Highlights
-
•
The methanolic extract of Enterobacter hormaechei showed leishmanicidal activity against promastigotes of Leishmania braziliensis (strain UA301).
-
•
P. ananatis, E. cloacae, A. gyllenbergii, O. anthropi and P. otitidis are promising bacteria in the biotechnology field as well as for biological control due to several enzymatic and antimicrobial bioactivities.
-
•
Differential production of enzymes and secondary metabolites by Gram-negative bacteria could contribute to physiological and metabolic processes in the intestine of Lutzomyia evansi.
Keywords: Bacteria-intestine interaction, Le. braziliensisenzymatic bioactivity, Antimicrobial screening, Leishmanicidal activity
Abstract
Knowledge regarding new compounds, peptides, and/or secondary metabolites secreted by bacteria isolated from the intestine of phebotominae has the potential to control insect vectors and pathogens (viruses, bacteria, and parasites) transmitted by them. In this respect, twelve Gram-negative bacteria isolated from the intestine of Lutzomyia evansi were selected and screened for their enzymatic, antimicrobial, and leishmanicidal activity. E. cancerogenus, E. aerogenes, P. otitidis, E. cloacae, L. soli, and P. ananatis exhibited enzymatic activity. 83.3% of the isolates displayed lipolytic and nitrate reductase activity and 58.3% of the isolates displayed protease activity. Hemolytic activity (17%) was identified only in E. hormaechei, and P. ananatis. E. cancerogenus, A. calcoaceticus, and P. otitidis showed cellulolytic activity. A. gyllenbergii, P. aeruginosa, and E. hormaechei showed amylolytic activity. In general, the totality of methanolic extracts exhibited antimicrobial activity, where E. hormaechei, A. calcoaceticus, and E. cancerogenus presented the highest activity against the evaluated reference bacteria strains. Cell-free supernatants (CFSS) of the Gram-negative bacteria showed higher growth inhibitory activity against the reference Gram-positive bacteria. The CFS of A. gyllenbergii was the most active antimicrobial in this study, against S. aureus (AAODs = 95.12%) and E. faecalis (AAODs = 86.90%). The inhibition percentages of CFS against Gram-positive bacteria showed statistically significant differences (repeated measure ANOVA df= 2; F= 6.095; P= 0.007832). The E. hormaechei methanolic extract showed leishmanicidal activity (CE-50 μg/ml = 47.7 + 3.8) against metacyclic promastigotes of Leishmania braziliensis (UA301). Based on this finding, we discuss the possible implications of these bacteria in digestion and physiological processes in the Lu. evansi intestine. P. ananatis, E. cloacae, E. hormaechei, and P. otitidis were considered the most promising bacteria in this study and they could potentially be used for biological control.
1. Introduction
The control of insect vectors and etiological agents that cause tropical illnesses needs alternative strategies that impact transmission and propagation in endemic areas in a more significant way [1], [2]. A determining source of natural products for biologic control is the intestinal lumen of some insect vectors. Recent studies have established that entire bacteria or their secondary metabolite molecules, as well as peptides secreted by intestinal tract bacteria, may possess the ability to modulate vector competence, impacting the life cycle of the vector species, generating reproductive alterations, and altering the immune system due to various antimicrobial activities. Some species have even demonstrated leishmanicidal activity [3], [4], [5], [6].
During interactions such as symbiosis, mutualism, and parasitism between the microbiota and the intestine of the insect vector, biologically active substances may be generated. Among these, proteolytic enzymes can be found (e.g., aminopeptidases), cytotoxins, thioesters, galectins, antimicrobial peptides, and signaling factors [3], [5], [7], [8], [9]. The insect also possesses an innate immune defense system against pathogens (defensins, antioxidant enzymes, caspases, peroxidases) which maintains the balance of this complex behavior [10], [11]. Additionally, it has been established that Gram-negative bacteria are responsible for activation via Toll, IMD, and Jak-STAT to control Gram-positive bacteria, fungi, viruses, and some parasites [12], [13].
Furthering the study of the insect intestinal microbiota is of the utmost importance due to the fact that it has been described that they can accomplish functions associated with physiological and/or metabolic processes, and in the production of amino acids and in the secretion of inhibiting substances that prevent pathogen bacteria colonization [6]. Nevertheless, few studies have described their enzymatic, antimicrobial or leishmanicidal bioactivities. For this reason, it is necessary to generate relevant information that allows us to understand the microorganism interactions in the intestinal lumen of phlebotominae and to design control strategies to interrupt the transmission cycle of the disease [5], [14], [15].
Recent studies have identified bacterial genera such as Serratia, Enterobacter, and Pseudomonas, with the ability to synthesize molecules with excellent antiparasitic potential; cytotoxic metalloproteases, hemolysins, antibiotics, prodigiosin, and hemagglutinins have been widely described [7]. Additionally, leishmanicidal activity against Le. infantum and Le. mexicana has been shown with entire bacteria such as Ochrobactrum intermedium, Asaia spp., Pantoea ananatis, Enterobacter cloacae, and O. anthropi [4], [16].
To summarize, some substances synthesized by Gram-negative bacteria have been characterized for being biologically active and functionally important compounds with metabolic activities as well as defensive roles against pathogens [6], [12]. Increased knowledge of the extracellular substances produced by these bacteria represents an important source for antibiotic and enzyme production.
In this study, bacterial strains isolated from the intestinal microbiota of Lu. evansi were evaluated for their antimicrobial activity from methanolic extracts and cell-free supernatants against pathogenic bacterial strains. Extracellular hydrolase activity (protease, lipase, hemolysis, amylase, cellulase, nitrate reduction) was qualitatively evaluated using entire bacteria. Finally, leishmanicidal activity was estimated against promastigotes of L. infantum and L. braziliensis. Increased knowledge of the bioactivities of the intestinal microbiota of phlebotominae could be useful to understand their interactions and lead to the formulation of new compounds with biotechnological potential.
2. Materials and methods
2.1. Gram-negative bacteria selection
In this study, we used 12 Gram-negative bacteria isolates previously obtained from the intestinal microbiota of wild populations of Lu. evansi collected in Sucre, Colombia [4] (Table 1). These isolates were selected for having leishmanicidal activity, and for previously reported biotechnological potential from other environmental sources. Taxonomical and molecular identification was carried out [4].
Table 1.
Gram-negative bacterial selected and isolated from the digestive tract of Lutzomyia evansi for enzymatic, antimicrobial and leishmanicidal bioactivity assays.
| Isolation source [4] |
Bacterial isolate code (Accession number_GenBank) | Strain |
|---|---|---|
| Unfed female | Isolate 41 (KU134743) | Enterobacter cancerogenus |
| Unfed female | Isolate 43 (KU134745) | Pseudomonas putida |
| Fed Female | Isolate 44 (KU134746) | Enterobacter aerogenes |
| Fed Female | Isolate 64 (KU134748) | Acinetobacter calcoaceticus |
| Male | Isolate 70 (KU134777) | Pseudomonas otitidis |
| Unfed female | Isolate 71 (KU134741) | Enterobacter cloacae |
| Larvae L4 | Isolate 102 (KU134766) |
Ochrobactrum anthropi |
| Unfed female | Isolate 140 (KU134754) |
Pantoea ananatis |
| Larvae L4 | Isolate 154 (KU134779) |
Acinetobacter gyllenbergii |
| Larvae L4 Larvae L4 Larvae L4 |
Isolate 157 (KU134771) Isolate 173 (KU134783) Isolate 188 KU134784) |
Enterobacter hormaechei Pseudomonas aeruginosa Lysobacter soli |
2.2. Bacteria and culture conditions
Isolates were reactivated using 100 μL of the cellular suspension, in LB liquid medium (Luria-Bertoni, LAB, United Kingdom) and subsequent growth in an LB-solid medium. Pure colonies were selected and incubated all night at 37 °C with shaking at 180 rpm, until a final concentration of 1.2 × 108 UFC/mL (OD600 = 0.125) [4].
