Abstract
Background
Up to 30–40% of Ewing sarcoma (EwS) patients with non-metastatic disease develop local or metastatic relapse within a time span of 2–10 years. This is in part caused by the absence of prognostic biomarkers that can identify high-risk patients and thus assign them to risk-adapted monitoring and treatment regimens. Since cancer stemness has been associated with tumour relapse and poor patient outcomes, we investigated in the current study the prognostic potential SOX2 (sex determining region Y box 2) – a major transcription factor involved in development and stemness – which was previously described to contribute to the undifferentiated phenotype of EwS.
Methods
Two independent patient cohorts, one consisting of 189 retrospectively collected EwS tumours with corresponding mRNA expression data (test-cohort) and the other consisting of 141 prospectively collected formalin-fixed and paraffin-embedded resected tumours (validation and cohort), were employed to analyse SOX2 expression levels through DNA microarrays or immunohistochemistry, respectively, and to compare them with clinical parameters and patient outcomes. Two methods were employed to test the validity of the results at both the mRNA and protein levels.
Findings
Both cohorts showed that only a subset of EwS patients (16–20%) expressed high SOX2 mRNA or protein levels, which significantly correlated with poor overall survival. Multivariate analyses of our validation-cohort revealed that high SOX2 expression represents a major risk-factor for poor survival (HR = 3·19; 95%CI 1·74–5·84; p < 0·01) that is independent from metastasis and other known clinical risk-factors at the time of diagnosis. Univariate analyses demonstrated that SOX2-high expression was correlated with tumour relapse (p = 0·002). The median first relapse was at 14·7 months (range: 3·5–180·7).
Interpretation
High SOX2 expression constitutes an independent prognostic biomarker for EwS patients with poor outcomes. This may help to identify patients with localised disease who are at high risk for tumour relapse within the first two years after diagnosis.
Funding
The laboratory of T. G. P. Grünewald is supported by grants from the ‘Verein zur Förderung von Wissenschaft und Forschung an der Medizinischen Fakultät der LMU München (WiFoMed)’, by LMU Munich's Institutional Strategy LMUexcellent within the framework of the German Excellence Initiative, the ‘Mehr LEBEN für krebskranke Kinder – Bettina-Bräu-Stiftung’, the Walter Schulz Foundation, the Wilhelm Sander-Foundation (2016.167.1), the Friedrich-Baur foundation, the Matthias-Lackas foundation, the Barbara & Hubertus Trettner foundation, the Dr. Leopold & Carmen Ellinger foundation, the Gert & Susanna Mayer foundation, the Deutsche Forschungsgemeinschaft (DFG 391665916), and by the German Cancer Aid (DKH-111886 and DKH-70112257). J. Li was supported by a scholarship of the China Scholarship Council (CSC), J. Musa was supported by a scholarship of the Kind-Philipp foundation, and T. L. B. Hölting by a scholarship of the German Cancer Aid. M. F. Orth and M. M. L. Knott were supported by scholarships of the German National Academic Foundation. G. Sannino was supported by a scholarship from the Fritz-Thyssen Foundation (FTF-40.15.0.030MN). The work of U. Dirksen is supported by grants from the German Cancer Aid (DKH-108128, DKH-70112018, and DKH-70113419), the ERA-Net-TRANSCAN consortium (project number 01KT1310), and Euro Ewing Consortium (EEC, project number EU-FP7 602,856), both funded under the European Commission Seventh Framework Program FP7-HEALTH (http://cordis.europa.eu/), the Barbara & Hubertus Trettner foundation, and the Gert & Susanna Mayer foundation. G. Hardiman was supported by grants from the National Science Foundation (SC EPSCoR) and National Institutes of Health (U01-DA045300). The laboratory of J. Alonso was supported by Instituto de Salud Carlos III (PI12/00816; PI16CIII/00026); Asociación Pablo Ugarte (TPY-M 1149/13; TRPV 205/18), ASION (TVP 141/17), Fundación Sonrisa de Alex & Todos somos Iván (TVP 1324/15).
Keywords: Ewing sarcoma, Biomarker, SOX2, Patient outcome, Risk-stratification
Research in context.
Evidence before this study
One third of EwS patients with localised disease experiences relapse during follow-up. So far, no study has succeeded in identifying a broadly available bona fide stratification biomarker to predict which patients will develop EwS recurrence. SOX2 was studied in EwS by two different groups and linked to stemness and tumourigenicity in in vitro and in vivo models. However, the expression of SOX2 in both studies was either not analysed in tumour material derived from EwS patients or was analysed only in small series and not associated with clinicopathological features, such as overall survival (OS), tumour volume, metastasis at diagnosis, relapse, and histological response.
Added value of this study
This is the first study to evaluate the prognostic potential of SOX2 at the mRNA and protein levels in EwS patients. Here, we report, based on the analysis of two independent cohorts (a retrospective mRNA-cohort and a prospective TMA-cohort) consisting of 189 and 141 cases, respectively, that high SOX2 is significantly correlated with poorer patient outcome. In both cohorts, the association of high SOX2 expression with OS was independent from the presence of metastasis at diagnosis. A univariate analysis showed in the TMA-cohort that SOX2-high expression is correlated with tumour relapse (p = 0·002), providing, for the first time, a promising and widely available prognostic biomarker to stratify high-risk patients with localised disease.
Implications of all the available evidence
A pivotal role of SOX2 in tumourigenesis has been consolidated in several tumours, including EwS. Although SOX2 does not represent a druggable target, in this study we suggest employing SOX2 as a stratification marker to identify high-risk patients and initiate molecular studies and clinical trials aimed at developing more personalised therapies for these patients. Moreover, patients with SOX2-high should be monitored more closely during the follow-up so that timely intervention can be made in cases of relapse.
