Abstract
Glioblastoma (GBM) is the most aggressive primary brain tumor in adults. Despite progress in surgical and medical neuro-oncology, prognosis for GBM patients remains dismal, with a median survival of only 14–15 months. The modest benefit of conventional therapies is due to the presence of GBM stem cells (GSCs) that cause tumor relapse and chemoresistance and, therefore, that play a key role in GBM aggressiveness and recurrence. So far, strategies to identify and target GSCs have been unsuccessful. Thus, the development of an approach for GSC detection and targeting would be fundamental for improving the survival of GBM patients. Here, using the cell-systematic evolution of ligand by exponential (SELEX) methodology on human primary GSCs, we generated and characterized RNA aptamers that selectively bind GSCs versus undifferentiated GBM cells. We found that the shortened version of the aptamer 40L, which we have called A40s, costained with CD133-labeled cells in human GBM tissue, suggestive of an ability to specifically recognize GSCs in fixed human tissues. Of note, both 40L and A40s were rapidly internalized by cells, allowing for the delivery of the microRNA miR-34c and the anti-microRNA anti-miR-10b, demonstrating that these aptamers can serve as selective vehicles for therapeutics. In conclusion, the aptamers 40L and A40s can selectively target GSCs. Given the crucial role of GSCs in GBM recurrence and therapy resistance, these aptamers represent innovative drug delivery candidates with a great potential in the treatment of GBM.
Keywords: aptamer, cancer stem cell, glioblastoma, microRNA, delivery
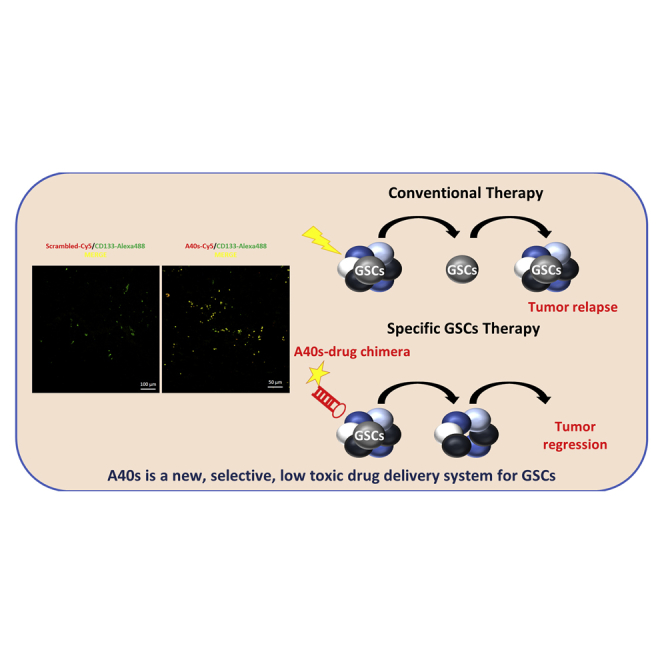
Graphical Abstract
Introduction
Glioblastoma (GBM) is the most common and aggressive primary brain tumor in adults.1, 2 A small population of cancer stem cells (GBM stem cells [GSCs]) that retains stem cell properties, including self-renewal and multipotency, are deemed responsible for the frequent relapse of GBM and its resistance to conventional therapeutics.3, 4 In contrast with highly proliferating cells from the tumor bulk, this rare quiescent cell population has the potential to reconstitute the intrinsic heterogeneity of the tumor mass and to spread into the brain.4, 5 Therefore, the development of highly specific and safe molecules able to selectively target and eradicate the GSC population represents a timely and important challenge for the treatment of brain tumors.
Aptamers are short, single-stranded oligonucleotides that act as high-affinity ligands and that can be potential antagonists of disease-associated proteins.6 The advantages of aptamers include low toxicity, easy penetration into tumors, and the ability to cross the blood-brain barrier (BBB).7 Due to their ability to diffuse throughout the tumor area in the intracranial cavity, aptamers have a great potential to be used as diagnostic and therapeutic tools, such as carriers for therapeutic drugs. By adopting an unbiased cell-based variant of the original combinatorial systematic evolution of ligand by exponential (SELEX) enrichment procedure, it is possible to generate aptamers that target specific cell surface-binding biomarkers.6, 8, 9
Here, in order to isolate GSC-specific aptamers, we took advantage of a panel of primary cultures of GSCs isolated from GBM patient tumors. For initial selection, we used two patient-derived cell lines characterized by different GBM subtypes as a complex target to generate a panel of aptamers able to distinguish stem-like cells from adherent, differentiated counterparts. From selected aptamers, we focused our investigation on one, named 40L, that resulted already enriched from early selection steps. Both the 40L aptamer and its short form, A40s, appeared to bind with high affinity to GSCs, but not to differentiated counterpart cells, both in in vitro assays and in fixed human GBM histological specimens. More interestingly, both aptamers were quickly internalized by GSCs. Therefore, they may represent a strategic diagnostic tool as well as a selective vehicle for therapeutics. Our findings provide a blueprint for the isolation of highly selective reagents as imaging tools for the clinical management of GBM.
Results
Aptamer Selection
To isolate aptamers that identify the rare population of GBM cells growing in the tumor mass as stem-like, non-adherent spheres, we adopted a differential cell-SELEX approach, using primary GBM stem cell lines derived from two patients. The GSC 1 line was derived from a patient diagnosed with neural GBM, whereas the GSC 83 line was from a patient diagnosed with mesenchymal GBM.1 The cell lines were propagated as non-adherent spheres in minimal F12 medium supplemented with cell growth factors (epidermal growth factor [EGF] and basic fibroblast growth factor [FGF]), as previously described,10 and alternately used as targets in the SELEX process.
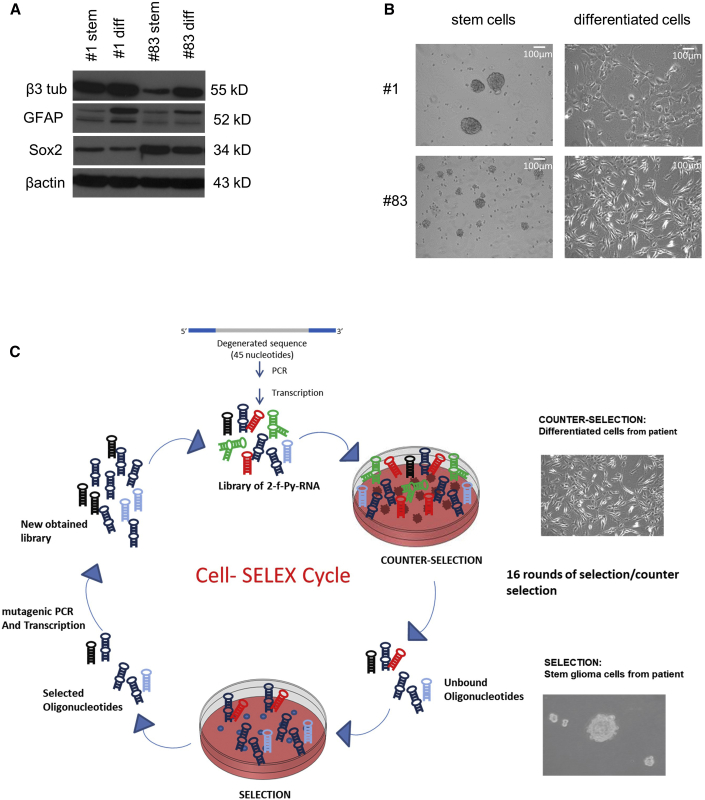
Stem features were evaluated by assessing the reduction of differentiation markers Glial fibrillary acidic protein (GFAP) and β-Tubulin-3 (β3 tub) and the increase in the stem cell-associated factor SRY-Box 2 (Sox-2) (Figure 1A) in the stem cell population as compared to the differentiated counterparts. At each round, selection was preceded by one or two counterselection steps, incubating the pool with adherent GSC 1 or GSC 83 cells. For counterselections, GSC 1 or GSC 83 cells were grown as adherent cells on a matrigel substrate for 2 weeks in serum-containing F12 medium to induce differentiation (Figure 1B). For the selection steps, spheres were dissociated and then incubated with the aptamer pool. The SELEX procedure is summarized in Figure 1C. During the selection process, we progressively increased the selective pressure by changing: (1) the incubation time (30 min up to cycle 14 and 15 min in the last 2 cycles); (2) the number of couterselections (1 up to cycle 13 and 2 in the last 3 cycles); and (3) the washing conditions (from 1 wash to 2 washes from cycle 3) and the addition of polyinosic acid (100 ng/μL) as non-specific competitor in the last cycle.
