Abstract
Background
Whole genome sequencing (WGS) has enabled the development of new approaches to track Mycobacterium tuberculosis (Mtb) transmission between tuberculosis (TB) cases but its utility may be challenged by the discovery that Mtb diversifies within hosts. Nevertheless, there is limited data on the presence and degree of within-host evolution.
Methods
We profiled a well-documented Mtb transmission cluster with three pulmonary TB cases to investigate within-host evolution and describe its impact on recent transmission estimates. We used deep sequencing to track minority allele frequencies (<50·0% abundance) during transmission and standard treatment.
Findings
Pre-treatment (n = 3) and serial samples collected over 2 months of antibiotic treatment (n = 16) from all three cases were analysed. Consistent with the epidemiological data, zero fixed SNP separated all genomes. However, we identified six subclones between the three cases with an allele frequency ranging from 35·0% to 100·0% across sampling intervals. Five subclones were identified within the index case pre-treatment and shared with one secondary case, while only the dominant clone was observed in the other secondary case. By tracking the frequency of these heterogeneous alleles over the two-month therapy, we observed distinct signatures of drift and negative selection, but limited evidence for de novo mutations, even under drug pressure.
Interpretation
We document within-host Mtb diversity in an index case, which led to transmission of minority alleles to a secondary case. Incorporating data on heterogeneous alleles may refine our understanding of Mtb transmission dynamics. However, more evidence is needed on the role of transmission bottleneck on observed heterogeneity between cases.
Keywords: Mycobacterium tuberculosis, Within-host evolution, Transmission dynamics, Rare variants, Transmission bottleneck
Research in context.
Evidence before this study
Whole genome sequencing (WGS) has greatly advanced our ability to trace direct Mycobacterium tuberculosis (Mtb) transmission, but this is increasingly being challenged by recent findings that within-host populations of pathogens are often heterogeneous. With current approaches, we implicitly assume one Mtb clone per tuberculosis (TB) patient and visualize only the predominant bacterial population to infer transmission links/clusters. A proposed cut off of ≤5 single nucleotide polymorphisms (SNPs) is conventionally used to infer direct transmission between cases. However, epidemiological analyses of serial clinical isolates have reported within-host genetic variability that exceeds that of linked cases, thereby challenging the convenience of a universal SNP-cut off to delineate Mtb transmission. While a number of studies have reported on within-host diversity and the impact on direct transmission estimates, there has been less focus on the precise mechanisms that drive within-host microevolution. The available evidence suggest two evolutionary scenarios to explain the observed within-host Mtb diversity in TB cases: The first proposes the idea of a dominant clone from which minority variants evolve continuously but are selected against by adequate drug pressure or the host immune system. The second suggests the presence of segregated but related within-host variant populations structurally shaped by drift and adaptation. Nevertheless, epidemiological evidence to support either scenario in Mtb remain limited.
Added value of this study
Although prior studies have used deep sequencing and profiled low frequency SNPs in serial isolates to investigate within-host evolution, they typically have applied this technique to unlinked cases, and cases with variable drug resistance profiles. In this study, we instead focus on a single discrete transmission cluster, documented through reverse contact tracing and traditional genotyping over 2 years, to investigate within-host evolution and describe its impact on SNP-based recent transmission estimates in more detail. We analyse pre-treatment and serial isolates from three human TB cases under standard treatment, with all cases receiving the same initial therapy. Thus, we specifically analysed the dynamics of the same strain within and between host(s) and over time.
Implications of all the available evidence
Our study expands on the available evidence that Mtb within-host heterogeneity is in the form of rare variants that seldom reach fixation. We also demonstrate that these rare variants are seeded during transmission but can be selected against during latency. More importantly, we show that over time, the within-host population dynamics can shift, with the potential to bias the reconstruction of transmission links in outbreaks that span years. Our report highlights the need to incorporate minority alleles in addition to consensus fixed SNPs when estimating recent transmission links. In addition, we provide the epidemiological impetus for public health officials to incorporate the identification of TB patients that may harbour polyclonal infections, as they may require more aggressive treatment. It is now accepted that the one genome/sample approach biases the reconstruction of Mtb transmission links and may even confound phenotypic drug susceptibility results. Consequently, we provide important evidence to support the integration of novel sampling approaches to capture the full spectrum of within-host diversity.
Alt-text: Unlabelled Box
1. Introduction
Tuberculosis (TB), predominantly caused by Mycobacterium tuberculosis, is currently the number one cause of death by an infectious agent globally [1]. In 2017, an estimated 10·0 million people developed active disease and 1·3 million died [1]. Rapid diagnosis and effective treatment of active cases are among the most effective ways to control TB and stop transmission in communities [2]. In low incidence settings, investigation of outbreaks aided by traditional molecular surveillance tools is a cornerstone of TB control efforts [3]. Conventional genotyping techniques were developed based on the fundamental assumption that Mtb transmission and infection is clonal [3]. Thus, cases with identical genotype patterns can be grouped into clusters, i.e. cases that share transmission links that can be further explored by contact investigation [3]. Mycobacterial interspersed repetitive unit – variable number tandem repeat (MIRU-VNTR) and spacer-oligonucleotide typing (spoligotyping) techniques have, however, been found to overestimate transmission based on clustering. Whole genome sequencing (WGS) has shown to be a promising technology that has higher resolution and can more accurately determine transmission dynamics and identify clusters more in agreement with the contact investigation data [4]. WGS is able to identify the direction and temporal sequence of transmission down to a single nucleotide polymorphism (SNP). This is especially useful in outbreaks that span years or for Mtb prospective genomic surveillance at the local or national level. WGS is revolutionizing TB outbreak investigations by helping to more efficiently identify recent transmission. Currently, by applying WGS, genotype clusters can be further classified into cases linked to the closest SNP, with a proposed ≤5 SNPs cut off to define recent outbreaks [5]; although larger SNP cut offs have been proposed and applied in diverse TB epidemiological settings [6]. WGS has also replaced traditional methods for routine strain typing [7] and drug susceptibility testing [8] for certain TB programs in low incidence settings.
