Abstract
Background
Early detection of influenza activity followed by timely response is a critical component of preparedness for seasonal influenza epidemic and influenza pandemic. However, most relevant studies were conducted at the regional or national level with regular seasonal influenza trends. There are few feasible strategies to forecast influenza activity at the local level with irregular trends.
Methods
Multi-source electronic data, including historical percentage of influenza-like illness (ILI%), weather data, Baidu search index and Sina Weibo data of Chongqing, China, were collected and integrated into an innovative Self-adaptive AI Model (SAAIM), which was constructed by integrating Seasonal Autoregressive Integrated Moving Average model and XGBoost model using a self-adaptive weight adjustment mechanism. SAAIM was applied to ILI% forecast in Chongqing from 2017 to 2018, of which the performance was compared with three previously available models on forecasting.
Findings
ILI% showed an irregular seasonal trend from 2012 to 2018 in Chongqing. Compared with three reference models, SAAIM achieved the best performance on forecasting ILI% of Chongqing with the mean absolute percentage error (MAPE) of 11·9%, 7·5%, and 11·9% during the periods of the year 2014–2016, 2017, and 2018 respectively. Among the three categories of source data, historical influenza activity contributed the most to the forecast accuracy by decreasing the MAPE by 19·6%, 43·1%, and 11·1%, followed by weather information (MAPE reduced by 3·3%, 17·1%, and 2·2%), and Internet-related public sentiment data (MAPE reduced by 1·1%, 0·9%, and 1·3%).
Interpretation
Accurate influenza forecast in areas with irregular seasonal influenza trends can be made by SAAIM with multi-source electronic data.
Keywords: Influenza, Influenza-like illness, Forecast, AI, Multi-source electronic data
Research in context.
Evidence before this study
Influenza is an acute respiratory infection caused by influenza viruses circulating in the world, which poses a significant threat to public health and causes around 290,000–650,000 deaths worldwide each year. There is an urgent need for accurate and prompt forecasts of an impending influenza emergency. Traditionally, in epidemiology, influenza prediction is conducted through transmission mechanism models. The generation and collection of big data from different sources provide new features and approaches for influenza prediction. We searched Web of Science, PubMed, and IEEE Xplore Digital Library with the combination of terms: “influenza OR flu”, “prediction OR forecast”, “epidemic OR infectious disease”, “AI OR artificial intelligence”, and “big data”, in English and Chinese. To summarize the evidence, different sources of data have been collected and diverse algorithms have been utilized for tracking influenza activities and predicting the outbreak. Ginsberg et al. first used Google search queries to detect influenza epidemics in the United States from 2007 to 2008. Since then, various sources of electronic data, especially the Internet-based data, have been harnessed to track illness activities. Paul et al. developed an influenza forecast model by combining ILI data with Twitter data, and a reduction of 17–30% in forecast errors was observed when compared to the baseline model with historical ILI data solely. Wikipedia access logs are another tentative data source for optimizing influenza predictions. Electronic health record data and participatory disease reports are also utilized to broaden the range and depth of surveillance information. A variety of machine learning algorithms have also been applied to ILI estimation, including the autoregressive integrated moving average (ARIMA), Lasso, Support Vector Regression (SVR), AdaBoost, and deep learning techniques. However, only limited methods adaptive to influenza forecast could capture both the seasonal pattern and the irregular variations of ILI efficiently. Additionally, few studies have focused on improving influenza forecast accuracy by taking advantage of multi-source data.
Added value of this study
Based on the feasibility of novel Internet data and big data modelling techniques, we developed an approach that accurately estimates the influenza activity one week ahead of official public health reports with data from multiple sources, and applied the developed machine learning-based methodology in Chongqing to validate our model. Here, we provided a feasible alternative for forecasting ILI in regions with irregular seasonal trends and named it Self-Adaptive AI Model (SAAIM). Trained with multi-source data, our model was able to forecast the influenza epidemics of Chongqing retrospectively in an out-of-sample way, and achieved a Mean Absolute Percentage Error (MAPE) of 11·9% between 2014 and 2016. Furthermore, the model continued to perform well on the test set of the year 2017 (MAPE 7·5%). To be noted, real-time ILI forecasting was conducted in Chongqing in 2018 and validated the accuracy and efficiency of the model in practice (MAPE 11·9%). To our knowledge, this is the first real-time AI-based influenza forecast model in China, which can provide accurate real-time influenza estimates in a city with irregular seasonal influenza trends.
Implications of the available evidence
SAAIM not only retains the periodic pattern of the ILI% time series, but also captures the incidental fluctuations through modelling with exogenous multi-source predictors. Our study indicates that the combination of XGBoost and seasonal autoregressive integrated moving average (SARIMA) models with the proposed self-adaptive method could accurately forecast influenza activities. As indicated by the real-time application in Chongqing, our improved influenza prediction model could better prepare the authorities and public for the upcoming influenza epidemics and limit the catastrophic consequences of the epidemics.
Improvements on influenza forecast could arise from further analysis of influenza-related features which were identified by SAAIM. Besides, differences in geographical patterns, the spatial distribution of population density, and population migration may also affect how influenza spreads in a specific region. Thus, the accuracy of the forecast is expected to be improved with the input of geographic and demographic data.
Alt-text: Unlabelled Box
1. Introduction
Influenza epidemic is a persistent threat to global public health [[1], [2], [3]]. Seasonal influenza epidemic results in millions of respiratory illness and 290,000–650,000 deaths worldwide each year [3,4]. In addition, the risk of influenza pandemic persists because the constant genetic mutating may produce new strains of influenza virus against which no previous immunity exists in the population [2,4]. Therefore, surveillance and early detection of influenza activity followed by timely response are essential for preparedness for seasonal influenza epidemic and influenza pandemic [[5], [6], [7], [8], [9]]. However, owing to the time needed for processing data, traditional influenza surveillance publishes influenza-like illness (ILI) report with one to two week lag behind real time, which is far from optimal for decision making [5,6,10].
To alleviate this information gap, lots of attempts have been made for real-time estimation of influenza activity in the past decade. Google Flu Trends created a new era which used Google search data to predict ILI in the US [5,6]. Thereafter, multi-source electronic data including Internet search data [[10], [11], [12]], influenza surveillance data [10], influenza-related posts on Twitter [[13], [14], [15], [16]], Wikipedia access logs [17] and electronic health records [5,18] were integrated with mathematical models to track illness activities with very good predictive results.
