Abstract
Here we describe the basic characteristics of Corynebacterium pacaense strain Marseille-P2417T (= CSUR P2417), Alistipes megaguti strain Marseille-P5997T (= CSUR P5997) and Alistipes provencensis strain Marseille-P2431T (= CSUR P2431 = DSM 102308). The phenotypic criteria, the 16S ribosomal RNA sequencing and MALDI-TOF MS spectra analysis were used to identify and characterize these new bacteria species, which were isolated from fresh human stool specimens.
Keywords: Alistipes megaguti, Alistipes provencensis, Corynebacterium pacaense, culturomics, taxonogenomics
Introduction
Culturomics is the concept of developing different culture conditions in order to enlarge our knowledge of the human microbiota through the discovery of previously uncultured bacteria [1], [2], [3], [4]. However, after bacterial strain isolation, we used a taxonogenomics approach, including MALDI-TOF MS, phylogenetic analysis, main phenotypic description and genome sequencing, to describe the bacterium [5], [6]. In this study, we describe three new species isolated for the first time from human stool samples.
Isolation and growth conditions
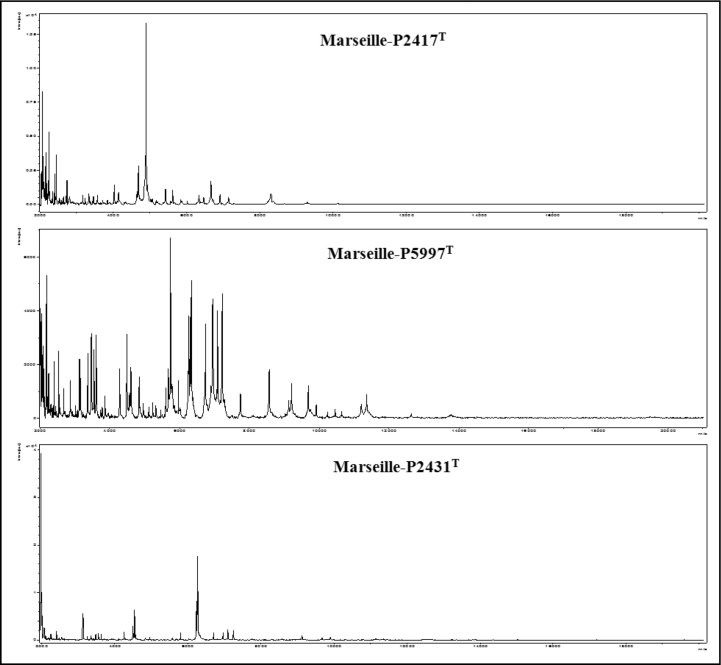
In 2016, strain Marseille-P2417T was isolated from a fresh stool sample of a 24-year-old healthy woman. In 2017, strain Marseille-P5997T was isolated from a fresh stool sample of a 25-year-old healthy woman. In 2015, strain Marseille-P2431Twas isolated from a 66-year-old man with hypertension and diabetes. Screening was performed by MALDI-TOF MS on a Microflex LT spectrometer (Bruker Daltonics, Bremen, Germany) as previously described [7]. The spectra generated from each strain (Fig. 1) were imported and analysed by Biotyper 3.0 software against the Bruker database, which is constantly updated with the Microbes Evolution Phylogeny and Infections (MEPHI) database.
Fig. 1.
MALDI-TOF MS reference mass spectrum. Spectra from 12 individual colonies were compared and reference spectrum generated.
The initial growth of strain Marseille-P2417T was obtained after 24 hours of culture in Colombia agar enriched with 5% sheep's blood (bioMérieux, Marcy l’Etoile, France) in aerobic conditions at 37°C and pH 7.5. The initial growth of strains Marseille-P5997T and Marseille-P2431T was obtained after 48 hours of culture in a Colombia agar enriched with 5% sheep's blood (bioMérieux) in anaerobic conditions at 37°C and pH 7.5.
