Abstract
Background
CpG island methylator phenotype (CIMP), a common biological phenomenon characterized by a subset of concurrently methylated genes, can have an influence on the progression of multiple cancers. However, the potential mechanism of CIMP in hepatocarcinogenesis and its clinical relevance remains only partially understood.
Methods: We used a methylation array from the cancer genome atlas (TCGA) to stratify HCC patients into different CIMP subtypes, and evaluated their correlation with clinical characteristics. In addition, mutation, CNV, and transcriptome profiles were also utilized to evaluate the distinctive genomic patterns correlated with CIMP. Finally, a CIMP-associated prognostic model (CPM) was trained and validated using four independent datasets.
Findings
A subgroup of patients was identified as having CIMP-H, which was associated with worse OS and DFS. Gene enrichment analysis indicated that the terms “liver cancer with EPCAM up”, “tumor invasiveness up”, “methyltransferase complex”, and “translational initiation” were enriched in CIMP-H subgroup. Notably, somatic mutation analysis indicated that CIMP-H patients presented with a higher mutation burden of BRD4, DDIAS and NOX1. Moreover, four CPM associated genes could significantly categorize patients into low- and high-risk groups in the training dataset and another 3 independent validation datasets. Finally, a nomogram incorporating a classifier based on four mRNAs, pathological M stage and CIMP status was established, which showed a favorable discriminating ability and might contribute to clinical decision-making for HCC.
Interpretation
Our work highlights the potential clinical application value of CPM in predicting the overall survival of HCC patients and the mechanisms underlying the role of CIMP in hepatocarcinogenesis.
Fund
This work was supported by the State Key Project on Infectious Diseases of China (2018ZX10723204-003), the National Nature Science Foundation of China (Nos. 81874065, 81500565, 81874149, 81572427, and 81401997), the Hepato-Biliary-Pancreatic Malignant Tumor Investigation Fund of Chen Xiao-ping Foundation for the Development of Science and Technology of Hubei Province (CXPJJH11800001-2018356).
Keywords: Hepatocellular carcinoma, Methylation, CpG island methylator phenotype, Prognostic signature
Research in context.
Evidence before this study
By searching PubMed on Jul 13, 2019, for original articles containing the terms “CpG island methylator phenotype AND hepatocellular carcinoma” without language or date restrictions, we find that the links between the molecular traits and CIMP have not been fully unveiled. In addition, this search also did not identify any studied, based on the high-throughput profiles, which had evaluated the potential prognostic role of CIMP-associated models in HCC.
Added value of this study
To our knowledge, our study is the first one to use clinical information and genomic profiles from TCGA to investigate the association between CIMP phenotype and genomic aberrations, immune infiltration. In addition, we identify the CIMP-associated prognostic model (CPM) in HCC, which was trained and validated using four independent datasets. This model is on the basis of four genes that could screen out the HCC patients with high risk of poor prognosis in both the training and validation cohorts. Our results indicate that this CPM is more accurate than conventional clinical characteristics alone, and a nomogram was also constructed for clinical practice to predict HCC prognosis.
Implication of all the available evidence
The CIMP-related prognostic model based on four genes was constructed and validated. It was found to act as an independent prognostic factor for HCC and reflects the overall epigenetic alterations in the whole genome. To our best knowledge, this is the first report of a prognostic model incorporating CIMP status and it could be utilized as a reference to understand the relevance of CIMP in other malignancies. Notably, the CPM provides epigenetic insights into the main mechanisms that potentially influence the prognosis of HCC.
Alt-text: Unlabelled Box
1. Introduction
Liver cancer ranks fifth among the most common malignancies and is the second leading cause of tumor-related death with an increasing global incidence in recent years [1,2]. Hepatocellular carcinoma (HCC), the predominant type of liver cancer, is correlated with well-known underlying etiologies, including chronic hepatitis (B and C) virus infections, alcohol abuse and aflatoxin exposure [3]. Under the influence of these risk factors, both genetic and epigenetic alterations will progressively accumulate and might contribute to activate oncogenes, leading to the inactivation of tumor suppressor genes and subsequently resulting in hepatocarcinogenesis [4]. Presently, the main treatments for HCC include hepatectomy, targeted therapy with sorafenib, thermal ablation, immunotherapy, transcatheter arterial chemoembolization (TACE) and liver transplantation [[5], [6], [7]]. However, in spite of great advances over the past decades, the prognosis of HCC patients remains poor due to high rate of recurrence [2]. Therefore, it is urgent to identify robust prognostic biomarkers for HCC patients who might benefit from curative therapy.
DNA methylation plays a crucial role in both physiologic and pathological cellular fate commitment [8]. During oncogenesis, aberrant DNA methylation mostly presents as focal hypermethylation surrounding the promoters of specific genes, as well as global hypomethylation in non-promoter regions [9,10]. Hypermethylation of promoter region is a crucial process that can lead to the epigenetic silencing of tumor suppressor genes [11,12]. At the same time, aberrant DNA methylation of non-promoter elements is an important contributor to intra-tumoral heterogeneity [13]. By contrast, global hypomethylation might contribute to chromosomal instability by affecting the intergenic regions of the whole genome.
CIMP is characterized by concurrent and widespread hypermethylation of a subset of CpG sites in clinically distinct cancer subtypes, and it plays a crucial role in chromosomal instability during carcinogenesis [14,15]. This aberrant methylation phenomenon was first discovered and validated in colorectal cancer, where it was termed colorectal CIMP [16,17]. From then on, the CIMP phenotype has been evaluated in a wide variety of other tumor types, including HCC [18]. The CIMP positive subgroup has a number of distinct epidemiological, clinicopathological, and genomic characteristics compared with its CIMP negative counterpart [19,20]. Since previous studies indicated that there is a lack of a pan-cancer overlap of specific gene, it appears that a tissue-specific CIMP pattern exists for each tumor [21]. However, the clinical relevance of CIMP in HCC still remains controversial, and the potential biological mechanism mediating its involvement in hepatocarcinogenesis is also only partially understood.
In the present study, we firstly clustered HCC patients into three distinct methylation subgroups, named CIMP-H, CIMP-M, and CIMP-L, and analyzed the diverse clinicopathological features correlated with CIMP status. The results indicated that patients with CIMP-H status had a worse prognosis. In addition, the correlations of CIMP status with RNA-seq datasets, somatic mutations, and copy number variations (CNVs) in HCC were also evaluated. Importantly, our robust CPM, consisting of four genes whose expression levels are influenced by CIMP status, was demonstrated as a robust prognostic model with favorable predictive performance. Therefore, CPM can greatly contribute to the decision-making process of clinicians, and related genes might act as promising therapeutic biomarkers for HCC.
2. Materials and methods
2.1. Data acquisition from TCGA
Level 3 DNA methylation profiles based on the Illumina HumanMethylation450 BeadChip Assay, including 377 HCC patients, were obtained from the TCGA using the TCGA-Assembler 2 R package [22], and the genomic annotation of each CpG site was conducted using the IlluminaHumanMethylation450kanno.ilmn12.hg19 R package (Version: 3.9; http://www.bioconductor.org/packages/release/data/annotation/html/IlluminaHumanMethylation450kanno.ilmn12.hg19.html). For individual CpG site, two measurements were accepted: a methylated intensity (denoted by M) and an unmethylated intensity (denoted by U). The methylation status of each CpG was expressed as a beta-value (β = M/(M + U), ranging from 0 to 1) [23]. We used the minfi R package [24] to remove low-quality probes based on the following criteria: first, the methylation of CpG site not available in any sample; second, single-nucleotide polymorphisms (SNPs) located in the assayed CpG dinucleotide [25]; third, not uniquely mapped to the human reference genome (hg19) [26]; fourth, locating in sex chromosomes [27].
Gene expression profiles obtained using the Illumina HiSeq RNA-Seq platform and the corresponding clinical information of 371 HCC patients were downloaded from the TCGA website (https://portal.gdc.cancer.gov/repository) (up to May 15, 2019). Gene symbols corresponding to ensemble IDs were obtained using the Homo_sapiens.GRCh38.91.chr.gtf file (http://asia.ensembl.org/index.html). The gene expression profiles were normalized using the scale method provided in the limma R package [28]. The highest RNA expression level was accepted in case of duplicates. In addition, genes with an average expression value of more than one were extracted, and low-abundance profiles were eliminated.
The somatic mutation profiles of 364 patients based on the whole exome sequencing platform were downloaded using the TCGAbiolinks [29]. They were summarized and analyzed using the maftools package [30]. Samples with missense mutations, nonsense mutations, multiple hits, splice-site mutations, frameshift insertions, frameshift deletions, in-frame insertions or in-frame deletions were regarded as positive for a mutation. Significant somatic mutated genes (SMGs) in three distinct subtype of HCC were identified using the Mutational Significance in Cancer (MuSiC) Genome Suite, (http://gmt.genome.wustl.edu/packages/genome-music/index.html). Tumor mutational burden (TMB), an emerging biomarker of immunotherapy responses, was calculated by the number of somatic, coding, base substitution, and indel mutations per megabase within the whole genome. In addition, 35 Mb was used as the estimated size of the whole human exome in [31].
Level 4 copy number variation (CNV) profiles of HCC were downloaded from GDAC Firehose (http://gdac.broadinstitute.org) and classified into three distinct subtypes according to the status of CIMP. Significant amplification or deletion alterations among the whole genome were identified using GISTIC 2.0, a biological program based on robust computational algorithm to detect recurrent somatic CNVs by evaluating the frequency and amplitude of corresponding events [32].
Complete clinical information of HCC patients in the TCGA cohort was collected, including gender, Body Mass Index (BMI), age, AFP (alpha fetoprotein) level, hepatitis C (HCV) status, hepatitis B (HBV) status, pathologic stage, vascular invasion, histologic grade, pathologic TNM stage, family, alcohol consumption and non-alcoholic fatty liver disease history (NAFLD), and used for the subsequent analyses.
This study fully complies with the TCGA publication requirements (http://cancergenome.nih.gov/publications/publicationguidelines).
2.2. Acquisition of gene expression matrix from ICGC and GEO
The gene expression files (ICGC-LIRI-JP) of HCC based on the Illumina HiSeq RNA Seq platform, including 212 Japanese patients, were obtained from the international cancer genomics consortium (ICGC, https://icgc.org/) [33]. The GSE14520 gene expression matrix files based on platform GPL571, including 225 Chinese patients were obtained from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). Among the two datasets, the highest RNA expression value was accepted when encountering duplicated data. Genes with an average expression value more than one were extracted, and low-abundance profiles were eliminated. Only patients in the two datasets with >90 days of overall survival (OS) (ICGC LIRI JP (n = 197), and GSE14520 (n = 216)) were extracted to validate CPM in the survival analysis. The downloaded profiles fully complied with the ICGC and GEO data access policies.