WGS has exposed a breadth of within-host genetic diversity in Mtb infection. A number of studies have reported on the within-host heterogeneity in Mtb and its implications for TB control [6,9]. Particularly, some studies have reported that the genomic variability within a TB patient may be greater than the variability observed between any two epidemiologically linked cases [6]. The consequence of this finding may be that we are unable to resolve transmission events using the standard ≤5 SNP cut off [6]. Other studies have suggested that within-host diversity may confound both phenotypic and rapid molecular diagnostics for drug resistance, severely impacting treatment efficacy, and leading to the selection of drug resistant clones during treatment [10] Nevertheless, there is limited data on the mechanisms of within-host evolution in M. tuberculosis [12,17]. Two evolutionary scenarios have been proposed to reconcile this seeming genomic clonality with the ability to diversify within hosts. The first proposes the idea of a dominant clone from which minority variants evolve continuously but are selected against by adequate drug pressure [11], or the host immune system [12]. The second scenario suggests a predominance of spatially separated but related variant populations within the host that are structurally shaped by genetic drift and adaptation [9,11,13,14]. However, most of the data supporting these evolutionary scenarios have come from animal studies, or epidemiological studies of unlinked TB cases [9,11,12,15] In addition, most of the epidemiology studies profiled SNPs that had become fixed in the sampled population, and as such did not explore the full spectrum of diversity due to the presence of minority variants [12,17].
In this study we investigated within-host Mtb evolution and describe its impact on recent transmission estimates. What sets our study apart is that we profiled a well-documented Mtb transmission cluster involving three cases linked by reverse contact tracing and conventional genotyping by MIRU-VNTR/spoligotyping. The availability of pre-treatment and serial isolates that were obtained during the standard antibiotic treatment course allowed us to investigate within-host evolution during transmission and under drug pressure within the same cluster. In addition, we used very deep (330× – 1500×) sequencing to track the population turnover of minority alleles with less than 50% abundance [11]. A secondary case was diagnosed soon after the index was reported, and a third case was identified 22 months later. We are thus able to account for disease latency in our estimates of the within-host evolution. We provide further evidence that Mtb within-host diversity is largely composed of minority alleles, with clonal evolution driven by negative selection at transmission and during latency. Interestingly, we observed little measureable microevolution under drug pressure. Our results may have important implications for how we estimate recent transmission.
2. Materials and methods
2.1. Cluster description
The cluster included three patients. The index case (P1) was a male in his early 50s. Originally from South America, P1 had lived in the United States (U.S.) for over a decade before he was diagnosed with advanced cavitary pulmonary TB in late 2016. The first secondary case (P2), who was confirmed to have TB by a positive culture 17 days after P1 was reported, was a long-term household contact born in the U.S. Patient three (P3), also born in the U.S., was diagnosed in early 2018 and linked to P1 by reverse contact tracing and traditional genotyping. All three cases were HIV-negative and shared identical S spoligotype (776377377760771) [16] and 24-locus MIRU-VNTR (213325153324 / 14143422332%) profiles. The MIRU loci are ordered as reported in Mazars et al., 2001, with CDC notations used for ambiguous and indeterminate sites [17]. The Florida Department of Health Bureau of Public Health Laboratories (BPHL) performed phenotypic drug susceptibility testing (DST) by broth microdilution method (Sensititre™; Thermo Fisher Scientific, Cleveland, OH, USA) on all pre-treatment isolates and molecular testing for the detection of multidrug resistance TB (MDR-TB) by MTBDRplus assay (Hain Lifescience GmbH, Nehren, Germany). DST was repeated on the last culture positive samples for P1 and P3.
2.2. Ethics statement
The Florida Department of Health TB control program managed the diagnosis and treatment of the patients, after obtaining their informed consents for services. Data from the Florida Department of Health TB Program and the Bureau of Public Health Laboratories were collected and shared anonymously. The use of the data for this study was approved by the Institutional Review Boards (IRB) of the University of Florida (IRB201600135, IRB201700445, and IRB201901133) and the Florida Department of Health (2013-05-UFL).
2.3. Whole genome sequencing and assembly
Details of sample preparation for genomic DNA extraction, library preparation, and sequencing using the Nextera XT library construction kit and Illumina MiSeq system (Illumina, Inc., San Diego, CA, USA) are described in the supplementary file. The metagenomics composition of individual paired-end libraries was assessed using kraken (v2·0·7), followed by bracken (v2·2), against the MiniKraken2 reference database (downloaded on March 7, 2019) to identify and remove contaminated reads [18,19].
2.4. Variant calling
The filtered mapped reads (in BAM-format) from the three libraries re-sequenced at high depth were combined with their respective normal-depth libraries. We called high-confidence fixed SNPs with samtools (v1·4) and bcftools (v1·4), consecutively. We retained all homozygous SNPs meeting a minimum phred quality score of 20, sequencing depth of 10, four forward and four reverse read support in non-repetitive regions of the H37Rv reference genome (excluding all transposases, pe-, ppe- and pe_pgrs-genes). To identify and track low-frequency SNPs, we used lofreq (v2·1·2) to call SNPs at previously confirmed high-confidence variant position [20]. We retained all low frequency SNPs having a minimum of one forward, one reverse read support, and a phred quality score >20. After confirming the absence of minority variants in these genomic positions, we excluded an additional 25 SNPs in or between genes belonging to the ESAT-6 and polyketide synthetases families to avoid underestimation of allele frequencies due to inconsistent read mapping.
