Abstract
Accumulating evidence has uncovered long non-coding RNAs (lncRNAs) as central regulators in the pathogenesis of diverse human cancers including colorectal cancer (CRC). The present study discovered that a novel lncRNA ITIH4 antisense RNA 1 (ITHI4-AS1) was frequently under-expressed in most normal human tissues, including colon tissues. Therefore, we aimed to investigate the role of ITHI4-AS1 in CRC. Interestingly, a significant overexpression of ITIH4-AS1 was observed in CRC cell lines relative to normal NCM460 cells. Also, we investigated the facilitating role of ITIH4-AS1 in CRC cell growth and metastasis both in vitro and in vivo. Additionally, we explained that ITIH4-AS1 upregulation in CRC was attributed to downregulation or even depletion of RE1 silencing transcription factor (REST), a presently identified transcriptional repressor for ITIH4-AS1. Meanwhile, the contribution of ITIH4-AS1 to CRC development was validated to rely on the activation of the JAK/STAT3 pathway. More importantly, we verified that FUS interacted with both ITIH4-AS1 and STAT3, and that ITIH4-AS1 evoked nuclear translocation of phosphorylated (p)-STAT3 in CRC through recruiting FUS. In summary, our findings unveiled for the first time that REST downregulation-enhanced ITIH4-AS1 exerts pro-tumor functions in CRC through FUS-dependent activation of the JAK/STAT3 pathway, implying that targeting ITIH4-AS1 may be a novel effective strategy for CRC therapy.
Keywords: ITIH4-AS1, colorectal cancer, REST, FUS, STAT3
Introduction
Colorectal cancer (CRC) is one of the most prevalent malignancies and the main cause of cancer-related death all over the world.1, 2 More than 10% of CRC patients are diagnosed at advanced stages, and nearly 30% of CRC cases that were diagnosed at an early stage eventually develop into metastases.3 Nowadays, CRC has become a serious hazard to public health because of high incidence and mortality, although great improvements have been made in diagnostic and therapeutic strategies for CRC.4, 5, 6, 7 Therefore, key biomarkers in the initiation and development of CRC need to be discovered to exploit more effective targets for the diagnosis and treatment of CRC.
Long non-coding RNAs (lncRNAs) are a class of endogenous transcripts larger than 200 bases without open reading frames (ORFs).8 Recently, the involvement of lncRNAs in tumorigenesis and cancer development has been increasingly illustrated,9, 10 including in CRC.11, 12 For instance, SNHG5 contributes to CRC cell survival via countervailing STAU1-induced mRNA destabilization.13 lncRNA UPAT facilitates colon tumorigenesis by regulating UHRF1.14 lncRNA PVT1-214 serves as an oncogene in promoting CRC cell proliferation and invasion.15
ITIH4 antisense RNA 1 (ITIH4-AS1) is a novel lncRNA that has never been studied until this research. The online databases including University of California, Santa Cruz (UCSC), NCBI, and NONCODE revealed the general low expression of ITIH4-AS1 in most of the normal human tissues including colon tissues. Meanwhile, the significant upregulation of ITIH4-AS1 in all five CRC cell lines was subsequently validated. Thus, we suspected that ITIH4-AS1 might function in CRC development. Herein, we investigated the carcinogenic role of ITIH4-AS1 in CRC through in vitro and in vivo experiments. In addition, we demonstrated that RE1 silencing transcription factor (REST), known as a transcription repressor,16 was downregulated in CRC and resulted in the transcriptional upregulation of ITIH4-AS1 in CRC. Moreover, ITIH4-AS1 facilitated the nuclear translocation of p-STAT3 homodimer and activated JAK/STAT3 signaling through an FUS-mediated manner so as to facilitate CRC progression.
All in all, our study first suggested that ITIH4-AS1 plays a contributing role in CRC through activating the JAK/STAT3 pathway by recruiting FUS, indicating ITIH4-AS1 as a promising target for CRC diagnosis and treatment.