However, most of these studies were done at the regional or national level with regular seasonal influenza trends, of which the estimates were hardly translated into actionable information for local health officials to make better decisions in cities [5]. Local influenza activities are not equivalent to those at the regional or national level, and local influenza epidemics exhibit more diverse seasonal patterns [19] and are more likely to fluctuate with local influenza-related factors, such as weather [20,21], economic and social activity [22,23], population immunity [24,25] and individual habits [26,27]. Although some studies had constructed models to improve forecasting accuracy in cities such as New York [28], Melbourne [29], and Hong Kong [30], efficient methods with great accuracy are still lacking.
Chongqing is one of the leading economic centres in south-western China, which covers approximately 82,400 km2 and has a population of about 30 million. It is a typical city that experiences highly irregular influenza epidemics annually, and the irregularity poses challenges for influenza forecasts. Additionally, the public health in Chongqing has been heavily affected by seasonal influenza A (H3N2) and A (H1N1) pdm09 in recent years [31]. It is notable that an avian influenza A (H7N9) virus emerged and resulted in human infections in Chongqing in 2017 [32]. The control and prevention of influenza is of great importance to the public health. Therefore, an applicable and suitable influenza forecasting method is in urgent need for real-time influenza epidemic response.
To address this critical public health issue, we developed a new Self-Adaptive AI Model (SAAIM) by integrating a time-series model and a nonlinear model through a self-adaptive AI weight adjustment mechanism. SAAIM can track seasonal patterns and irregular variations of ILI activity in Chongqing with local multi-source data including official influenza surveillance reports, weather information and Internet-based data. To our knowledge, this is the first real-time AI-based influenza forecast model in China, which can provide accurate real-time influenza estimates in a city with irregular seasonal influenza trends.
2. Materials and methods
2.1. Multi-source data collection
In this study, the forecasting target is the real-time weekly percentage of influenza-like illness (ILI%) in Chongqing, which is defined as the percentage of outpatients diagnosed with influenza-like illnesses among all outpatients each week. Based on literature review [5,6,[10], [11], [12],14,[17], [18], [19], [20], [21],29,30,[33], [34], [35]] and expert consultant, we collected multi-source data including historical ILI%, weather data, Baidu search index, and Sina Weibo data of Chongqing for model construction.
The ILI data of Chongqing was collected from Chinese surveillance system for influenza. ILI surveillance has been conducted in seven sentinel hospitals since October 2009, which were selected based on accessibility to patients, qualifications of medical staff, adequate specimen storage capacity, and the willingness of the physicians and nurses to participate voluntarily in the surveillance program. The hospitals included two urban comprehensive medical institutions (The First Affiliated Hospital of Chongqing Medical University and Banan Central Hospital), one urban paediatric hospital (Children's Hospital of Chongqing Medical University) and four rural comprehensive medical institutions (Qianjiang Central Hospital, Chongqing Three Gorges Central Hospital, Fuling Central Hospital and Yongchuan Hospital of Chongqing Medical University). ILI surveillance methods were described in a previous study [31].
Real-time weather and weather forecast data were collected from China Meteorological Administration (https://tianqi.911cha.com/). The Baidu Index is a statistical indicator that represents the search volume of demanding keywords or phrases based on Baidu's search query logs (https://index.baidu.com), which is the largest search engine in China. We summarized 81 influenza-related keywords that might correlate with the trend of influenza epidemics (Appendix Table S1). The keywords include early influenza symptoms, such as ‘sore throat’ and ‘dizzy’, relevant diagnoses, medicines, disease prevention and treatment, as well as phrases closely related to influenza, such as ‘immunity’ and ‘body temperature’. The Baidu Indexes used in this study were restricted to Chongqing municipality.
Sina Weibo is one of the most popular microblogging services in China. We tracked the number of daily tweets containing influenza-related keywords (63 in total) published from Chongqing municipality on Sina Weibo. The keywords were adapted from those we developed for Baidu Index, with some phrases infrequently seen on Weibo removed.
All the exogenous data were collected on a daily basis from 2012 to 2018 and included as input data. ILI% values of the previous three weeks, their averages, standard deviations, as well as the ILI% values in corresponding historical weeks of the past three years were also included in our model. Each kind of input data was converted to features for ILI forecasts (Appendix Table S1). We calculated the weekly maximums, minimums, averages, and variations for every numeric weather variable, and comparatively counted the weekly frequency of every categorical weather variable as weather features. In addition, we included the difference of weather features between the real-time week and previous weeks. For Baidu Index and Sina Weibo tweet counts, the weekly aggregate for each keyword was considered respectively. Moreover, the feature list included the year, month and week of the forecast week.
2.2. Model construction
SAAIM was constructed by integrating a time-series model and a nonlinear model. The time-series model is the SARIMA model, and the non-linear model is an optimized tree model called XGBoost. The final output of SAAIM is the self-adaptive weighted sum of base model results, and the weights were updated dynamically in the light of their historical forecast performance with the concept of Kalman Filter [36], as shown in Eq. (1). Thus, the output of SAAIM, or the posteriori estimate of the process state, was inclined to the base model that is currently more accurate.
| (1) |
In the equation, yA is the prediction of the SARIMA model at the week k, which is considered as a measurement of the state; yx is the prediction of XGBoost model at the week k, which is thought to be a prior estimate of the state; H represents the measurement gain in the Kalman Filter. The noisy measurement, i.e. the prediction of SARIMA in our study, is of the state itself, so H equals one [36]. Kk is the Kalman gain, which determines the weights of SARIMA and XGBoost in SAAIM. The iterative formula of Kk, which minimizes the posteriori estimate error covariance in Kalman Filter, is
| (2) |
where Pk− represents the covariance of the priori estimate calculated from
| (3) |
The historical prediction error variances of SARIMA and XGBoost were assigned to the measurement noise variance R and the process noise variance Q respectively. As the predictions of SARIMA performed more stable than XGBoost on time series data in our study, the priori estimate error covariance at the week k Pk− was estimated from incorporating the historical prediction error variance of XGBoost and the covariance of posteriori estimate (i.e. the prediction of the ensemble model SAAIM) error at previous moment Pk−1, which increased the weight of SARIMA to allow for the recent historical performance of the ensemble model. SAAIM iteratively updates the Kalman gain Kk using Eq. (2) to adjust the weights of base models on the basis of the historical performance (Appendix Page 1).