Alt-text: Unlabelled Box
1. Introduction
Ewing sarcoma (EwS), the second most common bone and soft tissue cancer in children and adolescents, potentially arises from neuroectodermal or mesodermal mesenchymal stem cells [1]. In fact, EwS tumours display a largely undifferentiated and ‘stemness’ phenotype, which is believed to contribute to its clinical aggressiveness [1]. Genetically, the hallmark of EwS is chromosomal translocations that generate chimeric proteins through fusion of the EWSR1 (Ewing sarcoma breakpoint region 1) gene to various members of the ETS (E26 transformation specific) family of transcription factors, in 85% FLI1 (Friend leukemia virus integration 1) [[2], [3], [4]]. These EWSR1-ETS fusion oncoproteins act as aberrant transcription factors that promote tumour initiation and progression by massively rewiring the cellular transcriptome and spliceosome [1].
Of EwS patients, 60–75% benefit from multimodal therapy [5,6]. Consequently, >30% of patients show a limited response to treatment, which is often first noted based on assessment of their histological response following neoadjuvant chemotherapy [7]. Since limited treatment response may be associated with early relapse [8], upfront identification of high-risk patients is essential to assigning them to adequate treatment regimens. Despite several clinicopathological features (such as tumour volume, histological response, tumour site and age at diagnosis) have been associated with high-risk disease, the presence of metastasis at diagnosis remains, thus far, the major factor for risk-stratification in EwS patients [9]. However, for patients with localised disease, risk prediction remains difficult, as no bona fide prognostic biomarkers independent from metastasis are available [1,10].
A previous report has indicated that stemness in EwS may be mediated via EWSR1-FLI-induced expression of SOX2 (sex determining region Y box 2) [11] – a well-known stemness gene overexpressed in many cancers [12]. We therefore explored, in the current study, the expression pattern of SOX2 in two independent EwS cohorts and investigated whether SOX2 may serve as a biomarker for outcome prediction in EwS.
We show that SOX2 is expressed only in ~16–20% of EwS tumours and is associated with very poor patient outcome independent of metastasis at time of diagnosis. Moreover, high expression of SOX2 is significantly correlated with local and/or metastatic tumour relapse. Our findings suggest that detection of high SOX2 expression through immunohistochemistry or RNA-based techniques may constitute a broadly available biomarker for risk-stratification even for patients with localised disease.
2. Materials and methods
2.1. Study populations
For this study, a retrospective test-cohort and a prospective validation-cohort were analysed. Patient characteristics of both cohorts are listed in Table 1.
Table 1.
Patients characteristics (n = 189;141).
| Variable | Test-cohort (mRNA) n (%) | Validation-cohort (TMA) n (%) |
|---|---|---|
| Sex | ||
| Male | 86 (57·7) | 87 (61·7) |
| Female | 63 (42·3) (missing: 47) |
54 (38·3) |
| Age at diagnosis | ||
| <15 years | 112 (59·3) | 70 (49·7) |
| ≥15 years | 77 (40·7) | 71 (50·3) |
| Metastasis at diagnosis | ||
| Absent | 53 (69·7) | 87 (61·7) |
| Present | 23 (30·3) (missing: 113) |
54 (38·3) |
| Sitea | ||
| Axial | NA | 86 (61·9) |
| Non-axial | NA | 53 (38·1) (missing: 2) |
| Tumour volume | ||
| <200 ml | NA | 76 (55·9) |
| <200 ml | NA | 60 (44·1) (missing: 5) |
| Histological responseb | ||
| Good | NA | 67 (79·8) |
| Poor | NA | 17 (20·2) (missing: 57) |
| Status | ||
| Alive | 113 (59·8) | 84 (59·6) |
| Dead | 76 (40·2) | 57 (40·4) |
| SOX2 expression | ||
| High | 38 (20·1) | 22 (15·6) |
| Low | 151 (79·9) | 119 (84·4) |
All extremity localization are non-axial.
NA = not available.
The study population of the test-cohort consisted of 189 EwS patients whose molecularly confirmed and retrospectively collected primary tumours were profiled at the mRNA level by gene expression microarrays in previous studies [[13], [14], [15], [16]]. To assemble this mRNA-cohort, microarray data generated on Affymetrix HG-U133Plus2.0, Affymetrix HuEx-1.0-st or Amersham/GE Healthcare CodeLink microarrays of the 189 EwS tumours (Gene Expression Omnibus (GEO) accession codes: GSE63157 [13], GSE12102 [14], GSE17618 [15], GSE34620 [16], and unpublished data) provided with clinical annotations were normalised separately as previously described [17]. Based on specific gene expressions, we predicted for each sample the tumour purity via the ESTIMATE algorithm [18], which revealed that all samples had a consensus purity estimation of >60%, corresponding to The Cancer Genome Atlas (TCGA) standard tissue sample requirements (http://cancergenome.nih.gov/cancersselected/biospeccriteria). Only genes that were represented on all microarray platforms were kept for further analysis. Batch effects were removed using the ComBat algorithm [19]. The removal of batch effects was demonstrated by t-SNE analysis of gene expression data before and after the batch correction (appendix fig. 1). This yielded a dataset comprising 189 EwS samples and 13,253 genes. Data processing was done in the statistical language R.
The validation-cohort was composed of 141 EwS patients treated with first-line therapy according to the successive phase III EwS protocols of European Intergroup Cooperative Ewing's Sarcoma Study (EICESS) 92 (1992 to 1998), and EURO-EWING 99 (1999 to 2009) run by the German Society for Paediatric Oncology and Haematology (GPOH). These studies were registered under ClinicalTrials.gov and approved by the appropriate ethics committees. The corresponding patient tumours were prospectively collected and tissue microarrays (TMAs) were constructed as detailed below. This study population included 87 males and 54 females.
