Abstract
Introduction
Asian Indians have a propensity for premature, severe, and diffuse coronary artery disease (CAD). Several single-nucleotide polymorphisms (SNPs) in the ‘core CAD’ region of the chromosomal region 9p21.3 are known to be strongly associated with CAD.
Objectives
We aimed to study SNPs in the 9p21.3 region associated with CAD and premature CAD and identify their association with demographic and clinical characteristics in an Asian Indian population.
Methods
SNP genotyping was performed for 30 SNPs of the 9p21.3 region using MassARRAY® technology. Along with demographic and SNP data analysis, we also performed multivariate logistic regression analysis and multifactor dimensionality reduction analysis to study SNP–SNP and SNP–demographic/clinical variable interactions.
Results
Our results suggest that females are at a higher risk of premature CAD. We found that SNPs rs1333045 (CC), rs16905599 (AA), rs2383206 (GG), rs2383208 (AG), and rs4977574 (GG) were significantly associated with premature CAD. When adjusted for covariates/confounders, we found that rs2383206 showed the strongest risk association with CAD followed by rs16905599 and rs2383208. Further, SNPs rs1333049 (CC) and rs4977574 (GG) were found to be exclusively associated with premature CAD cases, suggesting their potential as genetic markers for premature CAD in the local population. Upon gender-based stratification, it was found that rs10757272 (TT and TC) is significantly associated with eightfold to ninefold CAD risk specifically among females. SNP rs7865618 (GG) is significantly associated with more than 2.5-fold CAD risk specifically among males.
Conclusion
Our study suggests that SNPs at the 9p21 risk locus may be used to generate a reliable genetic risk score along with markers at other loci.
Keywords: Coronary artery disease, Single-nucleotide polymorphism, Asian Indian population, Genotype, 9p21.3 chromosomal region
1. Introduction
Coronary artery disease (CAD) has reached epidemic proportions in the Asian Indian population. Asian Indians have a propensity for premature, diffuse, and severe CAD.1, 2 Genome-wide association studies starting from 20073, 4, 5, 6, 7 identified the 9p21.3 chromosomal region as being strongly associated with CAD and myocardial infarction (MI). This region has also been associated with stroke,8, 9 aortic,10 abdominal,11 intracranial aneurysms,10 and several types of cancers.12, 13, 14
The chromosomal region 9p21.3 has been strongly associated with CAD in Caucasian, Italian,15 US Hispanic,16 Chinese,17 Japanese,18 Korean,19 Asian Indian,20 and Pakistani21 populations. This region is sparse in genes and also called ‘gene desert’. The nearest genes, about 100 kb upstream of the core CAD region, are two tumor suppressor genes called CDKN2A and CDKN2B that constitute the INK4/ARF locus. This locus encodes cyclin-dependent kinase inhibitors (p16INK4A and p14ARF from CDKN2A and p15INK4B from CDKN2B) that cause arrest of cell cycle in the G1 phase.22, 23 Further away is another gene MTAP that encodes methylthioadenosine phosphorylase, that is involved in the polyamine pathway (salvage of adenine and methionine).24 Most of the single-nucleotide polymorphisms (SNPs) associated with CAD are present in a 58 Kb region called the ‘core CAD’ region. The core CAD region overlaps CDKN2B and also contains a long noncoding RNA of about 126 kb named ANRIL for antisense noncoding RNA in the INK4 locus, hence also called CDKN2B-AS. ANRIL is known to have several linear25, 26, 27 and circular isoforms.27
The association of the 9p21.3 risk locus with CAD is believed to be mediated via ANRIL.28, 29, 30 Genetic variants associated with CAD are located within intronic and 3′ flanking sequences of ANRIL. ANRIL is known to be involved in the regulation of CDKN2A, CDKN2B,25, 30, 31, 32, 33, 34 possibly MTAP, and several other genes involved in the cardiovascular pathway.35 Repression of CDKN2A and CDKN2B is known to cause smooth muscle cell proliferation that occurs at the coronary artery wall during the initial stages of atherosclerosis.36
To understand the association of the 9p21 risk locus with CAD and premature CAD, we aimed to study 30 SNPs in this region, their association with demographic, clinical characteristics, and interferon alpha 21 (IFNA21) levels in an Asian Indian population.
2. Materials and methods
The study was conducted in the south Indian state of Telangana, in the twin cities of Hyderabad and Secunderabad. The study population consisted of 661 individuals including 443 controls and 218 angiographically documented CAD cases (recruited from June 2015 to June 2017). The controls were recruited from various parts of the twin cities of Hyderabad and Secunderabad. We have followed the guidelines of the 1975 Declaration of Helsinki in the recruitment of study subjects, sample, and data collection. The CAD cases were recruited from Krishna Institute of Medical Sciences (KIMS), Secunderabad, after the approval of the Ethics Committee of KIMS Foundation and Research Centre. Written informed consent was taken from all subjects prior to sample and data collection.
2.1. Selection criteria of study subjects
Inclusion criteria for controls include healthy individuals from the age of 40 to 85 years.
Exclusion criteria for controls include individuals with CAD, stroke, and peripheral artery disease.
Inclusion criteria for cases include angiographically documented CAD cases who showed at presentation unstable angina/ST segment elevation myocardial Infarction (STEMI)/Non-ST segment Elevation Myocardial Infarction (NSTEMI), aged 30–85 years.
Exclusion criteria for cases include individuals with liver, kidney, and gastrointestinal disorders and individuals with infectious diseases such as hepatitis, HIV, TB, and so on.
2.2. Methodology
The CAD cases were recruited in consultation with the hospital cardiologist. Peripheral blood sample of 5 ml was taken from each subject and equally dispensed in a vacutainer tube coated with EDTA for plasma and vacutainer tube with clot accelerator for serum separation. DNA extraction from plasma was performed using the kit manufactured by Epicentre (an Illumina company). The DNA samples were estimated qualitatively by electrophoresis on 0.8% agarose gels stained with ethidium bromide, and the bands were viewed in a UV transilluminator. The quantitative estimation of DNA was performed using a Nanodrop Spectrophotometer and taking the ratio of absorbance at 260/280 nm.
