Abstract
Background
Current fMRI-based classification approaches mostly use functional connectivity or spatial maps as input, instead of exploring the dynamic time courses directly, which does not leverage the full temporal information.
Methods
Motivated by the ability of recurrent neural networks (RNN) in capturing dynamic information of time sequences, we propose a multi-scale RNN model, which enables classification between 558 schizophrenia and 542 healthy controls by using time courses of fMRI independent components (ICs) directly. To increase interpretability, we also propose a leave-one-IC-out looping strategy for estimating the top contributing ICs.
Findings
Accuracies of 83·2% and 80·2% were obtained respectively for the multi-site pooling and leave-one-site-out transfer classification. Subsequently, dorsal striatum and cerebellum components contribute the top two group-discriminative time courses, which is true even when adopting different brain atlases to extract time series.
Interpretation
This is the first attempt to apply a multi-scale RNN model directly on fMRI time courses for classification of mental disorders, and shows the potential for multi-scale RNN-based neuroimaging classifications.
Fund
Natural Science Foundation of China, the Strategic Priority Research Program of the Chinese Academy of Sciences, National Institutes of Health Grants, National Science Foundation.
Keywords: Recurrent neural network (RNN), Schizophrenia, Multi-site classification, fMRI, Striatum, Cerebellum, Deep learning
Highlights
-
•
A deep learning method which combines CNN and RNN is developed for multi-site schizophrenia classification.
-
•
A leave-one-feature-out method is first proposed to discover the most discriminative biomarkers for schizophrenia classification.
-
•
Dorsal striatum and cerebellum components contribute the top two group-discriminative time courses.
Research in context.
Evidence before this study
Current fMRI-based classification approaches mostly use functional connectivity or spatial maps as input, instead of exploring the dynamic time courses directly, which does not leverage the full temporal information. In addition, the excellent feature-representation ability of deep learning methods provides us a way to capture spatiotemporal information from time courses.
Added value of this study
In the present study, we contributed a new deep learning-based framework which can directly work on fMRI time courses for identifying brain disorders. In addition, by using our proposed deep learning-interpretation method, dorsal striatum and cerebellum are discovered as the top two discriminative brain regions.
Implications of all the available evidence
To the best of our knowledge, this is the first attempt to enable deep learning directly to work on time courses of fMRI components in schizophrenia classification, which promise great potentials of deep-chronnectome-learning and a broad utility on neuroimaging applications, e.g., the extension to MEG, EEG learning.
Alt-text: Unlabelled Box
1. Introduction
Functional magnetic resonance imaging (fMRI), as a non-invasive imaging technique, has been extensively applied to study psychiatric disorders [1]. Due to the high-dimensional and low signal-to-noise ratio properties of the fMRI data, efficient feature selection procedures are usually required to reduce the redundancy before modeling. Two types of approaches, data-driven [2] and seed-based [3], have been extensively applied to decompose 4D fMRI data, resulting in spatial brain regions/independent components (ICs) and their corresponding time courses (TCs). Currently, existing fMRI-based classification models mostly adopt either subject-specific spatial maps [4] or functional (network) connectivity calculated by TC correlations as input features [5,6], though have achieved substantial progress, the sequential temporal dynamics were generally missed. The field is still striving to understand how to diagnose and discriminate complex mental illness, e.g., schizophrenia versus bipolar disorder, while ignoring the temporal information time-point by time-point is likely missing a critical, but available, part of the puzzle.
The power of deep learning models lies in enabling automatic discovery of latent or abstract higher-level information from high-dimensional neuroimaging data, which can be an important step to understand complex mental disorders [[7], [8], [9], [10], [11], [12], [13], [14]]. Specifically, convolutional neural network (CNN) which is “deep in space” and recurrent neural network (RNN) which is “deep in time” are two classic deep learning branches. It is natural to use CNN as an ‘encoder’ for obtaining correlations between brain regions and simultaneously employ RNN for sequence classification. RNN models such as long short-term memory (LSTM) [15] and gated recurrent unit (GRU) [16] have been firmly established as state-of-the-art approaches in sequence modeling, such as identifying autism using fMRI [17], diagnosing brain disorder by analyzing electroencephalograms [18], detecting temporally dynamic functional state translations [13,14,19].
In particular, GRU is a particular RNN-based model which can effectively solve the long-term dependency problem by controlling information flow with several gates, which may fit the fMRI brain voxel-wise changes along with time series. Moreover, multi-scale convolution layers can be complementary for CNN feature extraction, because it can account for different temporal scales (from seconds to minutes) of brain activity. Therefore, we combine the strengths of CNN and RNN models and develop a Multi-scale RNN (MsRNN) model, which can directly work on fMRI time courses for classifying brain disorders, thus avoids the second-level calculation (e.g., correlation analysis) of time courses and takes advantage of the high-level spatiotemporal information of fMRI data. Such a design of classification framework relies on two assumptions: 1) underlying dynamics of fMRI data, i.e., rules by which neural activities involved in time; 2) brain disorders may have different patterns of temporal changes recorded by fMRI.
In this work, based on a large-scale Chinese Han resting-state fMRI data consisting of 558 schizophrenia patients (SZ) and 542 healthy controls (HC) that were recruited from seven sites with compatible MRI scanning parameters and imaging quality, we tested the power of the proposed MsRNN model for deep chronnectome learning on multiple facets, with comparison of three classic classification algorithms and eight varietal deep-learning models. Furthermore, to improve the result's interpretability, which is the most challenging issue of deep learning in neuroimaging applications, we propose a leave-one-IC-out strategy for estimating the contribution of each IC on classifying schizophrenia. Subsequently, components of dorsal striatum and cerebellum contributed the top two group-discriminating time courses. Finally, the time courses extracted by using seed-based strategies, e.g., using brain atlases such as AAL [20] or Brainnetome Atlas [21], were compared further with ICA results. To the best of our knowledge, this is the first attempt to enable CNN + RNN directly to work on time courses of fMRI components in mental disorder classification, which promise great potentials of deep-chronnectome-learning and a broad utility on neuroimaging applications, e.g., the extension to MEG, EEG learning.
2. Materials and methods
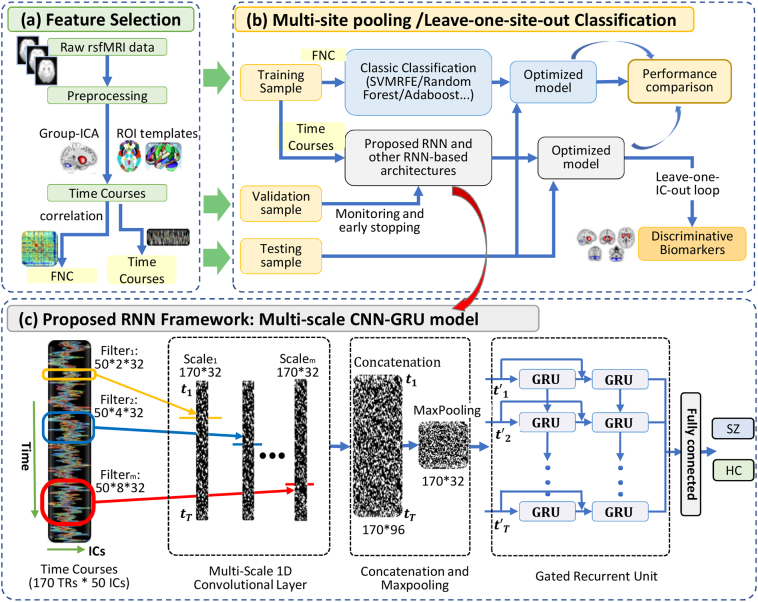
Fig. 1 presents an overview framework of the MsRNN classification method. Resting-state fMRI data from 1100 Chinese subjects (558 SZs, 542 HCs, from 7 sites) were used, which were preprocessed using the standard procedure [6]. Details of the demographic information are shown in Table S1. Time courses were extracted using group ICA [2]. Each subject was then represented with the TC features (No. time points × No. ICs, Fig. 1a, c). The proposed MsRNN model was directly applied on TCs of the selected non-artificial ICs to identify SZs from HCs using two types of classification strategies (Fig. 1b): 1) Multi-site pooling classification, in which all 1100 subjects from seven sites were pooled together, which were split into training set, validation set and testing set. Moreover, the classification performance was measured using k-fold cross-validation strategy; 2) Leave-one-site-out transfer classification, in which the subjects of a given site were left for testing, and the samples of all other sites were used for training and validation. These two types of classification strategies were independent of each other [9]. We trained the MsRNN using the TCs in training and validation sets with their corresponding labels (Fig. 1c). The learnable parameters of the MsRNN were iteratively adjusted using the error backpropagation algorithm. The validation samples were simultaneously used for monitoring the training process and avoid overfitting. The performance of the trained MsRNN was finally tested using held out TCs.