2.2. TMA establishment and immunohistochemistry (IHC)
Formalin-fixed paraffin-embedded (FFPE) EwS samples were retrieved from the archives of the Institute of Pathology of the LMU Munich (Germany) and the Gerhard-Domagk-Institute for Pathology of the University of Münster (Germany) with approval from the corresponding institutional review boards. All EwS samples were collected at diagnosis and reviewed by a reference pathologist, and diagnoses were confirmed by the detection of pathognomonic EWSR1-ETS fusion oncogenes by qRT-PCR or the detection of an EWSR1 break-apart by fluorescence-in-situ-hybridisation (FISH). TMAs were constructed as previously described [17]. Each EwS case was represented by at least two cores (each 1 mm in diameter).
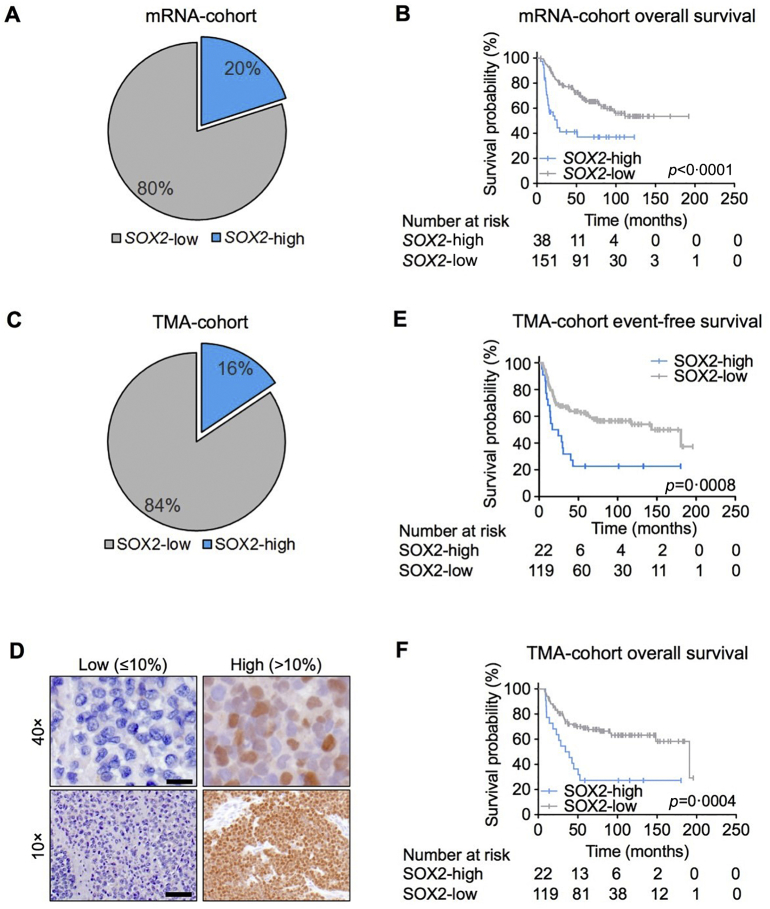
For IHC, 4 μm sections were cut, and antigen retrieval was performed using microwave treatment. Slides were incubated with a primary monoclonal rabbit anti-SOX2-antibody (1:100 dilution, D6D9 XP Cell Signaling) for 60 min. Then, the slides were incubated with a secondary anti-rabbit IgG antibody (ImmPress Reagent Kit; peroxidase-conjugated) followed by target detection using AECplus chromogen for 10 min (Dako, K3461). The specificity of the anti-SOX2-antibody used was confirmed in xenografts of the EwS cell line POE in NOD/Scid/gamma (NSG) mice, in which SOX2 expression was silenced by a doxycycline (dox-)inducible shRNA against the 3’UTR of SOX2 (appendix fig. 4B). For quantification of SOX2 immunoreactivity, the average percentage of SOX2-positive nuclei was evaluated by a data-blinded pathologist who examined at least five high-power fields per case. Examples of nuclear SOX2 immunoreactivity are given in Fig. 1D.
Fig. 1.
SOX2 is expressed in a subset of EwS patients and correlates with event-free survival (EFS) and overall survival (OS).
(A). Microarray analysis of SOX2 mRNA expression in 189 primary EwS cases.
(B). Kaplan-Meier analysis of OS in 189 EwS patients stratified by their SOX2 mRNA expression level (cut-off 80th percentile).
(C). Analysis of SOX2 protein expression levels in an independent cohort of 141 EwS patients in a TMA by IHC.
(D). Representative images for SOX2 IHC (nuclear staining) in EwS patients with different percentages of nuclear SOX2 immunoreactivity. Scale bars = 50 μM (40×) and 200 μM (10×).
(E). Kaplan-Meier analysis of EFS in 141 EwS patients stratified by their SOX2 protein expression level (cut-off >10% positive nuclei).
(F). Kaplan-Meier analysis of OS in 141 EwS patients stratified by their SOX2 protein expression level (cut-off >10% positive nuclei).
2.3. Statistical analyses
In the mRNA-cohort, the optimal cut-off for stratifying patients by SOX2 expression levels was identified by in-house software (GenEx), which tested all possible cut-offs between the 20th and 80th expression percentiles for the largest difference in area under the curves in the Kaplan-Meier analyses. This analysis identified the 80th percentile of SOX2 expression as the optimal cut-off for stratification of the mRNA-cohort in SOX2-high and -low. Statistical analyses were carried out with SPSS 19 (IBM Corporation, Armonk, NY) and SAS 9.2 (SAS Institute, Cary, NC) as described [20]. Event-free survival (EFS) and overall survival (OS) were estimated using the Kaplan-Meier method. OS time was defined as the interval between the date of diagnosis and the date of the patient's final follow-up or death. Group comparisons were calculated using the log-rank test. Multivariate analyses were carried out by applying the Cox proportional hazard method. Differences in proportions between groups were evaluated using chi-square or Fisher's exact tests. The significance level was set at p < 0·05 for two-sided testing. No alpha corrections were carried out for multiple testing. Outcome was analysed on an exploratory basis.