Results
ITIH4-AS1 Is Apparently Upregulated in CRC Cell Lines
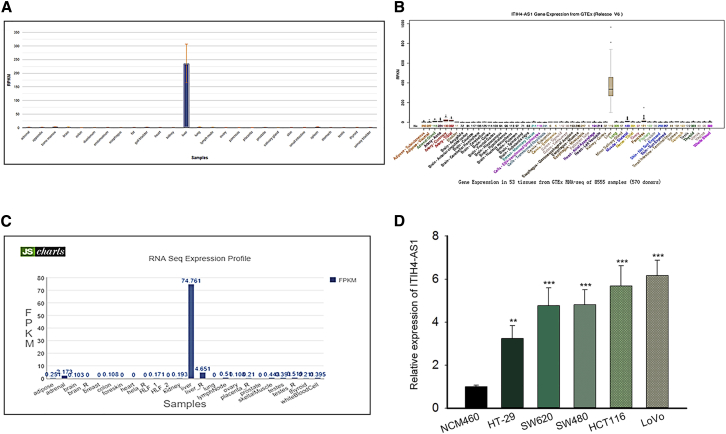
First of all, we browsed several online bioinformatics tools to find lncRNAs potentially implicated in the initiation and progression of CRC. As a result, the NCBI database (https://www.ncbi.nlm.nih.gov/gene/) showed that ITIH4-AS1 level was generally under-expressed in 26 of 27 different tissues (except liver tissues) from 95 healthy individuals, including normal colon tissues (Figure 1A). Consistently, the data from the UCSC database (http://genome.ucsc.edu/) suggested that ITIH4-AS1 was low expressed in colon tissues and 51 other normal tissues except liver tissues (Figure 1B). Similarly, data from the NONCODE database (http://www.noncode.org/) showed the under-expression of ITIH4-AS1 in normal colon tissues as well (Figure 1C). Conversely, we found an observable upregulation of ITIH4-AS1 expression in all five CRC cell lines (HT-29, SW620, SW480, HCT116 and LoVo) compared with the normal human intestinal epithelial cell line NCM460 (Figure 1D). Based on the above findings, we suspected that ITIH4-AS1 may play a role in CRC development.
Figure 1.
ITIH4-AS1 Was Highly Expressed in CRC Cell Lines
(A–C) The expression profiles of ITIH4-AS1 in human normal tissues were correspondingly obtained from NCBI (A), UCSC (B), and NONCODE (C). (D) Relative expression of ITIH4-AS1 in the five CRC cell lines and the normal NCM460 cells was tested by quantitative real-time PCR. All data were shown as mean ± SD, and error bars represent SD. **p < 0.01, ***p < 0.001.
ITIH4-AS1 Acts as a Facilitator of Cell Growth in CRC
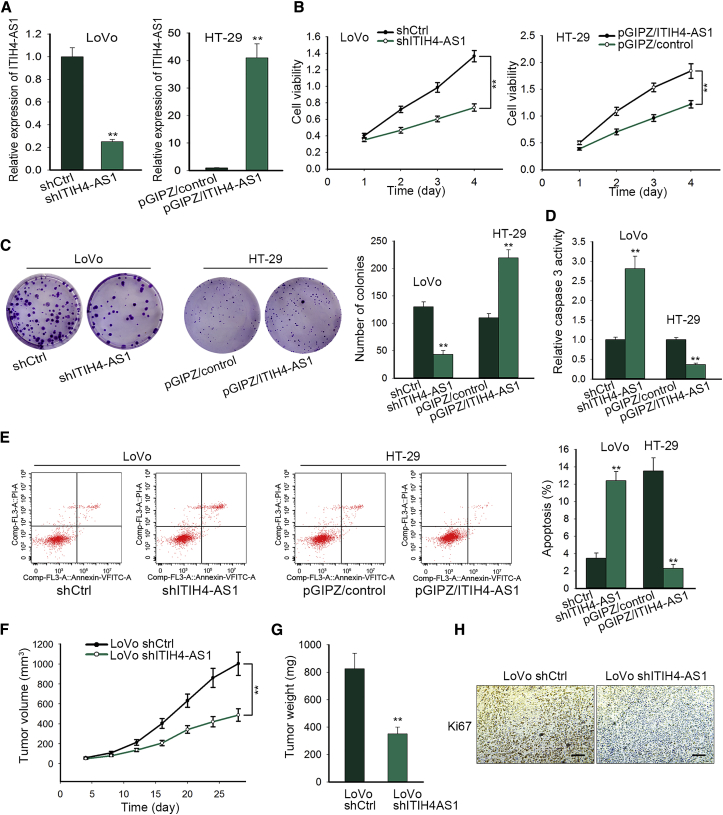
In order to confirm the role of ITIH4-AS1 in CRC, we investigated whether ITIH4-AS1 had an effect on cell proliferation in CRC. Therefore, loss-of-function assays were conducted in LoVo cells expressing the highest ITIH4-AS1 level, whereas gain-of-function assays were carried out in HT-29 cells with the lowest endogenous ITIH4-AS1 level. First, we proved that the expression of ITIH4-AS1 was indeed silenced in LoVo cells transfected with shITIH4-AS1 but successfully overexpressed in HT-29 cells transfected with pGIPZ/ITIH4-AS1 (Figure 2A). Then the Cell Counting Kit-8 (CCK-8) assays demonstrated that knockdown of ITIH4-AS1 markedly impaired the viability of LoVo cells, whereas overexpression of ITIH4-AS1 noticeably enhanced the viability of HT-29 cells (Figure 2B). Similarly, the colony formation capacity was restrained by ITIH4-AS1 silence but encouraged under ITIH4-AS1 upregulation (Figure 2C). In contrast, the caspase-3 activity was dramatically increased in response to ITIH4-AS1 inhibition but overtly decreased in the face of enforced expression of ITIH4-AS1 (Figure 2D). Likewise, the flow cytometry analysis further verified