2.5. Assessing composition of subclones in samples
Minimum inclusion criteria for considering a subclone detected within a given sample involved detection of at least two allelic SNPs from the respective subclone (except for subclone Sc6 which only had one identified allelic SNP). Subclone frequencies were then calculated based on pooled read depths of allele-specific nucleotide against pooled read depths supporting the respective reference nucleotides. The abundance of the primary clone (Pc) was calculated by subtracting the combined proportions of all detected subclones (Sc1-6) from 1. We used the Shannon Diversity index calculated from mapped files downsampled to a maximum 50× average coverage to track subclone abundance across sampling intervals [21].
2.6. Phylogenetic analyses
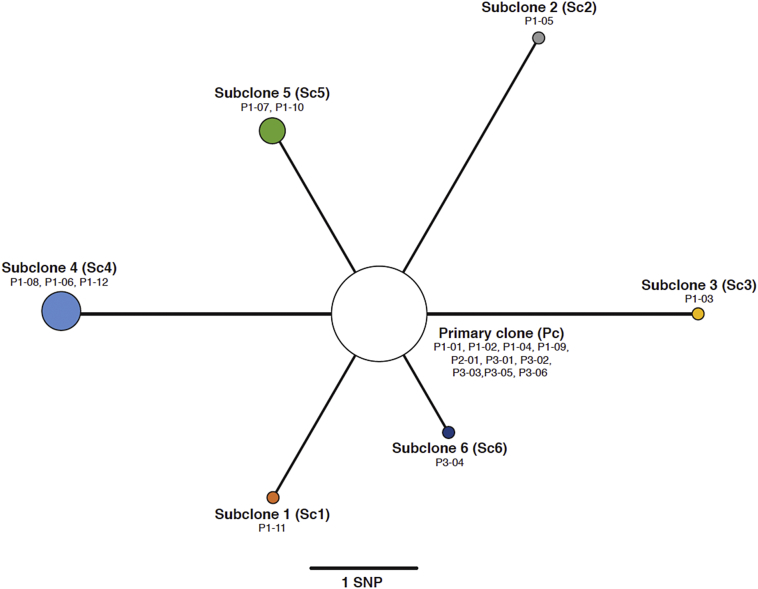
The phylogenetic relationship between the genomes was investigated using distance-based and maximum parsimony methods. The pairwise genetic distance between genomes (measured in SNPs) and a comparison between and within host(s) SNP differences were calculated using MEGA7 [22]. We used the PHYLIP/dnapars algorithm implemented in Seaview to infer a maximum parsimony phylogenetic tree, optimized by nearest neighbor interchange (NNI) [23], which formed the basis of the network shown in Fig. 1. Phylogenetic reconstruction of global diversity of the L4·4·1·1 Mycobacterium Tuberculosis Complex (MTBC)-lineage was carried out by first downloading currently publicly available datasets from the European Nucleotide Archive (ENA), from studies including >50 whole-genome sequenced Mtb genomes. MTBC-lineages were then identified by running TB-Profiler [24] on fastq-files and strains that identified as L4·4·1·1 were included in the subsequent analysis. Phylogenetic reconstruction was performed with RAxML (v7·2·8) implemented in the software package Geneious (v9·1·8) on an alignment consisting of 7554 SNPs with 100 bootstrap replicates to infer branch support.
Fig. 1.
Maximum parsimony phylogeny drawn using high confidence fixed SNPs. Node sizes correspond to the number of samples representing each subclone population. We chose the most dominant clone (primary or subclone) from each sample. This represents an allele frequency was below 50% in some samples (P1-04, P1-07, P1-09, and P2-01), while still being the most abundant clonal population.
2.7. Data deposition
Sequences are available in the EMBL-EBI European Nucleotide Archive (ENA) under study accession PRJEB30782 https://www.ebi.ac.uk/ena/data/view/PRJEB30782.
3. Results
3.1. Description of isolates
The initial pre-treatment isolates were available from all three cases (one each). In addition, 19 sputum samples were collected from P1 and seven from P3 over the course of 2-month intensive treatment phase to monitor progress. We only had the sputum sample collected at diagnosis for P2. Phenotypic drug susceptibility testing (DST) revealed full susceptibility towards the standard drugs rifampin, isoniazid, pyrazinamide, and ethambutol. Of the 26 original within-patient isolates, eight were later discarded from P1 due to very low sequencing coverage resulting from significant nontuberculous mycobacterial (NTM) contamination (Supplementary file). Furthermore, two samples from P3 were not sequenced due to lack of growth and substantial contamination, respectively. Overall, 19 isolates were included in the analyses, comprising twelve from P1, one from P2, and six from P3. Retained libraries comprised over 99% MTBC reads, except for one sample (UF01–18) comprising 98% MTBC reads and 1·9% NTM reads. Due to the detection of almost 2·0% contaminated reads in included samples, we also removed mapped reads that differed by >5% identity to the H37Rv reference genome as well as reads with less than half of their length mapped. All libraries included in final analyses had at least 99·9% of the mapped reads assigned to Mtb. We performed deep sequencing on first and last samples from P1 and P3 (and the single P2 isolate), to ensure at least 500× coverage, in order to track low-frequency SNPs following transmission and over the course of treatment. The time of isolation and sequencing coverage for all genomes are available in Supplementary Table 1.
3.2. Initial genotyping and isolates in a global context
We first determined that all sequenced isolates constituted a single genotype, namely MTBC L4·4·1·1, based on the confirmed presence, at >99% frequency, of all nine informative SNP markers specific to this particular sublineage [7]. Furthermore, consistent with the DST results, no well-known drug resistance conferring mutations were detected in any of the isolates.
Typically, in the absence of evidence of mixed-strain infection or contamination, SNPs are identified as present when their allele frequency exceeds 75%–90%. However, SNPs can justifiably be considered major alleles as long as they constitute more than half of the population (thus, a frequency > 50%), provided they are not located within or very near to repetitive- or inserted genome elements, in which case they are more likely a result of mis-mapped sequencing reads. In total, we identified 674 SNPs, universally conserved in all 19 isolates (99·8% of SNP-calls above >95% frequency, with all calls above 80% frequency), and 14 allelic SNPs present at >50% frequency in at least one isolate. From these 14 genome-positions, we were able to identify six distinct subclones (Sc1-Sc6) among the 19 isolates, each discernible through 1–3 allele-specific SNPs, which derived directly from a single, primary clone (Pc) lacking all 14 allele-specific SNPs (Supplementary Fig. 1). Thus, only 2% of the detected SNP positions contributed to the observed genetic heterogeneity.