The study of these microbes (strains Marseille-P2417, Marseille-P5997 and Marseille-P2431) was validated by the ethics committee of IHU–Méditerranée Infection under numbers 2016-010, 2016-011 and 09-022 respectively.
Strain identification
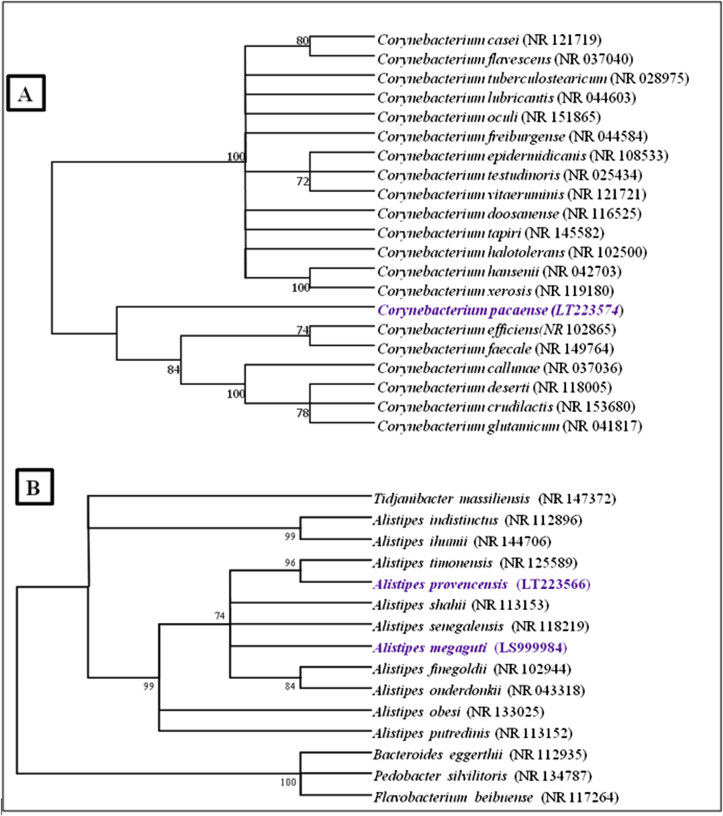
The 16S ribosomal RNA (rRNA) gene was sequenced in order to classify these bacteria. Amplification was performed using the primers fD1, rP2 and rpoB (Eurogentec, Angers, France) and was sequenced using the Big Dye Terminator v1.1 Cycle Sequencing Kit and the sequencer (Thermo Fisher Scientific, Waltham, MA, USA), as previously described [8]. The 16S rRNA nucleotide sequences were assembled and corrected using CodonCode Aligner software (https://www.codoncode.com/). The 16S rRNA gene (accession no. LT223574) of strain Marseille-P2417T exhibited a 97.7% sequence similarity with Corynebacterium efficiens strain YS-314T (GenBank accession no. NR_102865), the phylogenetically closest species with standing in nomenclature (Fig. 2(A)). Those of strain Marseille-P5997T (accession no. LS999984) and strain Marseille-P2431T (accession no. LT223566) showed a sequence similarity of 96.9% and 98.5% with Alistipes senegalensis strain JC50T (GenBank accession no. NR_118219) and Alistipes timonensis strain JC136T (GenBank accession no. NR_125589) respectively; these were the phylogenetically closest species with standing in nomenclature (Fig. 2(B)). These values were lower than the 98.7% 16S rRNA gene sequence threshold recommended by Meier-Kolthoff et al. [9] to delineate a new bacterial species without performing DNA–DNA hybridization.
Fig. 2.
Phylogenetic trees showing positions of Corynebacterium pacaense strain Marseille-P2417T (A), Alistipes megaguti strain Marseille-P5997T and Alistipes provencensis strain Marseille-P2431T (B) relative to other phylogenetically close neighbours. Respective GenBank accession numbers for 16S ribosomal RNA genes are indicated in parentheses. Sequences were aligned using Muscle v3.8.31 with default parameters, and phylogenetic inferences were obtained using maximum likelihood method within software. Numbers at nodes are percentages of bootstrap values obtained by repeating analysis 1000 times to generate majority consensus tree. Only bootstrap values > 71% were retained. Scale bar indicates 2% nucleotide sequence divergence.