that ITIH4-AS1 inhibition encouraged cell apoptosis, whereas its upregulation hampered apoptosis in CRC cells (Figure 2E). More importantly, we unveiled that LoVo cells with ITIH4-AS1 silence generated smaller and lighter tumors in vivo compared with the control cells (Figures 2F and 2G). Additionally, ITIH4-AS1 silence resulted in lower staining positivity of the proliferation marker Ki67 in xenografts (Figure 2H). Taken together, these data suggested that ITIH4-AS1 promotes CRC cell proliferation in vitro and CRC cell growth in vivo.
Figure 2.
ITIH4-AS1 Promoted CRC Cell Proliferation In Vitro and Accelerated Tumor Growth In Vivo
(A) Quantitative real-time PCR results of ITIH4-AS1 in LoVo cells transfected with shCtrl or shITIH4-AS1, and HT-29 cells transfected with pGIPZ/control or pGIPZ/ITIH4-AS1. (B and C) Cell viability (B) and proliferation ability (C) were evaluated by CCK-8 and colony formation assays, respectively. (D and E) The effect of ITIH4-AS1 depletion on CRC cell apoptosis was evaluated by conducting caspase-3 activity assays (D) and flow cytometry analysis (E). (F and G) The size (F) and weight (G) of tumors generated from LoVo cells with or without ITIH4-AS1 silence. (H) Ki67 expression in the above tumors was detected with IHC staining, with the scale bar indicating 200 μm. All data were shown as means ± SD, and error bars represent SD. *p < 0.05, **p < 0.01, ***p < 0.001.
ITIH4-AS1 Contributes to CRC Cell Metastasis
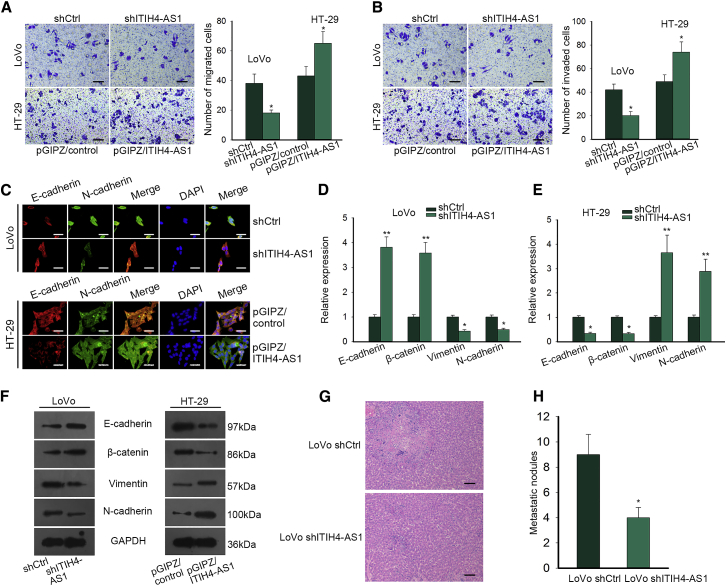
In view of the high rate of metastases in CRC patients, we also detected whether ITIH4-AS1 functioned in CRC metastasis. As displayed in Figures 3A and 3B, we observed that ITIH4-AS1 knockdown hindered cell migration and invasion in LoVo cells, whereas elevated expression of ITIH4-AS1 promoted migratory and invasive abilities in HT-29 cells. Besides, the immunofluorescence (IF) staining indicated that the expression of E-cadherin was enhanced, whereas that of N-cadherin was reduced in LoVo cells responding to ITIH4-AS1 silence; however, a reverse phenomenon was observed in ITIH4-AS1-overexpressed HT-29 cells (Figure 3C). Accordingly, inhibiting ITIH4-AS1 upregulated the expression of E-cadherin and β-catenin, and downregulated expression of Vimentin and N-cadherin, whereas overexpressing ITIH4-AS1 allowed the decreased expression of E-cadherin and β-catenin but increased expression of Vimentin and N-cadherin (Figures 3D–3F). Meanwhile, LoVo cells with ITIH4-AS1 silence resulted in less secondary tumor and metastatic nodules in liver than control, indicating ITIH4-AS1 accelerated CRC cell metastasis (Figures 3G and 3H). Collectively, it was conceivable that ITIH4-AS1 promotes CRC cell migration, invasion, and epithelial-mesenchymal transition (EMT) in vitro and facilitates tumor metastasis in vivo.
