Abstract
Background
Circular RNAs (circRNAs) have recently been identified as potential functional modulators of the cellular physiology processes. The study aims to uncover the potential clinical value and driving molecular mechanisms of circRNAs in gallbladder cancer (GBC).
Patients and methods
We performed RNA sequencing from four GBC and paired adjacent normal tissues to analyze the circRNA candidates. Quantitative real-time polymerase chain reaction (QRT-PCR) was used to measure the circFOXP1 expression from 40 patient tissue samples. Short hairpin RNA mediated knockdown or exogenous expression of circFOXP1 combined with in vitro and in vivo assays were performed to prove the functional significance of circFOXP1. Double luciferase reporter, RNA immunoprecipitation (RIP) and RNA pull-down assays were also performed.
Results
By performing RNA sequencing from GBC and paired adjacent normal tissues to analyze the circRNA candidates, we identified that circFOXP1 (hsa_circ_0008234) expression was significantly upregulated in GBC tissues and positively associated with lymph node metastasis, advanced TNM stage and poor prognosis in patients. Short hairpin RNA mediated knockdown or exogenous expression of circFOXP1 combined with in vitro assays demonstrated that circFOXP1 has pleiotropic effects, including promotion of cell proliferation, migration, invasion, and inhibition of cell apoptosis in GBC. In vivo, circFOXP1 promoted tumor growth. Mechanistically, double luciferase reporter, RNA immunoprecipitation (RIP) and biotin-labeled RNA pull-down assays clarified that circFOXP1 interacted with PTBP1 that could bind to the 3’UTR region and coding region (CDS) of enzyme pyruvate kinase, liver and RBC (PKLR) mRNA (UCUU binding bites) to protect PKLR mRNA from decay. Additionally, circFOXP1 acted as the sponge of miR-370 to regulate PKLR, resulting in promoting Warburg effect in GBC progression.
Conclusions
These results demonstrated that circFOXP1 serve as a prognostic biomarker and critical regulator in GBC progression and Warburg effect, suggesting a potential target for GBC treatment.
Keywords: Gallbladder cancer, circFOXP1, PTBP1, PKLR, miR-370
Background
Gallbladder cancer (GBC) is the most common and leading cause of cancer-associated mortality among biliary tract carcinomas worldwide. Complete surgical resection is the only potentially curative choice for GBC, but due to lack of specific signs, symptoms or reliably sensitive disease markers, many patients are diagnosed at advanced stages [1, 2]. Conventional chemotherapy, radiotherapy, and molecular targeted therapy provide palliative relief for GBC patients; however, in the past few decades, there has been no definitive improvement in overall survival (OS) [3–5]. Therefore, improving molecular understanding of the underlying pathogenesis in GBC and investigating mechanism-based therapeutic strategies for patients is essential.
Circular RNAs (circRNAs), covalently closed RNAs formed from both exonic and intronic sequences, are highly stable RNA molecules [6, 7]. CircRNAs exert certain functions, including as intermediates in RNA alternative splicing (AS), as regulators of transcription in cis, and as miRNA sponges [8]. Some of circRNAs are identified as potential functional modulators of cellular physiology processes involved in cell proliferation, differentiation, apoptosis and tumor progression [9, 10]. For example, CiRS-7 (also termed CDR1as) harbors more than 70 conventional miR-7 binding sites and is involved in cancer-related pathways [11]. CircHIPK3 but not HIPK3 mRNA regulates human cell growth in hepatocellular carcinoma [12]. Ectopic expression of circ-foxo3 repressed cell cycle progression by binding to cyclin-dependent kinase 2 (CDK2) and cyclin-dependent kinase inhibitor 1 (or p21) [13].
The Warburg effect (or aerobic glycolysis) is a well-characterized metabolic alteration that involved in cancer phenomenon including rapid cell proliferation, invasion, and migration in tumors [14]. enzyme pyruvate kinase, liver and RBC (PKLR) is identified as a pivotal regulator of glycolytic reprogramming and affect tumor cells. Nie et al. showed that mineralocorticoid receptor (MR) inhibits the Warburg effect and cancer progression via the miR-338-3p-PKLR axis in hepatocellular carcinoma [15]. Yang et al. demonstrated the NQO1/PKLR axis promotes lymph node metastasis and breast cancer progression by activating the AMPK and AKT/mTOR signaling pathway and consequently induced glycolytic reprogramming [16]. The association of between circRNAs and Warburg effect in gallbladder cancer remains unknown.
Here, we identified a circular FOXP1 RNA (circFOXP1, hsa_circ_0008234), derived from the exon region of the FOXP1 gene [17, 18], which is significantly upregulated in GBC tissues. CircFOXP1 interacted with PTBP1 or sponged miR-370, which promoted tumor progression and the Warburg effect in GBC by directly targeting the PKLR. Our results indicate that circFOXP1 may be a novel potential target for GBC treatment.
Methods
RNA-sequencing analysis
A detailed description of RNA-sequencing analysis was provided in Additional file 1: Supplementary Materials and Methods section. The GEO Accession number is GSE100363.
Patient tissue samples
Forty human GBC and adjacent normal tissue specimens were collected from patients who underwent radical resection at Xinhua Hospital between March 2009 and January 2013. The eleven patients were male and twenty-nine were female. Ages ranged from 35 to 82 years (the mean value = 54.85 years). Each tissue sample was snap-frozen in liquid nitrogen for further analysis. All of patients in this study belonged to the same ethnic group. The patients were selected according to the criteria: (1) All clinicopathological diagnoses were confirmed by two pathologists. (2) None of the patients received any treatments before surgery. (3) None of the patients received radiotherapy or chemotherapy during follow-up period. (4) availability of complete follow-up data and not lost follow-up. (5) no death in the perioperative period. (6) no history of other synchronous malignancies. (7) For patients undergoing radical resection of gallbladder cancer. The study procedure was approved by the Human Ethics Committee of Xinhua Hospital. All patients signed consent forms. Follow-ups after surgery were performed according to patient survival time until March 8, 2016. The clinicopathological data were shown in Table 1.
Table 1.
Correlation between circFOXP1 expression and clinicopathological characteristics in 40 cases GBC patients
| CircFOXP1 expression | P-value | |||
|---|---|---|---|---|
| Clinicopathological characteristics | The number of patients | Lower (n = 20) |
Higher (n = 20) |
|
| Age | ||||
| ≤ 60 | 25 | 10 | 15 | 0.102 |
| > 60 | 15 | 10 | 5 | |
| Gender | ||||
| Male | 11 | 6 | 5 | 0.723 |
| Female | 29 | 14 | 15 | |
| Tumor size | ||||
| < 5 cm | 16 | 11 | 5 | 0.053 |
| ≥ 5 cm | 24 | 9 | 15 | |
| Histological grade | ||||
| well and moderately | 21 | 10 | 11 | 0.752 |
| Poorly and others | 19 | 10 | 9 | |
| Lymph node metastasis | ||||
| N0 | 18 | 13 | 5 | 0.011* |
| N1/2 | 22 | 7 | 15 | |
| TNM stage | 0.027* | |||
| I-II | 19 | 13 | 6 | |
| III-IV | 21 | 7 | 14 | |
| Adjacent organ invasion | 0.110 | |||
| No | 17 | 11 | 6 | |
| Yes | 23 | 9 | 14 | |
*P < 0.05.TNM tumor-node-metastasis
Cell culture
Human GBC cell lines NOZ, GBC-SD, EHGB-1, SGC-996 and OCUG-1 and the human intrahepatic biliary epithelial cell line H69 and another normal biliary epithelia cell line HIBEC were used in the present study. GBC-SD and OCUG-1 cell lines were purchased from Cell Bank of the Chinese Academy of Science (Shanghai, China). The NOZ cell line was purchased from the Health Science Research Resources Bank (Osaka, Japan). EHGB-1 and SGC-996 cells were a generous gift from Eastern Hepatobiliary Surgical Hospital and Institute, The Second Military University, Shanghai, China. The cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco BRL, Grand Island, NY, USA); the cell media contained 10% fetal bovine serum (FBS, HyClone, Invitrogen). Cells were maintained in a humidified incubator at 37 °C in the presence of 5% CO2.
RNAi, plasmid construction and cell transfection
Cells were transfected using lipofectamine 2000 according to the manufacturer’s instructions. Additional information about the performed experiments can be found in the Additional file 1: Supplementary Materials and Methods. The sequences of all oligonucleotide used in the study were shown in Additional file 2: Table S1.
Cell proliferation, migration and invasion assays and cell cycle and cell apoptosis analysis
Cell proliferation was assessed using Cell Counting Kit 8 (Dojindo, Japan); cell migration and invasion was assessed with transwell assays. Cell cycle distribution and cell apoptosis rate analysis was analyzed using flow cytometry. Detailed descriptions of experiments can be found in Additional file 1: Supplementary Materials and Methods.