We consequently proposed to classify strains Marseille-P2417T, Marseille-P5997T and Marseille-P2431T as new species within the genera Corynebacterium and Alistipes respectively.
Phenotypic characteristics
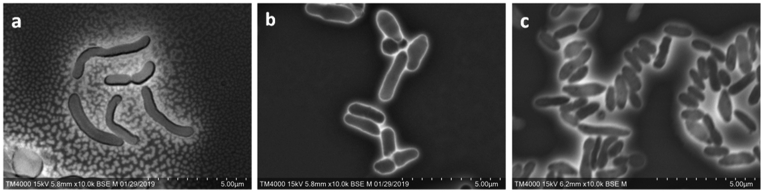
Strain Marseille-P2417T formed circular white colonies with a mean diameter of 1.15 mm. Bacterial cells of strain Marseille-P2417T were Gram-positive and rod shaped, and they ranged in length from 1.29 to 1.5 μm and in width from 0.5 to μm (Fig. 3(a)). Strain Marseille-P2417T showed catalase-positive and oxidase-negative activities. Colonies of the strain Marseille-P5997T were 0.2 to 0.9 mm in diameter on blood-enriched Columbia agar. Bacterial cells of strain Marseille-P5997T were Gram negative and rod shaped, and they ranged in length from 1.36 to 3.57 μm and in width from 0.45 to 0.6 μm (Fig. 3(b)). Strain Marseille-P5997T showed catalase-negative and oxidase-negative activities. Colonies of the strain Marseille-P2431T were 0.4 to 0.64 mm in diameter on blood-enriched Columbia agar. Bacterial cells were Gram negative and rod shaped, and they ranged in length from 0.9 to 1.43 μm and in width from 0.4 to 0.5 μm (Fig. 3(c)). Strain Marseille-P2431T showed catalase-positive and oxidase-negative activities.
Fig. 3.
Scanning electron micrograph of (a) Corynebacterium pacaense, (b) Alistipes megaguti and (c) Alistipes provencensis using TM4000Plus Tabletop microscope (Hitachi, Yokohama, Japan). Scale bar and acquisition settings are shown.
The main phenotypic properties of the three strains were studied by using API 50 CH strips (Table 1), API ZYM strips (Table 2), API Coryne strips (Table 3) and API 20A strips (Table 4). The main characteristics of strains Marseille-P2417T, Marseille-P5997T and Marseille-P2431T are respectively summarized at the Digital Protologue website (http://imedea.uib-csic.es/dprotologue/) under the following numbers: TA00965, TA00955 and TA00966. A comparative study of the main biochemical and phenotypic features of the closest Corynebacterium species and Alistipes species is presented in Table 5.
Table 1.