By comparing the seven clones identified among the 19 samples included in this study to publicly available whole-genome sequenced Mtb isolates belonging to the same MTBC sublineage (Supplementary Fig. 2), we saw that they formed a deeply branched monophyletic clade within L4·4·1·1. Eighty-five out of the 674 conserved SNPs were clade-specific (i.e unique to our isolates), while the majority (84%) of all other SNPs appeared universally conserved throughout L4·4·1·1. Despite 144 of the global isolates being from the American continent (South America, Canada and Greenland), our clones grouped among isolates of African and European origin. Furthermore, based on global genome typing studies, the L4·4 sublineage is not normally associated with South America or the USA, which sees a significantly higher abundance of generalist sublineages such as L4·1·2/Harlem, L4·3/LAM and L4·10/PGGE [25,26]. However, presence of this sublineage in this part of the world is still consistent with recent analysis suggesting a strong temporal correlation between European colonialization and the spread of L4 sublineages to Africa and the Americas [26].
3.3. Within-host genetic diversity and impact on observed heterogeneity between cases
As most SNP-based transmission analyses operate with a single clone per isolate, we traced the frequencies of each of the allele-specific SNPs across all isolates, to assess the proportion of individual subclones against the primary clone (Supplementary Fig. 1). Nine isolates contained the appropriate subclone-specific SNPs at >50% frequency, and thus appeared to be comprised of one of the six subclones, while the 14 SNPs were absent or below this threshold in ten isolates and were therefore initially identified as Pc isolates (Fig. 1). Six of the isolates were largely dominated (>80% abundance) by a single subclone (Sc1 and Sc3-Sc6, respectively), while the Pc completely dominated five isolates, all from patient P3. The Pc had the highest abundance in ten samples (35% – 100%), although three of these (P2–01, P1–04 and P1–09) were in reality dominated by mixtures of different subclones (see below).
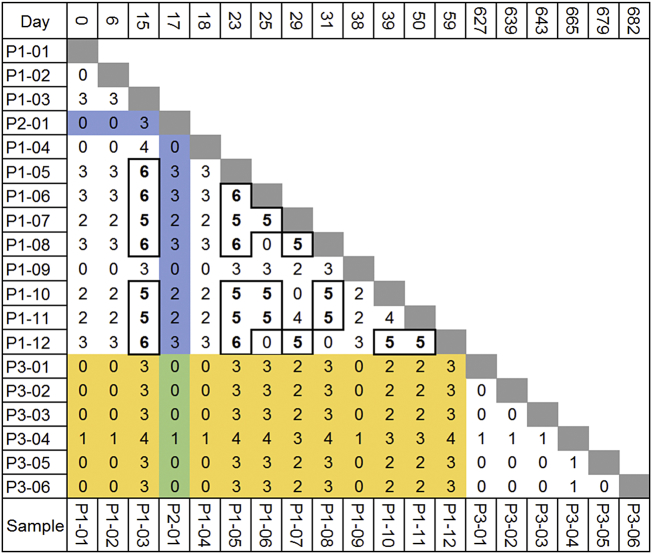
The pairwise genetic distance between any two isolates, when only considering their respective most abundant subclone is shown in Fig. 2. Consistent with the epidemiology and genotyping data, pre-treatment samples from all three patients (P1-01, P2-01and P3-01) were separated by zero SNPs, thus strongly supporting direct transmission. Interestingly, however, five of the six subclones (Sc1-Sc5) were detected in the index case (P1), demonstrating a significant within-host diversity, while only one (Sc6) was found exclusively in a secondary case (P3). Furthermore, while all isolates dominated by Pc were within three SNPs of all other isolates, the genetic distances were five or six SNPs between isolates in which Sc2, Sc3 or Sc4 were the most abundant, thereby exceeding the conventional threshold for recent transmission (Fig. 2). However, because the five subclones in P1 were demonstrably related directly to the primary clone, we could confidently rule out mixed infection (i.e transmission to P1 from multiple sources). On average, we measured a mean SNP-distance of 3·2 (n = 66) within P1, while P3 only had a distance of 0·33 (n = 15). Overall, we measured a mean distance of 2·19 SNPs between all isolates (n = 177). The mean between-host SNP-distance from P1 to P2 and P3 was 1·8 and 1·9, respectively, while we only saw a mean distance of 0·17 SNPs between P2 and P3. On the whole, we therefore saw between-host SNP distances within the conventionally defined limit of direct transmission (<2 SNPs) [5].
Fig. 2.