RNA isolation, quantitative real-time PCR (qRT-PCR) and western blot analysis
RNA isolation, qRT-PCR and western blot analysis were performed as described previously [19]. Detailed descriptions of experiments can be found in Additional file 1: Supplementary Materials and Methods. The sequences of all primers used in the study were shown in Additional file 2: Table S1.
Xenograft mouse model
NOZ cells (1 × 106) stably expressing sh-NC, sh-circFOXP1–1, sh-circFOXP1–2 or GBC-SD cells (1 × 106) stably expressing pLCDH-vector or pLCDH-circFOXP1 were subcutaneously injected into either side of the flank area of 3-week-old male nude mice (n = 5 mice per group). Tumor volumes were measured (0.5 × length × width2) and tumor weights were evaluated in mice on a weekly basis. After 4 weeks, mice were sacrificed, and the tumors were excised. The study protocol was approved by the Animal Care and Use committee of Xinhua Hospital (approval ID: 2014041). All animal experiments were performed in the animal laboratory center at Xinhua Hospital and conformed to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH publication number 85–23, revised 1996).
RNA pull-down assay
RNA pull-down assays were performed as described previously [20, 21]. Briefly, cells extract (2 μg) was mixed with biotinylated RNA (100 pmol). Washed streptavidin agarose beads (100 ml) were added to each binding reaction and further incubated at room temperature for 1 h. Beads were washed briefly three times and boiled in SDS buffer, and the retrieved protein was determined by western blot analysis.
RNA immunoprecipitation (RIP) assay
RIP assays were performed using an EZ-Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA) according to the manufacturer’s instructions. Cells at approximately 90% confluence was lysed using complete RIP lysis buffer containing RNase Inhibitor (Millipore) and protease inhibitor and then 100 μl of whole cell extract was incubated with RIP buffer containing magnetic beads conjugated to specific antibodies. The negative control was normal mouse anti-IgG antibody (Cell Signaling Technology, USA), and the positive control was anti-SNRNP70 antibody (Millipore, USA).
Statistical analysis
All experiments were independently repeated at least three times. Statistical analyses were performed using SPSS 20.0 (SPSS, Chicago, IL, USA). The data are expressed as the mean ± standard error of the mean (SEM). OS was estimated using the Kaplan-Meier method and a log-rank test. The difference between groups was analyzed using Student’s t-test. Differences were statistically significant at P < 0.05.
Results
Expression of circFOXP1 is significantly upregulated in GBC tissues and cells
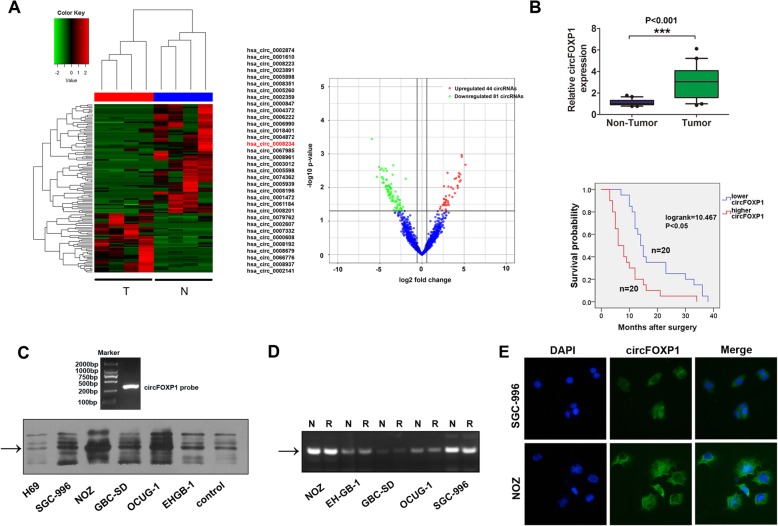
To explore circRNAs expression profile in GBC, we performed RNA sequencing analyses of ribosomal RNA-depleted total RNA from four pairs of GBC tissues and adjacent normal tissues. The cluster heat map demonstrated the differentially expressed circRNAs over 3.5-fold change (Fig. 1a, left). The scatter plots showed that upregulated 44 circRNAs and downregulated 81 circRNAs in GBC tissues compared with adjacent normal tissues (Fig. 1a, right). Compared with previously reported database obtained from circBase [18], 34 of the top 44 upregulated circRNAs were overlapped circRNAs and were listed in Fig. 1a, left. Several circRNAs including circXPO1 (chr2_61710091_61717911_-, hsa_circ_0002607), circFOXP1 (chr3_71090478_71102924_-,hsa_circ_0008234),circMAPK1(chr22_22153300_22162135_-, hsa_circ_0004872), circSENP1 (chr12_48477373_48491907_-, hsa_circ_0006222) and circSMAD2 (chr18_45391429_45423180_-, hsa_circ_0000847) were chosen for further investigation because their parental genes are involved in cell proliferation, invasion and cell cycle regulation in some tumors [22–26]. We examined these investigated circRNA candidates in GBC tissues and paired normal tissue samples from 40 patients. The results showed that expression of circFOXP1, circMAPK1 and circSAMD2 were upregulated in GBC tissues (Fig. 1b, top and Additional file 3: Figure S1A). GBC patients were classified into two groups: high-circFOXP1 group (n = 20, circFOXP1 expression ratio ≥ median ratio) and low-circFOXP1 group (n = 20, circFOXP1 expression ratio < median ratio). Kaplan-Meier analysis and log rank tests indicated that higher circFOXP1 expression levels were positively correlated with a shorter OS in patients (Fig. 1b, low), while circSMAD2 and circMAPK1 expression showed no statistical significance with OS in patients (Additional file 3: Figure S1B). CircFOXP1 expression was upregulated in several tumors compared with normal tissues through RNA sequencing data in a previous study [12]. However, the clinical significance and functional role of circFOXP1 remains unknown.
Fig. 1.
circFOXP1 is upregulated in GBC tissues. a The heat map showed that the upregulated 44 circRNAs and downregulated 81 circRNAs in GBC tissues when compared with adjacent non tumor tissues analyzed by RNA sequencing. The 34 of the upregulated 44 circRNAs were overlapped in database obtained from circBase. b Top, circFOXP1 expression levels were evaluated with qRT-PCR in tissues from 40 cases of GBC compared with adjacent normal tissues. The expression of circFOXP1 was normalized to GADPH. Significant differences between groups were analyzed with a paired samples t-test, ***P < 0.001. Low, Kaplan-Meier analysis and log-rank tests were performed to analyze the association between the expression of circFOXP1 and OS time of GBC patients. CircFOXP1 expression levels were classified as higher expression (circFOXP1 expression rate > median expression rate) and lower expression (circFOXP1 expression rate < median expression rate). c RNAs were isolated from five GBC cell lines and the H69 cell line and subjected to a northern blot analysis using a probe specific for circFOXP1 (top). Arrow, circFOXP1 expression was shown. d Standard PCR was performed to detect the circFOXP1 from control RNA or digested RNAs using RNase R exonuclease in five GBC cell lines. Arrow, circFOXP1 expression was shown. e RNA fluorescence in situ hybridization (FISH) was performed for circFOXP1. Nuclei were stained with 4, 6-diamidino-2-phenylindole (DAPI); the original magnification was 400×
As seen in Table 1, the statistical analysis revealed that high circFOXP1 expression levels were strongly correlated with lymph node metastasis (P = 0.011) and advanced TNM stage (III-IV) (P = 0.027) in GBC patients, but no correlation was found with the other factors (Table 1). Univariate and multivariate Cox analysis revealed that lymph node metastasis (HR = 3.749, 95% CI: 1.578–8.905, P < 0.05) and high circFOXP1 expression (HR = 3.658, 95% CI: 1.587–8.431, P < 0.05) were independent risk factors for the OS of GBC patients (Table 2). As shown in Additional file 4: Figure S2A, the transcript levels of circFOXP1 were also significantly higher in several GBC cells than in the bile duct epithelial cell line H69 or another normal biliary epithelia cell line HIBEC. RNA was isolated and RT-PCR with Sanger sequencing confirmed the circular form in GBC cells (Additional file 4: Figure S2B). RNA isolated from the above cell lines was subjected to northern blot using a probe specific for circFOXP1, confirming that circFOXP1 was present in these cells (Fig. 1c). In addition, we confirmed that circFOXP1 was resistant to RNase R after digestion of RNA using RNase R exonuclease (Fig. 1d and Additional file 4: Figure S2C), which was consistent with a previous study [27], the circHIPK3 was used as the control, which was reported as a circRNA in previous study [12]. Using fluorescence in situ hybridization (FISH) and qRT-PCR analysis, we also noted that circFOXP1 localized in the cytoplasm and nucleus, but was predominately enriched in the cytoplasm in GBC cells (Fig. 1e and Additional file 4: Figure S2D). Thus, the above results indicated that circFOXP1 may serve as a prognostic biomarker for GBC.