Biochemical tests of Corynebacterium pacaense, Alistipes megaguti and Alistipes provencensis (API 50 CH strips)
| Test | Result |
||
|---|---|---|---|
| C. pacaense | A. megaguti | A. provencensis | |
| Glycerol | − | + | − |
| Erythrol | − | − | − |
| d-Arabinose | − | − | w |
| l-Arabinose | − | + | − |
| d-Ribose | − | + | − |
| d-Xylose | − | + | + |
| l-Xylose | − | − | w |
| d-Adonitol | − | w | − |
| Methyl-βd-xylopyranoside | − | − | − |
| d-Galactose | − | + | − |
| d-Glucose | + | + | + |
| d-Fructose | + | + | − |
| d-Mannose | + | + | + |
| l-Sorbose | − | − | w |
| l-Rhammose | − | − | w |
| Dulcitol | − | w | − |
| Inositol | w | w | w |
| d-Mannitol | + | + | − |
| d-Sorbitol | − | − | − |
| Methyl-αd-mannopyranoside | − | − | w |
| Methyl-αd-glucopyranoside | − | − | − |
| N-Acetylglucosamine | − | + | − |
| Amygdalin | − | + | − |
| Arbutin | w | + | − |
| Esculin | + | + | + |
| Salicin | + | + | − |
| d-Cellobiose | − | + | − |
| D-Maltose | − | + | − |
| D-Lactose | − | + | + |
| d-Melibiose | − | + | − |
| d-Sucrose | + | − | − |
| d-Trehalose | w | + | − |
| Inulin | − | − | − |
| d-Melezitose | − | − | + |
| d-Raffinose | − | − | + |
| Starch | − | w | − |
| Glycogen | − | − | − |
| Xylitol | − | − | − |
| Gentiobiose | − | + | − |
| d-Turanose | − | − | − |
| d-Lyxose | − | w | − |
| d-Tagatose | − | + | − |
| d-Fucose | − | w | − |
| l-Fucose | − | w | − |
| d-Arabitol | − | − | − |
| l-Arabitol | − | − | w |
| Potassium gluconate | − | − | − |
| Potassium 2-ketogluconate | − | − | − |
| Potassium 5-ketogluconate | − | w | w |
+, positive result; −, negative result; w, weakly positive result.
Table 2.
Biochemical tests of Corynebacterium pacaense, Alistipes megaguti and Alistipes provencensis (API ZYM strips)
| Test | Result |
||
|---|---|---|---|
| Corynebacterium pacaense | Alistipes megaguti | Alistipes provencensis | |
| Alkaline phosphatase | − | − | − |
| Esterase (C4) | + | + | + |
| Esterase lipase (C8) | − | + | − |
| Lipase (C14) | − | + | + |
| Leucine arylamidase | + | − | − |
| Valine arylamidase | − | + | + |
| Cystine arylamidase | − | + | − |
| Trypsin | − | + | + |
| α-Chymotrypsin | − | + | − |
| Phosphatase acid | + | + | − |
| Naphthol-AS-BI-phosphohydrolase | + | + | + |
| α-Galactosidase | − | − | − |
| β-Galactosidase | − | + | + |
| β-Glucuronidase | − | + | + |
| α-Glucosidase | − | − | + |
| β-Glucosidase | + | − | + |
| N-Acetyl-β-glucosaminidase | − | − | − |
| α-Mannosidase | − | + | − |
| α-Fucosidase | − | + | − |
+, positive result; −, negative result.
Table 3.
Biochemical tests of Corynebacterium pacaense (API Coryne strips)
| Test | Result |
|---|---|
| Potassium nitrate | − |
| Pyrazine carboxamide | − |
| Pyroglutamic acid–β-naphthylamide | − |
| 2-Naphtyl-phosphate | − |
| Naphthol-ASBI-glucuronic acid | − |
| 2-Naphtyl-βd-galactopyranoside | − |
| 2-Naphtyl-αd-glucopyranoside | − |
| 1-Naphtyl-N-acetyl-βd-glucosaminide | − |
| Esculin ferric citrate | + |
| Urea | − |
| Gelatin (bovine origin) | − |
| d-Glucose | + |
| d-Ribose | − |
| d-Xylose | − |
| d-Mannitol | + |
| d-Maltose | − |
| d-Lactose (bovine origin) | − |
| d-Sucrose | + |
| Glycogen | − |
+, positive result; −, negative result.
Table 4.
Biochemical tests of Alistipes megaguti and Alistipes provencensis (API 20A strips)
| Test | Result |
|
|---|---|---|
| Alistipes megaguti | Alistipes provencensis | |
| l-Tryptophane | + | − |
| Urea | − | − |
| d-Glucose | + | + |
| d-Mannitol | + | − |
| d-Lactose | + | + |
| d-Saccharose | − | − |
| d-Maltose | + | + |
| Salicin | + | − |
| d-Xylose | + | + |
| l-Arabinose | + | − |
| Gelatin (bovine origin) | + | − |
| Esculin ferric citrate | + | + |
| Glycerol | − | − |
| d-Cellobiose | + | + |
| d-Mannose | + | + |
| d-Melezitose | − | + |
| d-Raffinose | − | + |
| d-Sorbitol | − | − |
| l-Rhamnose | + | − |
| d-Trehalose | + | − |
+, positive result; −, negative result.