Figure 1.
Cell Characterization and SELEX Conditions
(A) Growth pattern of glioblastoma cultures established under different cell conditions. Western blot (WB) analysis illustrating different expression markers in stem cell cultures and adherent cell cultures obtained after 2 weeks of culturing in 10% serum. Regarding the other promising sequences, aptamer 5 was not evaluated further since it was not among the most-enriched aptamers of the last six SELEX round media. (B) Panels show representative pictures of primary GBM cells growing as suspension or adherent cell culture. (C) Schematic summary of the selective approach of cell-SELEX.
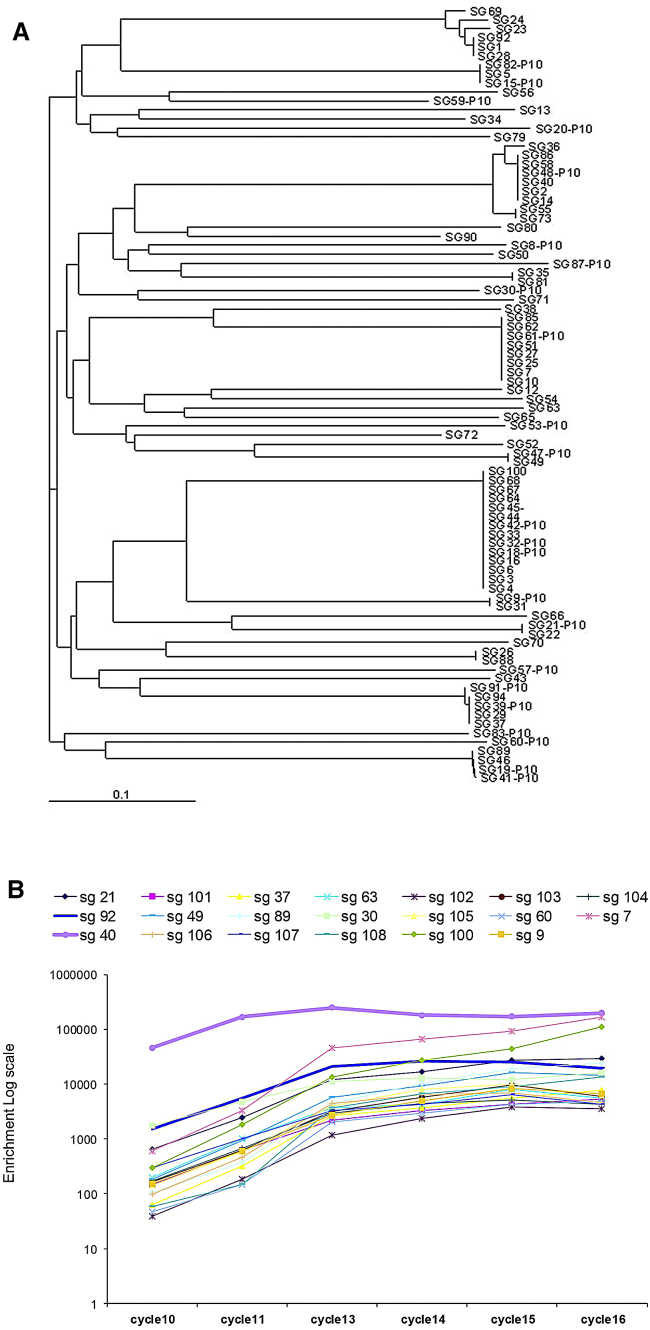
Upon 16 SELEX rounds, the final pool was cloned and 100 clones were sequenced and aligned for homology within their variable core region (Figure 2A). Four families dominated the pool, together covering approximately 30% of sequences. To validate the information obtained by clustering, the enriched pools from rounds 10, 11, 13, 14, 15, and 16 were sequenced by high-throughput sequencing (HTS) (Figure 2B). The enrichment of the top 20 sequences from HTS analyses of the last cycles is shown in Figure S1 and Table S1. The information obtained by coupling the two sequencing approaches provided a reliable way to identify the most promising sequences. Indeed, the most enriched sequences identified by HTS belonged to the four large clusters identified by conventional Sanger sequencing, on which we focused further analyses.
Figure 2.
GSC Aptamer Selection
(A) The random regions of all the sequenced aptamers were aligned using ClustalW. The dendrogram shows visual clasterization of similarity among 100 individual sequences cloned after 16 rounds of selection. (B) The enriched pools from rounds 10, 11, 13, 14, 15, and 16 were sequenced by high-throughput sequencing (HTS). sg, SELEX on GBM.