2.3. Collection of HCC patients in the Tongji cohort
From 2013 to 2017, surgical biopsies were collected from a total of 100 patients who underwent curative hepatic resection for HCC without preoperative therapy at Tongji Hospital (Wuhan, China). OS information was collected through electronic medical records or telephone follow-up. Informed consent forms to donate their tissue samples for biomedical research, which were approved by the ethics committee of Tongji Hospital, were signed by all patients.
2.4. Identification and validation of the CpG island methylator phenotype (CIMP)
To evaluate the CIMP phenomenon in HCC, CpG sites with a relatively high standard deviation of beta-values in 377 HCC tissues (SD > 0.2) and relatively low beta-values in 50 paracancerous tissues (mean β value <0.05), were selected as the most variable CpG sites for further survival analysis. The remaining CpG sites, which were considered to be significantly associated with OS based on the threshold of P < .05, were extracted for further cluster analysis. The ConsensusClusterPlus R package was used to conduct consensus cluster analysis based on the final 95 probes was on the basis of the K-means algorithm [34]. The correlations between clinical features and each cluster were evaluated using the chi-square or Fisher's exact test.
To validate the robustness of our 95 probes in clustering HCC patients into different CIMP-related subtypes, the GSE56588 DNA methylation profiles based on the Illumina HumanMethylation450 BeadChip, including 224 HCC samples and 10 normal tissues, was downloaded from the GEO database [35]. The method of cluster analysis in the GSE56588 cohort was the same as in the TCGA cohort.
2.5. Gene set enrichment analysis (GSEA)
In order to evaluate the potential mechanism underlying the involvement of CIMP in hepatocarcinogenesis, we performed GSEA analysis (Version: 3.0; http://software.broadinstitute.org/gsea/index.jsp) to identify the difference of the pathways and corresponding biomarkers between HCC patients with distinct CIMP statuses in the TCGA cohort [36]. The annotated gene set file (msigdb.v6.2.symbols.gmt) was accepted for our analysis as the reference. The significance was also based on the threshold of P < .05.
2.6. Differentially expressed gene (DEG) analysis
Analysis of differentially expressed genes among the three CIMP-related subtypes in the TCGA HCC cohort was performed using the DESeq2 R package [37]. We calculated the adjusted P-values of each gene using False-discovery rate (FDR) method. An FDR of <0.05 and absolute log2-fold change of >1 was set as the cut-off to detect DEGs.
2.7. Functional enrichment analysis
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses based on the CIMP-related DEGs were conducted using the clusterProfiler R package to evaluate the potential biological mechanisms mediating CIMP-related hepatocarcinogenesis [38]. We visualized significant biological pathways and processes using the GOplot R package (Version: 1.0.2; https://cran.r-project.org/web/packages/GOplot/index.html).
2.8. Construction and validation of a CIMP-related prognostic model
A total of 371 samples, including 109 CIMP-M, 185 CIMP-L and 77 CIMP-H patients, with both RNA-sequencing profiles and clinical patterns were extracted for further analyses. We evaluated the prognostic significance of 1179 DEGs based on the univariate Cox regression analysis, and those with P values <.05 were selected for further analyses. Importantly, the least absolute shrinkage and selection operator (LASSO) with L1-penalty, a widespread machine learning algorithm adopting explicable prediction rules that can solve the collinearity problem by dimension reduction, was utilized in our study [39]. Based on the prognostic CIMP-related DEGs, which were significant in the univariate Cox regression analysis, key CIMP-related genes were extracted using the LASSO algorithm. In this method, a sub-selection of CIMP-related biomarkers involved in hepatocarcinogenesis was extracted by shrinking the regression coefficient through using a penalty proportional to their size. Finally, we retained a relatively small group of biomarkers with nonzero regression coefficient. Conversely, the majority of the potential biomarkers were eliminated, with zero regression coefficients. Consequently, LASSO-penalized Cox regression analysis was performed to further narrow the range of candidate CIMP-related prognostic biomarkers. In our LASSO analysis performed using the glmnet R package (version: 2.0-16; https://cran.r-project.org/web/packages/glmnet/index.html), the dataset was sub-sampled 1000 times and the biomarkers that were repeated >900 times were selected as CIMP-related biomarkers. In the end, a CIMP-related prognostic signature was identified by extracting the regression coefficients from multivariate Cox regression analysis, and the risk score of each patient was calculated by multiplying the normalized expression level of each CIMP-related biomarker with its corresponding regression coefficients. The optimal cut-off was determined using the surv_cutpoint function of the survminer R package (Version: 0.4.3, https://cran.r-project.org/web/packages/survminer/index.html) to separate the patients into high- and low- risk subgroups. The OS and RFS rates of these subgroups were compared using Kaplan–Meier analysis based on the log-rank test. Multivariate Cox regression analyses were then implemented to detect independent risk factors correlated with the OS and RFS. Hazard ratios and corresponding 95% confidence intervals (95% CI) were also calculated in both univariate and multivariate Cox regression analyses. Finally, receiver operating characteristic (ROC) curve analyses were conducted using the survivalROC R package to investigate the prognostic performance of the model in four independent cohorts [40].
2.9. qRT-PCR
Total RNA from 100 fresh-frozen HCC tissue samples was extracted by TRIzol Reagent (Life Technologies, Carlsbad, CA, USA) and reverse transcribed using the PrimeScript® RT reagent Kit (Takara Bio, Dalian, China) according to the manufacturer's protocol. Further, quantitative real-time PCR was performed using the CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) and the SYBR Green Supermix kit (Takara Bio) according to the manufacturers' instructions. The expression levels of each genes were analyzed using the 2-ΔCT method with Homo sapiens GAPDH as the control housekeeping gene. We normalized the expression of the four genes using the scale function in R software, a generic function whose default method centered and scaled the data. The primers used in this study are summarized in Table S1.
2.10. 5-Aza-2′-deoxycytidine treatment
For methylation regulation analysis, HCC cell lines (LM3 and Huh7 cells) were split to low density (30% confluence) 12 h before treatment. Cells were treated with 5-Aza-2′-deoxycytidine (DAC, Sigma, St. Louis, MO, USA) at a concentration of 15 μM in the growth medium, which was exchanged every 24 h for a total of 48 h and cultured at 37 °C in a 5% CO2 incubator. At the end of the treatment period, cells were prepared for extraction of total RNA. The change of four genes expression was identified by qRT-PCR analysis and each sample makes three replicates.
2.11. Quantification of genome-wide DNA methylation
Genomic DNA from LM3 and Huh7 cells with or without 5-Aza-2′-deoxycytidine treatment was extracted using a QIAamp DNA Mini Kit (Qiagen, Germany) and each sample makes three replicates. Extracted DNA 500 ng from each sample was treated with sodium bisulfite using an EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA). Genome-wide DNA methylation of two HCC cell lines was quantified in bisulfite-converted genomic DNA at single-base resolution using the MethylationEPIC BeadChip (Illumina, San Diego, CA).
2.12. Single-sample gene set enrichment analysis (ssGSEA)
The tumor-infiltrating fraction of diverse immune cell subtypes was calculated using ssGSEA in the gsva R package (Version 1.32.0, http://www.bioconductor.org/packages/release/bioc/html/GSVA.html). The ssGSEA transforms specific gene expression patterns into quantities of immune cell populations in individual tumor samples [41]. The deconvolution algorithm utilized in the present study could distinguish 24 immune cell subtypes, including natural killer (NK) cells involved in innate immunity, as well as several B and T cell types involved in adaptive immunity. Kruskal-Wallis analysis was implemented to evaluate the differences in the tumor-infiltrating fractions of 24 human immune cell phenotypes between HCC patients with distinct CIMP statuses.
2.13. Construction and evaluation of the nomogram
The independent risk factors, identified by multivariate Cox regression analysis, were selected to construct a nomogram for the prediction of the likelihood of OS. Additionally, calibration plots were drawn to investigate the performance of the nomogram. The concordance index (C-index) was utilized to assess the consistency between the frequencies of the actual outcomes and probabilities of the model prediction. The nomograms and calibration plots were produced using the rms R package (Version 5.1-3.1, https://cran.r-project.org/web/packages/rms/). All statistical analyses were two-tailed, with a statistical significance level set at 0.05.
3. Results
3.1. Identification of the CpG island methylator phenotype in HCC
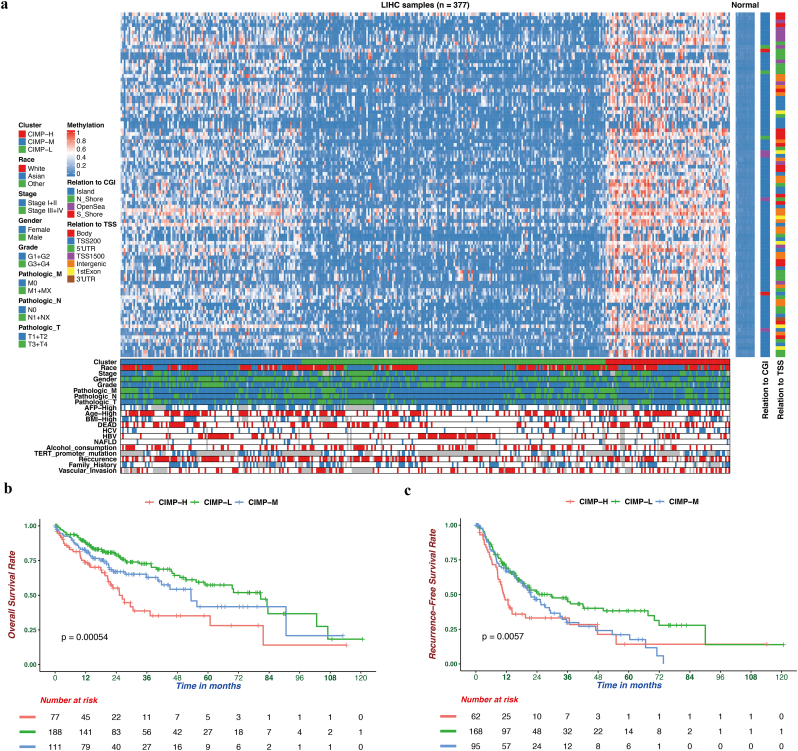
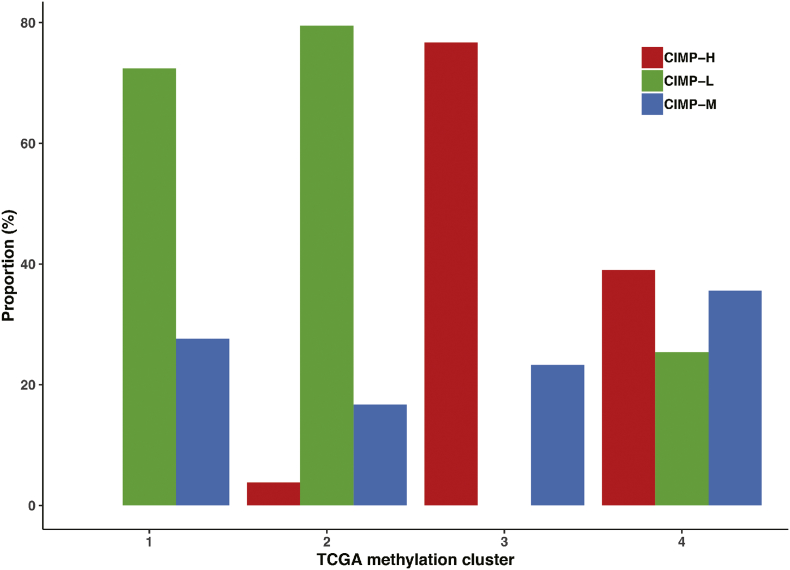
Based on the DNA methylation profiles of 377 HCC and 50 normal samples downloaded from TCGA, 95 most variable CpG sites (Table S2), which were correlated with OS, were extracted for unsupervised consensus clustering analysis. As a result, HCC patients were separated into three distinct groups (Fig. 1a). The methylation level of CIMP-L was the lowest and the patients in the CIMP-H subgroup had widespread hypermethylation among these variable CpG site. Furthermore, the robustness of these 95 most variable CpG sites was also conformed in an independent GSE56588 cohort from GEO (Fig. S1). Moreover, to compare the CIMP classification with previous four HCC hypermethylation clusters, we assigned each of HCC patients to one of the four DNA methylation-based subclasses from The Cancer Genome Atlas Research Network [42]. We found correspondence between the CIMP subtype and TCGA hypermethylation clusters. TCGA Cluster 1 and 2, presented degraded hypomethylation, consisted predominantly of CIMP-L patients, whereas TCGA Cluster 3, exhibited elevated hypermethylation, consisted predominantly of CIMP-H patients (Fig. S2).