2.3. Feature selection
Primary feature selection included two steps. Before model training, features with only a single unique value (zero variation features) in the training dataset were identified and removed. The correlation analysis between individual features and ILI% was then conducted, and features with no significant correlation were further eliminated.
After the primary screening, different strategies of feature selection were used for XGBoost and SARIMA individually according to the principles of the models. Given that feature subsampling was used to prevent over-fitting in XGboost [37], we counted on the XGBoost model itself to select the important features during the training process. The importance threshold for selecting features in XGBoost was considered as a hyper-parameter which was determined by cross-validation on the training dataset.
Exogenous features were selected for SARIMA. Firstly, all retained features were fed to a LASSO regression model, and the features with the absolute value of average coefficients larger than 0·01 were kept. Then, the final exogenous features used in SARIMA model were determined by stepwise regression with the evaluation metric of the Akaike information criterion (AIC).
2.4. Model assessment
To validate the effectiveness of SAAIM for influenza forecasting, three additional models were constructed for comparison: (a) Lasso (Baidu_index), a Lasso regression model built with Baidu Index features solely, which was inspired by the idea of Google Flu Trends [6]; (b) Lasso (ILI + Baidu_index), a Lasso regression model that used historical ILI% values and Baidu Index features, which was derived from ARGO [10]; (c) Long Short-term Memory (LSTM), a state-of-the-art tool for long sequence modelling [38]. The related model parameters were described in the Appendix.
Furthermore, the estimates of SAAIM were compared with those generated by modified SAAIM with individual feature groups left out separately, including historical ILI% values, weather, and Internet-based public sentiment data (Baidu Index and Sina Weibo tweets), to validate the effectiveness of different data sources.
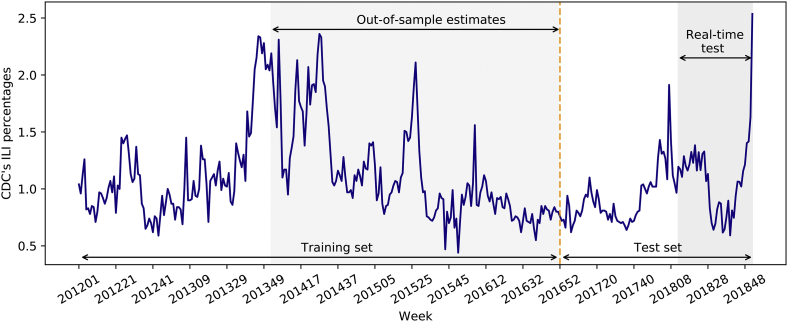
As for time series forecast, one-step-ahead rolling-origin-recalibration evaluation [39] was adopted in this study, so all the models were dynamically retrained weekly with updated data. Data from 2012 to 2016 were used as the training set. Retrospective estimates of influenza activity were performed between 2014 and 2016 in an out-of-sample fashion. In order to determine the optimized training strategy for each model, we tested all models with both a two-year rolling window and a fixed-origin expanding window in the light of a previous study [10]. For each model, the predictive performance was better when the MAPE value was smaller, and the corresponding training strategy was adopted. Based on the test results (Appendix Table S2), it turned out that LASSO and XGBoost models were more suitable to be trained with a two-year rolling window (i.e. data from the most recent 104 weeks) and a step size of one week, while LSTM and SARIMA models performed better with data from the first week of 2012 to the previous week of estimation. All the models were tested on a holdout validation period from 2017 to 2018. To be noted, SAAIM was applied to real-time forecast since the 12th week of 2018.
Four accuracy metrics were applied to evaluate the performance of the models: root mean square error (RMSE), mean absolute error (MAE), mean absolute percentage error (MAPE) and Pearson correlation coefficient (CORR).
In these equations, represents the estimation of the models at ith week and yi is the true value. As RMSE, MAE and MAPE approach zero and CORR approaches one, the result becomes more accurate.
2.5. Ethical considerations
The ILI surveillance protocol was approved by the National Health Commission (Previously called Ministry of Health) of the People's Republic of China as part of the monitoring of disease with epidemics. The study was approved by the Ethics Committee of the Chongqing Municipal Centre for Disease Control and Prevention. No experiment was done on humans or animals. All data was desensitized and would not be associated with any individual.
3. Results
3.1. Influenza activity in Chongqing
During the surveillance from 2012 to 2018, a total of 17,813,114 patient visits were recorded in the selected outpatient departments of the seven sentinel hospitals, with an average of 2,544,731 patient visits every year. Among these visits, 189,831 (1·1%) were ILI patients. ILI cases were reported throughout the year in Chongqing. However, ILI% showed an irregular seasonal trend (Fig. 1). Although ILI cases frequently peaked at the beginning and the middle of the year, they might peak twice within a short period of time. Moreover, there were shifts in peak times, outbreak intensity, duration, and time from onset to peak from year to year.
Fig. 1.
Time series of influenza-like illness percentages in Chongqing, China, 2012–2018.