Table 2.
Univariate and multivariate Cox analysis of the overall survival (OS) in 40 GBC patients
| Factors | Univariate Cox analysis | Multivariate Cox analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value | |
| Age | 1.005 | 0.974–1.037 | 0.754 | |||
| Gender | 0.796 | 0.390–1.624 | 0.530 | |||
| Tumor size | 0.917 | 0.484–1.738 | 0.790 | |||
| Histological grade | 1.426 | 0.755–2.693 | 0.274 | |||
| Lymph node metastasis | 2.938 | 1.476–5.848 | 0.002* | 3.749 | 1.578–8.905 | 0.003* |
| TNM stage | 2.105 | 1.064–4.164 | 0.033* | 1.123 | 0.372–2.032 | 0.747 |
| Adjacent organ invasion | 1.985 | 1.003–3.930 | 0.049* | 0.968 | 0.447–2.317 | 0.896 |
| Higher circFOXP1 | 2.762 | 1.422–5.362 | 0.003* | 3.658 | 1.587–8.431 | 0.002* |
*P < 0.05, HR Hazard Ratio, CI Confidence intervals
Overexpression of circFOXP1 promotes GBC cell growth in vitro and in vivo
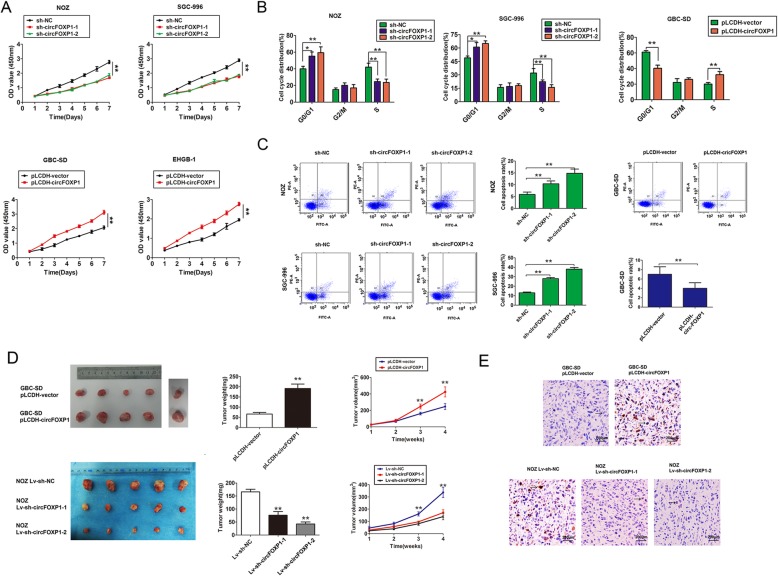
To further explore the biological significance of circFOXP1 in GBC progression, gain-and loss-of-function studies were performed. Based on endogenous expression of circFOXP1 in several GBC cell lines, we developed NOZ and SGC-996 cells with circFOXP1 stably silenced by shRNA-circFOXP1 and EHGB1 or GBC-SD cells with circFOXP1 stably overexpressed using pLCDH-circFOXP1. Meanwhile, to confirm the specificity of circFOXP1 silencing or overexpression, we showed that the cells transfection had no effects on mRNA expression of FOXP1 in GBC cells. In addition, circFOXP1 was also overexpressed after digestion of RNA using RNase R by using pLCDH-circFOXP1 (Additional file 4: Figure S2E-2F). By performing CCK8 assays and flow cytometry analysis in vitro, we observed that knockdown of endogenous circFOXP1 significantly suppressed cell proliferation capacity, G1-S arrest, and increased the cell apoptosis rate in NOZ and SGC-996 cells, but upregulated expression of circFOXP1 dramatically promoted the cell proliferation capacity, beyond the G1-S transition, and reduced the cell apoptosis rate in GBC-SD cells (Fig. 2a-c). Consistent with the decreased cell proliferation capacity, NOZ and SGC-996 cells exhibited reduced expression levels of proliferating cell nuclear antigen (PCNA), MMP9 and AKT, but increased Caspase-3 expression after knockdown of endogenous circFOXP1, and conversely, upregulated expression of circFOXP1 had the opposite effects in GBC cells (Additional file 5: Figure S3A). As shown in Additional file 5: Figure S3B-3C, migration and invasion capacities were impaired in NOZ and SGC-996 cells after knockdown of endogenous circFOXP1, but were dramatically promoted in GBC-SD cells after circFOXP1 overexpression.
Fig. 2.
circFOXP1 promotes cell proliferation in vitro and in vivo. a Cell proliferation capacity was evaluated with CCK8 assays. Briefly, 2000 cells/well were plated in triplicate and cell proliferation was detected at days 1, 2, 3, 4, 5, 6 and 7 days after transfection of NOZ and SGC-996 cells (top) with sh-NC, sh-circFOXP1–1 or sh-circFOXP1–2 or of GBC-SD and EHGB-1 cells (low) with pLCDH-vector and pLCDH-circFOXP1. The experiment was repeated three times, **P < 0.01. b Data are presented as the percentage cell phase distribution including G0/G1, S and G2/M phases after transfection of NOZ and SGC-996 cells with sh-NC, sh-circFOXP1–1 or sh-circFOXP1–2 or GBC-SD cells with pLCDH-vector and pLCDH-circFOXP1, *P < 0.05; **P < 0.01. c The data are presented as cell apoptosis rates after transfection of NOZ and SGC-996 cells (left) with sh-NC, sh-circFOXP1–1 or sh-circFOXP1–2 or GBC-SD cells (right) with pLCDH-vector and pLCDH-circFOXP1. **P < 0.01. d Tumor weight and volume were detected to monitor tumor growth in subcutaneous implantation mouse models; mice were implanted with GBC-SD cells (top) transfected with pLCDH-vector or pLCDH-circFOXP1 and NOZ cells (low) transfected with lv-sh-NC, lv-sh-circFOXP1–1 or lv-sh-circFOXP1–2. e Immunohistochemical staining of Ki67 expression was shown in tumor tissues, as indicated by the number of Ki67-positive cells with GBC-SD cells (top) transfected with pLCDH-vector or pLCDH-circFOXP1 and NOZ cells (low) transfected with lv-sh-NC, lv-sh-circFOXP1–1 or lv-sh-circFOXP1–2, (original magnification, 200×)
To evaluate the biological function of circFOXP1 in vivo, a xenograft tumor model was constructed by inoculating different clones of NOZ and GBC-SD cells subcutaneously into nude mice. The results confirmed that the mean tumor volumes and weights were larger and tumor growth was rapid in the upregulated expression of circFOXP1 group, compared with the control group. However, the tumor growth was inhibited, as shown by decreasing mean volumes and weights, in the knockdown of endogenous circFOXP1 group compared with the control group (Fig. 2d). Immunohistochemical staining of tumor tissues revealed an increased proportion of proliferating cells (Ki67+) in the upregulated expression of circFOXP1 group compared with the control group. In contrast, knockdown of endogenous circFOXP1 led to reduced expression of Ki67 compared with the control group (Fig. 2e). These results indicated that circFOXP1 promoted GBC growth in vitro and in vivo.
