0.323 | – |
| 157 | HvRaf-like103 | MAPKKK | Raf-like | HORVU7Hr1G003630.2 | chr7H | 433 | 2 | 6.54 | 48,300.04 | Nuclear | −0.295 | – |
| 158 | HvRaf-like104 | MAPKKK | Raf-like | HORVU7Hr1G021350.1 | chr7H | 371 | 2 | 6.01 | 40,657.44 | Nuclear | −0.126 | – |
| 159 | HvRaf-like105 | MAPKKK | Raf-like | HORVU7Hr1G029750.1 | chr7H | 1288 | 8 | 5.54 | 137,637.12 | Nuclear | −0.287 | AtRaf-like42 |
| 160 | HvRaf-like106 | MAPKKK | Raf-like | HORVU7Hr1G030370.10 | chr7H | 1151 | 1 | 8.24 | 124,540.57 | PlasmaMembrane | 0.059 | – |
| 161 | HvRaf-like107 | MAPKKK | Raf-like | HORVU7Hr1G031210.83 | chr7H | 823 | – | 6.12 | 90,280.99 | PlasmaMembrane | −0.1 | – |
| 162 | HvRaf-like108 | MAPKKK | Raf-like | HORVU7Hr1G038650.5 | chr7H | 964 | – | 5.59 | 106,329.97 | Chloroplast | −0.196 | AtRaf-like4 |
| 163 | HvRaf-like109 | MAPKKK | Raf-like | HORVU7Hr1G041430.2 | chr7H | 1115 | – | 6.74 | 121,239.44 | Extracellular | 0.038 | – |
| 164 | HvRaf-like110 | MAPKKK | Raf-like | HORVU7Hr1G044510.5 | chr7H | 598 | – | 6.71 | 65,728.91 | Cytoplasmic | −0.31 | – |
| 165 | HvRaf-like111 | MAPKKK | Raf-like | HORVU7Hr1G068410.1 | chr7H | 417 | 4 | 8.39 | 46,070.12 | Chloroplast | −0.146 | AtRaf-like28 |
| 166 | HvRaf-like112 | MAPKKK | Raf-like | HORVU7Hr1G078170.32 | chr7H | 567 | – | 5.63 | 63,786.57 | Cytoplasmic | −0.343 | – |
| 167 | HvRaf-like113 | MAPKKK | Raf-like | HORVU7Hr1G087320.1 | chr7H | 548 | – | 6.08 | 62,329.43 | Nuclear | −0.235 | – |
| 168 | HvRaf-like114 | MAPKKK | Raf-like | HORVU7Hr1G088430.1 | chr7H | 1055 | 3 | 5.76 | 114,538 | PlasmaMembrane | −0.101 | – |
| 169 | HvRaf-like115 | MAPKKK | Raf-like | HORVU7Hr1G092030.2 | chr7H | 397 | – | 9.19 | 44,690.52 | Nuclear | −0.359 | AtRaf-like19 |
| 170 | HvRaf-like116 | MAPKKK | Raf-like | HORVU7Hr1G098030.1 | chr7H | 694 | – | 6.95 | 77,384.09 | PlasmaMembrane | −0.147 | – |
| 171 | HvRaf-like117 | MAPKKK | Raf-like | HORVU7Hr1G109290.2 | chr7H | 575 | 1 | 5.37 | 62,356.03 | Nuclear | −0.371 | – |
| 172 | HvRaf-like118 | MAPKKK | Raf-like | HORVU7Hr1G109640.2 | chr7H | 426 | 7 | 6.28 | 47,228.18 | Chloroplast | −0.188 | – |
| 173 | HvRaf-like119 | MAPKKK | Raf-like | HORVU7Hr1G114620.5 | chr7H | 1106 | 9 | 5.63 | 118,989.35 | Nuclear | −0.508 | – |
| 174 | HvRaf-like120 | MAPKKK | Raf-like | HORVU7Hr1G116190.3 | chr7H | 632 | 2 | 6.04 | 70,455.32 | Cytoplasmic | −0.226 | – |
| 175 | HvRaf-like121 | MAPKKK | Raf-like | HORVU7Hr1G119100.1 | chr7H | 779 | 1 | 7.02 | 87,009.02 | PlasmaMembrane | −0.153 | – |
| 176 | HvRaf-like122 | MAPKKK | Raf-like | HORVU0Hr1G011480.3 | chrUn | 707 | 2 | 5.97 | 78,559.18 | PlasmaMembrane | −0.199 | – |
| 177 | HvRaf-like123 | MAPKKK | Raf-like | HORVU0Hr1G014630.8 | chrUn | 842 | – | 5.77 | 92,484.32 | PlasmaMembrane | −0.107 | – |
| 178 | HvRaf-like124 | MAPKKK | Raf-like | HORVU0Hr1G015980.4 | chrUn | 397 | – | 8.85 | 43,665.02 | Nuclear | −0.28 | – |
| 179 | HvZIK1 | MAPKKK | ZIK | HORVU2Hr1G036210.2 | chr2H | 352 | – | 6.6 | 39,258.55 | Nuclear | −0.448 | AtZIK8 |
| 180 | HvZIK2 | MAPKKK | ZIK | HORVU2Hr1G037990.1 | chr2H | 679 | 6 | 5.57 | 76,149.66 | Nuclear | −0.515 | AtZIK4 |
| 181 | HvZIK3 | MAPKKK | ZIK | HORVU5Hr1G046590.3 | chr5H | 461 | 1 | 4.91 | 51,074.45 | Chloroplast | −0.291 | AtZIK2 |
| 182 | HvZIK4 | MAPKKK | ZIK | HORVU6Hr1G065020.2 | chr6H | 619 | 2 | 4.78 | 69,307.43 | Nuclear | −0.365 | AtZIK5 |
Furthermore, the physical and chemical properties of these genes were investigated and compared. The length of MAPK cascade related proteins varied from 100 to 1332 amino acids, with an average of 596 in length. The putative molecular mass ranged from 11.2 kDa to 147.1 kDa, and the isoelectric points varied from 4.22 to 9.73, respectively (Table 1), which is similar to that of wheat and Brachypodium [29, 30]. The significance difference of physical and chemistry properties between the members of barley MAPK genes suggested that the subfunctionalization and neofunctionalization may have occurred among the MAPK cascade genes in barley [29]. Analysis of subcellular location showed that 52 (30%) HvMAPK cascade genes were predicted to be located in nuclear, followed by PlasmaMembrane (45) and Cytoplasmic (43), while the remaining ones were predicted to be located in chloroplast, mitochondrial and extra-cellular.