Binding Assay
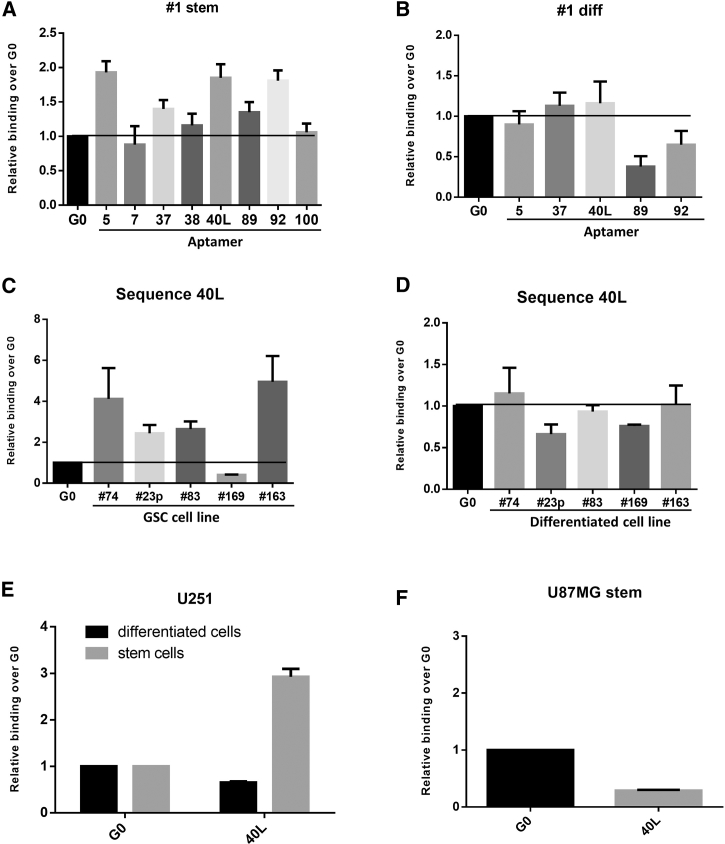
Given the good correlation between the two sequencing approaches, we determined the sequences that preferentially bound GSC spheres rather than differentiated, adherent cells. To this end, we used qRT-PCR to analyze aptamer binding at 200 nM in GSC 1 cells, the line used for the majority of selection rounds. Analysis was first performed for those aptamers belonging to the major clusters, i.e., aptamers 5, 7, 37, 38, 40L, 89, 92, and 100 (Figure 3A). Most of the sequences bound to GSC 1 stem cells rather than to the starting pool of the SELEX used as a negative control (G0) and, thus, specifically distinguished them, as shown by the relatively lower affinity of binding to the differentiated counterparts (Figure 3B). The best binding of GSCs as compared to the G0 unselected pool was detected for aptamers 5, 40L, and 92. Among the sequences with the best binding, we decided to further characterize 40L that from the HTS analyses was the most rapidly enriched during the SELEX rounds (from round 10).
Figure 3.
40L Binds GBM Primary Cell Cultures and Cell Lines
(A–D) Binding was performed with the most enriched sequences obtained from SELEX at 200 nM on GSC 1 stem (A) or adherent (diff) (B) cells from the same patient and on several GSC lines grown in suspension (C) or under adherent conditions (D). Representative experiments are shown and results are expressed relative to the background binding detected with the starting pool of sequences used for selection. (E and F) Binding of 40L was performed at 200 nM on U251-MG (E) and U87-MG (F) cell line cells in suspension or in adherent culturing conditions. The ability of 40L to be internalized into GSCs is shown. Results are expressed as percentages of the total bound after 30 min of incubation. Representative experiments are shown as mean (±SD) and results are expressed relative to the background binding detected with the starting pool of sequences used for selection.
Regarding the other promising sequences, aptamer 5 was not evaluated further since it was not among the most-enriched aptamers of the last six SELEX rounds, even if it showed a slight increased affinity to GSC compared to 40L (fold over G0 1.93 versus 1.85, respectively) (Figure 2B).
We validated 40L binding with a panel of five different GSCs (lines 74, 23p, 83, 169, and 163). 40L bound to almost all the GSCs analyzed (Figure 3C) at different extent; moreover, it showed no detectable binding to the differentiated counterparts of any of the cell lines (Figure 3D). We also tested 40L binding to the GSCs obtained from the U251 and U87MG cell lines, which, as previously demonstrated by Adamo et al.,11 acquire a stem phenotype when in suspension culture, with the upregulation of several stemness markers. We found that 40L bound to U251-derived stem-like cells, and not to the adherent counterpart (Figure 3E), but it did not bind to U87MG stem-like cells (Figure 3F). Due to the good specificity of 40L, we restricted our further functional analyses to this sequence.
Truncation of the 40L Sequence
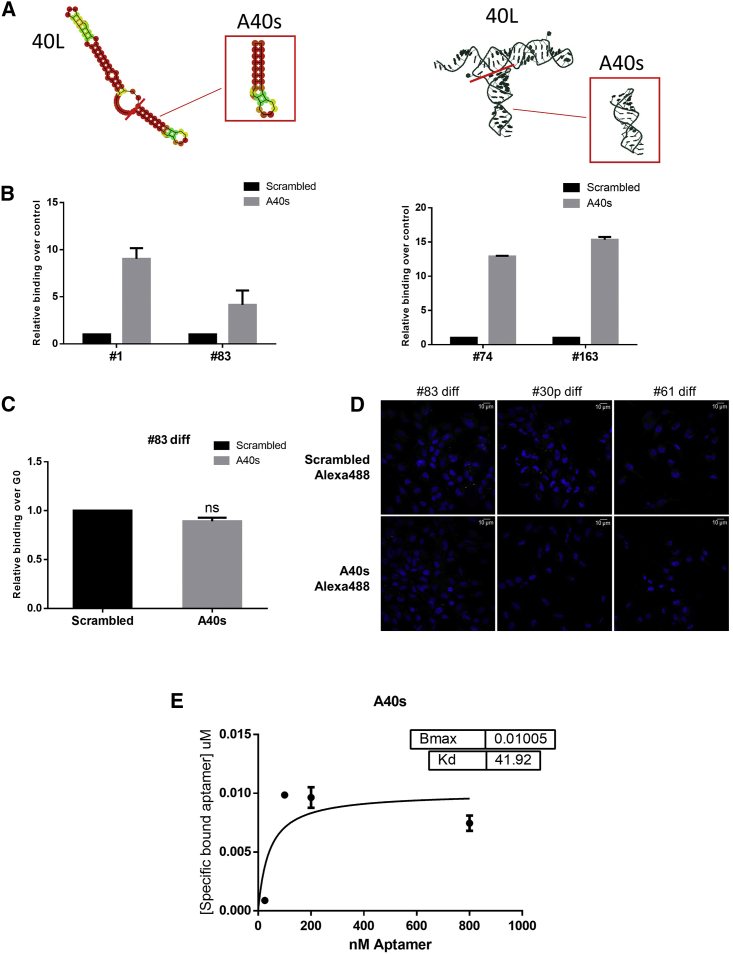
To identify a shorter aptamer with better properties, we first determined the 2D structure of the 40L aptamer. According to the structures predicted by software and online tools (RNAstructure version [v.]5.7, RNAcomposer,12 and RNAfold; http://rna.tbi.univie.ac.at/), we identified and tested two portions. One, a 30-bp sequence that we named A40s (Figure 4A), still had the three-dimensional shape of 40L. Importantly, A40s was still able to bind to the GSC 1, 83, 74, and 163 cell lines (Figure 4B), but no binding to GSC 83, 30p, and 61 cells grown in adherent, differentiated conditions was observed, either with qRT-PCR or fluorescence assay (for the latter assay, A40s and negative control aptamer were labeled with Alexa488) (Figures 4C and 4D).
Figure 4.
A40s Aptamer Binding
(A) Bidimensional and tridimensional shape predictions of the 40L aptamer are shown; 40L aptamer sequence was shortened in order to have a smaller aptamer with the best properties. The selected portion is shown in the squares. (B and C) Binding assay was performed at 200 nM on GSC lines 83, 1, 74, and 163 grown as stem (B) or as differentiated (C) cells. Representative experiments are shown and results are expressed as mean (±SD) relative to the background binding, detected with an unrelated aptamer of a length similar to that of A40s. (D) Immunofluorescence staining was performed by treating 83, 30p, or 61 differentiated cells with A40s-Alexa488 or scrambled-Alexa488 at 500 nM for 30 min. All images were captured at the same settings, enabling direct comparison of staining patterns. (E) Kd calculation is shown as mean (±SD). Nonlinear binding curve was obtained by treating GSCs with increasing amounts of A40s.