2.4. Role of the funding source
The funders of the study had no role in the study design, data collection, data analysis, data interpretation, or writing of the report. Both corresponding authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
3. Results
3.1. SOX2 is expressed in a subset of EwS patients and is correlated with OS
SOX2 is a well-known transcription factor involved in stemness during normal development and cancer [12]. In 2010, Riggi et al. proposed that EWSR1-FLI1 confers stemness features to EwS cells via up-regulation of SOX2 [11]. We therefore reasoned that SOX2 expression levels might be connected to patient outcome.
To test this hypothesis, a test-cohort obtained from a large EwS gene expression dataset comprising 189 samples, for which matched clinical data were available (Table 1), was employed to evaluate mRNA levels of SOX2. In contrast to previous suggestions that SOX2 overexpression might constitute a general feature of EwS [11], we found, using a microarray analysis, that SOX2 mRNA was not or was only minimally expressed in 79·9% (151/189) of tumours. Indeed, only 20·1% (38/189) of the samples exhibited moderate to strong SOX2 expression levels (Fig. 1A). Stratifying our mRNA-cohort by a cut-off of the 80th percentile of SOX2 expression in SOX2-low or -high cases, we noted that patients with high intratumoural SOX2 expression had worse OS than patients with low SOX2 expression (p < 0·0001) (Fig. 1B).
To validate this finding at the protein level, we stained an independent validation TMA-cohort comprising 141 EwS cases with an anti-SOX2-antibody with proven specificity for EwS (appendix fig. 4B). Patient characteristics are reported in Table 1. Scoring of the percentage of nuclear SOX2 positive cells confirmed that the majority of EwS cases (84·4%; 119/141) showed ≤10% positive nuclei (classified as SOX2-low) while only a small subset of patients (15·6%; 22/141) had >10% positive nuclei (classified as SOX2-high) (Fig. 1C,D). Strikingly, applying the cut-off of >10% SOX2 positive nuclei fully confirmed the strong association of SOX2 expression with poor event-free survival (EFS) (p = 0·0008) and OS of EwS patients (p = 0·0004) (Figs. 1E,F). In both the mRNA-test and TMA-validation cohorts, EwS samples were reviewed by a reference pathologist and diagnosis was confirmed either by the detection of pathognomonic EWSR1-ETS fusion oncogenes by qRT-PCR or the detection of an EWSR1 break-apart by fluorescence-in-situ-hybridisation (FISH). To investigate whether the fusion type (EWSR1-FLI1 type 1 and type 2 and EWSR1-ERG) affects SOX2 expression in EwS, we analysed data obtained from 18 EwS cell lines with different fusion types. These data demonstrated that there is no significant difference in SOX2 expression between EwS cell lines with different fusion types (appendix fig. 2).
Collectively, these results demonstrated, for the first time, that high SOX2 expression is not a common feature of EwS, but its high mRNA and protein levels may serve as a biomarker for outcome prediction.
3.2. High SOX2 expression is a major risk-factor for tumour associated-death independent from metastasis at time of diagnosis in EwS
To identify factors that may influence patient prognosis, we performed a multivariate analysis in our validation TMA-cohort, as here, additional clinicopathological information beyond OS was available. Interestingly, the multivariate analysis revealed that the major risk-factors were metastatic disease at time of diagnosis at the M2 stage and SOX2-high expression with hazard ratios (HRs) of 4·86 and 3·19 (both p < 0.01), respectively. Instead, M0 (HR = R; p < 0·01), M1 (n = 24; HR = 1·76; p = 0·13), age (≥15 years; HR = 1·34; p = 0·28) and primary axial tumour site (HR = 1·64; p = 0·11) did not show a significant impact on survival (n = 141; Table 2).
Table 2.
Multivariate analysis in all patients (n = 141).
| Variable | HR (95%CI) | p |
|---|---|---|
| Risk group | ||
| M0 (no met; n = 87) | Ra | <0·01 |
| M1 (lung met; n = 24) | 1·76 (0·85–3·62) | 0·13 |
| M2 (other +/− lung met; n = 30) | 4·86 (2·65–8·91) | <0·01 |
| SOX2 | ||
| High (>10%) | 3·19 (1·74–5·84) | <0·01 |
| Age | ||
| ≥15 years | 1·34 (0·79–2·27) | 0·28 |
| Site | ||
| Axial | 1·64 (0·90–2·96) | 0·11 |
R = Reference.
To exclude an interdependency of metastasis at time of diagnosis and SOX2 expression status, we repeated the multivariate analysis considering only patients with localised disease (M0, n = 87). In this analysis, SOX2-high expression (HR = 3·22; p < 0·01) represented the main risk-factor for survival, followed by age (≥15 years; HR = 2·43; p = 0·04), whereas primary axial tumour site (HR = 1·66; P = 0·25) showed only a tendency to affect survival (Table 3; n = 87). In agreement with these findings, in both validation- and test-cohorts, univariate analysis did not show a correlation between SOX2-high and metastasis at time of diagnosis (Table 4, n = 141, p = 0·8; and Table 5, n = 76, p 0·5). Taken together, these findings demonstrate that SOX2-high expression constitutes an independent risk-factor for survival of EwS patients, even in patients with localised disease.
Table 3.
Multivariate analysis in patients with localised disease (n = 87).
| Variable | HR (95%CI) | p |
|---|---|---|
| SOX2 | ||
| High (>10%) | 3·22 (1·36–7·65) | <0·01 |
| Age | ||
| ≥15 years | 2·43 (1·05–5·61) | 0·04 |
| Site | ||
| Axial | 1·66 (0·70–3·92) | 0·25 |
Table 4.