Upregulation of circFOXP1 promotes the Warburg effect in GBC cells
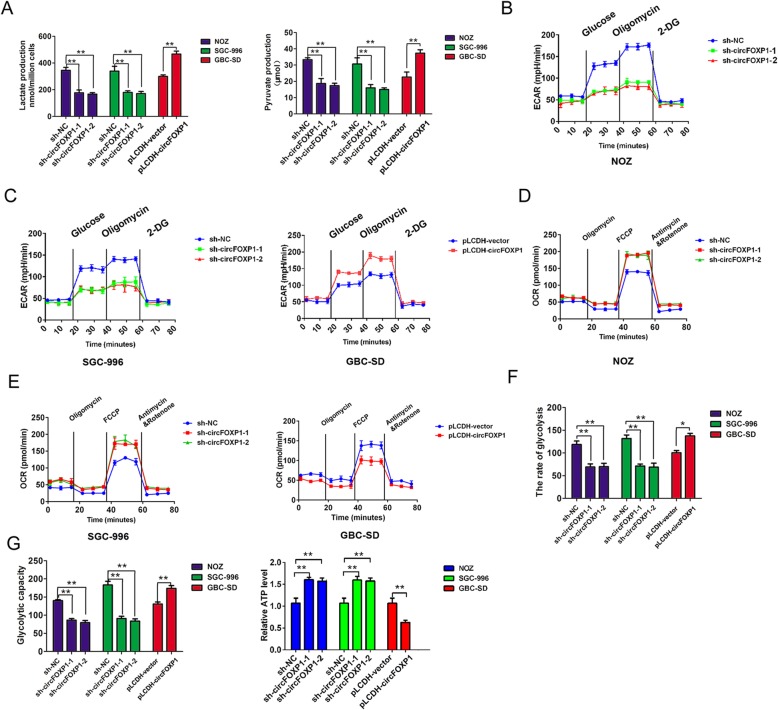
The Warburg effect, characterized by abnormal metabolic phenomena that enhance glycolysis and reduce oxidative phosphorylation, induces significant differences between cancer cells and normal cells and affects tumor progression [28]. Based on the above findings, we investigated whether the expression of circFOXP1 affected the Warburg effect by measuring the levels of pyruvate production and lactate production and the glycolytic rate in GBC cells. The statistical analysis revealed a dramatically lower level of lactate production and pyruvate production after circFOXP1 knockdown in NOZ and SGC-996 cells, conversely, were significantly higher after circFOXP1 overexpression in GBC-SD cells (Fig. 3a). Furthermore, glycolysis and extracellular acidification rates (ECAR) were analyzed using an XF24 Extracellular Flux analyzer (Seahorse), The results confirmed that circFOXP1 knockdown reduced the ECAR in NOZ and SGC-996 cells, while upregulated circFOXP1 expression increased ECAR in GBC-SD cells (Fig. 3b-c). we also measured oxygen consumption rate (OCR, a marker of OXPHOS) and found that knockdown of circFOXP1 enhanced OCR ability in NOZ and SGC-996 cells, in contrast, upregulated circFOXP1 expression reduced OCR ability in GBC-SD cells (Fig. 3d-e). The results confirmed that circFOXP1 knockdown impaired the glycolysis rate and glycolytic capacity in NOZ and SGC-996 cells. On the other hand, upregulated circFOXP1 expression significantly enhanced the rate of glycolysis and glycolytic capacity in GBC-SD cells (Fig. 3f-g). In addition, knockdown of circFOXP1 in NOZ and SGC-996 cells also elevated the formation of ATP produced by OXPHOS, but upregulated circFOXP1 has an decreased formation of ATP in GBC-SD cells (Fig. 3g). These results indicated that upregulation of circFOXP1 promoted the Warburg effect in GBC cells.
Fig. 3.
circFOXP1 promotes the Warburg effect in GBC cells. a Alterations in lactate production (left) and pyruvate production (right) levels were analyzed after transfection of NOZ and SGC-996 cells with sh-NC, sh-circFOXP1–1 or sh-circFOXP1–2 or GBC-SD cells with pLCDH-vector and pLCDH-circFOXP1, **P < 0.01. b, c The extracellular acidification rate was analyzed after transfection of NOZ and SGC-996 cells with sh-NC, sh-circFOXP1–1 or sh-circFOXP1–2 or GBC-SD cells with pLCDH-vector and pLCDH-circFOXP1. d, e The oxygen consumption rate was analyzed after transfection of NOZ and SGC-996 cells with sh-NC, sh-circFOXP1–1 or sh-circFOXP1–2 or GBC-SD cells with pLCDH-vector and pLCDH-circFOXP1. f, g The glycolysis rate, glycolysis capacity and ATP level was detected after transfection of NOZ and SGC-996 cells with sh-NC, sh-circFOXP1–1 or sh-circFOXP1–2 or GBC-SD cells with pLCDH-vector and pLCDH-circFOXP1. The extracellular acidification rate after glucose treatment indicates the glycolysis rate. The extracellular acidification rate after oligomycin treatment indicates the glycolysis capacity. Abbreviations: 2-DG, 2-deoxy-d-glucose; ECAR, extracellular acidification rate, **P < 0.01
The RNA-binding protein PTBP1 binds to circFOXP1 in GBC cells
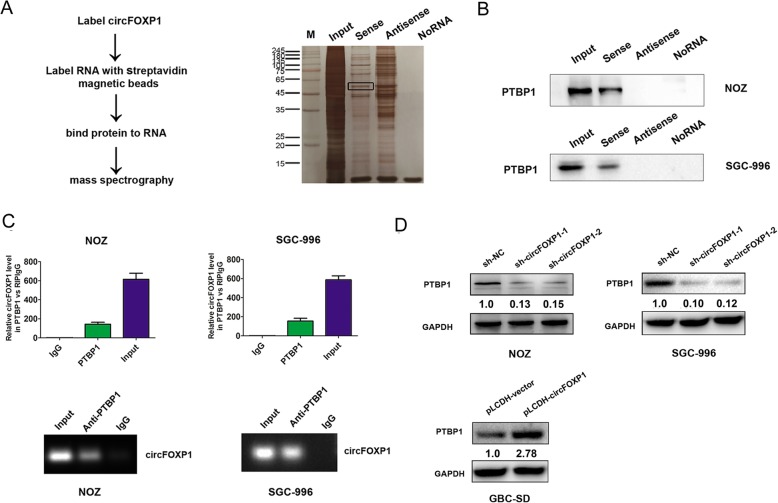
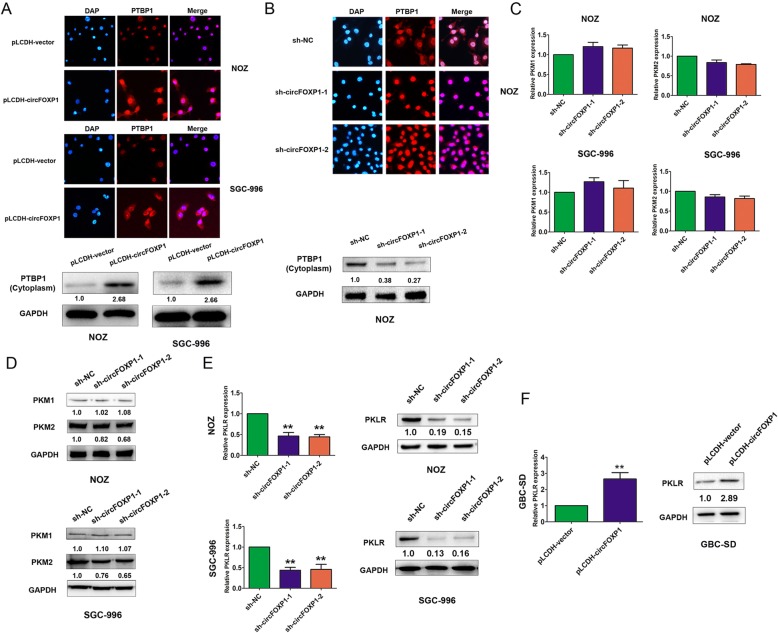
We further investigated the mechanisms by which circFOXP1 promoted the Warburg effect in GBC cells. Recent evidence has indicated that circRNAs participate in molecular regulation by interacting with several proteins [13]. Based on this hypothesis, we identified circFOXP1-interacting proteins by performing an RNA pull-down assay combined with Liquid Chromatography-Mass Spectrometry (LC-MS) in NOZ cells. The RNA-related proteins were determined using SDS-polyacrylamide gel electrophoresis (SDS/PAGE) and silver staining (Fig. 4a). By using LC-MS and comparing the results with those from antisense circFOXP1 experiments, proteins were identified by LC-MS that specifically associated with circFOXP1 (Additional file 6: Table S2). PTBP1 was further detected due to higher expression in the circFOXP1-sense probe compared with the circFOXP1-antisense sample or no RNA sample, and previous studies have verified that PTBP1 is associated with the Warburg effect in cancer cells through regulation of the PKM1/PKM2 ratio [29]. Western blot analysis with anti-PTBP1 antibody indicated the existence of PTBP1 within the circFOXP1 sense RNA probe pull-down samples in NOZ and SGC-996 cells (Fig. 4b). Meanwhile, a RIP assay with PTBP1 antibody showed that endogenous PTBP1 directly bound to circFOXP1 in NOZ and SGC-996 cells (Fig. 4c). Moreover, we observed that knockdown of endogenous circFOXP1 decreased the protein expression of PTBP1 in NOZ and SGC-996 cells, but upregulated expression of circFOXP1 upregulated the expression of PTBP1 in GBC-SD cells (Fig. 4d). By Immunofluorescence and western blot analysis, we further demonstrated that exogenous expression of circFOXP1 enhanced expression of PTBP1 by increased transportation levels of PTBP1 from the nucleus to the cytoplasm, but knockdown of circFOXP1 also revert PTBP1 back to the nucleus (Fig. 5a-b).