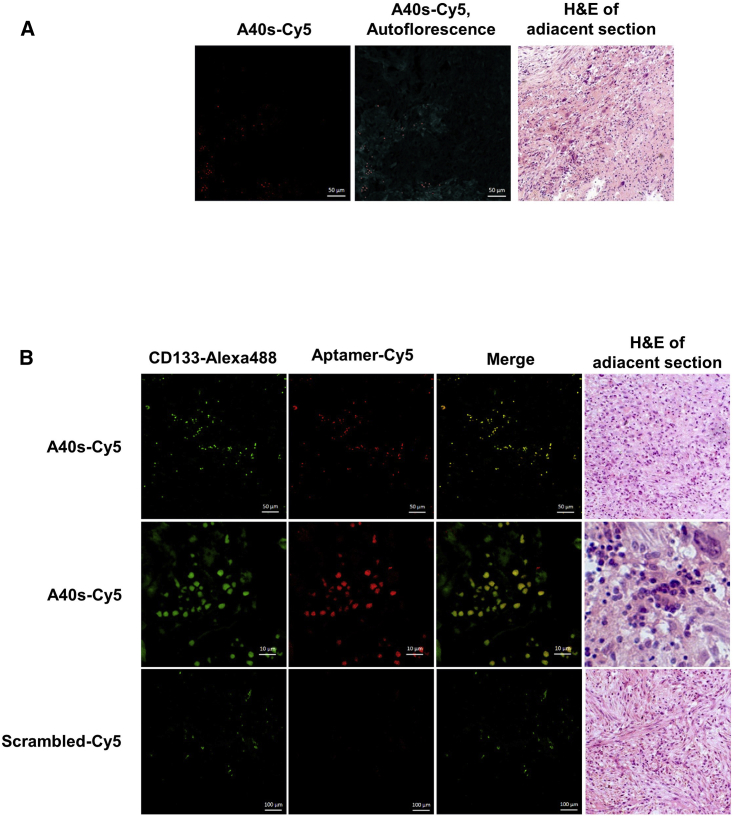
Low aptamer affinity and sensitivity for target cells can be a major limitation to applicability. Thus, we tested A40s’s binding kinetic for the GSC 1 line: it resulted in a very low affinity (about 42 nM) perfectly comparable with the range of other aptamers13 (Figure 4E). Furthermore, we evaluated the distribution of A40s-positive cells in GBM tissues obtained from patients that had undergone craniotomy (Figure 5A). The signals for A40s and CD133 were co-localized, proving specific recognition of GSCs in GBM tissues (Figure 5B). These results confirmed that the shortened sequence retains the binding properties of 40L and that it is a promising diagnostic tool, allowing a reduction in the cost of chemical synthesis.
Figure 5.
Discrimination of GSCs by A40s in Fixed Human Tissues
(A) Distribution of A40s-positive cells in human tissues. (B) The A40s signal perfectly co-localizes with CD133-labeled cells in human tissues. Images are accompanied by H&E staining of the adjacent section.
Internalization of 40L and A40s and Their Use as a Delivery System
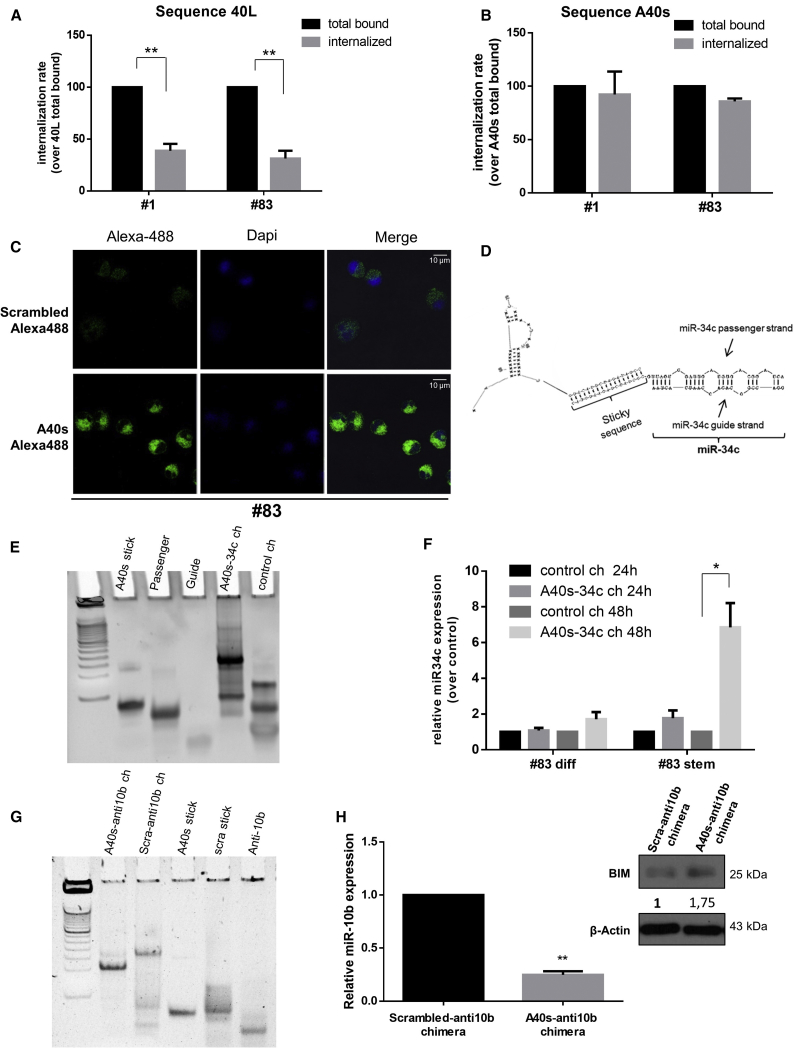
As previously reported, aptamer sequences for transmembrane cell surface receptors can be internalized in a receptor-mediated manner.14, 15 To test whether 40L is rapidly internalized, the GSC 1 line was incubated with the aptamer for 30 min, washed with 0.5 M NaCl in PBS to remove aptamers still exposed on the cell surface, then RNA extracted. As determined by qRT-PCR, approximately 40% of total bound 40L was not affected by the NaCl wash, indicative of intracellular uptake (Figure 6A). We then assessed the ability of A40s to be internalized by GSCs. After 30 min of exposure, almost 100% was internalized by the cells, suggestive of a better internalization rate than 40L (Figure 6B). Internalization of A40s was confirmed with immunofluorescence on the GSC 83 line (Figure 6C).
Figure 6.
