Abstract
Background
Transmission of Mycobacterium leprae, the pathogen causing leprosy, is still persistent. To facilitate timely (prophylactic) treatment and reduce transmission it is vital to both early diagnose leprosy, and identify infected individuals lacking clinical symptoms. However, leprosy-specific biomarkers are limited, particularly for paucibacillary disease. Therefore, our objective was to identify new biomarkers for leprosy and assess their applicability in point-of-care (POC) tests.
Methods
Using multiplex-bead-arrays, 60 host-proteins were measured in a cross-sectional approach in 24-h whole blood assays (WBAs) collected in Bangladesh (79 patients; 54 contacts; 51 endemic controls (EC)). Next, 17 promising biomarkers were validated in WBAs of a separate cohort (55 patients; 27 EC). Finally, in a third cohort (36 patients; 20 EC), five candidate markers detectable in plasma were assessed for application in POC tests.
Findings
This study identified three new biomarkers for leprosy (ApoA1, IL-1Ra, S100A12), and confirmed five previously described biomarkers (CCL4, CRP, IL-10, IP-10, αPGL-I IgM). Overnight stimulation in WBAs provided increased specificity for leprosy and was required for IL-10, IL-1Ra and CCL4. The remaining five biomarkers were directly detectable in plasma, hence suitable for rapid POC tests. Indeed, lateral flow assays (LFAs) utilizing this five-marker profile detected both multi- and paucibacillary leprosy patients with variable immune responses.
Interpretation
Application of novel host-biomarker profiles to rapid, quantitative LFAs improves leprosy diagnosis and allows POC testing in low-resource settings. This platform can thus aid diagnosis and classification of leprosy and also provides a tool to detect M.leprae infection in large-scale contact screening in the field.
Keywords: Biomarkers, Early diagnosis, Immune profiling, Lateral flow (LF), M.leprae, Upconverting reporter particles (UCP), User-friendly rapid test
Research in context.
Evidence before this study
The annual number of new leprosy cases has been stable for the past decade, indicating that despite the availability of effective multidrug treatment (MDT) transmission of Mycobacterium leprae (M.leprae) is still ongoing.
Leprosy can lead to life-long handicaps and patients frequently experience stigma, posing a severe burden. Diagnosis of leprosy relies on clinical symptoms, leaving asymptomatic M.leprae infected individuals undetected by the currently available diagnostic methods. As it can take up to 5–10 years before clinical symptoms arise, it is vital that a diagnostic tool is developed to detect M. leprae infection during this time period.
In leprosy endemic areas there is limited access to advanced healthcare institutes and specialized clinical expertise for leprosy has decreased enormously. This urges for newly developed point-of-care diagnostic tests applicable in resource-limited settings. Diagnostic tests to facilitate the detection of leprosy patients at an early disease stage as well as M.leprae infected individuals will enable timely therapeutic or prophylactic treatment, respectively. Overall, this will help prevent permanent leprosy-associated disabilities.
Added value of this study
We identified a novel five-biomarker signature using a funnel approach by extensive proteomic profiling in leprosy patients, household and endemic controls in Bangladesh. Subsequently, the five-marker signature validated in three independent cohorts was applied to a low-complexity lateral flow format; this point-of-care test sensitively detected not only multibacillary leprosy patients but also paucibacillary patients and thereby outperformed previously defined signatures. As M.leprae bacilli are difficult to detect in paucibacillary patients as well as in most infected household contacts, and detection requires advanced laboratory equipment, this field-friendly host biomarker-based test is well-suited for leprosy diagnostics in endemic areas covering multiple aspects of the diverse host immune response to M.leprae.
Implications of all the available evidence
This study shows the potential of biomarker profiles to identify M. leprae infected individuals and leprosy patients. To ensure global application validation of this five-marker signature in different populations is required. Early diagnosis of leprosy patients and identification of M.leprae infected individuals is vital to reduce the unremitting transmission. The low complexity format of the lateral flow strips would make large-scale contact screening field trials feasible in low resource settings, providing a diagnostic tool that can accurately detect leprosy patients. In this respect, the described five-marker test can aid in decision making for the WHO endorsed targeted prophylactic treatment in M. leprae infected contacts with single dose rifampicin. The objective read-out of the lateral flow strips will help guide decisions on which individuals are candidates for (prophylactic) treatment and facilitate leprosy diagnosis, thereby significantly improving the current diagnostic method that relies on detection of clinical symptoms.
Alt-text: Unlabelled Box
1. Introduction
Despite decades of control programs using multidrug therapy (MDT), leprosy still poses a public health problem in low and middle income countries affecting the poorest, most vulnerable people in their productive stage of life [1]. This does not only have impact on affected individuals, but also imposes a significant social and financial burden on society [2].
Key to leprosy control is the reduction of transmission of Mycobacterium leprae (M.leprae), the causative agent of leprosy, to breach the number of new cases which has stagnated around 200,000 annually for over a decade [2]. Development of methods and tools to early diagnose disease and detect infection to direct (prophylactic) treatment in leprosy healthcare programs therefore has a high priority on the leprosy research agenda.
Current diagnosis of leprosy relies on clinical symptoms requiring well-trained clinicians. However, due to decreased clinical expertise for leprosy in the field [3], delayed diagnosis occurs frequently which increases the risk of severe disabilities. M.leprae infected individuals lacking clinical symptoms who are at risk of developing leprosy disease are even more difficult to identify. A diagnostic test detecting leprosy disease as well as M.leprae infection would be a valuable tool for health care workers.
Leprosy is a spectral disease for which the clinical outcome after M.leprae infection is determined by host factors. The spectrum spans from anti-inflammatory T helper-2 (Th2) immunity concomitant with large numbers of bacteria as well as antibodies against M.leprae antigens in multibacillary (MB) leprosy, to paucibacillary (PB) leprosy characterised by strong pro-inflammatory, T helper-1 (Th1) as well as T helper 17 (Th17) immunity [4]. The pro-inflammatory response in PB patients leads to bacterial control, but also to collateral damage in the form of destruction of the body's own cells by the vigorous T cell response, mimicking autoimmunity.