Table 5.
Differential characteristics of Corynebacterium pacaense sp. nov. (1), Corynebacterium efficiens (2), Corynebacterium humireducens (3), Alistipes megaguti sp. nov. (4), Alistipes provencensis sp. nov. (5), Alistipes ihumii (6), Alistipes senegalensis (7) and Alistipes timonensis (8)
| Property | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| Cell diameter (μm) | 0.5–0.8 | 0.8–1.1 | 0.5–0.7 | 0.45–0.6 | 0.4–0.5 | 0.72 | 0.56 | 0.62 |
| Oxygen requirement | Facultatively anaerobic | Facultatively anaerobic | Facultatively anaerobic | Anaerobic | Anaerobic | Anaerobic | Anaerobic | Anaerobic |
| Gram stain | + | + | + | − | − | − | − | − |
| Motility | − | − | − | − | − | − | − | − |
| Endospore formation | − | − | − | − | − | − | − | − |
| Production of: | ||||||||
| Alkaline phosphatase | − | − | + | − | − | NA | NA | NA |
| Catalase | + | + | + | − | + | − | + | + |
| Oxidase | − | − | − | − | − | + | − | − |
| Urease | − | v | − | − | − | − | NA | NA |
| β-Galactosidase | − | − | − | − | − | − | w | + |
| N-Acetyl-glucosamine | − | − | − | − | − | + | NA | w |
| Acid from: | ||||||||
| l-Arabinose | − | − | − | + | − | NA | NA | NA |
| Ribose | − | + | + | + | − | NA | NA | NA |
| Mannose | + | + | + | + | + | + | + | − |
| Mannitol | + | − | − | + | − | NA | NA | NA |
| Sucrose | + | + | + | − | − | NA | NA | NA |
| d-Glucose | + | + | + | + | + | NA | NA | NA |
| d-Fructose | + | + | − | + | − | NA | NA | NA |
| d-Maltose | − | + | + | + | − | NA | NA | NA |
| d-Lactose | − | − | − | + | + | NA | NA | NA |
| G+C content (mol%) | 63.7 | 59.0 | 59.0 | 58.6 | 58.3 | 57.9 | 58.4 | 58.8 |
| Source | Human gut | Soil | Microbial fuel cell | Human gut | Human gut | Human gut | Human gut | Human gut |
+, positive result; −, negative result; w, weakly positive result; NA, data not available.
Genome sequencing
Genomic DNA was extracted using the EZ1 biorobot (Qiagen, Courtaboeuf, France) with the EZ1 DNA tissue kit, then was sequenced using with the), as previously described [10]. The assembly was performed with a pipeline incorporating different softwares (Velvet [11], Spades [12] and Soap Denovo [13], and trimmed (MiSeq and Trimmomatic [14] softwares) or untrimmed data (only MiSeq software). GapCloser was used to reduce assembly gaps. Scaffolds <800 bp and scaffolds with a depth value lower than 25% of the mean depth was removed. The best assembly was selected by using different criteria (number of scaffolds, N50, number of N).