Summary of the results of the correlation of SOX2 immunoreactivity with metastasis, histological response, tumour volume and relapse.
| Metastasis at diagnosis (n = 141) |
Histological response (n = 84) |
Tumour volume (n = 136) |
Relapse (n = 141) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No | Yes | p | Good | Poor | p | <200 ml | ≥200 ml | p | No | Yes | p | |
| SOX2 | ||||||||||||
| Low | 74 (62·2%) | 45 (37·8%) | 0·8 | 57 (79·2%) | 15 (20·8%) | 1·0 | 67 (58·3%) | 48 (41·7%) | 0·23 | 71 (59·7%) | 48 (40·3%) | 0·002 |
| High | 13 (59·1%) | 9 (40·9%) | 10 (83·3%) | 2 (16·7%) | 9 (42·9%) | 12 (57·1%) | 5 (22·7%) | 17 (77·3%) | ||||
Table 5.
Summary of the results of the correlation of SOX2 mRNA with metastasis.
| Metastasis at diagnosis |
|||
|---|---|---|---|
| No | Yes | p | |
| SOX2 | |||
| Low | 45 (71·4%) | 18 (28·6%) | 0·5 |
| High | 8 (61·5%) | 5 (38·5%) | |
3.3. High SOX2 expression is significantly correlated with tumour relapse
Next, we investigated whether the correlation between SOX2-high expression and OS is associated with histological response, high tumour volume (≥200 ml), and relapse, which are risk-factors for worse outcomes [10,[21], [22], [23]].
In our TMA-cohort, data on histological response, tumour volume, and tumour relapse were available for 84, 136 and 141 patients, respectively (Table 4). Our data did not show an association between high SOX2 expression and histological response (Table 4). However, we observed a tendency for an association between SOX2-high and tumour volume. In fact, 58·3% of patients with high SOX2 expression had a tumour volume ≥ 200 ml compared to 41·7% patients with low SOX2 expression (p = 0·23) (Table 4). Since the association of SOX2 and higher tumour volume suggested a role for SOX2 in EwS growth, we performed functional in vitro and in vivo experiments using an established EwS cell line model (POE) for which we generated a derivate with a doxycycline (dox-)inducible shRNA against SOX2. The results showed that the knockdown of SOX2 in POE cells led to strong reduction in cell proliferation, clonogenic growth and anchorage-independent growth compared to cells that expressed a non-targeting control shRNA (appendix fig. 4). Consistently, dox-induced silencing of SOX2 in POE cells xenografted in immunocompromised NSG mice significantly reduced tumour growth (appendix fig. 4).
In addition, we noted a striking correlation between SOX2-high expression and tumour relapse (p = 0·002) (Table 4), comprising in 86% of cases metastatic relapse (n = 39) or combined metastatic and local relapse (n = 17), and in 14% of cases exclusively local relapse (n = 9). While only 40·3% of patients with SOX2-low expression had relapses, recurrence occurred in 77·3% of patients with SOX2-high expression (Table 4), which may explain the poor outcome that was observed for these patients. Indeed, immunohistological assessment of 12 available pairs of primary EwS and relapse samples showed that while SOX2-high expression was only observed in one case at the time of diagnosis four cases exhibited SOX2-high expression at relapse (average percentage of SOX2 expression: 2·75 in primary EwS vs. 15·5 in relapse tumours, p = 0·219; Wilcoxon matched-pairs signed rank test). In agreement with these observations, gene set enrichment analysis (GSEA) in our mRNA cohort showed that genes co-expressed with SOX2 in primary EwS tumours are involved in stemness, proliferation, dedifferentiation, and cancer relapse (appendix fig. 5A, appendix table 1).
Collectively, these results suggest that SOX2-high expression may confer worse outcomes for EwS patients by contributing to tumour growth and relapse.
4. Discussion
Up to 30–40% of EwS patients with non-metastatic disease develop local or metastatic relapse within a time span of 2–10 years [24]. Clinical prognostic markers, such as primary dissemination, tumour site, size, age, and histological response to chemotherapy, are established risk-factors used for therapeutic stratification [24]. However, especially for patients with localised disease and otherwise no apparent risk-factors, risk prediction is challenging [25]. Although there is broad consensus that clinical management will benefit from prognostic or predictive biomarkers that can guide therapeutic decisions, there are currently no bona fide or broadly available biomarkers that may help predict tumour relapse and outcome among patients with EwS [1,10]. A recent retrospective study on a single cohort of 63 EwS patients with localised disease suggested that detection of a higher tumour-mutational-burden (TMB) in EwS may help to identify patients at risk for relapse among patients with localised disease [26]. However, TMB detection requires sophisticated sequencing technologies that may not be technically applicable in all EwS cases.
In this study, we reported, for the first time, that high mRNA or protein expression of SOX2, which can be determined with routine technologies such as immunohistochemical stains, is a stratification risk-factor for ~16–20% of EwS patients with poor outcomes. Notably, multivariate analyses that included either all or only patients with localised disease have demonstrated that SOX2-high expression represents an independent and strong risk-factor for EwS patients. In fact, SOX2-high expression has been more frequently observed in relapsed tumours, and SOX2-high expression in primary tumours is significantly associated with early relapse in patients with localised disease and no other apparent clinical risk-factors. In line with our findings in EwS, high expression or amplification of SOX2 has been reported to correlate with poor survival in breast, colorectal, esophageal, laryngeal, endometrial, and ovarian carcinomas [12]. Like EwS, SOX2 is highly expressed in a subset of patients with lung adenocarcinomas and was found to be an independent predictor of poor survival [27].
