Abstract
Background
Ankylosing spondylitis (AS) is a chronic inflammatory disease with worldwide high prevalence. Although AS is strongly associated with HLA-B27 MHC-I antigen presentation, the role played by αβ T cells in AS remains elusive.
Methods
Utilizing TCRβ repertoire sequencing and bioinformatics tools developed in house, we analyzed overall TCR repertoire structures and antigen-recognizing CDR3 motifs in AS patients with different disease activities.
Findings
We found that disease progression is associated with both CD4+ and CD8+ T cell oligo-clonal expansion, which suggests that αβ T cell activation may mediate AS disease progression. By developing a bioinformatics platform to dissect antigen-specific responses, we discovered a cell population consisting of both CD4+ and CD8+ T cells expressing identical TCRs, herein termed CD4/8 T cells. CD4/8 clonotypes were highly enriched in the spondyloarthritic joint fluid of patients, and their expansion correlated with the activity of disease.
Interpretation
These results provide evidence on the T cell clone side to reveal the potential role of CD4/8 T cells in the etiology of AS development.
Keywords: Ankylosing spondylitis, Autoimmune disease, T cells, TCR repertoire, Human, Complementarity determining region 3
Abbreviations: AS, ankylosing spondylitis; SpA, spondyloarthropathies; HLA, human leukocyte antigen; GWASs, genome-wide association studies; ERAP1, endoplasmic reticulum aminopeptidase 1; PBMC, peripheral blood mononuclear cells; CDR3, complementarity determining region 3; TCR, T cell receptor; JIA, Juvenile Idiopathic Arthritis; MS, Multiple Sclerosis; SJF, Spondyloarthritic Joint Fluid; OOF, out-of-frame
Research in context.
Evidence before this study
Ankylosing spondylitis (AS) is a chronic autoinflammatory disease that is highly associated with the MHC-I HLA-B27 allele. Although human genetic studies have suggested a strong relationship between AS and MHC-I antigen presentation, attributing disease etiology of AS remains elusive. There are two major enigmas: 1) disease manifestation remains nearly intact when CD8+ T cells are depleted in animal models whereas CD4+ T cells can transfer the disease; 2) if CD4+ T cells drive pathogenesis, it is unclear how HLA-B27 activates these cells.
Added value of this study
Aided by the analytical power of next-generation deep sequencing, we analyzed the TCR repertoires of T cell sub-populations from AS patients with varying disease activities. Using this approach, we have obtained direct evidence to support three major conclusions: 1) for individual patients, expansion of CD4 + CD45RA+ T cells is associated with the activity of AS; 2) for highly active disease, CD4 + CD45RO+ and CD8+ T cells are likely to be stimulated by certain shared autoantigens; 3) pathological CD4+ and CD8+ T cells share identical TCRs. Together, these findings reconcile the major controversy existing in the AS field: both CD4+ and CD8+ T cells are critical for disease manifestation; and, pathological CD4+ T cells might recognize HLA-B27 because they carry the exactly same TCR as CD8+ T cells.
Implications of all the available evidence
We developed several bioinformatics tools to dissect meaningful immunological information. Specifically, the “Motif Analysis” algorithm was critical for us to identify TCRs sharing the same antigen specificity without sequence homology. This method, which was generated and validated in 4 independent TCR datasets with over 400 repertoires, allowed us to determine that AS patients have an antigen profile that is distinct from that of other T cell-mediated autoimmune diseases. It also enabled the breakthrough discovery that pathological CD4+ and CD8+ T cells might share the same antigen specificity by virtue of expressing the same AS specific CDR3 motif. This tool enables the reliable and unbiased dissection of antigen-specific T cell responses from the global repertoire, which can be generally applied to and empower various repertoire-based clinical immune monitoring, such as those for cancer immunotherapies.
Alt-text: Unlabelled Box
1. Introduction
Ankylosing spondylitis (AS), the prototypic phenotype of chronic spondyloarthropathies (SpA), prevails in approximately 1.30–1.56 million Europeans and 4.63–4.98 million Asians, with an even higher estimated frequency in the United States [1]. Since 1973, the prevalence of AS has been associated with the expression of a human leukocyte antigen (HLA) class I surface molecule, HLA-B27 [2,3], and >90% of AS patients carry at least one HLA-B27 allele [4]. While association between AS and HLA-B27 represents one of the strongest genetic susceptibilities in any diseases, the etiology of AS remains elusive [5].
The central debate concerns whether AS is an αβ T cell-mediated autoimmune disease. Besides HLA-B27, genome-wide association studies (GWASs) identified polymorphism in endoplasmic reticulum aminopeptidase 1 (ERAP1) as a risk factor for AS, and ERAP1 variants only influence the susceptibility of patients carrying the HLA-B27 allele [6,7]. Since ERAP1 functions to process peptides for class I MHC antigen presentation, this linkage strongly suggests that AS is an autoimmune diseases driven by arthritogenic peptide-HLA-B27 activated CD8+ T cells. However, in patients, no arthritogenic peptides has been validated and no significant CD8+ T cell activity was observed [5]. Moreover, while the AS phenotype can be recapitulated using HLA-B27 transgenic rats, antibody-mediated [8] or genetic [9] depletion of CD8+ T cells in these animals is not sufficient to ameliorate the disease. Therefore, the role of CD8+ T cells in AS remains elusive.
Recent clinical success using an antibody against cytokine IL-17A [10,11], an inflammatory cytokine usually secreted by CD4+ T cells, triggered widespread interest in investigating the role of Th17 CD4+ T cells in AS. Indeed, an earlier study showed that transferring purified CD4+ T cells into adult euthymic HLA-B27 transgenic rat is sufficient to trigger SpA-like inflammatory disease [12]. Moreover, the percentage of IL-17A secreting CD4+ T cells is elevated in peripheral blood mononuclear cells (PBMCs) in two independent AS cohorts [13,14]. These evidences led the field to suspect that AS might be a Th17 CD4 T cell driven disease. However, there is no evidence from clinical trials that CD4+ T cells are the major source of pathological IL-17A secretion. Our own clinical studies failed to show any significant increase in effector/memory Th17 cells in patients' PBMCs, and CD4+ T cells from patients are not biased for Th17 differentiation. Other IL-17A-producing cells, such as γδ T-cells [15], natural killer cells [16], Type 3 innate lymphoid cells [17], neutrophils [[18], [19], [20], [21]], and mast cells [22], have been reported to be enriched in AS patients. Nevertheless, it remains an enigma how CD4+ T cells or these innate immune cell populations [[16], [17], [18], [19], [20], [21], [22], [23]] could drive AS disease manifestation through an MHC-I molecule, HLA-B27.
HLA-B27 has the capacity to form β2 microglobulin-free homodimers in solution [24] and on the cell surface [25], which can engage a range of innate killer immunoglobulin receptors such as KIR3DL2 [26]. T cells may utilize KIR3DL2 to engage HLA-B27 and trigger T cell expansion [27]. While this provides a plausible model to explain how CD4+ T cells isolated from AS patients can be activated by HLA-B27-expressing but antigen processing-defective cell lines [28], the clinical relevance of this model is unclear.
We reasoned that the best approach to demystify the etiology of AS is to monitor patients' T cell responses comprehensively and directly. This can be achieved using next-generation sequencing-aided high-throughput T cell receptor (TCR) repertoire analysis (Rep-seq) [29]. The diversity of the TCR repertoire is generated by somatic recombination of the TCRA and TCRB loci during early development in the thymus [30], and the junctional diversity generated during recombination [31] is responsible for shaping the spectrum of TCR antigen recognition [32]. The sequence of this joint region, complementarity determining region 3 (CDR3), determines the specificity and affinity of antigen recognition [32,33]. Therefore, the central benchmark of the T cell antigen response – oligo-clonal expansion of antigen-specific T cells – can be reflected by CDR3 clonotypic enrichment and diversity contraction [34]. While Rep-seq has emerged as a popular technology in recent years, its clinical application is limited by the complexity of the human TCR repertoire. Due to the life-long battle with environmental pathogens, meaningful information is usually muffled by noise from irrelevant clonal changes of T cells. Hence, the quantification of T cell responses against specific antigens is the central urgency facing the application of Rep-seq to clinical monitoring, especially for antigen-unknown conditions such as AS.
To address this challenge, we developed a new bioinformatics strategy attached to our validated Rep-seq platform [34], which allowed us to dissect antigen-specific T cell responses from global repertoire changes. Using this approach, we investigated TCR repertoires in sorted CD8+, CD4 + CD45RA+, and CD4 + CD45RO+ T cells from a cohort of AS patients with different pathological activities. Although our analyses do not have the power to suggest any characteristics of arthritogenic antigen, it provides direct evidence that AS progression is associated with both CD4+ and CD8+ T cell expansion in patients. Furthermore, we discovered that these pathology-associated CD4+ and CD8+ T cells share identical CDR3 and TCR sequences.
2. Materials and methods
2.1. Patients and sample collection
We enrolled 21 HLA-B27 positive patients with AS who fulfilled the SpondyloArthritis International Society (ASAS) classification criteria for axial spondylarthritis [35] or the modified New York criteria (1984) for AS [36]. Patients were enrolled to the rheumatology clinic at Xijing Hospital in Xi'an. Patients were ineligible to participate in this study if any of the following exclusion criteria were met: (1) currently receiving, or have previous use of corticosteroids, and synthetic or biological disease-modifying antirheumatic drugs (DMARD; e.g., methotrexate, sulfasalazine, tumor necrosis factor inhibitor); (2) currently receiving any medicine that could perturb the peripheral hemogram; (3) any infectious diseases (e.g., immunodeficiency virus, hepatitis B virus, hepatitis C virus or any chronic infection); (4) history of any other autoimmune rheumatic disease; (5) history of an infected joint prosthesis at any time; (6) history of any lymphoproliferative disorders, such as an Epstein Barr Virus-related lymphoproliferative disorder, history of lymphoma, leukemia, or signs and symptoms suggestive of current lymphatic disease; (7) vaccines given within the previous 6 weeks; (8) any known immunodeficiency disorder or a first-degree relative with a hereditary immunodeficiency; (9) significant trauma or surgical procedure within 1 month; (10) pregnant or lactating females, or females planning pregnancy.