Aptamer Internalization in GSCs
(A and B) 40L (A) and A40s (B) are internalized into GSCs. Results are expressed as percentages of the total bound after 30 min of incubation. (C) Immunofluorescence of 83 stem cells treated with A40s-Alexa488 or scrambled-Alexa488 at 500 nM for 30 min. All images were captured at the same settings, enabling direct comparison of the staining patterns. (D) Secondary structure of the A40s-miR34c chimera as predicted by RNA Structure v.5.7. (E) Non-denaturating gel electrophoresis analysis reveals correct A40s/miR-34c chimera conjugate annealing. (F) Stem or differentiated GBM cells were incubated with A40s/miR-34c chimera for 24 and 48 h. Relative miR-34c levels were assessed by using qRT-PCR. (G) Non-denaturating gel electrophoresis analysis reveals correct A40s-anti-miR-10b chimera conjugate annealing. (H) GSCs were incubated with A40s-anti-miR-10b chimera or negative control for 48 h. Relative miR-10b levels were assessed by qRT-PCR, and the upregulation of one of miR-10b’s targets, BIM, was assessed by WB. In (A), (F), and (H), vertical bars indicate SD values. *p ≤ 0.05, **p ≤ 0.01.
It is well established that aptamers function as highly selective vehicles for therapeutic substances, such as noncoding RNAs (ncRNAs),16, 17, 18, 19 to a specific target cell. Because of the significant internalization of 40L and A40s by target cells, both represent good candidates for selective delivery of therapeutic molecules to GSCs. Given the very rapid internalization of A40s, we tested its ability to function as a vehicle. To this end, as previously described,20, 21 we used sticky-end annealing to generate a molecular duplex chimera (termed A40s-miR-34c) (Figure 6D), consisting of a microRNA cargo and A40s as a carrier. We verified the correct annealing of the conjugate by non-denaturating gel electrophoresis analysis (Figure 6E). Treatment with A40s-miR34c increased miR-34c levels, as assessed by qRT-PCR, in GSCs, but not in differentiated cells (Figure 6F). Moreover, given the important role of miR-10b as a prognostic and predictive GBM biomarker (it is upregulated in GBM cells22), a chimera conjugate was generated to target this microRNA by annealing the sticky A40s aptamer to an anti-miR-10b sequence (Figure 6G). A40s-anti-miR10b was able not only to be internalized by GSCs but also to decrease miR-10b levels and cause upregulation of a validated miR-10b target, BCL-like protein 11 (BIM) (Figure 6H). This finding demonstrates that A40s may function as a selective therapeutic carrier for GSC targeting.
Discussion
GBM is a heterogeneous tumor consisting of differentiated cells and a small population of cancer stem cells.5, 10, 23 GSCs are responsible for tumor initiation, growth, and recurrence and, thus, represent an ideal target to increase the overall survival of GBM patients. In the present work, we addressed GSC targeting using an aptamer. Indeed, aptamers are excellent candidates because of their cell-specific recognition ability and other characteristics (e.g., short development and synthesis time, low size and cost, ease of modification, good tissue penetration, and high affinity and specificity); they represent a new class of therapeutic, diagnostic, and delivery molecules comparable to, or even better than, monoclonal antibodies.24
By developing an innovative cell-based selection strategy using primary patient-derived GSCs, we identified several sequences that effectively discriminate stem cells from their differentiated counterparts. Among the identified aptamers, we characterized in-depth a sequence (40L) that demonstrated a high selectivity for GSCs isolated from GBM patients. Given that long RNA sequences (>60–70 nt) have high manufacturing costs, in order to improve the potential use of this aptamer as a therapeutic molecule, we optimized it by identifying a shorter form (30-mer A40s) able to bind GSCs like the longer 40L. Interestingly, we found that, similar to 40L, A40s discriminated between GSCs and differentiated glioma cells.
More importantly, given that one emerging application of aptamers is as delivery tools, we demonstrate that 40L and, much more so, A40s show a high internalization rate in GSCs and, thus, may be used as specific carriers. This was confirmed by the ability of A40s to specifically deliver miR-34c and anti-miR10b to a stem population and not to differentiated cells.
Recently, we reported the use of GL21.T and Gint4.T aptamers as carriers for miR-137 and anti-miR-10b to target GSCs. Both bind Axl and PDGFRβ,21, 25, 26 respectively, two tyrosine kinase receptors commonly expressed on GBM cells. These aptamers bind to targets shared between stem and differentiated cells. Thus, it could be useful to compare different aptamers’ effects or, better yet, evaluate their synergistic activity. That study provided a strictly defined approach to GSC targeting, identifying an aptamer that specifically targets this cell population. Another attempt to identify aptamers targeting GSCs was previously reported by Kim et al.,27 who described a pool of DNA sequences able to discriminate between CD133-positive and CD133-negative cells derived from human GBM xenografts. In our selection, we isolated and amplified cells directly from human specimens without animal cell amplification using the already reported methods of Pallini and colleagues.10 Our RNA aptamer is the first reported aptamer able to identify GSCs directly in fixed human GBM tissue. In fact, we demonstrate that A40s and CD133 co-localize in sections from human brain tumor, indicating that the A40s signal localizes specifically to human brain cancer stem cells. Further, the selection process described here gave several additional promising sequences that could be further characterized for GSC targeting.
An important impediment for therapeutic compounds targeting GBM is the presence of the BBB, which limits the passage of large molecules to the tumor area. The ability of A40s to successful penetrate intracranial models has not been investigated yet. Nevertheless, recent evidence supports the ability of aptamers to cross the BBB,7, 26 and several strategies have been developed to transport therapeutics across the BBB28, 29 that could be easily combined with aptamers.30
Although A40s’s target remains to be determined, our aptamer is a good candidate to target GSCs selectively, and it shows great potential as a diagnostic as well as a therapeutic tool. The delivery properties of the aptamer further enhance its potential, opening the additional possibility of developing bifunctional conjugates for an effective, combined GBM therapy. Our study represents a proof of principle for the development of a novel tool to identify and target the GSC population.
Materials and Methods
Patient-Derived Tumor Samples
GBM tissue samples for selection were obtained from the Institute of Neurosurgery, School of Medicine, Università Cattolica, Rome, Italy, after craniotomy of adult patients from which, before surgery, informed consent was obtained. Mechanical dissociation of GBM tumor specimens allowed stem cell isolation, as previously described.10, 31 Not immortalized GSCs derived from patient specimens were then cultured in a serum-free medium supplemented with EGF and fibroblast growth factor beta (bFGF) (Sigma-Aldrich, Milan, Italy). The GSCs used were derived from human tissues of different GBM subtypes: neuronal subtype, 1; mesenchymal subtype, 23p and 83; and classical subtype, 61, 74, 163, and 169. Differentiation was induced by plating cells on flasks coated with BD Matrigel Basement Membrane Matrix (BD Biosciences, Milan, Italy) in the presence of 10% serum and absence of EGF and bFGF for 2 weeks. For human tissue immunostaining, samples were obtained from 3 patients with GBM who had undergone complete tumor resection at Krasnoyarsk Inter-District Ambulance Hospital. Tumor specimens were collected with the written informed consent of patients. The human studies were approved by the Local Committee on Ethics of the Krasnoyarsk Regional Clinical Cancer Center and Krasnoyarsk State Medical University, Krasnoyarsk, Russia. GBM tissue was removed during standard neurosurgery and immediately immersed in ice-cold RPMI-1640 medium supplemented with 1,000 UmL−1 penicillin G and 1,000 mgL−1 streptomycin. Samples were transported on ice to a laboratory within 4 h of resection.