Fig. 1.
The framework of the Multi-scale RNN model in distinguishing schizophrenia patients from healthy controls. (a) Data preprocessing and feature selection. All rsfMRI data were preprocessed using the standard procedure. Time courses were then extracted using group-ICA/AAL/Brainnetome Atlas respectively. (b) The TCs/FNC data were randomly split into training, validation and testing sets. In multi-site pooling classification, all seven datasets were pooled together, and then k-fold cross-validation strategies were used for evaluating classification performance. In leave-one-site-out transfer prediction, the samples of a given imaging site were left for testing, and the samples of other sites were used for training. The performance of conventional methods (including Adaboost, Random Forest and SVM) and various RNN-based models were used for comparison. The most discriminative components were found by using leave-one-IC-out method. (c) Details of the MsRNN classification model. Three different scales convolutional filters were used for extracting of spatial features from time courses. The extracted features were then concatenated, pooling, and sent to stacked GRU module.
2.1. Participants and demographics
Table S1 lists the demographic and clinical information of all 1100 participants (558 SZs and 542 age and gender-matched HCs) in this study. The subjects were within the 18–45 age range, right-handed who were screened for ethical clearance, with only Chinese Han people recruited from seven sites in China with the same recruitment criterion, including Peking University Sixth Hospital (Site 1); Beijing Huilongguan Hospital (Site 2); Xinxiang Hospital Simens (Site 3); Xinxiang Hospital GE (Site 4); Xijing Hospital (Site 5); Renmin Hospital of Wuhan University (Site 6); Zhumadian Psychiatric Hospital (Site 7). Each site received approval from their respective research ethics boards and written informed consents were obtained from all study participants. All the SZ patients were evaluated based on the Structured Clinical Interview for DSM disorders (SCID) and diagnosed by experienced psychiatrists according to the criteria of DSM-IV-TR. All the HCs were recruited from the same local geographical areas as the patients cohort through local advertisement and were free of Axis I or II disorders (SCID-Nonpatient) Additional exclusion criteria include factors such as current or past neurological illness, substance abuse or dependence, pregnancy, and prior electroconvulsive therapy or head injury resulting in loss of consciousness.
2.2. Image acquisition
The resting state fMRI data were collected with the following three different types of scanners: 3·0 T Siemens Trio Tim Scanner (Siemens; Site 1, 2 & 5), 3·0 T Siemens Verio Scanner (Siemens; Site 3), and 3·0 T Signa HDx GE Scanner (General Electric; Site 4, 6 & 7). To ensure equivalent, coincident and high-quality data acquisition, the scanning protocols for all the seven sites were set up by the same experienced experts [6]. Subjects were instructed to relax and lie still in the scanner while remaining calm and awake. More details of scanning parameters are listed in Supplementary Table S4.
2.3. Data preprocessing and IC extraction
The rsfMRI data were preprocessed according to the procedures which were the same as we did in [6] using SPM8 software (http://www.fil.ion.ucl.ac.uk/spm/). For each participant, the first ten volumes of each scan time series were discarded to ensure magnetization equilibrium. The remaining resting state volumes were first corrected by the acquisition time delay of different slices and then realigned to the first volume for head-motion correction [22]. For each subject, the translation of head motion was <3 mm and the rotation of head motion did not exceed 3° in all axes through the whole scanning process. Subsequently, the images were spatially normalized to EPI template conforming to the Montreal Neurological Institute (MNI) space. The data (originally collected at 3·44 mm × 3·44 mm × 4·60 mm) were then resliced to a voxel size of 3 mm × 3 mm × 3 mm, resulting in 53 × 63 × 46 voxels for each image. Subsequently, group ICA toolbox (GIFT, http://mialab.mrn.org/software/gift) was used to perform GIG-ICA [23] on the preprocessed fMRI data. 50 ICs were characterized as intrinsic connectivity networks (ICNs) after removing those ICs corresponding to physiological, movement-related or imaging artifacts, and their spatial maps (SMs) are listed in the Supplementary file Fig. S3. According to previous work [24,25], the control of movement-related artifacts should be stringent for the analysis of time courses of fMRI data. We compared the mean of framewise displacement (FD) for HC and SZ groups. The mean FD for HC and SZ are 0·137±0·071 and 0·142±0·085 respectively, with no significant group differences (P = .98, two-sample t-tests) existing. In our preprocessing, as did in previous work, nuisance covariates including six head motion parameters, mean FD, white matter signal, cerebrospinal fluid signal, and global mean signal were all regressed out [24,26,27]. Two covariants (age and gender) which may have potential confounding effects were also regressed out. Then the time courses were stacked to form a matrix with dimensions of [No. Subjects] × [No. Time courses] × [No. Independent components or ROIs)] which was then used to calculate the FNC matrix or to train the MsRNN model directly.
2.4. Multi-scale CNN-GRU (MsRNN)
As shown in Fig. 1c, MsRNN consists of 3 different scales of 1D convolutional filters (2TR, 4TR and 8TR, TR = 2 s), one concatenation layer, one max-pooling layer, a two-layer stacked gated recurrent unit (GRU) which are densely connected in a feed-forward manner, and an averaged layer which integrate the whole sequence. The time courses were fed into the proposed MsRNN model for parameter optimization. After optimizing the parameters, the model was saved for testing and comparison. Equations are listed in the Supplementary files for a precise definition of the MsRNN model.
2.4.1. Multi-scale convolutional layer
Multi-scale convolution layers may be helpful in feature extraction because it can account for different scales (from seconds to minutes) of brain activity. Inspired by 1D convolution (Conv1D) layers [28], we designed an architecture which expands upon simple convolutional layers by including multiple filters of varying sizes in each Conv1D layer. This architecture allows the network to extract information over multiple time scales. The filter lengths used in the Conv1D were drawn from a logarithmic instead of a linear scale, leading to exponentially varying filter lengths (2TR, 4TR, and 8TR). Therefore, the size of 3 different scales of convolutional filters are 50 (ICs) × 2 × 32 (number of filters), 50 × 4 × 32, 50 × 8 × 32 in our experiment. A concatenation layer then concatenates the incoming features among the depth axis, resulting in feature maps whose size are 170 (time points) ×96 (feature dimension). Whereafter, a max-pooling layer performs downsampling operation along the time dimensions with filter size 3, resulting in feature such as 57(time points) × 96(feature dimension).The downsampled features are as the input of the following GRU layers.