Fig. 1.
The DNA methylation landscape of hepatocellular carcinoma. (a) Three methylation clusters were generated via k-means consensus clustering. The rows represent 95 CpGs that had high variation (SD > 0.2) in tumor tissues and low methylation levels (β value <0.05) in normal tissues. CIMP-H (red) presented a hypermethylation pattern in nearly all CpG sites and was regarded as the CpG island methylator phenotype. (b-c) Kaplan-Meier survival curves of each subtypes. The CIMP-H subgroup had a worse OS (b) and RFS (c) than the other groups. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. S1.
The DNA methylation landscape of hepatocellular carcinoma in the GSE56588 cohort. Three methylation clusters were generated via k-means consensus clustering based on the 95 variable CpG sites. CIMP-H (red) showed a hypermethylation pattern in nearly all CpG sites and was regarded as the CpG island methylator phenotype.
Fig. S2.
Relative proportions in each TCGA methylation cluster defined by CIMP phenotype.
To investigate whether the CIMP status was associated with OS, survival analysis was performed using the Kaplan-Meier method, and the P-value calculated using the log-rank analysis was approximately 0.0005, which indicated that there were highly significant differences in OS among the subtypes with different CIMP status (Fig. 1b). In addition, the relationship between CIMP status and relapse free survival (RFS) was also investigated, and the result indicated that significant differences (P = .0057) in RFS also existed among the subtypes with different CIMP status (Fig. 1c). In conclusion, the patients in the CIMP-H subgroup had the worst OS and RFS, while the CIMP-L subgroup had the best OS and RFS.
The associations between CIMP and clinical characteristics were also investigated. The demographic and clinicopathological characteristics of HCC patients in TCGA cohort are summarized in Table 1. There were no statistically significant differences in the majority of indices, with the exception of age, AFP level and HCV, HBV infection. There were significantly more patients with higher level of serum AFP (P = .003) in the CIMP-H subgroup. Conversely, significantly fewer patients in the CIMP-L subgroup were infected with HCV (P = .048) and this subgroup had a significantly lower age (P < .001).
Table 1.
Demographic and clinical characteristics of HCC patients from the TCGA cohort in different CIMP-related subgroups.
| CIMP-M | CIMP-L | CIMP-H | P-value | |
|---|---|---|---|---|
| Number of patients | 112 | 188 | 77 | |
| Age (mean (sd)) | 62.06 (11.18) | 56.68 (14.46) | 62.47 (12.92) | <.001 |
| AFP (median [IQR]) | 14.00 [4.75, 136.00] | 11.00 [3.00, 126.00] | 43.00 [7.50, 2599.00] | .003 |
| Pathologic_M (%) | .642 | |||
| M0 | 79 (70.5) | 138 (73.4) | 55 (71.4) | |
| M1 | 1 (0.9) | 1 (0.5) | 2 (2.6) | |
| MX | 32 (28.6) | 49 (26.1) | 20 (26.0) | |
| Pathologic_N (%) | .691 | |||
| N0 | 73 (65.2) | 129 (69.0) | 55 (71.4) | |
| N1 | 1 (0.9) | 3 (1.6) | 0 (0.0) | |
| NX | 38 (33.9) | 55 (29.4) | 22 (28.6) | |
| Pathologic_Stage (%) | .675 | |||
| Stage I | 54 (50.5) | 90 (51.4) | 31 (43.7) | |
| Stage II | 29 (27.1) | 42 (24.0) | 16 (22.5) | |
| Stage III | 23 (21.5) | 41 (23.4) | 22 (31.0) | |
| Stage IV | 1 (0.9) | 2 (1.1) | 2 (2.8) | |
| Pathologic_T (%) | .86 | |||
| T1 | 57 (50.9) | 95 (50.8) | 33 (43.4) | |
| T2 | 30 (26.8) | 46 (24.6) | 19 (25.0) | |
| T3 | 21 (18.8) | 39 (20.9) | 21 (27.6) | |
| T4 | 4 (3.6) | 7 (3.7) | 3 (3.9) | |
| Family_History (Yes %) | 36 (36.7) | 56 (33.3) | 22 (36.7) | .815 |
| Pathologic_Grade (%) | .712 | |||
| G1 | 17 (15.3) | 31 (16.6) | 7 (9.5) | |
| G2 | 53 (47.7) | 93 (49.7) | 34 (45.9) | |
| G3 | 37 (33.3) | 57 (30.5) | 30 (40.5) | |
| G4 | 4 (3.6) | 6 (3.2) | 3 (4.1) | |
| Race (%) | .488 | |||
| ASIAN | 47 (43.5) | 85 (46.4) | 29 (38.2) | |
| Other | 3 (2.8) | 11 (6.0) | 5 (6.6) | |
| WHITE | 58 (53.7) | 87 (47.5) | 42 (55.3) | |
| Vascular_Invasion (%) | .542 | |||
| Macro | 6 (6.6) | 6 (3.7) | 5 (7.4) | |
| Micro | 22 (24.2) | 51 (31.5) | 21 (30.9) | |
| None | 63 (69.2) | 105 (64.8) | 42 (61.8) | |
| Gender (Male %) | 82 (73.2) | 129 (68.6) | 44 (57.1) | .062 |
| NAFLD (Yes %) | 8 (7.3) | 9 (5.0) | 3 (4.3) | .617 |
| HBV = (Yes %) | 18 (25.7) | 56 (31.3) | 33 (30.3) | .685 |
| HCV (Yes %) | 20 (18.3) | 20 (11.2) | 16 (22.9) | .048 |
| Alcohol_consumption (Yes %) | 33 (30.3) | 65 (36.3) | 20 (28.6) | .391 |
| BMI (Low %) | 68 (68.0) | 129 (75.9) | 49 (69.0) | .304 |
3.2. Potential mechanism underlying the role of CIMP in hepatocarcinogenesis
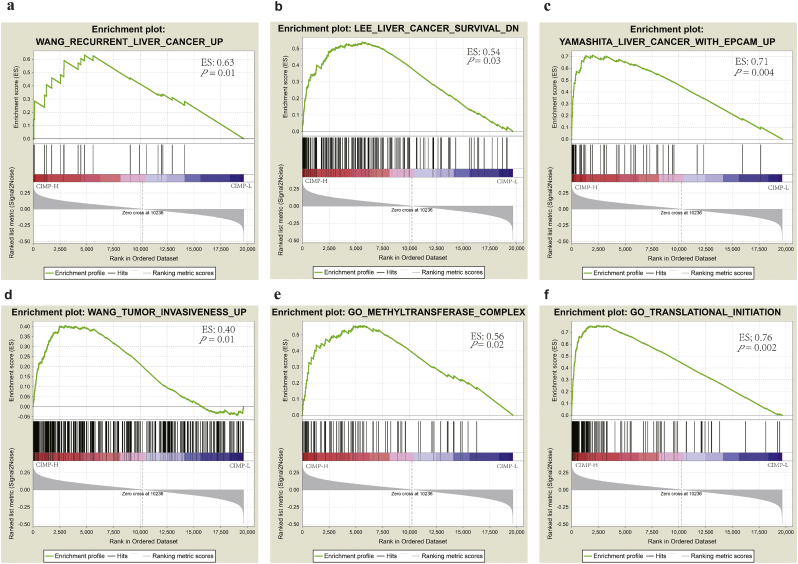
GSEA analysis of HCC patients with different CIMP status based on the RNA-seq profiles was performed to evaluate the potential biological mechanism by which CIMP influences hepatocarcinogenesis and the results indicated that the gene signatures of “Recurrent liver cancer up”, “Liver cancer survival down”, “Liver cancer with EPCAM up”, “Tumor invasiveness up”, “Methyltransferase complex”, and “Translational initiation” were enriched in patients with CIMP-H status (Fig. 2). Importantly, the negative relationships between CIMP and prognosis were also confirmed in the GSEA analysis, which were consistent with the results obtained from the TCGA cohort. In addition, it can be concluded that the signatures involved in the aforementioned biological process play an important role in the role of CIMP in hepatocarcinogenesis. In conjunction with the association between the expression of EPCAM and CIMP [43], these facts strongly indicate the potential role of CIMP as a tumor activator in hepatocarcinogenesis, and HCC in different CIMP subtypes originated in precursor cells might have a different epigenetic background of the cell of origin.
Fig. 2.
Gene set enrichment analysis of CIMP status in the TCGA dataset. (a-f) Significant enrichment in CIMP-H compared with the other groups in HCC.