The mean age of the patients was 24.86 (SD, ± 5.50) years (range 17–34 years) (online supplementary table 2a) with a disease duration of 6.48 ± 3.92 years. Clinicopathological measurements including back pain, peripheral pain with swelling, duration of morning stiffness, patient global assessment, Ankylosing Spondylitis Disease Activity Score (ASDAS), erythrocyte sedimentation rate (ESR), and C-reactive protein (CRP) levels are summarized in online supplementary table 2a. The disease activity of AS was graded by the ASDAS.CRP and ASDAS.ESR score, the most commonly used disease activity scores according to standard criteria [37,38]. Based on the ASDAS.CRP score, patients were divided into high disease (AS-hi) and low disease activity groups (AS-lo) (online supplementary table 2b). With informed consent, twenty milliliters of peripheral blood were obtained from patients with active AS. Synovial fluid samples were collected from the affected knee joints when available. For comparative analysis, twenty milliliters of peripheral blood from healthy controls was also obtained. The mean age of healthy controls was 24.29 (SD, ± 3.35) years (range 21–29 years).
Peripheral blood and synovial fluid was collected with a syringe and needle into EDTA-treated Vacutainer tubes (BD Biosciences, Franklin Lakes, NJ, USA). Synovial fluid was diluted with two volumes of phosphate-buffered saline (PBS) and treated with bovine testicular hyaluronidase (10 mg/ml; Sigma-Aldrich, St Louis, MO, USA) for 30 min at 37 °C. Cells were washed twice with PBS and filtered through a 40-μm nylon filter (Becton Dickinson, Franklin Lakes, NJ, USA) to remove debris. PBMCs and spondyloarthritic joint fluid mononuclear cell (SFMCs) (at least 7 × 10 [6]/sample) were isolated by Ficoll-Paque (Paneco, Russia) density-gradient centrifugation. Total RNA (at least 6 μg/sample) was isolated using Trizol reagent (Invitrogen, Carlsbad, CA), according to the manufacturer's protocol.
Ethics approval was granted for this study and all individuals provided their informed consent. All donors were recruited into a clinical trial at Xijing Hospital (Chinese Clinical Trial Registry; Registration number: ChiCTR-ONRC-11001565).
2.2. Antibodies and flow cytometry
For CD4 + CD8-CD45RA + CD45RO−, CD4 + CD8-CD45RA–CD45RO+ and CD4-CD8+ T-cell separation, isolated mononuclear cells were stained with antibody to CD45RA (clone HI100; BD Pharmingen), CD45RO (clone UCHL1; BD Pharmingen), CD4 (clone SK3; BD Pharmingen) and CD8 (clone RPA-T8; BD Pharmingen). Cells were sorted using a FACSAria cell sorter (BD Biosciences). Recovered cells were lysed in Trizol reagent (Life Technologies, Inc., Gaithersburg, MD) and stored at −80 °C. Data was analyzed using FlowJo software (TreeStar).
2.3. Sequencing of the TCR-β repertoire
The TCR-β repertoire sequencing process has been previously published in detail [34]. All raw data generated for this study was uploaded to the Sequence Read Archive (SRA) under BioProject ID PRJNA378893 (https://www.ncbi.nlm.nih.gov/bioproject/378893).
2.4. Processing of raw reads
We conducted data pre-processing by using Ion Torrent Suite software filters to exclude low quality reads and erroneous sequences derived from unrecognized multiplex barcodes. Next, through the Ion Torrent PGM built-in plugin, raw sequences with recognizable barcode identities were converted to a FASTQ format. For TCR clonotype identification and clustering, the FASTQ files were imported to the MiTCR software [39]. This software was further used for correcting reverse transcription, PCR, and sequencing errors. The resulting TCR clonotype data are summarized in online supplementary table 1. Further statistical analysis of the TCR-β repertoire was performed using the tcR R package [40], Excel (Microsoft Office 2013), Prism 5 (GraphPad) software and the R statistical programming language via manual scripts.
2.5. Asymptotic diversity profile analysis
Diversity indices such as richness, Shannon index and Simpson index are frequently used to evaluate the clonal frequency distributions in the repertoire and represent the state of clonal expansion and selection. However, these types of analyses based on different diversity indices can yield qualitatively different results. In order to guarantee the reliability of diversity analysis, we introduced diversity indices based on Hill-based diversity which was calculated as follows:
where n is the number of unique sampling clones and pi is the abundance of sampling clonotypes in its repertoire. The α values represent weights on rare and abundant TCR clonotypes differently. Hence the diversity of different alpha value focuses on a different part of relatively dominant TCR clonotypes. Since we didn't know which part of the dominant TCR clonotypes was pathological, unbiased, asymptotic diversity indices of continuum alpha were used to evaluate the structure of the TCR repertoire comprehensively.
To compare the diversity of each group, we plotted the asymptotic diversity profiles of alpha between 0 and 4, the 95% confidence intervals (shaded graphs), and the mean (solid lines) for diversity profiles based on the CDR3 amino acid sequence frequency. To evaluate the clinical relevance of TCR repertoire clonal expansion, we calculated the Pearson correlation coefficients between diversity and disease activity score ASDAS.CRP, and plotted alpha against correlation coefficients. Significant correlation positions were indicated by the corresponding color shades.
2.6. Similarity profile analysis
To quantitatively depict similarities in the entire repertoire level, we used custom R scripts and the tcR R package [40] to calculate the Morisita index, which is frequently used in similarity analysis. In addition, we developed the Top[N] index to evaluate the similarity between the dominant clonotypes in both samples. Top[N] index indicates that the most abundant shared CDR3 between repertoires A and B is ranked at least as “N” in these two repertoires. To calculate the Top[N] index, first, CDR3s in each repertoire were ranked based on their frequency. Second, starting from the top 1 CDR3 of sample A, we search for shared CDR3s in other samples. If found, we determine the rank of that CDR3 (N) in sample B as “top N CDR3s” can be identified as shared CDR3. If not, we will move down to lower ranked CDR3s in sample A and repeats the search. After all, the “Top[N] index” was defined as the “Top N CDR3s” in sample 1 and sample 2, from which the very first shared CDR3 could be found in both samples.
2.7. Statistical analysis
TCR diversity and similarity was compared using an unpaired Student's t-test (two groups) or one-way ANOVA with Tukey's HSD post-test (three groups or more). A p value of <0.05 was considered statistically significant; ∗p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001 and n.s. (or lack of indicated p value) denotes not significant (p > 0.05).
2.8. Clustering of TCR repertoires through CDR3 motif usage
In each sample, we acquired a defined number of Top N most abundant clones (“Top N CDR3s”). “Top N CDR3s” were deconstructed into connected amino acid motifs with various length.
All the possible a.a. motifs used by the Top[N] CDR3s were merged into a list of CDR3 motifs and calculated for their frequencies. The frequencies of motifs used by Top[N] CDR3s in each sample were used for pairwise Manhattan distance calculation and applied Ward.D2 clustering [41] (Illustration of workflow in online supplementary Fig. 3b). In order to evaluate the discriminating ability of the motif analysis, we introduced the analysis of similarities (ANOSIM). ANOSIM was performed on Manhattan distance matrices with 1000 permutations. Circular dendrogram visualization and ANOSIM was performed using the R via manual scripts.
2.9. CDR3 motif similarity network construction, clustering and consensus motif summary
CDR3s were deconstructed into all possible M a.a. connected motifs (Illustration in online supplementary Fig. 3a), and merged into a list of CDR3 motifs. The corresponding frequencies of the merged motif list were used for calculating the pairwise CDR3 similarity matrix using Jaccard similarities. CDR3 motif similarity networks were constructed through connecting CDR3 pairs with Jaccard similarities of motif usage >0.5 and graphed using a Fruchterman-Reingold force-directed layout algorithm. CDR3s sharing similar motif usage are positioned proximal to each other, whereas CDR3s with less similarity are positioned farther apart (illustration of workflow in online supplementary Fig. 8).
For the display of CDR3s from the specific network cluster, CDR3 similarity matrices were calculated and used for applied Ward.D2 clustering. Next, a sequence logo was generated for consensus motif summary (online supplementary Fig. 8). Network, circular dendrograms and sequence logo visualizations were performed in R.
3. Results
3.1. TCR repertoire analysis links AS disease activity to CD4+ T cell clonal expansion
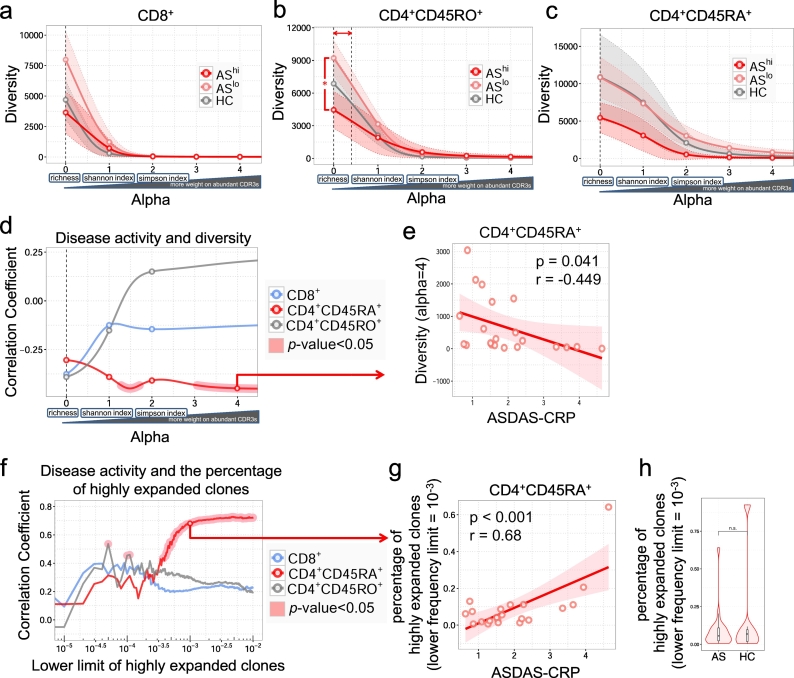
To directly investigate whether T cells play a role in the onset or progression of AS, we utilized TCRβ Rep-seq to comprehensively characterize the clonal distribution of T cells from a cohort of 21 early diagnosed AS patients. Ankylosing Spondylitis Disease Activity Scores (ASDAS) for these patients ranged from 0.6 to 4.6 (Supplementary Table 2). Blood was collected from age-matched healthy controls or AS patients before any steroid or antibody treatments, and FACS was employed to purify CD3 + CD4 + CD45RA+, CD3 + CD4 + CD45RO+, and CD3 + CD8+ T cells. ~500,000 cells from each T cell population of individual patients were subjected to Rep-seq. In total, 34 million effective reads were generated, which mapped to ~700,000 TCR clonotypes (Supplementary Table 1). To detect oligo-clonal expansion, we first compared TCR repertoire diversity among subjects. Since various diversity measurements emphasize different aspects (e.g. clonotype numbers, relative size, evenness, etc.) of a repertoire, to avoid bias, we employed Hill-based Diversity [42,43]. Unlike those, Hill indices enable us to search through a large range of continuum weight (alpha) values and generate a diversity index profile, which provides a continuum of angles to inspect the same repertoire structure. As alpha value increases, highly expanded clones have more weight on the value of indices. As depicted by the 95% confidence intervals of different groups, for most diversity measurements, we failed to distinguish repertories between healthy individual and AS patients, or between patients with different activities (Fig. 1a-c). However, when diversity indices were calculated with alpha value close to richness (α = 0.0–0.4), which emphasize the number of different clonotypes a specific repertoire contains, diversity of CD4 + CD45RO+ T cells was reduced in PBMCs from patients with highly active AS disease (Fig. 1b). This suggests that CD4+ memory T cells in highly active AS patients might be moderately restricted to fewer clonotypes.