3.2. Performance on influenza forecast by using SAAIM
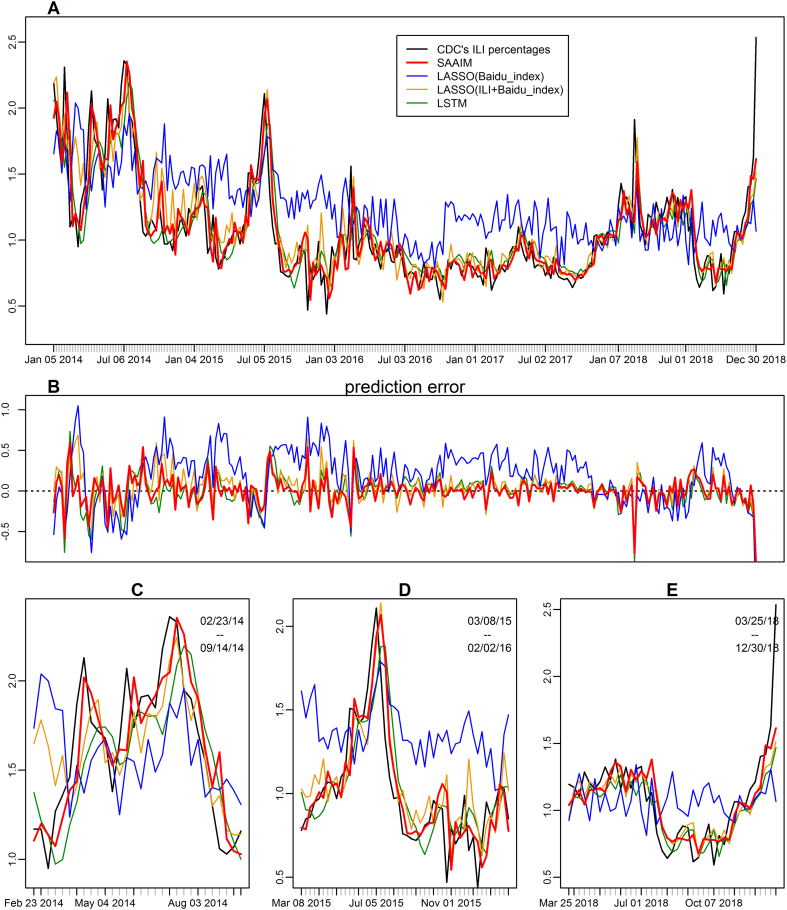
Overall, SAAIM produced smaller prediction error and less lag between the prediction and the true value (Fig. 2). Moreover, the estimates of SAAIM fitted the CDC-reported values better than other currently available models during influenza epidemic periods. SAAIM showed good approximate epidemic peak values (Fig. 2C) and the onset and end of the epidemic period (Fig. 2D) as well as the real-time forecasting period (Fig. 2E).
Fig. 2.
Estimation results of SAAIM in comparison of reference models. (A) The estimated ILI% values from SAAIM (thick red), comparing with the true CDC's ILI percentages (thick black) as well as the estimates from Lasso model with Baidu Index (blue), Lasso model with Baidu Index plus historical ILI% values(orange) and LSTM model (green) between the first week of 2014 and the last week of 2018. (B) The estimation error, defined as estimated value minus the CDC's ILI activity level. (C-E) Zoomed-in plots for estimation results in different study periods. (C) The 2014 flu season. (D) The 2015 flu season. (E) The real-time prediction of ILI percentages 1 week before official publication from March 25th, 2018 to December 30th, 2018. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
SAAIM uniformly outperformed all other tested models in the performance metrics consisting of RMSE, MAPE, MAE, and correlation (Table 1). During the period from 2014 to 2018, SAAIM (RMSE = 0·175, MAPE = 0·110, MAE = 0·117) reduced 17% RMSE, 26% MAPE, 23% MAE compared to the LASSO regression with ILI% values and Baidu index (RMSE = 0·211, MAPE = 0·149, MAE = 0·152), and 15% RMSE, 18% MAPE, 18% MAE compared to LSTM model (RMSE = 0·206, MAPE = 0·134, MAE = 0·143). Meanwhile, the influenza forecast of SAAIM achieved the highest correlation coefficient with the reported ILI activities. It should be noted that in the real-time influenza forecast application, SAAIM still had good accuracy metrics and performed best among all the tested models.
Table 1.
Performance metrics of SAAIM compared to reference models.
| 2014–2018 | 2014–2016 | 2017–2018 | 2014 | 2015 | 2016 | 2017 | 2018 | |
|---|---|---|---|---|---|---|---|---|
| RMSE | ||||||||
| SAAIM | 0.175 | 0.183 | 0.161 | 0.221 | 0.188 | 0.130 | 0.083 | 0.212 |
| LASSO (ILI + Baidu_index) | 0.211 | 0.230 | 0.179 | 0.286 | 0.223 | 0.163 | 0.101 | 0.232 |
| LASSO (Baidu_index) | 0.381 | 0.416 | 0.322 | 0.437 | 0.478 | 0.314 | 0.313 | 0.330 |
| LSTM | 0.206 | 0.218 | 0.187 | 0.273 | 0.219 | 0.142 | 0.098 | 0.246 |
| MAPE | ||||||||
| SAAIM | 0.110 | 0.119 | 0.097 | 0.114 | 0.144 | 0.099 | 0.075 | 0.119 |
| LASSO (ILI + Baidu_index) | 0.149 | 0.171 | 0.115 | 0.169 | 0.201 | 0.143 | 0.100 | 0.131 |
| LASSO (Baidu_index) | 0.340 | 0.375 | 0.288 | 0.283 | 0.508 | 0.331 | 0.351 | 0.225 |
| LSTM | 0.134 | 0.144 | 0.119 | 0.136 | 0.179 | 0.116 | 0.100 | 0.138 |
| MAE | ||||||||
| SAAIM | 0.117 | 0.128 | 0.101 | 0.165 | 0.134 | 0.086 | 0.062 | 0.140 |
| LASSO (ILI + Baidu_index) | 0.152 | 0.175 | 0.117 | 0.224 | 0.179 | 0.122 | 0.081 | 0.152 |
| LASSO (Baidu_index) | 0.314 | 0.356 | 0.251 | 0.373 | 0.427 | 0.268 | 0.268 | 0.234 |
| LSTM | 0.143 | 0.157 | 0.122 | 0.205 | 0.169 | 0.097 | 0.079 | 0.165 |
| Correlation | ||||||||
| SAAIM | 0.892 | 0.905 | 0.845 | 0.860 | 0.845 | 0.652 | 0.770 | 0.788 |
| LASSO (ILI + Baidu_index) | 0.843 | 0.855 | 0.799 | 0.762 | 0.792 | 0.475 | 0.684 | 0.732 |
| LASSO (Baidu_index) | 0.509 | 0.579 | 0.132 | 0.217 | 0.376 | 0.273 | −0.075 | 0.245 |
| LSTM | 0.843 | 0.858 | 0.789 | 0.784 | 0.773 | 0.552 | 0.735 | 0.729 |
Boldface highlights the best performance for each metric in each study period.
The prediction delay was quantified by the combination of the time-shift parameter, which describes how much the prediction curve slides along the time axis comparing with the true value curve, and the RMSE associated with the time-shift parameter (Appendix Page 1). LASSO (Baidu_index) model showed less time-shift compared to SAAIM on the training set, but a higher delay score due to the high RMSE of the model (Appendix Table S3 and Fig. S1). Collectively, SAAIM showed the smallest prediction delay score during the period of 2014–2016 (delay score = 0·136) and 2017–2018 (delay score = 0·052) respectively, and reached the overall delay score 0·122 from 2014 to 2018 (Appendix Table S3).