Since antibodies against M.leprae phenolic glycolipid I (PGL—I) indicate infection and are associated with bacillary load [5] rapid diagnostic tests detecting anti-PGL-I antibodies have been developed [5,6]. However, these are still not yet widely implemented in the field due to limited availability. Moreover, to capture the different clinical outcomes of M.leprae infection across the leprosy disease spectrum we have shown that both cellular and humoral markers should be included in diagnostic tests: biomarker profiles including cellular and/or inflammatory biomarkers such as CCL4, IL-10, IP-10, CRP combined with M.leprae specific anti-PGL-I antibodies, increased sensitivity for leprosy [7,8]. In this respect, IL-10 discriminated disease and infection from healthy status, whereas CCL4 was particularly informative for PB patients. On one hand, for classification and confirmation of leprosy diagnosis 24 h incubation with M. leprae antigens in WBAs represents a specific approach, similar to the application of the Quantiferon® test for TB diagnosis [9]. On the other hand, a triage for rapid identification of infection/disease (e.g. in large-scale contact screening efforts) must rely on biomarkers detectable in samples directly, without stimulation. To allow improved diagnosis and classification of leprosy patients as well as detection of infection by triage, we thus used a funnel approach assessing additional host proteins for their diagnostic performance in both rapid tests and 24 h WBAs, including cytokines, chemokines and growth factors (CCGF). First, we applied high throughput multiplex bead arrays (WBAs) and ELISAs (WBA and plasma) of samples from leprosy patients, household contacts (HC) and endemic controls (EC) from Bangladesh. Appropriate biomarkers were subsequently validated in low complexity, quantitative up converting phosphor lateral flow assays (UCP-LFAs) [7].
2. Materials and methods
2.1. Study setting
During this study the prevalence in the four districts (Nilphamari, Rangpur, Panchagar and Thakurgaon; population 8,190,035) was 0.9 per 10,000 and the new case detection rate 1.18 per 10,000 (Rural health program, the leprosy mission Bangladesh, yearly district activity report 2018).
2.2. Study participants
Participants were recruited on a voluntary basis between January 2013 and 2018 in leprosy endemic areas in Bangladesh as described previously [10]. Leprosy was diagnosed based on clinical and bacteriological observations and classified as MB or PB as described by the WHO [11]. Clinical and demographic data were collected in a database. As a reference group healthy individuals without known contact to leprosy patients in their village or at work from the same area (EC) were assessed for the absence of clinical signs and symptoms of leprosy and TB at intake, and after 2 and 4 years. Samples were collected from 8 villages spread randomly across the study area (2 representative villages for each of the 4 districts).
2.3. Inclusion/exclusion criteria
Patients of the Rural Health program and their contacts who were willing to participate were included in the study [10]. Contacts were either living in the same house (household members) or in a house on the same compound, sharing the same kitchen or direct neighbors (first neighbors). The following exclusion criteria were applied to patients: refusal of examination of contacts, suffering from the pure neural form of leprosy, residing only temporarily in the study area, new patients found during contact examination of the index case, living <100 m away from a patient already included in the study or first and second degree relatives of a patient already included in the study. Contacts who refused informed consent were also excluded, as well as any woman indicating to be pregnant, any person on TB or leprosy treatment, children below 5 years of age, contacts known to suffer from liver disease or jaundice, residing temporarily in the area, suffering from leprosy at the initial survey (these patients were referred to the clinic for leprosy treatment) and contacts who were already enrolled in the contact group of another patient. Staff of leprosy or TB clinics were excluded as EC.
2.4. Study cohorts
Three different cohorts were tested: a discovery cohort, including age and gender matched [7] leprosy patients (n = 79; 34 MB; 45 PB), HC (n = 54) and EC (n = 51) from Bangladesh for biomarker discovery; two validation cohorts, cohort I for biomarker validation in WBA including leprosy patients (n = 55; 27 MB; 28 PB) and EC (n = 27) and cohort II for biomarker validation in plasma consisting of leprosy patients (n = 36; 21 MB; 15 PB) and EC (n = 20). For age and gender matching a 50/50 male/female ratio and a 1:1:1 ratio of three age groups (0–14, 15–29 and 30+) was aimed at [7].
2.5. Samples
For discovery cohort and validation cohort I WBA samples, 4 ml venous blood was drawn and 1 ml applied directly to a microtube pre-coated with 10 μg M.leprae whole cell sonicate (WCS), 10 μg ML2478 and 10 μg ML0840 recombinant proteins (combined designated as Mlep) [3] or without stimulus (Med). Pre-coating of the tubes was done by lyophilizing the material. After 24 h incubation at 37 °C the microtube was frozen at −20 °C, shipped to the LUMC and stored at −80 °C until further analysis. For validation cohort II, plasma was collected as described previously [12].
2.6. Ethics
This study was performed according to the Helsinki Declaration (2008 revision) and the study protocol was approved by the National Research Ethics Committee (Bangladesh Medical Research Council) (Ref no. BMRC/NREC/2010–2013/1534). Participants were informed about the study objectives, the samples and their right to refuse to take part or withdraw from the study without consequences for their treatment. Written informed consent was obtained before enrolment. All patients received treatment according to national guidelines.
2.7. Multiplex bead arrays (MBA)
BCA-1 (CXCL13), CCL17, CTACK (CCL27), sCD40L, EGF, ENA-78 (CXCL5), Eotaxin (CCL11), FGF, Flt3L, Fraktalkine (CX3CL1), G-CSF, GM-CSF, GRO, I309, IFN-α2, IFN-γ, IL-1α, IL-1β, IL-1ra, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12(p40), IL-12(p70), IL-13, IL-15, IL-16, IL-17A, IL17F, IL-20, IL-21, IL-22, IL-23, IL-27, IL28A, IL-33, IP-10, MCP-1 (CCL2), MCP-3 (CCL7), MDC (CCL22), MIP-1α (CCL3), MIP-1β (CCL4), PDGF-AA, PDGF-AB/BB, RANTES (CCL5), SCF, SDF-1, TGF-α, TNF-α, TNF-β, TPO, TRAIL, TSLP and VEGF were measured in the discovery cohort using the Milliplex magnetic bead kit (Merck, USA) as described previously [13].