Whole-Cell SELEX
The SELEX cycle was performed essentially as described elsewhere.9 GSCs and differentiated GBM cells were used respectively as selection and counterselection counterparts. Given the resistance to degradation against serum nucleases provided by fluoropyrimidine, transcription was performed in the presence of 1 mM 2′-F pyrimidines and a mutant form of T7 RNA polymerase (2.5 U/μL T7 R&DNA polymerase, Epicenter Biotechnologies, WI, USA) was used to improve yields. The complexity of the starting pool was roughly 1,014. Before each incubation with the cells, the 2′F-Py RNAs were heated at 85°C for 5 min, snap-cooled on ice for 2 min, and allowed to warm up to 37°C. The library was purchased from Eurogentec (Seraing, Belgium). It was composed of a central degenerated sequence of 45 nt flanked by two fixed regions.
Selection Step
To sort aptamers able to selectively bind GSCs, a selection step was performed, incubating the pool with 106 GSCs at 37°C for 30 min up to the 14th round or for 15 min in the last two rounds of SELEX. The bound aptamers were recovered after washings (one for the first two cycles and two for the others cycles) with 5 mL serum-free DMEM-F12 (Sigma-Aldrich).
Counterselection Step
To select sequences specifically recognizing GSCs, counterselection against differentiated GBM cells was performed before the selection step (as previously described). The pool was first incubated for 30 min (one time for rounds 1–13 and two times in the last three rounds) with 106 GSCs grown under adherent conditions (150-mm cell plate), and then unbound sequences were recovered for the selection phase.
During the selection process, we increased the number of counterselections or of washings and decreased incubation time to progressively raise the SELEX selective pressure. The use of polyinosinic acid (Sigma-Aldrich) as a competitor was introduced to minimize non-specific binding. At the end of SELEX, sequences of the pools were cloned with TOPO-TA cloning kit (Thermo Fisher Scientific, Milan, Italy) before sequencing. Afterward, they were compared by Clustal, and their structure predictions were obtained with RNAstructure v.5.7 software, RNAcomposer12 and RNAfold (http://rna.tbi.univie.ac.at/).
Immortalized Cell Culture
Human U251 and U87MG cell lines were purchased from the ATCC (LG Standards, Milan, Italy). Cells were grown in DMEM (Sigma-Aldrich, Milan, Italy) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich, Milan, Italy).
Binding and Internalization Analysis
Binding assays were performed as described.32 The amount of bound RNAs was determined by performing qRT-PCR as reported or by performing Custom TaqMan Small RNA Assays (Thermo Fisher Scientific) for aptamer sequences, following the manufacturer’s recommendations. 2 × 105 cells were treated with 200 nM individual aptamers (or the starting pool as a negative control) for 30 min at 37°C in the presence of 100 μg/mL polyinosine (Sigma-Aldrich) used as a nonspecific competitor. Following two washes with PBS to remove unbound RNA, bound RNA was recovered by TRIzol (Thermo Fisher Scientific) containing 0.5 pmol/mL non-related aptamer used as a reference control (for each experiment, data were normalized to the reference control). The amount of bound RNA was determined by performing qRT-PCR, as reported, with the following primers for the long sequences: P10 (forward), 5′-TAATACGACTCACTATAGGGAGACAAGAATAAACGCTCAA-3′; P20 (reverse), 5′-GCCTGTTGTGAGCCTCCTGTCGAA-3′. Otherwise, the amount of bound RNA was determined by performing Custom TaqMan Small RNA Assays (Thermo Fisher Scientific) for aptamer sequences, following the manufacturer’s recommendations. To check internalization, cell surface-bound aptamers were removed by washing the cells three times before recovering with PBS/0.5 M NaCl. Internalization rate is expressed as percentage of internalized aptamer compared to total bound aptamer.
Western Blot Analysis
Western blot analysis was performed as described previously.33 After washing cells twice in ice-cold PBS, protein extracts were prepared by incubating cell pellets in JS buffer (50 mM HEPES [pH 7.5] containing 150 mM NaCl, 1% glycerol, 1% Triton X-100, 1.5 mM MgCl2, 5 mM EGTA, 1 mM Na3VO4, and 1X protease inhibitor cocktail). Protein concentrations were determined with Bio-Rad Protein Assay, and equal amounts of proteins were separated by SDS-PAGE (10% polyacrylamide gel). The separated proteins were transferred to nitrocellulose membranes (GE Healthcare, Milan, Italy). Membranes were blocked for 1 h with 5% non-fat dry milk in Tris-buffered saline (TBS) containing 0.1% Tween-20. Primary antibodies were incubated at 4°C overnight, and peroxidase-conjugated secondary antibodies were used to perform an enhanced chemiluminescence (ECL Star, Euroclone, Milan, Italy) reaction, according to the manufacturer’s protocol, in order to identify target proteins. Primary antibodies used were as follows: anti-β3-tubulin, anti-Sox2, anti-GFAP, anti-Nanog, and anti-β-actin (Sigma-Aldrich).
RNA Extraction and Real-Time PCR
Total RNA (microRNA and mRNA) were extracted using EuroGOLDTriFast (EuroClone, Milan, Italy), according to the manufacturer’s protocol. All the RNAs were reverse transcribed as described by Iaboni and colleagues.25 Reverse transcription of total microRNA was performed starting from equal amounts of total RNA per sample (500 ng) using miScript reverse Transcription Kit (QIAGEN, Hilden, Germany). Quantitative analysis of microRNAs and RNU6B (as an internal reference) was performed by real-time PCR using specific primers (QIAGEN) and miScript SYBR Green PCR Kit (QIAGEN Hilden, Germany). All reactions were run in duplicate.
Immunofluorescence Analysis
Assay was performed as previously described.34 Spheres were mechanically disaggregated and cells were treated with Alexa488-A40s or Alexa488-unrelated aptamer (scrambled) at 37°C for 30 min. Subsequently, cells were washed two times with PBS and forced to adhere to polylysine-coated glass coverslips for 15 min. Alternatively, differentiated cells were directly seeded on coverslips and treated with 500 nM Alexa488-A40s or Alexa488-unrelated aptamer (scrambled) at 37°C for 30 min. In both cases, cells were fixed with 4% paraformaldehyde in PBS for 20 min at room temperature. Coverslips were washed three times in PBS, mounted with Invitrogen Gold antifade reagent with DAPI (Thermo Fisher Scientific), and finally visualized by confocal microscopy (LSM700, Zeiss, Milan, Italy).