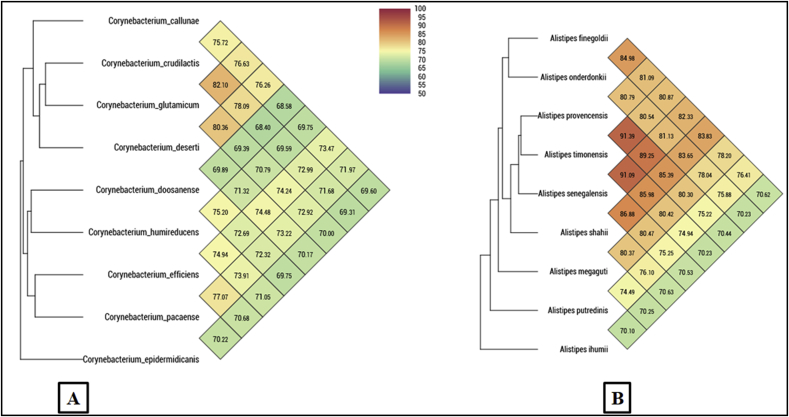
The genome of strain Marseille-P2417T was 3 030 201 bp long (13 scaffolds, 16 N50) with a 63.75 mol% G+C content and contains 2820 predicted genes. The degree of genomic similarity of Marseille-P2417T with closely related species was estimated using the OrthoANI software [15]. Values among closely related species (Fig. 4(A)) ranged from 68.40% between Corynebacterium crudilactis and Corynebacterium doosanense to 82.10% between C. crudilactis and Corynebacterium gultamicum. When the isolate was compared to these closely related species, values ranged from 70.22% with Corynebacterium epidermidicanis to 77.07% with Corynebacterium efficiens. Likewise, the genomes of strains Marseille-P5997T and Marseille-P2431T measured about 3 270 862 and 3 805 103 bp long respectively. They contain 2727 and 2973 predicted genes, with 58.6 and 58.3 mol% G+C content respectively. A genomic similarity analysis of Alistipes species was performed by OrthoANI software. Values among closely related species (Fig. 4(B)) ranged from 70.10% between Alistipes putredinis and Alistipes ihumii to 91.39% between Alistipes provencensis and Alistipes timonensis. When Marseille-P5997T was compared to these closely related species, values ranged from 70.25% with Alistipes ihumii to 80.47% with Alistipes senegalensis. When Marseille-P2431T was compared to these closely related species, values ranged from 70.44% with Alistipes ihumii to 91.39% with Alistipes timonensis.
Fig. 4.
Heat maps generated with OrthoANI values calculated using OAT software for Corynebacterium pacaense sp. nov. (A), Alistipes megaguti sp. nov., and Alistipes provencensis sp. nov. (B) with other closely related species with standing in nomenclature.
Conclusion
Strains Marseille-P2417T, Marseille-P5997T and Marseille-P2431T exhibited a 16S rRNA sequence similarity of <98.65% and an OrthoANI value < 95% with its phylogenetically closest species with standing in nomenclature, and we consequently propose them to be the type strains of the new Corynebacterium pacaense sp. nov., Alistipes megaguti sp. nov. and Alistipes provencensis sp. nov. respectively.
Description of Corynebacterium pacaense sp. nov.
Corynebacterium pacaense (pa.ca.en'se, L. adj. fem., ‘PACA,’ the name of the region Provence-Alpes-Côte d’Azur, France, where the strain was isolated). Cells were aerobic, Gram positive, rod shaped, nonmotile and non–spore forming, and catalase and oxidase. Cells had a length of 1.29 to 1.5 μm and a width of 0.5 to 0.8 μm. Colonies growing on 5% sheep's blood–enriched Columbia agar (bioMérieux) were circular and white after 72 hours of incubation in aerobic atmosphere at 37°C, and they showed a mean diameter of 1.15 mm. Positive reactions were observed for d-glucose, d-mannitol, d-sucrose, d-fructose, d-mannose, esculin, salicin, esterase (C4), leucine arylamidase, phosphatase acid, naphtalo-AS-BI-phosphohydrolase and β-glucosidase. Negative reactions were obtained with glycerol, d-maltose, d-ribose, d-xylose, glycogen, amygdalin, d-adonitol, d-arabinose, d-arabitol, d-cellobiose, d-fucose, d-galactose, d-lactose, d-lyxose, d-melezitose, d-melibiose, d-raffinose, d-sorbitol, d-tagatose, d-turanose, dulcitol, erythrol, gentiobiose, inulin, l-arabinose, l-arabitol, l-fucose, l-rhammose, l-sorbose, l-xylose, methyl-αd-glucopyranoside, methyl-αd-mannopyranoside, methyl-βd-xylopyranoside, N-acetylglucosamine, potassium 2-ketogluconate, potassium 5-ketogluconate, potassium gluconate, starch, xylitol, alkaline phosphatase, esterase lipase (C8), lipase (C14), valine arylamidase, cystine arylamidase, trypsin, α-chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase and α-fucosidase.