2.3. Detection of hydrolyzing extracellular enzymes
2.3.1. Assay for proteolytic activity
Proteolytic activity was determined using skim milk-supplemented media (100 g L−1), according to [17] and Hossain (2015). Pure isolates were grown on agar milk plates and incubated at 30 °C for 48 h. The presence of a clear zone around the colony indicated casein hydrolysis [18]. Additionally, proteolytic activity was evaluated by gelatin liquefaction (120 g L−1, in distilled H2O; Merck), incubated at room temperature for 24 h. For this assay, a deep culture of each bacterial isolate was carried out. Their activity was evaluated in triplicate. Staphylococcus aureus ATCC 29213 and Bacillus cereus ATCC 14579™ were used as the positive and negative controls, respectively.
2.3.2. Assay for lipase activity
Bacteria were screened using medium supplemented with Tween 20 at 1% (w/v), which contained peptone 1% (10 g L−1), NaCl 0.5% (5 g L−1), and CaCl2 0.01% (0.1 g L−1) (El-bestawy and El-masry, 2005). Isolates were streaked onto media plates and incubated at 30 °C. Each sample was evaluated in triplicate. After seven days, the activity was confirmed by the appearance precipitation zone around the colony [19]. Serratia sp. B006 and Escherichia coli ATCC 8739™strains were used as the positive and negative controls, respectively [20].
2.3.3. Assay for amylolytic activity
Amylolytic activity was evaluated following a previously described protocol [21]. Pure colonies of each bacterium were grown in medium supplemented with starch 1% (w/v), yeast extract 0.3% (3 g L−1), and agar 1.5% (15 g L−1). The isolates were streaked and incubated at 35 °C ± 2 °C for 48 h. After incubation, iodine solution (3 mL) was flooded with a dropper for five minutes onto the starch agar plate. The clear zone of hydrolysis around the colony indicates a positive result. Escherichia coli ATCC 9029™ and the Bacillus cereus ATCC 14579™ strains were used as the negative and positive controls, respectively.
2.3.4. Assay for cellulolytic activity
Pure colonies were grown in medium supplemented with carboxymethylcellulose 1% (w/v), yeast extract 0.25% (2.5 g L−1), peptone 0.25% (2.5 g L−1), (NH4)2SO4 0.05% (0.5 g L−1), CaCl2 0.05% (0.5 g L−1), KH2PO4 0.01% (0.1 g L−1), K2HPO4 0.01% (0.1 g L−1), and 1.5% agar (15 g L−1). Subsequently, cellulolytic activity was assessed by means of a qualitative evaluation, adding 3 ml of Congo red reactive at 1% for 15 min, used to illustrate the clear the zone around the colony [22]. Afterwards, 2 mL of 0.1 M NaCl was added for 15 min and the excess was decanted. The Petri plates with growth were incubated at 4 °C for 24 h. After that time, the corresponding readings of the degradation halos were conducted. Bacillus cereus ATCC 14579™ strain was used as the positive control [23]. PBS was used as the negative control.
2.3.5. Assay for hemolytic activity
Hemolysis was determined according to [24]. Pure isolates were evaluated on blood agar plates for 30 °C for 72 h. S. aureus (clinical isolate), E. coli ATCC 9029™ and E. faecalis (clinical isolate) were used as controls to determine complete hemolysis (β), partial hemolysis (α), and non-hemolysis (γ), respectively [25], [26], [27].
2.3.6. Nitrate reductase and nitrite reductase activity
Nitrate reduction was evaluated following a previously described protocol [21]. Pure colonies of each bacterial isolate were grown in medium supplemented with potassium nitrate 1% (w/v) broth, meat extract 0.3% (3 g L−1), and peptone 0.5 (5 g L−1), and incubated at 35 °C ± 2 °C for 24 h [21]. Nitrate and nitrite reduction was assessed by adding Griess reagent. Escherichia coli ATCC 9029™ and Acinoteobacter baumanni ATCC 17978™were used as the positive and negative controls, respectively [21].
2.4. Extraction of secondary metabolites and antimicrobial activity
Secondary metabolite extraction was performed according to the described method [28]. An Erlenmeyer with 50 mL of LB broth was inoculated with 0.5 mL of each of the selected strains culture for 18 h at 30 °C, then amberlite XAD-16 (AMRESCO®) 2% (w/v) was added. After seven days of incubation at 30 °C and 180 rpm, the metabolites absorbed by the amberlite were eluted with 20 mL of methanol (100%) for 60 min [29]. The methanolic fraction of each extract was concentrated up to 1.0 mL in a rotary evaporator at 40 °C (Heidolph Laborota 4001 Efficient Rotary Evaporator) [4].
Antimicrobial screening was carried out by means of the agar diffusion test on Mueller-Hinton medium (Becton Dickinson) [30] with some modifications. Sterile Whatman filter paper discs (No. 1, 6 mm in diameter) were impregnated with each extract. The discs were placed over the Mueller-Hinton agar (Becton Dickison) in Petri dishes, evenly inoculating a liquid culture of the reference Pseudomonas aeruginosa ATCC 9027, E. coli ATCC 8739, Enterococcus faecalis ATCC 51299, and S. aureus ATCC 29213 strains and two clinical isolates, which corresponded to S. marcescens and B. cereus. The determination of antimicrobial activity was evaluated following the procedure described by the Clinical and Laboratory Standards Institute (CLSI, 2014). The dishes were incubated at 37 °C for 18 h, and then the diameter of the inhibition halo of the growth surrounding each of the discs was measured. The antibacterial activity assays were performed in triplicate in six independent experiments.
2.5. Assessment of antimicrobial activity of cell-free supernatants
Strains were inoculated in an LB broth until reaching their exponential growth. The supernatants were recovered by centrifugation at 10,000 g, at 4 °C for 10 min, and were neutralized by pH (7.0) modification. Finally, they were filtered through a 0.2 μm membrane [31].
Antimicrobial activity of cell-free supernatants was done by means of an agar diffusion test and serial microdilution [30]. This assay was done using 96-well microplates on the previously described reference strains (1.0 × 106 CFU/ml with serial dilutions) [32]. As negative control, 100 μL of BMH along with 100 μL of reference strain were added to each well. The positive control was evaluated with 20 μL of chloramphenicol, 80 μL of BMH in 100 μL of the pathogen. Additionally, cyclophenicol (30 μg/mL) in BMH was used as a negative control. Each assay was carried out in triplicate, and the microplates were incubated at 37 °C for 18 h with 600 nm photometric readings every hour [30]. Bacterial inhibition percentages based on absorbance were calculated using the formula: antibacterial activity (% OD) = ((DC-Ds)/DC) x 100, where DC is the control (cellular concentration of reference strains more LB broth) and Ds is the final growth of reference strains with CFS in the plate.
2.6. Leishmanicidal activity of secondary metabolites
Roto-evaporation was used to eliminate the solvent from the methanolic extracts. Afterwards, they were weighed and solubilized with DMSO. The assay for the effectiveness on metacyclic promastigotes of L. infantum, (BCN- strain) and L. braziliensis (UA301 strain) promastigotes was done in 96-well plates with six serial dilutions to a maximum concentration of 400 μg/mL. Subsequently, the promastigotes were added at 1,200,000 parasites/mL and incubated at 26 °C for 72 h. Amphotericin B was used as the control. Each concentration of the compound (the extracts), as well as the compound-free control, were tested in triplicate in two different experiments. The viability of the parasites was determined by measuring mitochondrial dehydrogenase activity by the MTT method [33].