Aptamer-miRNA Chimera
2′-fluoropyrimidine RNAs synthesized by TriLink Biotechnologies (San Diego, CA, USA) were used for chimera production according to previously described protocols.26 The sequences used for the chimera conjugates were as follows: miR-34c passenger strand sticky, 5′-ACU AGGCAGUGUAGUUAGCUGAUUGC2′OMe(GG)CU2′OMe(A)UCU2′OMe(AGAA)U2′OMe(G)U2′OMe(A)C-3′; miR-34c guide strand, 5′-AAUCACUAACCACACGGCCAGG-3′; antimiR-10b sticky (indicated as anti10b), 5′-CACAAAUUCGGUUCUACAGGGUA2′OMe(GG)CU2′OMe(A)UCU2′OMe(AGAA)U2′OMe(G)U2′OMe(A)C-3′; and scrambled sticky, 5′-UUCGUACCGGGUAGGUUGGCUUGCACAUAGAACGUGUCA2′OMe(G)U2′OMe(A)C 2′OMe(A)UUCU2′OMe(AGA)U2′OMe(AG)CC-3′. All RNAs have 2′-fluoropyrimidine. For the A40s-34c chimera, the negative control was made from the unannealed single portions of the chimera (miR-34c guide strand, miR-34c passenger strand, and sticky and not sticky A40s). To prepare A40s/miR-34c, 10 μM passenger RNA strand and 10 μM guide strand, in the appropriate 10× binding buffer (200 mM N-2-Hydroxyethylpiperazine-N’-2-Ethanesulfonic Acid [pH 7.4], 1.5 M NaCl, and 20 mM CaCl2), were first denatured at 95°C for 15 min, subsequently brought to 55°C for 10 min, and finally warmed up to 37°C for 20 min. The annealed passenger and guide strand thereby obtained was combined with sticky A40s and kept 30 min at 37°C. For the A40s-anti-miR-10b chimera, the negative control was made by using a scrambled sticky aptamer in place of sticky A40s. To prepare A40s-anti-miR-10b and scrambled-anti-10b chimera, aptamers sticky and anti-10b sticky were annealed for 30 min at 37°C in 10× binding buffer.
Determination of Binding Kinetic
Binding kinetic for the A40s-GSC complex was determined by performing Custom TaqMan Small RNA Assays (Thermo Fisher Scientific). Fitting curves were designed by using GraphPad Prism 6 software.
Immunohistochemistry
For immunostaining of human tissues, paraffin-immersed tissue slices were prepared using a standard procedure. Adjacent sections were stained with H&E dye and/or antibodies. Nonspecific binding of the antibodies was blocked by incubation of the sections with 10% BSA followed by incubation with primary anti-CD-133 and then staining with secondary antibody (2 ng μL−1 goat anti-rabbit labeled with fluorescein amidites [FAMs]). Biotinylated A40s aptamer was linked with Cy5-labeled streptavidin. Nonspecific binding of the aptamers with tissue slices was blocked with yeast tRNA (1 ng μL−1) (Sigma-Aldrich, USA) and then incubated with 100 nM Cy5-labeled aptamers in Dulbecco’s PBS (DPBS) for 2 h at 6°С. Scrambled aptamer was used as a control oligonucleotide. Bio Mount mounting medium (Bio-Optica, Italy) was used to fix the sections. The sections were analyzed with a laser-scanning fluorescence microscope (Carl Zeiss LSM780).
Statistical Analysis
Continuous variables are given as mean ± 1 SD. Statistical values were defined using GraphPad Prism 6 (San Diego, CA, USA) software, by Student’s t test (two variables) or one-way ANOVA (more than two variables). p value ≤ 0.05 was considered significant for all analyses.
Ethics Statement
Informed consents were obtained from all the patients according to the protocols approved by the Ethical Committee of the Catholic University School of Medicine. Experiments involving animals were approved by the Ethical Committee of the Instituto Superiore di Sanitá and by the Ministry of Health (Aut. N. 259/2017).
Author Contributions
Experiment Design and Performance, C.Q. and A.A. (selection and bindings) and A.A. (all the other experiments); Technical Support, C.V.; Data Interpretation, S.N., A.A., C.Q., C.L.E., and G.R.; Cellular Samples, L.R.-V., G.D.L., and R.P.; Immunofluorescence on Tissue Samples, A.S.K. and I.N.L.; Manuscript Preparation, A.A., G.C., and V.d.F.; Figure Assembly, A.A. and S.N.; Financial Support, G.C. All the authors reviewed the manuscript.
Conflicts of Interest
All authors have read and approved the manuscript, its contents, and its submission and disclose no potential conflicts of interest.
Acknowledgments
This work was partially supported by Associazione Italiana Ricerca sul Cancro (AIRC) (IG 2013 N.14046 and IG 2016 N.18473 to G.C., IG 2014 N.15584 to L.R.-V., and N.13345 and 9980 to V.d.F.); Fondazione Berlucchi to G.C.; Italian Ministry of Health GR-2011-02352546 to C.L.E.; MSCA IF N. 838988 to C.Q.; and POR Campania FESR 2014-2020 “SATIN” to G.C. G.R. is a recipient of a Fondazione Umberto Veronesi individual fellowship. We are thankful to Andrey Narodov (V.F. Voino-Yasenecki, Krasnoyarsk, Russia) for providing tissue samples for immunofluorescence and Fortunato Moscato and Deborah Rotoli (CNR-IEOS, Naples, Italy) for technical support.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.08.015.
Contributor Information
Cristina Quintavalle, Email: cristina.quintavalle@gmail.com.
Gerolama Condorelli, Email: gecondor@unina.it.
Supplemental Information
References
- 1.Louis D.N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W.K., Ohgaki H., Wiestler O.D., Kleihues P., Ellison D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.Sant M., Minicozzi P., Lagorio S., Børge Johannesen T., Marcos-Gragera R., Francisci S., EUROCARE Working Group Survival of European patients with central nervous system tumors. Int. J. Cancer. 2012;131:173–185. doi: 10.1002/ijc.26335. [DOI] [PubMed] [Google Scholar]
- 3.Bao S., Wu Q., McLendon R.E., Hao Y., Shi Q., Hjelmeland A.B., Dewhirst M.W., Bigner D.D., Rich J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 4.Bovenberg M.S., Degeling M.H., Tannous B.A. Advances in stem cell therapy against gliomas. Trends Mol. Med. 2013;19:281–291. doi: 10.1016/j.molmed.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Wang K., Wu X., Wang J., Huang J. Cancer stem cell theory: therapeutic implications for nanomedicine. Int. J. Nanomedicine. 2013;8:899–908. doi: 10.2147/IJN.S38641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 7.Cheng C., Chen Y.H., Lennox K.A., Behlke M.A., Davidson B.L. In vivo SELEX for Identification of Brain-penetrating Aptamers. Mol. Ther. Nucleic Acids. 2013;2:e67. doi: 10.1038/mtna.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Catuogno S., Esposito C.L., de Franciscis V. Developing Aptamers by Cell-Based SELEX. Methods Mol. Biol. 2016;1380:33–46. doi: 10.1007/978-1-4939-3197-2_3. [DOI] [PubMed] [Google Scholar]
- 9.Fitzwater T., Polisky B. A SELEX primer. Methods Enzymol. 1996;267:275–301. doi: 10.1016/s0076-6879(96)67019-0. [DOI] [PubMed] [Google Scholar]