2.4.2. Densely connected GRU layer
A two-layer stacked GRU may capture higher-level dynamic information than single-layer GRU model. The size of the GRU's hidden state was set as 32. However, one of the central challenges of training a deep GRU-based network the gradient exploding/vanishing problem. It is worthy to note that the densely-connected structure may effectively prohibit the “gradient exploding/vanishing” problem by connecting each layer to every other layer in a feed-forward manner [29].
2.4.3. Averaged layer
Even with the best experimental fMRI design, it is infeasible to control the random thoughts of the subjects during the resting-state fMRI scanning because they depend on too many subject-specific factors. Also, it is not possible to label the beginning and the end of brain activities. Hence combining all fMRI steps by averaging all of the GRU outputs is a compromised solution [10]. In this way, all activities of the brain during scanning may be leveraged for obtaining better classification performance.
In summary, the proposed MsRNN classification model consists of multiple-scale Conv1D layers, stacked GRU layers which are densely connected in a feed-forward manner, an averaged layer which integrates the context of the whole sequence, and fully-connected layers. More details of the model can be found in Supplementary Fig. S2.
2.5. MsRNN model implementation
The time courses of ICs described above were used as the inputs for training the Multi-scale RNN model. The model was trained by minimizing the cross-entropy loss using Adam optimizer. The training batch size was set as 64. The learning rate started from 0.001 and decayed after each epoch with the decay rate of 10−210−2. To improve the generalization performance of the model and overcome overfitting, dropout(dropout = 0.5) and L1,2-norm regularization (L1 = 0.0005, L2 = 0.0005) were also applied for regulating the model parameters. The training process was stopped when the validation loss stopped decreasing for 50 epochs or when the maximum epochs (1000 epochs) had been executed. In our experiment, the training time for MsRNN was around five minutes, while the testing time for a new subject is <0.01 s. The intermediate model which achieved the highest accuracy on the validation dataset was reserved for testing. Also, the proposed models were implemented on the platform of Keras (https://keras.io/) and ScikitLearn (https://scikit-learn.org/).
The visualization of MsRNN codes was performed by the unsupervised dimensionality reduction technique t-SNE, which embeds high-dimensional data into a low-dimensional space while preserving the pairwise distances of the data points, implemented in MATLAB. The activation strengths of individual neurons at the last hidden layer by the training and testing samples were used as the raw variables. The parameters for the stochastic optimization for t-SNE [30] were as follows [31]: The perplexity was 30, and the dimension for initial principal components analysis was 30.
2.6. Estimating the discriminative power of independent components (leave-one-IC-out)
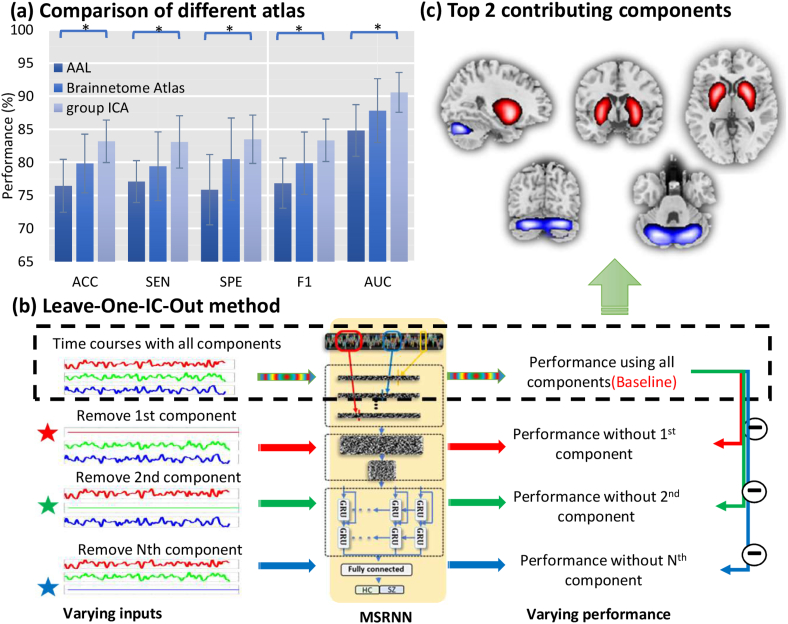
The basic idea is that the feature whose elimination lead to the most significant damage of classification performance should be regarded as the top contributing features. More specifically, as shown in Fig. 3b, each subject is represented with a T × D matrix, where T is the length of time courses and D is the number of independent components (ICs). A specific element in the matrix can be denoted by vtd. To quantify the classification contribution of the dth IC, we replace the time courses of dth IC with its averaged value while keep other ICs' time courses as they were. This is equivalent to eliminating the contribution of dth component. All the testing samples are processed in the same way and subsequently fed to the trained MsRNN model. The classification performance of the trained model which is fed with reduced features may decrease compared to that using all features. The variation of the classification performance (i.e., accuracy, sensitivity, specificity) when removing dth dimension are recorded and sorted. The features which maximize the decrease of the classification performance are further selected as the most discriminative features. Specifically, the 1100 samples were randomly split into five folds. 880 samples (four folds) were used for optimizing the parameters of MsRNN, and 220 samples (one-fold) were used for further finding the contribution of each IC during each cross-validation. The specific procedures are as follow: 1) After optimizing the trained model with 880 samples, the parameters of the trained model were saved; 2) The time courses of 220 subjects without removing any component were fed to the model to obtain a baseline classification performance; 3) The 220 subjects which have removed the contribution of one specific IC were fed to the model to obtain the classification performance repeatedly. The decrease of sensitivity/specificity when removing a specific component was recorded and sorted; 4) Repeat step 3 until each IC has been removed once.
Fig. 3.
Comparison of different atlas and Leave-One-IC-Out method. (a) The MsRNN classification results using three different feature selection methods. * P < .05 (two-sample t-test). (b) Leave-one-IC-out method for estimating the contribution of each IC. (c) Top two discriminative independent components discovered using the leave-one-IC-out method.
2.7. Statistics
The performance of identifying schizophrenia from normal controls was evaluated by five metrics including accuracy (ACC), sensitivity (SEN), specificity (SPE), F-score (F1) and area under curve (AUC) based on the results of cross-validation (k-fold or leave-one-site-out). They are defined as below:
where TP, TN, FP, FN, PPV denote true positive, true negative, false positive, false negative and positive predictive value respectively, e.g., SEN represents the percentage of SZ are classified as SZ correctly. The full k-fold cross-validation procedure was repeated ten times to generate the means and standard deviations of accuracy, sensitivity, and specificity. We used two-sample t-test to compare classification performances between different algorithms and hyperparameter settings.
2.8. Data availability
All data needed to evaluate the conclusions are present in the paper and/or the supplementary materials. Additional data related to this paper may be requested from the authors.