3.3. The landscape of tumor-infiltrating immune cells in patients with different CIMP status
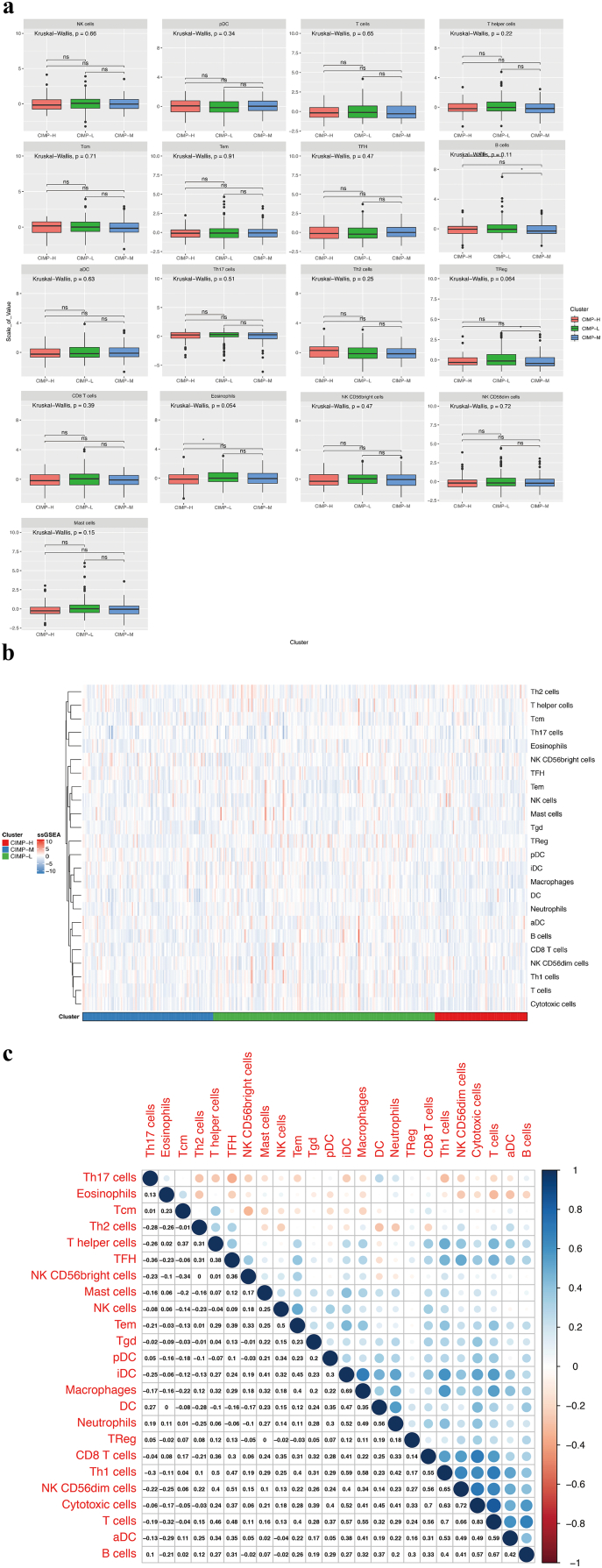
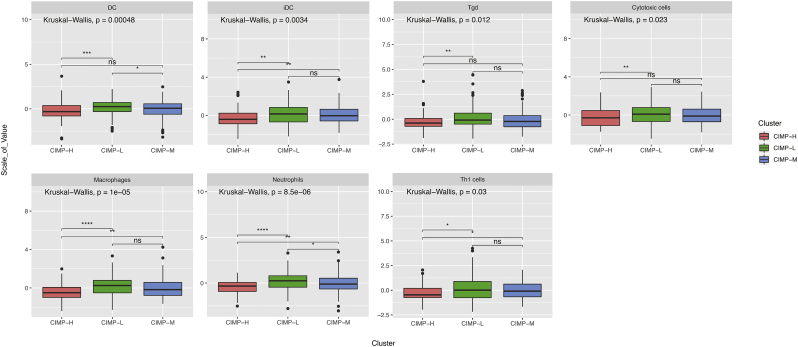
The ssGSEA function in gsva R package was used in combination with a signature matrix of 24 immune cell types to calculate the differences in the proportions of different infiltrating immune cells among patients with different CIMP status. The resulting heatmap summarizes the tumor-infiltrating immune-cells landscape of 371 HCC patients in the TCGA cohort (Fig. S3b). However, there were no statistically significant differences in the majority of immune-cell subtypes. In addition, the correlations among the tumor-infiltrating immune cell types were only weak or moderate (Fig. S3c). Nevertheless, the CIMP-H subgroup had significantly lower proportions of cytotoxic cells, dendritic cells, interdigitating cells, macrophages, neutrophils, gamma delta T cell (Tgd), and type 1 T. helper cell (Th1) cells than the other groups (P < .05, Figs. 3 and S3a). These results indicated that compared with its counterparts, patients in CIMP-H subgroup have a distinct immune phenotype, characterized by less immune infiltration, lower cytotoxic potential and immune activation. Since the T-cell immune response in antitumor immunity is the central event, effective immunotherapy of immune checkpoint inhibitors depends on the generation of neoantigen-specific T-cells and its penetration into the tumor microenvrironment. Consequently, immunotherapies are more likely to be less efficacious in CIMP-H phenotype with less immune infiltration, lower cytotoxic potential and immune activation.
Fig. S3.
The landscape of tumor-infiltrating immune cells between CIMP-H and the other groups in HCC. (a) No significant differences in the abundance of tumor-infiltrating immune cells between CIMP-H and the other groups in HCC. (b) Relative proportions of infiltrating immune cells between CIMP-H and the other groups in HCC. (c) Correlation matrix of all the proportions of24 detected immune cell types.
Fig. 3.
The significant differences in the abundance of tumor-infiltrating immune cells between CIMP-H and the other groups in HCC.
3.4. Analysis of mutations and CNVs in patients with distinct CIMP statuses
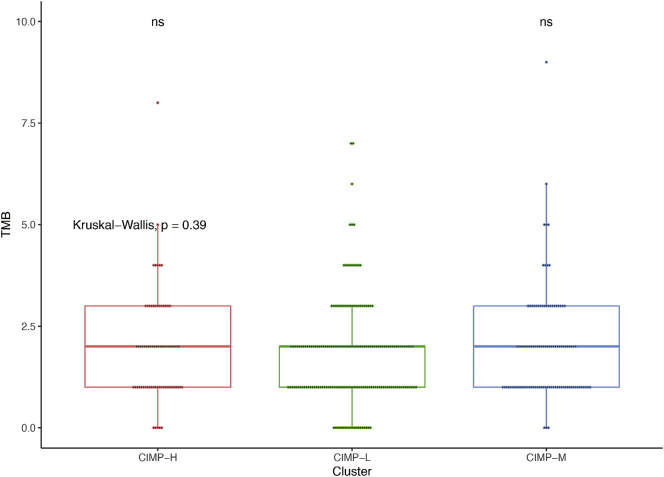
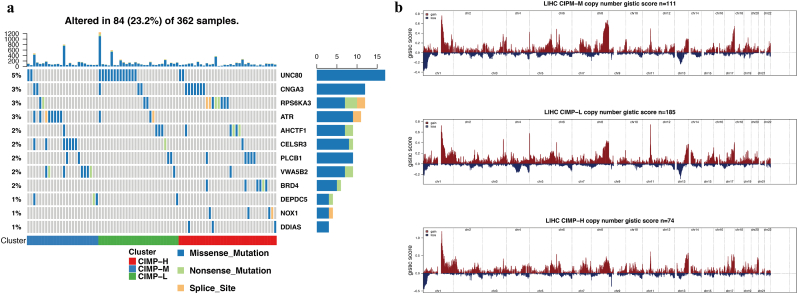
In previous studies, the precise landscape of the driver gene mutations of HCC was illustrated based on whole genomic profiles [42], Consequently, the correlation of CIMP with multiple omics mutational profiles was investigated in present study. There was no significant difference of tumor mutational burden (TMB), an emerging biomarker of immunotherapy responses, among the patients in the different CIMP-related subgroups (Fig. S4). However, patients in the CIMP-H group had significantly higher somatic mutation burdens in DDIAS and NOX1 (Fig. 4a), which have been shown to play an important role in the carcinogenesis of multiple tumors [[44], [45], [46], [47]]. At the same time, somatic mutations in BRD4 (Fig. 4a), which is considered a key oncogene and promising therapeutic target, were also significantly enriched in the CIMP-H subtype [48,49].
Fig. S4.
Tumor mutation burden in the three clusters with different methylation statuses. The Mann–Whitney test was applied to evaluate the significant difference of means between each two subgroups and Kruskal-Wallis test for all three subgroups. (ns: not significant).
Fig. 4.
Association between CIMP and mutational signatures and CNV in HCC. (a) Significantly mutated genes in the HCC subsets stratified by CIMP status. (b) Composite copy number profiles of CIMP-H compared with the other HCC groups with gains shown in red and losses in blue. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Furthermore, differences in somatic copy number alternations between patients with different CIMP statuses were evaluated using GISTIC 2.0 and a total of 158 genes were within the chromosome regions with copy number significantly amplified or deleted in CIMP-H subgroup (Table S3). As illustrated, amplifications on chromosomes 2 accompanied with a deletion on chromosome 9 were enriched in the CIMP-H subgroup. Focal amplification peaks, including well-studied drivers such as CRIM1 (2p22), NBAS (2p24) were identified in the patients with CIMP-H, along with a focal deletion peak at 9p21.3 (C9orf53) (Fig. 4b).