- 10.Pallini R., Ricci-Vitiani L., Banna G.L., Signore M., Lombardi D., Todaro M., Stassi G., Martini M., Maira G., Larocca L.M., De Maria R. Cancer stem cell analysis and clinical outcome in patients with glioblastoma multiforme. Clin. Cancer Res. 2008;14:8205–8212. doi: 10.1158/1078-0432.CCR-08-0644. [DOI] [PubMed] [Google Scholar]
- 11.Adamo A., Fiore D., De Martino F., Roscigno G., Affinito A., Donnarumma E., Puoti I., Ricci Vitiani L., Pallini R., Quintavalle C., Condorelli G. RYK promotes the stemness of glioblastoma cells via the WNT/ β-catenin pathway. Oncotarget. 2017;8:13476–13487. doi: 10.18632/oncotarget.14564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Popenda M., Szachniuk M., Antczak M., Purzycka K.J., Lukasiak P., Bartol N., Blazewicz J., Adamiak R.W. Automated 3D structure composition for large RNAs. Nucleic Acids Res. 2012;40:e112. doi: 10.1093/nar/gks339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mercier M.C., Dontenwill M., Choulier L. Selection of Nucleic Acid Aptamers Targeting Tumor Cell-Surface Protein Biomarkers. Cancers (Basel) 2017;9:E69. doi: 10.3390/cancers9060069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lao Y.H., Phua K.K., Leong K.W. Aptamer nanomedicine for cancer therapeutics: barriers and potential for translation. ACS Nano. 2015;9:2235–2254. doi: 10.1021/nn507494p. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y., Hong H., Cai W. Tumor-targeted drug delivery with aptamers. Curr. Med. Chem. 2011;18:4185–4194. doi: 10.2174/092986711797189547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Catuogno S., Esposito C.L., de Franciscis V. Aptamer-Mediated Targeted Delivery of Therapeutics: An Update. Pharmaceuticals (Basel) 2016;9:E69. doi: 10.3390/ph9040069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iaboni M., Russo V., Fontanella R., Roscigno G., Fiore D., Donnarumma E., Esposito C.L., Quintavalle C., Giangrande P.H., de Franciscis V., Condorelli G. Aptamer-miRNA-212 Conjugate Sensitizes NSCLC Cells to TRAIL. Mol. Ther. Nucleic Acids. 2016;5:e289. doi: 10.1038/mtna.2016.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esposito C.L., Catuogno S., Condorelli G., Ungaro P., de Franciscis V. Aptamer Chimeras for Therapeutic Delivery: The Challenging Perspectives. Genes (Basel) 2018;9:E529. doi: 10.3390/genes9110529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Catuogno S., Esposito C.L., Condorelli G., de Franciscis V. Nucleic acids delivering nucleic acids. Adv. Drug Deliv. Rev. 2018;134:79–93. doi: 10.1016/j.addr.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 20.Russo V., Paciocco A., Affinito A., Roscigno G., Fiore D., Palma F., Galasso M., Volinia S., Fiorelli A., Esposito C.L. Aptamer-miR-34c Conjugate Affects Cell Proliferation of Non-Small-Cell Lung Cancer Cells. Mol. Ther. Nucleic Acids. 2018;13:334–346. doi: 10.1016/j.omtn.2018.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esposito C.L., Cerchia L., Catuogno S., De Vita G., Dassie J.P., Santamaria G., Swiderski P., Condorelli G., Giangrande P.H., de Franciscis V. Multifunctional aptamer-miRNA conjugates for targeted cancer therapy. Mol. Ther. 2014;22:1151–1163. doi: 10.1038/mt.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sasmita A.O., Wong Y.P., Ling A.P.K. Biomarkers and therapeutic advances in glioblastoma multiforme. Asia Pac. J. Clin. Oncol. 2018;14:40–51. doi: 10.1111/ajco.12756. [DOI] [PubMed] [Google Scholar]
- 23.Singh S.K., Clarke I.D., Terasaki M., Bonn V.E., Hawkins C., Squire J., Dirks P.B. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 24.Jayasena S.D. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- 25.Romanelli A., Affinito A., Avitabile C., Catuogno S., Ceriotti P., Iaboni M., Modica J., Condorelli G., Catalucci D. An anti-PDGFRβ aptamer for selective delivery of small therapeutic peptide to cardiac cells. PLoS ONE. 2018;13:e0193392. doi: 10.1371/journal.pone.0193392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Esposito C.L., Nuzzo S., Kumar S.A., Rienzo A., Lawrence C.L., Pallini R., Shaw L., Alder J.E., Ricci-Vitiani L., Catuogno S., de Franciscis V. A combined microRNA-based targeted therapeutic approach to eradicate glioblastoma stem-like cells. J. Control. Release. 2016;238:43–57. doi: 10.1016/j.jconrel.2016.07.032. [DOI] [PubMed] [Google Scholar]
- 27.Kim Y., Wu Q., Hamerlik P., Hitomi M., Sloan A.E., Barnett G.H., Weil R.J., Leahy P., Hjelmeland A.B., Rich J.N. Aptamer identification of brain tumor-initiating cells. Cancer Res. 2013;73:4923–4936. doi: 10.1158/0008-5472.CAN-12-4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abbott N.J., Romero I.A. Transporting therapeutics across the blood-brain barrier. Mol. Med. Today. 1996;2:106–113. doi: 10.1016/1357-4310(96)88720-x. [DOI] [PubMed] [Google Scholar]
- 29.Azad T.D., Pan J., Connolly I.D., Remington A., Wilson C.M., Grant G.A. Therapeutic strategies to improve drug delivery across the blood-brain barrier. Neurosurg. Focus. 2015;38:E9. doi: 10.3171/2014.12.FOCUS14758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monaco I., Camorani S., Colecchia D., Locatelli E., Calandro P., Oudin A., Niclou S., Arra C., Chiariello M., Cerchia L., Comes Franchini M. Aptamer Functionalization of Nanosystems for Glioblastoma Targeting through the Blood-Brain Barrier. J. Med. Chem. 2017;60:4510–4516. doi: 10.1021/acs.jmedchem.7b00527. [DOI] [PubMed] [Google Scholar]
- 31.Ricci-Vitiani L., Pallini R., Biffoni M., Todaro M., Invernici G., Cenci T., Maira G., Parati E.A., Stassi G., Larocca L.M., De Maria R. Tumour vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature. 2010;468:824–828. doi: 10.1038/nature09557. [DOI] [PubMed] [Google Scholar]
- 32.Catuogno S., Rienzo A., Di Vito A., Esposito C.L., de Franciscis V. Selective delivery of therapeutic single strand antimiRs by aptamer-based conjugates. J. Control. Release. 2015;210:147–159. doi: 10.1016/j.jconrel.2015.05.276. [DOI] [PubMed] [Google Scholar]
- 33.Roscigno G., Puoti I., Giordano I., Donnarumma E., Russo V., Affinito A., Adamo A., Quintavalle C., Todaro M., Vivanco M.D., Condorelli G. MiR-24 induces chemotherapy resistance and hypoxic advantage in breast cancer. Oncotarget. 2017;8:19507–19521. doi: 10.18632/oncotarget.14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iaboni M., Fontanella R., Rienzo A., Capuozzo M., Nuzzo S., Santamaria G., Catuogno S., Condorelli G., de Franciscis V., Esposito C.L. Targeting Insulin Receptor with a Novel Internalizing Aptamer. Mol. Ther. Nucleic Acids. 2016;5:e365. doi: 10.1038/mtna.2016.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.