Within and between host pairwise genetic distance. Genomes are ordered by the date of isolation. Genetic distance is measured in SNP. Between-host SNP distances are highlighted in blue, green and yellow, respectively. Bold numbers/boxes indicate pairwise SNP distance above the conventional ≤5 SNP cut off to define direct transmission. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
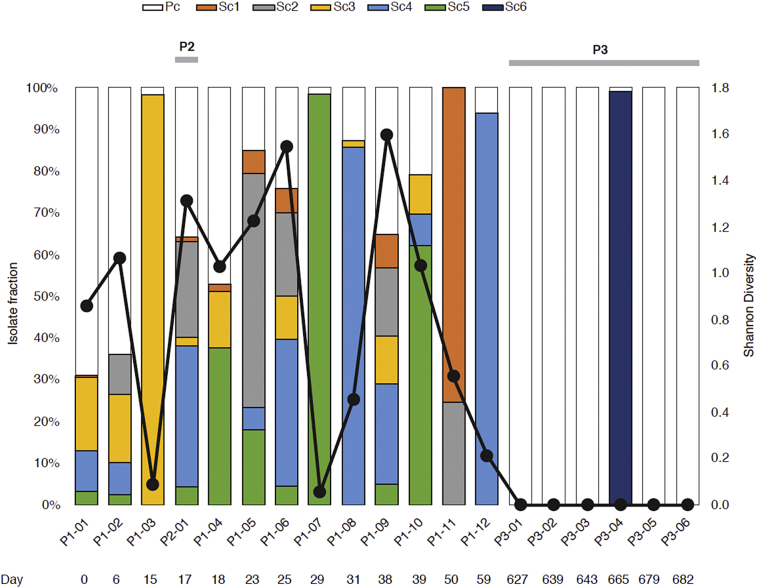
Intriguingly, eight of the isolates were revealed to be comprised of complex mixtures of three or more clones (Fig. 3). Isolates consisting almost entirely of single subclones appeared on days 15, 29, 31 and 59, while within-host heterogeneity, expressed via the Shannon diversity index, was the highest at days 6, 23, 25 and 38 of treatment in P1. Surprisingly, we also saw a high degree of heterogeneity, comparable to P1, in the pre-treatment sample of P2. Most importantly, all five subclones (Sc1-Sc5) detected in P1 were represented in the P2 pre-treatment sample. We also note that diversity seemed to decrease after 1 month of treatment (days 38–59), and that all P3 isolates, apart from P3-01, appeared to consist mostly of pure clones (Pc or Sc6, respectively).
Fig. 3.
Subclone population and diversity across sampling intervals. Genomes are ordered by the date of isolation. Colours represent each of the six subclone populations, while the black line represents the Shannon diversity index across sampling interval. The diversity index was normalized for sequencing depth by downsampling each mapped genome to an average of 50×.
3.4. Deep sequencing for detection of ultra-low-abundance subclones in heterogeneous sputum isolates
Initially, our analyses of the isolates seemed to suggest evidence of significant within-host microevolution occurring in P1, as multiple subclones, all deriving directly from a single clone (Pc), appeared as pure isolates at various times over the course of treatment. However, as we later observed, subclone-mixtures also appeared in subsequent isolates, suggesting a more dynamic picture than simple purifying selection on emerging de novo mutations. Conversely, from days 6–59 of treatment in P1, we saw a more or less steady increase in the abundance of Sc4, which increased from 7·7% to 94·1%, while Sc3 and Sc5 were undetected after day 39 of treatment, which also saw an overall decrease in sample heterogeneity (Fig. 3). This bore a distinct signature of purifying selection, possibly influenced by drug pressure. We therefore became interested at the realistic detection limit of subclones by supplementing selected isolates, the pre-treatment- (P1-01, P2-01, P3-01) and final (P1-12 and P3-06) isolates of the three patients with deep sequencing, corresponding to 1–4 isolates per MiSeq flow cell. This allowed the detection of subclones at as low as 0·2% allele frequency, although this corresponded to only nine sequencing reads over two allelic SNP positions (Supplementary Table 3). This level of sensitivity was sufficient to confirm the presence of all five subclones in the P1 and P2 pre-treatment isolates.
3.5. Functional consequences of within-host heterogeneity
We looked at the genes affected by mutations in these 14 variable regions (Supplemental Table S2). All but one occurred in protein-coding regions, with seven leading to amino acid changes (missense). The mutations were mainly in genes required for Mtb replication, lipid synthesis, or cellular metabolism [27]. For example, we observed a missense variant (Thr128Met) in the lipoprotein gene lppE in subclone-4, while subclone-3 had a synonymous variant in LppF at position 2,172,722 in LppF. The lipoprotein genes LppE and LppF encode membrane-anchored proteins that are involved in virulence and immunoregulatory processes [28,29]. Another interesting missense mutation (Asp422Ala) was in the phoR gene, which codes for the sensor histidine kinase PhoR, part of the two-component regulatory system PhoPR. PhoPR has a regulatory role in virulence and cell wall composition and is believed to control the expression of approximately 2% of the Mtb genome [30]. These regulatory systems alter gene expression in response to stimuli such as external stress. The response regulator PhoP regulates processes such as lipid metabolism, stress responses, and persistence, but the action of the sensor histidine kinase PhoR is less understood [31,32]. The presence of these mutations in genes required for membrane and cell-wall synthesis and the absence of mutations in drug targets, supports our hypothesis that these mutations did not occur over the study sampling period.
4. Discussion
In this study we confirm the findings of a number of epidemiological studies that Mtb is variable within-host [9,11,[33], [34], [35], [36]]. Within our index case sampled at eleven different time points, we identified five subclones over the course of the TB infection and treatment separated by up to 6 SNPs. This observation may have profound implications for transmission dynamics studies [37]. In addition, we show that the bulk of the diversity is in the form of minority alleles, which are often not taken into consideration by most within-host epidemiological studies. Within-host heterogeneity in Mtb and its impact on transmission and drug susceptibility testing has been documented [38]. Nevertheless, our understanding of the mechanisms of within-host diversification remains limited [11]. Recently, Herranz et al. 2018 reported that Mtb acquires limited genetic diversity during prolonged infection, reactivation and transmission involving multiple hosts [34]. This is in line with our own observations. It took 2 months of combination therapy with four effective drugs for P1 and P3 to successfully eradicate their Mtb infection. This gave us the unique opportunity to track the fate of the minority allele populations identified at baseline under drug pressure and between hosts. We observed that all six subclones were present in pre-treatment samples and in sputa collected at subsequent time points, demonstrating that none of these subclones became fixed over time, with the primary clone remaining dominant in all three patients. Given that at the estimated mutation rate of 0·25–0·30 SNP/genome/year during latent infection [39,40] three SNPs would take on average 10–12 years to develop, we would argue that part of the diversity observed in P1 could have been transmitted to him as we observed in the case of P2. However, more experimental data that include the date of infection are needed to confirm these findings.