Fig. 1.
Asymptotic diversity profiles reveal disease activity related diversity and clonal expansion in CD4 + CD45RA+ T cells. Asymptotic diversity profiles and 95% confidence intervals of different groups when alpha is between 0 and 4 in CD8+ (a), CD4 + CD45RO+ (b) and CD4 + CD45RA+ (c) T cells. The 95% CI (shaded) and mean (solid lines) for diversity profiles are based on the CDR3 amino acid sequence frequency counts in each repertoire. One-way ANOVA and Tukey's test were used to compare diversities across different groups of different alpha. No significant alpha interval was observed in CD8+ (a) or CD4 + CD45RA+ (c) T cells. Significant alpha interval in CD4 + CD45RO+ (b) is indicated with a double-headed arrow. (d) Asymptotic correlation between disease activity ASDAS.CRP score and diversity when alpha is between 0 and 4. Correlation coefficient (r) and P-values were calculated based on two-tailed Pearson's correlation. Significant correlation is indicated by red shade. (e) Representative correlation between ASDAS.CRP score and CD4 + CD45RA+ T cell diversity (alpha = 4). (f) Asymptotic correlation between the ASDAS.CRP score and the percentage of dominant T cells with CDR3 clonal proportion exceeding certain lower limit. Significant correlation is indicated by red shade. (g) Representative correlation between ASDAS.CRP score and the percentage of relatively dominant CD4 + CD45RA+ T cell clones (lower frequency limit = 10−3). (h) Percentages of relatively dominant CD4 + CD45RA+ T cell clones (lower frequency limit = 10−3) in AS patients and health controls (HC). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To further assess the clinical relevance of T cell diversity in individual AS patients, we performed correlation analysis between diversity indices and ASDAS.CRP scores. Across a large range of alpha values, no correlation was identified between CD8+ or CD4 + CD45RO+ T cell diversity and AS activity. However, for CD4 + CD45RA+ cells that contains terminally differentiated effector cells (TEMRA, Supplementary Fig. 1), we found a range of diversity indices (when 1.3 < α < 1.7 and α > 3.1) were inversely correlated with disease activity (Fig. 1d, red shade). Specifically, when the unevenness of clone size within a repertoire was emphasized in the diversity measurements (alpha>3.1), higher disease scores were associated with lower diversity, which indicates potential progression-associated oligo-clonal T cell expansion (p < 0.05, r = −0.449, Fig. 1e). To directly evaluate the impact of highly expanded CD4 + CD45RA+ T cell clones, which are most likely differentiated effector T cells, we ranked clonotypes according to their dominancy within each T cell repertoire, measured their collective space occupancy in that repertoire, and performed association analysis against disease activity. This revealed that, when the dominancy cutoff was set to 0.037% and above, the collective size of dominant clonotypes is strongly correlated with disease severity (Fig. 1f-g, when dominancy cutoff = 0.001, r = 0.680, p < .001). However, no significant difference was observed regarding the collective size of dominant CD4 + CD45RA+ clonotypes between AS patients and HCs (Fig. 1h). Based on these results, we reasoned that the progression of AS is accompanied by a mild expansion of CD4 + CD45RA+ T cells, which is likely autoantigen-driven TEMRA cell expansion. Analyzing the antigen specificity of T cells should be important to investigate the role of T cells in disease progression.
3.2. TCR repertoire analysis reveals that CD8+ T cells are stimulated by shared antigens among AS patients with active disease
Since all patients in this cohort are HLA-B27+, we reasoned that certain CDR3 sequences, which code for the major structural component of a TCR involved in antigen recognition, should be enriched among different patients if there are shared autoantigens. To investigate whether any dominant autoantigens were shared among individual AS patients, and whether T cell responses against these autoantigens played any role in disease progression, we performed bioinformatics analysis to quantify the inter-patient T cell repertoire similarity.
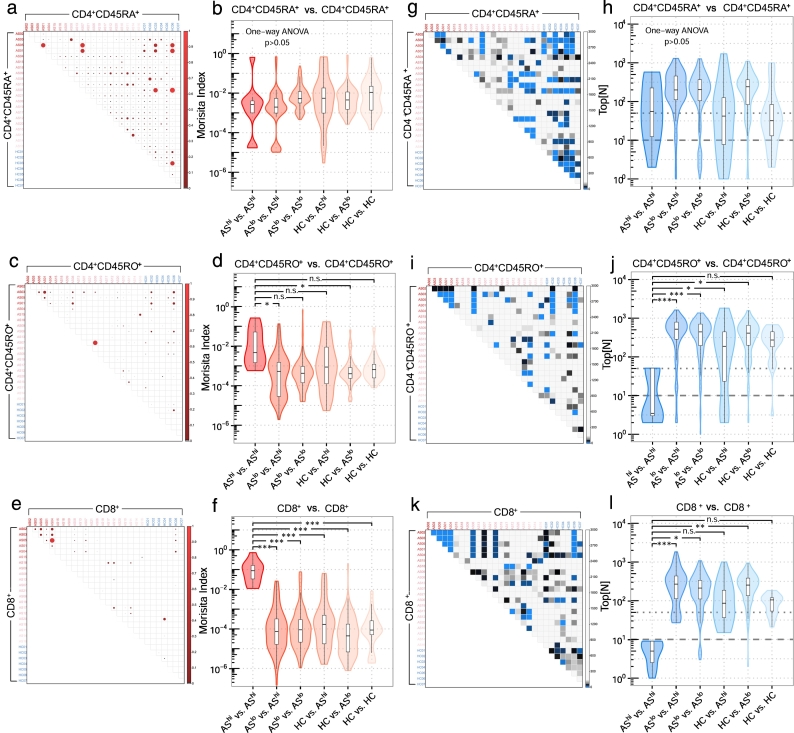
To quantitatively assess similarity for the entire repertoire, we calculated Morisita indices for each pairwise combination within T cell sub-populations. Although disease activity was associated with CD4 + CD45RA+ cell expansion, it did not correlate with inter-repertoire CDR3 overlapping (Fig. 2a, b). This could be explained by inter-patient diversity of MHC-II. However, we found that CDR3 sharing was enriched in the CD4 + CD45RO+ population from patients with active disease (Fig. 2c, d). This activity-associated similarity enrichment was even more dramatic for CD8+ T cells (Fig. 2e, f). Thus, at the advanced disease stage, CD8+ T cells in different patients might be stimulated by shared autoantigens.
Fig. 2.
Similarity profiles reveal shared CDR3 amino acids among high disease activity AS patients in CD8+ T cells. Morisita similarity indices for each pairwise combination of CD4 + CD45RA+ (a), CD4 + CD45RO+ (c) and CD8+ (e) samples plotted as heat maps (upper triangles). Red gradients denote MH values, which range from 0.0 (No shared clonotypes between repertoires) to 1.0 (identical repertoires: same clonotypes and clonotype frequencies). Violin plots indicate the spread of Morisita similarity indices for CD4 + CD45RA+ (b), CD4 + CD45RO+ (d) and CD8+ (f). One-way ANOVA and Tukey's test were used to compare similarities across different groups. Top[N] analysis for each pairwise combination of CD4 + CD45RA+ (g), CD4 + CD45RO+ (i) and CD8+ (k) samples plotted as heat maps (upper triangles). Blue gradients denote Top[N] values, which range from 1 (the first shared clonotype was from Top 1 clone between repertoires) to Max (no shared clonotype). Violin plots indicate the spread of Top[N] indices for CD4 + CD45RA+ (h), CD4 + CD45RO+ (j) and CD8+ (l) samples. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
We also developed Top[N] index analysis (Fig. 2g-l), which measures the dominancy of shared TCR clonotypes between two subjects and therefore indirectly reflects the strength of shared autoantigens (Supplementary Fig. 2). Similar to the Morisita index measurement, CDR3 sharing between the CD4 + CD45RA+ pools was not significant in any disease stage (Fig. 2g, h); however, shared CDR3s were highly enriched in the most abundant CD4 + CD45RO+ (Fig. 2i, j) and CD8+ T cells (Fig. 2k, l) in active patients. In this group, every patient has at least one top50 CD4 + CD45RO+ clonotype that could be identified in at least one other patient and ranked also within the top50, and, at least one top10 CD8+ clonotype that was shared by at least one other patient within the top10. This result strongly suggested that, at the active disease stage, shared autoantigen between AS patients could drive dominant CD8+ T cell responses.