2.7. Statistical analysis
The statistical analysis of the antibacterial activity of the extracts was estimated by repeated measures ANOVA using XLSTAT software (version 2018.5), considering the inhibition-halo diameter comparison. The same analysis was done for the cell-free supernatants in terms of the inhibition percentages. Antimicrobial activity is shown in a graph with the standard error using PAST (Version 3.21) software.
3. Results
3.1. Extracellular hydrolase detection
Out of the 12 Gram-negative isolates, seven showed light halo formation around the colony in the supplemented milk medium (Table 2). The isolates with the highest activity proteolytic in milk were E. hormaechei and P. ananatis (Appendix A). Proteolytic activity by gelatin liquefaction was also positive for these two bacteria (Table 2). A wide number of isolates (n = 10; 83.3%) that showed lipolytic activity were found (Table 2, Appendix A). P. ananatis and E. hormaechei were the only bacterial isolates with complete hemolytic activity (hemolysis β) (Table 2, Appendix A).
Table 2.
Enzymatic activity of Gram-negative bacteria isolated from the intestinal tract of Lutzomyia evansi.
| Gram-negative bacteria - Code | Enzymatic activity |
||||||
|---|---|---|---|---|---|---|---|
| Proteolytic * | Proteolytic ** | Lipolytic | Hemolytic | Cellulolytic | Amylolytic | Nitrate reductase and nitrite reductase | |
| E. cancerogenus_41 | + | – | ++ | – | + | – | + |
| P. putida_43 | – | – | + | – | – | – | – |
| E. aerogenes_44* | ++ | – | ++ | – | – | – | + |
| A. calcoaceticus_64* | – | – | ++ | – | + | – | + |
| P. otitidis_70 | + | – | ++ | – | + | + | + |
| E. cloacae_71* | + | – | + | – | – | – | + |
| O. anthropi_102* | – | – | – | – | – | – | + |
| P. ananatis_140* | +++ | + | +++ | + | – | – | – |
| A. gyllenbergii_154 | – | – | + | – | – | – | + |
| E. hormaechei_157* | +++ | + | – | ++ | . | + | + |
| P. aeruginosa_173** | – | – | +++ | – | . | + | + |
| L. soli_188 | ++ | – | + | – | – | – | + |
Proteolytic activity of Gram-negative bacteria on agar supplemented with milk * and gelatin**.-: no activity (absence of clear areas around the colony); +: positive activity, presence of clear areas around the colony. Degradation zone on specific supplements (cm): - (0); + (0.1-0.3); ++ (0.4-0.6); +++ (0.7-1.0). * Reported positive for nitrate reduction (Gitaitis et al., 2003; Villalobo et al., 1977; Brenner et al., 2015); * Reported positive for amylase (genome.jp/kegg-bin/show_pathway?pae00500).
One of the enzymatic tests of great interest, and which is associated with complex carbohydrate hydrolysis, was the amylolytic activity test where E. hormaechei, P. aeruginosa and P. otitidis isolates were positive (Table 2, Appendix A). With regard to the qualitative test of cellulolytic activity, P. otitidis, A. calcoaceticus, and E. cancerogenus were biologically active (Table 2, Appendix A). Finally, P. putida and P. ananatis isolates were the only species in which nitrate reductase activity and molecular nitrogen production were not evidenced (Table 1, Appendix A).
3.2. Antimicrobial activity of secondary metabolites extracts
Antibacterial activity was showed by all strains tested against at least six strains used as the target (Table 3). E. hormaechei, A. calcoaceticus, and E. cancerogenus showed the greatest inhibitory activity with a standard inhibition halo average of 19 mm in diameter (Table 3). Statistically significant differences were found when comparing the measurements of the inhibition zone produced against S. marcescens (Repeated Measure ANOVA df= 2; F= 12.18 P= 0.01432) with regards to E. coli and P. aeruginosa. In contrast to this, the antimicrobial activity does not reflect statistically significant differences in the size of the halos generated by the methanolic extracts activities against Gram-positive bacteria (Repeated Measure ANOVA df= 2; F= 2.598 P= 0.08157). No antimicrobial activity of bacterial isolates was determined for L. soli against some bacteria like E. coli (ATCC 8739TM) and E. faecalis (ATCC 51299TM).
Table 3.
Evaluation of antimicrobial activity using metabolic extract of 12 Gram-negative isolates of gut microbiota of Lu. evansi against Gram negative y Gram positive bacteria.
| metabolic extracts of bacteria isolates_Code | Diameter of growth or zone of inhibition |
|||||
|---|---|---|---|---|---|---|
|
Gram-negative bacteria |
Gram-positive bacteria |
|||||
|
E. coli (ATCC 8739TM) |
P. aeruginosa (ATCC 9027TM) |
S. marcescens (Clinical isolated) |
B. cereus (Enviromental) |
E. faecalis (ATCC 51299TM) |
S. aureus subsp aureus (ATCC 29213 TM) |
|
| E. cancerogenus_41 | ++ | +++ | ++ | ++ | ++ | ++ |
| P. putida_43 | + | ++ | + | ++ | + | + |
| E. aerogenes_44 | ++ | ++ | ++ | ++ | ++ | ++ |
| A. calcoaceticus_64 | + | +++ | + | ++ | +++ | ++ |
| P. otitidis_70 | ++ | ++ | ++ | ++ | ++ | + |
| E. cloacae_71 | + | ++ | ++ | ++ | + | +++ |
| O. anthropi_102 | ++ | ++ | + | ++ | ++ | ++ |
| P. ananatis_140 | ++ | ++ | + | ++ | ++ | ++ |
| A. gyllenbergii_154 | ++ | ++ | ++ | ++ | ++ | ++ |
| E. hormaechei_157 | ++ | +++ | ++ | +++ | +++ | ++ |
| P. aeruginosa_173 | ++ | ++ | + | ++ | + | ++ |
| L. soli_188 | – | ++ | ++ | + | – | + |
| E. cloacae (C+) | 11 ± 2 | 12 ± 1 | 12 ± 2 | 12 ± 2 | 12 ± 3 | 13 ± 2 |
| Cloranfenicol (C+) | 22 ± 2 | 8 ± 1 | 25 ± 2 | 23 ± 1 | 12 ± 2 | 23 ± 2 |
| Metanol (C-) | 0 | 0 | 0 | 0 | 0 | 0 |
Determination of diameter of growth (measured in mm) for the antimicrobial activity showing different degrees of inhibition for the twelve metabolic extracts. (-): Absence zone of inhibition, (+): zone of inhibition between 8–11 mm, (++): zone of inhibition between 12–15 mm, (+++): zone of inhibition ≥16 mm. C +: Positive control, C-: Negative control. The results represent the average of the triplicates.
3.3. Antimicrobial activity of cell-free supernatants (CFS)
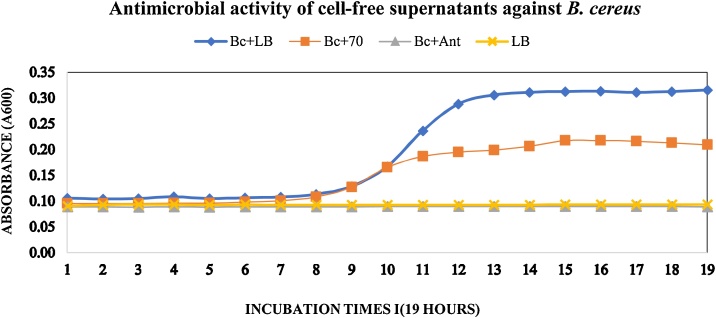
The inhibition percentage of CFS against three Gram-negative and three-Gram positive reference strains are listed in Table 4. An example of the bioassay is shown in Fig. 1 (a representation of the growth of B. cereus against the CFS of P. otitidis). P. aeruginosa, P. ananatis, and E. aerogenes CFSs showed strong antimicrobial activity against S. marcescens (inhibition of bacterial growth > 60%), while O. anthropi showed the lowest activity against this bacterium (Table 4). In general, most of the CFS shows less activity with regard to the three Gram-negative reference strains isolate (Table 4). The inhibition percentages of CFS against Gram-negative bacteria do not show any statistically significant differences (repeated measured ANOVA df= 2; F= 0.7936; P= 0.4647).