3.3. Contribution of different data sources to SAAIM
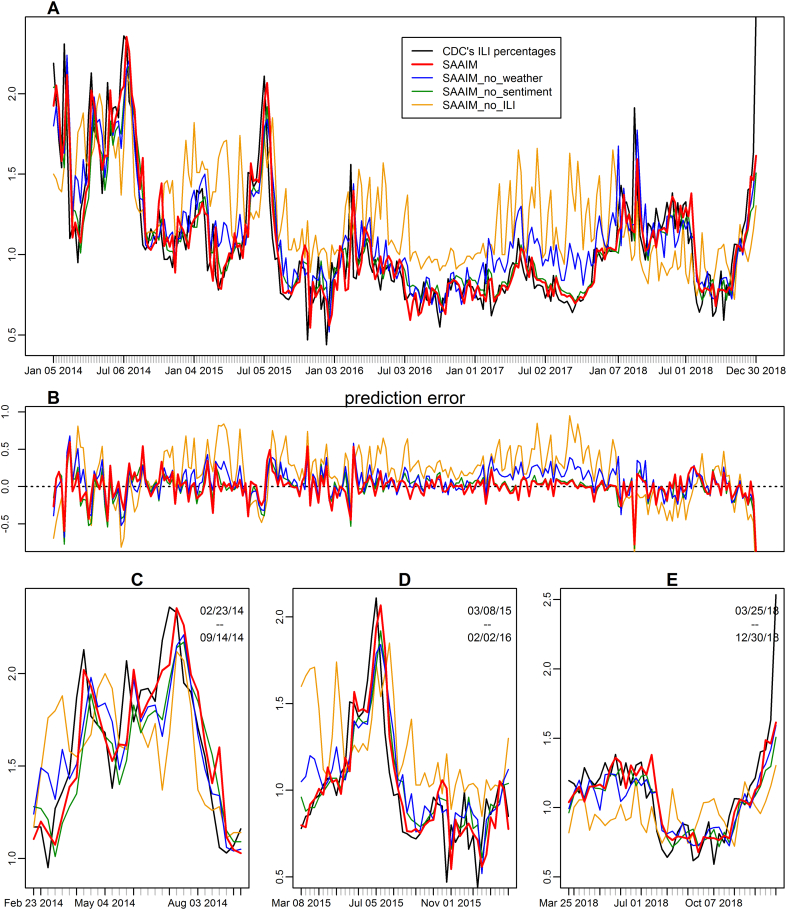
We evaluated the contribution of different data sources to SAAIM. Individual features were categorised into three feature groups, including ILI, weather, and public sentiment. Each group was removed respectively from SAAIM and the outputs were compared to original SAAIM (Table 2; Appendix Table S4 and Fig. S2). Time-series information (i.e. the historical ILI data) contributed the most to the prediction. Removal of historical ILI data resulted in a 119% increase in RMSE from 0·175 to 0·384 and 204% increase in MAPE from 0·110 to 0·334. Removal of weather features increased 25% and 53% respectively in terms of RMSE and MAPE, indicating that weather features, such as humidity and temperature variation (Appendix Fig. S3), are essential for accurate estimates of the subtle changes of influenza epidemics (Fig. 3). And a majority of weather features have been selected by XGBoost in our model as important features. The average temperature of the previous week, in particular, ranked second in terms of the contribution to the model performance (Appendix Fig. S3).
Table 2.
Performance metrics of prediction from SAAIM with different groups of features as input or not.
| 2014–2018 | 2014–2016 | 2017–2018 | 2014 | 2015 | 2016 | 2017 | 2018 | |
|---|---|---|---|---|---|---|---|---|
| RMSE | ||||||||
| SAAIM | 0.175 | 0.183 | 0.161 | 0.221 | 0.188 | 0.130 | 0.083 | 0.212 |
| SAAIM _no_weather | 0.218 | 0.214 | 0.214 | 0.260 | 0.214 | 0.154 | 0.213 | 0.236 |
| SAAIM _no_sentiment | 0.190 | 0.198 | 0.177 | 0.250 | 0.191 | 0.138 | 0.086 | 0.236 |
| SAAIM _no_ILI | 0.384 | 0.375 | 0.396 | 0.378 | 0.423 | 0.315 | 0.454 | 0.330 |
| MAPE | ||||||||
| SAAIM | 0.110 | 0.119 | 0.097 | 0.114 | 0.144 | 0.099 | 0.075 | 0.119 |
| SAAIM _no_weather | 0.169 | 0.152 | 0.194 | 0.135 | 0.193 | 0.126 | 0.246 | 0.141 |
| SAAIM _no_sentiment | 0.122 | 0.130 | 0.108 | 0.123 | 0.157 | 0.112 | 0.084 | 0.132 |
| SAAIM _no_ILI | 0.334 | 0.315 | 0.363 | 0.213 | 0.397 | 0.332 | 0.506 | 0.220 |
| MAE | ||||||||
| SAAIM | 0.117 | 0.128 | 0.101 | 0.165 | 0.134 | 0.086 | 0.062 | 0.140 |
| SAAIM _no_weather | 0.168 | 0.160 | 0.180 | 0.197 | 0.174 | 0.108 | 0.194 | 0.165 |
| SAAIM _no_sentiment | 0.130 | 0.141 | 0.112 | 0.186 | 0.144 | 0.094 | 0.067 | 0.156 |
| SAAIM _no_ILI | 0.316 | 0.309 | 0.326 | 0.301 | 0.352 | 0.273 | 0.400 | 0.253 |
| Correlation | ||||||||
| SAAIM | 0.892 | 0.905 | 0.845 | 0.860 | 0.845 | 0.652 | 0.770 | 0.788 |
| SAAIM _no_weather | 0.837 | 0.871 | 0.694 | 0.794 | 0.811 | 0.599 | 0.645 | 0.719 |
| SAAIM _no_sentiment | 0.870 | 0.885 | 0.817 | 0.813 | 0.831 | 0.567 | 0.768 | 0.746 |
| SAAIM _no_ILI | 0.488 | 0.635 | −0.063 | 0.497 | 0.411 | 0.370 | 0.266 | 0.488 |
Boldface highlights the best performance for each metric in each study period.