Figure 3.
ITIH4-AS1 Contributed to CRC Cell Migration, Invasion, and EMT In Vitro and Facilitated Tumor Metastasis In Vivo
(A and B) The migratory (A) and invasive (B) capacities of LoVo and HT-29 cells in different conditions were detected by Transwell assays. Scale bar, 100 μm. (C) The expression of E-cadherin and N-cadherin in indicated CRC cells was evaluated by IF staining. Scale bar, 50 μm. (D–F) The mRNA and protein levels of epithelial markers (E-cadherin and β-catenin) and mesenchymal markers (Vimentin and N-cadherin) were assessed by quantitative real-time PCR (D and E) and western blotting (F), respectively. (G and H) H&E-stained images of metastatic nodules in the liver of mice injected with shCtrl or shITIH4-AS1-transfected LoVo cells (G; scale bar, 200 μm) and corresponding quantitative graph (H). All data were shown as means ± SD, and error bars represent SD. *p < 0.05, **p < 0.01.
ITIH4-AS1 Is Transcriptionally Regulated by REST
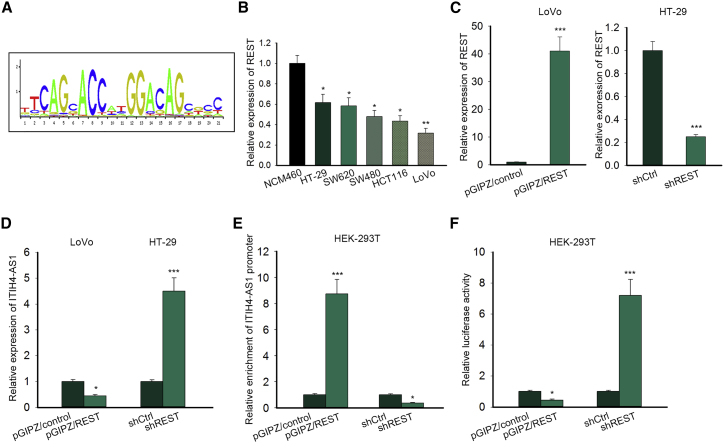
Given that ITIH4-AS1 was upregulated in CRC progression, we next probed the upstream regulator of ITIH4-AS1 in CRC. The UCSC tool exhibited that RE1 silencing transcription factor (REST) was the only transcription factor (TF) potently modulating ITIH4-AS1 transcription. The binding motif obtained from JASPAR (http://jaspardev.genereg.net/) is exhibited in Figure 4A. Previously, REST was proved as a tumor-suppressive gene in the colon.17 Here, we unveiled that REST was low expressed in CRC cell lines relative to the normal NCM460 cells (Figure 4B). Interestingly, REST overexpression in LoVo cells erased the expression of ITIH4-AS1, and converse results were observed when silencing REST in HT-29 cells (Figures 4C and 4D). Furthermore, we discovered that the enrichment of the ITIH4-AS1 promoter in immunoprecipitated products of REST was robustly strengthened upon REST overexpression but distinctly abolished under REST inhibition (Figure 4E). Nevertheless, overexpressing REST significantly reduced the luciferase activity of the ITIH4-AS1 promoter, whereas REST silence led to the opposite result (Figure 4F), indicating that REST negatively regulated ITIH4-AS1 transcription. Altogether, these results illustrated that REST is a transcription repressor of ITIH4-AS1 in CRC.