The genome of strain Marseille-P2417T was 3 030 201 bp long, and its G+C content was 63.75 mol%. The strain Marseille-P2417T was isolated from a fresh stool sample from a 24-year-old healthy woman. It was deposited in the Collection de Souches de l’Unité des Rickettsies (CSUR) collection under accession number CSUR P2417. The 16S rRNA, rpoB genes and genome sequences are available in GenBank under accession numbers LT223574, LT223580 and FWCI00000000 respectively.
Description of Alistipes megaguti sp. nov.
Alistipes megaguti (mega.gu'ti, N.L. masc. adj. megaguti, ‘megagut,’ the name of a project aiming to individually cultivate all species from the digestive tract of healthy humans). Cells were anaerobic, Gram-negative, nonmotile and asporogenous rods. Catalase and oxidase activities were negative. Bacterial cells had a length of 1.36 to 3.57 μm and a width of 0.4 to 0.6 μm. Colonies of the strain Marseille-P2431T were 0.2 to 0.9 mm in diameter on blood-enriched Columbia agar. Growth occurred at 37°C under anaerobic conditions. Positive reactions were observed for esterase (C4), esterase lipase (C8), cystine arylamidase, trypsin, α-chymotrypsin, phosphatase acid, naphtalo-AS-BI-phosphohydrolase, β-galactosodase, α-mannosidase, l-arabinose, d-xylose, glucose, fructose, mannose, d-mannitol, esculin, d-maltose, d-lactose, d-tagatose and α-fucosidase. Negative reactions were observed for alkaline phosphatase, leucine arylamidase, α-galactosidase, α-glucosidase, β-glucosidase, N-acetyl-β glucosaminidase, d-arabinose, l-xylose, methyl β-d-xylopyranoside, l-rhammose, d-sorbitol, methyl α-d-mannopyranoside, sucrose, glycogen and potassium 2-ketogluconate. The most abundant fatty acid by far was hexadecanoic acid (53%), followed by 13-methyltetradecanoic acid (16%) and 9-octadecenoic acid (8%).
The genome of strain Marseille-P5997T was 3 270 862 bp long, and its G+C content was 58.6 mol%. Strain Marseille-P5997T was isolated from a fresh stool sample from a 25-year-old healthy woman and was deposited in the CSUR collection under accession number CSUR P5997. The 16S rRNA and genome sequences are available in GenBank under accession numbers LS999984 and LR027382 respectively.
Description of Alistipes provencensis sp. nov.
Alistipes provencensis (pro.ven.cen'sis, N.L. adj. neut., from Provence, the region in France, where the strain was isolated). Cells were anaerobic, Gram-negative, nonmotile, asporogenous rods that were catalase positive and oxidase negative. Cells had a length of 0.9 to 1.43 μm and a width of 0.4 to 0.5 μm. Colonies of the strain Marseille-P2431T were 0.4 to 0.64 mm in diameter on blood-enriched Columbia agar. Growth occurred at 37°C under anaerobic conditions. Positive reactions were observed for esterase (C4), lipase (C14), β-galactosodase, β-glucuronidase, α-glucosidase, β-glucosidase, d-xylose, d-glucose, d-mannose, esculin, d-lactose, d-melezitose and d-raffinose. Negative reactions were observed with alkaline phosphatase, esterase lipase (C8), α-chymotrypsin, phosphatase acid, α-galactosidase, N-acetyl-β-glucosaminidase, glycerol, erythrol, l-arabinose, d-ribose, d-galactose, d-fructose, dulcitol, d-mannitol, amygdalin, arbutin, d-cellobiose, d-maltose, d-sucrose, d-trehalose, inulin, starch, glycogen, d-fucose and potassium 2-ketogluconate.