Previous reports have suggested that SOX2 may constitute a direct EWSR1-FLI1 target gene [11,28]. Surprisingly, we found that the vast majority of molecularly confirmed EwS cases do not express SOX2 neither at the mRNA nor the protein level, suggesting that the mode of regulation and the functional role of SOX2 may be more complex than previously thought. Recently, Boulay et al. identified a polymorphic EWSR1-FLI1-bound enhancer-like GGAA-microsatellite near SOX2, and demonstrated that epigenetic silencing of this DNA sequence strongly reduced SOX2 expression in EwS cells [28]. In fact, EWSR1-FLI1 encodes an aberrant transcription factor, which induces many of its target genes by binding to enhancer-like GGAA-microsatellites, whose activity increases with the number of consecutive GGAA-repeats [1,29]. Since the number of GGAA-repeats at such EWSR1-FLI1-bound GGAA-microsatellites is highly variable across individuals [30,31], it is tempting to speculate that the number of GGAA-repeats at the SOX2-associated GGAA-microsatellite may contribute to the strong heterogeneity of SOX2 expression in EwS patients.
In our TMA-cohort, SOX2-high expression showed a correlation with tumour relapse (p = 0·002), which likely explains the poor outcome of these patients. In support of our findings, it has been shown in sinonasal carcinomas that patients with SOX2 amplification have a significantly higher rate of tumour recurrence than those without SOX2 amplification [32]. However, in our TMA-cohort 38·7% of patients with SOX2-low expression showed relapses, suggesting that in this subset of patients, a different driver might mediate tumour relapse.
Although the correlation between high SOX2 expression and tumour volume was not significant in our TMA-cohort, in line with previous reports regarding EwS [11,33], our in vitro and in vivo results confirmed a functional role of SOX2 in EwS cell proliferation and tumour growth suggesting that a larger cohort might enable validation of this correlation in EwS patients. In agreement with this, our GSEA in primary EwS indicated that SOX2-high tumours are enriched for gene signatures involved in dedifferentiation, proliferation, and stemness, which may promote tumour relapse. Interestingly, GSEA also revealed that SOX2 co-expressed genes overlap with genes involved in relapse of malignant melanoma – a tumour of neuroectodermal origin [34] that has also been proposed for EwS [1]. In support of these data, GSEA of differentially expressed genes upon SOX2 knockdown in three SOX2-high EwS cell lines demonstrated a significant upregulation of gene signatures involved in differentiation-related processes, such as neurite outgrowth, integrin cell surface interaction, and components of the basement membrane (appendix fig. 5B, appendix table 2).
Despite the exploratory nature of this study, SOX2-high expression may constitute the first promising biomarker for outcome prediction and stratification of high-risk EwS patients with localised disease, which is readily available due to standardised assessments by qRT-PCR and/or IHC. Early identification of such high-risk patients may enable clinicians to apply a closer monitoring and eventually intervene earlier in cases of incipient relapse. In addition, translational and clinical studies aimed at discovering specific therapeutic approaches for this high-risk sub-group of EwS patients could be initiated. Therefore, we recommend validation of these observations in additional prospective studies and experimental elucidation of the precise molecular role of SOX2 in EwS. For prospective studies, we recommend to evaluate the SOX2 expression status by IHC following our staining protocol during routine pathology work-up on diagnostic specimens and to apply the same cut-off as described here (>10% positive nuclei of tumour cells) for classifying samples in SOX2-high and -low, respectively. Since immunohistochemical stains for SOX2 are relatively easy to establish, inexpensive, and straight-forward to evaluate from a pathologist's perspective, we anticipate that this biomarker can be widely used in the clinics. For these reasons, SOX2 will be assessed prospectively in the new iEuroEwing trial led by the Cooperative Ewing Sarcoma Study (CESS) group of the German Society for Paediatric Oncology and Haematology (GPOH) (U. Dirksen personal communication).
Declartation of Competing Interest
The authors declare no conflict of interest.
Author contributions
Conception and design: Giuseppina Sannino and Thomas G. P. Grünewald.
Provision of study material and patients: Uta Dirksen, Heribert Jürgens, Wolfgang Hartmann, Thomas Kirchner, Ana Sastre, Javier Alonso.
Financial and administrative support: Thomas Kirchner and Thomas G. P. Grünewald.
Data analysis and interpretation: Giuseppina Sannino, Andreas Ranft, Susanne Jabar, Uta Dirksen, Aruna Marchetto, Martin F. Orth, Julia S. Gerke, Willian De Silveira and Gary Hardiman, Fabienne S. Wehweck, Merve M. Kiran, Thomas G. P. Grünewald.
Experimental support: Shunya Ohmura, Constanze Zacherl, Rebeca Alba-Rubio, Stefanie Stein, Tilman L. B. Hölting, Julian Musa, Laura Romero-Pérez, Florencia Cidre-Aranaz, Maximilian M. L. Knott, Jing Li.
Manuscript writing: Giuseppina Sannino, Uta Dirksen and Thomas G. P. Grünewald.
Final approval of the manuscript: All the authors.