3. Results
3.1. Multi-site pooling classification
We compared the MsRNN with three traditional popular classifiers (SVM [32], Adaboost [33], Random Forest [34]), one multi-layer perception model, and seven RNN-based alternative deep learning models (Table 1). The detailed hyperparameters and the time complexity of these methods can be found in Supplementary Table S5. All the above models were implemented on a desktop computer (Intel(R) Xeon(R) CPU E5–1650 v4 @ 3.60GHz, 6 CPU cores) with a single GPU (12GB NVIDIA GTX TITAN 12GB), and can be trained within five minutes. Note that the three conventional classification methods usually work on the FNC matrix that was computed using the correlation of TCs of selected components instead of the TCs themselves. Therefore, in performance comparison, FNCs were used as the input of conventional methods while TCs were used as the input of MsRNN, multi-layer perception, and other RNN-based deep learning methods. All models were trained using the training dataset and tested using testing dataset, embedded in nested five-fold cross-validation cycles. Fig. 1c shows the architecture of the proposed MsRNN model.
Table 1.
Performance comparison in multi-site pooling classification.
| Methods | ACC | SEN | SPE | F1 | AUC | |
|---|---|---|---|---|---|---|
| CON | Adaboost | 75.6(3.8)** | 77.0(4.4)** | 74.2(4.4)** | 76.2(3.8)** | 84.2(3.6)** |
| CON | Random Forest | 76.0(3.5)** | 81.0(3.9)o | 71.4(5.5)** | 77.4(3.5)** | 84.0(3.4)** |
| CON | SVM | 79.4(3.1)* | 80.4(3.5)o | 78.4(3.9)* | 79.6(3.3)* | 86.8(3.2)* |
| RNN | GRU_1_last | 51.6(3.6)** | 52.0(5.3)** | 51.2(4.3)** | 52.0(3.8)** | 51.2(3.6)** |
| RNN | GRU_1_ave | 77.8(3.4)** | 78.4(3.8)** | 77.0(3.5)** | 78.2(3.4)** | 86.8(3.5)* |
| RNN | GRU_2_ave | 78.0(3.9)** | 80.8(5.1)o | 76.0(4.2)** | 78.8(3.9)* | 86.8(4.1)* |
| CMLP | Multi_CNN_MLP | 77.8(3.4)** | 76.2(4.0)** | 79.2(4.8)○ | 77.2(3.4)** | 86.4(3.1)** |
| CRNN | Simple_CNN_GRU_2_ave | 80.8(3.0)○ | 80.2(4.3)○ | 82.0(3.5)○ | 80.8(3.1)○ | 89.2(2.8)○ |
| CRNN | Multi_CNN_GRU_1_ave | 80.6(3.5)○ | 80.8(4.1)○ | 80.6(4.3)○ | 80.8(3.3)○ | 88.2(3.6)○ |
| CRNN | Multi_CNN_GRU_2_ave | 81.2(3.4)○ | 81.4(4.1)○ | 81.0(4.9)○ | 81.0(3.5)○ | 88.6(3.7)○ |
| CRNN | Multi_CNN_LSTM_2_ave | 81.6(2.9)○ | 82.6(3.6)○ | 80.4(3.8)○ | 82.0(2.7)○ | 89.4(2.8)○ |
| CRNN | MsRNN(Proposed) | 83.2(3.2) | 83.1(3.7) | 83.5(3.7) | 83.3(3.2) | 90.6(3.0) |
CON: conventional classification methods; RNN: RNN-based methods; CMLP: CNN linked with multi-layer perception; CRNN: CNN-RNN based methods; SVM: Support vector machine with Gaussian kernel; LSTM: Long short-term memory network; GRU: gated recurrent unit. GRU_1: one layer of GRU; GRU_2: two-layer stacked GRU; #_last: the output of the last GRU step is connected to the next layer. #_ave: the average of the outputs of all GRU steps is connected to the next layer; SimpleCNN: Convolutional layer has fixed kernel size; Multi_CNN: Convolutional layer has different kernel size; ○ denotes that the methods have no significant difference (two-sample t-test) with the proposed. */** denote respectively that the methods are significantly worse than the proposed model with P value = .05/0.01. Details of all these mentioned architectures are shown in Supplementary file Fig. S2. The last row is our proposed method.
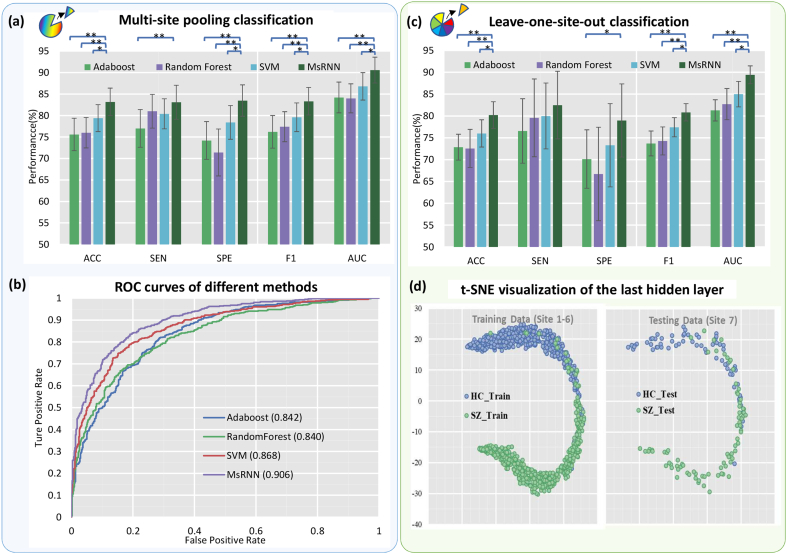
Table 1 and Fig. 2a listed the averaged accuracy and variance of classification performance achieved by all 11 methods in multi-site pooling condition. In the deep learning classification frameworks (including MsRNN, multi-layer perception, and other RNN-based architectures), we used four folds as the training set (10% samples of the training set were further randomly selected as validation dataset), and one-fold as the testing dataset. As for conventional classification models (Adaboost, Random Forest and SVM), four folds were used for training and one-fold for testing.
Fig. 2.
Classification results of multi-site pooling and leave-one-site-out transfer classification. (a) 5-fold multi-site pooling classification results. ** P < .01(two-sample t-test), * P < .05 (two-sample t-test). (b) The comparison of receiver operating characteristic curves of different methods. (c) Leave-one-site-out transfer classification results. (d) t-SNE visualization of the last hidden layer representation in the MsRNN for SZ/HC classification. Here we show the MsRNN's internal representation of SZ and HC by applying t-SNE, a method for visualizing high-dimensional data, to the last hidden layer in the MsRNN of training (Site 1–6: 951 subjects) and testing (Site 7: 149 subjects) samples.
The accuracy of 83·2 ± 3·2% was obtained by using the MsRNN method, which is significantly higher than those obtained by using the Adaboost, Random Forest and SVM (P = 2·1e-4, 1·9e-4, 1.1e-2, two-sample t-tests, df = 18). Also, the ROC curves of these methods are shown in Fig. 2b. The proposed MsRNN achieved an AUC of 0.906, while the AUC of Adaboost, Random Forest and SVM ranges from 0·840–0·868. To validate the advantage of the proposed model, other RNN architectures based on GRU and one similar network architecture based on LSTM were also compared with MsRNN. As shown in Table 1, a single layer GRU model can easily reach a higher classification performance than the classic FNC-based methods. The improvement may be due to the ability of GRU in extracting dynamic information from time sequences. In addition, the performance of GRU_1_ave is better than GRU_1_last because the former one made full use of temporal information at every time point. Furthermore, combining the GRU layer with Conv1D layer is a remedy for improving the classification performance because CNN-GRU model is “double deep” which include both spatial and temporal layers. Thus it can be jointly trained to learn convolutional perceptual representations and temporal dynamics simultaneously.