Table 4.
Growth bacterial inhibition of reference strains using cell-free supernatants (CFS) of Gram-negative bacteria by serial microdilution test.
| CFS of bacteria isolates_Code | Inhibition of bacterial growth (% OD600) |
|||||
|---|---|---|---|---|---|---|
| Bacterial reference strains | ||||||
| E. coli ATCC 8739 | P. aeruginosa ATCC 9027 | S. marcescens aislado clínico | B. cereus | S. aureus | E. faecalis | |
| E. cancerogenus_41 | 35.11 ± 0.56 | 26.16 ± 1.18 | 17.76 ± 1.18 | 34,37 0,12 | 31,460,06 | 54,55 0,03 |
| P. putida_43 | 2.08 ± 0.94 | 11.88 ± 1.19 | 44.66 ± 0.92 | 63,330,19* | 28,010,14 | 31,860,07 |
| E. aerogenes_44 | 28.39 ± 0.72 | 19.89 ± 1.0 | 61.34 ± 0.97* | 50,00 0,07 | 55,50 0,08 | 78,050,03* |
| A. calcoaceticus_64 | 3.92 ± 0.81 | 25.62 ± 1.29 | 16.22 ± 1.59 | 9,49 0,13 | 26,46 0,14 | 45,46 0,05 |
| P. otitidis_70 | 34.41 ± 0.57 | 25.37 ± 1.19 | 18.02 ± 1.80 | 53,72 0,06 | 53,63 0,09 | 67,100,04* |
| E. cloacae_71 | 24.85 ± 0.83 | 16.12 ± 1.33 | 9.41 ± 2.16 | 17,65 0,12 | 44,28 0,06 | 76,890,03* |
| O. anthropi_102 | 17.48 ± 0.87 | 20.25 ± 1.11 | 0.28 ± 2.14 | 30,81 0,09 | 86,530,01* | 73,110,04* |
| P. ananatis_140 | 26.43 ± 0. 70 | 27.68 ± 0.98 | 75.89 ± 0.56* | 23,66 012 | 46,28 0,05 | 81,240,03* |
| A. gyllenbergii_154 | 32.02 ± 0.67 | 7.87 ± 1.24 | 13.54 ± 1.96 | 50,50 0,06 | 95,120,01* | 86,900,04* |
| P. aeruginosa_173 | 3.02 ± 0.97 | 18.03 ± 1.18 | 69.71 ± 0.81* | 7,89 0,08 | 45,90 0,07 | 52,36 0,04 |
| L. soli_188 | 36.41 ± 0.54 | 25.16 ± 1.17 | 16.22 ± 1.75 | 61,820,05* | 48,610,05 | 70,230,04* |
Antibacterial activity (% OD) = ((DC-Ds)/DC) x 100, where DC is the control (cellular concentration of reference strains more LB broth) and Ds is the final growth of reference strains with CFS in the plate. *: Inhibition of CFS > 60%. The standard deviation (SD) is represented as the mean of the triplicates. Inhibitory activity above 60% are showed in bold.
Fig. 1.
Growth curves of B. cereus (clinical isolate) against the exposure of cell-free supernatants (CFSs) of Pseudomonas otitidis.Bc + LB:B. cereus in LB culture medium; Bc+70: B. cereus in presence of CFSs of Pseudomonas otitidis (isolate 70); Bc + Ant:B. cereus in LB medium supplemented with antibiotic (Chloramphenicol 30 μg/ml).
In contrast, the CFS of the Gram negative bacteria showed greater growth inhibitory activity against the reference Gram positive bacteria, specifically against E. faecalis (Table 4). The most active CFS were from E. aerogenes (which also showed activity against S. marcescens), P. otitidis, E. cloacae, O. anthropi, A. gyllenbergii, P. ananatis and L. soli (Table 4). The A. gyllenbergii CFS was the most active antimicrobial in this study against S. aureus (AAODs = 95.12%) and E. faecalis (AAODs = 86.90%) (Table 4). The inhibition percentages of CFS against Gram-positive bacteria show statistically significant differences (repeated measures ANOVA df= 2; F= 6,095; P= 0.007832). Only Tukey’s paired test showed differences between the CFS of S. aureus (P= 0,0364) and E. faecalis (0,0096), when contrasted against the CFSs of B. cereus.
3.4. Leishmanicidal activity of secondary metabolites in the extracts
The E. hormaechei metabolic extract showed activity (CE-50**μg/ml = 47.7 + 3.8) against Le. braziliensis metacyclic promastigotes (UA301), using the MTT method to determine the viability of the parasites (Table 5). None of the evaluated extracts and drugs used in the effectiveness evaluation showed any activity against metacyclic promastigotes of the Le. infantum (BCN) strain, indicating that the half maximal effective concentration (EC50) of these extracts is higher than 400 μg/mL against the evaluated strain, i.e., it is higher than the maximum evaluated concentration (Table 5).
Table 5.
The median effective concentration (EC-50) of the bacterial extracts on metacyclic promastigotes of the Leishmania infantum (BCN-GFP)* and Leishmania braziliensis (UA301-GFP)** strains.
| Leishmanicidal activity of methanolic bacteria extracts | ||
|---|---|---|
| methanol crude extract_Code | EC-50* (μg/ml) | EC-50** (μg/ml) |
| O. anthropi_102 | >400 | >400 |
| E. aerogenes_44 | >400 | >400 |
| P. otitidis_70 | >400 | >400 |
| E. hormaechei_157 | >400 | 47.7 + 3.8 |
| E. cloacae_71 | >400 | >400 |
| A. calcoaceticus_64 | >400 | >400 |
| P. ananatis_140 | >400 | >400 |
| A. gyllenberguii_ | >400 | >400 |
| P. aeruginosa_173 | >400 | >400 |
| E. cancerogenus_41 | >400 | >400 |
| L. soli_188 | >400 | >400 |
| P. putida_43 | >400 | >400 |
(μg/ml): micrograms per milliliter; EC-50: The median effective concentration. Leishmanicidal activity are showed in bold.
4. Discussion
In recent years, it has been reported an increase of microbial community studies based on sequenced amplicons from 16S rRNA gene on insect gut communities [34]. However, the knowledge of the intestinal microbiota function and its biologic potential could be useful for understand the role on metabolism, physiology, defense against pathogens, or in vector competence modulation.
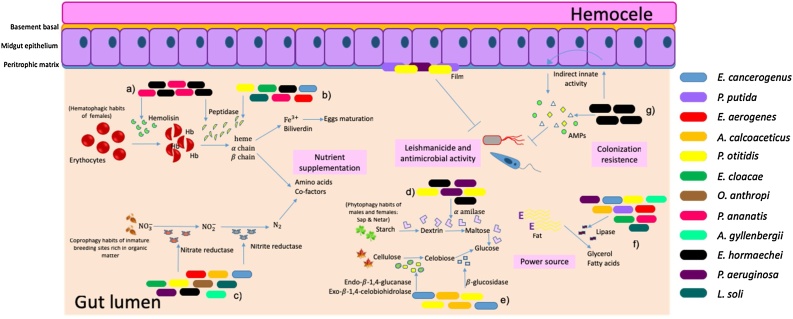
In this study, we report the antimicrobial, enzymatic, and leishmanicidal activities of 12 Gram-negative isolates, obtained from the intestine of wild populations of Lu. evansi, a known vector of visceral leishmaniasis in Colombia. The resulting information was used to infer probable functions of these bacteria in the Lu. evansi gut compartment (Fig. 2), besides providing aggregate value to secondary metabolite production, due the great biotechnological and pharmacological potential use.