The genome of strain Marseille-P2431T was 3 805 103 bp long, and its G+C content was 58.3 mol%. Strain Marseille-P2431T was isolated from a 66-year-old man with diabetes and hypertension, and was deposited in the CSUR collection under accession number CSUR P2431. The 16S rRNA and genome sequences are available in GenBank under accession numbers LT223566 and FKYL00000000 respectively.
Nucleotide sequence accession number
The 16S rRNA gene, the rpoB gene and genome sequences of strain Marseille-P2417T were deposited in GenBank under accession numbers LT223574, LT223580 and FWCI00000000 respectively. The 16S rRNA gene and genome sequences of strain Marseille-P5997T and strain Marseille-P2431T were deposited in GenBank under accession numbers LS999984 and LT223566 respectively, and NR027382 and FKYL00000000 respectively.
Deposit in culture collection
Strain Marseille-P2417T, strain Marseille-P5997T and strain Marseille-P2431T were respectively deposited in the CSUR collection under the following numbers: CSUR 2417T, CSUR P5997T and CSUR 2431T = DSM 102308.
Acknowledgements
This work has received financial support from the French Government through the Agence Nationale pour la Recherche, including the “Programme d'Investissement d'Avenir” under the reference Méditerranée Infection 10-IAHU-03. This work was supported by the Région Provence-Alpes-Côte d'Azur and European funding FEDER PRIMI.
Conflict of Interest
None declared.
References
- 1.Lagier J.C., Armougom F., Million M., Hugon P., Pagnier I., Robert C. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012;18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 2.Lagier J.C., Hugon P., Khelaifia S., Fournier P.E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lagier J.C., Khelaifia S., Alou M.T., Ndongo S., Dione N., Hugon P. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Lagier J.C., Edouard S., Pagnier I., Mediannikov O., Drancourt M., Raoult D. Current and past strategies for bacterial culture in clinical microbiology. Clin Microbiol Rev. 2015;28:208–236. doi: 10.1128/CMR.00110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fournier P.E., Lagier J.C., Dubourg G., Raoult D. From culturomics to taxonomogenomics: a need to change the taxonomy of prokaryotes in clinical microbiology. Anaerobe. 2015;36:73–78. doi: 10.1016/j.anaerobe.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 6.Ramasamy D., Mishra A.K., Lagier J.C., Padhmanabhan R., Rossi M., Sentausa E. A polyphasic strategy incorporating genomic data for the taxonomic description of novel bacterial species. Int J Syst Evol Microbiol. 2014;64:384–391. doi: 10.1099/ijs.0.057091-0. [DOI] [PubMed] [Google Scholar]
- 7.Seng P., Drancourt M., Gouriet F., La Scola B., Fournier P.E., Rolain J.M. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 8.Morel A.S., Dubourg G., Prudent E., Edouard S., Gouriet F., Casalta J.P. Complementarity between targeted real-time specific PCR and conventional broad-range 16S rDNA PCR in the syndrome-driven diagnosis of infectious diseases. Eur J Clin Microbiol Infect Dis. 2015;34:561–570. doi: 10.1007/s10096-014-2263-z. [DOI] [PubMed] [Google Scholar]
- 9.Meier-Kolthoff J.P., Göker M., Spröer C., Klenk H.P. When should a DDH experiment be mandatory in microbial taxonomy? Arch Microbiol. 2013;195:413–418. doi: 10.1007/s00203-013-0888-4. [DOI] [PubMed] [Google Scholar]
- 10.Diop A., Khelaifia S., Armstrong N., Labas N., Fournier P.E., Raoult D., Million M. Microbial culturomics unravels the halophilic microbiota repertoire of table salt: description of Gracilibacillus massiliensis sp. nov. Microb Ecol Health Dis. 2016;27:32049. doi: 10.3402/mehd.v27.32049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J. SOAPdenov02: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee I., Ouk Kim Y., Park S.C., Chun J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016;66:1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]