Finally, the proposed multi-scale convolution is even better than a single-scale convolution layer because it can extract dynamics from a variety of timescales. In summary, multi-site pooling results indicated that our proposed MsRNN model achieved the best performance by smartly integrating the advantages of CNN and RNN, while the LSTM-based model can reach competitive performance compared with the GRU-based model.
3.2. Leave-one-site-out transfer classification
In the leave-one-site-out classification, we left each of the seven sites as the testing data and used the other six sites for training and validation, in which 10% samples were randomly selected as validation dataset and the other 90% were used for training MsRNN or other deep learning architectures. In the Adaboost, Random Forest and SVM classification frameworks, we used the samples of the given imaging site for testing and the samples of other sites for training. The leave-one-site-out transfer classification results are shown in Table 2 and Table S2. The averaged classification performance of the seven sites was used to represent the overall performance of cross-site prediction. The accuracy of 80·2% was achieved by using the MsRNN method, which was significantly higher than the accuracies obtained by using the Adaboost, Random Forest and SVM (P = 6·2e-4, 7.0e-3, 1.9e-2, two-sample t-test, df = 12) (Fig. 2c). To visualize the performance of MsRNN classifier, we used t-Distributed Stochastic Neighbor Embedding (t-SNE) to project the 32-dimensional representations of subjects extracted from the hidden layer of the trained MsRNN model to a 2D plane. As shown in Fig. 2d, samples from six sites (951 subjects, site 1–6) were used as the training/validation set, and the samples from site 7 (149 subjects) were used for testing. The tSNE result indicates that the proposed MsRNN model can successfully distill features and separate the SZ and HC apart.
Table 2.
Performance comparison in leave-one-site-out classification.
| Methods | ACC | SEN | SPE | F1 | AUC | |
|---|---|---|---|---|---|---|
| CON | Adaboost | 72.9(3.0)** | 76.6(7.4)○ | 70.1(6.7)* | 73.7(2.8)** | 81.3(2.4)** |
| CON | Random Forest | 72.6(4.4)** | 79.6(8.9)○ | 66.7(10.7○ | 74.3(3.2)** | 82.7(3.6)** |
| CON | SVM | 76.0(3.1)* | 80.0(7.5)○ | 73.3(9.5)○ | 77.4(2.2)* | 85.0(2.9)* |
| RNN | GRU_1_last | 47.7(3.2)** | 50.6(6.8)** | 44.7(7.1)** | 49.3(3.7)** | 46.7(2.4)** |
| RNN | GRU_1_ave | 78.7(2.8)○ | 80.9(7.3)○ | 77.4(7.4)○ | 79.4(1.9)○ | 86.9(2.3)* |
| RNN | GRU_2_ave | 77.9(3.9)○ | 79.0(9.2)○ | 77.9(7.5)○ | 78.1(2.7)○ | 87.7(3.0)○ |
| CMLP | Multi_CNN_MLP | 76.1(3.2)* | 79.7(8.2)○ | 73.4(9.8)○ | 77.0(2.1)** | 85.4(2.7)* |
| CRNN | Simple_CNN_GRU_2_ave | 79.1(3.7)○ | 82.4(7.9)○ | 76.7(10.7)○ | 80.1(2.3)○ | 89.1(2.3)○ |
| CRNN | Multi_CNN_GRU_1_ave | 80.3(3.0)○ | 82.9(7.3)○ | 79.0(9.4)○ | 81.1(1.8)○ | 88.7(2.3)○ |
| CRNN | Multi_CNN_GRU_2_ave | 79.7(3.0)○ | 80.4(7.2)○ | 79.6(7.7)○ | 79.9(2.7)○ | 88.6(2.3)○ |
| CRNN | Multi_CNN_LSTM_2_ave | 78.7(3.9)○ | 83.1(8.3)○ | 75.3(9.7)○ | 79.7(2.6)○ | 89.6(3.0)○ |
| CRNN | MsRNN(Proposed) | 80.2(3.0) | 82.5(7.7) | 79.0(8.4) | 80.8(2.0) | 89.4(2.1) |
CON: conventional classification methods; RNN: RNN-based methods; CMLP: CNN linked with multi-layer perception; CRNN: CNN-RNN based methods; SVM: Support vector machine with Gaussian kernel; LSTM: Long short-term memory network; GRU: gated recurrent unit. GRU_1: one layer of GRU; GRU_2: two-layer stacked GRU; #_last: the output of the last GRU step is connected to the next layer. #_ave: the average of the outputs of all GRU steps is connected to the next layer; SimpleCNN: Convolutional layer has fixed kernel size; Multi_CNN: Convolutional layer has different kernel size; Details of all these mentioned architectures are shown in Supplementary file Fig. S2. The last row is our proposed method. ○ denotes that the methods have no significant difference (two-sample t-test) with the proposed. */** denote respectively that the methods are significantly worse than the proposed model with P =.05/0.01.
3.3. Comparison of TC-extracting strategies
Besides using ICA to extract TCs, we further tested the performance of the MsRNN by using TCs obtained from brain parcellation using both AAL template and Brainnetome Atlas, where the TCs of each brain regions of interests (ROI) were calculated by averaging the voxel-wise time series within each ROI. The dimension of TCs for AAL atlas is 170(time points) × 116(ROIs) andª 170(time points) × 273(ROIs) for Brainnetome Atlas. MsRNN models were separately trained and evaluated, as shown in Fig. 3a and Table S3, the TCs generated from ICA achieved the best performance, surpassing the AAL feature extraction strategies by at least 7% on AUC (P = 3·0e-2, two-sample t-test). This is likely due to the ability of ICA to capture variability in the components among subjects and is also consistent with earlier work showing that ICA time courses show better performance than fixed ROIs for graph theory metrics [35].
3.4. Estimating the most discriminating ICs
The ultimate goal of fMRI classification studies is to identify a collection of statistical features that can serve as reliable imaging biomarkers for disease diagnosis and are reproducible across multiple datasets. Despite extraordinary classification performance, the lack of interpretability often restricts the application of deep learning methods. Some previous work tried to open the black box of deep learning by analyzing the weight matrix of the trained model [9,12]. Generally speaking, the most important features are those whose removal can cause the most significant performance decrease compared to other features. Here we proposed a leave-one-IC-out method to leave one IC's time course out, and used the remaining 49 ICs' time course to train the model. After that, we compared the alteration of classification performances by looping all 50 ICs (shown in Fig. 3b). As a result, TCs from two components: 1) putamen and caudate which are parts of striatum; 2) declive and uvula which are parts of the cerebellum (Fig. 3c), contributed the top 2 group-discriminating time courses. Table 3 listed the Talairach labels of the two components. Note that similar findings of the most group-discriminating ROIs were obtained from both AAL and Brainnetome atlas.
Table 3.