Figure 4.
ITIH4-AS1 Was Transcriptionally Regulated by REST in CRC
(A) The binding motif of REST to the promoter region of its targets was obtained from JASPAR. (B) The expression of REST in normal NCM460 cells and five CRC cell lines was examined using quantitative real-time PCR. (C and D) The levels of REST (C) and ITIH4-AS1 (D) in pGIPZ/control or pGIPZ/REST-transfected LoVo cells, as well as those in shCtrl or shREST-transfected HT-29 cells, were determined by quantitative real-time PCR. (E and F) The effect of REST on ITIH4-AS1 transcription was confirmed via ChIP (E) and luciferase reporter assay (F). All data were shown as means ± SD, and error bars represent SD. *p < 0.05, **p < 0.01, ***p < 0.001.
ITIH4-AS1 Aggravates CRC Progression through the JAK/STAT3 Signaling Pathway
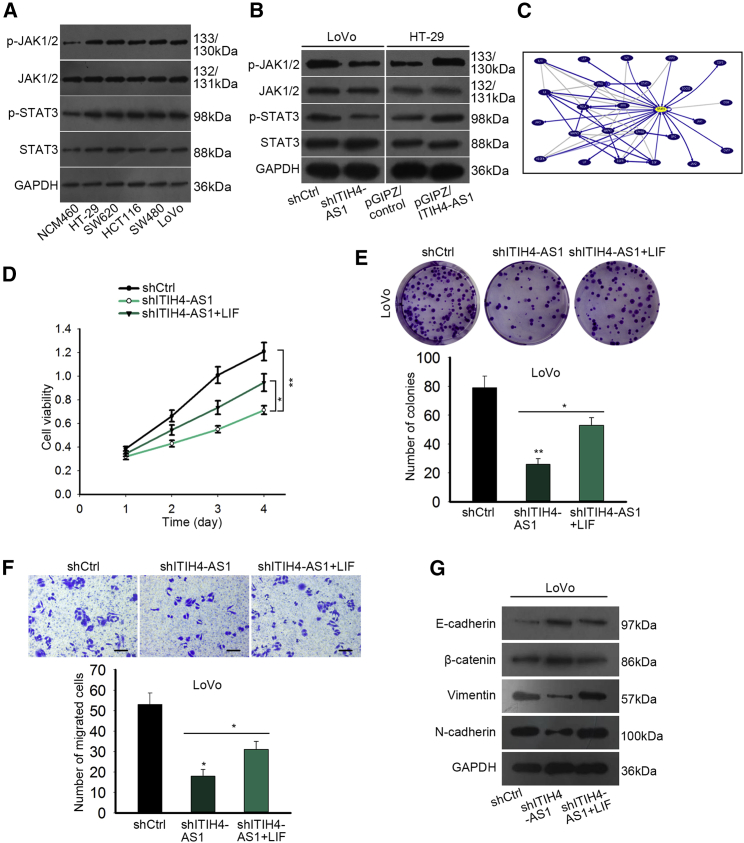
Previously, the important role of JAK/STAT3 signaling in CRC tumorigenesis has been already reported.18, 19 In this study, we validated the notable activation of JAK/STAT3 signaling in CRC cells because the phosphorylation levels of JAK1/2 and STAT3 were strikingly upregulated in all five CRC cell lines in comparison with the normal NCM460 cells (Figure 5A). Besides, the levels of phosphorylated JAK1/2 and STAT3 were abated under depletion of ITIH4-AS1 but were stimulated in the face of upregulation of ITIH4-AS1 (Figure 5B), suggesting ITIH4-AS1 activated the JAK/STAT3 pathway in CRC. Additionally, the UCSC database revealed the co-expression of STAT3 with multiple oncogenes, such as EGFR and MAPK1 (Figure 5C). To make clear whether the JAK/STAT3 pathway mediated the regulation of ITIH4-AS1 on CRC progression, we applied LIF, a key activator of JAK/STAT3 signaling, to conduct rescue assays in ITIH4-AS1-silenced LoVo cells. As a result, the repressive effect of ITIH4-AS1 silence on viability and the proliferative ability of CRC cells were pronouncedly normalized in the context of LIF upregulation (Figures 5D and 5E). Also, the processes of cell migration and EMT abrogated by ITIH4-AS1 downregulation were both disinhibited after activating the JAK/ATAT3 pathway (Figures 5F and 5G). By and large, JAK/STAT3 signaling mediates the facilitating effect of ITIH4-AS1 on CRC development.