Another observation in our study is that the five subclones identified in P1 were recovered in P2, diagnosed within a couple of weeks, while only the primary clone was observed in P3 diagnosed almost 2 years later. The role of purifying selection in Mtb within-host clonal evolution has previously been reported [9,11,12]. However, there is as of yet little known about the role of transmission bottleneck in curtailing within-host diversity in Mtb [12]. Based on our data, we would hypothesize that the mechanisms of within-host clonal evolution in Mtb are likely ambiguous and largely dependent on host and environmental factors. Some transmission events are a mixture of clones while in others, one dominant clone is transmitted. Perhaps this transmitted diversity is necessary to guarantee infection and, ultimately, the survival of this obligate pathogen and this warrants further investigation [41].
We did not identify mutations conferring resistance to the anti-tuberculosis drugs in any isolates from P1 and phenotypic drug resistance conducted at baseline and 2 months after treatment initiation. However, we did not explore alternative drug resistance mechanisms, such as antibiotic tolerance, as explanation for why P1 took so long to eradicate the infection without observed de novo mutation in any of the serial isolates collected while under therapy [[42], [43], [44]]. Among the four antibiotics to treat drug susceptible TB, rifampin (RIF) is the most important and targets the RNA polymerase, beta subunit to inhibit bacterial transcription [45]. Recent data suggest RIF preferentially inhibits one of the two rpoB promotor regions, resulting in increased expression from the second promotor and increased bacterial growth under drug pressure [44]. This hypothesis should be addressed in future investigations.
There are some limitations to our study. First, we only had one genome for P2, which limited our ability to explore the full spectrum of within-host clonal evolution during transmission and under drug pressure. In addition, several of the within-host isolates were too contaminated to be useable. This may have limited our ability to track the full dynamics of the within-host minority allele turn over in these discarded time-intervals. In addition, we selectively deep sequenced a limited number of isolates and had uneven genome coverage across sampling intervals, which meant that some low frequency SNPs were likely not observed. We subcultured the isolates, first on LJ and then MGIT, to generate enough genomic DNA for sequencing. It is possible that these serial subculture steps resulted in a loss of genetic heterogeneity, which would partially explain the observed changes in genetic diversity [46,47]. One approach to circumvent this bias is direct sequencing from sputum sample [47]. However, the design of our study limited us to working with isolates. All three patients in our study initially provided sputum samples following standard procedures for TB diagnosis and treatment management at the clinic. Two patients in particular, P1 and P3 had several isolates collected over the course of therapy. We tracked subclonal populations across these sampling intervals and observed distinct signatures of drift and negative selection under drug pressure (Fig. 3). Nevertheless, we must acknowledge ascertainment bias in the underlying within-host diversity. Indeed sampling effects whereby subclonal populations are differentially captured would also present as purifying selection. These data raise major concerns that even when multiple samples are analysed, we do not capture the underlying within-host heterogeneity in clinical practice [11, 48]. Our study, however, has several strengths in that it provides an in-depth look at the variability within and between a cluster of patients who are pan-susceptible to first-line therapy and thus avoids variability introduced due to differing history of acquired drug resistance. Additionally, because we have an epidemiologically well linked TB cluster, we are able to compare the intra- and inter-patient variability without ambiguity as to the cases' relatedness.
WGS is becoming the new “gold standard” in molecular epidemiology of TB. Mtb is exceptionally slow-growing and slow to mutate, and the level of genetic diversity is very low compared to other pathogens. Nevertheless, WGS has shown that there is more diversity than previously recognized. WGS provides unparalleled sensitivity to detect small genetic changes, enabling the determination of directionality of transmission by comparing numbers of SNPs between cases and over time. However, in order to effectively utilize WGS in public health, clinical practice, and research into Mtb, we must understand the extent of intra-patient variability. More analyses are needed to elucidate the mechanism and functionality of this variability. Future studies on this subject should include greater numbers of patients (both index and secondary cases) and more in-depth sequencing of isolates. As more TB programs transition to WGS for routine strain surveillance and outbreak investigation, more data is needed on the role of transmission bottleneck in observed heterogeneity between cases. Ultimately, these data may help guide the integration of minority allele frequency to refine transmission estimates [49].
Acknowledgments
Acknowledgements
We thank Pia N. Kristiansen from the Statens Serum Institut for technical assistance with DNA isolation and sequencing. We thank Dr. Matthias Merker at the German Tuberculosis Reference Center in Borstel for providing the metadata used for the global phylogenetic tree in Fig. S1. We thank the staff of the Florida Department of Health (FDOH), Section of Tuberculosis Control, for providing the de-identified surveillance data, with special acknowledgment of Thomas Privett, Jose Zabala, and Lori Johnston. We thank the FDOH Bureau of Public Health Laboratories for performing patient testing, including sequencing of P3 isolates. Finally, we acknowledge the tireless work of the Florida Department of Health Staff throughout the 67 counties in Florida, especially those in the county where these cases were diagnosed and received care.
Funding sources
This research was supported by the NIH/NCATS Clinical and Translational Science Award to the University of Florida KL2TR001429 and a Department of Medicine Gatorade start-up research grant (MNS). The funding organizations had no role in the design, conduct, collection, analysis, and interpretation of the data and no role in the preparation, review, or approval of the manuscript. The corresponding author had full access to all data in the study and had final responsibility for the decision to submit for publication.
Declaration of Competing Interests
We declare no competing interests.
Author contributions
MNS, EMR, ML and TL conceived and designed the study. CC, MR, and EMR collected the data, performed the laboratory experiments and sequencing. AN, MNS, TL, and ML developed, performed, and interpreted the analyses. MNS and AN wrote the first draft of the manuscript. All authors critically reviewed and approved the final version of the report.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.08.010.