Talairach labels of the peak activations in spatial maps of selected ICs.
| Area | Brodmann area | Volume (cc) | Random effects: Max Value (x, y, z) |
|---|---|---|---|
| IC_4 | |||
| Putamen | 4.2/4.9 | 1.4 (−24, 12, 15)/1.4 (29, −8, 14) | |
| Lentiform nucleus | 1.6/1.3 | 1.4 (−28, −17, 13)/1.4 (14, −1, −2) | |
| Parahippocampal Gyrus | 34 | 0.8/0.8 | 1.4 (−23, −8, −16)/1.4 (32, −10, −13) |
| Claustrum | 0.8/1.0 | 1.4 (−36, −13, 2)/1.4 (34, 1, 9) | |
| Inferior Frontal Gyrus | 13, 47 | 0.6/0.1 | 1.4 (−32, 10, −15)/1.4 (30, 13, −12) |
| Caudate | 1.7/1.8 | 1.4 (−11, 17, 7)/1.4 (16, −8, 19) | |
| IC_2 | |||
| Declive | 2.9/3.0 | 1.9 (−27, −71, −22)/1.9 (21, −71, −22) | |
| Uvula | 0.5/0.8 | 1.6 (−27, −71, −25)/1.8 (24, −71, 24) | |
| Pyramis | 0.0/0.1 | NA/1.6 (27, −71, −27) |
4. Discussion
As known, the current clinical diagnosis of schizophrenia is based solely on clinical manifestations. In recent years, many studies attempted to find stable neuroimaging-based biomarkers by machine learning techniques. To the best of our knowledge, this is the first attempt to apply an RNN model directly on fMRI time courses for schizophrenia diagnosis, which avoids second-level correlation analysis and make full use of time-varying functional network information. Accuracies of 83·2% and 80·2% were obtained in the multi-site pooling classification and leave-one-site-out transfer prediction between schizophrenia patients and healthy controls respectively, yielding 4% improvement of accuracy compared to conventional approaches, suggesting a remarkable increase of the discriminative power via deep learning in neuroimaging predictions. The promising results may benefit from the following two aspects: 1) the proposed MsRNN can learn both temporal and spatial information simultaneously based on time courses rather than the second-level FNC features. Specifically, the multi-scale CNN module can capture the spatial correlation of components from different time scales (2TR~8TR), and the RNN module can leverage temporal information; 2) the large-scale dataset (1100 subjects) provide us the opportunity to train the deep learning model sufficiently. From this view of point, the present study may mark a significant breakthrough for enhancing the capabilities of psychiatrists by bringing RNN-based deep learning method to the task of diagnosing brain disorders across sites. Such applications would be critical and useful in clinical practice to predict for the new imaging sites or subjects. We also noticed a recently published multi-center study using deep learning method to diagnose schizophrenia [9]. The deep discriminant autoencoder network proposed by Zeng et al., aiming at learning imaging site-shared functional connectivity features, achieved desirable discrimination of schizophrenia across multiple independent imaging sites. To clarify, the current study used an entirely different deep learning architecture (AutoEncoder [Zeng et al.] vs. MsRNN [ours]) and different input features for classification (functional connectivity [Zeng et al.] vs. time courses [ours]), which avoid the second-level computation of fMRI data.
As to the identified brain regions, the dominating component is related to the dorsal striatum in the classification of schizophrenia. The dorsal striatum, comprising caudate and putamen, primarily mediates cognition involving motor function, certain executive functions (e.g., inhibitory control), and stimulus-response learning. It receives input from cortex, thalamus, hippocampus and amygdala, then projects its output information to thalamus. The thalamus, which projects back to the cortex, thereby completing the circuit is also a component of the reward system that may suffer severely in SZ [[36], [37], [38]]. A similar impairment in SZ was verified in multiple resting-state fMRI studies [39] and cognitive studies [40]. For example, Yoon et al. [41] observed a link between impaired prefrontal-basal ganglia functional connectivity and the severity of psychosis, and Sarpal et al. [42] found a negative relationship between the functional connectivity of striatal regions and reduction in psychosis.
Another cerebellum component consist of declive, uvula and pyramis. The cerebellum is engaged in basic cognitive function such as attention, working memory, verbal learning and sensory discrimination, has led to an emerging interest in the role of the cerebellum in schizophrenia [43]. Structural and functional cerebellar abnormalities have been observed in schizophrenia, with evidence the impairment in white matter integrity in specific cerebellar lobes [44], as well as the abnormal size and a significant decrease in cerebral blood flow during a broad range of cognitive tasks [43,45]. Besides, researchers have posited the role of the cerebellum in reinforcement learning, allowing for more direct convergence between the theories of cognitive dysmetria and impaired reinforcement learning in schizophrenia [46].
Across several studies, altered connectivity patterns between the striatum and cerebellum have been frequently found in schizophrenia. Abnormalities in the relationship between cortical and sub-cortical regions, in particular, the prefrontal cortex, thalamus, basal ganglia, and cerebellum, were observed in patients with schizophrenia and correlated primarily with deficits in executive functioning, as well as deficits in processing speed and working memory [45]. Su et al. [47] and Repovs et al. [48] provided evidence that the connectivity strength between cerebellum and caudate is associated with executive functioning loss in schizophrenia. Also, reduced functional connectivity between the cerebellum and medial dorsal nucleus of the thalamus in schizophrenia providing evidence of abnormalities in this portion of the cortico-cerebellar-thalamic-cortico circuit [9,12,45,49]. Our results suggest that the temporal dynamics in the two identified brain regions and their connectivity are highly different between HC and SZ, which may serve as potential biomarkers for SZ discrimination.
The proposed model is stable and robust. Fig. S1 shows the learning curves on training and validation data while optimizing the parameters of MsRNN. The model convergent quickly during the first 100 epochs and reached a steady point after around 300 epochs. Since the number of hidden nodes in GRU layer may directly affect the learning capacity of a GRU model., we compared the performance of MsRNN model with a varying number of hidden units (i.e. [21, 22, 23 …, 210]) to validate the influence of the number of hidden notes in GRU layer. The statistical results indicate that our proposed model is not sensitive to the number of hidden units (Fig. S1b). The model can reach an over 80% classification accuracy with a range of 23~29 GRU hidden nodes. More hyperparameters about MsRNN including batch size, number of filters, scales of filters were analyzed thoroughly (Table S6-S8). The results show that the proposed MsRNN model is quite robust and not sensitive to these hyperparameters. Moreover, the hyperparameters combination we used in this work is close to an optimal solution. We also compared the influence of multiple training-testing ratios (Table S9), results show that the higher training-testing ratio is, the better performance MsRNN model achieves, which is consistent with the previous finding [9], suggesting further potential improvement of our proposed method when gathering more samples for modeling. Finally, to study the influence of the number of ICs, we further compared four different ICA component settings (Table S10). The two-sample t-test results show that only when the number of ICs is 16, the classification accuracy is less attractive than using 50 ICs(proposed), however, using more ICs does not show significant improvement, and many previous studies use a similar number of ICs as we did [50,51].
The current study has a few limitations. One is that information on antipsychotic or mood stabilizing medications for part of the patients were unavailable, which makes it difficult to assess the medication effect that may result in specific functional changes [6]. Secondly, the time courses were filtered within the range of 0·01–0·1HZ during the preprocessing step. However, the discriminative functional activity in the human brain may occur in a higher frequency range. Since the proposed MsRNN model can be applied to classify using either magnetoencephalogram (MEG) or electroencephalography (EEG) data due to its feasibility to higher temporal resolution data [6], therefore, a more stable and generative deep learning classification model may be designed by fusing multi-modalities to extract fused features and apply them to the RNN classification model in the future [52]. Another limitation is that even though head motion effect has been substantially attenuated through preprocessing procedures, it may not be completely removed and may remain certain influences. Moreover, the fMRI data acquisition protocols for all sites were set up by the same experienced experts and more harmonized in our study. Therefore the classification performance of the proposed model may be weighted down a bit if the data acquisition protocols in new sites are very different from each other. We admit that the proposed MsRNN model is still a preliminary model which did not give each hidden state a specific weight. One complementary strategy which may enhance GRU's performance is “attention” mechanism that can learn the weight of each hidden state automatically [53]. Furthermore, interpretation of deep learning networks remains an emerging but key field of research, our future work will focus more on a better interpretation of deep learning results, which would provide us with more clues on identifying potential biomarkers.