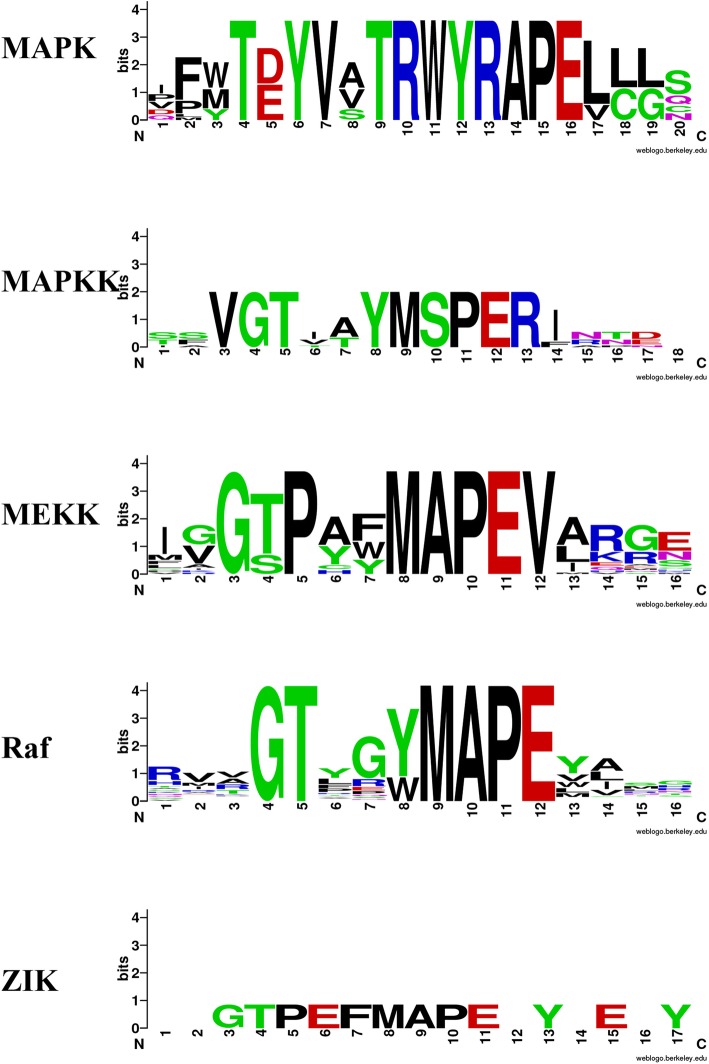
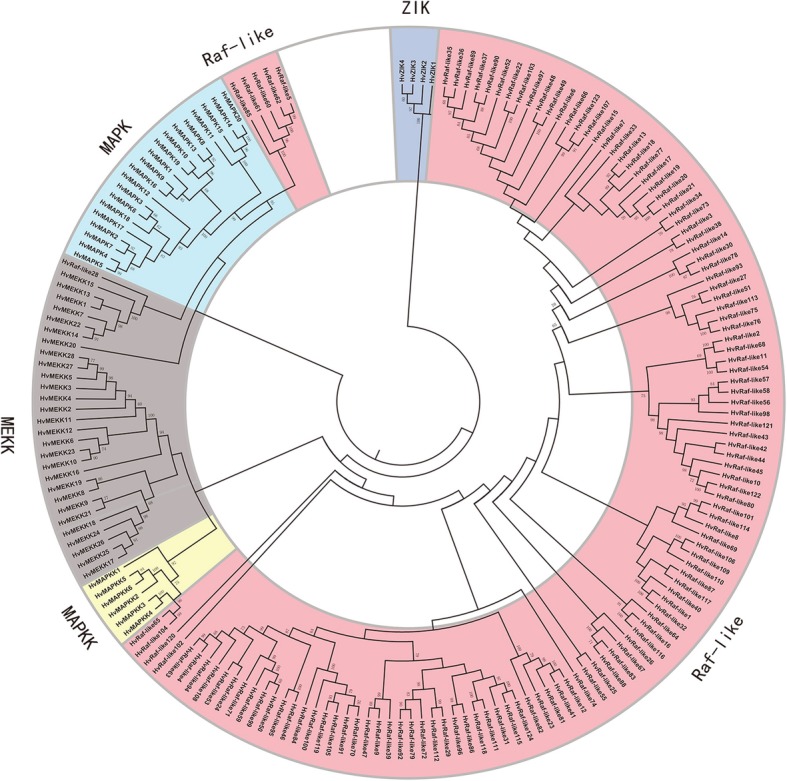
These 182 barley MAPK cascade genes can be classified into three major clades in coordination to MAPK, MAPKK and MAPKKK with the specific conserved signature motifs, respectively (Fig. 1). Among them, 20 genes harboring the specific conserved signature motifs of T(E/D)YVxTRWYRAPE(L/V), and 6 genes possessing the VGTxxYMSPER conserved signature, which were categorized into MAPK and MAPKK subfamilies, respectively [3, 31]. Consistent with the other species [3, 10], these HvMAPKs could be assigned into the 10 TDY- and 10 TEY-subtype members (Fig. 2a and Additional file 1: Figure S1). We also investigated the docking site CD (Common docking) domain in HvMAPKs. Results showed that the TDY-subtype HvMAPKs lacked this domain (Fig. 2c and Additional file 2: Figure S2), which was the same as that of Arabidopsis [3]. All of MAPKK members contained the VGTxxYMSPER motif and the putative MAPK docking sites [K/R][K/R][K/R]x(1–5)[L/I]x[L/I] (Additional file 3: Figure S3). The remaining 156 genes belonged to MAPKKK subfamily. The barley MAPKKK genes could be further divided into three groups, which owned the conserved motifs of G(T/S)Px(W/Y/F)MAPEV, GTxx(W/Y)MAPE and GTPEFMAPE(L/V)Y for MEKK, Raf-like and ZIK subfamilies, respectively (Additional file 4: Figure S4). Remarkably, the Raf-like subfamily had 124 members, ranking the largest group of MAPKKK in barley, whereas the ZIK subfamilies possessed only 4 members as the smallest group, which was consistent with the abundance and composition of MAPKKK genes in other species, especially in wheat [29, 30] (Table 2).
Fig. 1.
List of barley MAPK signalling components
Fig. 2.
The subfamily organizations based on phylogenetic relationships (a), intron-exon structure structures (b) and protein structures (c) analysis of MAPK cascade genes in barley
Table 2.
Comparison of the abundance of MAPK cascade gene family in different plant species
| Hordeum vulgare | Triticum aestivum | Oryza sativa | Zea mays | Brachypodium distachyon | Arabidopsis thaliana | Lycopersicon esculentum | Glycine max | Vitis vinifera | |
|---|---|---|---|---|---|---|---|---|---|
| MAPK | 20 | 54 | 17 | 19 | 16 | 20 | 16 | 38 | 14 |
| MAPKK | 6 | 18 | 8 | 9 | 12 | 10 | 6 | 11 | 5 |
| MAPKKK | 156 | 155 | 75 | 74 | 75 | 80 | 89 | 150 | 45 |
| RAF | 124 | 115 | 43 | 46 | 45 | 48 | 40 | 92 | 27 |
| MEKK | 28 | 29 | 22 | 22 | 24 | 21 | 33 | 34 | 9 |
| ZIK | 4 | 11 | 10 | 6 | 6 | 11 | 16 | 24 | 9 |
Phylogenetic relationship, gene structure and motifs analysis
To further support the subfamily grouping, phylogenetic analysis were performed using the full-length protein sequences of these barley MAPK cascade genes (Fig. 3). Consistent with specific conserved signature motifs [3], the MEKK, Raf-like and ZIK subfamilies belonging to MAPKKK family were also clustered into independent sub-clade, respectively. For MAPK, it could be further divided into TDY and TEY two sub-clades, and TEY sub-clade was further assigned into A to C subgroups. We further performed phylogenetic analysis of these HvMAPK and the reported rice and Arabidopsis MAPKs. Results found they could clustered into different groups and the orthology pairs of them were obtained depending on phylogenetic relationship (Additional file 5: Figure S5). These results could provide some clues for candidate selection for further functional study as some orthologous genes in rice and Arabidopsis has been extensively functionally characterized [16, 18].
Fig. 3.