2.8. ELISAs
Validation cohort I was assessed by ELISA for ApoA1, CCL4, CFH, CRP, CCL27, CXCL9, IL-1Ra, IL-19, IL-32, MMP9, PDGF-BB, PTX3, S100A12, SAA1 (R&D systems, Minneapolis, USA), IP-10 and IL-10 (Diaclone Research, Besancon, France) and TTR (Abcam, Cambridge, UK). To detect anti-PGL-I IgM the ELISA was performed as previously described [5]. Validation cohort II was assessed by ELISA for anti-PGL-I IgM, ApoA1, CCL4, CRP, IL-1Ra, IP-10 and S100A12.
2.9. Lateral flow assays (LFA)
LFAs for IP-10, CRP and αPGL-I IgM strips were produced as described earlier [3]. ApoA1 and S100A12 strips were produced similarly with 200 ng goat-anti-S100A12 pAb (AF1052; R&D systems, Minneapolis, USA) and Goat-anti-ApoA1 pAb (AF3664; R&D systems, Minneapolis, USA) on the test lines. The respective flow control lines comprised 100 ng Goat-anti-Rabbit or Rabbit-anti-Goat antibody. Conjugates of UCP particles were applied to the sample/conjugate pad at a density of 200 ng per 4 mm. UCP conjugates were prepared according to a previously described protocol [14] with Rabbit-anti-ApoA1 (Clone # 2083A; R&D systems, Minneapolis, USA) or goat-anti-S100A12 pAb (AF1052; R&D systems, Minneapolis, USA) at a concentration of 50 μg antibody per mg UCP.
10 μl, 1 μl, 0.1 μl and 0,01 μl plasma was diluted in high salt lateral flow (HSLF) buffer (100 mM HEPES pH 7.5, 270 mM NaCl, 1% (w/v) BSA, 0.5% (v/v) Tween-20). 50 ul of diluted sample was added to microtiter plate wells before target-specific LF strips were placed in the corresponding wells. Immunochromatography was allowed to continue for at least 30 min until dry.
2.10. Statistical analysis
Statistical analysis was performed using GraphPad Prism version 7 (GraphPad Software, San Diego,CA, USA; http://www.graphpad.com), SPSS Statistics 24 (http://www.spss.com.hk) and R Version 3.3.0 (R, Vienna, Austria; http://www.R-project.org).
Hierarchical clustering of the CCGF based on absolute correlation difference and average linkage was performed using the global test [15]. Log2 fold changes were calculated for MB, PB and HC compared to EC. Volcano plots were computed using R, by plotting the log2 fold change against the -log10 (p-value) of each marker (p-values calculated by global test). Radar plots showing the log2 fold change were generated using Excel 2016. Receiver operating characteristic (ROC) curves were computed in Graphpad Prism and the respective area under the curve (AUC) was calculated. Cut-offs were determined by calculating the Youden's index [16]. To determine the optimal classification method three approaches (logistic regression, random forest classification and classification tree) were computed using Orange data mining version 3.3.9 [17], comparing the AUC after 10-fold stratified cross-validation for each method.
3. Results
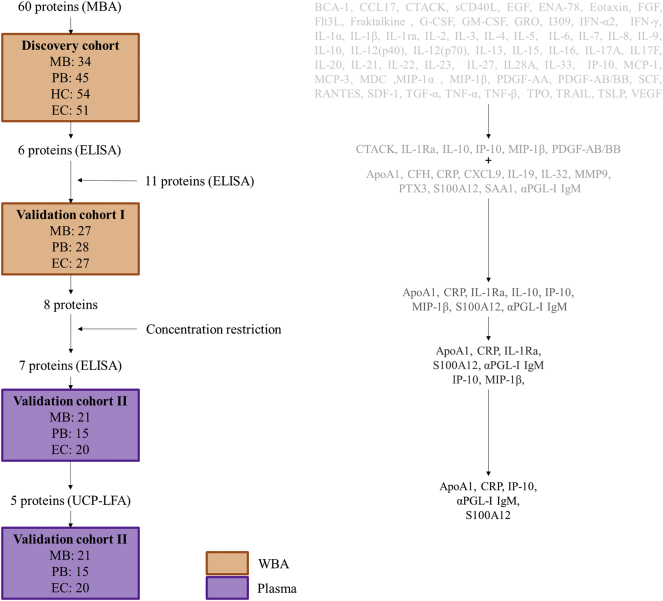
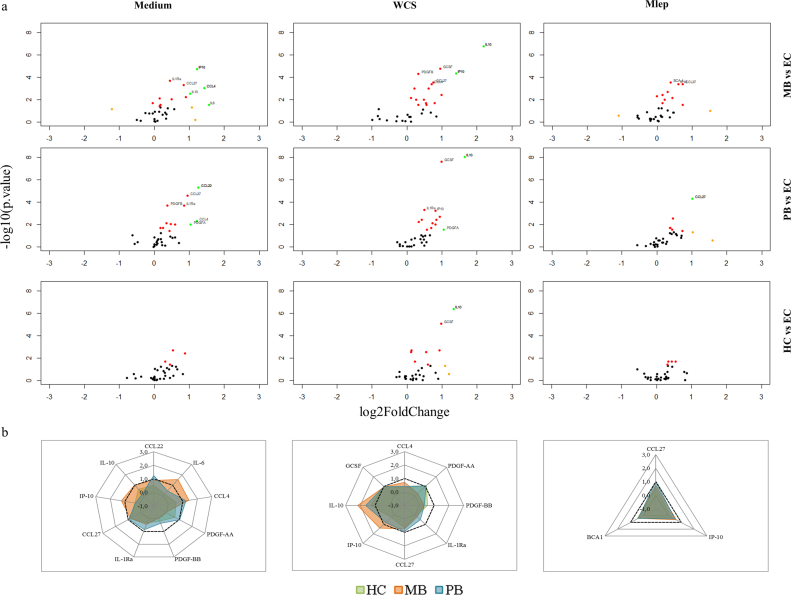
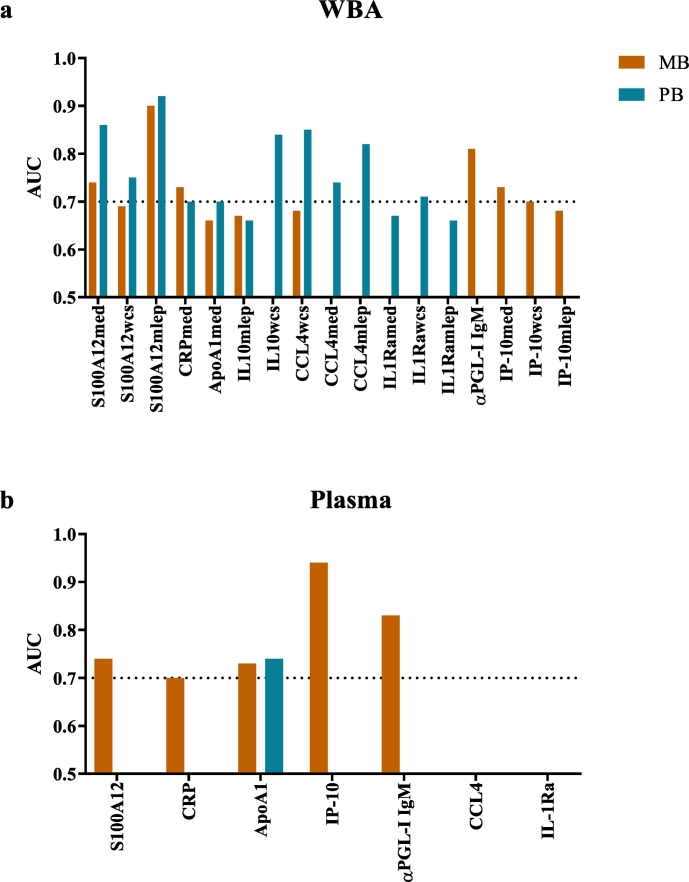
To obtain new biomarkers for leprosy with high potential for user-friendly POC applications, we applied a funnel approach using discovery and validation cohorts (Fig. 1). First, in a discovery cohort of MB (n = 34) and PB (n = 45) patients, HC (n = 54) and EC (n = 51), 60 CCGFs were measured in WBA supernatant using high throughput multiplex bead arrays (Fig. 2; Supplementary Table S1–4): in 24 h whole blood samples without stimulus IL-1Ra, CCL27 and CCL4 identified both MB and PB patients. IL-6, IL-10 and IP-10 levels were significantly different from EC in MB patients only, whereas CCL22, PDGF-AA and PDGF-BB identified PB patients (Fig. 2A, left column). In samples stimulated with M. leprae WCS IL-10 and GCSF levels were higher in both leprosy