3.3. CD4+ and CD8+ T cells with identical TCR sequences are pathological for AS
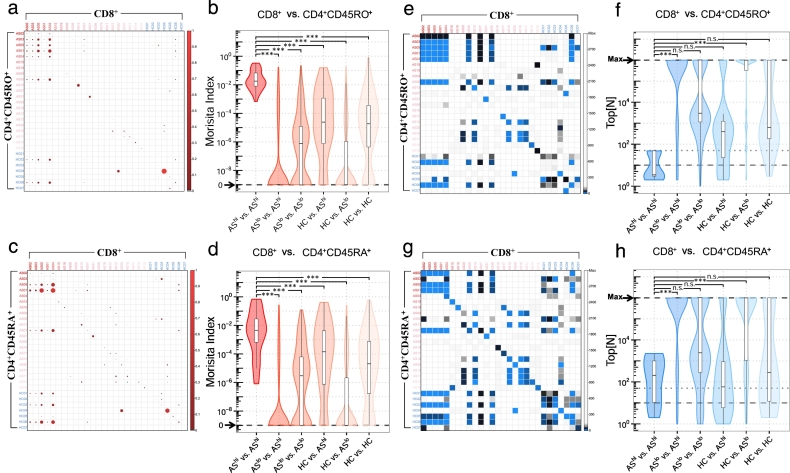
Previous study with HLA-B27 transgenic mice suggested that both HLA-B27-restricted CD4+ and CD8+ T cells were fully functional in vitro and in vivo [44]. This indicated that dominant autoantigens from HLA-B27+ AS patients may also trigger CD4+ T cell responses. Thus, we evaluated similarities of the TCR sequence between CD8+ and CD4 + CD45RO+ or CD4 + CD45RA+ TCR repertoires. Morisita similarity analysis quantitatively showed that a significant amount of complete CDR3 amino acid sequences (Supplementary Fig. 5) and full TCR sequences identified in CD8+ repertoires can be found in both CD4 + CD45RO+ (Fig. 3a, b) and CD4 + CD45RA+ (Fig. 3c, d) T cells from the same patient or other patients when AS is highly active. Specifically, for CD8+ and CD4 + CD45RO+ T cells, the Morisita index is multi-logs higher within this group of patients (Fig. 3a, b). The identification of these common TCRs was not a result of contamination during sample sorting or processing because, at the nucleic acid level, the majority of these common TCRs were derived from distinct mother clones (Supplementary Fig. 6). In addition, Top[N] analysis showed that, for highly active AS patients, common TCRs between CD8+ and CD4 + CD45RO+ repertoires could be identified within their top 80 clones (Fig. 3e, f). To a lesser extent, identical CD8+ and CD4 + CD45RA+ TCRs were also significantly enriched in highly ranked clonotypes (Fig. 3g, h). We hereafter refer to these T cells with CD4+ or CD8+ surface expression but an identical TCR sequence as “CD4/8” T cells.
Fig. 3.
Similarity profiling reveals shared total TCRs (CDR3 nucleotides in combination with their corresponding V and J genes) between CD4+ and CD8+ in high disease activity AS patients. Morisita similarity indices for each pairwise combination of CD8+ and CD4 + CD45RO+ (a) or CD4 + CD45RA+ (c) samples plotted as heat maps. Red gradients denote MH values, which range from 0.0 (no shared clonotype between repertoires) to 1.0 (identical repertoires). Violin plots indicate the spread of Morisita similarity indices between CD8+ and CD4 + CD45RO+ (b) or CD4 + CD45RA+ (d). One-way ANOVA and Tukey's test were used to compare similarities across different groups. Top[N] index for each pairwise combination of CD8+ and CD4 + CD45RO+ (e) or CD4 + CD45RA+ (g) T cell samples plotted as heat maps. Blue gradients denote Top[N] values, which range from 1 (the very first shared clonotype from Top 1 clone between repertoires) to Max (no shared clonotype). Violin plots indicate the spread of Top[N] indices between CD8+ and CD4 + CD45RO+ (f) or CD4 + CD45RA+ (h) samples. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
We previously developed a strategy to detect antigen-expanded T cells in the repertoire, which are highly expanded clonotypes with multiple ways of nucleotide coding [34]. We examined the collective CD4+ or CD8+ TCR repertoire from AS patients and focused on clonotypes with this pattern. In both CD4+ and CD8+ repertoires, a large majority of these expanded clones with high codon degeneracy were indeed CD4/8 cells (Supplementary Fig. 7a, b). Compared to conventional non-CD4/8 T cells, highly expanded clonotypes with codon degeneracy were significantly enriched in the CD4/8 population by 7.45- and 1.43-folds compared to conventional CD8+ and CD4+ T cells, respectively (Supplementary Fig. 7c, d). Based on these results, we reached two conclusions: 1) CD4+ T cell expansion in AS patients is likely due to sharing identical TCR sequences with CD8+ T cells, which can recognize HLA-B27-presented autoantigens; 2) for patients with active disease, the inter-patient sharing of CD4+ and CD8+ TCR sequences suggests that certain common autoantigens are responsible for driving disease progression.
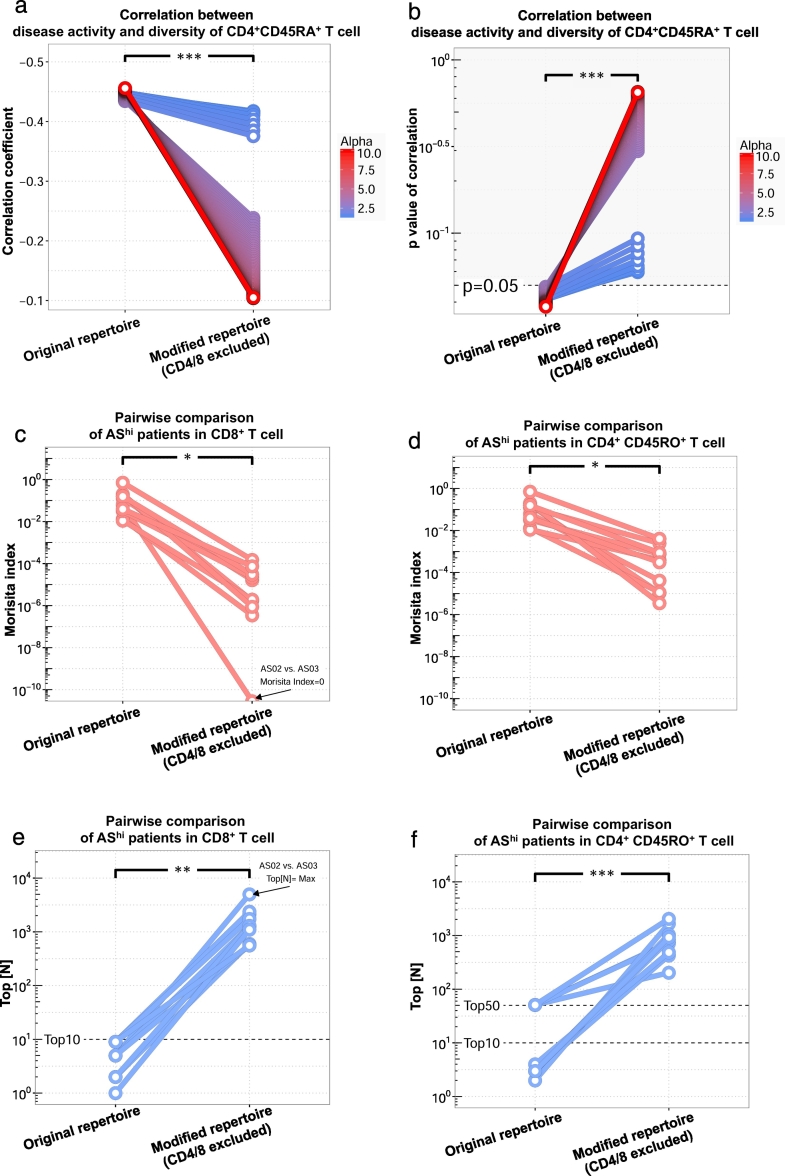
To assess the pathological impact of these TCRs, we performed subtraction analysis. From 21 AS patients subjected to repertoire sequencing, 376 clonotypes were identified to be CD4/8. By subtracting these clonotypes, we built a “modified repertoire” and re-evaluated its correlation with disease severity. As shown in Fig. 1d, the oligo-clonal expansion of CD4 + CD45RA+ T cells was correlated to the active disease at the individual patient level. After we excluded these CD4/8 cells, this correlation was greatly weakened (Fig. 4a) to be statistically insignificant (Fig. 4b). This loss-of-correlation was observed when diversity indices were calculated across a large range of alpha values and was most significant when highly expanded T cell clones carried more weight in diversity calculation, which suggested that the major CD4 + CD45RA+ expansion during disease progression was constituted by CD4/8 T cells.
Fig. 4.
Common TCRs shared between CD4+ and CD8+ T cells are pathological for AS. To evaluate the pathological power of CD4/8 T cells, we recalculated the previous significant clinical relevance of the CD4 + CD45RA+ T cell clonal expansion (Fig. 1d) and the significantly elevated similarity between high disease activity patients in either CD8+ (Fig. 2e,f,k,l) or CD4 + CD45RO+ T cells (Fig. 2c,d,i,j) in the corresponding repertoires without CD4/8 T cells (“Modified repertoire”), and compared the clinically relevant features between the original repertoire and the modified repertoire. (a) Correlation coefficients and (b)p-values of Pearson correlation between ASDAS.CRP scores and the CD4 + CD45RA+ T cell diversity indices for alpha between 1.3 and 1.7 or beyond 3.1. Statistics are based on paired t-test. (c) Morisita and (e) Top[N] similarity indices between CD8+ samples from each pairwise combination of high disease activity AS patients. After depletion of CD4/8 T cells, there was no shared CDR3 between one pair of AS-high patients (arrow pointed). (d) Morisita and (f) Top[N] similarity indices between CD4CD45RO+ samples from each pairwise combination of high disease activity AS patients.
Our analysis also showed that, among the most severe AS patients, shared CDR3s are highly enriched in the CD8+ population (Fig. 2e, f), and, to a lesser extent, in the CD4 + CD45RO+ population (Fig. 2c, d). After depleting CD4/8 CDR3s, we observed log-wide reductions of Morisita indices for both CD8+ (Fig. 4c) and CD4 + CD45RO+ populations (Fig. 4d). For two patients, depletion of CD4/8 results in the total loss of CD8+ TCR similarity to any other patients (Fig. 4c, arrow). The most significant inter-patient TCR sharing was observed in the group of clones with the most dominant size (Fig. 2 i-l). Accordingly, by depleting CD4/8 clones, we observed that the clones shared among patients were dramatically less dominant for both CD8+ (Fig. 4e) and CD4 + CD45RO+ (Fig. 4f) cell populations.