Fig. 2.
Predictive model of the interactions and functions of Gram-negative bacteria in the intestine of Lutzomyia evansi, based on in vitro evaluation of enzymatic, antimicrobial, and leishmanicidal activity. a) Intestinal bacteria such as P. ananatis and E. hormaechei display hemolytic activity, suggesting that they might be related to erythrocyte lysis. b) Hemoglobin could be released and used by P. ananatis, E. hormaechei, P. otitidis, E. cloacae, E. cancerogenus, L. soli, and E. aerogenes for nutrient supplementation. Erythrocytes are also an useful source of iron for egg maturation. c) E. aerogenes, A. calcoaceticus, E. cancerogenus, E. cloacae, P. otitidis, O. anthropi, L. soli, E. hormaechei, and A. gyllenbergii are able to reduce nitrates to nitrites and/or molecular nitrogen, from decomposing organic matter, possibly during the development of the immature states of Lu. evansi. This process allows for the synthesis of some necessary amino acids for protein and glycoprotein synthesis, which could also permit formation of the peritrophic matrix in the adult phase. d) P. aeruginosa, E. hormaechei, and P. otitidis are responsible for carbohydrate hydrolysis, processing starch to obtain glucose and produce energy. e) Cellulose is hydrolyzed by P. otitidis, E. cancerogenus, and A. calcoaceticus as an additional source of glucose.) L. soli, P. aeruginosa, E. cancerogenus, P. putida, E. aerogenes, P. otitidis, and A. gyllenbergii showed lipolytic activity that might be associated with fat degradation from the fatty abdominal body, which is filled with nutrients and allows for development and/or reproduction, as fats are the precursors of the synthesis of most proteins and metabolites secreted into the hemolymph. g) E. hormaechei showed activity against metacyclic promastigotes of Leishmania braziliensis. The majority of Gram-negative bacteria show antimicrobial activity, which is a determining aspect in intestinal microbiota dynamics.
Proteases have a number of key roles in bacteria viability, stress response and pathogenicity., which provides them benefits over other microorganisms and enables them to survive in hostile or high pH environments into the insect intestine [35], [36]. In this study, P. ananatis, E. cancerogenus, E. cloacae, E. aerogenes, and E. hormaechei displayed proteolytic activity, which suggests a possible role in the degradation of protein into amino acids or small peptides during nutrition events or cell division processes [8] (Fig. 2). The secretion into the extracellular medium of a high variety of proteolytic enzymes such as serine proteases, metalloproteases, and bacteriocins has been reported by Enterobacter genera [37].
It was also shown a lipolytic activity associated to P. otitidis, P. aeruginosa, P. putida, E. cancerogenus, E. aerogenes, E. cloacae, and P. ananatis. In insects, the fat body carries out multiple metabolic activities, such as storage, energy production, circulating metabolite synthesis, and protein synthesis according to requirements (Arrase and Soulages, 2010; [36]. It has been established that bacteria from Pseudomonaceae and Enterobacteriaceae families act directly on lipid hydrolysis and glycolipid degradation in insects [38]; Anand et al., 2009; [36], as well as the degradation of microbial components (lipopolysaccharides, peptidoglycans) inside the digestive tract [9], [39].
A common physiological event of females insect vectors is erythrocyte lysis into the midgut that is required to produce eggs. Hematophagous arthropods are faced with a very particular situation concerned to heme metabolism, as well as the fact that they ingest many times their own mass in vertebrate blood [40]. gut isolates described in the study could participate in blood digestion, acting on haemoglobin degradation pathway, as demonstrated for P. ananatis and E. hormaechei that have shown complete hemolytic activity and probably increasing their abundance after the blood intake.
During this process, massive liberation of the heme group can occur (Fig. 2), a known pre-oxidant molecule, whose internal regulation can produce reactive oxygen species (ROS) and concomitant Fe2+ liberation during this process [41], [42]. Reactive oxygen species are beneficial in low and moderate conditions within cellular processes [42]. Heme to biliverdin and carbon monoxide degradation (CO) definitely has a crucial role in pigmentation and can protect insects against the oxidative damage induced by light [43]. The amount and composition of blood nourishment is the main factor that affects vitellogenesis and egg production in sand flies [9], [44], [45]. However, feeding preferences and digestive patterns of the blood of each species can depend on the bacterial microbiota.
All isolated strains were tested for cellulolytic and aminolytic activities (Fig. 2), as shown in Table 2. However, it is necessary to highlight P. otitidis for having these two enzymatic activities and the reductase nitrate capacity, which suggests that it is useful in nutrient digestion in Lu. evansi. In general, phlebotominae have coprophagous habits, i.e. larvae forms of Lu. evansi develop in places with abundant decomposing organic matter [46]. We also found that most of the cellulolytic bacteria belonging to the family Enterobacteriaceae. In some insects, like Bombyx mori, it has been concluded that the number of cellulolytic bacteria increases with each urge [47].
Both males and females Phlebotomine in their adult state have phytophagous habits, associated with the ingestion of sugar from sap, plant phylloplanes, or from leaves that are rich in protein and water [36], [48], [49], [50], [51]. This suggests that amylolytic bacteria play a role only in the initial stages of digestion, which includes the decomposition of complex sugar polymers into dimers or oligomers (Fig. 2).
Maladjustment in the content of nitrogen in phytophagous insects and their host plants has been recognized for many years as a critical factor that influences hematophagous insect success in their adult state when blood sources are insufficient [52], [53]. In the majority of plants, nitrogen, from the sap of both the xylem and the phloem, is dominated by non-essential amino acids; essential amino acids represent less than 20% of the total. The majority of Gram-negative bacteria were nitrate reductase positive, and some generated molecular nitrogen (Fig. 2), which is important on many occasions for the formation of amino acids. Various ecological and evolutionary factors (trophic level, feeding style, body size, and phylogeny) may contribute to variations in the N content in insects that spend part of their life cycle in terrestrial substrates [54]. However, many insects obtain a nutritional advantage from persistent associations with the intestinal microbiota, which synthesizes various nutrients, digest and detoxify ingested foods, and provide essential amino acids [8], [36].
A suggested hypothesis is that the nutritional contributions of the intestinal microbiota represented by Gram-negative bacteria in Lu. evansi might occur in different ways: 1) improved digestion efficiency, 2) optimized ability to live with a suboptimal diet, 3) acquisition of digestive enzymes, and 4) vitamin supply. Secondary metabolites extracts derived from Gram-negative bacteria exhibited similar antimicrobial activity patterns against reference bacteria. The diameters of the inhibitory halos were similar to the activity detected for complete bacteria of P. ananatis, E. cloacae, and O. anthropi isolated from Lu. evansi [4]. E. hormaechei, P. ananatis, A. calcoaceticus, and E. cancerogenus bacteria presented the strongest antimicrobial activity. It has been suggested that these bacteria might use a type 4 secretion system (T4SS) that gives them advantages in terms of microbial competence, antagonist effectors, and antibiotic resistance [55], [56]. Unlike other Gram-negative isolates found in the Pseudomonaceae family that showed antimicrobial activity, they may use a type 2 (T2SS) or type 6 secretion system (T6SS) [57], [58], [59].