The DNA samples were then genotyped for 30 SNPs of the 9p21 region (Table 1). The 9p21 SNPs were chosen based on 2 criteria: (A) extensive literature survey and (B) minor allele frequency (MAF) taken from the dbSNP site of the National Center for Biotechnology Information. We have chosen SNPs having MAF >0.15. The SNP genotyping was performed by the MassARRAY® technology (Sequenom platform) using the AGENA protocol.
Table 1.
SNPs studied in the 9p21.3 region.
| S. No. | SNP ID | Chromosomal positiona | Gene view | Functional consequence | Minor allele and global frequency |
|---|---|---|---|---|---|
| 1 | rs1004638 (T/A) | 22115590 | CDKN2B-AS1 | Intron variant | A- 0.31 |
| 2 | rs10116277 (T/G) | 22081398 | CDKN2B-AS1 | Intron variant | G- 0.323 |
| 3 | rs1011970 (G/T) | 22062135 | CDKN2B-AS1 | Intron variant | T- 0.247 |
| 4 | rs1063192 (A/G) | 22003368 | CDKN2B-AS1 | Intron variant, UTR variant 3′ | G- 0.205 |
| 5 | rs10757272 (C/T) | 22088261 | CDKN2B-AS1 | Intron variant | T- 0.45 |
| 6 | rs10757274 (A/G) | 22096056 | CDKN2B-AS1 | Intron variant | G- 0.404 |
| 7 | rs10757278 (A/G) | 22124478 | Near CDKN2B-AS1 | Intron variant | G- 0.408 |
| 8 | rs10757283 (C/T) | 22134173 | Near CDKN2B-AS1 | Intron variant | T- 0.497 |
| 9 | rs10811661 (T/C) | 22134095 | Near CDKN2B-AS1 | Intron variant | C- 0.176 |
| 10 | rs1333040 (T/C) | 22083405 | CDKN2B-AS1 | Intron variant | C- 0.383 |
| 11 | rs1333042 (G/A) | 22103814 | CDKN2B-AS1 | Intron variant | A- 0.321 |
| 12 | rs1333045 (T/C) | 22119196 | CDKN2B-AS1 | Intron variant | C- 0.498 |
| 13 | rs1333048 (A/C) | 22125348 | Near CDKN2B-AS1 | Intron variant | C- 0.442 |
| 14 | rs1333049 (G/C) | 22125504 | Near CDKN2B-AS1 | Intron variant | C- 0.418 |
| 15 | rs16905599 (G/A) | 22069145 | CDKN2B-AS1 | Intron variant | A- 0.190 |
| 16 | rs2383206 (A/G) | 22115027 | CDKN2B-AS1 | Intron variant | G- 0.487 |
| 17 | rs2383207 (G/A) | 22115960 | CDKN2B-AS1 | Intron variant | A- 0.310 |
| 18 | rs2383208 (A/G) | 22132077 | Near CDKN2B-AS1 | Intron variant | G- 0.210 |
| 19 | rs2811712 (A/G) | 21998036 | CDKN2B-AS1 | Intron variant | G- 0.160 |
| 20 | rs2891169 (A/G) | 22131826 | Near CDKN2B-AS1 | Intron variant | G- 0.493 |
| 21 | rs3731239 (A/G) | 21974219 | CDKN2A | Intron variant | G- 0.175 |
| 22 | rs4977574 (A/G) | 22098575 | CDKN2B-AS1 | Intron variant | G- 0.395 |
| 23 | rs4977756 (A/G) | 22068653 | CDKN2B-AS1 | Intron variant | G- 0.288 |
| 24 | rs564398 (T/C) | 22029548 | CDKN2B-AS1 | Intron variant, nc transcript variant | C- 0.184 |
| 25 | rs615552 (T/C) | 22026078 | CDKN2B-AS1 | Intron variant | C- 0.195 |
| 26 | rs6475606 (T/C) | 22081851 | CDKN2B-AS1 | Intron variant | C-0.322 |
| 27 | rs7023329 (A/G) | 21816529 | MTAP | Intron variant | G- 0.449 |
| 28 | rs7865618 (A/G) | 22031006 | CDKN2B-AS1 | Intron variant | G- 0.188 |
| 29 | rs944797 (T/C) | 22115287 | CDKN2B-AS1 | Intron variant | C- 0.487 |
| 30 | rs9632884 (C/G) | 22072302 | CDKN2B-AS1 | Intron variant | G- 0.304 |
SNP, single-nucleotide polymorphism.
Genome build is GRCh38.p12 (taken from dbSNP site).
IFNA21 is a proinflammatory cytokine, and elevated serum IFNA21 levels have been associated with the 9p21 risk locus.37 To examine the association between 9p21 SNPs and the expression of IFNA21 gene, the serum circulating levels of the cytokine were estimated using sandwich enzyme linked immuno sorbent assay (ELISA) (Wuhan Fine Biotech kit method) in a random subset of the entire sample that included 184 controls and 167 CAD cases. The IFNA21 gene is located 946,000 base pairs downstream of “core CAD” region in the IFNA gene cluster.
2.3. Statistical analyses
Demographic data were analyzed using SPSS software (Statistical Package for Social Sciences, version 21). The SNP data were analyzed using SPSS (version 21), SNPStats38 (available online), Haploview39 (available online), and multifactor dimensionality reduction (MDR)40 software (version 3.2). Two-tailed P values were considered to assess the significance of association in Chi-square tests used in all the statistical analyses. The power of the study was estimated by using online available software M GAS (Genetic Association Study) Power Calculator (version 3) and found to be 0.966.
3. Results and discussion
Demographic data analysis revealed a higher frequency of conventional risk factors such as diabetes, hypertension, hyperlipidemia, smoking, alcohol, nonvegetarian diet, low fruit intake, lack of physical exercise, and family history (affected first-degree relative) in CAD cases as compared to controls (Table 2). We compared demographic and clinical characteristics between premature CAD cases (age at presentation <55 years in men and <65 years in women) and nonpremature CAD cases (Table 3). Analysis revealed that frequency of females and individuals with diabetes, hypertension, and nonvegetarian diet is significantly higher in premature cases as compared to nonpremature cases. Premature cases did not score well with respect to lifestyle habits, except for regular fruit intake. Our results suggest that lifestyle habits and an increased genetic predisposition may play an important role in the etiopathology of premature CAD.