Phylogenetic analysis of barley MAPK cascade proteins
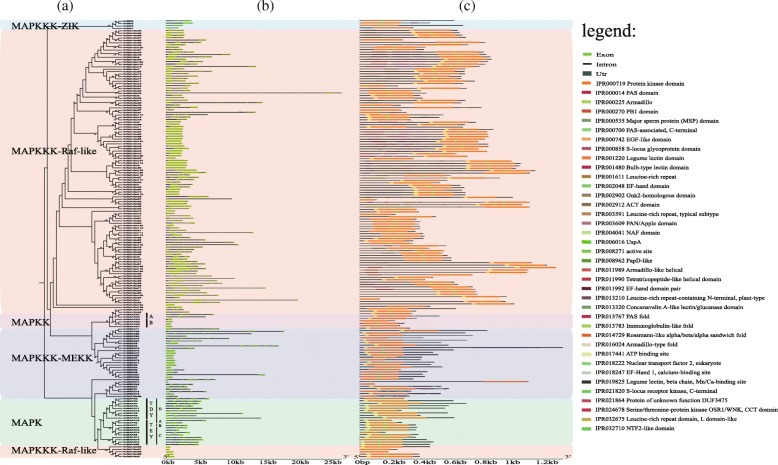
Gene structure played vital roles in the evolution of gene families and provided extra evidence to estimate the functional diversifications [32]. Thus, the exon-intron organization of these barley MAPK cascade genes was further analyzed (Fig. 2b). Result found that there were significant intron abundance variations between these genes. It is reported that C- and D-group of MAPKKs tend to have no introns in Arabidopsis [3]. The C-group of HvMAPKKs also showed intron-less while D-group have abundant introns. For instance, HvMAPKK3 and HvMAPKK4, which assigned into D subgroup, possessed 7 and 9 introns, respectively. Furthermore, the intron count of HvMAPKKK gene family ranged from 1 to 24, showing obviously variations even in the same subgroup. For the MEKK subfamily, more than half (54.2%) of the genes possessed no or one intron, while the other MEKK members had 6 to 24 introns. The intron number of the ZIK subfamily varied from 2 to 5, whereas the RAF genes with the intron number ranged from 1 to 20 and presented the highest level of variation among them.
Additionally, the conserved protein domains in the barley MAPK cascade genes were identified and compared. A total of 32 conserved motifs were detected (Fig. 2c). The protein kinase domain was found in each member of the MAPK cascade proteins. A certain degree of conservation could be observed in the HvMAPK and HvMAPKK genes that almost all of them harbored the ATP (Adenosine triphosphate) binding site and serine/threonine-protein kinase active site. Similar to the intron/exon structure, the composition of conserved motifs was also highly variable in HvMAPKKK family. Apart from the protein kinase and its related domains, a series of other functional motifs was widely distributed, such as Bulb-type lectin domain, S-locus glycoprotein domain and PAN/Apple domain, suggested they are widely involved in growth and development as well as signaling transduction [33]. The PAS domain, S-locus glycoprotein domain and Concanavalin A-like lectin/glucanase domain were possessed by 4, 1 and 3 Raf subfamily members. The EF-hand domain pair, EF-Hand 1, calcium-binding site and EF-hand domain were uniquely found in MEKK subfamily, whereas no domains were specific to the ZIK subfamily. On the whole, the MAPK cascade proteins clustered into the same group phylogenetically tended to share similar motifs composition.
Finally, the 1.5 kb genomic sequences upstream of the transcriptional start sites of HvMAPK genes were extracted and used to identify the cis-regulatory elements. Totally, 27 cis-elements were obtained, of which SARE(salicylic acid responsiveness) domain and the TGA(auxin-responsive) domain were found to be present only in 3 and 7 genes respectively, whereas the Skn-1 motif was shared by 159 genes, which ranked the least and most abundant motifs (Additional file 7: Table S2). Skn-1 motif is reported to be a cis-acting regulatory element required for endosperm expression and oxidative stress response in eukaryotes [34], suggesting the MAPK cascades played the important role in regulating the barley development and stress response. In addition, a large amount of plant growth and development (including circadian, meristem and endosperm), hormone-related (e.g., abscisic acid, auxin, MeJA, ethylene, gibberellin) cis-elements were found in these promoter regions, suggesting that MAPK cascade genes widely involved in regulating the signal transduction network of diverse developmental processes. Meanwhile, the cis-element related to biotic (e.g. fungal and wound) and abiotic stress response (e.g. salt, extreme temperature, dehydration) were also identified in the promoter region of the HvMAPK cascade genes, which suggested that these MAPK cascade genes might have potential functions in stress adaptation and signaling pathways [33].
Gene duplication and synteny analysis
In order to investigate the mechanism of expansion of the MAPK cascade genes in barley, we further investigated the segmental and tandem duplication events by genome synteny analysis. Results showed that 13 paralogs composed of 26 HvMAPK cascade genes were identified, of which 5 were segmental duplications and 8 were tandem duplication events (Fig. 4 and Additional file 7: Table S3). In detail, 3 and 2 segmental events were found in HvMAPKs and HvMAPKKKs, as well as 8 tandem repeats events in HvMAPKKKs, suggesting that segmental duplication played important roles in the expansion of MAPKs while tandem repeat duplication was the driven force for HvMAPKKK gene family expansion. It is noteworthy that the segmental events mainly occurred at chromosome 1 and chromosome 3, whereas the tandem duplication blocks distributed throughout the whole genome, of which 1, 1, 4, 1, 1 paralogous pairs were mapped to chromosome 1, 2, 3, 4 and 5, respectively (Fig. 4). In order to detect the selection effect during gene divergence after duplication, the Ka/Ks substitution ratio of the duplicated pairs were further calculated. Result showed that Ka/Ks ratios of MAPK cascade genes ranged from 0.001 to 0.4727, with an average of 0.1964, suggesting that they have undergone purifying selection pressure during the process of evolution in barley [35].
Fig. 4.
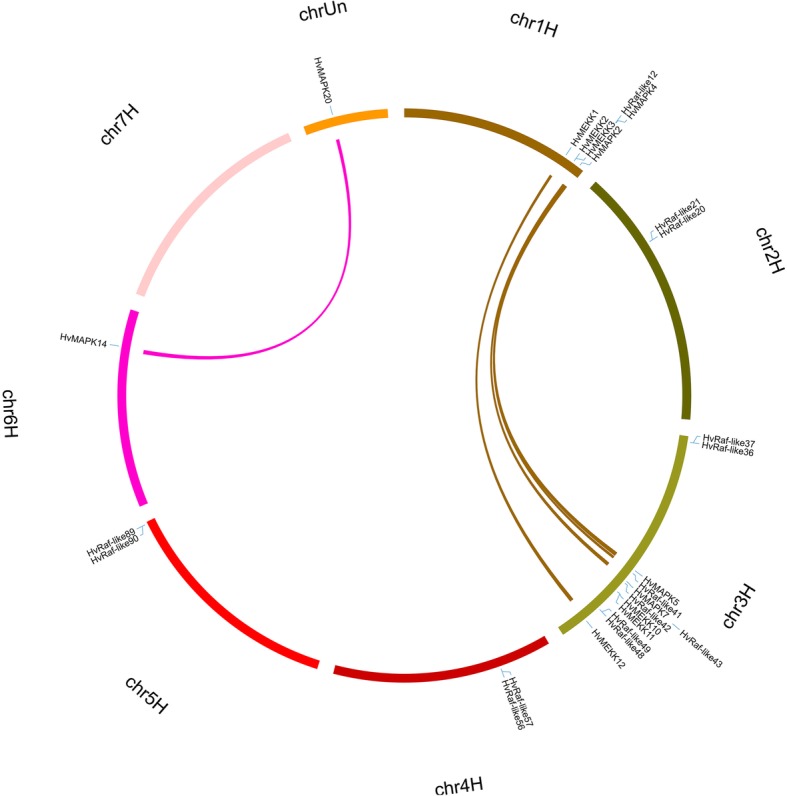
Chromosome locations and duplicated genes pairs of MAPK cascade genes in the barley genome. Each barley chromosome is displayed in different color. Duplicated gene pairs are displayed in corresponding color and linked using lines with the same color
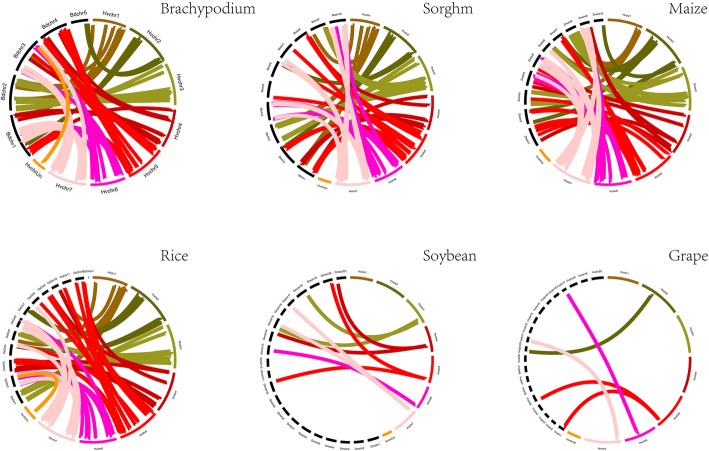
Furthermore, the comparative analysis between barley with other six species (Brachypodium, sorghum, maize, rice, soybean and grape) was performed to determine the origin and evolutionary relationships of MAPK cascade genes (Fig. 5). Through whole genome-wide syntenic analysis, a total of 84, 80, 77, 67, 5 and 7 barely MAPK cascade genes were identified to have orthologous counterpart in Brachypodium, rice, sorghum, maize, grape and soybean (Additional file 7: Table S4 to S9). The average Ka/Ks value was maximum between barley and Brachypodium (0.1641), followed by rice and sorghum (0.1544) as well as maize (0.43), suggesting the genes pairs between barley and those species appeared to have undergone extensive intense purifying selection. Besides, we found that most of MAPK cascade genes showed syntenic bias towards particular chromosomes of sorghum, maize, rice, which indicated that the chromosomal rearrangement events like duplication and inversion may predominantly shape the distribution and organization of MAPK genes in these genomes [35].