As far as we know, this is the first time that the antimicrobial activity of cell-free supernatants (CFS) of bacteria associated to the intestinal tract of phlebotominae has been evaluated. This characterization allows for a more complete spectrum of bioactive molecules produced by Gram-negative bacteria [60]. The antimicrobials produced in the CFS, presumably at variable levels and at different times depending on factors such as cellular density, intercellular signaling, and the variable composition of the intestine environments of the insect [61]. Significant antimicrobial activity was obtained regarding P. aeruginosa, P. ananatis, and E. aerogenes over S. marcescens. The inhibiting action of the CFS produced by bacteria tends to be very stable against the action of proteases [62] and some metabolites are resistant to heat degradation, i.e. non-protein molecules can also exist. This activity may confer to Lu. evansi the ability to adapt to several ecological niches. In general, supernatant could comprise bacteriocins, which may have inhibitory action against groups of Gram-positive and Gram-negative bacteria [62], [63].
In our study, the E. hormaechei metabolic extract showed leishmanicidal activity against Le. braziliensis. This bacterium is found within the E. cloacae complex and is spread by horizontal transference. E. hormaechei metabolomics approach to identify secondary metabolites that causes leishmanicidal activity is justified as a technological prospective. E. hormaechei may have enzymes such as phospholipase C, cytolysins, or type IV (T4SS) and VI (T6SS) secretion systems [55], [59], [64], [65] linked to virulence factors, with direct effects on the surface of some prokaryotes (Telleira et al., 2018). Recently, it has been found that the use of complete Gram-negative bacteria of the P. ananatis, E. cloacae, and O. antropi species may modulate the development of procyclic and metacyclic promastigotes of Le. infantum [4] in in vitro assays. [16], found that Pseudozyma sp., Asaia sp., and O. intermedium reduced the number of females infected with Le. mexicana promastigotes using an experimental infection model in adults of L. longipalpis. This effect was possibly caused by the secretion of extracellular metabolites [16], [66], [67].
Unfortunately, the results from methanolic extracts of the twelve Gram-negative isolates indicated that neither of these showed inhibitory activity on Le. infantum, which suggests that the antiparasitic mechanisms of these bacteria against this parasite species might be associated with the action of complete cells, as evidenced by [4] in a previous study. It has also been described that L. infantum is highly resistant to various antimonial compounds and/or pharmaceutical derivates [68], [69]. In many cases, antimicrobials are also believed to be produced in subinhibitory concentrations in natural environments or used in relatively high concentrations to study the activity of antimicrobials in the laboratory [33]. A complementary analysis of cytotoxicity will be developed as a step the in vitro test.
Finally, we can conclude that intestinal bacteria associated with insect vectors have the ability to adapt to changes in the insect’s diet due to the induction of enzymes or changes in the microbial community population. In this study, we suggest Gram-negative bacteria may provide digestive enzymes in synergy, and they may contribute to larvae growth, ingested blood digestion, complex carbohydrate breakdown, and the generation of the components required for protein synthesis, among other activities. Nevertheless, it is unclear how in vitro results obtained here are related to in vivo situation. Thus, the relative functions of the enzymes produced endogenously by Lu. evansi need to be studied further because they might be targets for the strategic design of control measures against pathogenic bacteria, the Leishmania parasite, or the insect vector. Subsequently, it may be possible to conduct studies investigating antibacterial synergy. Lastly, P. ananatis and E. hormaechei are promising strains with great biotechnological potential due to their excellent antimicrobial and enzymatic activity.
Funding information
The present study was funded by the Foundation for the Promotion of Research and Technology, Bank of the Republic of Colombia Project No. 4.124; the Administrative Department of Science, Technology and Innovation (COLCIENCIAS code FP44842-128-2018; Quipu Code 201010021937; HERMES 40051 Postdoctoral call; National Call 528 for doctoral studies in Colombia, 2011); Strengthening of Research, Creation and Innovation 2016-2018, Universidad Nacional de Colombia (Project Code: 36012; 2010100-Basic Research).
Author’s contributions
RJV, CXMH, GECR, SMR: Designed the study and contributed to writing the manuscript.
RJV, GBD: Analyzed the data, performed the experiments and contributed to writing the manuscript.
Declaration of Competing Interest
Authors have no conflict of interest to declare
Acknowledgements
Foundation for the promotion of research and technology, Bank of the Republic of Colombia; Project No. 4.124; Administrative Department of Science, Technology and Innovation (COLCIENCIAS code FP44842-128-2018; Quipu Code 201010021937; HERMES 40051 Postdoctoral call); Strengthening of the research, creation and Innovation 2016-2018, Universidad Nacional de Colombia (Projet Code: 36012; 2010100-Basic Research). Special recognition to Tatiana Pineda, Eliany Yised Henao and Daniel Lozada, for their technical collaboration.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.btre.2019.e00379.
Contributor Information
Rafael J. Vivero, Email: rjviverog@unal.edu.co.
Gustavo Bedoya Mesa, Email: guabedoyame@unal.edu.co.
Sara M. Robledo, Email: sara.robledo@udea.edu.co.
Claudia Ximena Moreno Herrera, Email: cxmoreno@unal.edu.co.
Gloria Cadavid-Restrepo, Email: gecadavi@unal.edu.co.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Overral L., Rebek E. Insect vectors and current management strategies for diseases caused by Xylella fastidiosa in the Southern United States. J. Integr. Pest Manag. 2017;8(1):1–12. [Google Scholar]
- 2.Wilke A., Toledo M. Paratransgenesis: a promising new strategy for mosquito vector control. Parasit. Vectors. 2015;8:342. doi: 10.1186/s13071-015-0959-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelly P., Bahr S., Serafim T., Ajami N., Petrosino J., Meneses C. The gut microbiome of the vector Lutzomyia longipalpis is essential for survival of leishmania infantum. MBio. 2017;8:e01121–16. doi: 10.1128/mBio.01121-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vivero R., Restrepo G.E., Herrera C., Ospina V., Uribe S.I., Robledo S. Antagonistic effect of Bacteria Isolated from the digestive tract of Lutzomyia evansi against promastigotes of leishmania infantum, antimicrobial activities. Adv. Microbiol. 2016;6(10):760–775. [Google Scholar]
- 5.Heerman M., Weng J.L., Hurwitz I., Durvasula R., Ramalho-Ortigao M. Bacterial infection and immune responses in Lutzomyia longipalpis sand fly larvae midgut. PLoS Negl. Trop. Dis. 2015;9(7) doi: 10.1371/journal.pntd.0003923. e0003923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McQuade R., Stock S.P. Secretion systems and secreted proteins in gram-negative entomopathogenic Bacteria: their roles in insect virulence and beyond. Insects. 2018;9(2):68. doi: 10.3390/insects9020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Azambuja P., Garcia E.S., Ratcliffe N.A. Gut microbiota and parasite transmission by insect vectors. Trends Parasitol. 2005;21:568–572. doi: 10.1016/j.pt.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 8.Douglas The microbial dimension in insect nutritional ecology. Funct. Ecol. 2009;58:38–47. [Google Scholar]