Figure 5.
ITIH4-AS1-Aggravated CRC Progression Was Mediated by JAK/STAT3 Signaling
(A) Western blotting results of the total and phosphorylated levels of proteins involved in the JAK/STAT3 pathway in NCM460 cells and CRC cell lines. (B) The levels of proteins participating in the JAK/STAT3 pathway in LoVo and HT-29 cells with specific transfections were assayed using western blotting. (C) The co-expression network of STAT3 was provided by UCSC. (D–G) The impact of the activated JAK/STAT3 pathway on the cellular processes of ITIH4-AS1-silenced LoVo cells was estimated by performing CCK-8 (D), colony formation assay (E), Transwell assay (F; scale bar, 100 μm), and western blotting (G). All data were shown as mean ± SD, and error bars represent SD. *p < 0.05, **p < 0.01.
ITIH4-AS1 Facilitates the Translocation of STAT3 into the Nucleus of CRC Cells through Recruiting FUS
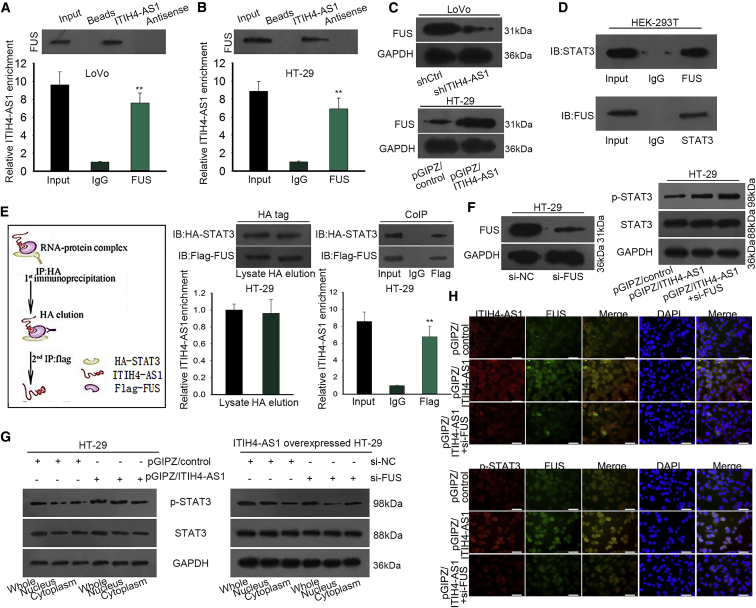
Furthermore, we aimed to explore the in-depth mechanism underlying the activation of the JAK/STAT3 pathway by ITIH4-AS1. A recent report disclosed that the binding of FUS to AKT contributes to AKT nuclear translocation and activation.20 Meanwhile, the online starBase (http://starbase.sysu.edu.cn/starbase2/rbpLncRNA.php) predicted the interaction between ITIH4-AS1 and FUS, and such interaction was further validated in CRC cells by performing RNA pull-down and RIP assays (Figures 6A and 6B). Also, we found that FUS was positively regulated by ITIH4-AS1 in CRC cells, because its protein level was decreased under ITIH4-AS1 inhibition but increased upon ITIH4-AS1 stimulation (Figure 6C). Furthermore, the interaction of FUS with STAT3 in CRC cells was also proved by the co-immunoprecipitation (coIP) assay (Figure 6D). Thus, we speculated that FUS was required for the activation of JAK/STAT3 signaling by ITIH4-AS1.
Figure 6.
ITIH4-AS1 Intensified the Nuclear Translocation of STAT3 in CRC via Recruiting FUS
(A and B) RNA pull-down and RIP assays were conducted to prove the interaction between ITIH4-AS1 and FUS in LoVo (A) and HT-29 (B) cells. (C) The effect of ITIH4-AS1 on FUS protein in two CRC cells was explained using western blotting. (D) The interplay between FUS and STAT3 was confirmed by coIP assay. (E) The existence of the complex containing ITIH4-AS1, FUS, and STAT3 was validated using two-step coIP assay. (F and G) The influence of FUS knockdown on STAT3 protein level (F) and distribution (G) in ITIH4-ASI1-overexpressed HT-29 cells was determined by western blotting. (H) The co-localization of ITIH4-AS1 and FUS, as well as that of FUS and STAT3, was validated by IF analysis (scale bar, 50 μm). All data were shown as means ± SD, and error bars represent SD. **p < 0.01.