Table 2.
Epidemiological characteristics of the study population.
| Demographic/clinical parameter | Controls | Cases | P value |
|---|---|---|---|
| Age in years (mean ± SD) | 51.8 ± 9.8 | 55.9 ± 10.7 | 1 × 10−6 |
| Percentage of males/females | 36.6/63.4 | 81.2/18.8 | <1 × 10−7 |
| Height in cms (mean ± SD) | 157.61 ± 10 | 163.49 ± 8.1 | 3 × 10−3 |
| Weight in Kg (mean ± SD) | 67.5 ± 11.6 | 67.8 ± 11.7 | 0.603 |
| BMI (mean ± SD) | 26.1 ± 4.9 | 25.4 ± 3.8 | 0.044 |
| Systolic BP (mean ± SD) | 121.4 ± 6.7 | 125.7 ± 18.1 | 8 × 10−4 |
| Diastolic BP (mean ± SD) | 80.8 ± 5.9 | 76.6 ± 10.7 | 1 × 10−7 |
| Percentage of diabetics/normal | 15.8/84.2 | 50.5/49.5 | <1 × 10−7 |
| Duration of diabetes in years (mean ± SD) | 0.431 ± 1.13 | 1.50 ± 1.80 | <1 × 10−7 |
| Percentage of hypertensives/normal | 24.6/75.4 | 58.7/41.3 | <1 × 10−7 |
| Duration of hypertension in years (mean ± SD) | 0.76 ± 1.55 | 1.74 ± 1.82 | <1 × 10−6 |
| Percentage having affected first-degree relative/no affected first-degree relative | 30/70 | 39.4/60.6 | 0.015 |
| Percentage of cigarette smokers (ever)/nonsmokers | 4.7/95.3 | 43.6/56.4 | <1 × 10−7 |
| No. of cigarettes per day (mean ± SD) | 0.13 ± 0.92 | 4.21 ± 7.30 | <1 × 10−7 |
| Percentage of alcoholics (ever)/nonalcoholics | 9.5/90.5 | 39.5/60.5 | <1 × 10−7 |
| Number of pegs of alcohol (mean ± SD) | 1.85 ± 1.10 | 1.01 ± 1.40 | <1 × 10−7 |
| Number of tea/coffee cups per day (mean ± SD) | 2.37 ± 1.12 | 2.69 ± 1.86 | 0.021 |
| Percentage of individuals with vegetarian diet/mixed diet | 51.9/48.1 | 17.9/82.1 | <1 × 10−7 |
| Percentage of individuals with regular fruit intake/rare fruit intake | 75/25 | 56.9/43.1 | 2 × 10−6 |
| Percentage of individuals with regular physical exercise/no physical exercise | 78.3/21.7 | 45.5/54.5 | <1 × 10−7 |
SD, standard deviation; BMI, body mass index; BP, blood pressure.
Table 3.
Comparison of epidemiological and clinical variables between nonpremature and premature cases.
| S. No. | Variable | % of Nonpremature cases (n = 109) | % of Premature cases (n = 109) | P value |
|---|---|---|---|---|
| 1. | Gender | |||
| Male | 86.7 | 75.2 | 0.030 | |
| Female | 13.3 | 24.8 | ||
| 2. | Diabetes | |||
| Yes | 58.4 | 41.9 | 0.015 | |
| No | 41.6 | 58.1 | ||
| 3. | Hypertension | |||
| Yes | 70.8 | 45.7 | 0.0002 | |
| No | 29.2 | 54.3 | ||
| 4. | Hyperlipidemia | |||
| Yes | 10.6 | 13.3 | 0.404 | |
| No | 55.8 | 46.7 | ||
| No information | 33.6 | 40.0 | ||
| 5. | Alcohol | |||
| Yes | 36.3 | 42.9 | 0.321 | |
| No | 63.7 | 57.1 | ||
| 6. | Smoking habit | |||
| Yes | 40.7 | 46.7 | 0.375 | |
| No | 59.3 | 53.3 | ||
| 7. | Exercise | |||
| Yes | 49.6 | 41 | 0.202 | |
| No | 50.4 | 59 | ||
| 8. | Food habit | |||
| Vegetarian | 23.9 | 11.4 | 0.016 | |
| Mixed | 76.1 | 88.6 | ||
| 9. | Fruits intake | |||
| Daily | 38.1 | 37.1 | 0.592 | |
| 1–2 times weekly | 5.3 | 6.7 | ||
| 3–4 times weekly | 10.6 | 16.2 | ||
| Occasionally/rarely | 46.0 | 40.0 | ||
| 10. | Family history | |||
| Yes | 37.6 | 41.6 | 0.597 | |
| No | 62.4 | 58.4 | ||
3.1. SNP data analysis
The genotypic and allele frequency distribution of the 30 SNPs among the controls and CAD and premature CAD cases revealed that the CC genotype of rs1333045 (T/C) showed more than 1.4-fold risk for CAD (P = 0.046 in the recessive model). We found more than 1.6-fold risk for premature CAD (P = 0.051 in the recessive model). Our study is the first to report an association of rs1333045 with CAD risk in an Asian Indian population.
The CC genotype of rs1333049 (G/C), a highly replicated SNP, showed a trend toward risk association with premature CAD in the study population (P = 0.061 in the recessive model).41, 42
Further, we found that the AA genotype of rs16905599 (G/A) is associated with more than 2.4-fold risk for CAD in the study population (P = 0.025 in the codominant model and P = 0.0069 in the recessive model). The AA genotype of rs16905599 showed about threefold risk for premature CAD (P = 0.018 in the codominant model and P = 0.0081 in the recessive model). This is a first ever report of an association of rs16905599 (AA) with CAD indicating its potential as a useful genetic marker for CAD in the local population.