In summary, to the best of our knowledge, this is the first attempt to enable RNN directly to work on time courses of fMRI components in schizophrenia classification. The model takes advantage of high-level spatiotemporal information of fMRI data, and the high classification performances indicate the advantages of the proposed model. Also, the proposed leave-one-IC-out strategy provides a potential solution for increasing the clinical interpretability of the deep learning-based methods. Our work promises great potentials of deep-chronnectome-learning and a broad utility on neuroimaging applications, e.g., the extension to MEG, EEG learning.
Funding sources
This work was supported by the Natural Science Foundation of China(No. 61773380), the Strategic Priority Research Program of the Chinese Academy of Sciences (No.XDB32040100), Beijing Municipal Science and Technology Commission (Z181100001518005), the National Institute of Health (1R56MH117107, R01EB005846, R01MH094524, and P20GM103472) and the National Science Foundation (1539067). The authors report no financial interests or potential conflicts of interest. No funders played a role in the study. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
Declaration of Competing Interest
The authors report no biomedical financial interests or potential conflicts of interest.
Acknowledgments
The current research was designed by WZY and JS. Acquisition of data was performed by TZJ, MS, YC, LZF, ZYY, NMZ, KBX, LXL, JC, YCC, HG, PL, LL, PW, HNW, HLW, HY, JY, YFY, HXZ, DZ. The data was analyzed by WZY, SFL, VDC, JS. The draft was written by WZY, VDC and JS. All authors contribute to the discussion of the results and have approved the final manuscript to be published.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.08.023.
Contributor Information
Tianzi Jiang, Email: jiangtz@nlpr.ia.ac.cn.
Jing Sui, Email: jing.sui@nlpr.ia.ac.cn.
Appendix A. Supplementary data
Supplementary material
References
- 1.Calhoun V.D., Miller R., Pearlson G., Adali T. The chronnectome: time-varying connectivity networks as the next frontier in fMRI data discovery. Neuron. 2014;84(2):262–274. doi: 10.1016/j.neuron.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calhoun V.D., Adali T., Pearlson G.D., Pekar J.J. A method for making group inferences from functional MRI data using independent component analysis. Hum Brain Mapp. 2001;14(3):140–151. doi: 10.1002/hbm.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lynall M.-E., Bassett D.S., Kerwin R., McKenna P.J., Kitzbichler M., Müller U. Functional connectivity and brain networks in schizophrenia. J Neurosci. 2010;30(28):9477–9487. doi: 10.1523/JNEUROSCI.0333-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sui J., Qi S., van Erp T.G.M., Bustillo J., Jiang R., Lin D. Multimodal neuromarkers in schizophrenia via cognition-guided MRI fusion. Nat Commun. 2018;9(1):3028. doi: 10.1038/s41467-018-05432-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rashid B., Arbabshirani M.R., Damaraju E., Cetin M.S., Miller R., Pearlson G.D. Classification of schizophrenia and bipolar patients using static and dynamic resting-state fMRI brain connectivity. Neuroimage. 2016;134:645–657. doi: 10.1016/j.neuroimage.2016.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu S., Wang H., Song M., Lv L., Cui Y., Liu Y. Linked 4-way multimodal brain differences in schizophrenia in a large Chinese Han population. Schizophr Bull. 2018;45(2):436–449. doi: 10.1093/schbul/sby045. [:sby045-sby] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan W., Plis S., Calhoun V.D., Liu S., Jiang R., Jiang T.Z. 2017 IEEE 27th international workshop on machine learning for signal processing (MLSP) 2017. Discriminating schizophrenia from normal controls using resting state functional network connectivity: A deep neural network and layer-wise relevance propagation method. [25-28 Sept. 2017] [Google Scholar]
- 8.Yan W., Zhang H., Sui J., Shen D. International conference on medical image computing and computer-assisted intervention. Springer; 2018. Deep chronnectome learning via full bidirectional long short-term memory networks for MCI diagnosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng L.-L., Wang H., Hu P., Yang B., Pu W., Shen H. Multi-site diagnostic classification of schizophrenia using discriminant deep learning with functional connectivity MRI. EBioMedicine. 2018;30:74–85. doi: 10.1016/j.ebiom.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dvornek N.C., Ventola P., Pelphrey K.A., Duncan J.S. Identifying autism from resting-state fMRI using long short-term memory networks. In: Wang Q., Shi Y., Suk H.-I., Suzuki K., editors. Machine learning in medical imaging: 8th international workshop, MLMI 2017, held in conjunction with MICCAI 2017, Quebec City, QC, Canada, September 10, 2017, proceedings. Springer International Publishing; Cham: 2017. pp. 362–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durstewitz D., Koppe G., Meyer-Lindenberg A. Deep neural networks in psychiatry. Mol Psychiatry. 2019 doi: 10.1038/s41380-019-0365-9. https://www.nature.com/articles/s41380-019-0365-9 [DOI] [PubMed] [Google Scholar]
- 12.Kim J., Calhoun V.D., Shim E., Lee J.H. Deep neural network with weight sparsity control and pre-training extracts hierarchical features and enhances classification performance: evidence from whole-brain resting-state functional connectivity patterns of schizophrenia. Neuroimage. 2015;124:127–146. doi: 10.1016/j.neuroimage.2015.05.018. Pt A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davatzikos C., Sotiras A., Fan Y., Habes M., Erus G., Rathore S. Precision diagnostics based on machine learning-derived imaging signatures. Magn Reson Imaging. 2019 doi: 10.1016/j.mri.2019.04.012. https://www.sciencedirect.com/science/article/abs/pii/S0730725X18306301?via%3Dihub [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H., Fan Y., editors. Medical image computing and computer assisted intervention – MICCAI 2018. Springer International Publishing; Cham: 2018. Identification of temporal transition of functional states using recurrent neural networks from functional MRI. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hochreiter S., Schmidhuber J. Long short-term memory. J Neural Comput. 1997;9(8):1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 16.Chung J., Gulcehre C., Cho K., Bengio Y. 2014. Empirical Evaluation of Gated Recurrent Neural Networks on Sequence Modeling. arXiv preprint arXiv:14123555. [Google Scholar]
- 17.Dvornek N.C., Ventola P., Pelphrey K.A., Duncan J.S. Machine learning in medical imaging MLMI (Workshop) Vol. 10541. 2017. Identifying autism from resting-state fMRI using long short-term memory networks; pp. 362–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roy S., Kiral-Kornek I., Harrer S. 2018. ChronoNet: A deep recurrent neural network for abnormal EEG identification. arXiv preprint arXiv:180200308. [DOI] [PubMed] [Google Scholar]