Fig. 5.
Comparative physical mapping showing the degree of orthologous relationships of MAPK cascade genes with Brachypodium, Sorghum, Maize, Rice, Soybean and Grape
Comprehensive analysis of the expression profiles of barley MAPK cascade genes
To preliminarily predict the biological function of these barley MAPK cascade genes, gene ontology (GO) analysis was firstly performed (Additional file 6: Figure S6) and they could be annotated into 40 GO terms, including 9 terms of molecular function, 19 of biological processes and 11 of cellular components, respectively. In the cellular components category, cell and cell part were main annotation terms, whereas binding, catalytic nucleoside and transferase were the most presented function in the molecular function category. In the biological process category, cellular metabolic, cellular, metabolic and macromolecule metabolic process occupied most of the proportion. By employing the fisher statistical test method, a total of 17 terms were significant enriched (P < 0.05 and Q < 0.05) when taking the whole barley genome as customized backgrounds, including 5 biological process categories, 6 molecular function categories and 6 cellular component categories (Additional file 7: Table S10). These results revealed that the MAPK cascade genes played diverse roles in diverse development and stress response pathways in barley.
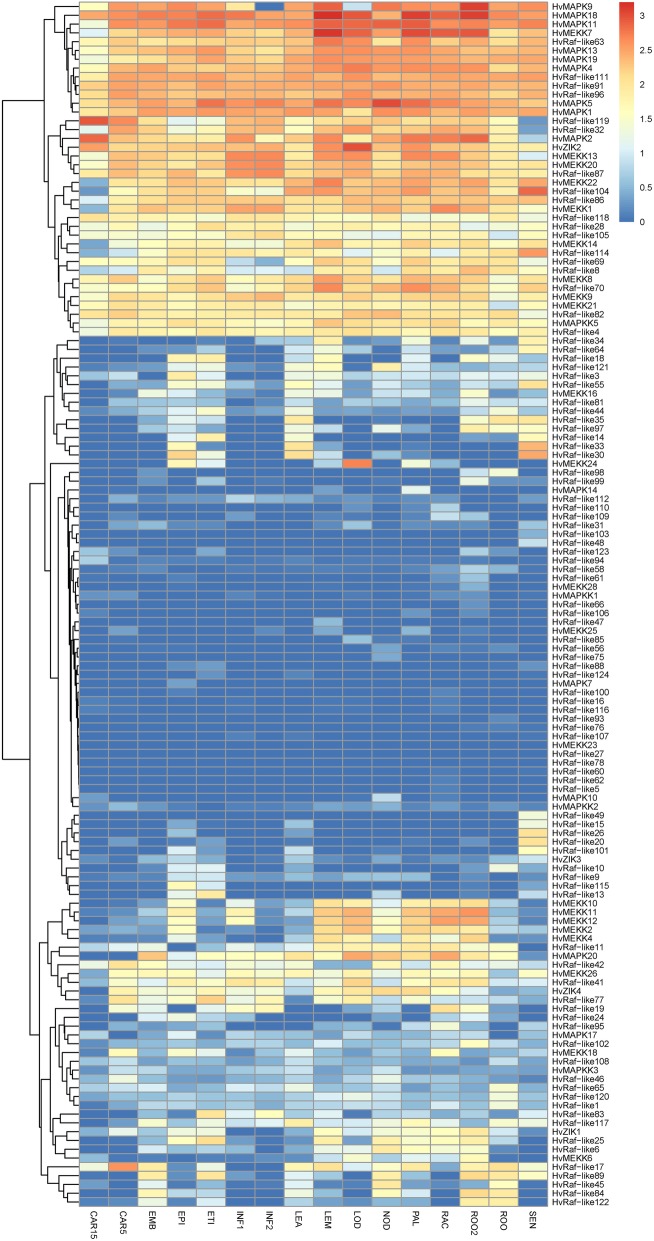
Furthermore, the expression profiles of MAPK cascade genes at 16 developmental stages were investigated using RNA-Seq data. A total of 75 genes were found to be expressed in at least one organ or stage (Fig. 6). A high variance in the expression levels among these MPAK cascade genes was observed, of which a series of them showed relatively high expression in all the tested tissues, such as HvMAPK1, HvMAPK4, HvRaf-like63, HvRaf-like87 and HvZIK2, The ortholog of HvZIK2 in Arabidopsis is AtZIK4(WNK1), which is found to regulating internal circadian rhythm and flowering time [36]. It highly expressed in different organs, suggesting it also played the indispensable role in organ formation and development. Additionally, the tissue- and stage-specific MAPK cascade genes were also identified. HvRaf-like103 and HvRaf-like49 were found to be predominantly expressed in senescing leaf, whereas HvRaf-like66, HvRaf-like47, HvRaf-like93 and HvMAPK7 showed preferential expression in the root, lemma, seedling root and epidermis, respectively, suggesting that these genes may mainly involve into organ- or tissue-specific development in barley.
Fig. 6.