In addition, rs2383206 (A/G) showed robust association with CAD in the study population where the GG genotype of rs2383206 conferred about twofold risk for CAD (P = 0.0004 in the codominant model and P = 0.0001 in the recessive model)42, 43 and 2.4-fold risk for Premature CAD (P = 0.0002 in Recessive model).
An interesting case of under dominance (risk associated with only heterozygote) was found in case of rs2383208 (A/G) where the AG genotype showed about 1.7-fold risk for premature CAD (P = 0.021 in the codominant model and P = 0.034 in the overdominant model). We found a similar trend in all CAD cases (P = 0.051 in the overdominant model). The GG genotype of rs4977574 (A/G) was also associated with more than 1.7-fold risk for premature CAD (P = 0.025 in the recessive model).42
Thus, the present study identified SNPs rs1333049 (CC) and rs4977574 (GG)41, 42 to be associated exclusively with premature CAD in the local population, suggesting that these have the potential to be used as markers to identify asymptomatic individuals at a higher risk of premature CAD. However, these results require further validation in larger cohorts.
We performed association analysis using SNPStats by adjustment for the following covariates: age, gender, diabetes, hypertension, hyperlipidemia, smoking, alcohol, and family history. The SNPs that showed significant risk association despite adjustment for the covariates were rs2383206, rs16905599, and rs2383208. Of the three SNPs, rs2383206 consistently showed the strongest CAD risk association with odds ratio greater than 2 in our study population. Hence, rs2383206, rs16905599, and rs2383208 have the potential to be used as 9p21 markers along with markers at other loci to generate a more reliable genetic risk score (GRS) for CAD in the local population.
We used the Bonferroni correction to reduce the probability of false-positive results (type I errors) and found that only SNP rs2383206 showed significant risk association with CAD (P < 0.0016) after doing multiple testing corrections for the 30 9p21 SNPs studied.
Upon gender stratification of the SNP genotyping data, we found that the TT and TC genotypes of SNP rs10757272 (C/T) showed an eightfold to ninefold significant risk association (P = 0.0032 in the dominant model and P = 0.012 in the codominant model) with CAD specifically among females.
The GG genotype of SNP rs7865618 (A/G) showed more than 2.5-fold risk association (P = 0.037 in the recessive model) with CAD specifically among males.
3.2. Multivariate logistic regression, MDR, and Haploview analysis
We performed multivariate logistic regression analysis of 30 9p21 SNPs with 12 demographic/clinical variables (Table 4). Our analysis revealed association of several 9p21 SNPs with demographic and clinical variables such as age, diabetes, hyperlipidemia, Rh blood groups, ABO blood groups, food habit, and fruit intake.
Table 4.
Association of SNPs with demographic and clinical parameters using multivariate logistic regression analysis.
| S. No. | SNP ID/variablea | Wild-type genotype/reference value | Risk genotype/OR with 95% CI | P value | Heterozygous genotype/OR with 95% CI | P value |
|---|---|---|---|---|---|---|
| 1. | rs1004638 | TT(1) | AA | AT | ||
|
0.676 (0.469–0.974) | 0.036 | 0.767 (0.576–1.022) | 0.070 | ||
| 2. | rs10116277 | TT(1) | GG | GT | ||
|
3.394 (0.917–12.566) | 0.067 | 3.137 (1.044–9.429) | 0.042 | ||
|
0.329 (0.097–1.115) | 0.074 | ||||
|
1.930 (0.910–4.096) | 0.087 | ||||
| 3. | rs1011970 | GG(1) | TT | TG | ||
|
0.294 (0.070–1.235) | 0.095 | ||||
|
0.555 (0.278–1.110) | 0.096 | ||||
| 4. | rs1063192 | AA(1) | GG | GA | ||
|
0.205 (0.040–1.053) | 0.058 | ||||
|
0.638 (0.376–1.084) | 0.096 | ||||
| 5. | rs10757272 | CC(1) | TT | CT | ||
|
2.703 (0.918–7.958) | 0.071 | ||||
| 6. | rs10757274 | AA(1) | GG | AG | ||
|
0.216 (0.059–0.785) | 0.020 | ||||
|
1.388 (0.998–1.932) | 0.051 | ||||
|
2.656 (0.971–7.266) | 0.057 | ||||
| 7. | rs10757578 | AA(1) | GG | AG | ||
|
0.354 (0.108–1.157) | 0.086 | ||||
|
0.565 (0.307–1.040) | 0.067 | ||||
|
2.801 (0.898–8.736) | 0.076 | ||||
| 8. | rs10811661 | TT(1) | CC | CT | ||
|
2.739 (0.920–8.154) | 0.070 | ||||