Fig. 4.
circFOXP1 interacts with PTBP1 in GBC cells. a A RNA pull-down assay was performed (left) and the RNA-related proteins were determined with SDS-polyacrylamide gel electrophoresis (SDS/PAGE) and silver staining (right). b PTBP1 was pulled down by a circFOXP1 sense RNA probe but not by an antisense RNA probe or no RNA in NOZ and SGC-996 cells. Western blot analysis was performed to detect the specific association of PTBP1 and circFOXP1 (n = 3). c RIP assays with qRT-PCR (top) or RT-PCR (low) showed that circFOXP1 was pulled down by an anti-PTBP1 antibody in NOZ and SGC-996 cells (n = 3). d The relative protein expression of PTBP1 after transfected with sh-NC, sh-circFOXP1–1 or sh-circFOXP1–2 in NOZ and SGC-996 cells and transfected with pLCDH-vector or pLCDH-circFOXP1 in GBC-SD cells were determined by western blot analysis (n = 3). All data are shown as mean ± S.E.M., n = 3 or more, **P < 0.01
Fig. 5.
Effects of circFOXP1 on PTBP1, PKM1, PKM2 and PKLR expression in GBC cells. a, b The relative protein expression of PTBP1 after transfected with pLCDH-vector and pLCDH-circFOXP1 in NOZ and SGC-996 cells or sh-NC, sh-circFOXP1–1 and sh-circFOXP1–2 in NOZ cell were determined by immunofluorescence assays, original magnification, 200×. c The mRNA expression of PKM1 and PKM2 were determined with qRT-PCR after transfection of NOZ (top) and SGC-996 cells (low) with sh-NC, sh-circFOXP1–1 or sh-circFOXP1–2. d The relative protein expression of PKM1 or PKM2 after transfected with sh-NC, sh-circFOXP1–1 or sh-circFOXP1–2 in NOZ and SGC-996 cells was determined by western blot analysis (n = 3). e, f The mRNA and protein expression of PKLR were determined with qRT-PCR and western blot analysis after transfection of NOZ and SGC-996 cells with sh-NC, sh-circFOXP1–1 or sh-circFOXP1–2 or GBC-SD cells with pLCDH-vector and pLCDH-circFOXP1. All data are shown as mean ± S.E.M., n = 3 or more, **P < 0.01
PTBP1 promotes PKLR mRNA expression in GBC cells
In mammalian cells, pyruvate kinase is encoded by 4 isozymes: M1, M2, liver (PKL), and red blood cell (PKR). While the M1, PKL, and PKR isozymes are described to exhibit tissue-specific expression, the pyruvate kinase M2 isoform is highly expressed across cancer types. Glycolytic deregulation, including activation of upregulated PKM2 and liver and RBC (PKLR), is a driver that promotes cancer progression [15, 30, 31]. Previous reports confirmed that PTBP1 regulates AS of the pyruvate kinase gene (PKM) in the acquisition of oncogenic features by increasing recruitment to the PKM pre-mRNA to promote PKM2 splicing, which contributes to the Warburg effect [32]. First, we speculated whether decreased circFOXP1 led to exertion of the function of PTBP1 to regulate AS. The results confirmed that knockdown of endogenous circFOXP1 resulted in no significant change in the mRNA and protein levels of PKM1 in NOZ and SGC-996 cells but had a partial effect on the PKM2 protein level (Fig. 5c-d). PKLR is also known as a promoter in modulation of the Warburg effect [15]. Interestingly, dramatically decreased PKLR expression levels were observed after downregulation of circFOXP1 in NOZ and SGC-996 cells or in NOZ cells in vivo, but increased expression of PKLR was observed after upregulation of circFOXP1 in GBC-SD cells or in vivo (Fig. 5e-f and Additional file 7: Figure S4A). In addition, we showed that PKLR silencing impaired the glycolysis rate and glycolytic capacity in NOZ and SGC-996 cells, which inhibited the Warburg effect in GBC (Additional file 7: Figure S4B-4F).
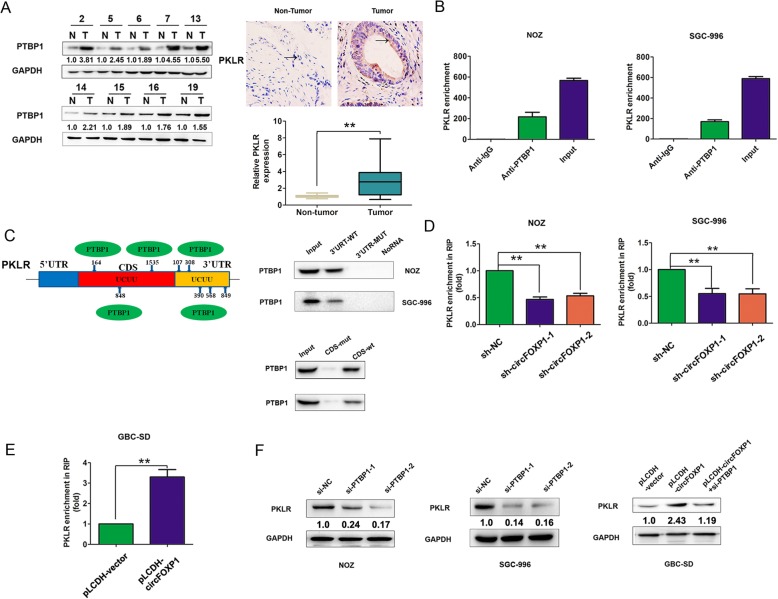
PKLR acts as a driver of tumor growth and metastasis [15, 33]. We sought to elucidate the underlying mechanism by which circFOXP1 affect PKLR expression. PTBP1, an RNA-binding protein, exerts various molecular functions, including RNA metabolism, for example, control of mRNA stability or degradation [34, 35], determination of mRNA localization [36], and protection of mRNAs from decay [37]. Compared with normal tissues, PTBP1 was upregulated in gallbladder cancer tissues and the protein and mRNA of PKLR was also upregulated in gallbladder cancer tissues (Fig. 6a). Furthermore, we showed that the proteasome inhibitor MG-132 could increase PKLR expression and abolished the reduction in PKLR protein levels in circFOXP1-knockdown NOZ cells (Additional file 8: Figure S5A). The protein synthesis inhibitor cycloheximide (CHX) decreased the expression of PKLR proteins by inhibiting protein synthesis, but upregulation of PKLR in circFOXP1-overexpressing SGC-996 and GBC-SD cells was not abolished by treatment with CHX (Additional file 8: Figure S5B). These results showed that circFOXP1 may affect PKLR degradation in GBC.
Fig. 6.
circFOXP1 promotes PKLR expression by interacting with PTBP1 in GBC cells. a left, the protein level of PTBP1 was detected using western blot analysis in GBC tissues compared with adjacent normal tissues. Right, immunohistochemical staining and mRNA expression of PKLR in GBC tissues compared with adjacent normal tissues. Arrow, PKLR expression was shown. b RIP assays with PTBP1 antibody were performed to detect PKLR enrichment in NOZ and SGC-996 cells. All data are shown as mean ± S.E.M., n = 3. c A RBP map was used to analyze the binding sites between PTBP1 and the 3’UTR and CDS region of PKLR. PTBP1 was pulled down by the 3’UTR and CDS wt RNA probe but not by the mut RNA probe in NOZ and SGC-996 cells. d, e The mRNA enrichment of PKLR was determined using RIP-qRT-PCR after silencing of circFOXP1 in NOZ and SGC-996 cells or overexpression of circFOXP1 in GBC-SD cells. All data are shown as mean ± S.E.M., n = 3, **P < 0.01. f The expression levels of circFOXP1 were determined using western blot analysis after transfection of NOZ and SGC-996 cells with sh-NC, sh-circFOXP1–1 or sh-circFOXP1–2 or GBC-SD cells with pLCDH-vector, pLCDH-circFOXP1, pLCDH-circFOXP1+ si-PTBP1, n = 3 or more
Based on these findings, we hypothesized that circFOXP1 may enhance the capacity of PTBP1 to bind to PKLR mRNA and protect it from mRNA decay and consequently increasing the PKLR protein levels. To test this hypothesis, we utilized a RIP assay to detect the association between PKLR mRNA and PTBP1. The results demonstrated that PTBP1 could bind to PKLR mRNA in NOZ and SGC-996 cells (Fig. 6b). Using the online prediction software RBP map (http://rbpmap.technion.ac.il/), we found that PTBP1 could bind to the 3’UTR region and coding region (CDS) of PKLR mRNA (UCUU binding bites) (Additional file 9: Figure S6A-6B). Furthermore, an RNA pull-down assay with the 3’UTR region or CDS region of PKLR biotin-labeled RNA probe confirmed that PTBP1 directly bound to PKLR mRNA in NOZ and SGC-996 cells (Fig. 6c). A RIP assay with PTBP1 antibody demonstrated that downregulation of circFOXP1 reduced the enrichment of PKLR mRNA in NOZ and SGC-996 cells, but enhanced the enrichment of PKLR mRNA after overexpression of circFOXP1 in GBC-SD cells (Fig. 6d-e). We further detected the effects of PTBP1 knockdown on PKLR expression. Downregulation of PTBP1 reduced both PKLR mRNA and protein in NOZ and SGC-996 cells, and knockdown of PTBP1 abrogated the effect of overexpressed circFOXP1 on PKLR expression in GBC-SD cells (Additional file 10: Figure S7A and Fig. 6f). These results revealed that circFOXP1 promoted PKLR mRNA expression by interacting with PTBP1 in GBC cells.