Hierarchical clustering of expression profiles of barley MAPKKK cascade genes across different stages. CAR15: bracts removed grains at 15DPA; CAR5: bracts removed grains at 5DPA; EMB: embryos dissected from 4d-old germinating grains; EPI: epidermis with 4 weeks old; ETI: etiolated from 10-day old seedling; INF1: young inflorescences with 5 mm; INF2: young inflorescences with 1–1.5 cm; LEA: shoot with the size of 10 cm from the seedlings; LEM: lemma with 6 weeks after anthesis; LOD: lodicule with 6 weeks after anthesis; NOD: developing tillers at six-leaf stage; PAL: 6-week old palea; RAC: rachis with 5 weeks after anthesis; ROO2: root from 4-week old seedlings; ROO: Roots from the seedlings at 10 cm shoot stage; SEN: senescing leaf
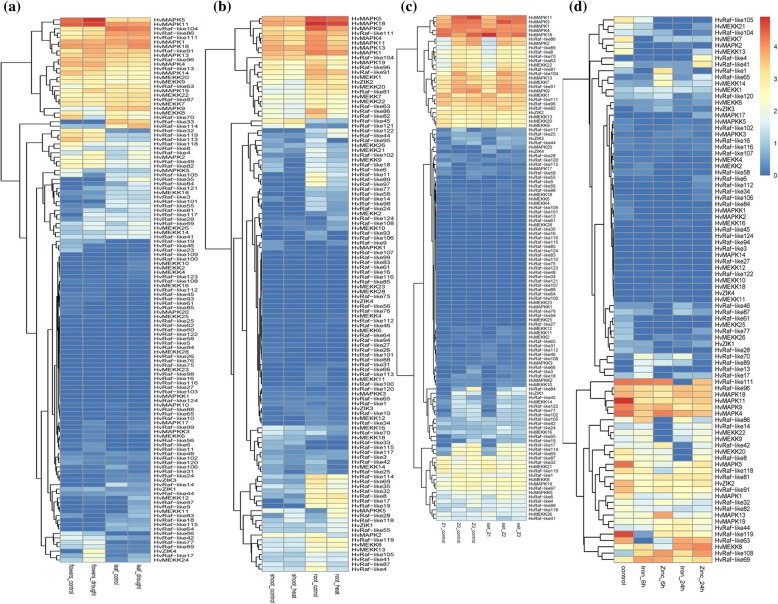
To get insight into the roles of MAPK cascade genes in response to abiotic stresses, the expression profiles of them under drought, heat, salt were investigated to discover the abiotic stress-responsive candidates. Results showed that a total of 123 genes were detected to be expressed under drought stress (Fig. 7a). Among them, 10 and 24 genes were significantly up-regulated, whereas 5 and 19 MAPK cascade genes were significantly down-regulated in flowers and leaves when subjecting to drought. Meanwhile, 114 MAPK cascade genes were found to express under heat stress (Fig. 7b). Remarkably, HvRaf-like124 and HvMAPKK5 presented about 62 and 21 times higher expression level under heat stress compared to control. Previous study found the MPK20 have the defense function in cotton, while its ortholog HvMAPKK5 involved in regulating heat stress adaptation in barley, suggesting it might have divergent function in different species [37]. The expression patterns of MAPK cascades genes under salt stress were also examined (Fig. 7c). Totally, 5, 7 and 9 genes showed up-regulated in the root Z1, Z2 and Z3 respectively, of which the expression level of HvRaf-like28 and Hv-Raf-like113 were up-regulated with more than 10 fold at the Z1 zone and HvMAPKK1 showed 34-fold change at the Z2 zone. Besides, a total of 7, 11 and 4 genes were identified to be down-regulated at root Z1, Z2 and Z3 zone respectively. HvZIK4 and HvRaf-like56 was 862 and 558 time lower expression at Z1 and Z2 zone of root under salt stress than that of control.
Fig. 7.
Hierarchical clustering of expression profiles of barley MAPKKK cascade genes under five stressed conditions. a: Drought stress; b: Heat stress; c: Salt stress; d: Zinc and Iron stress
Finally, the expression profiles of these genes under zinc metal poisoning and iron were investigated (Fig. 7d). When in response to iron stress, 9 genes showed up-regulated and 7 showed down-regulated after 6 h treatment. Furthermore, 8 up-regulated and 11 down-regulated genes were found after 24 h treatment. Among them, HvMAPK17, HvRaf-like4, HvRaf-like70, HvRaf-like109 HvZIK3and HvRafZIK4 all presented up-regulated under iron stress after both 6 h and 24 h treatment, whereas HvMAPK2 and HvRaf-like41 showed down-regulated. Under zinc stress, a total of 13 and 12 up-regulated genes as well as 14 and 16 down-regulated genes were found after 6 h and 24 h treatment, respectively. Among them HvMAPKK5, HvMEKK7, HvMEKK26, HvRaf-like28 and HvRaf-like58 were all down-regulated at all treatment, whereas HvZIK3, HvRaf-like65, HvRaf-like4, HvRaf-like108, HvMEKK14, HvMEKK10 and HvRaf-like108 displayed up-regulated after both 6 h and 24 h treatment. Obviously, HvZIK3, HvRaf-like4, HvRaf-108 showed up-regulated expression under both iron and zinc treatment, which might play the important roles in regulating signal transduction process for metal poisoning response and detoxification.
Network construction of HvMAPK cascade genes
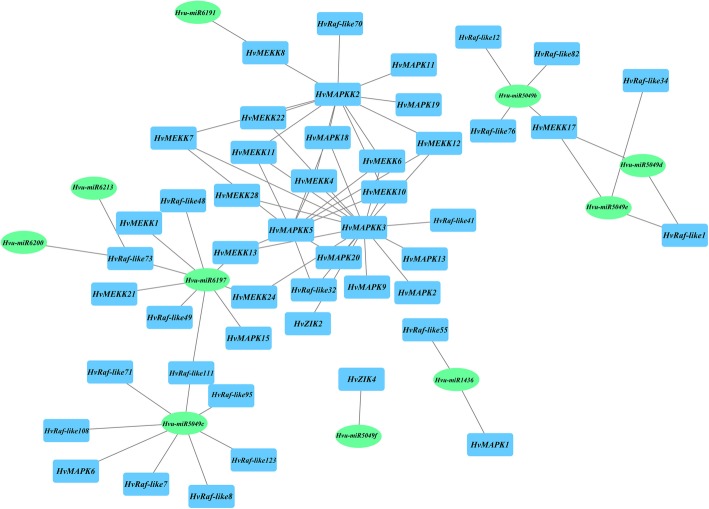
To get the network of miRNA targeting on MAPK cascade genes, the putative miRNAs targeted HvMAPK cascade genes were analyzed. Results found that 26 MAPK cascade genes including 3 MAPKs and 23 MAPKKK genes were predicted to be targeted by 11 miRNAs, while no miRNA target was found for HvMAPKK genes, which might be due to the limited barley miRNA reported at present (Additional file 7: Table S11). Totally, 36 miRNA-MAPK interactions were constructed based on the target relationship. The barley cascade genes were mainly inhibited by miRNAs through transcript cleavage (94.44%), while HvRaf-like12 and HvRaf-like12 and HvRaf-like76 were inhibited to translation by miRNAs. Additionally, miRNAs mainly targeted on the CDS region but behind the protein kinase domain of these MAPK cascade genes to function gene silence.
The co-expression regulatory network was further constructed to detect the interaction among these barley MAPK cascade genes based on weighted correlation of their expressions using a big datasets of 173 RNA-seq data. Only the relations between MAPKKK and MAPKK as well as MAPKK and MAPK were presented. A total of 40 interactions composed of 25 genes were constructed, including 7 MAPK, 3 MAPKK and 15 MAPKKK genes respectively (Fig. 8). Among them, some MAPK cascade modules has been verified in model plants, such as MKK3-MPK6 in Arabidopsis [38]and MAPK18-MAPKK2-MEKK4 in Brachypodium [30]. Furthermore, a total of 18 genes including 2 MAPK, 10 MEKK, 2 HvRaf-like and one ZIK gene were predicted to be interacted with HvMAPKK3, suggesting that it may be the hub gene of the co-expression regulatory network, playing the key role in barley MAPK cascade signaling pathway. In Arabidopsis, MAPKK3 is found to be expressed in all organs, and plays a vital role in photomorphogenesis to regulate gene expression under various light conditions, as well as involved in cell expansion, pathogen signaling and jasmonate signaling pathway, indicating it is critical for development and signaling transduction [39, 40]. Thus, the barley ortholog HvMAPKK3 might also play the hub role in co-expression network in barley response to development and stresses. In addition, there was 10 MAPK-MAPKK, 30 MAPKK-MAPKKK interactions were also obtained to use to subsequently experimental validation. Combined with miRNA-target interaction mentioned above, the regulatory network containing a total of 46 HvMAPK cascade genes and 46 miRNAs were constructed and 72 branches were linked for each other, which provided the indispensable resource to facilitate the MAPK pathway and signal transduction mechanism studies in barley and beyond.
Fig. 8.