3.5. Construction of a CIMP-based prognostic model
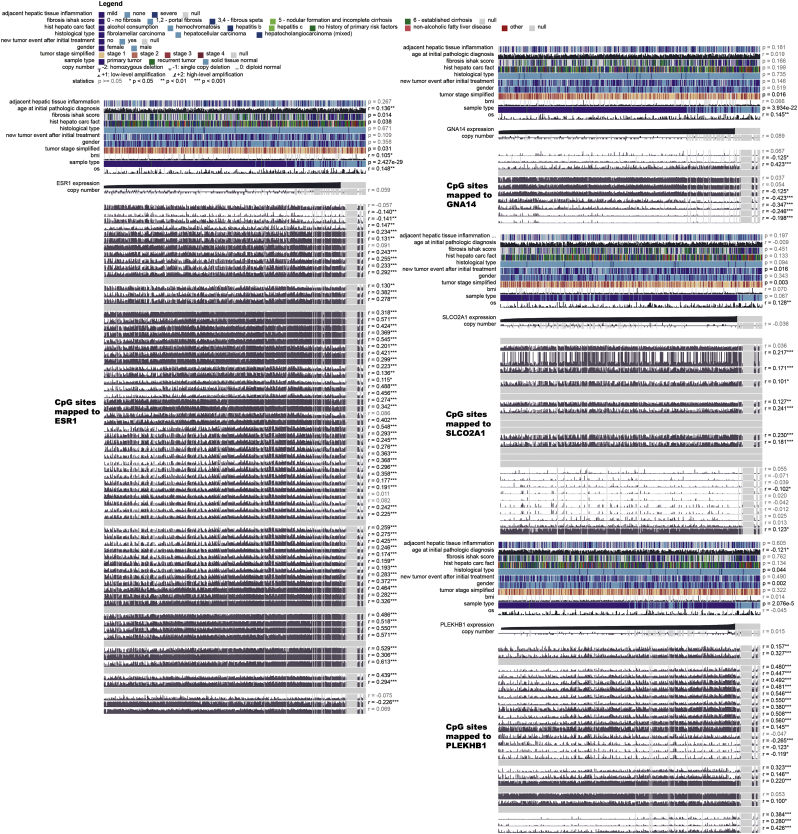
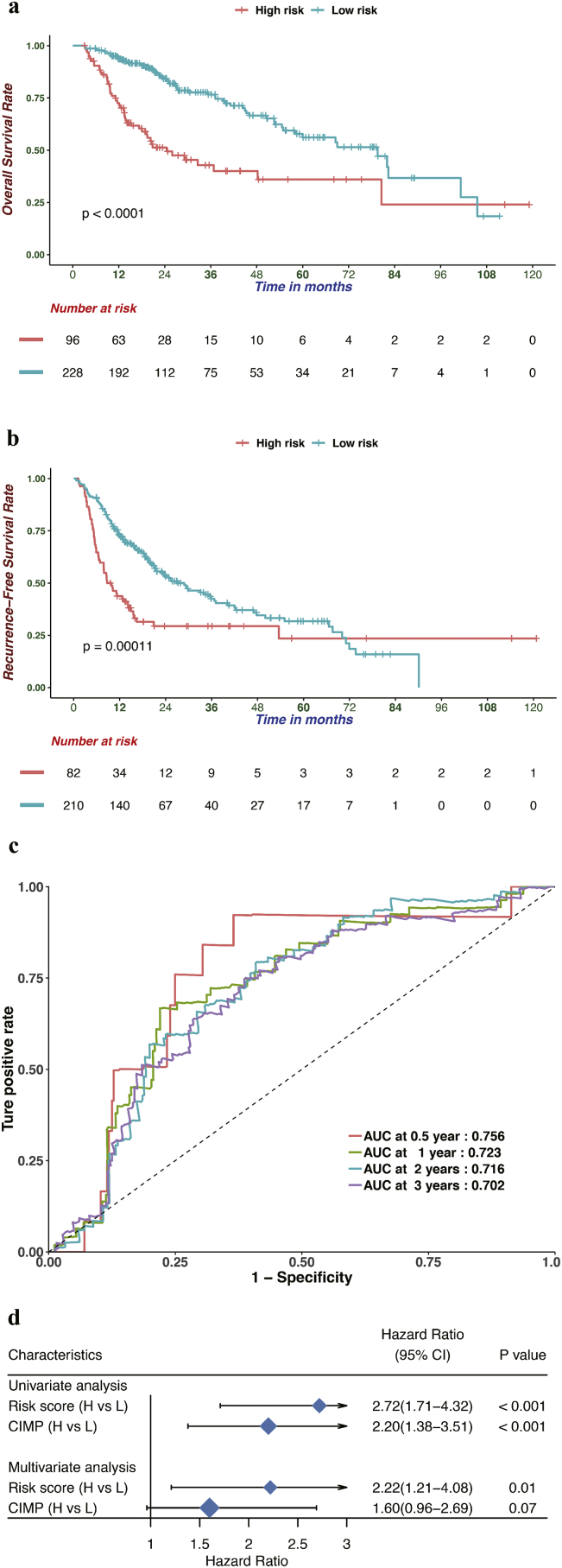
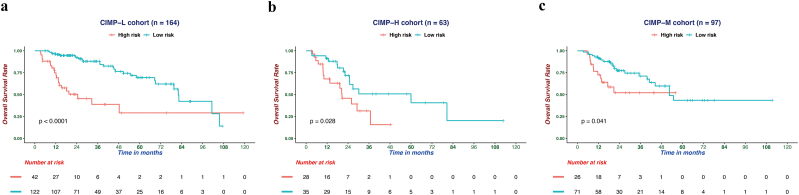
To identify differentially expressed genes between HCC patients with different CIMP statuses, differential expression analysis was conducted by using the DEseq2 R package [37], which revealed and 1180 differentially expression expressed genes were identified with FDR values of <0.05 and the absolute log2-fold changes of >1. Then, trans-regulation analysis, which was defined as the correlation of one gene's methylation and another gene's expression, was performed based on differentially expressed genes and differentially methylated CpG sites among different CIMP subtypes, according to previous study [50]. As a result, numerous differentially expressed genes were predominantly trans-regulated by differentially methylated CpGs (Fig. S5). By performing univariate Cox regression analysis, 137 differentially expressed genes were identified to be significantly correlated with OS (Table S4). To obtain the genes with the greatest potential prognostic values, LASSO regression analysis was performed, and the four genes PLEKHB1, ESR1, SLCO2A1, and GNA14 were finally selected. In addition, we have analyzed the correlation between the expression level of four genes and methylation levels of corresponding CpG sites, and found that the expression of GNA14 were significantly negatively correlated with the methylation level of most CpG sites mapped to it and the other three genes showed significant positive correlation (Fig. S6) [51]. Upon treatment with 5-Aza-2-deoxycytidine, re-expression of PLEKHB1, ESR1, SLCO2A1 and GNA14 was found and 39 of 95 most variable CpG sites presented elevated DNA methylation in LM3 and Huh7 cells. It is generally believed that distal regulatory DNA methylation can also affect dysregulation of cancer genes by influencing transcription factor regulatory networks. The results indicated that 39 of 95 most variable CpG sites, especially 7 CpG sites with the most change of methylation, might affect the expressions of PLEKHB1, ESR1, SLCO2A1, and GNA14 (Table S5, Table S6). Our studies also suggested that the expression of four genes could be regulated by specific DNA methylation in HCC cells (Fig. S7). Then, we normalized the expression of these four major marker genes using scale, a generic R function whose default method centered and scaled the data. The regression coefficient of each gene was calculated using multivariate Cox regression. Finally, the CPM (risk score = 0.16 × normalized expression level of PLEKHB1–0.08 × normalized expression level of ESR1–0.13 × normalized expression level of SLCO2A1–0.21 × normalized expression level of GNA14) was constructed as the predictive prognostic model. The optimal cutoff point (0.23) was calculated using the surv_cutpoint function from the survminer R package, and the patients in the TCGA cohort were categorized into high and low-risk subgroups. As illustrated in Fig. 5a, the patients in the high-risk subgroup had a worse OS than their low-risk counterparts (HR, 2.77; 95% CI, 1.89–4.05; P < .001). In addition, survival plots for the CPM in different CIMP subgroups were drawn and the results indicated that the CPM was also significantly associated with OS in CIMP-L, CIMP-M, and CIMP-H subgroups (Fig. S8). Furthermore, we also investigated the performance of the CPM in predicting RFS, and the result indicated that it could also distinguish high-risk from low-risk patients (HR, 1.87; 95% CI, 1.36–2.59; P < .001, Fig. 5b). The predictive performance of the CPM was evaluated using time-dependent ROC curves, and the area under the ROC curve (AUC) for OS was 0.756 at 0.5 years, 0.723 at 1 year, 0.716 at 2 years and 0.702 at 3 years (Fig. 5c). Finally, the results of uni- and multivariate Cox regression analyses indicated that the predictive performance of CPM for OS is independent of CIMP status (Fig. 5d).
Fig. S5.
Correlation between differentially expressed genes and differentially methylated CpG sites. The coefficients were calculated by Pearson's correlation analysis. Positive and negative correlations are shown in red and blue, respectively.
Fig. S6.
Correlation between the expression level of four genes and methylation levels of corresponding CpG sites. The coefficients were calculated by Pearson's correlation analysis.
Fig. S7.
Relative PLEKHB1, ESR1, SLCO2A1 and GNA14 mRNA levels with or without 5-Aza treatment in LM3 (A), and Huh7 (B) cell lines.
Fig. 5.
Prognostic analysis of the CPM in the TCGA cohort. (a, b) Based on the Kaplan-Meier survival analysis, OS (a) and RFS (b) was significantly higher in the low-risk score group than the other groups. (c) Time-dependent receiver operating characteristic analysis was utilized to evaluate the predictive performance of the CPM. (d) Univariate and multivariate regression analysis of the correlation between the CPM and CIMP status regarding the prognostic value.
Fig. S8.
Prognostic analysis of the CPM in CIMP-L (a), CIMP-H (b), and CIMP-M (c) cohort. Based on the Kaplan-Meier survival analysis, OS was significantly higher in the low-risk score group than its counterparts.
3.6. Validation and evaluation the CPM in the GEO, ICGC, and Tongji cohorts
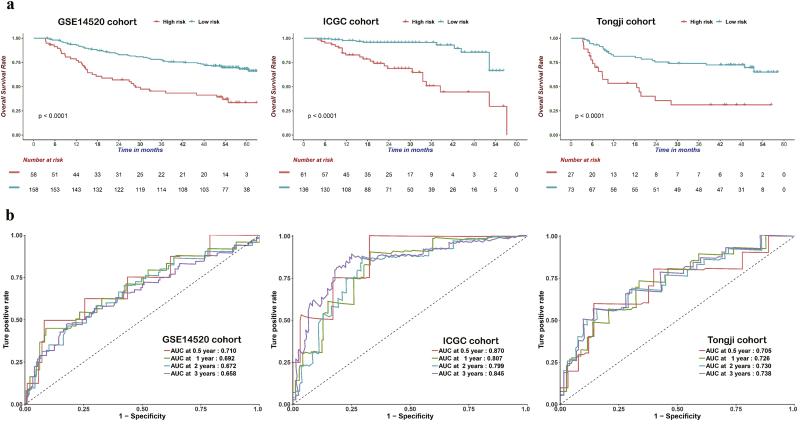
In order to evaluate the robustness of the CPM constructed using data from the TCGA cohort, its performance was also assessed using the GEO, ICGC, and Tongji cohorts, respectively including 216, 197, and 100 HCC patients. The patients in the different cohort were categorized into a high- and low-risk groups using the same risk formula and cutoff obtained using the TCGA cohort. Consistent with the results generated obtained for the TCGA cohort, patients in the high-risk group had worse OS than those assigned to the low-risk group (GEO: HR: 2.95; 95% CI: 1.90–4.58; P < .001; ICGC: HR: 6.27; 95% CI: 2.95–13.33, P < .001; Tongji: HR: 3.42; 95% CI: 1.80–6.54, P < .001 Fig. 6a). In addition, the CPM reached an AUC of 0.710 at 0.5 years, 0.692 at 1 year, 0.672 at 2 years, and 0.658 at 3 years in the GEO cohort, as well as an AUC of 0.870 at 0.5 years, 0.807 at 1 year, 0.799 at 2 years, and 0.845 at 3 years in the ICGC cohort, and an AUC of 0.705 at 0.5 years, 0.726 at 1 year, 0.730 at 2 years, and 0.738 at 3 years in the Tongji cohort (Fig. 6b). These results demonstrate the robust performance of the CPM in the distinct cohorts.