patients and their contacts. Elevated levels of IP-10 were observed in both MB and PB patients, whereas PDGF-BB, CCL4 and CCL27 levels were significantly higher for MB patients and IL-1Ra and PDGF-AA for PB patients in response to WCS (Fig. 2A, middle column). In response to 2 M.leprae specific proteins (Mlep) CCL27 was identified as a marker for both types of leprosy disease, IP-10 and BCA-1 for MB leprosy only (Fig. 2A, right column). Thus, in this discovery cohort IP-10, IL-1Ra, CCL4, CCL27 and PDGF-BB enabled the distinction of leprosy patients from EC irrespective of leprosy classification (Fig. 2B) and were used for further evaluation by ELISAs in validation cohort I consisting of 27 MB patients, 28 PB patients and 27 EC. The WCS-induced levels of IL-10 and GCSF, discriminating both patients and HC from EC significantly, correlated in the discovery cohort. Therefore, only IL-10 was included as a marker for infection as these data confirm previous reports on IL-10 as an infection marker [7]. Additionally, 11 markers with potential for diagnosis of mycobacterial diseases in earlier reports [8,[18], [19], [20], [21], [22]] (not available in the multiplex bead assay) were also included (Fig. 1; Supplementary Table S1). AUCs were calculated to assess the potential of the markers tested to discriminate the test groups from EC, demonstrating significance for S100A12, CRP, ApoA1, IL-10 in response to M.leprae specific proteins and CCL4 in response to M. leprae WCS for both leprosy types. Furthermore, MB patients could be discriminated from EC based on αPGL-I IgM and IP-10 as well, whereas for PB patients this was feasible based on IL-10WCS, CCL4Med/CCL4Mlep and IL-1Ra (Fig. 3A). Thus, this validation cohort confirmed diagnostic potential for leprosy of 8 markers.
Fig. 1.
Funnel approach workflow. Three different cohorts including samples originating from Bangladesh (multibacillary (MB) and paucibacillary (PB) leprosy patients, household contacts (HC), healthy endemic controls (EC)) were used. Both whole blood assays (WBA) samples (orange; unstimulated and stimulated with M. leprae whole cell sonicate or M. leprae specific proteins (ML0840, ML2478)) and plasma samples (purple) were analyzed using multiplex bead assays (MBA), ELISA or up-converting phosphor lateral flow assays (UCP-LFA). The markers tested in each step are displayed in the right column. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 2.
Production of cytokines, chemokines and growth factors (CCGFs) in leprosy patients and household contacts compared to endemic controls. 60 CCGFs were detected in whole blood assay (WBA) supernatant of multibacillary (MB) and paucibacillary (PB) leprosy patients, household contacts (HC) and endemic controls (EC). (a) Volcano plots show the log2 fold change compared to EC (x-axis) and the -log10 (p-value) (y-axis) in unstimulated WBA supernatant (Medium; left column), in response to M.leprae whole cell sonicate (WCS; middle column) and two specific M.leprae proteins (Mlep; ML0840, ML2478; right column). The markers in either of the three groups with both a log2 fold change of 1 (−1) and a p-value <0,05 or markers with a p-value <0,001 are indicated (P-value <0,05 = red dot, log2 fold change of 1(−1) = orange dot, P-value <0,05 & log2 fold change of 1(−1) = green dot). (b) Summary of the markers indicated in the volcano plots per stimulus (Medium = left, WCS = middle and Mlep = right). Radar plots show the log2 FC of the markers indicated in the volcano plots for MB (orange), PB (blue) and HC (green) compared to the levels in EC. Dotted lines indicated a log2 FC of 1. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 3.