Expectedly, a two-step coIP assay demonstrated that both the tagged STAT3 and FUS could be easily observed in anti-hemagglutinin (HA)-precipitated complex with enriched ITIH4-AS1; also, HA-STAT3 and FLAG-FUS, as well as the concentration of ITIH4-AS1, were validated to be precipitated by FLAG, but not immunoglobulin G (IgG) (Figure 6E), suggesting that ITIH4-AS1, FUS, and STAT3 formed a complex. In addition, we revealed that FUS knockdown abrogated the upregulation of nuclear phosphorylated (p)-STAT3, but not total activated STAT3, in ITIH4-AS1-overexpressing CRC cells, with the level of total STAT3 unchanged all the way (Figures 6F and 6G). At length, the IF staining testified that forced ITIH4-AS1 overexpression accelerated the nuclear translocation of p-STAT3 in HT-29 cells, whereas such acceleration was abrogated in the context of si-FUS co-transfection (Figure 6H). On the whole, we concluded that ITIH4-AS1 helped the nuclear translocation of activated STAT3 by an FUS-mediated way.
Discussion
In the past decades, the role of lncRNAs in human cancers has become an attractive research spot. Mounting evidence has recognized lncRNAs as oncogenes or tumor suppressors that affect the onset and development of a large number of human cancers through regulating multiple cellular processes.21, 22 Meanwhile, the implication of lncRNAs in CRC progression has also been uncovered recently. For example, lncRNA CCAL is verified as a contributor in CRC development.23 lncRNA ASBEL promotes the tumorigenicity of CRC cells by inhibiting ATF3 via interacting with TCF3.24 Also, lncRNA OCC-1 acts as a tumor suppressor in CRC through impairing HuR stabilization.25 In the present study, we identified a novel lncRNA ITIH4-AS1 as a key regulator in CRC. Bioinformatics data revealed that ITIH4-AS1 was usually expressed at low level in almost all kinds of normal tissues except liver tissues, and quantitative real-time PCR data validated that ITIH4-AS1 was dramatically upregulated in CRC cell lines. Of note, this study was the first one to unveil ITIH4-AS1 as a facilitator in CRC cell growth and metastasis both in vitro and in vivo. Moreover, we validated that ITIH4-AS1 in CRC was transcriptionally repressed by REST, a transcription repressor to control gene expression in human diseases.26, 27 A previous study also proved that REST was downregulated in CRC and functioned as an anti-tumor gene.28 These findings suggested that upregulation of ITIH4-AS1 was partially attributed to the low expression of REST in CRC.
JAK/STAT3 signaling is a well-identified oncogenic pathway that plays a pivotal part in various biological processes, including cell proliferation, survival, migration, invasion, and EMT in multiple cancers,29 and the activation of the JAK/STAT3 pathway in CRC has been largely indicated.19, 30, 31 As an example, herein we elucidated that the contribution of ITIH4-AS1 to CRC tumorigenesis was partly mediated by the activated JAK/STAT3 signaling pathway. FUS is a multi-function RNA-binding protein that interacts with both DNA and RNA.32, 33 A recent report elucidated that LINC00470 regulates AKT activation and nuclear translocation via interacting with FUS.20 In the current study, using online starBase 2.0, we found that FUS was the only RNA-binding protein (RBP) that binds with ITIH4-AS1, and then confirmed the interaction of FUS with both ITIH4-AS1 and STAT3 in CRC cells. Intriguingly, this study elucidated the mechanism that ITIH4-AS1 formed a complex with FUS and STAT3, and helped STAT3 nuclear translocation to activate JAK/STAT3 signaling in CRC cells, opposite to a previous finding that lncRNA OLA1P2 confines the nucleus import of STAT3 in CRC.34