Acknowledgements
The laboratory of T. G. P. Grünewald is supported by grants from the ‘Verein zur Förderung von Wissenschaft und Forschung an der Medizinischen Fakultät der LMU München (WiFoMed)’, by LMU Munich's Institutional Strategy LMUexcellent within the framework of the German Excellence Initiative, the ‘Mehr LEBEN für krebskranke Kinder – Bettina-Bräu-Stiftung’, the Walter Schulz Foundation, the Wilhelm Sander-Foundation (2016.167.1), the Friedrich-Baur foundation, the Matthias-Lackas foundation, the Barbara & Hubertus Trettner foundation, the Dr. Leopold und Carmen Ellinger foundation, the Gert & Susanna Mayer foundation, the Rolf M. Schwiete foundation, the Deutsche Forschungsgemeinschaft (DFG 391665916), and by the German Cancer Aid (DKH-111886 and DKH-70112257). J. Li was supported by a scholarship of the China Scholarship Council (CSC), J. Musa was supported by a scholarship of the Kind-Philipp foundation, and T. L. B. Hölting by a scholarship of the German Cancer Aid. M. F. Orth and M. M. L. Knott were supported by scholarships of the German National Academic Foundation. G. Sannino was supported from a scholarship from the Fritz-Thyssen Foundation (FTF-40.15.0.030MN). The work of U. Dirksen is supported by grants from the German Cancerr Aid (DKH-108128, DKH-70112018, and DKH-70113419), the ERA-Net-TRANSCAN consortium (project number 01KT1310), and Euro Ewing Consortium (EEC, project number EU-FP7 602856), both funded under the European Commission Seventh Framework Program FP7-HEALTH (http://cordis.europa.eu/), the Barbara & Hubertus Trettner foundation, and the Gert & Susanna Mayer foundation. G. Hardiman was supported by grants from the National Science Foundation (SC EPSCoR) and National Institutes of Health (U01-DA045300). The laboratory of J. Alonso was supported by Instituto de Salud Carlos III (PI12/00816; PI16CIII/00026); Asociación Pablo Ugarte (TPY-M 1149/13; TRPV 205/18), ASION (TVP 141/17), Fundación Sonrisa de Alex & Todos somos Iván (TVP 1324/15).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.08.002.
Contributor Information
Uta Dirksen, Email: uta.dirksen@uk-essen.de.
Thomas G.P. Grünewald, Email: thomas.gruenewald@med.uni-muenchen.de.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
Supplementary material 3
References
- 1.Grünewald T.G.P., Cidre-Aranaz F., Surdez D., Tomazou E.M., de Álava E., Kovar H. Ewing sarcoma. Nat Rev Dis Primer. 2018;4:5. doi: 10.1038/s41572-018-0003-x. [DOI] [PubMed] [Google Scholar]
- 2.Delattre O., Zucman J., Melot T., Garau X.S., Zucker J.M., Lenoir G.M. The Ewing family of tumours--a subgroup of small-round-cell tumours defined by specific chimeric transcripts. N Engl J Med. 1994;331:294–299. doi: 10.1056/NEJM199408043310503. [DOI] [PubMed] [Google Scholar]
- 3.Delattre O., Zucman J., Plougastel B., Desmaze C., Melot T., Peter M. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 4.Anderson N.D., de Borja R., Young M.D., Fuligni F., Rosic A., Roberts N.D. Rearrangement bursts generate canonical gene fusions in bone and soft tissue tumours. Science. 2018;361 doi: 10.1126/science.aam8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Deley M.-C., Paulussen M., Lewis I., Brennan B., Ranft A., Whelan J. Cyclophosphamide compared with ifosfamide in consolidation treatment of standard-risk Ewing sarcoma: results of the randomized noninferiority Euro-EWING99-R1 trial. J Clin Oncol. 2014;32:2440–2448. doi: 10.1200/JCO.2013.54.4833. [DOI] [PubMed] [Google Scholar]
- 6.Gaspar N., Hawkins D.S., Dirksen U., Lewis I.J., Ferrari S., Le Deley M.-C. Ewing sarcoma: current management and future approaches through collaboration. J Clin Oncol. 2015;33:3036–3046. doi: 10.1200/JCO.2014.59.5256. [DOI] [PubMed] [Google Scholar]
- 7.Whelan J., Le Deley M.-C., Dirksen U., Le Teuff G., Brennan B., Gaspar N. High-dose chemotherapy and blood autologous stem-cell rescue compared with standard chemotherapy in localized high-risk Ewing sarcoma: results of Euro-E.W.I.N.G.99 and Ewing-2008. J Clin Oncol. 2018 doi: 10.1200/JCO.2018.78.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biswas B., Bakhshi S. Management of Ewing sarcoma family of tumors: current scenario and unmet need. World J Orthop. 2016;7:527–538. doi: 10.5312/wjo.v7.i9.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shukla N., Schiffman J., Reed D., Davis I.J., Womer R.B., Lessnick S.L. Biomarkers in Ewing sarcoma: the promise and challenge of personalized medicine. A report from the children's oncology group. Front Oncol. 2013;3:141. doi: 10.3389/fonc.2013.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinto A., Dickman P., Parham D. Pathobiologic markers of the ewing sarcoma family of tumors: state of the art and prediction of behaviour. Sarcoma. 2011;2011 doi: 10.1155/2011/856190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riggi N., Suvà M.-L., De Vito C., Provero P., Stehle J.-C., Baumer K. EWS-FLI-1 modulates miRNA145 and SOX2 expression to initiate mesenchymal stem cell reprogramming toward Ewing sarcoma cancer stem cells. Genes Dev. 2010;24:916–932. doi: 10.1101/gad.1899710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wuebben E.L., Rizzino A. The dark side of SOX2: cancer - a comprehensive overview. Oncotarget. 2017;8:44917–44943. doi: 10.18632/oncotarget.16570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Volchenboum S.L., Andrade J., Huang L., Barkauskas D.A., Krailo M., Womer R.B. Gene expression profiling of Ewing sarcoma tumors reveals the prognostic importance of tumor-stromal interactions: a report from the children's oncology group. J Pathol Clin Res. 2015;1:83–94. doi: 10.1002/cjp2.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scotlandi K., Remondini D., Castellani G., Manara M.C., Nardi F., Cantiani L. Overcoming resistance to conventional drugs in Ewing sarcoma and identification of molecular predictors of outcome. J Clin Oncol. 2009;27:2209–2216. doi: 10.1200/JCO.2008.19.2542. [DOI] [PubMed] [Google Scholar]