Fig. 6.
Prognostic analysis of the CPM in three independent cohorts. (a) Based on the Kaplan-Meier survival analysis, OS was significantly higher in the low-risk score group than in the other groups from the GSE14520, ICGC, and Tongji cohorts. (b) Time-dependent receiver operating characteristic analysis was performed to compare the performance of CPM in the GSE14520, ICGC, and Tongji cohorts.
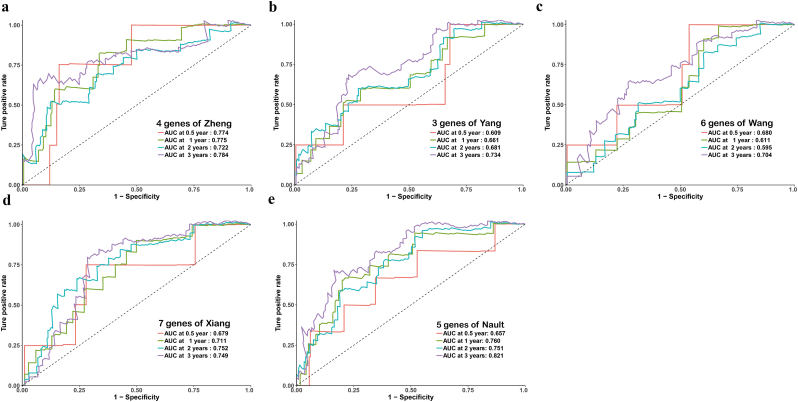
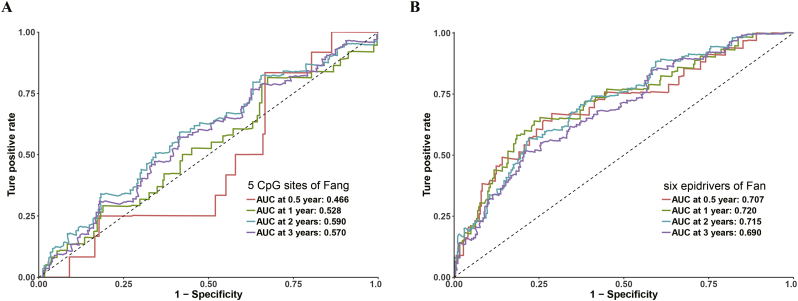
Next, CPM was compared with HCC-related prognostic biomarkers published in the literature [[52], [53], [54], [55], [56], [57], [58]]. The formulae were extracted from each publication, and the results of comparative time-dependent ROC curve analysis indicated that the CPM as best at predicting the prognosis in the ICGC or TCGA cohort (Figs. 7 and S9). Therefore, our results indicate that CPM is relatively robust across distinct molecular levels, platforms and datasets.
Fig. 7.
ROC curves showing the sensitivity and specificity of the CPM and other known biomarkers reported in previous studies in the prediction of OS for the ICGC cohort.
Fig. S9.
ROC curves showing the sensitivity and specificity of other known methylation-related biomarkers reported in previous studies in the prediction of OS for the TCGA cohort.
3.7. Altered biological processes and pathways in high- and low-risk subgroups
We performed GO and KEGG analysis to investigate the biological mechanisms revealed by CPM. Gene, highly associated with risk scores (absolute Pearson correlation coefficient > 0.5 and P < .05) were regarded as risk score-related biomarkers. Genes correlated with the risk score in the TCGA cohort were significantly enriched in the “protein targeting to membrane”, “ribonucleoprotein complex biogenesis”, “nuclear transcribed mRNA catabolic process”, “regulation of ubiquitin protein ligase activity”, “positive regulation of PI3K signaling”, “condensed chromosome”, and “structural constituent of ribosome” terms based on GO analysis, as well as “Ribosome tryptophan metabolism”, “Valine, leucine and isoleucine degradation”, “beta Alanine metabolism”, and “Propanoate metabolism” according to KEGG analysis (Fig. 8a, b).
Fig. 8.
Circular plot of the biological processes (a) and KEGG pathways (b) enriched for the risk score associated genes.
3.8. The CPM is independent of frequently used clinical characteristics
Univariate and multivariate Cox regression analyses were performed to investigate whether CPM was an independent predictive factor for the prognosis of HCC patients from the TCGA cohort. The results of the adjustment for conventional clinical patterns, including gender, diagnostic age, pathologic TNM, pathologic grade, pathologic stage, vascular invasion, and serum AFP level, indicated that CPM also acted as an independent prognostic factor, which confirmed its robust predictive ability for the OS of HCC patients (OS: HR, 2.82; 95% CI:, 1.85–4.32; P < .001, Fig. 9a). Furthermore, the C-index between CPM and common clinical patterns, including 15 prognosis-predictive factors, were compared. The results showed that CPM had the highest C-index (0.68, Table S7). Finally, CPM was also found to act as an independent prognostic factor for predicting the RFS in the TCGA cohort (HR, 1.61; 95% CI:, 1.08–2.40; P = .019, Fig. 9b). In conclusion, the results demonstrated that CPM possesses an optimal predictive ability for prognosis independent of common clinical patterns.
Fig. 9.
Univariate and multivariate regression analysis of the relation between the CPM and clinicopathological characteristics regarding OS (a) and RFS (b) in the TCGA cohort.
3.9. Establishment of a nomogram based on the CPM
In order to establish a quantitative approach for HCC prognosis, which might contribute to the clinical decision-making of practitioners, we integrated the CPM and independent clinical risk factors (CIMP type and Pathologic M) to construct a nomogram (Fig. 10a). On the basis of multivariate Cox analysis, a point scale of the nomogram was utilized to dispense points to respective variables. We drew a horizontal straight line to ascertain the points for each variable, and the total points of each patient were calculated by adding the points of all variables together, which were normalized to a distribution from 0 to 100. The estimated survival rates at 1, 3, and 5 years of HCC patients were calculated by drawing a vertical line between the total point coordinate axis and each prognostic coordinate axis. The results of the nomogram indicated that CPM had the greatest weight among the total points, consistent with the previous multivariate regression analysis. The C-index of our nomogram reached 0.71 with 1000 bootstrap iterations (95% CI: 0.68–0.74). The results of the calibration plots indicated that there was good consistency between the predicted and the actually observed outcomes (Fig. 10b). The predictive performance of our nomogram was also compared with that of CPM, CIMP type and Pathologic M, and the results indicated that the nomogram performance was better than that of CPM (C-index: 0.68), CIMP type (C-index: 0.59) and Pathologic M (C-index: 0.51). Consequently, our results suggest that the nomogram is an optimal model for the prediction of HCC prognosis comparing with individual risk factors.
Fig. 10.
Integration of CPM and clinical characteristics (a) Nomogram constructed to predict the 1-, 3-, and 5-year OS in the TCGA cohort. (b) Calibration curve of the nomogram for predicting the probability of OS at 1, 2, and 3 years.
4. Discussion
As changeable and possibly heritable genetic alterations, epigenetic mechanisms offer promising clues for the treatment of various diseases, including many cancers [59]. The results of previous studies indicated that a number of epigenome-targeting therapies in hematological malignancies seem to be advantageous and safe [60]. While investigations in solid malignancies had several limitations, painstaking explorations of epigenetic targeting therapies, including clinical trials, are nevertheless under way [61,62]. To our best knowledge, there are no registered clinical trials of targeted drugs for CIMP-H subtypes in HCC.
DNMT inhibitors, such as 5-azacytidine, can result in dose-dependent global demethylation through the degradation of DNA methyltransferases, which can restore the expression of aberrantly methylated oncogenesis-related genes. In addition, the results of previous studies indicated that DNMT inhibitors can contribute to the reversion of neoplastic immune evasion by activating important defense pathways. For instance, in a HepG2 xenograft model of HCC, guadecitabine, a second-generation demethylating agent, can inhibit the growth of HCC cells by restoring the DNA methylation levels of carcinogenesis-related genes, which might act as a promising therapy in advanced HCC [63]. Although drugs aimed at epigenetic targets showed promising results in previous studies, a related clinical investigation utilizing guadecitabine against HCC has not been registered to date. Taking the potential unfavorable influence of reversion of DNA methylation in hepatocarcinogenesis into account, which was reported in previous studies [64], it was reasonable to consider the CIMP-H subtypes in when designing clinical trials.
CIMP is characterized by simultaneous and general hypermethylation of specific CpG sites, which plays a vital role in chromosomal instability in diverse cancer subtypes [14,15,65]. However, the potential mechanism by which CIMP impacts hepatocacinogenesis are still only partially understood. Furthermore, it is necessary to construct potential CIMP-related prognostic models to categorize patients, which would increase the effectiveness of epigenetic therapy. To our best knowledge, this is the first attempt to investigate the potential mechanism linking CIMP with HCC. In agreement with previous studies, we found that CIMP is significant associated with worse OS and RFS in HCC, and the CIMP-H group tended to have relative higher serum AFP level [19,50,66]. Based on the gene set enrichment analysis between CIMP-H and the other groups, we identified that the CIMP-H subtype was enriched with biomarkers associated with the terms in “Recurrent liver cancer up”, “Liver cancer survival down”, “Liver cancer with EPCAM up”, “Tumor invasiveness up”, “methyltransferase complex”, and “Translational initiation”. It could be speculated that CIMP-related biomarkers might play a vital role in hepatocarcinogenesis through the aforementioned biological process. In view of the correlation between the expression of EPCAM and CIMP in colorectal carcinoma [43], our results predominantly indicate that the CIMP might act as a tumor promoter in hepatocarcinogenesis by inducing the formation of the methyltransferase complex to translationally regulate corresponding genes, and HCC in different CIMP subtypes originated in precursor cells might have a different epigenetic background of the cell of origin.
In addition, the landscape of tumor-infiltrating immune cells among patients with different CIMP statuses was also investigated in our study, and the results indicated that there are no statistically significant differences in the abundance of the majority of immune cell subtypes. Nevertheless, the CIMP-H subgroup presented with significantly less proportions of cytotoxic cells, dendritic cell, interdigitating cell, macrophages, neutrophils, Tgd, and Th1 cells than the other groups, which indicated that immunotherapies are more likely to be less efficacious in CIMP-H phenotype with less immune infiltration, lower cytotoxic potential and immune activation.
Based on the mutation analysis, the patients in CIMP-H had significantly higher somatic mutation burdens in the DDIAS, NOX1, and BRD4. DDIAS, a DNA damage-induced apoptosis suppressor, can exert an anti-apoptotic activity when DNA damage occurs, and has been identified as a promising therapeutic target in HCC [44]. NOX1 (NADPH oxidase 1), was found to be able to increase the stemness of CD133+ thyroid cancer cells by activating the Akt signaling pathway [47]. BRD4 (bromodomain containing 4) may act as an epigenetic reader and transcriptional coactivator to participate in carcinogenesis [48]. To date, a few selective small-molecule inhibitors, including JQ1 and I-BET762, have already been developed to target BRD4, and studies indicated that they may have anti-proliferative activity in various tumors [67]. Our study sheds new light on the potential mechanisms by which a high mutation burden in DDIAS, NOX1, and BRD4 in the CIMP-H subgroup might contribute to hepatocarcinogenesis by reducing cancer cell apoptosis, as well as the regulation of stemness and epigenomic signals.