The circFOXP1 binds to miR-370 targeting PKLR in GBC cells
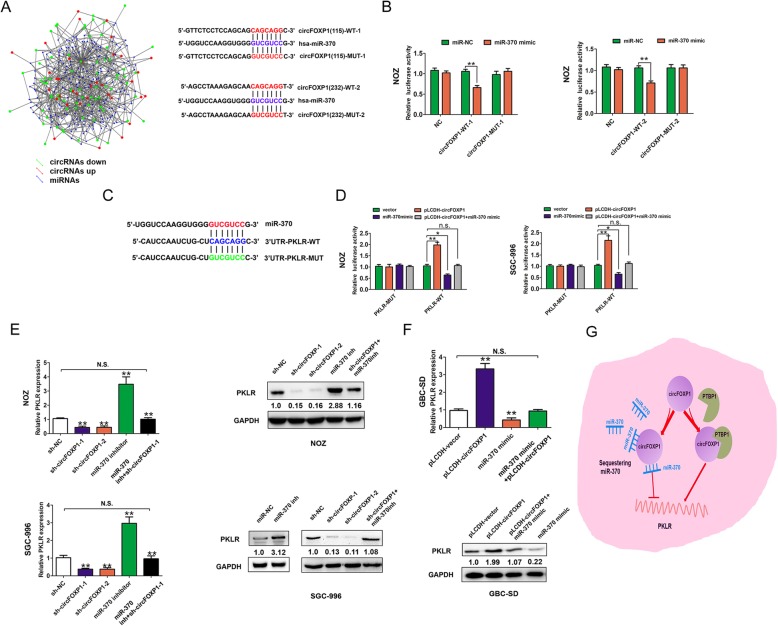
Endogenous circRNAs have been found to act as microRNA (miRNA) sponges in human cancers [38]. As circFOXP1 was predominantly localized in the cytoplasm, we hypothesized that circFOXP1 could also regulate PKLR expression by binding to specific miRNAs. To confirm this hypothesis, we performed a search for miRNAs that have complementary base pairing with circFOXP1 using the online software tools circinteractome (http://circinteractome.nia.nih.gov) and circRNAs from RNA sequencing targeted miRNAs predicted analysis by miRanda (www.microrna.org) (Fig. 7a, left, Additional file 11: Table S3). The results from RNA sequencing showed that miR-370 could form complementary base pairing with circFOXP1 and PKLR, respectively (Fig. 7a, right, and Fig. 7c). To verify that circFOXP1 could bind to miR-370, the wild-type (WT) and two mutant-type (MUT) circFOXP1 reporter vectors were constructed. We observed that a miR-370 mimic significantly reduced the luciferase activity of the circFOXP1-WT-1 (37%) and circFOXP1-WT-2 (33%) reporter vectors but not that of circFOXP1-MUT-1 or circFOXP1-MUT-2, which suggested that miR-370 was a target of circFOXP1 in a sequence-specific manner (Fig. 7b). To further clarify the regulatory association between circFOXP1 and the 3’UTR of PKLR, we constructed wild-type (WT) and mutant-type (MUT) PKLR 3’UTR reporter vectors (Fig. 7c). The results revealed that the miR-370 mimic significantly reduced the luciferase activity of the WT PKLR 3’UTR reporter vector but not that of the mutant PKLR 3’UTR reporter. Co-transfection with the WT PKLR 3’UTR and miR-370 mimic and pLCDH-circFOXP1 antagonized the effects (Fig. 7d). Furthermore, we confirmed that miR-370 expression was significantly downregulated in GBC tissues and cells, compared with adjacent normal tissues and H69 cell, respectively (Additional file 10: Figure S7B-7C). Moreover, an inverse correlation was observed between circFOXP1 and miR-370 expression in GBC tissues (r = − 0.45, P < 0.05, Additional file 10: Figure S7D). The PKLR mRNA and protein expression was downregulated after transfection of NOZ and SGC-996 cells with sh-circFOXP1, but was rescued by co-transfection with miR-370 inhibitor and sh-circFOXP1 (Fig. 7e). Conversely, the PKLR mRNA and protein expression was increased after transfection of GBC-SD cells with a pLCDH-circFOXP1, but was rescued by co-transfection with pLCDH-circFOXP1 and miR-370 mimic (Fig. 7f). These results showed that circFOXP1 promoted PKLR expression by sponging miR-370 in GBC cells.
Fig. 7.
circFOXP1 promotes PKLR expression by binding to miR-370 in GBC cells. a MiR-370 have complementary base pairing with circFOXP1 using circRNAs from RNA sequencing targeted miRNAs predicted by miRanda (www.microrna.org) and the online software tools circinteractome (http://circinteractome.nia.nih.gov) (left). The wild type and mutant type complementary sequences of the circFOXP1 and miR-370 binding sequence are shown (right). b Luciferase reporter assays were performed in NOZ cells co-transfected with miR-370 mimic or miR-NC and circFOXP1-WT1/2 or circFOXP1-MUT1/2 reporter plasmids. Data are shown as mean ± S.E.M., n = 3, **P < 0.01. c The wild-type and mutant-type complementary sequences of the PKLR and miR-370 binding sequence are shown. d Luciferase reporter assays were performed in NOZ and SGC-996 cells transfected with pLCDH-vector, pLCDH-circFOXP1, miR-370 mimic or co-transfected with miR-370 mimic+pLCDH-circFOXP1 and PKLR-WT or PKLR-MUT reporter vector. Data are shown as mean ± S.E.M., n = 3, *P < 0.05, **P < 0.01, n.s., not significant. e The relative expression of PKLR was detected after transfection of NOZ and SGC-996 cells with sh-NC, sh-circFOXP1–1 or sh-circFOXP1–2, miR-370 inhibitor or cotransfection with miR-370 inhibitor and sh-circFOXP1–1. f The relative expression of PKLR was detected after transfection of GBC cells with pLCDH-vector, pLCDH-circFOXP1 or cotransfection with pLCDH-circFOXP1 and miR-370 mimic. g Proposed mechanism by which circFOXP1 promoted GBC progression by interacting with PTBP1 or sponging miR-370 targeting PKLR. Data are shown as mean ± S.E.M., n = 3 or more, **P < 0.01, n.s., not significant
Discussion
The annotations of circRNAs by transcriptome-wide sequencing in various fundamental cellular biological processes including tumor development and progression have been thrust into the spotlight [39]. Fusion circRNAs (f-circRNAs) produced from transcribed exons of distinct genes were upregulated by cancer-associated chromosomal translocations and subsequently contributed to cellular transformation and enhanced cell viability and resistance upon tumor therapy [40]. In clear cell renal cell carcinoma, AR suppressed circHIAT1 expression by regulating the expression of its host gene (HIAT1) at the transcriptional level, which resulted in deregulation of miR-195-5p/29a-3p/29c-3p, and increased CDC42 to enhance cell migration and invasion [41]. Silencing of circZKSCAN1 promoted cell proliferation, migration and invasion in hepatocellular carcinoma [42]. The circRNA CCDC66 promotes colon cancer growth and metastasis [43]. In spite of some advanced findings, the expression and possible carcinogenic involvement of circRNAs in GBC remains unknown. In this study, we first determined that circFOXP1 was highly upregulated in GBC tissues and cells, and higher circFOXP1 expression was implicated as an independent prognostic marker for OS in patients. Furthermore, upregulation of circFOXP1 expression had pleiotropic effects, including promotion of cell proliferation, migration, invasion, and cell cycle progression and inhibition of cell apoptosis in GBC. These results suggested that circFOXP1 acts as an oncogene and may serve as a prognostic biological marker in GBC.