Biomarkers validated by ELISA in whole blood assay supernatant and plasma of leprosy patients. Markers showing significant areas under the curve (AUC) for multibacillary (MB; orange) and/or paucibacillary (PB; blue) leprosy patients in unstimulated whole blood assay (WBA) supernatant (med), in response to M.leprae whole cell sonicate (WCS) and two specific M.leprae proteins (Mlep) (A) or plasma samples (B). Biomarkers levels were compared to those of endemic controls. Values for AUC can range from 0.5 to 1, the dotted line indicates an AUC of 0.7. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fingerstick blood (FSB) is an easy to use sample, requiring no phlebotomist or overnight stimulation, making it suitable for rapid testing using field friendly LFAs. As a proxy for FSB [3] we here tested plasma samples from Bangladeshi leprosy patients and EC (validation cohort II) for the seven markers that were significantly different in unstimulated WBA samples (Fig. 3A). Since stimulation is required for detection of IL-10 we did not further include this marker for analysis of plasma samples. Without antigen stimulation, anti-PGL-I IgM antibodies, IP-10, CRP and S100A12 remained valuable markers in plasma for MB patients and ApoA1 for both MB and PB (Fig. 3B), whereas IL-1Ra and CCL4 levels could not be detected in these plasma samples.
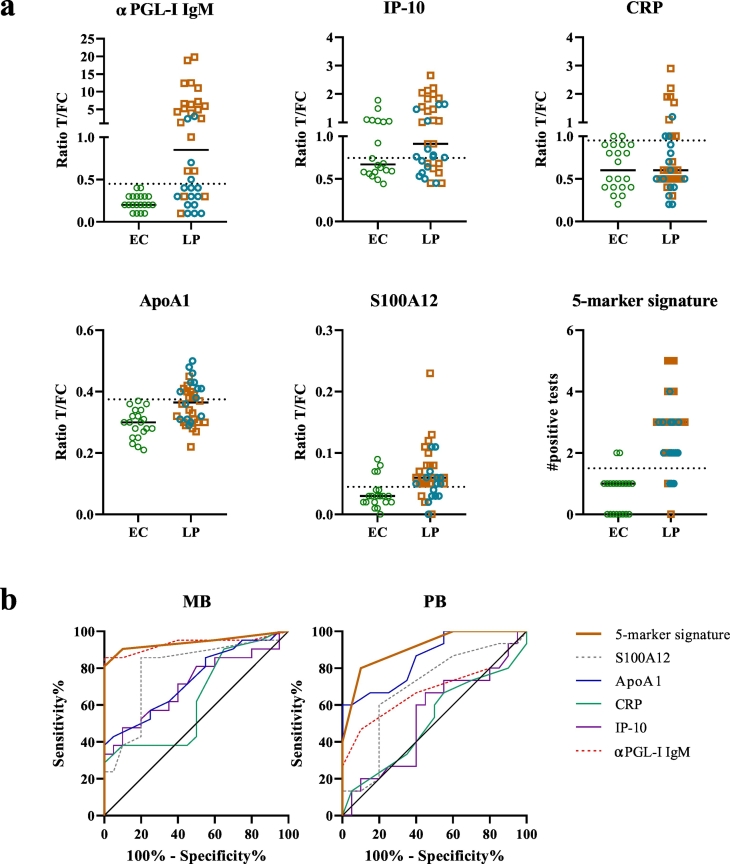
To assess the potential of the five-marker plasma signature for POC/field applications, quantitative UCP-LFAs specific for ApoA1, CRP, IP-10, αPGL-IgM and S100A12 were tested in validation cohort II. Data obtained by the UCP-LFAs are in line with the ELISA data for plasma samples, with ApoA1 being the most optimal marker to discriminate PB patients from EC, whereas the other four markers are elevated especially in MB patients, but also discriminate some PB patients from EC (Fig. 4). To optimally identify leprosy across the disease spectrum, cut-offs were determined comparing patients irrespective of leprosy type to EC (Supplementary Table S5). Based on the cut-off values, the number of positive tests was determined per individual resulting in a five-marker signature. A sum of positive test results is a practical way to apply biomarker signatures in the field. This signature (AUC: 0.93, p < 0,0001) identified 86% of the leprosy patients, with a specificity of 90% (cut-off >2 tests positive). Moreover, in contrast to single markers, the five-marker signature showed similar AUCs for MB (AUC:0,94) and PB (AUC:0,91) patients (Fig. 4B).
Fig. 4.
Five-marker plasma signature assessed by up-converting phosphor lateral flow assays (UCP-LFA). Levels of αPGL-I IgM, IP-10, CRP, ApoA1 and S100A12 were measured by UCP-LFAs comparing 36 leprosy patients (LP, orange squares = multibacillary (MB) patients and blue dots = paucibacillary (PB) patients) to 20 endemic controls (EC = green dots). (a) Ratio values for the 5 markers tested were calculated by dividing the relative fluorescence units (RFU) from the test line (T) by the RFU from the flow controls (FC). The dotted line indicates the cut-off value for each markers as calculated by the Youden's index. Values above the cut-off line are considered a positive test result, the sum of all positive tests results in the values displayed for the five-marker signature. Cut-offs are shown in supplementary Table S5. (b) receiver operating characteristic curves (ROC) for MB and PB patients compared to EC showing all 5 markers tested (αPGL-I IgM (red), IP-10 (purple), CRP (green), ApoA1 (blue), S100A12 (grey)) and the five-marker signature (orange).