Fig. 3.
Importance analyses of different feature groups. SAAIM was constructed with four kinds of features: historical ILI, weather, sentiment and time. (A) The estimates of SAAIM without the climate features (blue), the public sentiment features containing Baidu Index and Weibo (green), without the historical ILI features (orange) are drawn. The estimated ILI% values of SAAIM with all features (red) and the true CDC's ILI activity level (black) are shown as references. (B) The estimation error, defined as estimated value minus the CDC's ILI activity level. (C-E) Zoomed-in plots for estimation results in different study periods. (C) The 2014 flu season. (D) The 2015 flu season. (E) The real-time prediction of ILI percentages 1 week before official publication from March 25th, 2018 to December 30th, 2018. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Similarly, removal of either historical ILI data or weather features led to a drastic increase in prediction delay. The delay scores amplified around 2-fold and 6-fold in the SAAIM trained without historical ILI data compared to the original SAAIM during the periods of 2014–2016 and 2017–2018, and amplified around 6-fold and 15-fold in the SAAIM trained without weather features compared to the original SAAIM during the periods of 2014–2016 and 2017–2018 (Appendix Table S4).
Internet-based public sentiment data, including Baidu Index and Sina Weibo, had the least impact on SAAIM. This observation is consistent with our finding that the LASSO model constructed with Baidu Index alone was not sufficient to predict ILI activity efficiently (Fig. 2). However, removal of web search and social media features did increase the prediction delay (Appendix Table S4) and marginally decreased the accuracy of SAAIM (Table 2), suggesting that the Internet-based public sentiment data also contributed to the forecast.
3.4. Statistical significance test
According to previous researches [40,41], computing prediction intervals is important to indicate the likely uncertainty in point forecasts. Using the bootstrap strategy specialized for time series proposed by Lorenzo Pascual [42] (Appendix Page 2), we obtained the 95% prediction intervals of SAAIM compared with all other models, which were constructed by the 2·5% and 97·5% percentage points of the bootstrap distribution function of the one-step-ahead forecast. The results indicated that 96·2% of the true ILI% data points were enclosed within the prediction intervals from 2017 to 2018 (Appendix Fig. S4), which was very close to the desired value of 95%. In addition, we compared the average margin of the 95% prediction intervals of SAAIM on the test dataset with all other reference models. As SAAIM shows an error reduction of at least 15% over other models, the average margin of the prediction intervals confirms the statistical significance of these results, with a minimum margin reduction of 15·57% (Appendix Table S5).
4. Discussion
Real-time prediction of influenza epidemics has been a great challenge in areas with irregular influenza activities, such as epidemic peak shift and reoccurring peaking in short periods, which hindered timely influenza epidemic response. In this study, we constructed an innovative AI model (SAAIM) with multi-source data to forecast the influenza activity in Chongqing, south-western China, which is a representative of cities with irregular influenza activities.
We (i) collected multi-source data based on literature review and expert consultant, (ii) derived features that could contribute to the prediction based on multiple statistic values from various data sources, (iii) extracted valid features via the feature selection methods in the proposed methodology, (v) merged the SARIMA model and the XGBoost model into an integrated model with the concept of Kalman Filter, an algorithm that dynamically updates the weights of the predictions from different base models.
SARIMA is a prevalent time-series forecasting model and can uncover the autocorrelation in time series, while XGBoost model is an optimized tree-based model and can display the nonlinear relationship between features and ILI%. As a result, our method not only retains the irregular trends of the ILI% time series but also captures the incidental fluctuations. In addition, we incorporated multi-source data including historical ILI%, weather data, Baidu search index, and Sina Weibo data of Chongqing into our model based on previous studies [5,6,[10], [11], [12],14,[17], [18], [19], [20], [21],29,30,[33], [34], [35]]. The diverse data contributed to retrieving miscellaneous influenza-related features and thus accurately constructing the model.
Ensemble approaches have been adopted to improve model performance in many studies. While simple averaging methods have been widely applied [33,[43], [44], [45], [46]], methods involving performance-based weighting system have also been proposed [47]. Our self-adaptive ensemble approach integrated the advantages of single models by dynamically updating the weights of the predictions from different base models, and outperformed the simple averaging method (Appendix Table S6).
SAAIM has been applied to real-time forecast since the 12th week of 2018 in Chongqing and reached a MAPE of 11·9%, which validated the forecast capacity of SAAIM in practice. The estimates by SAAIM have provided guidance on real-time influenza prevention and control to Chongqing health authorities. The reliable estimates and guidance have enabled authorities to make timely and scientific decisions on public health resources allocation and to prepare hospitals for the massive influx of influenza patients during flu season. Furthermore, the service of providing influenza activity forecast by the intelligent model could improve public health in the long run by arousing public awareness of infectious disease prevention and control.
Moreover, SAAIM could enlighten both theoretical and operational influenza forecasting studies by evaluating feature importance in the forecast and revealing novel factors that related to influenza epidemics. The features in the model could provide public health workers with some clues for investigating the potential reasons for the epidemic increase. The results showed that historical ILI activity one week prior to real time had the highest ranking score compared with all other features, suggesting that the ILI activity is highly autoregressive.
Interestingly, we identified three features that contributed significantly to the ILI prediction, including the foggy day counts of the prediction week, the overcast day counts of the prediction week, and the difference in average temperature between the prediction week and the prior week. Although the weather could affect influenza virus viability and the spread of influenza [20,21], to our knowledge, this is the first influenza forecast study that connects foggy and overcast weather with influenza activity, of which the mechanism remains to be studied.
SAAIM could be further improved by incorporating data sources with higher granularity, such as personal electronic medical records and demographic surveillance system. As in metropolises such as Chongqing, the spatial distribution of population density, population migration, and geographical features may affect the spread of influenza. Therefore, SAAIM is being expanded to and will be further tested in more major cities in China. In conclusion, our study provides a feasible methodology for irregular influenza activity forecast.