The co-expression regulatory network of MAPK cascade genes in barley. Box colour: blue, MAPK gene in barley; green, miRNA s found in barley
Conclusion
This is the first study to identify the MAPK cascade genes in barley at genomic level. Totally, 20 HvMAPKs, 6 HvMAPKKs and 156 HvMAPKKKs were obtained, which was further supported the existence by EST or full-length cDNA sequences. The phylogenetic relationships, intron-exon structure as well as conserved motif analysis all strongly supported the prediction. Furthermore, both segmental and tandem duplication events contributed to the expansion of the MAPK cascade genes in barley. The expression profiles of these MAPK cascade genes during development and under abiotic stresses were investigated and the tissue-specific or stress-responsive genes were identified, which could be considered as the candidates for further functional studies. Finally, the co-expression regulatory network of the MAPK cascade genes was constructed using WGCNA tool based on a total of 174 RNA-seq data. A total of 30 MAPKKK-MAPKK, 10 MAPKK-MAPK potential interactions were identified, which contributed to better understanding the MAPK signal transduction pathway in barely.
Methods
Identification of MAPK cascade genes in barley
The protein sequences of the latest updated barley genome Morex v2.0 [26] were retrieved from the IPK website (http://webblast.ipk-gatersleben.de/barley_ibsc/). Then, the MAPK cascade proteins of Arabidopsis from the TAIR database, were used as queries to search against the barley proteins using BLASTP program with an e-value of 1e-5 and identity of 50% as the threshold. The HMMER 3.0 program was employed to conduct for Hidden Markov Model (HMM) algorithm search using the serine/threonine-protein kinase-like domain (PF00069) as the query with the threshold of E < 1e− 5. The HMMER hits were further integrated with the BLASTP results and parsed by manual editing to remove redundant. Those genes displayed the consensus sequences as Jonak et al described were considered as the potential MAPK cascade genes [3]. The candidates were subsequently submitted to SMART and PFAM web tool to verify the kinase domain. Additionally, the putative MAPK cascade genes were further verified through searching against the barely ESTs by BLASTN tool. The theoretical isoelectric point (pI), molecular weight (MW) and gravy of the identified barley MAPK cascade genes were evaluated using ProtParam tool (http://web.expasy.org/protparam/) integrated in ExPASy database. The cello online server (http://cello.life.nctu.edu.tw/) was used to detect the subcellular localization and protein solubility was predicted by PROSOII tool (http://mips.helmholtz-muenchen.de/prosoII).
Phylogenetic relationship and conserved motif analysis
Multiple sequence alignment were performed using ClustalX v2.0 with default parameter [41]. A neighbor–joining (NJ) phylogenetic tree was constructed based on the full-length protein sequences using the MEGA software with a bootstrap of 1000 replications [42]. The gene structures were obtained from the GTF annotation file of barley genome and then were displayed by Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/index.php). Furthermore, the protein domain and conserved motifs of barley MAPK cascade genes were predicted using InterProScan tool. Finally, the upstream 1.5 kb genomic DNA sequences of each gene were extracted from barley genome, and then submitted to PlantCARE database to detect the putative cis-regulatory elements [43].
Gene duplication and molecular selection analysis
Gene duplication events were defined based on the following three criteria: 1) the alignment should cover more than 70% of the longer gene; (b) the identity of the aligned region should be more than 70%; 3) for tightly linked genes only one duplication event was counted [44]. The gene synteny between barley and other species, including Brachypodium distachyon, Sorghum bicolor, Zea mays, Oryza sativa, Vitis vinifera and Glycine max was conducted using the MCScanX toolkit [45]. The linked genes pairs were displayed using the Circus tool. The rate of Ka (non-synonymous substitution)/Ks (synonymous substitution) was employed to assess the codon evolutionary rate between the synteny genes using the codeml program embedded in the PAML package [46]. The formula T = Ks/2λ was employed to calculate the duplication and divergence time, where λ referred to the mutation rate, was considered as 6.5 × 10− 9 synonymous substitutions per site per year.
Expression profiles and co-expression networks construction
The MAPK cascade genes were firstly searched against the NR protein database using the local BLASTx with an E-value cut off of 10–5. Based on the Nr annotation, Blast2GO [47] program was used to retrieved the GO (gene ontology) annotation. AgriGO v2 (http://systemsbiology.cau.edu.cn/agriGOv2/index.php) was applied to conduct the singular enrichment analysis. Furthermore, a total of 172 public available RNA-seqs (Additional file 7: Table S12) including multiple tissues and developmental stages as well as biotic and abiotic stresses were downloaded from the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) database to investigate the expression profiles of these genes. The FPKM (fragments per kilobase of transcript per million fragments mapped reads) value were calculated by Hisat2 and Stringtie software [48]. Then, differentially expressed genes were identified with the following threshold values: fold change≥2, FDR(false discovery rate) ≤ 0.01, and the absolute ratio of log2 ≥ 1. All FPKM data was finally reported by log2 counts and the heat map was visualized using pheatmap package in R. WGCNA was used to construct the co-expression network based on all of the downloaded transcriptome data [49]. Besides, all the identified MAPK cascade transcripts were submitted to the psRNATarget tool [50] to search the barley miRNAs targets in the miRBase. The regulatory network of Hvu-miRNA and HvMAPK cascade genes were visualized using cytoscape tool (http://www.cytoscape.org/).
Supplementary information
Additional file 1: Figure S1. Multiple sequence alignment of the partial sequences of 20 HvMAPK proteins to identify the TDY and TEY motif. The red color marked sequence is the TDY or TEY motif.
Additional file 2: Figure S2. Multiple sequence alignment of the full length sequence of 20 HvMAPK proteins to identify the conserved kinase motifs. The color marked indicated the conserved motifs found.
Additional file 3: Figure S3. Multiple sequence alignment of the HvMAPKK to identify the conserved kinase motifs. The red color marked are the signature motif of MAPKK proteins.
Additional file 4: Figure S4. Multiple sequence alignment of the HvMAPKKK to identify the conserved kinase motifs. The red color marked are the signature motif of MEKK, Raf and ZIK three sub family.
Additional file 5: Figure S5. Evolutionary relationships and grouping among barley, rice and Arabidopsis MAPKs.
Additional file 6: Figure S6. GO annotation of these identified barley MAPK cascade genes.
Additional file 7: Table S1. Motif identification based on PFAM database. Table S2. Characteristics of cis-acting regulatory elements in the promoter region of these identified barley MAPK cascade genes. Table S3. Duplicated MAPK cascade gene pairs identified in barley. Table S4. The Ka/Ks ratios for orthologous MAPK cascade proteins between barley and brachypodium. Table S5. The Ka/Ks ratios for orthologous HvMAPK cascade proteins between barley and rice sorghum. Table S6. The Ka/Ks ratios for orthologous MAPK cascade proteins between barley and maize. Table S7. The Ka/Ks ratios for orthologous MAPK cascade proteins between barley and sorghum. Table S8. The Ka/Ks ratios for orthologous MAPK cascade proteins between barley and soybean. Table S9. The Ka/Ks ratios for orthologous MAPK cascade proteins between barley and grape. Table S10. GO annotation of the identified barley MAPK cascade genes. Table S11. List of the putative miRNAs targeted on HvMAPK cascade genes identified by psRNATarget online tool. Table S12. Accession number and sample information of RNA-seq data using in this study.
Acknowledgments
We thank the High Performance Computing center of Northwest A&F University for providing computational resources in this work.