Additionally, three different classification methods (logistic regression, classification tree and random forest classification) were applied to the two validation cohorts to assess the performance of the POC five-marker signature. In general, ten-fold stratified cross-validation showed the most optimal AUC and classification accuracy for the classification tree algorithm (Supplementary Table S6). The cross-validated sensitivity and specificity of this algorithm for WBA and plasma as assessed by ELISA was comparable to that assessed by UCP-LFA, indicating that the signature can also be accurately detected in POC-tests (Supplementary Fig. S1). Moreover, cross-validated signatures showed only a minor decrease in sensitivity (12%)/specificity (16%) compared to the POC signature, indicating the robustness of this signature. The here described “funnel- approach” thus identified biomarker signatures, applicable to either WBAs and plasma, that sensitively detect MB as well as PB leprosy patients.
4. Discussion
Tools that detect disease at an early state and identify M.leprae infection are eminent to interrupt transmission. Previous reports showed that the combined detection of humoral markers capturing MB leprosy and cellular markers detecting PB, significantly improved the detection of leprosy patients [7,8]. However, PB patients and HC could not be distinguished as these markers showed similar responses for these cellular markers, especially in highly endemic areas [7,8]. In this study, using a wide array of CCGFs, five markers differentiated PB patients from HC (Supplementary Table S2–4), whereas 18 makers were different in PB patients compared to EC in WBA samples. These included markers previously tested in the UCP-LFA format such as CCL4, CRP and IL-10 [7,8], as well as the newly identified markers ApoA1, IL-1Ra and S100A12.
Apolipoprotein A1 (ApoA1) is a negative acute phase protein which is suggested to bind to stimulated T-cells thereby inhibiting contact-mediated activation of monocytes [23] and reported to be decreased during inflammation [24] and active tuberculosis [18]. Indeed, in WBA samples both MB and PB patients showed decreased levels of anti-inflammatory ApoA1. IL-1Ra (Interleukin-1 receptor antagonist) also exerts anti-inflammatory functions by binding to the IL-1 receptor, thereby inhibiting the function of the proinflammatory IL-1α and IL-1β. M.leprae can induce high levels of IL-1Ra in monocytes, and high expression of IL-1Ra in skin lesions was associated with increased susceptibility to leprosy irrespective of polarity [25]. Both MB and PB patients showed elevated levels of IL-1Ra in WBA samples, supporting the use of IL-1Ra as a biomarker in leprosy diagnostics. S100A12 (calgranulin C) can induce proinflammatory cytokines and serum levels have been shown to correlate with disease activity in inflammatory disorders [26]. Interestingly, S100A12 has antimicrobial properties exerting direct effects on both M.leprae and M.tuberculosis and was more strongly expressed in skin lesions of PB leprosy patients [27]. Serum levels did, however, not significantly differ between MB and PB patients [21] in line with the data observed in this study. In response to M.leprae specific proteins, S100A12 showed the optimal AUC of all the markers tested in WBAs, both for MB and PB patients. For MB leprosy this study also confirmed the use of IP-10 as a biomarker in line with our previous studies [7,8].
In view of point-of-care (POC) test applicability (i.e. direct analysis of clinicals samples without antigen stimulation), biomarker levels were also assessed in plasma samples as a proxy for FSB collectable without venipuncture [3]. A plasma biomarker signature including αPGL-I IgM, IP-10, S100A12, ApoA1, CRP accurately detected leprosy patients irrespective of type with high sensitivity (86%) and specificity (90%) in the UCP-LFAs; indicating the diagnostic value of this signature in leprosy as it identifies both patients with high and low bacillary loads. The future detection of this signature in FSB by rapid POC testing can be useful for screening purposes in a triage approach: a FSB-based multi-biomarker LF strip rules out individuals who lack host biomarkers associated with leprosy, and individuals requiring further testing are selected for overnight incubation of whole blood with M.leprae specific antigens [7]. In the 24 h stimulated WBA samples a larger selection of (stimulated) discriminatory markers were identified, especially to detect PB patients. The levels of biomarkers in WBAs can thus be used for multiple applications, besides contact screening i.e. to help in classification of leprosy patients in referral hospitals or for monitoring of the development of complications such as leprosy reactions [12,28].
PB patients and HC show similar immune responses and often have undetectable loads of M.leprae bacilli. The infection status of HC is, however, largely unknown. Reports from Brazil and India indicate the presence of M.leprae DNA in nasal swabs and skins slit smears of HC ranging from 8,8% to 49% [29,30] or 21%, respectively [31]. Therefore, elaborate host immune profiling of HC stratified by M.leprae DNA presence in nasal swabs or slit skin slides may aid in identifying biomarkers associated with M.leprae exposure or infection without clinical symptoms. αPGL-I IgM levels have been measured in HC in order to predict the development of leprosy disease, but has so far proven insufficient for early detection of leprosy or onset of disease [32,33]. Longitudinal monitoring of the host biomarkers described in this study can provide more insight into the predictive capacity of this biomarker signature. Moreover, validation of this signature in different populations in leprosy endemic areas and validation in FSB is required for large numbers of samples before multi-biomarker testing can be implemented in leprosy healthcare. Translation of biomarkers into clinical practice is still challenging as evidenced by the low percentage of discovered biomarkers validated for routine clinical practice [34]. Identifying markers in three independent cohorts using a funnel approach ensure that the strongest biomarkers remain.
Application of biomarker signatures in rapid POC tests can not only facilitate leprosy diagnosis and classification but also aid decision making on which individuals are candidate for prophylactic treatment. Contacts of leprosy patients are 4 to 9 times more at risk of developing leprosy than the general population [35]. Therefore, these individuals are targeted for post-exposure prophylaxis. Large scale contact screening trials to select M.leprae infected individuals for post-exposure prophylaxis with single dose rifampicin (SDR) according to WHO guidelines [28] for leprosy control will thus contribute to decrease transmission and thereby prevent leprosy-associated irreversible nerve damage. Moreover, the quantitative LF test data enable the assessment of SDR efficacy and dosage regimens in infected individuals, as well as monitoring of treatment in leprosy patients. Importantly, the biomarker signature identified in this study, including novel biomarkers, accurately detected patients across the leprosy spectrum and was compatible with low-complexity lateral flow tests. Implementation of these host biomarker-based field tests can thus provide the urgently needed diagnostic tool for leprosy applicable in low-resource settings.
Author contributions
Designed research: AG, AH.