This suggests that the major CD8+ and CD4 + CD45RO+ expansions in active AS are driven by the expansion of CD4/8 T cells. Therefore, we hypothesized that CD4/8 T cells might drive the pathogenesis of ankylosing spondylitis.
3.4. Discriminating T cell repertoires of AS from other autoimmune diseases through Motif Analysis
Potential antigen specificity sharing among TCR repertoires of AS patients was revealed by similarity analyses focusing on the entire CDR3β sequence. However, recent comprehensive structural biology studies clearly suggest that not every amino acid of CDR3β is required to execute TCR-pMHC interaction [[45], [46], [47]]. Instead, antigen recognition relies on hotspots formed by only a few (3–4) amino acids in the CDR3β loop [48]. Hence, it is conceivable that traditional similarity analysis mistakenly excludes TCRs that recognize a specific autoantigen with the same hotspot but varied amino acids in other positions. To improve the accuracy of antigen specificity-based similarity measurement, we developed a new computational framework to search for TCRs sharing the same antigen specificity in repertoires – the Motif Analysis.
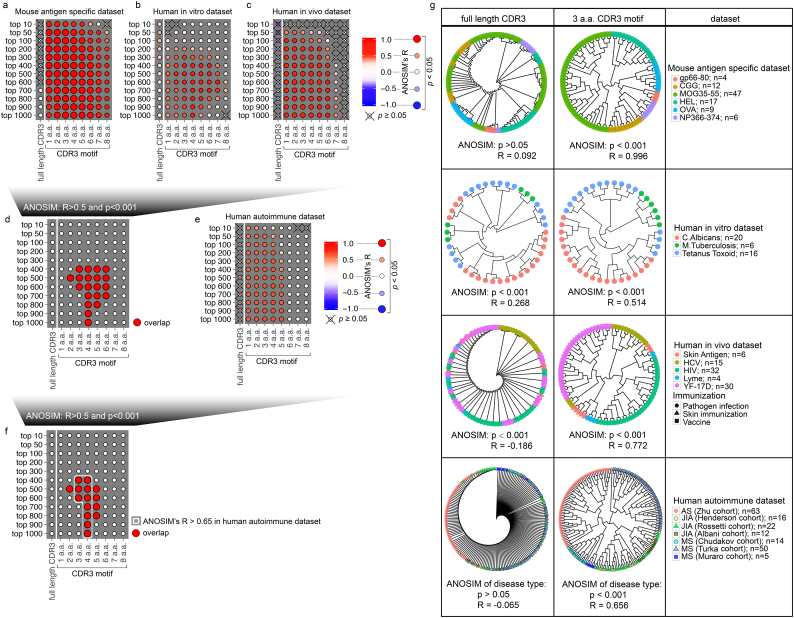
As a first step, in this study, we restricted our motif searching to the primary protein structure consisting of continuous amino acids within CDR3. The CDR3 sequence was then dissected into multiple motifs, such that the similarity between any two TCRs, or two TCR repertoires, can be digitalized by their collective motif sharing (Supplementary Fig. 3a-c). To develop our algorithm, we built a training data set consisting of 95 mouse TCR repertoires with defined and distinct antigen specificities (Supplementary Table 3). This artificial repertoire included LCMV gp66–80 I-Ab specific CD4+ T cells (unpublished), chicken gamma globulin specific CD4+ T cells (unpublished), myelin oligodendrocyte glycoprotein (MOG) a.a.35–55 I-Ab specific CD4+ T cells [49], hen egg lysozyme (HEL) specific CD4+ T cells (NCBI: SRP049789), ovalbumin (OVA) specific CD4+ T cells [50] and influenza NP366–374H-2b specific CD8+ T cells [51]. We used the Analysis Of Similarities (ANOSIM) to objectively assess the power of algorithm in distinguishing TCRs with different antigen specificity (Supplementary Fig. 3d). As expected, since antigen recognition does not require the complete CDR3, CDR3 sequences for a specific antigen were too diversified among individual repertoires (Supplementary Fig. 4a) to reflect the primary structural requirement for antigen recognition. Unsupervised hierarchical clustering based on traditional similarity calculations using full length CDR3s failed to distinguish T cell repertoires based on their antigen specificities (Fig. 5a, full length CDR3; corresponding to Supplementary Fig. 3c, d). However, consistent with structural data [[45], [46], [47]], our Motif Analysis indicated that CDR3 motifs were sufficient to group TCR repertoires based on antigen specificity among their most dominant CDR3s (Fig. 5a, CDR3 motif; corresponding to Supplementary Fig. 3c, d). Next, we validated this Motif Analysis algorithm using human TCR repertoires sequenced from PBMCs stimulated by in vitro pathogen infection. This validation data set was comprised of 42 TCR repertoires post C. albicans infection [52], Tetanus toxoid priming, and M. Tuberculosis challenge [52] (Supplementary Table 4). With the greatly increased antigen complexity, CDR3s were too diversified (Supplementary Fig. 4b) to sufficiently distinguish antigen specificity (Fig. 5b, full length CDR3). However, even without filtering HLA restriction, motif analysis could successfully identify infection types when using proper motif length and dominant CDR3s number (Fig. 5b, ANOSIM: R > 0.5).
Fig. 5.
Discriminating T-cell repertoires with different antigenic specificity through motif analysis.
(a-c) Antigen discrimination capacity of full length CDR3 and CDR3 motifs in 3 T-cell repertoire training datasets, including 95 mouse antigen-specific samples (a; corresponding to Supplementary Fig. 3c, d), 42 human in vitro antigen-stimulated samples (b), and 87 human in vivo antigen-challenged peripheral blood samples (c). The significance and quantitative evaluation of the antigen discrimination capacity were measured by the analysis of similarities (ANOSIM) p and R values. R values range from −1.0 (completely cross-reactive) to 1.0 (completely antigen-specific). The color gradient represents the analysis of similarities (ANOSIM) R values. (d) Overlap of the CDR3 motif length used by top[N] dominant CDR3s to reflect antigen specificity in 3 training datasets (ANOSIM: R>0.5; p < .001). (e) Significance and quantitative evaluation of disease discrimination capacity of full length CDR3 and CDR3 motif in 182 human autoimmune disease peripheral blood samples. (f) Overlap of CDR3 motif length used by top[N] dominant CDR3s to discriminate antigen specificity and T cell responses in autoimmune diseases (ANOSIM: R>0.5; p < .001). (g) Circular dendrogram representing unsupervised hierarchical clustering using frequencies of top 500 dominant CDR3s or 3 amino acid CDR3 motifs from top 500 dominant CDR3. Samples of different groups are indicated by corresponding color and shape.
We further tested our algorithm using 87 human PBMC TCR repertoires after in vivo antigen challenge (Supplementary Table 5), which demanded that the algorithm act on repertoire complexity of ex vivo PBMCs. While diversified CDR3s failed to discriminate antigen specificity (Fig. 5c, full length CDR3), the motif analysis using most of the various parameters successfully distinguished repertoires from human immunodeficiency virus (HIV) infection [53], hepatitis C virus (HCV) infection [54], Lyme disease [55], yellow fever vaccination [56], and diphencyprone-induced skin DTH responses [57] despite differing times of sample collection, Rep-seq methods, routes of antigen challenging, and HLA diversity (Fig. 5c, ANOSIM: R > 0.5). Furthermore, we identified overlapping, robust parameters for motif analysis (ANOSIM: R > 0.5, p < .001) across these 3 independent datasets (Fig. 5d). Based on these results, we conclude that Motif Analysis should be a robust assay with sufficient sensitivity to dissect antigen-specific T cell responses from a complex repertoire.
We next performed Motif Analysis with a data set mixing our 63 repertoires from AS patients with 50 repertoires from 3 independent cohorts of Juvenile Idiopathic Arthritis (JIA) patients [55,58,59] and 69 repertoires from 3 independent cohorts of Multiple Sclerosis (MS) patients [[60], [61], [62]] (Supplementary Table 6). We hypothesized that Motif Analysis should distinguish AS from these two well-known T cell-mediated autoimmune diseases if AS is also driven by pathological T cells. While full length CDR3 sequences were unsuccessful, motif analysis showed a promising ability to discriminate T cell responses in distinct autoimmune diseases (Fig. 5e). Moreover, motif analysis using most antigen specificity discriminating parameters successfully identified different autoimmune diseases (Fig. 5f).
When these 182 repertoires were subjected to Motif Analysis using 3 amino acid CDR3 motif and top 500 CDR3s (Fig. 5g, Human autoimmune dataset), we observed a few interesting clustering patterns: 1) motif similarity grouped patients based on their disease type, which strongly suggested that, like JIT and MS, AS is also an autoimmune disease mediated by T cell antigen responses against a specific set of autoantigens; 2) although JIA repertoires were collected from 3 cohorts in independent studies, the enriched motifs were not distinguishable between cohorts, which suggests that certain common autoantigens are dominant among JIA patients; 3) among MS repertoires, patients from two out of three independent studies shared highly similar collections of TCR motifs. Surprisingly, a distinct cluster of MS repertoires was constituted by samples from a third independent cohort. This cohort consisted entirely of patients with poor prognostic features including highly active disease, progressive disability, and deficiency in response to various treatments. This suggests that these MS patients may have pathological T cell activation stimulated by distinct autoantigens; 4) for AS patients, all TCR repertoires were explicitly clustered together without distinction between CD4+ and CD8+ T cells, which implies shared antigen specificity might exist between CD4+ and CD8+ T cells.