Funding sources
A donation from Medical Research Key Project of Chongqing Municipal Health Commission (20141026) funded this study. The funder had no role in the study design, data collection, data analysis, data interpretation, or writing of the manuscript. The corresponding authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Author contributions
KS led the preparation of all analyses and drafting of the paper. LX and XR led the execution of the study. GL, XL, and PD contributed to data analysis, data interpretation, and writing of the paper. XmL contributed to the literature search and project management. XC and SL contributed to data analysis and figure creation. QL, YX, LQ, WT, RR, CS, and ZY contributed to data collection. YN provided suggestions on paper writing. XS, QZ, BH, DL, JyX, and ZZ coordinated project management. YL, LZ, and JX designed and directed the project. YL, LZ, and JX, as corresponding authors, have confirmed that all authors have seen and approved the final text.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgments
We appreciate the funding from Chongqing Municipal Health Commission, and gratefully acknowledge the staff members of the seven sentinel hospitals (the First Affiliated Hospital of Chongqing Medical University, Banan Central Hospital, Children's Hospital of Chongqing Medical University, Qianjiang Central Hospital, Chongqing Three Gorges Central Hospital, Fuling Central Hospital and Yongchuan Hospital of Chongqing Medical University) and 6 district Centres for Disease Control and Prevention (Yuzhong, Wanzhou, Fuling, Yongchuan, Qianjiang and Banan district) for their assistance with data collection, quality control and management.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.08.024.
Contributor Information
Linqi Zhang, Email: zhanglinqi@mail.tsinghua.edu.cn.
Jing Xiao, Email: xiaojing661@pingan.com.cn.
Yafei Li, Email: liyafei2008@hotmail.com, liyafei2008@tmmu.edu.cn.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
Supplementary material 3
References
- 1.Dawood F.S., Iuliano A.D., Reed C., Meltzer M.I., Shay D.K., Cheng P.Y. Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: a modelling study. Lancet Infect Dis. 2012;12(9):687–695. doi: 10.1016/S1473-3099(12)70121-4. [DOI] [PubMed] [Google Scholar]
- 2.Taubenberger J.K., Kash J.C. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe. 2010;7(6):440–451. doi: 10.1016/j.chom.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iuliano A.D., Roguski K.M., Chang H.H., Muscatello D.J., Palekar R., Tempia S. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet. 2018;391(10127):1285–1300. doi: 10.1016/S0140-6736(17)33293-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nicholson K.G., Wood J.M., Zambon M. Influenza. Lancet. 2003;362(9397):1733–1745. doi: 10.1016/S0140-6736(03)14854-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santillana M., Nguyen A.T., Louie T., Zink A., Gray J., Sung I. Cloud-based electronic health records for real-time, region-specific influenza surveillance. Sci Rep. 2016;6 doi: 10.1038/srep25732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ginsberg J., Mohebbi M.H., Patel R.S., Brammer L., Smolinski M.S., Brilliant L. Detecting influenza epidemics using search engine query data. Nature. 2009;457(7232):1012–1014. doi: 10.1038/nature07634. [DOI] [PubMed] [Google Scholar]
- 7.Yang W., Lipsitch M., Shaman J. Inference of seasonal and pandemic influenza transmission dynamics. Proc Natl Acad Sci U S A. 2015;112(9):2723–2728. doi: 10.1073/pnas.1415012112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polansky L.S., Outin-Blenman S., Moen A.C. Improved global capacity for influenza surveillance. Emerg Infect Dis. 2016;22(6):993–1001. doi: 10.3201/eid2206.151521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Longini I.M., Jr., Nizam A., Xu S., Ungchusak K., Hanshaoworakul W., Cummings D.A. Containing pandemic influenza at the source. Science. 2005;309(5737):1083–1087. doi: 10.1126/science.1115717. [DOI] [PubMed] [Google Scholar]
- 10.Yang S., Santillana M., Kou S.C. Accurate estimation of influenza epidemics using Google search data via ARGO. Proc Natl Acad Sci U S A. 2015;112(47):14473–14478. doi: 10.1073/pnas.1515373112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan Q., Nsoesie E.O., Lv B., Peng G., Chunara R., Brownstein J.S. Monitoring influenza epidemics in China with search query from baidu. PLoS One. 2013;8(5) doi: 10.1371/journal.pone.0064323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santillana M., Nguyen A.T., Dredze M., Paul M.J., Nsoesie E.O., Brownstein J.S. Combining search, social media, and traditional data sources to improve influenza surveillance. PLoS Comput Biol. 2015;11(10) doi: 10.1371/journal.pcbi.1004513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paul M.J., Dredze M., Broniatowski D. Twitter improves influenza forecasting. PLoS Curr. 2014;6 doi: 10.1371/currents.outbreaks.90b9ed0f59bae4ccaa683a39865d9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J., Cardie C. 2013. Early stage influenza detection from twitter. arXiv preprint arXiv:13097340. [Google Scholar]
- 15.Hu H., Wang H., Wang F., Langley D., Avram A., Liu M. Prediction of influenza-like illness based on the improved artificial tree algorithm and artificial neural network. Sci Rep. 2018;8(1):4895. doi: 10.1038/s41598-018-23075-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alessa A., Faezipour M. A review of influenza detection and prediction through social networking sites. Theor Biol Med Model. 2018;15(1):2. doi: 10.1186/s12976-017-0074-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hickmann K.S., Fairchild G., Priedhorsky R., Generous N., Hyman J.M., Deshpande A. Forecasting the 2013-2014 influenza season using Wikipedia. PLoS Comput Biol. 2015;11(5) doi: 10.1371/journal.pcbi.1004239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang S., Santillana M., Brownstein J.S., Gray J., Richardson S., Kou S.C. Using electronic health records and internet search information for accurate influenza forecasting. BMC Infect Dis. 2017;17(1):332. doi: 10.1186/s12879-017-2424-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang W., Cowling B.J., Lau E.H., Shaman J. Forecasting influenza epidemics in Hong Kong. PLoS Comput Biol. 2015;11(7) doi: 10.1371/journal.pcbi.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roussel M., Pontier D., Cohen J.M., Lina B., Fouchet D. Quantifying the role of weather on seasonal influenza. BMC Public Health. 