Next, a CIMP-based prognostic model (CPM) was developed based on 4 genes, and trained using the TCGA cohort. It was able to screen out the HCC patients with poor prognosis. The robustness of CPM was validated in three independent cohorts. These results might contribute to a potential therapeutic strategy involving epigenetic regulation to improve the prognosis of HCC patients. The four genes used in CPM, PLEKHB1, ESR1, SLCO2A1, and GNA14, might act as individual targets, and it is definitely possible to combine these four targets could produce a more effective therapeutic approach.. However, the potential mechanisms by which these four genes contribute hepatocarcinogenesis remain poorly understood, and further evaluation of potential mechanisms may be worthwhile. In addition, our results indicate that the CPM could act as an independent prognostic factor after adjusting conventional clinical characteristics. It seemed that CPM might potentially have predictive power than traditional prognostic patterns. Subsequently, a comprehensive evaluation combining CPM with other important clinical patterns (CIMP status and pathologic M) was performed. Based on the calibration plot, there was a favorable consistency between the actual and predicted values for 1-, 2-, and 3-year OS. Our model was constructed based on the complementary perspective for respective tumors, and provides a personalized score for individual patients. Consequently, our nomogram could be a valuable new prognostic method for clinicians in the future.
There are a number of strengths in our study. Firstly, our robust CPM was based on four independent cohorts, which validated our prognostic signature sufficiently. Secondly, the CIMP-related biomarkers of CPM showed a significant biological background, which indicates that they can be potentially applied in clinical adjuvant therapies, which was not necessarily the case with previous studies. Thirdly, CPM showed a more robust predictive performance than conventional clinical prognostic features and other signatures reported in the literature.
Although our study sheds new light on the epigenomic microenvironment and possible CIMP-related therapies, it still has some limitations. Firstly, our studies was based on single-omics (DNA methylation), so that patients with same CIMP phenotype might possess different heterogeneity because of different characteristics in terms of other omics data platforms. Secondly, our attempts was based on a retrospective design, and prospective studies should be conducted to validate our results. Furthermore, the biological functions and molecular mechanisms of the four indicator genes alone and in combination should be evaluated to accelerate their clinical application in HCC. Thirdly, the rate of lacking data for some clinical features was relatively high, which might reduce the statistical reliability and validity of multivariable Cox regression analysis. Fourthly, it was reported that the quantitative signatures would be unsuitable for direct application to individual samples because their application needed pre-collecting a set of samples for normalization [68,69], the prognostic signature based on quantitative method of genes should be identified in the future.
5. Conclusion
The CIMP-related prognostic model based on four genes was constructed and validated. It was found to act as an independent prognostic factor for HCC and reflects the overall epigenetic alterations in the whole genome. To our best knowledge, this is the first report of a prognostic model incorporating CIMP status and it could be utilized as a reference to understand the relevance of CIMP in other malignancies. Notably, the CPM provides epigenetic insights into the main mechanisms that potentially influence the prognosis of HCC. Finally, it is urgent to design prospective clinical trials for further validation of its prognostic performance and evaluation of its clinical applicability in personalized management of HCC.
The following are the supplementary data related to this article.
Sequences of primers used for qRT-PCR.
Basic information on the 95 most variable CpG sites utilized for unsupervised consensus cluster analysis.
A total of 158 genes were within the chromosome regions with copy number significantly amplified or deleted in CIMP-H subgroup.
Complete list of 137 prognostic DEGs in the TCGA cohort.
Differentially DNA methylation analysis of 95 most variable CpG sites between Huh7 cells with and without 5-Aza-2′-deoxycytidine treatment.
Differentially DNA methylation analysis of 95 most variable CpG sites between LM3 cells with and without 5-Aza-2′-deoxycytidine treatment.
The C-indices of the nomogram, CPM and different conventional clinical characteristics.
Acknowledgments
Acknowledgements
Thank Dr. Chris Lou (Blogger, WeChat Official Accounts), Jiayun Chen and Xingjie Hao for the bioinformatics technology support. Thank Hajnal Ivan for English language editing.
Funding sources
This work was supported by the State Key Project on Infectious Diseases of China (2018ZX10723204-003), the National Nature Science Foundation of China (Nos. 81874065, 81500565, 81874149, 81572427, and 81401997), the Hepato-Biliary-Pancreatic Malignant Tumor Investigation Fund of Chen Xiao-ping Foundation for the Development of Science and Technology of Hubei Province (CXPJJH11800001-2018356). The funders did not play a role in manuscript design, data collection, data analysis, data interpretation or writing of the manuscript.
Declaration of competing interests
None.
Author contributions
All authors searched the literature, designed the study, interpreted the findings and revised the manuscript. Ganxun Li, Weiqi Xu, Lu Zhang and Tongtong Liu carried out data management and statistical analysis and drafted the manuscript. Guannan Jin, Jia Song, Jingjing Wu, Yuwei Wang, Weixun Chen, Chuanhan Zhang, and Xiaoping Chen helped with cohort identification and data management. Zeyang Ding, Peng Zhu, and Bixiang Zhang performed project administration.
Contributor Information
Zeyang Ding, Email: zyding@tjh.tjmu.edu.cn.
Peng Zhu, Email: zhupeng@tjh.tjmu.edu.cn.
Bixiang Zhang, Email: bixiangzhang@hust.edu.cn.
References
- 1.Global Burden of Disease Liver Cancer C, Akinyemiju T., Abera S., Ahmed M., Alam N., Alemayohu M.A. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level: results from the global burden of disease study 2015. JAMA Oncol. 2017;3(12):1683–1691. doi: 10.1001/jamaoncol.2017.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.European Association for the Study of the Liver. Electronic address eee and European Association for the Study of the L EASL clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2018;69(1):182–236. doi: 10.1016/j.jhep.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 3.Yang J.D., Roberts L.R. Hepatocellular carcinoma: a global view. Nat Rev Gastroenterol Hepatol. 2010;7(8):448–458. doi: 10.1038/nrgastro.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dhanasekaran R., Bandoh S., Roberts L.R. Molecular pathogenesis of hepatocellular carcinoma and impact of therapeutic advances. F1000Research. 2016;5 doi: 10.12688/f1000research.6946.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heimbach J.K., Kulik L.M., Finn R.S., Sirlin C.B., Abecassis M.M., Roberts L.R. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology. 2018;67(1):358–380. doi: 10.1002/hep.29086. [DOI] [PubMed] [Google Scholar]
- 6.Brown Z.J., Greten T.F., Heinrich B. Adjuvant treatment of hepatocellular carcinoma: prospect of immunotherapy. Hepatology. 2019 doi: 10.1002/hep.30633. [DOI] [PubMed] [Google Scholar]
- 7.Bruix J., Cheng A.L., Meinhardt G., Nakajima K., De Sanctis Y., Llovet J. Prognostic factors and predictors of sorafenib benefit in patients with hepatocellular carcinoma: analysis of two phase III studies. J Hepatol. 2017;67(5):999–1008. doi: 10.1016/j.jhep.2017.06.026. [DOI] [PubMed] [Google Scholar]
- 8.Portela A., Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28(10):1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 9.Lee S.T., Wiemels J.L. Genome-wide CpG island methylation and intergenic demethylation propensities vary among different tumor sites. Nucleic Acids Res. 2016;44(3):1105–1117. doi: 10.1093/nar/gkv1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baylin S.B., Esteller M., Rountree M.R., Bachman K.E., Schuebel K., Herman J.G. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum Mol Genet. 2001;10(7):687–692. doi: 10.1093/hmg/10.7.687. [DOI] [PubMed] [Google Scholar]
- 11.Berdasco M., Esteller M. Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Dev Cell. 2010;19(5):698–711. doi: 10.1016/j.devcel.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 12.Esteller M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum Mol Genet. 2007;16:R50–R59. doi: 10.1093/hmg/ddm018. Spec No 1. [DOI] [PubMed] [Google Scholar]
- 13.Hansen K.D., Timp W., Bravo H.C., Sabunciyan S., Langmead B., McDonald O.G. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43(8):768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noushmehr H., Weisenberger D.J., Diefes K., Phillips H.S., Pujara K., Berman B.P. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes L.A., Melotte V., de Schrijver J., de Maat M., Smit V.T., Bovee J.V. The CpG island methylator phenotype: what's in a name? Cancer Res. 2013;73(19):5858–5868. doi: 10.1158/0008-5472.CAN-12-4306. [DOI] [PubMed] [Google Scholar]
- 16.Toyota M., Ahuja N., Ohe-Toyota M., Herman J.G., Baylin S.B., Issa J.P. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96(15):8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weisenberger D.J. Characterizing DNA methylation alterations from The Cancer Genome Atlas. J Clin Invest. 2014;124(1):17–23. doi: 10.1172/JCI69740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang C., Guo X., Jiang G., Zhang L., Yang Y., Shen F. CpG island methylator phenotype association with upregulated telomerase activity in hepatocellular carcinoma. Int J Cancer. 2008;123(5):998–1004. doi: 10.1002/ijc.23650. [DOI] [PubMed] [Google Scholar]
- 19.Zhang C., Li Z., Cheng Y., Jia F., Li R., Wu M. CpG island methylator phenotype association with elevated serum alpha-fetoprotein level in hepatocellular carcinoma. Clin Cancer Res. 2007;13(3):944–952. doi: 10.1158/1078-0432.CCR-06-2268. [DOI] [PubMed] [Google Scholar]
- 20.Malta T.M., de Souza C.F., Sabedot T.S., Silva T.C., Mosella M.S., Kalkanis S.N. Glioma CpG island methylator phenotype (G-CIMP): biological and clinical implications. Neuro Oncol. 2018;20(5):608–620. doi: 10.1093/neuonc/nox183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez-Vega F., Gotea V., Margolin G., Elnitski L. Pan-cancer stratification of solid human epithelial tumors and cancer cell lines reveals commonalities and tissue-specific features of the CpG island methylator phenotype. Epigenetics Chromatin. 2015;8:14. doi: 10.1186/s13072-015-0007-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei L., Jin Z., Yang S., Xu Y., Zhu Y., Ji Y. TCGA-assembler 2: software pipeline for retrieval and processing of TCGA/CPTAC data. Bioinformatics. 2018;34(9):1615–1617. doi: 10.1093/bioinformatics/btx812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bibikova M., Barnes B., Tsan C., Ho V., Klotzle B., Le J.M. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98(4):288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Fortin J.P., Triche T.J., Jr., Hansen K.D. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics. 2017;33(4):558–560. doi: 10.1093/bioinformatics/btw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sandoval J., Mendez-Gonzalez J., Nadal E., Chen G., Carmona F.J., Sayols S. A prognostic DNA methylation signature for stage I non-small-cell lung cancer. J Clin Oncol. 2013;31(32):4140–4147. doi: 10.1200/JCO.2012.48.5516. [DOI] [PubMed] [Google Scholar]