3.5. Identification and validation of pathological AS specific CDR3 motifs
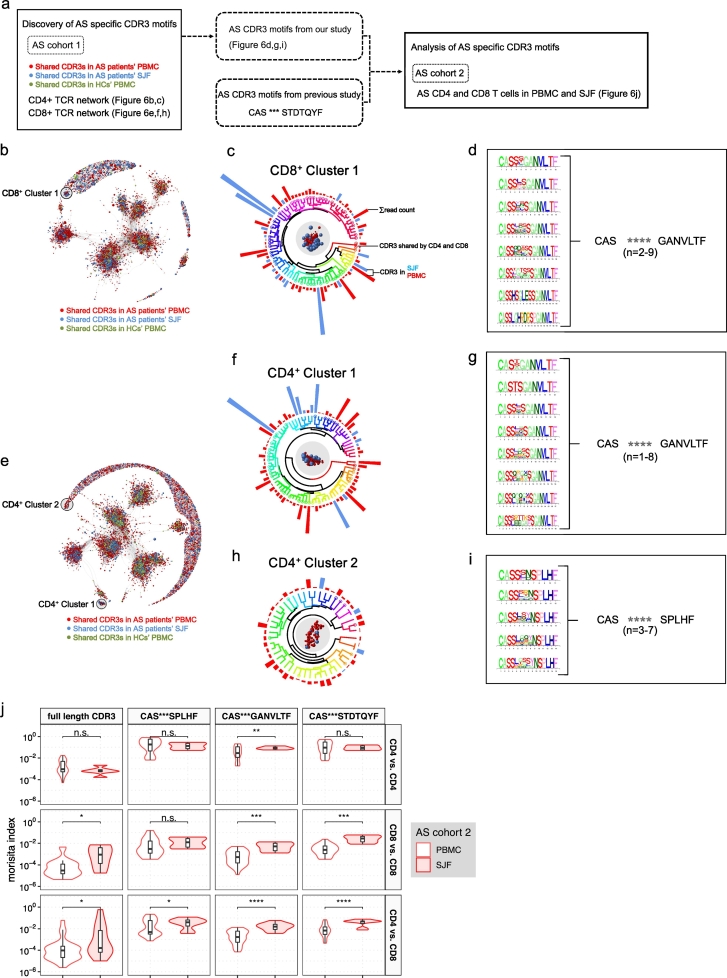
To assess the clinical relevance of CD4/8 cells, we undertook two independent approaches. Since our evidence so far strongly supports the notion that AS is a T cell-driven autoimmune disease, we reasoned that autoantigen-specific T cells should be enriched in the spondyloarthritic joint fluid (SJF), the site of autoinflammation. Consequently, AS autoantigen-specific motifs should also be enriched in SJF. To test this, we collected CD4+ and CD8+ T cells from SJF in 2 AS patients from our cohort, sequenced their TCR repertoires and performed Motif Analysis to identify AS specific motifs. Moreover, using another independent AS cohort [63], we validated AS specific motifs from both our AS cohort and previous studies [64,65] (Fig. 6a). Using our AS cohort, we combined all CD8+ TCR sequences from patients' SJF, patients' PBMCs, and healthy controls' PBMCs together to build the CD8+ data set. CDR3s were broken down into 3 a.a. motifs and Jaccard indices were calculated for every individual pair of CDR3s to measure CDR3-CDR3 similarities. Focusing on CDR3s with highly shared motif usage, we set up a high cutoff (Jaccard index ≥0.5) and constructed a network with qualified CDR3 pairs (Supplementary Fig. 8). Depicting CDR3-CDR3 similarities by distances, CDR3s were assembled into clusters (Fig. 6b). We identified an isolated cluster that consisted entirely of TCRs from patients' SJF or PBMCs, but completely excluded TCRs from PBMC of healthy controls (Fig. 6c). We found the selected TCR cluster was composed of TCRs with a universal TCR motif. From the selected cluster, we extracted one AS-specific CD8+ TCR motif (Fig. 6d) and two AS-specific CD4+ TCR motifs (Fig. 6e-i). From this stringent analysis, one of these two identified CD4+ motifs was, again, identical to the one from CD8+ T cells (Fig. 6d, g). This identification of a common unique-in-patients motif from both CD4+ and CD8+ T cells, but excluded from shared repertoire of healthy controls, supported a role of CD4/8 TCRs in AS pathogenesis.
Fig. 6.
Enrichment of AS specific CDR3 motifs in spondyloarthritic joint fluid from AS patients. (a) Schematic of analysis of AS specific CDR3 motifs. (b-i) Discovery of AS specific CDR3 motifs. CDR3 motif similarity network of shared CDR3s in CD8+ (b) or CD4+ (e) T cells from healthy control subjects' peripheral blood, AS patients' peripheral blood and spondyloarthritic joint fluid (SJF). To determine similarity, Jaccard similarities of motif usage were computed for all pairs of CDR3s. CDR3 pairs with Jaccard similarities >0.5 were connected and graphed using a Fruchterman-Reingold force-directed layout algorithm. CDR3s with similar motif usage were positioned proximal to each other, whereas CDR3s with less motif similarity were positioned farther apart. Nodes are color-coded according to 3 distinct samples and tissue compartments (CDR3s shared by AS patients' PBMC, red spheres; AS patients' SJF, blue spheres; control PBMC, green spheres). Sizes of nodes correspond to CDR3 frequency. The AS-specific cluster consisting only of CDR3s from patients' SJF and PBMCs were indicated by black shaded regions (CD8+ cluster 1; CD4+ cluster 1; CD4+ cluster 2). Circular dendrogram clustering of CDR3s from CD8+ cluster 1 (c), CD4+ cluster 1 (f) and CD4+ cluster 2 (h) based on motif usage similarity. Representations of sum of CDR3 read count (indicated by bar size), tissue compartments (indicated by bar color: red, SJF; blue, PBMC), CDR3s shared by CD4+ and CD8+ (denoted by circle with corresponding color). Sequence logo plot of CDR3s from CD8+ cluster 1 (d), CD4+ cluster 1 (g) and CD4+ cluster 2 (i) with different CDR3 length (left). For the ensemble of CDR3s with different amino acid length, (d, g, i) CDR3 motif summary indicated the consensus motif (right). (j) Overlap of full length CDR3s and AS specific CDR3 motifs within CD4+ T cells, within CD8+ T cells and between CD4+ and CD8+ T cells. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Next, using another AS validation cohort [63], we investigated a common T cell response using shared full length CDR3s and AS specific CDR3 motifs from our study and previous study [64,65] (Fig. 6j). This revealed significantly enriched common T cell responses in CD4/8 T cells from AS SJF, thereby providing further evidence to support the role of CD4/8 TCRs in AS pathogenesis. Moreover, the previously reported CDR3 motif “CAS***STDTQYF” was significantly enriched in SJF CD8 and CD4/8 T cells but not in CD4+ T cells. “CAS***SPLHF” was also a specific CDR3 motif enriched in SJF CD4/8 T cells, whereas “CAS***GANVLTF” was significantly enriched in all CD4, CD8 and CD4/8 T cell populations from SJF. Therefore, we discovered and validated 2 new AS specific CDR3 motifs which could depict the common T cell response of CD4/8 T cells enriched in the AS arthritogenic tissue compartment.
4. Discussion
Limited by the moderate nature of T cell activation and the limitation in sensitivity of old cellular immunology technology, the immunopathology of AS disease remains elusive. In this study, through high-throughput TCR Rep-seq and CDR3 Motif Analysis, our data provide direct evidence to support four major hypotheses related to the etiology of AS: 1) although perhaps not initiated by T cells, AS is driven by T cell activation for disease progression; 2) both CD4+ and CD8+ T cells contribute to the activity of disease; 3) a subset of CD4+ T cells sharing identical TCRs with CD8+ T cells are capable of being activated by HLA-B27; 4) pathology-associated CD4/8 T cells can be identified in human spondyloarthritic joint fluid.
Conventionally, CD4+ and CD8+ T cell antigen recognition is diametrically restricted by class II and class I MHC specificity. Data collected from this study does not challenge this dogma. Instead, we provide a novel mechanism to explain HLA-B27-associated CD4+ T cell activation – identical TCR usage for both CD4+ and CD8+ lineages. The enrichment of identical CDR3s between CD4+ and CD8+ T cells in patients could result from presentation of the same antigen epitopes by both MHC-II and MHC-I, respectively. However, since CDR1 and CDR2 regions of the TCRβ chain anchor on the MHC helix [32,66], the enrichment of common TCRs between CD4+ and CD8+ T cells prompted our hypothesis that TCR recognition in AS patients naturally crosses the MHC class boundary. A previous study indicates that CD4+ T cells isolated from PBMCs of AS patients can be expanded by cell lines expressing HLA-B27 [28]. Based on animal models, this was interpreted as stimulation through surface β2m-free HLA-B27 or HLA-B27 dimers [24] engaged by innate receptors, such as KIR3DL2, on CD4+ T cells [27]. However, TCR repertoire analysis provides three lines of evidence supporting an alternative model: 1) in each patient, CD4/8 T cells account for 30–95% of moderately-to-highly expanded clonotypes. These clonotypes share the same CDR3 amino acid sequences encoded with nucleic acid degeneracy. Most plausible explanation is that CD4/8 T cells were experienced antigen stimulation. 2) we were able to detect identical CD4/8 T cells in different patients, and, for each patient, these shared T cells can be found in the most dominant range of the repertoire. This inter-patient sharing of CD4/8 TCRs also argues for a mechanism of antigen stimulation, especially stimulation from common arthritogenic antigens. 3) CD4/8 T cells are a major contributor to the correlation between T cells and disease activity. Taken together, although we do not have data to exclude the possibility of innate receptor-driven expansion, although we do not have data to suggest any characteristics of arthritogenic antigens, the identified expansion of CD4/8 T cells strongly supports the presence of such arthritogenic antigens during AS progression.
What is the origin of these CD4/8 T cells? While we cannot exclude the possibility of lineage conversion between mature CD4+ and CD8+ T cells, more plausible model is that HLA-B27-recognizing CD4+ T cells were generated during thymic selection. Although the mechanisms determining thymic CD4+ versus CD8+ lineage choice remain to be determined, it is clear that, at the very beginning of positive selection, CD4 + CD8+ double positive thymocytes terminate Cd8 gene transcription but maintain Cd4 expression regardless of TCR specificity to MHC-I or –II67. At this transitional CD4 + CD8low stage, prolonged TCR signaling with positive selecting ligands [68] or impaired function of certain transcription factors such as MAZR [69] can promote MHC-I-restricted TCRs to become CD4+ T cells. These results from animal models suggest possible pathways through which AS patients might produce HLA-B27-restricted CD4+ T cells. Moreover, the pathogenic development of these cells might not be stochastic, but instead emerge from undiscovered fundamental characteristics of HLA-B27. In support of this, in HLA-B27 transgenic mice, fully functional CD4+ T cells were selected by HLA-B27 [44]. From our patient cohort, we have performed preliminary analysis with out-of-frame (OOF) TCR sequences, which fail to produce functional TCR proteins and consequently are not subject to any selection pressure. By comparing the identical OOF TCRs between CD4+ and CD8+ T cells and OOF repertoires among different patients (data not shown), we speculate that CD4/8 T cells likely originate from thymic development.