- 15.Savola S., Klami A., Myllykangas S., Manara C., Scotlandi K., Picci P. High expression of complement component 5 (C5) at tumor site associates with superior survival in Ewing's sarcoma family of tumour patients. ISRN Oncol. 2011;2011 doi: 10.5402/2011/168712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Postel-Vinay S., Véron A.S., Tirode F., Pierron G., Reynaud S., Kovar H. Common variants near TARDBP and EGR2 are associated with susceptibility to Ewing sarcoma. Nat Genet. 2012;44:323–327. doi: 10.1038/ng.1085. [DOI] [PubMed] [Google Scholar]
- 17.Baldauf M.C., Orth M.F., Dallmayer M., Marchetto A., Gerke J.S., Rubio R.A. Robust diagnosis of Ewing sarcoma by immunohistochemical detection of super-enhancer-driven EWSR1-ETS targets. Oncotarget. 2018;9:1587–1601. doi: 10.18632/oncotarget.20098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshihara K., Shahmoradgoli M., Martínez E., Vegesna R., Kim H., Torres-Garcia W. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stein C.K., Qu P., Epstein J., Buros A., Rosenthal A., Crowley J. Removing batch effects from purified plasma cell gene expression microarrays with modified ComBat. BMC Bioinformatics. 2015;16:63. doi: 10.1186/s12859-015-0478-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grunewald T.G.P., Ranft A., Esposito I., da Silva-Buttkus P., Aichler M., Baumhoer D. High STEAP1 expression is associated with improved outcome of Ewing's sarcoma patients. Ann Oncol. 2012;23:2185–2190. doi: 10.1093/annonc/mdr605. [DOI] [PubMed] [Google Scholar]
- 21.Haeusler J., Ranft A., Boelling T., Gosheger G., Braun-Munzinger G., Vieth V. The value of local treatment in patients with primary, disseminated, multifocal Ewing sarcoma (PDMES) Cancer. 2010;116:443–450. doi: 10.1002/cncr.24740. [DOI] [PubMed] [Google Scholar]
- 22.Cotterill S.J., Ahrens S., Paulussen M., Jürgens H.F., Voûte P.A., Gadner H. Prognostic factors in Ewing's tumor of bone: analysis of 975 patients from the European intergroup cooperative Ewing's sarcoma study group. J Clin Oncol. 2000;18:3108–3114. doi: 10.1200/JCO.2000.18.17.3108. [DOI] [PubMed] [Google Scholar]
- 23.Rodríguez-Galindo C., Liu T., Krasin M.J., Wu J., Billups C.A., Daw N.C. Analysis of prognostic factors in Ewing sarcoma family of tumors: review of St. Jude Children's Research Hospital Studies Cancer. 2007;110:375–384. doi: 10.1002/cncr.22821. [DOI] [PubMed] [Google Scholar]
- 24.Bosma S.E., Ayu O., Fiocco M., Gelderblom H., Dijkstra P.D.S. Prognostic factors for survival in Ewing sarcoma: a systematic review. Surg Oncol. 2018;27:603–610. doi: 10.1016/j.suronc.2018.07.016. [DOI] [PubMed] [Google Scholar]
- 25.Stahl M., Ranft A., Paulussen M., Bölling T., Vieth V., Bielack S. Risk of recurrence and survival after relapse in patients with Ewing sarcoma. Pediatr Blood Cancer. 2011;57:549–553. doi: 10.1002/pbc.23040. [DOI] [PubMed] [Google Scholar]
- 26.Liu K.X., Lamba N., Hwang W.L., Niemierko A., DuBois S.G., Haas-Kogan D.A. Risk stratification by somatic mutation burden in Ewing sarcoma. Cancer. 2019 doi: 10.1002/cncr.31919. [DOI] [PubMed] [Google Scholar]
- 27.Sholl L.M., Barletta J.A., Yeap B.Y., Chirieac L.R., Hornick J.L. Sox2 protein expression is an independent poor prognostic indicator in stage I lung adenocarcinoma. Am J Surg Pathol. 2010;34:1193–1198. doi: 10.1097/PAS.0b013e3181e5e024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boulay G., Volorio A., Iyer S., Broye L.C., Stamenkovic I., Riggi N. Epigenome editing of microsatellite repeats defines tumour-specific enhancer functions and dependencies. Genes Dev. 2018;32:1008–1019. doi: 10.1101/gad.315192.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gangwal K., Sankar S., Hollenhorst P.C., Kinsey M., Haroldsen S.C., Shah A.A. Microsatellites as EWS/FLI response elements in Ewing's sarcoma. Proc Natl Acad Sci U S A. 2008;105:10149–10154. doi: 10.1073/pnas.0801073105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monument M.J., Johnson K.M., McIlvaine E., Abegglen L., Watkins W.S., Jorde L.B. Clinical and biochemical function of polymorphic NR0B1 GGAA-microsatellites in Ewing sarcoma: a report from the Children's oncology group. PLoS One. 2014;9 doi: 10.1371/journal.pone.0104378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grünewald T.G.P., Bernard V., Gilardi-Hebenstreit P., Raynal V., Surdez D., Aynaud M.-M. Chimeric EWSR1-FLI1 regulates the Ewing sarcoma susceptibility gene EGR2 via a GGAA microsatellite. Nat Genet. 2015;47:1073–1078. doi: 10.1038/ng.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schröck A., Göke F., Wagner P., Bode M., Franzen A., Braun M. Sex determining region Y-box 2 (SOX2) amplification is an independent indicator of disease recurrence in sinonasal cancer. PLoS One. 2013;8 doi: 10.1371/journal.pone.0059201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren C., Ren T., Yang K., Wang S., Bao X., Zhang F. Inhibition of SOX2 induces cell apoptosis and G1/S arrest in Ewing's sarcoma through the PI3K/Akt pathway. J Exp Clin Cancer Res CR. 2016;35:44. doi: 10.1186/s13046-016-0321-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shain A.H., Bastian B.C. From melanocytes to melanomas. Nat Rev Cancer. 2016;16:345–358. doi: 10.1038/nrc.2016.37. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2
Supplementary material 3