- 19.Li H., Fan Y., editors. Medical image computing and computer assisted intervention – MICCAI 2018. Cham; Springer International Publishing: 2018. Brain decoding from functional mri using long short-term memory recurrent neural networks. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tzourio-Mazoyer N., Landeau B., Papathanassiou D., Crivello F., Etard O., Delcroix N. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15(1):273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- 21.Fan L., Li H., Zhuo J., Zhang Y., Wang J., Chen L. The human Brainnetome atlas: a new brain atlas based on connectional architecture. Cereb Cortex. 2016;26(8):3508–3526. doi: 10.1093/cercor/bhw157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He Y., A Evans. Magnetic resonance imaging of healthy and diseased brain networks. Front Hum Neurosci. 2014;vol. 8:890. doi: 10.3389/fnhum.2014.00890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du Y., Fan Y. Group information guided ICA for fMRI data analysis. Neuroimage. 2013;69:157–197. doi: 10.1016/j.neuroimage.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 24.Satterthwaite T.D., Elliott M.A., Gerraty R.T., Ruparel K., Loughead J., Calkins M.E. An improved framework for confound regression and filtering for control of motion artifact in the preprocessing of resting-state functional connectivity data. Neuroimage. 2013;64:240–256. doi: 10.1016/j.neuroimage.2012.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeng L.-L., Wang D., Fox M.D., Sabuncu M., Hu D., Ge M. Neurobiological basis of head motion in brain imaging. Proc Natl Acad Sci. 2014;111(16):6058. doi: 10.1073/pnas.1317424111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fox M.D., Snyder A.Z., Vincent J.L., Corbetta M., Van Essen D.C., Raichle M.E. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci U S A. 2005;102(27):9673. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan C.-G., Cheung B., Kelly C., Colcombe S., Craddock R.C., Di Martino A. A comprehensive assessment of regional variation in the impact of head micromovements on functional connectomics. Neuroimage. 2013;76:183–201. doi: 10.1016/j.neuroimage.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roy S., Kiral-Kornek I., S Harrer. 2018. ChronoNet: A deep recurrent neural network for abnormal EEG identification. [DOI] [PubMed] [Google Scholar]
- 29.Huang G., Liu Z., Van Der Maaten L., Weinberger K.Q. Proceedings of the IEEE conference on computer vision and pattern recognition. 2017. Densely connected convolutional networks. [Google Scholar]
- 30.Laurens van der Maaten G.H. Visualizing Data using t-SNE. J Mach Learn Res. 2008;9(Nov):2579–2605. [Google Scholar]
- 31.Jo Y., Park S., Jung J., Yoon J., Joo H., Kim M.H. Holographic deep learning for rapid optical screening of anthrax spores. Sci Adv. 2017;3(8) doi: 10.1126/sciadv.1700606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guyon I., Weston J., Barnhill S., Vapnik V. Gene selection for cancer classification using support vector machines. Mach Learn. 2002;46(1):389–422. [Google Scholar]
- 33.Zhu J., Zou H., Rosset S., Hastie T. Multi-class AdaBoost. Stat Interface. 2009;2(3):349–360. [Google Scholar]
- 34.Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. [Google Scholar]
- 35.Yu Q., Du Y., Chen J., He H., Sui J., Pearlson G. Comparing brain graphs in which nodes are regions of interest or independent components: a simulation study. J Neurosci Methods. 2017;291:61–68. doi: 10.1016/j.jneumeth.2017.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nestler E.J., Hyman S.E., Malenka R.C. Appleton & Lange East Norwalk; Conn: 2001. Molecular basis of neuropharmacology: a foundation for clinical neuroscience. [Google Scholar]
- 37.Yager L.M., Garcia A.F., Wunsch A.M., Ferguson S.M. The ins and outs of the striatum: role in drug addiction. Neuroscience. 2015;301:529–541. doi: 10.1016/j.neuroscience.2015.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferré S., Lluís C., Justinova Z., Quiroz C., Orru M., Navarro G. Adenosine-cannabinoid receptor interactions. Implications for striatal function. Br J Pharmacol. 2010;160(3):443–453. doi: 10.1111/j.1476-5381.2010.00723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sui J., Pearlson G.D., Du Y., Yu Q., Jones T.R., Chen J. In search of multimodal neuroimaging biomarkers of cognitive deficits in schizophrenia. Biol Psychiatry. 2015;78(11):794–804. doi: 10.1016/j.biopsych.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simpson E.H., Kellendonk C., Kandel E. A possible role for the striatum in the pathogenesis of the cognitive symptoms of schizophrenia. Neuron. 2010;65(5):585–596. doi: 10.1016/j.neuron.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoon J.H., Minzenberg M.J., Raouf S., D'Esposito M., Carter C.S. Impaired prefrontal-basal ganglia functional connectivity and substantia Nigra hyperactivity in schizophrenia. Biol Psychiatry. 2013;74(2):122–129. doi: 10.1016/j.biopsych.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarpal D.K., Robinson D.G., Lencz T. Antipsychotic treatment and functional connectivity of the striatum in first-episode schizophrenia. JAMA Psychiat. 2015;72(1):5–13. doi: 10.1001/jamapsychiatry.2014.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andreasen N.C., Pierson R. The role of the cerebellum in schizophrenia. Biol Psychiatry. 2008;64(2):81–88. doi: 10.1016/j.biopsych.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim D.-J., Kent J.S., Bolbecker A.R., Sporns O., Cheng H., Newman S.D. 40(6) 2014. Disrupted modular architecture of cerebellum in schizophrenia: a graph theoretic analysis; pp. 1216–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sheffield J.M., Barch D.M. Cognition and resting-state functional connectivity in schizophrenia. Neurosci Biobehav Rev. 2016;61:108–120. doi: 10.1016/j.neubiorev.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swain R.A., Kerr A.L., Thompson R.F. The cerebellum: a neural system for the study of reinforcement learning. Front Behav Neurosci. 2011;vol. 5:8. doi: 10.3389/fnbeh.2011.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Su T.-W., Lan T.-H., Hsu T.-W., Biswal B.B., Tsai P.-J., Lin W.-C. Reduced neuro-integration from the dorsolateral prefrontal cortex to the whole brain and executive dysfunction in schizophrenia patients and their relatives. Schizophr Res. 2013;148(1):50–58. doi: 10.1016/j.schres.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 48.Repovs G., Csernansky J.G., Barch D.M. Brain network connectivity in individuals with schizophrenia and their siblings. Biol Psychiatry. 2011;69(10):967–973. doi: 10.1016/j.biopsych.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anticevic A., Yang G., Savic A., Murray J.D., Cole M.W., Repovs G. Mediodorsal and visual thalamic connectivity differ in schizophrenia and bipolar disorder with and without psychosis history. Schizophr Bull. 2014;40(6):1227–1243. doi: 10.1093/schbul/sbu100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allen E.A., Erhardt E.B., Damaraju E., Gruner W., Segall J.M., Silva R.F. A baseline for the multivariate comparison of resting-state networks. Front Syst Neurosci. 2011;5:2. doi: 10.3389/fnsys.2011.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Du Y., Pearlson G.D., Liu J., Sui J., Yu Q., He H. A group ICA based framework for evaluating resting fMRI markers when disease categories are unclear: application to schizophrenia, bipolar, and schizoaffective disorders. Neuroimage. 2015;122:272–280. doi: 10.1016/j.neuroimage.2015.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Plis S.M., Amin M.F., Chekroud A., Hjelm D., Damaraju E., Lee H.J. Reading the (functional) writing on the (structural) wall: multimodal fusion of brain structure and function via a deep neural network based translation approach reveals novel impairments in schizophrenia. Neuroimage. 2018;181:734–747. doi: 10.1016/j.neuroimage.2018.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou P., Shi W., Tian J., Qi Z., Li B., Hao H. Proceedings of the 54th annual meeting of the association for computational linguistics (volume 2: Short papers) 2016. Attention-based bidirectional long short-term memory networks for relation classification. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
All data needed to evaluate the conclusions are present in the paper and/or the supplementary materials. Additional data related to this paper may be requested from the authors.