Data sharing
All raw data generated for this study was uploaded to the Sequence Read Archive (SRA) under BioProject ID PRJNA378893 (https://www.ncbi.nlm.nih.gov/bioproject/378893).
Acknowledgments
Acknowledgements
All patient samples were collected upon informed consent. All experiments were conducted according to the principles expressed in the Declaration of Helsinki. All donors were recruited into a clinical trial at Xijing Hospital (Chinese Clinical Trial Registry; Registration number: ChiCTR-ONRC-11001565). This study was approved by Ethics Committee of the first affiliated hospital of the Fourth Military Medical University (Approved No. of ethic committee:20110303-7). We thank the surgeons and research nurses for providing us with access to these samples.
Funding sources
This work was partially supported by the National Key Research and Development Program of China (2017YFC0909002), the National Basic Research Program of China (NO.2015CB553704) and the National Science and Technology Major Projects of New Drugs (2014ZX09508002–002). Qi-Jing Li is a Whitehead Family Foundation Scholar. Ying Wan is supported by National Natural Science Foundation of China (Grant No. 91642119). All the funders had no role in the study design, data collection, data analysis, interpretation, writing of the manuscript, or the decision to submit the paper for publication.
Declarations of Competing Interests
The authors declare no potential conflicts of interest.
Author contributions
Ming Zheng, Xin Zhang, Yinghui Zhou, Juan Tang, Qing Han and Yang Zhang performed most of the experiments with the help from other authors. Ying Wan, Qi-Jing Li, Zhi-Nan Chen and Ping Zhu designed and supervised the study. Ming Zheng, Ping Zhu and Qi-Jing Li wrote the manuscript with input from co-authors. Qingshan Ni, Gang Chen, Qingzhu Jia, Haili Yu, Siqi Liu and Elizabeth Robins performed TCRβ repertoire sequencing. Ning Jenny Jiang revised the manuscript. Ming Zheng and Yinghui Zhou conducted the data analysis.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.07.032.
Contributor Information
Ying Wan, Email: wanying.cn@gmail.com.
Qi-Jing Li, Email: qi-jing.li@duke.edu.
Zhi-Nan Chen, Email: znchen@fmmu.edu.cn.
Ping Zhu, Email: zhuping@fmmu.edu.cn.
Appendix A. Supplementary data
Supplementary material
References
- 1.Dean L.E., Jones G.T., MacDonald A.G., Downham C., Sturrock R.D., Macfarlane G.J. Global prevalence of ankylosing spondylitis. Rheumatology (Oxford) 2014;53(4):650–657. doi: 10.1093/rheumatology/ket387. [DOI] [PubMed] [Google Scholar]
- 2.Caffrey M.F., James D.C. Human lymphocyte antigen association in ankylosing spondylitis. Nature. 1973;242(5393):121. doi: 10.1038/242121a0. [DOI] [PubMed] [Google Scholar]
- 3.Schlosstein L., Terasaki P.I., Bluestone R., Pearson C.M. High association of an HL-A antigen, W27, with ankylosing spondylitis. N Engl J Med. 1973;288(14):704–706. doi: 10.1056/NEJM197304052881403. [DOI] [PubMed] [Google Scholar]
- 4.Brown M.A., Pile K.D., Kennedy L.G. HLA class I associations of ankylosing spondylitis in the white population in the United Kingdom. Ann Rheum Dis. 1996;55(4):268–270. doi: 10.1136/ard.55.4.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowness P. HLA-B27. Annu Rev Immunol. 2015;33:29–48. doi: 10.1146/annurev-immunol-032414-112110. [DOI] [PubMed] [Google Scholar]
- 6.Evans D.M., Spencer C.C., Pointon J.J. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat Genet. 2011;43(8):761–767. doi: 10.1038/ng.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cortes A., Hadler J., Pointon J.P. Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat Genet. 2013;45(7):730–738. doi: 10.1038/ng.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.May E., Dorris M.L., Satumtira N. CD8 alpha beta T cells are not essential to the pathogenesis of arthritis or colitis in HLA-B27 transgenic rats. J Immunol. 2003;170(2):1099–1105. doi: 10.4049/jimmunol.170.2.1099. [DOI] [PubMed] [Google Scholar]
- 9.Taurog J.D., Dorris M.L., Satumtira N. Spondylarthritis in HLA-B27/human beta2-microglobulin-transgenic rats is not prevented by lack of CD8. Arthritis Rheum. 2009;60(7):1977–1984. doi: 10.1002/art.24599. [DOI] [PubMed] [Google Scholar]
- 10.Baeten D., Sieper J., Braun J. Secukinumab, an interleukin-17A inhibitor, in Ankylosing spondylitis. N Engl J Med. 2015;373(26):2534–2548. doi: 10.1056/NEJMoa1505066. [DOI] [PubMed] [Google Scholar]
- 11.Baraliakos X., Borah B., Braun J. Long-term effects of secukinumab on MRI findings in relation to clinical efficacy in subjects with active ankylosing spondylitis: An Observational Study. Ann Rheum Dis. 2016;75(2):408–412. doi: 10.1136/annrheumdis-2015-207544. [DOI] [PubMed] [Google Scholar]
- 12.Breban M., Fernandez-Sueiro J.L., Richardson J.A. T cells, but not thymic exposure to HLA-B27, are required for the inflammatory disease of HLA-B27 transgenic rats. J Immunol. 1996;156(2):794–803. [PubMed] [Google Scholar]
- 13.Shen H., Goodall J.C., Hill G.J.S. Frequency and phenotype of peripheral blood Th17 cells in ankylosing spondylitis and rheumatoid arthritis. Arthritis Rheum. 2009;60(6):1647–1656. doi: 10.1002/art.24568. [DOI] [PubMed] [Google Scholar]
- 14.Zhang L., Li Y.G., Li Y.H. Increased frequencies of Th22 cells as well as Th17 cells in the peripheral blood of patients with ankylosing spondylitis and rheumatoid arthritis. PLoS One. 2012;7(4) doi: 10.1371/journal.pone.0031000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kenna T.J., Davidson S.I., Duan R. Enrichment of circulating interleukin-17-secreting interleukin-23 receptor-positive gamma/delta T cells in patients with active ankylosing spondylitis. Arthritis Rheum. 2012;64(5):1420–1429. doi: 10.1002/art.33507. [DOI] [PubMed] [Google Scholar]
- 16.Mousavi T., Poormoghim H., Moradi M., Tajik N., Shahsavar F., Soofi M. Phenotypic study of natural killer cell subsets in ankylosing spondylitis patients. Iran J Allergy Asthma Immunol. 2009;8(4):193–198. [PubMed] [Google Scholar]
- 17.Ciccia F., Guggino G., Rizzo A. Type 3 innate lymphoid cells producing IL-17 and IL-22 are expanded in the gut, in the peripheral blood, synovial fluid and bone marrow of patients with ankylosing spondylitis. Ann Rheum Dis. 2015;74(9):1739–1747. doi: 10.1136/annrheumdis-2014-206323. [DOI] [PubMed] [Google Scholar]
- 18.Gokmen F., Akbal A., Resorlu H. Neutrophil-lymphocyte ratio connected to treatment options and inflammation markers of Ankylosing spondylitis. J Clin Lab Anal. 2015;29(4):294–298. doi: 10.1002/jcla.21768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boyraz I., Onur C.S., Erdem F. Assessment of relation between neutrophil lympocyte, platelet lympocyte ratios and epicardial fat thickness in patients with ankylosing spondylitis. Med Glas (Zenica) 2016;13(1):14–17. doi: 10.17392/832-16. [DOI] [PubMed] [Google Scholar]
- 20.Mercan R., Bitik B., Tufan A. The association between neutrophil/lymphocyte ratio and disease activity in rheumatoid arthritis and Ankylosing spondylitis. J Clin Lab Anal. 2016;30(5):597–601. doi: 10.1002/jcla.21908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kucuk A., Uslu A.U., Ugan Y. Neutrophil-to-lymphocyte ratio is involved in the severity of ankylosing spondylitis. Bratisl Lek Listy. 2015;116(12):722–725. doi: 10.4149/bll_2015_142. [DOI] [PubMed] [Google Scholar]
- 22.Noordenbos T., Yeremenko N., Gofita I. Interleukin-17-positive mast cells contribute to synovial inflammation in spondylarthritis. Arthritis Rheum. 2012;64(1):99–109. doi: 10.1002/art.33396. [DOI] [PubMed] [Google Scholar]
- 23.Buckley M.G., Walters C., Wong W.M. Mast cell activation in arthritis: detection of alpha- and beta-tryptase, histamine and eosinophil cationic protein in synovial fluid. Clin Sci (Lond) 1997;93(4):363–370. doi: 10.1042/cs0930363. [DOI] [PubMed] [Google Scholar]
- 24.Allen R.L., O'Callaghan C.A., McMichael A.J., Bowness P. Cutting edge: HLA-B27 can form a novel beta 2-microglobulin-free heavy chain homodimer structure. J Immunol. 1999;162(9):5045–5048. [PubMed] [Google Scholar]
- 25.Bird L.A., Peh C.A., Kollnberger S., Elliott T., McMichael A.J., Bowness P. Lymphoblastoid cells express HLA-B27 homodimers both intracellularly and at the cell surface following endosomal recycling. Eur J Immunol. 2003;33(3):748–759. doi: 10.1002/eji.200323678. [DOI] [PubMed] [Google Scholar]
- 26.Kollnberger S., Bird L., Sun M.Y. Cell-surface expression and immune receptor recognition of HLA-B27 homodimers. Arthritis Rheum. 2002;46(11):2972–2982. doi: 10.1002/art.10605. [DOI] [PubMed] [Google Scholar]
- 27.Wong-Baeza I., Ridley A., Shaw J. KIR3DL2 binds to HLA-B27 dimers and free H chains more strongly than other HLA class I and promotes the expansion of T cells in ankylosing spondylitis. J Immunol. 2013;190(7):3216–3224. doi: 10.4049/jimmunol.1202926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyle L.H., Goodall J.C., Opat S.S., Gaston J.S. The recognition of HLA-B27 by human CD4(+) T lymphocytes. J Immunol. 2001;167(5):2619–2624. doi: 10.4049/jimmunol.167.5.2619. [DOI] [PubMed] [Google Scholar]
- 29.Freeman J.D., Warren R.L., Webb J.R., Nelson B.H., Holt R.A. Profiling the T-cell receptor beta-chain repertoire by massively parallel sequencing. Genome Res. 2009;19(10):1817–1824. doi: 10.1101/gr.092924.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chien Y.H., Gascoigne N.R., Kavaler J., Lee N.E., Davis M.M. Somatic recombination in a murine T-cell receptor gene. Nature. 1984;309(5966):322–326. doi: 10.1038/309322a0. [DOI] [PubMed] [Google Scholar]