| 9. | rs1333040 | TT(1) | CC | CT | ||
|
2.292 (1.034–5.079) | 0.041 | ||||
|
2.665 (0.998–7.117) | 0.050 | ||||
|
0.282 (0.082–0.975) | 0.045 | ||||
| 10. | rs1333042 | GG(1) | AA | AG | ||
|
2.461 (1.093–5.544) | 0.030 | ||||
|
3.045 (1.094–8.478) | 0.033 | ||||
|
0.720 (0.488–1.060) | 0.096 | ||||
| 11. | rs1333045 | TT(1) | CC | CT | ||
|
0.287 (0.085–0.972) | 0.045 | ||||
| 12. | rs1333048 | AA(1) | CC | CA | ||
|
0.263 (0.071–0.969) | 0.045 | ||||
|
3.385 (1.110–10.324) | 0.032 | 3.905 (1.462–10.430) | 0.007 | ||
| 13. | rs1333049 | GG(1) | CC | CG | ||
|
3.206 (1.029–9.991) | 0.045 | 3.222 (1.232–8.424) | 0.017 | ||
|
1.316 (0.951–1.821) | 0.097 | ||||
| 14. | rs16905599 | GG(1) | AA | AG | ||
|
0.273 (0.065–1.155) | 0.078 | ||||
|
4.052 (1.500–10.948) | 0.006 | ||||
|
1.678 (0.939–2.999) | 0.080 | ||||
| 15. | rs2383206 | AA(1) | GG | AG | ||
|
1.365 (0.986–1.891) | 0.061 | 1.424 (1.023–1.981) | 0.036 | ||
| 16. | rs2383207 | GG(1) | AA | AG | ||
|
2.430 (1.073–5.501) | 0.033 | ||||
|
2.887 (1.041–8.011) | 0.042 | ||||
| 17. | rs2383208 | AA(1) | GG | GA | ||
|
7.737E+136 (7.737E+136 to 7.737E+136) | 0.000 | ||||
|
2.349 (0.878–6.282) | 0.089 | ||||
|
1.285 (0.994–1.660) | 0.055 | ||||
| 18. | rs2811712 | AA(1) | GG | AG | ||
|
5455280.330 (5455280.330–5455280.330) | 0.000 | ||||
| 19. | rs2891169 | AA(1) | GG | AG | ||
|
0.241 (0.081–0.719) | 0.011 | 0.246 (0.092–0.656) | 0.005 | ||
|
0.328 (0.101–1.063) | 0.063 | ||||
|
0.523 (0.258–1.060) | 0.072 | ||||
| 20. | rs4977574 | AA(1) | GG | GA | ||
|
0.236 (0.065–0.855) | 0.028 | ||||
|
2.509 (0.832–7.565) | 0.102 | 2.675 (1.025–6.981) | 0.044 | ||
|
1.354 (0.980–1.871) | 0.066 | ||||
| 21. | rs4977756 | AA(1) | GG | GA | ||
|
2.089 (0.907–4.810) | 0.083 | ||||
|
0.432 (0.180–1.037) | 0.060 | ||||
|
0.603 (0.351–1.038) | 0.068 | ||||
| 22. | rs615552 | TT(1) | CC | CT | ||
|
2.319 (1.024–5.249) | 0.044 | ||||
|
0.542 (0.260–1.129) | 0.102 | ||||
|
0.331 (0.134–0.816) | 0.016 | ||||
|
0.521 (0.300–0.905) | 0.021 | ||||
| 23. | rs6475606 | TT(1) | CC | CT | ||
|
2.684 (1.150–6.265) | 0.022 | ||||
|
0.364 (0.148–0.892) | 0.027 | ||||
|
1.992 (0.934–4.252) | 0.075 | ||||
|
3.003 (1.046–8.623) | 0.041 | ||||
| 24. | rs7023329 | AA(1) | GG | GA | ||
|
1.383 (1.039–1.840) | 0.026 | ||||
|
6.096 (1.595–23.302) | 0.008 | ||||
| 25. | rs944797 | TT(1) | CC | CT | ||
|
0.711 (0.487–1.039) | 0.078 | ||||
|
0.369 (0.112–1.210) | 0.100 | ||||
|
3.066 (0.931–10.100) | 0.065 | ||||
|
1.562 (1.123–2.171) | 0.008 | 1.443 (1.038–2.005) | 0.029 | ||
| 26. | rs9632884 | CC(1) | GG | GC | ||
|
2.584 (1.149–5.815) | 0.022 | ||||
|
0.766 (0.569–1.029) | 0.077 | ||||
|
0.768 (0.594–0.992) | 0.043 |
SNP, single-nucleotide polymorphism; CAD, coronary artery disease; OR, odds ratio; CI, confidence interval.
The variables included for multivariate logistic regression analysis are age at onset, duration of CAD, diabetes, hypertension, hyperlipidemia, ABO blood groups, Rh blood groups, alcohol habit, smoking habit, physical exercise, food habit, and fruit intake. Only variables for which there is an association or trend towards an association have been shown in the table for each SNP.
Further, we examined SNP–SNP interactions and SNP–demographic/clinical variable interactions using MDR analysis for ten 9p21 SNPs showing good risk association trends with CAD: rs1011970, rs10757272, rs10757274, rs1333045, rs1333049, rs16905599, rs2383206, rs2383208, rs4977574, and rs7865618.
MDR risk prediction model analysis revealed that rs2383206 is the best one-locus model, and rs16905599 with rs2383206 is the best two-locus model in the study population.
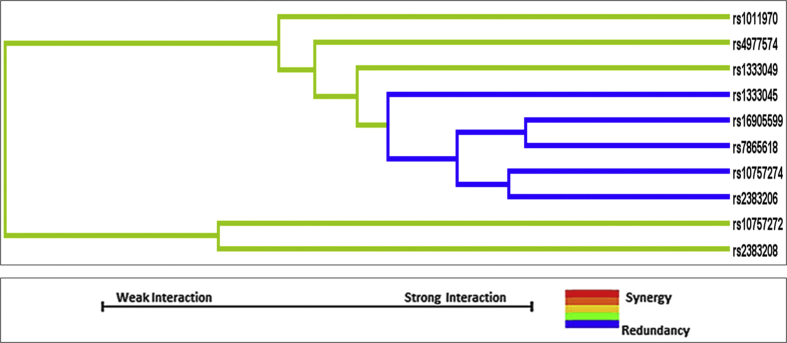
With respect to SNP–SNP interactions, MDR analysis revealed a mild interaction between rs10757272 and rs2383208.
SNPs rs1333045, rs16905599, rs7865618, rs10757274, and rs2383206 showed no interaction among themselves. Therefore, these SNPs (showing significant risk association with CAD in our study population) appear to be operating independent of each other (Fig. 1).
Fig. 1.
MDR analysis: SNP–SNP interactions. MDR, multifactor dimensionality reduction; SNP, single-nucleotide polymorphism.