Mechanistic studies confirmed that circFOXP1 exerted its tumor-promoting roles by modulating the Warburg effect through upregulation of PKLR expression by interacting with PTBP1, protecting PKLR from mRNA decay. PTBP1, an RNA-binding protein, exerts various molecular functions, including RNA metabolism, for example, by repressively regulating AS [44], controlling mRNA stability [34, 35], and determining mRNA localization [36]. The PKM1/M2 isoforms are generated through AS of two mutually exclusive exons, and this AS is controlled by PTBP1 [29]. Herein, we first detected the effects of circFOXP1 on PKM1 and PKM2 expression, and found that decreased circFOXP1 does not significantly change PKM1 expression and only partially affected the PKM2 protein levels, which had been reported in previous studies [32, 45]. Interestingly, we uncovered a novel regulation target of circFOXP1, PKLR, which was activated by upregulation of circFOXP1. We speculated that PTBP1 could affect PKLR mRNA levels by its typical characteristic nucleocytoplasmic shuttling. We demonstrated that circFOXP1 facilitated PTBP1 nuclear to cytoplasmic translocation, resulting in enhancement of the control of PKLR mRNA stability. PTBP1 is critical for post-transcriptional upregulation of proinsulin and other granule proteins biosynthesized shortly after glucose stimulation by stabilizing mRNAs encoding proteins of secretory granules [46, 47]. We found that PTBP1 bound to the 3’UTR and CDS region of PKLR mRNA. Silencing circFOXP1 decreased the ability of PTBP1 to bind PKLR mRNA, which significantly suppressed the Warburg effects in GBC. This is consistent with a previous study that reported PTBP1-mediated post-transcriptional upregulation of proinsulin [46]. Because PTBP1 is mainly located in the nucleus, but circFOXP1 is located in the cytoplasm, we speculated that circFOXP1 could serve as a protein scaffold to recruit not only PTBP1 but also other proteins to the circFOXP1-PTBP1 complex to affect PKLR mRNA expression. Future studies may be necessary to elucidate how circFOXP1 affects the ability of PTBP1 to bind target PKLR mRNA.
Several circRNAs have been found to function as miRNA “sponges” to counteract miRNA-mediated repression of mRNA. CiRS-7, which acts as a designated miR-7 inhibitor, harbors more than 70 conventional miR-7 binding sites and provides a conceptual mechanistic understanding of competing RNA (ceRNA) networks [11]. The circHIPK3 regulates cell growth by binding to miR-124 and inhibiting miR-124 activity [12]. circRNA HRCR functions as an endogenous miR-223 sponge to sequester and inhibit miR-223 activity, which protects the heart from pathological hypertrophy and heart failure [48]. The circPVT1 promotes gastric cancer cell proliferation by acting as a sponge for members of the miR-125 family [49]. Based on the findings, we demonstrated that miR-370 was a direct target of circFOXP1. Upregulation of circFOXP1 promoted PKLR expression by sponging miR-370 in GBC cells.
Conclusions
Together, our study is the first to reveal that circFOXP1 was upregulated in GBC and promoted the tumor progression in GBC cells by interacting with PTBP1 or sponging miR-370 targeting PKLR (Fig. 7g). These findings have significant implications for our understanding of GBC pathogenesis and provide a target for GBC treatment.
Supplementary information
Additional file 1. Supplementary materials and methods.
Additional file 2. The sequences of all primers and oligonucleotide used in the study.
Additional file 3. Relative expression levels of several circRNAs in GBC tissues are shown.
Additional file 4. CircFOXP1 is identified in GBC cells.
Additional file 5. CircFOXP1 promotes cell migration and invasion in GBC.
Additional file 6. Proteins are identified by LC-MS that specifically associated with circFOXP1.
Additional file 7. Effects of PKLR on Warburg effect in GBC cells.
Additional file 8. CircFOXP1 affects PKLR expression in GBC cells.
Additional file 9. PTBP1 binds to 3’UTR and CDS region of human PKLR mRNA.
Additional file 10. Expression of circFOXP1 is negatively associated with miR-370 in GBC tissues and cells.
Additional file 11. CircRNAs from RNA sequencing targeted miRNAs.
Acknowledgments
We thank for the members of Department of General Surgery, Xinhua Hospital, Shanghai Jiao Tong University School of Medicine for helpful discussions.
Abbreviations
- AS
Alternative splicing
- CDK2
cyclin-dependent kinase 2
- cDNA
complementary DNA
- ceRNA
competing endogenous RNA
- circFOXP1
circular RNA FOXP1
- circRNAs
circular RNAs
- FISH
Fluorescence in situ hybridization
- GBC
Gallbladder cancer
- LC-MS
Liquid Chromatography-Mass Spectrometry
- MUT
Mutant-type
- ncRNA
non-coding RNA
- OS
Overall survival
- PCNA
Proliferating cell nuclear antigen
- PKLR
pyruvate kinase, liver and RBC
- qRT-PCR
quantitative real-time PCR
- RIP
RNA immunoprecipitation
- SDS/PAGE
SDS-polyacrylamide gel electrophoresis
- shRNA
short hairpin RNA
- WT
wild-type
Authors’ contributions
SW and ZQ study concept and design, acquisition of data, analysis and interpretation of data, statistical analysis, project funding, and drafting of the manuscript. YZ, QC, MM, and LYJ acquisition of data, material support, and statistical analysis. SW, MW, DZ, ZT and JDW, study concept and design, critical revision of the manuscript for important intellectual content, analysis and interpretation of data, perform the experiments. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (grant number 81572297 and 81802315) and Sponsored by Shanghai Sailing Program (grant number18YF1416000).
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The study procedure was approved by the Human Ethics Committee of Xinhua Hospital and All patients signed consent forms. The study protocol was approved by the Animal Care and Use committee of Xinhua Hospital (approval ID: 2014041).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhaohui Tang, Phone: 86-20-25078999, Email: tangzhaohui@xinhuamed.com.cn.
Jian Dong Wang, Phone: 86-20-25078999, Email: wangjiandong@xinhuamed.com.cn.
Zhiwei Quan, Phone: 86-20-25078999, Email: quanzhiwei@xinhuamed.com.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12943-019-1078-z.
References
- 1.Kakaei F, Beheshtirouy S, Nejatollahi SM, Zarrintan S, Mafi MR. Surgical treatment of gallbladder carcinoma: a critical review. Updat Surg. 2015;67:339–351. doi: 10.1007/s13304-015-0328-x. [DOI] [PubMed] [Google Scholar]
- 2.Lazcano-Ponce EC, Miquel JF, Munoz N, Herrero R, Ferrecio C, Wistuba II, Alonso de Ruiz P, Aristi Urista G, Nervi F. Epidemiology and molecular pathology of gallbladder cancer. CA Cancer J Clin. 2001;51:349–364. doi: 10.3322/canjclin.51.6.349. [DOI] [PubMed] [Google Scholar]
- 3.Kanthan R, Senger JL, Ahmed S, Kanthan SC. Gallbladder Cancer in the 21st century. J Oncol. 2015;2015:967472. doi: 10.1155/2015/967472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caldow Pilgrim CH, Groeschl RT, Quebbeman EJ, Gamblin TC. Recent advances in systemic therapies and radiotherapy for gallbladder cancer. Surg Oncol. 2013;22:61–67. doi: 10.1016/j.suronc.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Horgan AM, Amir E, Walter T, Knox JJ. Adjuvant therapy in the treatment of biliary tract cancer: a systematic review and meta-analysis. J Clin Oncol. 2012;30:1934–1940. doi: 10.1200/JCO.2011.40.5381. [DOI] [PubMed] [Google Scholar]
- 6.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32:309–316. doi: 10.1016/j.tig.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lasda E, Parker R. Circular RNAs: diversity of form and function. Rna. 2014;20:1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, Yang J, Zhou P, Le Y, Zhou C, Wang S, Xu D, Lin HK, Gong Z. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am J Cancer Res. 2015;5:472–480. [PMC free article] [PubMed] [Google Scholar]
- 10.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 11.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 12.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathupala SP, Ko YH, Pedersen PL. Hexokinase-2 bound to mitochondria: cancer's stygian link to the “Warburg effect” and a pivotal target for effective therapy. Semin Cancer Biol. 2009;19:17–24. doi: 10.1016/j.semcancer.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nie H, Li J, Yang XM, Cao QZ, Feng MX, Xue F, Wei L, Qin W, Gu J, Xia Q, Zhang ZG. Mineralocorticoid receptor suppresses cancer progression and the Warburg effect by modulating the miR-338-3p-PKLR axis in hepatocellular carcinoma. Hepatology. 2015;62:1145–1159. doi: 10.1002/hep.27940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Yang, Zhu Guang, Dong Bing, Piao Junjie, Chen Liyan, Lin Zhenhua. The NQO1/PKLR axis promotes lymph node metastasis and breast cancer progression by modulating glycolytic reprogramming. Cancer Letters. 2019;453:170–183. doi: 10.1016/j.canlet.2019.03.054. [DOI] [PubMed] [Google Scholar]