- 31.Schatz D.G., Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nat Rev Immunol. 2011;11(4):251–263. doi: 10.1038/nri2941. [DOI] [PubMed] [Google Scholar]
- 32.Davis M.M., Boniface J.J., Reich Z. Ligand recognition by alpha beta T cell receptors. Annu Rev Immunol. 1998;16:523–544. doi: 10.1146/annurev.immunol.16.1.523. [DOI] [PubMed] [Google Scholar]
- 33.Garcia K.C., Gapin L., Adams J.J. A closer look at TCR germline recognition. Immunity. 2012;36(6):887–888. doi: 10.1016/j.immuni.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jia Q., Zhou J., Chen G. Diversity index of mucosal resident T lymphocyte repertoire predicts clinical prognosis in gastric cancer. Oncoimmunology. 2015;4(4) doi: 10.1080/2162402X.2014.1001230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rudwaleit M., van der Heijde D., Landewe R. The development of assessment of SpondyloArthritis international society classification criteria for axial spondyloarthritis (part II): validation and final selection. Ann Rheum Dis. 2009;68(6):777–783. doi: 10.1136/ard.2009.108233. [DOI] [PubMed] [Google Scholar]
- 36.van der Linden S., Valkenburg H.A., Cats A. Evaluation of diagnostic criteria for ankylosing spondylitis. A proposal for modification of the New York criteria. Arthritis Rheum. 1984;27(4):361–368. doi: 10.1002/art.1780270401. [DOI] [PubMed] [Google Scholar]
- 37.Lukas C., Landewe R., Sieper J. Development of an ASAS-endorsed disease activity score (ASDAS) in patients with ankylosing spondylitis. Ann Rheum Dis. 2009;68(1):18–24. doi: 10.1136/ard.2008.094870. [DOI] [PubMed] [Google Scholar]
- 38.van der Heijde D., Lie E., Kvien T.K. ASDAS, a highly discriminatory ASAS-endorsed disease activity score in patients with ankylosing spondylitis. Ann Rheum Dis. 2009;68(12):1811–1818. doi: 10.1136/ard.2008.100826. [DOI] [PubMed] [Google Scholar]
- 39.Bolotin D.A., Shugay M., Mamedov I.Z. MiTCR: software for T-cell receptor sequencing data analysis. Nat Methods. 2013;10(9):813–814. doi: 10.1038/nmeth.2555. [DOI] [PubMed] [Google Scholar]
- 40.Nazarov V.I., Pogorelyy M.V., Komech E.A. TCR: an R package for T cell receptor repertoire advanced data analysis. BMC bioinforma. 2015;16:175. doi: 10.1186/s12859-015-0613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Legendre P. Ward's hierarchical agglomerative clustering method: which algorithms implement Ward's criterion? J of Classif. 2014;31(3):274–295. [Google Scholar]
- 42.Greiff V., Bhat P., Cook S.C., Menzel U., Kang W., Reddy S.T. A bioinformatic framework for immune repertoire diversity profiling enables detection of immunological status. Genome Med. 2015;7(1):49. doi: 10.1186/s13073-015-0169-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hill M.O. Diversity and evenness: a unifying notation and its consequences. Ecology. 1973;54(2):427–432. [Google Scholar]
- 44.Roddis M., Carter R.W., Sun M.Y. Fully functional HLA B27-restricted CD4+ as well as CD8+ T cell responses in TCR transgenic mice. J Immunol. 2004;172(1):155–161. doi: 10.4049/jimmunol.172.1.155. [DOI] [PubMed] [Google Scholar]
- 45.Newell E.W., Ely L.K., Kruse A.C. Structural basis of specificity and cross-reactivity in T cell receptors specific for cytochrome c-I-E(k) J Immunol. 2011;186(10):5823–5832. doi: 10.4049/jimmunol.1100197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Birnbaum M.E., Mendoza J.L., Sethi D.K. Deconstructing the peptide-MHC specificity of T cell recognition. Cell. 2014;157(5):1073–1087. doi: 10.1016/j.cell.2014.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adams J.J., Narayanan S., Birnbaum M.E. Structural interplay between germline interactions and adaptive recognition determines the bandwidth of TCR-peptide-MHC cross-reactivity. Nat Immunol. 2016;17(1):87–94. doi: 10.1038/ni.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Glanville J., Huang H., Nau A. Identifying specificity groups in the T cell receptor repertoire. Nature. 2017;547(7661):94–98. doi: 10.1038/nature22976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao Y., Nguyen P., Ma J. Preferential use of public TCR during autoimmune encephalomyelitis. J Immunol. 2016;196(12):4905–4914. doi: 10.4049/jimmunol.1501029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Madi A., Shifrut E., Reich-Zeliger S. T-cell receptor repertoires share a restricted set of public and abundant CDR3 sequences that are associated with self-related immunity. Genome Res. 2014;24(10):1603–1612. doi: 10.1101/gr.170753.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carey A.J., Gracias D.T., Thayer J.L. Rapid evolution of the CD8+ TCR repertoire in neonatal mice. J Immunol. 2016;196(6):2602–2613. doi: 10.4049/jimmunol.1502126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Becattini S., Latorre D., Mele F. T cell immunity. Functional heterogeneity of human memory CD4(+) T cell clones primed by pathogens or vaccines. Science. 2015;347(6220):400–406. doi: 10.1126/science.1260668. [DOI] [PubMed] [Google Scholar]
- 53.Heather J.M., Best K., Oakes T. Dynamic perturbations of the T-cell receptor repertoire in chronic HIV infection and following antiretroviral therapy. Front Immunol. 2015;6:644. doi: 10.3389/fimmu.2015.00644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abdel-Hakeem M.S., Boisvert M., Bruneau J., Soudeyns H., Shoukry N.H. Selective expansion of high functional avidity memory CD8 T cell clonotypes during hepatitis C virus reinfection and clearance. PLoS Pathog. 2017;13(2) doi: 10.1371/journal.ppat.1006191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Henderson L.A., Volpi S., Frugoni F. Next-generation sequencing reveals restriction and Clonotypic expansion of Treg cells in juvenile idiopathic arthritis. Arthritis Rheumatol. 2016;68(7):1758–1768. doi: 10.1002/art.39606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DeWitt W.S., Emerson R.O., Lindau P. Dynamics of the cytotoxic T cell response to a model of acute viral infection. J Virol. 2015;89(8):4517–4526. doi: 10.1128/JVI.03474-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gaide O., Emerson R.O., Jiang X. Common clonal origin of central and resident memory T cells following skin immunization. Nat Med. 2015;21(6):647–653. doi: 10.1038/nm.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rossetti M., Spreafico R., Consolaro A. TCR repertoire sequencing identifies synovial Treg cell clonotypes in the bloodstream during active inflammation in human arthritis. Ann Rheum Dis. 2017;76(2):435–441. doi: 10.1136/annrheumdis-2015-208992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spreafico R., Rossetti M., van Loosdregt J. A circulating reservoir of pathogenic-like CD4+ T cells shares a genetic and phenotypic signature with the inflamed synovial micro-environment. Ann Rheum Dis. 2016;75(2):459–465. doi: 10.1136/annrheumdis-2014-206226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shugay M., Bagaev D.V., Turchaninova M.A. VDJtools: unifying post-analysis of T cell receptor repertoires. PLoS Comput Biol. 2015;11(11) doi: 10.1371/journal.pcbi.1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Muraro P.A., Robins H., Malhotra S. T cell repertoire following autologous stem cell transplantation for multiple sclerosis. J Clin Invest. 2014;124(3):1168–1172. doi: 10.1172/JCI71691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Paula A.S.A., Johnson K.R., Nicholas R. Intrathecal T-cell clonal expansions in patients with multiple sclerosis. Ann Clin Transl Neurol. 2016;3(6):422–433. doi: 10.1002/acn3.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Komech E.A., Pogorelyy M.V., Egorov E.S. CD8+ T cells with characteristic T cell receptor beta motif are detected in blood and expanded in synovial fluid of ankylosing spondylitis patients. Rheumatology (Oxford) 2018;57(6):1097–1104. doi: 10.1093/rheumatology/kex517. [DOI] [PubMed] [Google Scholar]
- 64.Dulphy N., Peyrat M.A., Tieng V. Common intra-articular T cell expansions in patients with reactive arthritis: identical beta-chain junctional sequences and cytotoxicity toward HLA-B27. J Immunol. 1999;162(7):3830–3839. [PubMed] [Google Scholar]
- 65.May E., Dulphy N., Frauendorf E. Conserved TCR beta chain usage in reactive arthritis; evidence for selection by a putative HLA-B27-associated autoantigen. Tissue Antigens. 2002;60(4):299–308. doi: 10.1034/j.1399-0039.2002.600404.x. [DOI] [PubMed] [Google Scholar]
- 66.Garcia K.C., Adams J.J., Feng D., Ely L.K. The molecular basis of TCR germline bias for MHC is surprisingly simple. Nat Immunol. 2009;10(2):143–147. doi: 10.1038/ni.f.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yasutomo K., Doyle C., Miele L., Fuchs C., Germain R.N. The duration of antigen receptor signalling determines CD4+ versus CD8+ T-cell lineage fate. Nature. 2000;404(6777):506–510. doi: 10.1038/35006664. [DOI] [PubMed] [Google Scholar]
- 69.Sakaguchi S., Hombauer M., Bilic I. The zinc-finger protein MAZR is part of the transcription factor network that controls the CD4 versus CD8 lineage fate of double-positive thymocytes. Nat Immunol. 2010;11(5):442–448. doi: 10.1038/ni.1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material