Abbreviations
- ABA
Abscisic acid
- ATP
Adenosine triphosphate
- CD
Common docking
- EST
Expressed sequence tag
- FDR
False discovery rate
- FPKM
Fragments per kilobase of transcript per million fragments mapped reads
- GO
Gene ontology
- HMM
Hidden Markov Model
- Ka
Non-synonymous substitution
- Ks
Synonymous substitution
- MAPK/MPK
Mitogen-activated protein kinase
- MAPKK/MKK
Mitogen-activated protein kinase kinase
- MAPKKK/MEKK
Mitogen-activated protein kinase kinase
- MeJA
Methyl jasmonate
- miRNA
MicroRNA
- MW
Molecular weight
- PAS
Per-Arnt-Sim
- pI
Isoelectric point
- SARE
Salicylic acid responsiveness
- TDY
Putative activation motif in MAPK gene TDY1
- TGA
Auxin-responsive element
Authors’ contributions
CLC collected data, perform analysis and also drafted the manuscript. YG contributed to data analysis. YJL and PY contributed to data collection. NXJ conceived this study and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was mainly supported by the Open Project Program of the State Key Laboratory of Crop Stress Biology in Arid Areas, Northwest A&F University (Grant No. CSBAA2019001) and National Natural Science Foundation of China (Grant No. 31771778). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its additional files.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Licao Cui, Email: juelianjunjie@foxmail.com.
Guang Yang, Email: drbiology@aliyun.com.
Jiali Yan, Email: 577891374@qq.com.
Yan Pan, Email: panya321@qq.com.
Xiaojun Nie, Phone: +86-29-87082984, Email: small@nwsuaf.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12864-019-6144-9.
References
- 1.Tena G, Asai T, Chiu WL, Sheen J. Plant mitogen-activated protein kinase signaling cascades. Curr Opin Plant Biol. 2001;4(5):392–400. doi: 10.1016/S1369-5266(00)00191-6. [DOI] [PubMed] [Google Scholar]
- 2.MAPK Group. Ichimura K, et al. Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci. 2002;7(7):301–308. doi: 10.1016/S1360-1385(02)02302-6. [DOI] [PubMed] [Google Scholar]
- 3.Jonak C, Okrész L, Bögre L, Hirt H. Complexity, cross talk and integration of plant MAP kinase signalling. Curr Opin Plant Biol. 2002;5(5):415. doi: 10.1016/S1369-5266(02)00285-6. [DOI] [PubMed] [Google Scholar]
- 4.Zhang T, Liu Y, Yang T, Zhang L, Xu S, Xue L, An L. Diverse signals converge at MAPK cascades in plant. Plant Physiol Biochem. 2006;44(5):274–283. doi: 10.1016/j.plaphy.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Hamel LP, Nicole MC, Sritubtim S, Morency MJ, Ellis M, et al. Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 2006;11:192–198. doi: 10.1016/j.tplants.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 6.Pitzschke A, Schikora A, Hirt H. MAPK cascade signalling networks in plant defence. Curr Opin Plant Biol. 2009;12(4):421–426. doi: 10.1016/j.pbi.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 7.Hao L, Wen Y, Zhao Y, Lu W, Xiao K. Wheat mitogen-activated protein kinase gene TaMPK4 improves plant tolerance to multiple stresses through modifying root growth, ROS metabolism, and nutrient acquisitions. Plant Cell Rep. 2015;34:2081–2097. doi: 10.1007/s00299-015-1853-2. [DOI] [PubMed] [Google Scholar]
- 8.Menzel Wilhelm, Stenzel Irene, Helbig Lisa‐Marie, Krishnamoorthy Praveen, Neumann Susanne, Eschen‐Lippold Lennart, Heilmann Mareike, Lee Justin, Heilmann Ingo. A PAMP ‐triggered MAPK cascade inhibits phosphatidylinositol 4,5‐bisphosphate production by PIP 5K6 in Arabidopsis thaliana. New Phytologist. 2019;224(2):833–847. doi: 10.1111/nph.16069. [DOI] [PubMed] [Google Scholar]
- 9.Rohila JS, Yang Y. Rice mitogen-activated protein kinase gene family and its role in biotic and abiotic stress response. J Integr Plant Biol. 2007;49(6):751–759. doi: 10.1111/j.1744-7909.2007.00501.x. [DOI] [Google Scholar]
- 10.Rao KP, Richa T, Kumar K, Raghuram B, Sinha AK. In silico analysis reveals 75 members of mitogen-activated protein kinase kinase kinase gene family in rice. DNA Res. 2010;17(3):139–153. doi: 10.1093/dnares/dsq011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wankhede DP, Misra M, Singh P, Sinha AK. Rice mitogen activated protein kinase kinase and mitogen activated protein kinase interaction network revealed by in-silico docking and yeast two-hybrid approaches. PLoS One. 2013;8(5):e65011. doi: 10.1371/journal.pone.0065011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen L, Hu W, Tan S, Wang M, Ma Z, Zhou S, Deng X, Zhang Y, Huang C, Yang G. Genome-wide identification and analysis of MAPK and MAPKK gene families in Brachypodium distachyon. PLoS One. 2012;7(10):e46744. doi: 10.1371/journal.pone.0046744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng K, Liu F, Zou J, Xing G, Deng P, Song W, Tong W, Nie X. Genome-wide identification, evolution and co-expression network analysis of mitogen-activated protein kinase kinase kinases in Brachypodium distachyon. Front Plant Sci. 2016;7(556):1400. doi: 10.3389/fpls.2016.01400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Zhang D, Wang L, Li D. Genome-wide analysis of mitogen-activated protein kinase gene family in maize. Plant Mol Biol Report. 2013;31(6):1446–1460. doi: 10.1007/s11105-013-0623-y. [DOI] [Google Scholar]
- 15.Kong X, Pan J, Zhang D, Jiang S, Cai G, Wang L, Li D. Identification of mitogen-activated protein kinase kinase gene family and MKK–MAPK interaction network in maize. Biochem Biophys Res Commun. 2013;441(4):964–969. doi: 10.1016/j.bbrc.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 16.Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomezgomez L, Boller T, Ausubel FM, Sheen J. MAP kinase signaling cascade in Arabidopsis innate immunity. Nature. 2002;415(6875):977. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- 17.Galletti R, Ferrari S, De LG. Arabidopsis MPK3 and MPK6 play different roles in basal and oligogalacturonide- or flagellin-induced resistance against Botrytis cinerea. Plant Physiol. 2011;157(2):804–814. doi: 10.1104/pp.111.174003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eckardt NA. A complete MAPK signaling cascade that functions in stomatal development and patterning in Arabidopsis. Plant Cell. 2007;19(1):7. doi: 10.1105/tpc.107.190110. [DOI] [Google Scholar]
- 19.Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE. Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell. 2000;103(7):1111–1120. doi: 10.1016/S0092-8674(00)00213-0. [DOI] [PubMed] [Google Scholar]
- 20.Qiu JL, Zhou L, Yun BW, Nielsen HB, Fiil BK, Petersen K, Mackinlay J, Loake GJ, Mundy J, Morris PC. Arabidopsis mtogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiol. 2008;148(1):212–222. doi: 10.1104/pp.108.120006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furuya T, Matsuoka D, Nanmori T. Membrane rigidification functions upstream of the MEKK1-MKK2-MPK4 cascade during cold acclimation in Arabidopsis thaliana. FEBS Lett. 2014;588(11):2025–2030. doi: 10.1016/j.febslet.2014.04.032. [DOI] [PubMed] [Google Scholar]
- 22.Danquah A, De ZA, Boudsocq M, Neubauer J, Frei DFN, Leonhardt N, Pateyron S, Gwinner F, Tamby JP, Ortizmasia D. Identification and characterization of an ABA-activated MAP kinase cascade in Arabidopsis thaliana. Plant J Cell Mol Biol. 2015;82(2):232–244. doi: 10.1111/tpj.12808. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Schiff M, Dineshkumar SP. Involvement of MEK1 MAPKK, NTF6 MAPK, WRKY/MYB transcription factors, COI1 and CTR1 in N-mediated resistance to tobacco mosaic virus. Plant J Cell Mol Biol. 2004;38(5):800–809. doi: 10.1111/j.1365-313X.2004.02085.x. [DOI] [PubMed] [Google Scholar]