- 17.Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 18.Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. Rna. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang SH, Zhang WJ, Wu XC, Weng MZ, Zhang MD, Cai Q, Zhou D, Wang JD, Quan ZW. The lncRNA MALAT1 functions as a competing endogenous RNA to regulate MCL-1 expression by sponging miR-363-3p in gallbladder cancer. J Cell Mol Med. 2016;20:2299–2308. doi: 10.1111/jcmm.12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, Jiang Z, Xu J, Liu Q, Cao X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 21.Wang SH, Ma F, Tang ZH, Wu XC, Cai Q, Zhang MD, Weng MZ, Zhou D, Wang JD, Quan ZW. Long non-coding RNA H19 regulates FOXM1 expression by competitively binding endogenous miR-342-3p in gallbladder cancer. J Exp Clin Cancer Res. 2016;35:160. doi: 10.1186/s13046-016-0436-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferreiro-Neira I, Torres NE, Liesenfeld LF, Chan CH, Penson T, Landesman Y, Senapedis W, Shacham S, Hong TS, Cusack JC. XPO1 inhibition enhances radiation response in preclinical models of rectal Cancer. Clin Cancer Res. 2016;22:1663–1673. doi: 10.1158/1078-0432.CCR-15-0978. [DOI] [PubMed] [Google Scholar]
- 23.Flori M, Schmid CA, Sumrall ET, Tzankov A, Law CW, Robinson MD, Muller A. The hematopoietic oncoprotein FOXP1 promotes tumor cell survival in diffuse large B-cell lymphoma by repressing S1PR2 signaling. Blood. 2016;127:1438–1448. doi: 10.1182/blood-2015-08-662635. [DOI] [PubMed] [Google Scholar]
- 24.Mebratu Y, Tesfaigzi Y. How ERK1/2 activation controls cell proliferation and cell death: is subcellular localization the answer? Cell Cycle. 2009;8:1168–1175. doi: 10.4161/cc.8.8.8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui Chun-Ping, Wong Carmen Chak-Lui, Kai Alan Ka-Lun, Ho Daniel Wai-Hung, Lau Eunice Yuen-Ting, Tsui Yu-Man, Chan Lo-Kong, Cheung Tan-To, Chok Kenneth Siu-Ho, Chan Albert C Y, Lo Regina Cheuk-Lam, Lee Joyce Man-Fong, Lee Terence Kin-Wah, Ng Irene Oi Lin. SENP1 promotes hypoxia-induced cancer stemness by HIF-1α deSUMOylation and SENP1/HIF-1α positive feedback loop. Gut. 2017;66(12):2149–2159. doi: 10.1136/gutjnl-2016-313264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ying Z, Tian H, Li Y, Lian R, Li W, Wu S, Zhang HZ, Wu J, Liu L, Song J, et al. CCT6A suppresses SMAD2 and promotes prometastatic TGF-beta signaling. J Clin Invest. 2017;127:1725–1740. doi: 10.1172/JCI90439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki H, Tsukahara T. A view of pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci. 2014;15:9331–9342. doi: 10.3390/ijms15069331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 29.Chen M, Zhang J, Manley JL. Turning on a fuel switch of cancer: hnRNP proteins regulate alternative splicing of pyruvate kinase mRNA. Cancer Res. 2010;70:8977–8980. doi: 10.1158/0008-5472.CAN-10-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452:181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 31.Lu W, Cao Y, Zhang Y, Li S, Gao J, Wang XA, Mu J, Hu YP, Jiang L, Dong P, et al. Up-regulation of PKM2 promote malignancy and related to adverse prognostic risk factor in human gallbladder cancer. Sci Rep. 2016;6:26351. doi: 10.1038/srep26351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calabretta S, Bielli P, Passacantilli I, Pilozzi E, Fendrich V, Capurso G, Fave GD, Sette C. Modulation of PKM alternative splicing by PTBP1 promotes gemcitabine resistance in pancreatic cancer cells. Oncogene. 2016;35:2031–2039. doi: 10.1038/onc.2015.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen A, Loo JM, Mital R, Weinberg EM, Man FY, Zeng Z, Paty PB, Saltz L, Janjigian YY, de Stanchina E, Tavazoie SF. PKLR promotes colorectal cancer liver colonization through induction of glutathione synthesis. J Clin Invest. 2016;126:681–694. doi: 10.1172/JCI83587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tillmar L, Carlsson C, Welsh N. Control of insulin mRNA stability in rat pancreatic islets. Regulatory role of a 3′-untranslated region pyrimidine-rich sequence. J Biol Chem. 2002;277:1099–1106. doi: 10.1074/jbc.M108340200. [DOI] [PubMed] [Google Scholar]
- 35.Cui J, Placzek WJ. PTBP1 modulation of MCL1 expression regulates cellular apoptosis induced by antitubulin chemotherapeutics. Cell Death Differ. 2016;23:1681–1690. doi: 10.1038/cdd.2016.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cote CA, Gautreau D, Denegre JM, Kress TL, Terry NA, Mowry KL. A Xenopus protein related to hnRNP I has a role in cytoplasmic RNA localization. Mol Cell. 1999;4:431–437. doi: 10.1016/S1097-2765(00)80345-7. [DOI] [PubMed] [Google Scholar]
- 37.Ge Z, Quek BL: Polypyrimidine tract binding protein 1 protects mRNAs from recognition by the nonsense-mediated mRNA decay pathway. 2016, 5. [DOI] [PMC free article] [PubMed]
- 38.Cortes-Lopez M, Miura P. Emerging functions of circular RNAs. Yale J Biol Med. 2016;89:527–537. [PMC free article] [PubMed] [Google Scholar]
- 39.Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143:1838–1847. doi: 10.1242/dev.128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH, Pandolfi PP. Oncogenic role of fusion-circRNAs derived from Cancer-associated chromosomal translocations. Cell. 2016;165:289–302. doi: 10.1016/j.cell.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 41.Wang K, Sun Y, Tao W, Fei X, Chang C. Androgen receptor (AR) promotes clear cell renal cell carcinoma (ccRCC) migration and invasion via altering the circHIAT1/miR-195-5p/29a-3p/29c-3p/CDC42 signals. Cancer Lett. 2017;394:1. doi: 10.1016/j.canlet.2016.12.036. [DOI] [PubMed] [Google Scholar]
- 42.Yao Zhicheng, Luo Jingyan, Hu Kunpeng, Lin Jizong, Huang He, Wang Qiangliang, Zhang Peng, Xiong Zhiyong, He Chonghua, Huang Zejian, Liu Bo, Yang Yang. ZKSCAN1gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Molecular Oncology. 2017;11(4):422–437. doi: 10.1002/1878-0261.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsiao Kuei-Yang, Lin Ya-Chi, Gupta Sachin Kumar, Chang Ning, Yen Laising, Sun H. Sunny, Tsai Shaw-Jenq. Noncoding Effects of Circular RNA CCDC66 Promote Colon Cancer Growth and Metastasis. Cancer Research. 2017;77(9):2339–2350. doi: 10.1158/0008-5472.CAN-16-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheung HC, Hai T, Zhu W, Baggerly KA, Tsavachidis S, Krahe R, Cote GJ. Splicing factors PTBP1 and PTBP2 promote proliferation and migration of glioma cell lines. Brain. 2009;132:2277–2288. doi: 10.1093/brain/awp153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dayton TL, Jacks T, Vander Heiden MG: PKM2, cancer metabolism, and the road ahead. 2016, 17:1721–1730. [DOI] [PMC free article] [PubMed]
- 46.Knoch KP, Bergert H, Borgonovo B, Saeger HD, Altkruger A, Verkade P, Solimena M. Polypyrimidine tract-binding protein promotes insulin secretory granule biogenesis. Nat Cell Biol. 2004;6:207–214. doi: 10.1038/ncb1099. [DOI] [PubMed] [Google Scholar]
- 47.Knoch KP, Nath-Sain S, Petzold A, Schneider H, Beck M, Wegbrod C, Sonmez A, Munster C, Friedrich A, Roivainen M, Solimena M. PTBP1 is required for glucose-stimulated cap-independent translation of insulin granule proteins and Coxsackieviruses in beta cells. Mol Metab. 2014;3:518–530. doi: 10.1016/j.molmet.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 49.Chen J, Li Y, Zheng Q, Bao C, He J, Chen B, Lyu D, Zheng B, Xu Y, Long Z, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–219. doi: 10.1016/j.canlet.2016.12.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplementary materials and methods.
Additional file 2. The sequences of all primers and oligonucleotide used in the study.
Additional file 3. Relative expression levels of several circRNAs in GBC tissues are shown.
Additional file 4. CircFOXP1 is identified in GBC cells.
Additional file 5. CircFOXP1 promotes cell migration and invasion in GBC.
Additional file 6. Proteins are identified by LC-MS that specifically associated with circFOXP1.
Additional file 7. Effects of PKLR on Warburg effect in GBC cells.
Additional file 8. CircFOXP1 affects PKLR expression in GBC cells.
Additional file 9. PTBP1 binds to 3’UTR and CDS region of human PKLR mRNA.
Additional file 10. Expression of circFOXP1 is negatively associated with miR-370 in GBC tissues and cells.
Additional file 11. CircRNAs from RNA sequencing targeted miRNAs.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.