2016;16:441. doi: 10.1186/s12889-016-3114-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis R.E., Rossier C.E., Enfield K.B. The impact of weather on influenza and pneumonia mortality in New York City, 1975-2002: a retrospective study. PLoS One. 2012;7(3) doi: 10.1371/journal.pone.0034091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ajelli M., Poletti P., Melegaro A., Merler S. The role of different social contexts in shaping influenza transmission during the 2009 pandemic. Sci Rep. 2014;4:7218. doi: 10.1038/srep07218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cauchemez S., Bhattarai A., Marchbanks T.L., Fagan R.P., Ostroff S., Ferguson N.M. Role of social networks in shaping disease transmission during a community outbreak of 2009 H1N1 pandemic influenza. Proc Natl Acad Sci U S A. 2011;108(7):2825–2830. doi: 10.1073/pnas.1008895108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kopf M., Brombacher F., Bachmann M.F. Role of IgM antibodies versus B cells in influenza virus-specific immunity. Eur J Immunol. 2002;32(8):2229–2236. doi: 10.1002/1521-4141(200208)32:8<2229::AID-IMMU2229>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 25.Kanai Y., Boonsathorn N., Chittaganpitch M., Bai G., Li Y., Kase T. The impact of antigenic drift of influenza A virus on human herd immunity: Sero-epidemiological study of H1N1 in healthy Thai population in 2009. Vaccine. 2010;28(33):5437–5444. doi: 10.1016/j.vaccine.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 26.Lin C.J., Nowalk M.P., Toback S.L., Rousculp M.D., Raymund M., Ambrose C.S. Importance of vaccination habit and vaccine choice on influenza vaccination among healthy working adults. Vaccine. 2010;28(48):7706–7712. doi: 10.1016/j.vaccine.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 27.Nowalk M.P., Lin C.J., Zimmerman R.K., Fox D.E., Raymund M., Tanis M.D. Establish the habit: influenza vaccination for health care personnel. J Healthc Qual. 2010;32(2):35–42. doi: 10.1111/j.1945-1474.2010.00073.x. [DOI] [PubMed] [Google Scholar]
- 28.Viboud C., Charu V., Olson D., Ballesteros S., Gog J., Khan F. Demonstrating the use of high-volume electronic medical claims data to monitor local and regional influenza activity in the US. PLoS One. 2014;9(7) doi: 10.1371/journal.pone.0102429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moss R., Zarebski A., Dawson P., McCaw J.M. Forecasting influenza outbreak dynamics in Melbourne from internet search query surveillance data. Influenza Other Respi Viruses. 2016;10(4):314–323. doi: 10.1111/irv.12376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Q., Gel Y.R., Ramirez Ramirez L.L., Nezafati K., Zhang Q., Tsui K.L. Forecasting influenza in Hong Kong with Google search queries and statistical model fusion. PLoS One. 2017;12(5) doi: 10.1371/journal.pone.0176690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi L., Xiong Y., Xiao B., Tang W., Ling H., Long J. Epidemiological and Virological characteristics of influenza in Chongqing, China, 2011-2015. PLoS One. 2016;11(12) doi: 10.1371/journal.pone.0167866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su K., Ye S., Li Q., Xie W., Yu H., Qi L. Influenza A(H7N9) virus emerged and resulted in human infections in Chongqing, southwestern China since 2017. Int J Infect Dis. 2019;81:244–250. doi: 10.1016/j.ijid.2019.02.011. [DOI] [PubMed] [Google Scholar]
- 33.Shaman J., Karspeck A. Forecasting seasonal outbreaks of influenza. Proc Natl Acad Sci U S A. 2012;109(50):20425–20430. doi: 10.1073/pnas.1208772109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen L., Hossain K.T., Butler P., Ramakrishnan N., Prakash B.A. Syndromic surveillance of flu on twitter using weakly supervised temporal topic models. Data Min Knowl Disc. 2016;30(3):681–710. [Google Scholar]
- 35.Axelsen J.B., Yaari R., Grenfell B.T., Stone L. Multiannual forecasting of seasonal influenza dynamics reveals climatic and evolutionary drivers. Proc Natl Acad Sci U S A. 2014;111(26):9538–9542. doi: 10.1073/pnas.1321656111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bishop G., Welch G. An introduction to the Kalman filter. Proc SIGGRAPH Course. 2001;8(27599–3175):59. [Google Scholar]
- 37.Chen T., Guestrin C. 2016. XGBoost: A scalable tree boosting system. Acm Sigkdd international conference on knowledge discovery & data mining. [Google Scholar]
- 38.Chae S., Kwon S., Lee D. Predicting infectious disease using deep learning and big data. Int J Environ Res Public Health. 2018;15(8) doi: 10.3390/ijerph15081596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bergmeir C., Benítez J.M. On the use of cross-validation for time series predictor evaluation. Inform Sci. 2012;191:192–213. (none. [Google Scholar]
- 40.Chatfield C. 2001. Prediction intervals for time-series forecasting. [Google Scholar]
- 41.Shrestha D.L., Solomatine D.P. Machine learning approaches for estimation of prediction interval for the model output. Neural Netw. 2006;19(2):225–235. doi: 10.1016/j.neunet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 42.Pascual L., Romo J., Ruiz E. Bootstrap predictive inference for ARIMA processes. J Time. 2010;25(4):449–465. [Google Scholar]
- 43.McGowan C.J., Biggerstaff M., Johansson M., Apfeldorf K.M., Ben-Nun M., Brooks L. Collaborative efforts to forecast seasonal influenza in the United States, 2015-2016. Sci Rep. 2019;9(1):683. doi: 10.1038/s41598-018-36361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang L., Ai H., Chen W., Yin Z., Hu H., Zhu J. CarcinoPred-EL: novel models for predicting the carcinogenicity of chemicals using molecular fingerprints and ensemble learning methods. Sci Rep. 2017;7(1):2118. doi: 10.1038/s41598-017-02365-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farrow D.C., Brooks L.C., Hyun S., Tibshirani R.J., Burke D.S., Rosenfeld R. A human judgment approach to epidemiological forecasting. PLoS Comput Biol. 2017;13(3) doi: 10.1371/journal.pcbi.1005248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shaman J., Karspeck A., Yang W., Tamerius J., Lipsitch M. Real-time influenza forecasts during the 2012-2013 season. Nat Commun. 2013;4:2837. doi: 10.1038/ncomms3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu F.S., Hou S., Baltrusaitis K., Shah M., Leskovec J., Sosic R. Accurate influenza monitoring and forecasting using novel internet data streams: a case study in the boston metropolis. JMIR Public Health Surveill. 2018;4(1) doi: 10.2196/publichealth.8950. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2
Supplementary material 3