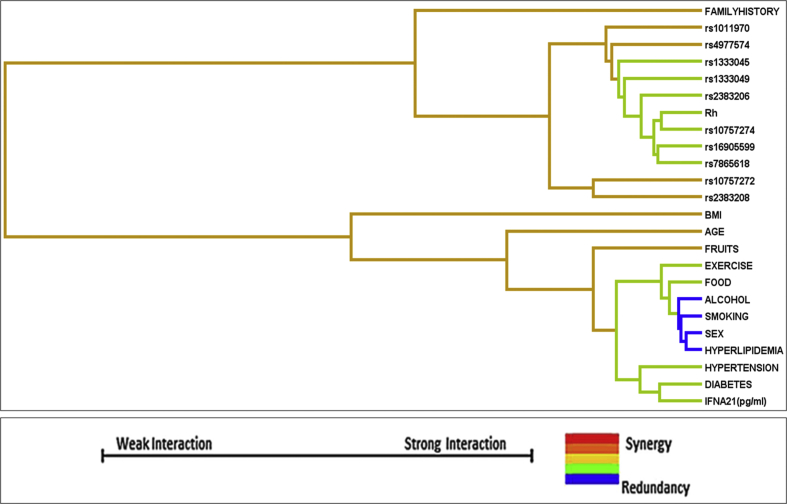
With respect to SNP–demographic/clinical variable interactions, a moderate interaction was observed among the variables body mass index, age, and fruit intake, whereas a mild interaction was observed among hypertension, diabetes, and serum IFNA21 levels. Similarly, there is a mild interaction between exercise and food habit. The variables alcohol, smoking, gender, and hyperlipidemia are found to be operating independently (Fig. 2). This corroborates the observation that the 9p21 locus confers CAD risk independent of classic risk factors.4, 7
Fig. 2.
MDR analysis: SNP–demographic/clinical variable interactions. MDR, multifactor dimensionality reduction; SNP, single-nucleotide polymorphism; BMI, body mass index.
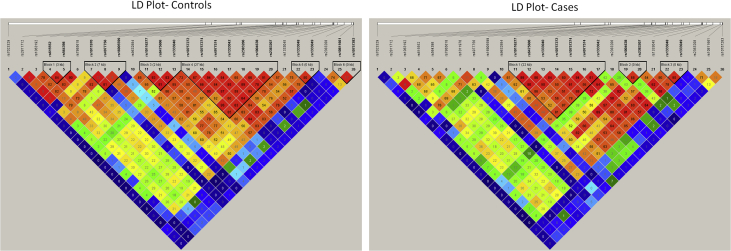
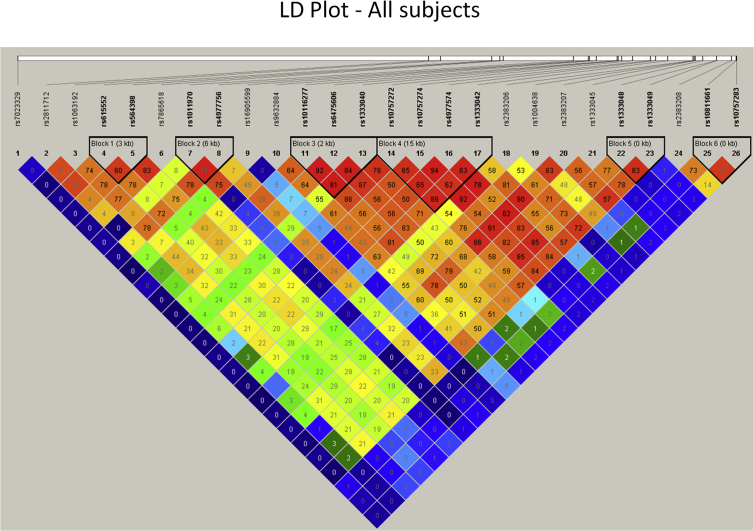
Linkage disequilibrium (LD) analysis was performed using Haploview software to generate LD plots for controls and cases. Our analysis has revealed six blocks (blocks 1, 2, 3, 4, 5, and 6) of linked SNP variants in controls and three blocks (blocks 1, 2, and 3) in cases (Fig. 3). Similarly, six blocks (blocks 1, 2, 3, 4, 5, and 6) of linked SNP variants were observed for the entire sample (Fig. 4).
Fig. 3.
LD plot for controls and cases. LD, linkage disequilibrium.
Fig. 4.
LD plot for all subjects. LD, linkage disequilibrium.
One limitation of our study is the limited size of CAD cases. Hence, the results need to be validated in larger CAD cohorts. Our study subjects have been taken from the twin cities of Hyderabad and Secunderabad, irrespective of the geographical region, language, caste group, and community they hail from. Most CAD cases have been taken from KIMS hospital, Secunderabad, which is centrally located and receives patients from different parts of the state and country. Despite these measures, there may be a possibility of a hidden population substructure that has not been considered in this study.
4. Conclusions
Premature CAD cases showed a significantly higher frequency of females, diabetics, hypertensives, and individuals with nonvegetarian food habit as compared to nonpremature cases. SNPs rs1333045 (CC), rs16905599 (AA), rs2383206 (GG), rs2383208 (AG), and rs4977574 (GG) showed significant risk association with premature CAD in our study population. We found rs1333049 (CC) and rs4977574 (GG) to be associated exclusively with premature CAD, suggesting their potential as genetic markers to predict premature CAD in the local population. Of all the 9p21 SNPs that showed significant CAD risk association, rs2383206 emerged as the strongest genetic risk factor followed by rs16905599 and rs2383208. SNP rs10757272 (TT and TC) has shown a robust female-specific risk association, and SNP rs7865618 (GG) has shown significant male-specific risk association with CAD in our study population.
To conclude, results of the present study suggest that SNPs at the 9p21 risk locus may be used to generate a reliable GRS along with markers at other loci. This could be used for presymptomatic diagnosis in high-risk families such that suitable therapeutic measures could be advised to prevent the serious outcomes of acute coronary syndromes.
Key messages
What is already known about this subject?
-
•
The 9p21 risk locus is independent of most traditional risk factors of CAD such as high lipids, hypertension, obesity, and diabetes. The association of SNPs at this locus with CAD has been replicated in many ethnicities. 9p21 is also known to be a predictor of severity of CAD based on the number of vessels involved. The 9p21 risk locus has shown about twofold greater risk in individuals who develop premature CAD (<55 years in men and <65 years in women). This locus mediates its risk at the vessel wall leading to the onset of atherosclerosis in coronary arteries, thereby resulting in CAD. The 9p21 locus may have clinical utility as an early marker for CAD risk.