- 24.Soyano T, Nishihama R, Morikiyo K, Ishikawa M, Machida Y. NQK1/NtMEK1 is a MAPKK that acts in the NPK1 MAPKKK-mediated MAPK cascade and is required for plant cytokinesis. Genes Dev. 2003;17(8):1055. doi: 10.1101/gad.1071103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayer KFX, Waugh R, Langridge P, Close TJ, Wise RP, Graner A, Matsumoto T, Sato K, Schulman A, Muehlbauer GJ. A physical, genetic and functional sequence assembly of the barley genome. Nature. 2012;491(7426):711–716. doi: 10.1038/nature11543. [DOI] [PubMed] [Google Scholar]
- 26.Mascher M, Gundlach H, Himmelbach A, Beier S, Twardziok SO, Wicker T, Radchuk V, Dockter C, Hedley PE, Russell J. A chromosome conformation capture ordered sequence of the barley genome. Nature. 2017;544(7651):427–433. doi: 10.1038/nature22043. [DOI] [PubMed] [Google Scholar]
- 27.Lv SZ, Nie XJ, Wang L, Biradar SB, Jia XO, Weining S. Identification and characterization of microRNAs from barley (Hordeum vulgare L.) by Solexa sequencing. Int J Mol Sci. 2012;13:2973–2984. doi: 10.3390/ijms13032973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Křenek P, Niks RE, Vels A, Vyplelová P, Šamaj J. Genome-wide analysis of the barley MAPK gene family and its expression patterns in relation to Pucciniahordei infection. Acta Physiol Plant. 2015;37(11):254. doi: 10.1007/s11738-015-2010-9. [DOI] [Google Scholar]
- 29.Wang M, Yue H, Feng K, Deng P, Song W, Nie X. Genome-wide identification, phylogeny and expressional profiles of mitogen activated protein kinase kinase kinase (MAPKKK) gene family in bread wheat (Triticum aestivum L.) BMC Genomics. 2016;17:668. doi: 10.1186/s12864-016-2993-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang M, Wen F, Cao J, Li P, She J, Chu Z. Genome-wide exploration of the molecular evolution and regulatory network of mitogen-activated protein kinase cascades upon multiple stresses in Brachypodium distachyon. BMC Genomics. 2015;16:228. [DOI] [PMC free article] [PubMed]
- 31.Lehti-Shiu MD, Shiu SH. Diversity, classification and function of the plant protein kinase superfamily. Philos Trans R Soc Lond Ser B Biol Sci. 2012;367(1602):2619–2639. doi: 10.1098/rstb.2012.0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye J, Yang H, Shi H, Wei Y, Tie W, Ding Z, Yan Y, Luo Y, Xia Z, Wang W, Peng M, Li K, Zhang H, Hu W. The MAPKKK gene family in cassava: genome-wide identification and expression analysis against drought stress. Sci Rep. 2017;7(1):14939. doi: 10.1038/s41598-017-13988-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Colcombet J, Hirt H. Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem J. 2008;413(2):217. doi: 10.1042/BJ20080625. [DOI] [PubMed] [Google Scholar]
- 34.Wilson MA, Iser WB, Son TG, Logie A, Cabral-Costa JV, Mattson MP, Camandola S. Skn-1 is required for interneuron sensory integration and foraging behavior in Caenorhabditis elegans. PLoS ONE. 2017;12(5):e0176798. doi: 10.1371/journal.pone.0176798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akhunov ED, Sunish S, Hanquan L, Shichen W, Akhunova AR, Gaganpreet K, Wanlong L, Forrest KL, Deven S, Hana S. Comparative analysis of syntenic genes in grass genomes reveals accelerated rates of gene structure and coding sequence evolution in polyploid wheat. Plant Physiol. 2013;161(1):9090–9100. doi: 10.1104/pp.112.205161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Liu K, Liao H, Zhuang C, Ma H, Yan X. The plant WNK gene family and regulation of flowering time in Arabidopsis. Plant Biol (Stuttg) 2008;10(5):548–562. doi: 10.1111/j.1438-8677.2008.00072.x. [DOI] [PubMed] [Google Scholar]
- 37.Wang C, He X, Li Y, Wang L, Guo X, Guo X. The cotton MAPK kinase GhMPK20 negatively regulates resistance to Fusarium oxysporum by mediating the MKK4-MPK20-WRKY40 cascade. Mol Plant Pathol. 2018;19(7):1624–1638. doi: 10.1111/mpp.12635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sethi V, Raghuram B, Sinha AK, Chattopadhyay S. A mitogen-activated protein kinase cascade module, MKK3-MPK6 and MYC2, is involved in blue light-mediated seedling development in Arabidopsis. Plant Cell. 2014;26(8):3343–3357. doi: 10.1105/tpc.114.128702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schikora A, Carreri A, Charpentier E, Hirt H. The dark side of the salad: Salmonella typhimurium overcomes the innate immune response of Arabidopsis thaliana and shows an endopathogenic lifestyle. PLoS One. 2008;3(5):e2279. doi: 10.1371/journal.pone.0002279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dóczi R, Brader G, Pettkó-Szandtner A, Rajh I, Djamei A, Pitzschke A, Teige M, Hirt H. The Arabidopsis mitogen-activated protein kinase kinase MKK3 is upstream of group C mitogen-activated protein kinases and participates in pathogen signaling. Plant Cell. 2007;19(10):3266–3279. doi: 10.1105/tpc.106.050039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larkin MA, Blackshields G, Brown N, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R. ClustalW and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 42.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gu Z, Cavalcanti A, Chen FC, Bouman P, Li WH. Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol Biol Evol. 2002;19(3):256–262. doi: 10.1093/oxfordjournals.molbev.a004079. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, Tang H, DeBarry JD, Tan X, Li J, Wang X, Lee T-h, Jin H, Marler B, Guo H. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40(7):e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 47.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 48.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4(1):17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 50.Dai X, Zhuang Z, Zhao PX. psRNATarget: a plant small RNA target analysis server (2017 release) Nucleic Acids Res. 2018;46(W1):W49–W54. doi: 10.1093/nar/gky316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Multiple sequence alignment of the partial sequences of 20 HvMAPK proteins to identify the TDY and TEY motif. The red color marked sequence is the TDY or TEY motif.
Additional file 2: Figure S2. Multiple sequence alignment of the full length sequence of 20 HvMAPK proteins to identify the conserved kinase motifs. The color marked indicated the conserved motifs found.
Additional file 3: Figure S3. Multiple sequence alignment of the HvMAPKK to identify the conserved kinase motifs. The red color marked are the signature motif of MAPKK proteins.
Additional file 4: Figure S4. Multiple sequence alignment of the HvMAPKKK to identify the conserved kinase motifs. The red color marked are the signature motif of MEKK, Raf and ZIK three sub family.
Additional file 5: Figure S5. Evolutionary relationships and grouping among barley, rice and Arabidopsis MAPKs.
Additional file 6: Figure S6. GO annotation of these identified barley MAPK cascade genes.
Additional file 7: Table S1. Motif identification based on PFAM database. Table S2. Characteristics of cis-acting regulatory elements in the promoter region of these identified barley MAPK cascade genes. Table S3. Duplicated MAPK cascade gene pairs identified in barley. Table S4. The Ka/Ks ratios for orthologous MAPK cascade proteins between barley and brachypodium. Table S5. The Ka/Ks ratios for orthologous HvMAPK cascade proteins between barley and rice sorghum. Table S6. The Ka/Ks ratios for orthologous MAPK cascade proteins between barley and maize. Table S7. The Ka/Ks ratios for orthologous MAPK cascade proteins between barley and sorghum. Table S8. The Ka/Ks ratios for orthologous MAPK cascade proteins between barley and soybean. Table S9. The Ka/Ks ratios for orthologous MAPK cascade proteins between barley and grape. Table S10. GO annotation of the identified barley MAPK cascade genes. Table S11. List of the putative miRNAs targeted on HvMAPK cascade genes identified by psRNATarget online tool. Table S12. Accession number and sample information of RNA-seq data using in this study.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its additional files.