Enrolled patients, performed and registered clinical diagnosis: KA, JR, RR, RF, MK, AC.
Performed research: AH, ET, LW, SE, MK.
Provided tools/technical expertise: PC, KF.
Analyzed the data: AG, AH, PC.
Wrote the paper: AG, AH.
Agree with manuscript results and conclusions: all authors.
Data sharing statement
All biomarker data used for analysis in this study is available upon request from the corresponding author.
Declaration of Competing Interest
The authors declare that they have no conflict of interest
Acknowledgements
The authors gratefully acknowledge all patients and blood donors. LUMC, EMC and TLMI, B are part of the IDEAL (Initiative for Diagnostic and Epidemiological Assays for Leprosy) Consortium. We thank the staff of the Rural Health Program, The Leprosy Mission International Bangladesh, Nilphamari, Bangladesh for recruitment of study participants and sample collection.
This study was supported by the Order of Malta-Grants-for-Leprosy-Research (MALTALEP), the Q.M. Gastmann-Wichers Foundation and the Leprosy Research Initiative (LRI) together with the Turing Foundation (ILEP#: 703.15.07). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.08.009.
Appendix A. Supplementary data
Supplementary material
References
- 1.Rao P.S., Darlong F., Timothy M., Kumar S., Abraham S., Kurian R. Disability adjusted working life years (DAWLYs) of leprosy affected persons in India. Indian J Med Res. 2013;137(5):907–910. [PMC free article] [PubMed] [Google Scholar]
- 2.Global leprosy update 2016: Accelerating reduction of disease burden. Wkly Epidemiol Rec. 2017;92(35):501–519. [PubMed] [Google Scholar]
- 3.Corstjens P., van Hooij A., Tjon Kon Fat E.M., Alam K., Vrolijk L.B., Dlamini S. Fingerstick test quantifying humoral and cellular biomarkers indicative for M. leprae infection. Clin Biochem. Apr 2019;66:76–82. doi: 10.1016/j.clinbiochem.2019.01.007. (Epub 2019 Jan 26) [DOI] [PubMed] [Google Scholar]
- 4.Saini C., Ramesh V., Nath I. CD4+ Th17 cells discriminate clinical types and constitute a third subset of non Th1, non Th2 T cells in human leprosy. PLoS Negl Trop Dis. 2013;7(7) doi: 10.1371/journal.pntd.0002338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Hooij A., Tjon Kon Fat E.M., van den Eeden S.J.F., Wilson L., Batista da Silva M., Salgado C.G. Field-friendly serological tests for determination of M. leprae-specific antibodies. Sci Rep. 2017;7(1):8868. doi: 10.1038/s41598-017-07803-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duthie M.S., Balagon M.F., Maghanoy A., Orcullo F.M., Cang M., Dias R.F. Rapid quantitative serological test for detection of infection with Mycobacterium leprae, the causative agent of leprosy. J Clin Microbiol. 2014;52(2):613–619. doi: 10.1128/JCM.02085-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Hooij A., Tjon Kon Fat E.M., Richardus R., van den Eeden S.J., Wilson L., de Dood C.J. Quantitative lateral flow strip assays as user-friendly tools to detect biomarker profiles for leprosy. Sci Rep. 2016;6 doi: 10.1038/srep34260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Hooij A., Tjon Kon Fat E.M., Batista da Silva M., Carvalho Bouth R., Cunha Messias A.C., Gobbo A.R. Evaluation of immunodiagnostic tests for leprosy in Brazil, China and Ethiopia. Sci Rep. 2018;8(1) doi: 10.1038/s41598-018-36323-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uzorka J.W., Bossink A.W.J., Franken W.P.J., Thijsen S.F.T., Leyten E.M.S., van Haeften A.C. Borderline QuantiFERON results and the distinction between specific responses and test variability. Tuberculosis (Edinb) 2018;111:102–108. doi: 10.1016/j.tube.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 10.Richardus R.A., Alam K., Pahan D., Feenstra S.G., Geluk A., Richardus J.H. The combined effect of chemoprophylaxis with single dose rifampicin and immunoprophylaxis with BCG to prevent leprosy in contacts of newly diagnosed leprosy cases: a cluster randomized controlled trial (MALTALEP study) BMC Infect Dis. 2013;13:456. doi: 10.1186/1471-2334-13-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Global leprosy update 2015: Time for action, accountability and inclusion. Wkly Epidemiol Rec. 2015;91(35):405–420. [PubMed] [Google Scholar]
- 12.Khadge S., Banu S., Bobosha K., van der Ploeg-van Schip J.J., Goulart I.M., Thapa P. Longitudinal immune profiles in type 1 leprosy reactions in Bangladesh, Brazil, Ethiopia and Nepal. BMC Infect Dis. 2015;15:477. doi: 10.1186/s12879-015-1128-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geluk A., Bobosha K., van der Ploeg-van Schip J.J., Spencer J.S., Banu S., Martins M.V. New biomarkers with relevance to leprosy diagnosis applicable in areas hyperendemic for leprosy. J Immunol. 2012;188(10):4782–4791. doi: 10.4049/jimmunol.1103452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corstjens P., Zuiderwijk M., Brink A., Li S., Feindt H., Niedbala R.S. Use of up-converting phosphor reporters in lateral-flow assays to detect specific nucleic acid sequences: a rapid, sensitive DNA test to identify human papillomavirus type 16 infection. Clin Chem. 2001;47(10):1885–1893. [PubMed] [Google Scholar]
- 15.Goeman J.J., van de Geer S.A., de Kort F., van Houwelingen H.C. A global test for groups of genes: testing association with a clinical outcome. Bioinformatics. 2004;20(1):93–99. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- 16.Fluss R., Faraggi D., Reiser B. Estimation of the Youden Index and its associated cutoff point. Biom J. 2005;47(4):458–472. doi: 10.1002/bimj.200410135. [DOI] [PubMed] [Google Scholar]