What does this study add?
-
•
Our study suggests that female gender may be at higher risk of premature CAD. Lifestyle factors and genetic predisposition may have a major role to play in the etiopathology of premature CAD.
-
•
Our study has reported a first ever risk association of SNPs rs1333045 (CC) and rs16905599 (AA) with CAD in an Asian Indian population.
-
•
SNPs rs1333049 (CC) and rs4977574 (GG) have potential as genetic markers to identify individuals at a higher risk of premature CAD.
-
•
Some demographic and clinical variables have shown significant association with 9p21 SNP genotypes.
-
•
SNP rs2383206 is the strongest genetic risk factor for CAD followed by rs16905599 and rs2383208 in the study population.
-
•
SNP rs10757274 (TT and TC) has shown a strong female-specific risk association and rs7865618 (GG) has shown a significant male-specific risk association with CAD in the study population.
How might this impact clinical practice?
This study aimed to identify and understand the unique pattern of risk factors, especially those at the chromosome 9p21.3 locus in our local population. This may contribute to presymptomatic diagnosis in high-risk families and development of preventive/therapeutic strategies most suited to our population in the near future. The 9p21.3 locus also holds good promise for pharmacogenetics research and clinical applications, so that medical treatment could be personalized to suit individuals based on their complex genetic and environmental backgrounds. However, these results require further validation in larger cohorts.
Author contributions
B.K. has contributed to the design of the work and acquisition of data. D.K.M. has contributed to the substantial revision of the manuscript. N.B. has contributed to the statistical analysis of the data. M.T.A. has contributed to the interpretation of the data and to the preparation of the final draft of the manuscript. All authors have read and approved the submitted version of the manuscript and have agreed to be personally accountable for their own contribution and all queries related to the study.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Conflicts of interest
All authors have none to declare.
Acknowledgments
The authors are extremely thankful to Dr. V. Dayasagar Rao, Senior Interventional Cardiologist, KIMS, and Ms. Apoorva Sharma, Clinical Research Coordinator, KIMS, for their kind help in recruitment of patients with CAD. They owe the success of this work to the willing participation and cooperation of all the study subjects.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ihj.2019.04.005.
Contributor Information
Bellary Kalpana, Email: kalpana_bellary@yahoo.com.
Dwarkanath K. Murthy, Email: dwarkanath49@yahoo.co.in.
Nagalla Balakrishna, Email: dr_nbk@yahoo.com.
Mohini T. Aiyengar, Email: mohini.aiyengar13@gmail.com.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Krishnaswamy S., Prasad N.K., Jose V.J. A study of lipid level in Indian patients with coronary artery disease. Int J Cardiol. 1989;4:337–345. doi: 10.1016/0167-5273(89)90013-2. [DOI] [PubMed] [Google Scholar]
- 2.Tewari S., Kumar S., Kapoor A. Premature coronary artery disease in North India: an angiography study of 1971 patients. Indian Heart J. 2005;57:311–318. [PubMed] [Google Scholar]
- 3.Helgadottir A., Thorleifsson G., Manolescu A. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 4.McPherson R., Pertsemlidis A., Kavaslar N. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samani N.J., Erdmann J., Hall A.S., WTCCC and the Cardiogenics Consortium Genome-wide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coronary Artery Disease (C4D) Genetics Consortium A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet. 2011;43(4):339–344. doi: 10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- 7.Broadbent H.M., Peden J.F., Lorkowski S. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet. 2008;17:806–814. doi: 10.1093/hmg/ddm352. [DOI] [PubMed] [Google Scholar]
- 8.Gschwendtner A., Bevan S., Cole J.W. Sequence variants on chromosome 9p21.3 confer risk for atherosclerotic stroke. Ann Neurol. 2009;65(5):531–539. doi: 10.1002/ana.21590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matarin M., Brown W.M., Singleton A., Hardy J.A., Meschia J.F. Whole genome analyses suggest ischemic stroke and heart disease share an association with polymorphisms on chromosome 9p21. Stroke. 2008;39:1586–1589. doi: 10.1161/STROKEAHA.107.502963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helgadottir A., Thorleifsson G., Magnusson K.P. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat Genet. 2008;40:217–224. doi: 10.1038/ng.72. [DOI] [PubMed] [Google Scholar]
- 11.Thompson A.R., Golledge J., Cooper J.A., Hafez H., Norman P.E., Humphries S.E. Sequence variant on 9p21 is associated with the presence of abdominal aortic aneurysm disease but does not have an impact on aneurysmal expansion. Eur J Hum Genet. 2008;17(3):391–394. doi: 10.1038/ejhg.2008.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherborne A.L., Hosking F.J., Prasad R.B. Variation in CDKN2A at 9p21.3 influences childhood acute lymphoblastic leukemia risk. Nat Genet. 2010;42:492–494. doi: 10.1038/ng.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turnbull C., Ahmed S., Morrison J. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42:504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stacey S.N., Sulem P., Masson G. New common variants affecting susceptibility to basal cell carcinoma. Nat Genet. 2009;41:909–914. doi: 10.1038/ng.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gori F., Specchia C., Pietri S. Common genetic variants on chromosome 9p21 are associated with myocardial infarction and type 2 diabetes in an Italian population. BMC Med Genet. 2010;11:60. doi: 10.1186/1471-2350-11-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Assimes T.L., Knowles J.W., Basu A. Susceptibility locus for clinical and subclinical coronary artery disease at chromosome 9p21 in the multi-ethnic ADVANCE study. Hum Mol Genet. 2008;17:2320–2328. doi: 10.1093/hmg/ddn132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding H., Xu Y., Wang X. 9p21 is a shared susceptibility locus strongly for coronary artery disease and weakly for ischemic stroke in Chinese Han population. Circ Cardiovasc Genet. 2009;2:338–346. doi: 10.1161/CIRCGENETICS.108.810226. [DOI] [PubMed] [Google Scholar]