- 9.Gaio A.D., Gusmão D.S., Santos A., Berbert-molina M.A., Pimenta P.F., Lemos F.J. Contribution of midgut bacteria to blood digestion and egg production in Aedes. Parasit. Vectors. 2011;14(4):105. doi: 10.1186/1756-3305-4-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dostálová A., Volf P. Leishmania development in sand flies : parasite-vector interactions overview. Parasit. Vectors. 2012;5:1–12. doi: 10.1186/1756-3305-5-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eleftherianos I., Atri J., Accetta J., Castillo J.C. Endosymbiotic bacteria in insects : guardians of the immune system? Front. Physiol. 2013;4:1–10. doi: 10.3389/fphys.2013.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Müller U., Vogel P., Alber G., Schaub G. The Innate Immune System of Mammals and Insects. Contrib. Microbiol. 2008;15:21–44. doi: 10.1159/000135684. [DOI] [PubMed] [Google Scholar]
- 13.Baxter R., Contet A., Krueger K. Arthropod innate immune systems and vector-borne diseases. Biochemistry. 2017;56:907–918. doi: 10.1021/acs.biochem.6b00870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weiss B., Aksoy S. Microbiome influences on insect host vector competence. Trends Parasitol. 2011;27:514–522. doi: 10.1016/j.pt.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krishnan M., Bharathiraja C., Pandiarajan J., Prasanna V., Rajendhran J., Gunasekaran P. P. Review Insect gut microbiome - An unexploited reserve for biotechnological application. Asian Pac. J. Trop. Biomed. 2014;4:16–21. doi: 10.12980/APJTB.4.2014C95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sant’Anna M., Diaz H., Aguiar K., Al S., Cavalcante R., Dillon M., Bates A., Genta A., Dillon J. Colonisation resistance in the sand fly gut: Leishmania protectsLutzomyia longipalpis from bacterial infection. Parasit. Vectors. 2014;23(7):329. doi: 10.1186/1756-3305-7-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kazanas N. Proteolytic activity of microorganisms isolated from freshwater fish. Appl. Microbiol. 1968;16:128–132. doi: 10.1128/am.16.1.128-132.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pereira M., Ribeiro D., Marques D., Freitas R. Proteolytic activities of bacteria, yeasts and filamentous fungi isolated from coffee fruit (Coffea arabica L.) Acta Sci. Agron. 2011;33:457–464. [Google Scholar]
- 19.Ramnath L., Sithole R. Govinden Identification of lipolytic enzymes isolated from bacteria indigenous to Eucalyptus wood species for application in the pulping industry. Biotechnol. Rep. 2017;15:114–124. doi: 10.1016/j.btre.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silva-Bedoya, Silva-Bedoya L.M., Sánchez-Pinzón M.S., Cadavid-Restrepo G.E., Moreno-Herrera C.X. Bacterial community analysis of an industrial wastewater treatment plant in Colombia with screening for lipid-degrading microorganisms. Microbiol. Res. (Pavia) 2016;192:313–325. doi: 10.1016/j.micres.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 21.MacFaddi J. Williams & Wilkins Co. Lippincott Williams and Wilkins; 2003. Biochemical Tests for Identification of Medical Bacteria. 912 p. [Google Scholar]
- 22.Teather R.M., Wood P.J. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl. Environ. Microbiol. 1982;43(4):777–780. doi: 10.1128/aem.43.4.777-780.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zamani H., Salehzadeh A., Abdolhosseini M. Isolation and molecular identification of a cellulotic bacterium from muncipal waste and investigation of its cellulase production. Biosci. J. 2018;34(3) [Google Scholar]
- 24.Lányi B. Classical and rapid identification methods for medically important Bacteria. Methods Microbiol. 1987;19:1–67. [Google Scholar]
- 25.Ali-Vehmas T., Vikerpuur M., Pyörälä S., Atroshi F. Characterization of hemolytic activity of Staphylococcus aureus strains isolated from bovine mastitic milk. Microbiol. Res. 2001;155:339–344. doi: 10.1016/S0944-5013(01)80013-6. [DOI] [PubMed] [Google Scholar]
- 26.Short E.C., Kurtz H.J. Properties of the hemolytic activities of Escherichia coli. Infect. Immun. 1971;3:678–687. doi: 10.1128/iai.3.5.678-687.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Semedo T., Santos M.A., Martins P., Lopes M.F., Figueiredo J.J., Tenreiro R., Barreto M.T. Comparative study using type strains and clinical and food isolates to examine hemolytic activity and occurrence of the cyl operon in enterococci. J. Clin. Microbiol. 2003;41:2569–2576. doi: 10.1128/JCM.41.6.2569-2576.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sierra-García N., Romero-Tabarez M., Orduz-Peralta S. Determinación de la actividad antimicrobiana e insecticida de extractos producidos por bacterias aisladas del suelo. Acta Biol. 2012;34:5–19. [Google Scholar]
- 29.Sangnoi Y., Srisukchayakul P., Arunpairojana V., Kanjana-Opas A. Diversity of marine gliding bacteria in Thailand and their cytotoxicity. Electron. J. Biotechnol. 2009;12:1–8. [Google Scholar]
- 30.Balouiri M., Sadiki M., Ibnsouda S.K. Methods for in vitro evaluating antimicrobial activity : A review. J. Pharm. Anal. 2016;6:71–79. doi: 10.1016/j.jpha.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leyton A., Urrutia H., Vidal J.M., Fuente M., Aroca G., González-Rocha G., Sossa K. Actividad inhibitoria del sobrenadante de la bacteria Antártica Pseudomonas sp. M19B en la formación de biopelículas de. Flavobacterium psychrophilum. 2015;50(2):375–381. 19749. [Google Scholar]
- 32.Elshikh M., Ahmed S., Mcgaw M., Marchant R., Funston S., Dunlop P., Banat I.M. Resazurin-based 96-well plate microdilution method for the determination of minimum inhibitory concentration of biosurfactants. Biotechnol. Lett. 2016;38:1015–1019. doi: 10.1007/s10529-016-2079-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez A., Robledo S., Muñoz D., Blair S., Higuita E., Echeverri E. Antiparasitic activity of methanol extracts and isolated fractions from Caribbean Sponges. Vitae. Rev. Fac. Cienc. Quim. Quim. Farm. 2001;8:71–81. [Google Scholar]
- 34.Engel P., Moran N. The gut microbiota of insects – diversity in structure and function. FEMS Microbiol. Rev. 2013;37(5):699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- 35.Arrese E., Soulages J. Insect fat body: energy, metabolism, and regulation. Annu. Rev. Entomol. 2010;55:207–225. doi: 10.1146/annurev-ento-112408-085356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Assis W., Malta J., Pimenta P., Ramalho-Ortigão J., Martins G. The characterization of the fat bodies and oenocytes in the adult females of the sand fly vectors Lutzomyia longipalpis and Phlebotomus papatasi. Arthropod Struct. Dev. 2014;43(5):501–509. doi: 10.1016/j.asd.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 37.Akhoundi M., Bakhtiari R., Guillard T., Baghaei A., Tolouei R. Diversity of the bacterial and fungal microflora from the midgut and cuticle of phlebotomine sand flies collected in North-Western Iran. PLoS One. 2012;7:1–10. doi: 10.1371/journal.pone.0050259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Azevedo T.R., Terra W.R., Ferreira C. Purification and characterization of three β- glycosidases from midgut of the sugar cane borer, Diatraea saccharalis. Insect Biochem. Mol. Biol. 2003;33:81–92. doi: 10.1016/s0965-1748(02)00179-0. [DOI] [PubMed] [Google Scholar]
- 39.Iliev I., Trifonova S. Lipolytic activity of genus Pseudomonas. J. Biosci. Biotechnol. Discov. 2012;1314:163–168. [Google Scholar]
- 40.Lara F., Lins U., Bechara G., Oliveira P. Tracing heme in a living cell: hemoglobin degradation and heme traffic in digest cells of the cattle tick Boophilus microplus. J. Exp. Biol. 2005;208:3093–3101. doi: 10.1242/jeb.01749. [DOI] [PubMed] [Google Scholar]