- 17.Demsar J.C.T., Erjavec A., Gorup C., Hocevar T., Milutinovic M., Mozina M. Orange: data mining toolbox in python. J Mach Learn Res. 2013;14(Aug):2349–2353. [Google Scholar]
- 18.Chegou N.N., Sutherland J.S., Malherbe S., Crampin A.C., Corstjens P.L., Geluk A. Diagnostic performance of a seven-marker serum protein biosignature for the diagnosis of active TB disease in African primary healthcare clinic attendees with signs and symptoms suggestive of TB. Thorax. 2016;71(9):785–794. doi: 10.1136/thoraxjnl-2015-207999. [DOI] [PubMed] [Google Scholar]
- 19.Jacobs R., Malherbe S., Loxton A.G., Stanley K., van der Spuy G., Walzl G. Identification of novel host biomarkers in plasma as candidates for the immunodiagnosis of tuberculosis disease and monitoring of tuberculosis treatment response. Oncotarget. 2016;7(36):57581–57592. doi: 10.18632/oncotarget.11420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mendes M.A., de Carvalho D.S., Amadeu T.P., Silva B.J.A., Prata R., da Silva C.O. Elevated Pentraxin-3 concentrations in patients with leprosy: potential biomarker of erythema nodosum leprosum. J Infect Dis. 2017;216(12):1635–1643. doi: 10.1093/infdis/jix267. [DOI] [PubMed] [Google Scholar]
- 21.Kim M.H., Choi Y.W., Choi H.Y., Myung K.B., Cho S.N. The expression of RAGE and EN-RAGE in leprosy. Br J Dermatol. 2006;154(4):594–601. doi: 10.1111/j.1365-2133.2005.07112.x. [DOI] [PubMed] [Google Scholar]
- 22.Montoya D., Inkeles M.S., Liu P.T., Realegeno S., Teles R.M., Vaidya P. IL-32 is a molecular marker of a host defense network in human tuberculosis. Sci Transl Med. 2014;6(250):250ra114. doi: 10.1126/scitranslmed.3009546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hyka N., Dayer J.M., Modoux C., Kohno T., Edwards C.K., 3rd, Roux-Lombard P. Apolipoprotein A-I inhibits the production of interleukin-1beta and tumor necrosis factor-alpha by blocking contact-mediated activation of monocytes by T lymphocytes. Blood. 2001;97(8):2381–2389. doi: 10.1182/blood.v97.8.2381. [DOI] [PubMed] [Google Scholar]
- 24.Montecucco F., Favari E., Norata G.D., Ronda N., Nofer J.R., Vuilleumier N. Impact of systemic inflammation and autoimmune diseases on apoA-I and HDL plasma levels and functions. Handb Exp Pharmacol. 2015;224:455–482. doi: 10.1007/978-3-319-09665-0_14. [DOI] [PubMed] [Google Scholar]
- 25.Shah J.A., Berrington W.R., Vary J.C., Jr., Wells R.D., Peterson G.J., Kunwar C.B. Genetic variation in toll-interacting protein is associated with leprosy susceptibility and cutaneous expression of interleukin 1 receptor antagonist. J Infect Dis. 2016;213(7):1189–1197. doi: 10.1093/infdis/jiv570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foell D., Wittkowski H., Vogl T., Roth J. S100 proteins expressed in phagocytes: a novel group of damage-associated molecular pattern molecules. J Leukoc Biol. 2007;81(1):28–37. doi: 10.1189/jlb.0306170. [DOI] [PubMed] [Google Scholar]
- 27.Realegeno S., Kelly-Scumpia K.M., Dang A.T., Lu J., Teles R., Liu P.T. S100A12 is part of the antimicrobial network against mycobacterium leprae in human macrophages. PLoS Pathog. 2016;12(6) doi: 10.1371/journal.ppat.1005705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hagge D.A., Parajuli P., Kunwar C.B., Rana D., Thapa R., Neupane K.D. Opening a can of worms: leprosy reactions and complicit soil-transmitted helminths. EBioMedicine. 2017;23:119–124. doi: 10.1016/j.ebiom.2017.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brito e Cabral P., Junior J.E., de Macedo A.C., Alves A.R., Goncalves T.B., Brito e Cabral T.C. Anti-PGL1 salivary IgA/IgM, serum IgG/IgM, and nasal Mycobacterium leprae DNA in individuals with household contact with leprosy. Int J Infect Dis. 2013;17(11):e1005–e1010. doi: 10.1016/j.ijid.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 30.Araujo S., Freitas L.O., Goulart L.R., Goulart I.M. Molecular evidence for the aerial route of infection of mycobacterium leprae and the role of asymptomatic carriers in the persistence of leprosy. Clin Infect Dis. 2016;63(11):1412–1420. doi: 10.1093/cid/ciw570. [DOI] [PubMed] [Google Scholar]
- 31.Turankar R.P., Lavania M., Chaitanya V.S., Sengupta U., Darlong J., Darlong F. Single nucleotide polymorphism-based molecular typing of M. leprae from multicase families of leprosy patients and their surroundings to understand the transmission of leprosy. Clin Microbiol Infect. 2014;20(3):O142–O149. doi: 10.1111/1469-0691.12365. [DOI] [PubMed] [Google Scholar]
- 32.Leturiondo A.L., Noronha A.B., do Nascimento M.O.O., Ferreira C.O., Rodrigues F.D.C., Moraes M.O. Performance of serological tests PGL1 and NDO-LID in the diagnosis of leprosy in a reference Center in Brazil. BMC Infect Dis. 2019;19(1):22. doi: 10.1186/s12879-018-3653-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richardus R.A., van der Zwet K., van Hooij A., Wilson L., Oskam L., Faber R. Longitudinal assessment of anti-PGL-I serology in contacts of leprosy patients in Bangladesh. PLoS Negl Trop Dis. 2017;11(12) doi: 10.1371/journal.pntd.0006083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poste G. Bring on the biomarkers. Nature. 2011;469(7329):156–157. doi: 10.1038/469156a. [DOI] [PubMed] [Google Scholar]
- 35.van Beers S.M., Hatta M., Klatser P.R. Patient contact is the major determinant in incident leprosy: implications for future control. Int J Lepr Other Mycobact Dis. 1999;67(2):119–128. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material