- 18.Hiura Y., Fukushima Y., Yuno M. Validation of the association of genetic variants on chromosome 9p21 and 1q41 with myocardial infarction in a Japanese population. Circ J. 2008;72:1213–1217. doi: 10.1253/circj.72.1213. [DOI] [PubMed] [Google Scholar]
- 19.Shen G.Q., Li L., Rao S. Four SNPs on chromosome 9p21 in a South Korean population implicate a genetic locus that confers high cross-race risk for development of coronary artery disease. Arterioscler Thromb Vasc Biol. 2008;28:360–365. doi: 10.1161/ATVBAHA.107.157248. [DOI] [PubMed] [Google Scholar]
- 20.Maitra A., Dash D., John S. A common variant in chromosome 9p21 associated with CAD in Asian Indians. J Genet. 2009;88(1):113–118. doi: 10.1007/s12041-009-0017-y. [DOI] [PubMed] [Google Scholar]
- 21.Saleheen D., Alexander M., Rasheed A. Association of the 9p21.3 locus with risk of first-ever myocardial infarction in Pakistanis: case-control study in South Asia and updated meta-analysis of Europeans. Arterioscler Thromb Vasc Biol. 2010;30:1467–1473. doi: 10.1161/ATVBAHA.109.197210. [DOI] [PubMed] [Google Scholar]
- 22.Quelle D.E., Zindy F., Ashmun R.A., Sherr C.J. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 23.Popov N., Gil J. Epigenetic regulation of the INK4b-ARF-INK4a locus: in sickness and in health. Epigenetics. 2010;5:685–690. doi: 10.4161/epi.5.8.12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas T., Thomas T.J. Polyamines in cell growth and cell death: molecular mechanisms and therapeutic applications. Cell Mol Life Sci. 2001;58:244–258. doi: 10.1007/PL00000852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pasmant E., Laurendeau I., Héron D., Vidaud M., Vidaud D., Bièche I. Characterization of a germ-line deletion including the entire INK4/ARF locus, in a melanoma-neural system tumor family. Identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963–3969. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- 26.Folkersen L., Kyriakou T., Goel A. PROCARDIS consortia. Relationship between CAD risk genotype in the chromosome 9p21 locus and gene expression. Identification of eight new ANRIL splice variants. PLoS One. 2009;4 doi: 10.1371/journal.pone.0007677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burd C.E., Jeck W.R., Liu Y., Sanoff H.K., Wang Z., Sharpless N.E. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Congrains A., Kamide K., Oguro R. Genetic variants at the 9p21 locus contribute to atherosclerosis through modulation of ANRIL and CDKN2A/B. Atherosclerosis. 2012;220:449–455. doi: 10.1016/j.atherosclerosis.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 29.Holdt L.M., Beutner F., Scholz M. ANRIL expression is associated with atherosclerosis risk at chromosome 9p21. Arterioscler Thromb Vasc Biol. 2010;30:620–627. doi: 10.1161/ATVBAHA.109.196832. [DOI] [PubMed] [Google Scholar]
- 30.Cunnington M.S., Santibanez Koref M., Mayosi B.M., Burn J., Keavney B. Chromosome 9p21 SNPs associated with multiple disease phenotypes correlate with ANRIL expression. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harismendy O., Notani D., Song X. 9p21 DNA variants associated with coronary artery disease impair interferon-γ signalling response. Nature. 2011;470:264–268. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y., Sanoff H.K., Cho H. INK4/ARF transcript expression is associated with chromosome 9p21 variants linked to atherosclerosis. PLoS One. 2009;4 doi: 10.1371/journal.pone.0005027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yap K.L., Li S., Muñoz-Cabello A.M. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu W., Gius D., Onyango P. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bochenek G., Hasler R., El Mokhtari N.E. The large non-coding RNA ANRIL, which is associated with atherosclerosis, periodontitis and several forms of cancer, regulates ADIPOR1, VAMP3 and C11ORF10. Hum Mol Genet. 2013;22(22):4516–4527. doi: 10.1093/hmg/ddt299. [DOI] [PubMed] [Google Scholar]
- 36.Motterle A., Pu X., Wood H. Functional analyses of coronary artery disease associated variation on chromosome 9p21 in vascular smooth muscle cells. Hum Mol Genet. 2012;21(18):4021–4029. doi: 10.1093/hmg/dds224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Almontashiri N.A., Fan M., Chen H.-H. Abstract 15730: serum interferon alpha 21 is a biomarker of the 9p21.3 risk locus for coronary artery disease. Circulation. 2011;124:A15730. [Google Scholar]
- 38.Solé X., Guinó E., Valls J., Iniesta R., Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006 Aug 1;22(15):1928–1929. doi: 10.1093/bioinformatics/btl268. Epub 2006 May 23. [DOI] [PubMed] [Google Scholar]
- 39.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005 Jan 15;21(2):263–265. doi: 10.1093/bioinformatics/bth457. Epub 2004 Aug 5. [DOI] [PubMed] [Google Scholar]
- 40.Motsinger A.A., Ritchie M.D. Multifactor dimensionality reduction: an analysis strategy for modelling and detecting gene-gene interactions in human genetics and pharmacogenomics studies. Hum Genomics. 2006;2(5):318–328. doi: 10.1186/1479-7364-2-5-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhanushali A.A., Contractor A., Das B.R. Variant on 9p21 rs 1333049 is associated with age of onset of CAD in a Western Indian population: a case control association study. Genet Res (Camb) 2013;95(5):138–145. doi: 10.1017/S0016672313000189. [DOI] [PubMed] [Google Scholar]
- 42.Shanker J., Arvind P., Jambunathan S., Nair J., Kakkar V. Genetic analysis of the 9p21.3 CAD risk locus in Asian Indians. Thromb Haemost. 2014;111(5):960–969. doi: 10.1160/TH13-08-0706. [DOI] [PubMed] [Google Scholar]
- 43.Kumar J., Yumnam S., Basu T. Association of polymorphisms in 9p21 region with CAD in North Indian population: replication of SNPs identified through GWAS. Clin Genet. 2011;79(6):588–593. doi: 10.1111/j.1399-0004.2010.01509.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.