- 41.Paiva-Silva G., Cruz-Oliveira C., Nakayasu E., Maya-Monteiro C., Dunkov B., Masuda H. A heme-degradation pathway in a blood-sucking insect. Proc. Natl. Acad. Sci. U.S.A. 2006;103(21):8030–8035. doi: 10.1073/pnas.0602224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ariza M.M. Estrés oxidativo: origen, evolución y consecuencias de la toxicidad del oxígeno. Nova. 2012;10:213–225. [Google Scholar]
- 43.Graça-Souza, Graça-Souza A.V., Maya-Monteiro C., Paiva-Silva G.O., Braz G.R., Paes M.C., Sorgine M.H., Oliveira P.L. Adaptations against heme toxicity in blood-feeding arthropods. Insect Biochem. Mol. Biol. 2006;36:322–335. doi: 10.1016/j.ibmb.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 44.Koller C.N., Raikhel A.S. Initiation of vitellogenin uptake and protein synthesis in the mosquito (Aedes aegypti) ovary in response to a blood meal. J. Insect Physiol. 1991;37:703–711. [Google Scholar]
- 45.Nieves E., Oliveros J., Rondon M. Impacto de Leishmania amazonensis e Sangue de Ave no potencial Biológico e fertilidade de Lutzomyia migonei e Lutzomyia ovallesi (Diptera: psychodidae) EntomoBrasilis. 2011;4(4):20–25. [Google Scholar]
- 46.Vivero R., Torres-Gutierrez C., Bejarano E., Cadena H., Estrada L., Florez F. Study on natural breeding sites of sand flies (Diptera: phlebotominae) in areas of Leishmania transmission in Colombia. Parasit. Vectors. 2015;8:116. doi: 10.1186/s13071-015-0711-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anand A., Vennison S., Sankar S., Gilwax D., Thirumalai P. Isolation and characterization of bacteria from the gut of Bombyx mori that degrade cellulose, xylan, pectin and starch and their impact on digestion. J. Insect Sci. 2010;10:107. doi: 10.1673/031.010.10701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murakami A. Genetic studies on tropical races of silkworm (Bombyx mori), with special reference to cross breeding strategy between tropical and temperate races. Jpn. Agric. Res. Q. 1989;23(1):127–133. [Google Scholar]
- 49.Cavalcante R., Pereira M., Freitas J., Gontijo N. Ingestion of saliva during carbohydrate feeding by Lutzomyia longipalpis (Diptera; Psychodidae. Mem. Inst. Oswaldo Cruz. 2006;101(1):85–87. doi: 10.1590/s0074-02762006000100016. [DOI] [PubMed] [Google Scholar]
- 50.Dillon R.J., Dillon V.M. The gut bacteria of insects: Nonpathogenic interactions. Annu. Rev. Entomol. 2004;49:71–92. doi: 10.1146/annurev.ento.49.061802.123416. [DOI] [PubMed] [Google Scholar]
- 51.Abbasi I., Queiroz A., Kirstein O., Nasereddin A., Horwitz B., Hailu A. Plant-feeding phlebotomine sand flies, vectors of leishmaniasis, prefer Cannabis sativa. PNAS. 2018;115:11790–11795. doi: 10.1073/pnas.1810435115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.White T. Springer-Verlag; Berlin: 1993. The Inadequate Environment: Nitrogen and the Abundance of Animals. pp 425, Google Scholar. [Google Scholar]
- 53.Broadway R., Duffey S. The effect of plant protein quality on insect digestive physiology and the toxicity of plant proteinase inhibitors. J. Insect Physiol. 1988;34:1111–1117. [Google Scholar]
- 54.Fagan W., Siemann E., Mitter C., Denno R., Huberty A., Woods H., Elser J. Nitrogen in insects: implications for trophic complexity and species diversification. Am. Nat. 2002;160(6):784–802. doi: 10.1086/343879. [DOI] [PubMed] [Google Scholar]
- 55.Liu W., Wong C., Chung K.M., Jiang J. F.C. Comparative genome analysis of bacter cloacae. PLoS One. 2013;8:1–15. doi: 10.1371/journal.pone.0074487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chudasama K.S., Thaker V.S. Genomics Data Genome sequence of Ochrobactrum anthropi strain SUBG007, a plant pathogen and potential xenobiotic compounds degradation bacterium. Genom. Data. 2017;11:116–117. doi: 10.1016/j.gdata.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Putker F., Boxtel R.T., Stork M., Rodríguez-herva J.J., Koster M., Tommassen J. The type II secretion system (Xcp) of Pseudomonas putida is active and involved in the secretion of phosphatases. Environ. Microbiol. 2013;15:2658–2671. doi: 10.1111/1462-2920.12115. [DOI] [PubMed] [Google Scholar]
- 58.Costa T.R., Felisberto-rodrigues C., Meir A., Prevost M.S., Redzej A., Trokter M., Waksman G. Secretion systems in Gram-negative insights. Nat. Publ. Gr. 2015;13:343–359. doi: 10.1038/nrmicro3456. [DOI] [PubMed] [Google Scholar]
- 59.Shyntum Y., Theron J., Venter S.N., Moleleki L.N., Toth I.K., Coutinho T.A. Pantoea ananatis utilizes a type VI secretion system for pathogenesis and bacterial competition. Mol. plan-Microbe Interact. 2015;28:420–431. doi: 10.1094/MPMI-07-14-0219-R. [DOI] [PubMed] [Google Scholar]
- 60.Makarova O., Rodriguez- Rojas A., Eravci M., Weise C., Dobson A., Johnston P., Rolff J. Antimicrobial defence and persistent infection in insects revisited. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 2016;26(371) doi: 10.1098/rstb.2015.0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Singh S., Orr D., Divinagracia E., McGraw J., Dorff K., Forst E. Role of secondary metabolites in establishment of the mutualistic partnership between Xenorhabdus nematophila and the entomopathogenic nematode Steinernema carpocapsae. Appl. Environ. Microbiol. 2015;81(2):754–764. doi: 10.1128/AEM.02650-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mariam S., Zegeye N., Tariku T., Andargie E., Endalafer N., Aseffa A. Potential of cell-free supernatants from cultures of selected lactic acid bacteria and yeast obtained from local fermented foods as inhibitors of Listeria monocytogenes, Salmonella spp. And Staphylococcus aureus. BMC Res. Notes. 2014;7:606. doi: 10.1186/1756-0500-7-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dobson A.J., Johnston P.R., Vilcinskas A., Rolff J. Identification of immunological expressed sequence tags in the mealworm beetle Tenebrio molitor. J. Insect Physiol. 2012;58:1556–1561. doi: 10.1016/j.jinsphys.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 64.Cox C.R., Coburn P.S., Gilmore M.S. Enterococcal Cytolysin : A Novel Two Component Peptide System that Serves as a Bacterial Defense Against Eukaryotic and Prokaryotic Cells. Curr. Protein Pept. Sci. 2005;6:77–84. doi: 10.2174/1389203053027557. [DOI] [PubMed] [Google Scholar]
- 65.Rossignol G., Merieau A., Guerillon J., Veron W., Lesouhaitier O., Feuilloley M.G.J., Orange N. Involvement of a phospholipase C in the hemolytic activity of a clinical strain of Pseudomonas fluorescens. BMC Microbiol. 2008;8:1–14. doi: 10.1186/1471-2180-8-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Diaz-Albiter H., Sant’Anna M., Genta F. R. Dillon Reactive Oxygen Species-mediated Immunity againstLeishmania mexicana and Serratia marcescens in the Phlebotomine Sand Fly Lutzomyia longipalpis. J. Biol. Chem. 2012;287(28):23995–24003. doi: 10.1074/jbc.M112.376095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Telleria E., Martins-da-Silva A., Tempone J., Traub-Csekö Y. Leishmania, microbiota and sand fly immunity. Parasitology. 2018;145(10):1336–1353. doi: 10.1017/S0031182018001014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ashutosh S., Sundar N. Goyal Molecular mechanisms of antimony resistance in Leishmania. J. Med. Microbiol. 2007;56:143–153. doi: 10.1099/jmm.0.46841-0. [DOI] [PubMed] [Google Scholar]
- 69.Mondelaers A., Sanchez-Cañete M.P., Hendrickx S., Eberhardt E., Garcia-Hernandez R., Lachaud L. Genomic and molecular characterization of miltefosine resistance in Leishmania infantum strains with either natural or acquired resistance through experimental selection of intracellular amastigotes. PLoS One. 2016;11 doi: 10.1371/journal.pone.0154101. e0154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.