- 26.Price M.E., Cotton A.M., Lam L.L., Farre P., Emberly E., Brown C.J. Additional annotation enhances potential for biologically-relevant analysis of the Illumina Infinium HumanMethylation450 BeadChip array. Epigenetics Chromatin. 2013;6(1):4. doi: 10.1186/1756-8935-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y.A., Lemire M., Choufani S., Butcher D.T., Grafodatskaya D., Zanke B.W. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8(2):203–209. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colaprico A., Silva T.C., Olsen C., Garofano L., Cava C., Garolini D. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44(8):e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayakonda A., Lin D.C., Assenov Y., Plass C., Koeffler H.P. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28(11):1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chalmers Z.R., Connelly C.F., Fabrizio D., Gay L., Ali S.M., Ennis R. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9(1):34. doi: 10.1186/s13073-017-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mermel C.H., Schumacher S.E., Hill B., Meyerson M.L., Beroukhim R., Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.International Cancer Genome C, Hudson T.J., Anderson W., Artez A., Barker A.D., Bell C. International network of cancer genome projects. Nature. 2010;464(7291):993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilkerson M.D., Hayes D.N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Villanueva A., Portela A., Sayols S., Battiston C., Hoshida Y., Mendez-Gonzalez J. DNA methylation-based prognosis and epidrivers in hepatocellular carcinoma. Hepatology. 2015;61(6):1945–1956. doi: 10.1002/hep.27732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu G.C., Wang L.G., Han Y.Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gui J., Li H. Penalized cox regression analysis in the high-dimensional and low-sample size settings, with applications to microarray gene expression data. Bioinformatics. 2005;21(13):3001–3008. doi: 10.1093/bioinformatics/bti422. [DOI] [PubMed] [Google Scholar]
- 40.Heagerty P.J., Lumley T., Pepe M.S. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56(2):337–344. doi: 10.1111/j.0006-341x.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 41.Rooney M.S., Shukla S.A., Wu C.J., Getz G., Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160(1–2):48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cancer Genome Atlas Research Network. Electronic address wbe and Cancer Genome Atlas Research N Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169(7):1327–1341 e1323. doi: 10.1016/j.cell.2017.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim J.H., Bae J.M., Song Y.S., Cho N.Y., Lee H.S., Kang G.H. Clinicopathologic, molecular, and prognostic implications of the loss of EPCAM expression in colorectal carcinoma. Oncotarget. 2016;7(12):13372–13387. doi: 10.18632/oncotarget.5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Im J.Y., Kim B.K., Lee J.Y., Park S.H., Ban H.S., Jung K.E. DDIAS suppresses TRAIL-mediated apoptosis by inhibiting DISC formation and destabilizing caspase-8 in cancer cells. Oncogene. 2018;37(9):1251–1262. doi: 10.1038/s41388-017-0025-y. [DOI] [PubMed] [Google Scholar]
- 45.Won K.J., Im J.Y., Kim B.K., Ban H.S., Jung Y.J., Jung K.E. Stability of the cancer target DDIAS is regulated by the CHIP/HSP70 pathway in lung cancer cells. Cell Death Dis. 2017;8(1):e2554. doi: 10.1038/cddis.2016.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Echizen K., Horiuchi K., Aoki Y., Yamada Y., Minamoto T., Oshima H. NF-kappaB-induced NOX1 activation promotes gastric tumorigenesis through the expansion of SOX2-positive epithelial cells. Oncogene. 2019;38(22):4250–4263. doi: 10.1038/s41388-019-0702-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang C., Wang Z., Liu W., Ai Z. ROS-generating oxidase NOX1 promotes the self-renewal activity of CD133+ thyroid cancer cells through activation of the Akt signaling. Cancer Lett. 2019;447:154–163. doi: 10.1016/j.canlet.2019.01.028. [DOI] [PubMed] [Google Scholar]
- 48.Wu Y., Wang Y., Diao P., Zhang W., Li J., Ge H. Therapeutic targeting of BRD4 in head neck squamous cell carcinoma. Theranostics. 2019;9(6):1777–1793. doi: 10.7150/thno.31581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu J., Duan Z., Guo W., Zeng L., Wu Y., Chen Y. Targeting the BRD4/FOXO3a/CDK6 axis sensitizes AKT inhibition in luminal breast cancer. Nat Commun. 2018;9(1):5200. doi: 10.1038/s41467-018-07258-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng J., Wei D., Ji Y., Chen L., Yang L., Li G. Integrative analysis of DNA methylation and gene expression reveals hepatocellular carcinoma-specific diagnostic biomarkers. Genome Med. 2018;10(1):42. doi: 10.1186/s13073-018-0548-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koch A., Jeschke J., Van Criekinge W., van Engeland M., De Meyer T. MEXPRESS update 2019. Nucleic Acids Res. 2019;47(W1):W561–W565. doi: 10.1093/nar/gkz445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng Y., Liu Y., Zhao S., Zheng Z., Shen C., An L. Large-scale analysis reveals a novel risk score to predict overall survival in hepatocellular carcinoma. Cancer Manag Res. 2018;10:6079–6096. doi: 10.2147/CMAR.S181396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Z., Teng D., Li Y., Hu Z., Liu L., Zheng H. A six-gene-based prognostic signature for hepatocellular carcinoma overall survival prediction. Life Sci. 2018;203:83–91. doi: 10.1016/j.lfs.2018.04.025. [DOI] [PubMed] [Google Scholar]
- 54.Xiang X.H., Yang L., Zhang X., Ma X.H., Miao R.C., Gu J.X. Seven-senescence-associated gene signature predicts overall survival for Asian patients with hepatocellular carcinoma. World J Gastroenterol. 2019;25(14):1715–1728. doi: 10.3748/wjg.v25.i14.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang Y., Lu Q., Shao X., Mo B., Nie X., Liu W. Development of a three-gene prognostic signature for hepatitis B virus associated hepatocellular carcinoma based on integrated transcriptomic analysis. J Cancer. 2018;9(11):1989–2002. doi: 10.7150/jca.23762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nault J.C., De Reynies A., Villanueva A., Calderaro J., Rebouissou S., Couchy G. A hepatocellular carcinoma 5-gene score associated with survival of patients after liver resection. Gastroenterology. 2013;145(1):176–187. doi: 10.1053/j.gastro.2013.03.051. [DOI] [PubMed] [Google Scholar]
- 57.Fang F., Wang X., Song T. Five-CpG-based prognostic signature for predicting survival in hepatocellular carcinoma patients. Cancer Biol Med. 2018;15(4):425–433. doi: 10.20892/j.issn.2095-3941.2018.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fan G., Tu Y., Chen C., Sun H., Wan C., Cai X. DNA methylation biomarkers for hepatocellular carcinoma. Cancer Cell Int. 2018;18:140. doi: 10.1186/s12935-018-0629-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jones P.A., Issa J.P., Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet. 2016;17(10):630–641. doi: 10.1038/nrg.2016.93. [DOI] [PubMed] [Google Scholar]
- 60.Azad N., Zahnow C.A., Rudin C.M., Baylin S.B. The future of epigenetic therapy in solid tumours – lessons from the past. Nat Rev Clin Oncol. 2013;10(5):256–266. doi: 10.1038/nrclinonc.2013.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reifenberger G., Wirsching H.G., Knobbe-Thomsen C.B., Weller M. Advances in the molecular genetics of gliomas – implications for classification and therapy. Nat Rev Clin Oncol. 2017;14(7):434–452. doi: 10.1038/nrclinonc.2016.204. [DOI] [PubMed] [Google Scholar]
- 62.Maio M., Covre A., Fratta E., Di Giacomo A.M., Taverna P., Natali P.G. Molecular pathways: at the crossroads of cancer epigenetics and immunotherapy. Clin Cancer Res. 2015;21(18):4040–4047. doi: 10.1158/1078-0432.CCR-14-2914. [DOI] [PubMed] [Google Scholar]
- 63.Jueliger S., Lyons J., Cannito S., Pata I., Pata P., Shkolnaya M. Efficacy and epigenetic interactions of novel DNA hypomethylating agent guadecitabine (SGI-110) in preclinical models of hepatocellular carcinoma. Epigenetics. 2016;11(10):709–720. doi: 10.1080/15592294.2016.1214781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mei Q., Chen M., Lu X., Li X., Duan F., Wang M. An open-label, single-arm, phase I/II study of lower-dose decitabine based therapy in patients with advanced hepatocellular carcinoma. Oncotarget. 2015;6(18):16698–16711. doi: 10.18632/oncotarget.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brennan K., Koenig J.L., Gentles A.J., Sunwoo J.B., Gevaert O. Identification of an atypical etiological head and neck squamous carcinoma subtype featuring the CpG island methylator phenotype. EBioMedicine. 2017;17:223–236. doi: 10.1016/j.ebiom.2017.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Q., Wang G., Liu C., He X. Prognostic value of CpG island methylator phenotype among hepatocellular carcinoma patients: a systematic review and meta-analysis. Int J Surg. 2018;54:92–99. doi: 10.1016/j.ijsu.2018.04.033. Pt A. [DOI] [PubMed] [Google Scholar]
- 67.Delmore J.E., Issa G.C., Lemieux M.E., Rahl P.B., Shi J., Jacobs H.M. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ao L., Zhang Z., Guan Q., Guo Y., Guo Y., Zhang J. A qualitative signature for early diagnosis of hepatocellular carcinoma based on relative expression orderings. Liver Int. 2018;38(10):1812–1819. doi: 10.1111/liv.13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qi L., Chen L., Li Y., Qin Y., Pan R., Zhao W. Critical limitations of prognostic signatures based on risk scores summarized from gene expression levels: a case study for resected stage I non-small-cell lung cancer. Brief Bioinform. 2016;17(2):233–242. doi: 10.1093/bib/bbv064. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequences of primers used for qRT-PCR.
Basic information on the 95 most variable CpG sites utilized for unsupervised consensus cluster analysis.
A total of 158 genes were within the chromosome regions with copy number significantly amplified or deleted in CIMP-H subgroup.
Complete list of 137 prognostic DEGs in the TCGA cohort.
Differentially DNA methylation analysis of 95 most variable CpG sites between Huh7 cells with and without 5-Aza-2′-deoxycytidine treatment.
Differentially DNA methylation analysis of 95 most variable CpG sites between LM3 cells with and without 5-Aza-2′-deoxycytidine treatment.
The C-indices of the nomogram, CPM and different conventional clinical characteristics.