Appendix A. Supplementary data
Supplementary material
References
- 1.World Health Organization . 2018. Global Tuberculosis Report 2018. Geneva. [Google Scholar]
- 2.Glaziou P., Floyd K., Raviglione M.C. Global epidemiology of tuberculosis. Semin. Respir. Crit. Care Med. 2018;39:271–285. doi: 10.1055/s-0038-1651492. [DOI] [PubMed] [Google Scholar]
- 3.Castro K.G., Jaffe H.W. Rationale and methods for the national tuberculosis genotyping and surveillance network. Emerg. Infect. Dis. 2002;8:1188–1191. doi: 10.3201/eid0811.020408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niemann S., Supply P. Diversity and evolution of Mycobacterium tuberculosis: moving to whole-genome-based approaches. Cold Spring Harb. Perspect. Med. 2014;4 doi: 10.1101/cshperspect.a021188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker T.M., Ip C.L.C., Harrell R.H., Evans J.T., Kapatai G., Dedicoat M.J. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect. Dis. 2013;13:137–146. doi: 10.1016/S1473-3099(12)70277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hatherell H.-A., Colijn C., Stagg H.R., Jackson C., Winter J.R., Abubakar I. Interpreting whole genome sequencing for investigating tuberculosis transmission: a systematic review. BMC Med. 2016;14:21. doi: 10.1186/s12916-016-0566-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coll F., McNerney R., Guerra-Assunção J.A., Glynn J.R., Perdigão J., Viveiros M. A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat. Commun. 2014;5 doi: 10.1038/ncomms5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Köser C.U., Bryant J.M., Becq J., Török M.E., Ellington M.J., Marti-Renom M.A. Whole-genome sequencing for rapid susceptibility testing of M. tuberculosis. N. Engl. J. Med. 2013;369:290–292. doi: 10.1056/NEJMc1215305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lieberman T.D., Wilson D., Misra R., Xiong L.L., Moodley P., Cohen T. Genomic diversity in autopsy samples reveals within-host dissemination of HIV-associated Mycobacterium tuberculosis. Nat. Med. 2016;22:1470–1474. doi: 10.1038/nm.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merker M., Kohl T.A., Roetzer A., Truebe L., Richter E., Rüsch-Gerdes S. Whole genome sequencing reveals complex evolution patterns of multidrug-resistant Mycobacterium tuberculosis Beijing strains in patients. PLoS One. 2013;8 doi: 10.1371/journal.pone.0082551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trauner A., Liu Q., Via L.E., Liu X., Ruan X., Liang L. The within-host population dynamics of Mycobacterium tuberculosis vary with treatment efficacy. Genome Biol. 2017;18:71. doi: 10.1186/s13059-017-1196-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Copin R., Wang X., Louie E., Escuyer V., Coscolla M., Gagneux S. Within host evolution selects for a dominant genotype of Mycobacterium tuberculosis while T cells increase pathogen genetic diversity. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1006111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dheda K., Lenders L., Magombedze G., Srivastava S., Raj P., Arning E. Drug-penetration gradients associated with acquired drug resistance in patients with tuberculosis. Am. J. Respir. Crit. Care Med. 2018;198:1208–1219. doi: 10.1164/rccm.201711-2333OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jorth P., Staudinger B.J., Wu X., Hisert K., Hayden H., Garudathri J. Regional isolation drives bacterial diversification within cystic fibrosis lungs. Cell Host Microbe. 2015;18:307–319. doi: 10.1016/j.chom.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun G., Luo T., Yang C., Dong X., Li J., Zhu Y. Dynamic population changes in Mycobacterium tuberculosis during acquisition and fixation of drug resistance in patients. J. Infect. Dis. 2012;206:1724–1733. doi: 10.1093/infdis/jis601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allix-Béguec C., Harmsen D., Weniger T., Supply P., Niemann S. Evaluation and strategy for use of MIRU-VNTRplus, a multifunctional database for online analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates. J. Clin. Microbiol. 2008;46:2692–2699. doi: 10.1128/JCM.00540-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mazars E., Lesjean S., Banuls A.L., Gilbert M., Vincent V., Gicquel B. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc. Natl. Acad. Sci. U. S. A. 2001;98:1901–1906. doi: 10.1073/pnas.98.4.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wood D.E., Salzberg S.L. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu J., Breitwieser F.P., Thielen P., Salzberg S.L. Bracken: estimating species abundance in metagenomics data. PeerJ Comput. Sci. 2017;3 [Google Scholar]
- 20.Wilm A., Aw P.P.K., Bertrand D., Yeo G.H.T., Ong S.H., Wong C.H. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012;40:11189–11201. doi: 10.1093/nar/gks918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Neill M.B., Mortimer T.D., Pepperell C.S. Diversity of Mycobacterium tuberculosis across evolutionary scales. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016 doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gouy M., Guindon S., Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 24.Coll F., McNerney R., Preston M.D., Guerra-Assunção J.A., Warry A., Hill-Cawthorne G. Rapid determination of anti-tuberculosis drug resistance from whole-genome sequences. Genome Med. 2015;7:51. doi: 10.1186/s13073-015-0164-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stucki D., Brites D., Jeljeli L., Coscolla M., Liu Q., Trauner A. Mycobacterium tuberculosis lineage 4 comprises globally distributed and geographically restricted sublineages. Nat. Genet. 2016 doi: 10.1038/ng.3704. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brynildsrud O.B., Pepperell C.S., Suffys P., Grandjean L., Monteserin J., Debech N. Global expansion of Mycobacterium tuberculosis lineage 4 shaped by colonial migration and local adaptation. Sci. Adv. 2018;4 doi: 10.1126/sciadv.aat5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamichhane G., Zignol M., Blades N.J., Geiman D.E., Dougherty A., Grosset J. A postgenomic method for predicting essential genes at subsaturation levels of mutagenesis: application to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 2003;100:7213–7218. doi: 10.1073/pnas.1231432100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sutcliffe I.C., Harrington D.J. Lipoproteins of Mycobacterium tuberculosis: an abundant and functionally diverse class of cell envelope components. FEMS Microbiol. Rev. 2004;28:645–659. doi: 10.1016/j.femsre.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 29.Becker K., Sander P. Mycobacterium tuberculosis lipoproteins in virulence and immunity – fighting with a double-edged sword. FEBS Lett. 2016;590:3800–3819. doi: 10.1002/1873-3468.12273. [DOI] [PubMed] [Google Scholar]
- 30.Cimino M., Thomas C., Namouchi A., Dubrac S., Gicquel B., Gopaul D.N. Identification of DNA binding motifs of the Mycobacterium tuberculosis PhoP/PhoR two-component signal transduction system. PLoS One. 2012;7 doi: 10.1371/journal.pone.0042876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schreuder L.J., Carroll P., Muwanguzi-Karugaba J., Kokoczka R., Brown A.C., Parish T. Mycobacterium tuberculosis H37Rv has a single nucleotide polymorphism in PhoR which affects cell wall hydrophobicity and gene expression. Microbiology. 2015;161:765–773. doi: 10.1099/mic.0.000036. [DOI] [PubMed] [Google Scholar]
- 32.Chiner-Oms Á., Sánchez-Busó L., Corander J., Gagneux S., Harris S.R., Young D. Genomic determinants of speciation and spread of the Mycobacterium tuberculosis complex. Sci. Adv. 2019;5:eaaw3307. doi: 10.1126/sciadv.aaw3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pérez-Lago L., Comas I., Navarro Y., González-Candelas F., Herranz M., Bouza E. Whole genome sequencing analysis of intrapatient microevolution in Mycobacterium tuberculosis: potential impact on the inference of tuberculosis transmission. J. Infect. Dis. 2014;209:98–108. doi: 10.1093/infdis/jit439. [DOI] [PubMed] [Google Scholar]
- 34.Herranz M., Pole I., Ozere I., Chiner-Oms Á., Martínez-Lirola M., Pérez-García F. Mycobacterium tuberculosis acquires limited genetic diversity in prolonged infections, reactivations and transmissions involving multiple hosts. Front. Microbiol. 2018;8 doi: 10.3389/fmicb.2017.02661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cohen T., Chindelevitch L., Misra R., Kempner M.E., Galea J., Moodley P. Within-host heterogeneity of Mycobacterium tuberculosis infection is associated with poor early treatment response: a prospective cohort study. J. Infect. Dis. 2016;213:1796–1799. doi: 10.1093/infdis/jiw014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Q., Via L.E., Luo T., Liang L., Liu X., Wu S. Within patient microevolution of Mycobacterium tuberculosis correlates with heterogeneous responses to treatment. Sci. Rep. 2015;5 doi: 10.1038/srep17507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Worby C.J., Lipsitch M., Hanage W.P. Within-host bacterial diversity hinders accurate reconstruction of transmission networks from genomic distance data. PLoS Comput. Biol. 2014;10 doi: 10.1371/journal.pcbi.1003549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alizon S., Luciani F., Regoes R.R. Epidemiological and clinical consequences of within-host evolution. Trends Microbiol. 2011;19:24–32. doi: 10.1016/j.tim.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 39.Lillebaek T., Norman A., Rasmussen E.M., Marvig R.L., Folkvardsen D.B., Andersen Å.B. Substantial molecular evolution and mutation rates in prolonged latent Mycobacterium tuberculosis infection in humans. Int. J. Med. Microbiol. 2016;306:580–585. doi: 10.1016/j.ijmm.2016.05.017. [DOI] [PubMed] [Google Scholar]
- 40.Ford C.B., Lin P.L., Chase M., Shah R.R., Iartchouk O., Galagan J. Use of whole genome sequencing to estimate the mutation rate of Mycobacterium tuberculosis during latent infection. Nat. Genet. 2011;43:482–486. doi: 10.1038/ng.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gagneux S. Host-pathogen coevolution in human tuberculosis. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2012;367:850–859. doi: 10.1098/rstb.2011.0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wiuff C., Zappala R.M., Regoes R.R., Garner K.N., Baquero F., Levin B.R. Phenotypic tolerance: antibiotic enrichment of noninherited resistance in bacterial populations. Antimicrob. Agents Chemother. 2005;49:1483–1494. doi: 10.1128/AAC.49.4.1483-1494.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matern W.M., Rifat D., Bader J.S., Karakousis P.C. Gene enrichment analysis reveals major regulators of Mycobacterium tuberculosis gene expression in two models of antibiotic tolerance. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.00610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu J.-H., Wang B.-W., Pan M., Zeng Y.-N., Rego H., Javid B. Rifampicin can induce antibiotic tolerance in mycobacteria via paradoxical changes in rpoB transcription. Nat. Commun. 2018;9 doi: 10.1038/s41467-018-06667-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarkar S., Ganguly A., Sunwoo H.H. Current overview of anti-tuberculosis drugs: metabolism and toxicities. Mycobact. Dis. 2016;6 [Google Scholar]
- 46.Metcalfe J.Z., Streicher E., Theron G., Colman R.E., Penaloza R., Allender C. Mycobacterium tuberculosis subculture results in loss of potentially clinically relevant heteroresistance. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.00888-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nimmo C., Shaw L.P., Doyle R., Williams R., Brien K., Burgess C. Correction to: whole genome sequencing Mycobacterium tuberculosis directly from sputum identifies more genetic diversity than sequencing from culture. BMC Genomics. 2019;20:433. doi: 10.1186/s12864-019-5841-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shockey A.C., Dabney J., Pepperell C.S. Effects of host, sample, and in vitro culture on genomic diversity of pathogenic mycobacteria. Front. Genet. 2019;10 doi: 10.3389/fgene.2019.00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin M.A., Lee R.S., Cowley L.A., Gardy J.L., Hanage W.P. Within-host Mycobacterium tuberculosis diversity and its utility for inferences of transmission. Microb. Genomics. 2018 doi: 10.1099/mgen.0.000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material