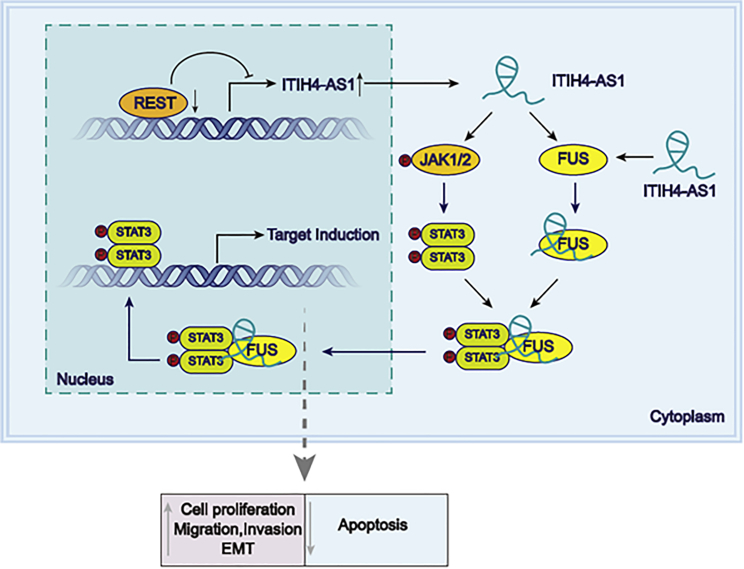
In conclusion, the observations in the present study unmasked that REST downregulation-stimulated lncRNA ITIH4-AS1 promoted CRC progression through interacting with FUS to activate the tumorigenic JAK/STAT3 signaling (Figure 7), suggesting ITIH4-AS1 as an important regulator in CRC progression, which may provide a new promising road to treating patients with CRC.
Figure 7.
REST Downregulation-Enhanced ITIH4-AS1 Acted as a Facilitator in CRC through Activating the Tumorigenic JAK/STAT3 Signaling by Stimulating STAT3 Translocation via Interacting with FUS
Materials and Methods
Cell Culture and Transfection
All five human CRC cell lines (HT-29, SW620, SW480, HCT116, and LoVo) and the human colonic epithelial cell line NCM460 were provided by American Type Culture Collection (ATCC; Manassas, VA, USA). Cells were all grown in RPMI 1640 (GE Healthcare, Chicago, IL, USA) containing 10% fetal bovine serum (FBS; GE Healthcare) at 37°C in a humidified incubator with 5% CO2. Short hairpin RNA (shRNA) targeting ITIH4-AS1 (shITIH4-AS1) or REST (shREST) and the negative control (shCtrl) were purchased from Sigma-Aldrich (USA) and then ligated into the pGIPZ lentiviral vector (Open Biosystems, Huntsville, AL, USA). In addition, pGIPZ vectors sub-cloned with full-length ITIH4-AS1 (pGIPZ/ITIH4-AS1) or REST cDNA (pGIPZ/REST) were respectively used to overexpress ITIH4-AS1 or REST, with the empty control (pGIPZ/control) serving as negative controls. Afterward, HEK293T cells were co-transfected with Lenti-Pac HIV Expression Packaging Mix and above lentiviral vectors by using Lipofectamine 2000 (Life Technologies, USA) according to the manufacturer’s guides. Subsequently, LoVo cells were transfected with lentivirus containing shITIH4-AS1 or shCtrl, whereas HT-29 cells were transfected with those containing pGIPZ/ITIH4-AS1 or pGIPZ/control. Finally, the stably transfected cells that were then applied in subsequent experiments were selected by puromycin (2 μg/mL) treatment for 2 weeks.
Quantitative Real-Time PCR
RNA isolation from cells was processed with TRIzol reagent (Invitrogen, USA). Then the reverse transcription of total RNA to cDNA was performed by the use of Moloney Murine Leukemia Virus Reverse Transcriptase (M-MLVRT; Promega, USA). Quantitative real-time PCR was conducted on Applied Biosystems 7500 Sequence Detection system (ABI, USA) using SYBR green real-time Master Mix (Takara, Japan). GAPDH was used as an internal control, and the relative expression of genes was calculated with the 2−ΔΔCt method. The sequences of primers used here were shown in Table 1.
Table 1.
Primer Sequences for Gene Amplification
| Gene | Strand | Sequences |
|---|---|---|
| ITIH4-AS1 | forward | 5′-GTACCGGCCACTTACAGCTT-3′ |
| reverse | 5′-TGAGAGTGCGTGTTCCTCAG-3′ | |
| E-cadherin | forward | 5′-GCCTCCTGAAAAGAGAGTGGAAG-3′ |
| reverse | 5′-TGGCAGTGTCTCTCCAAATCCG-3′ | |
| β-Catenin | forward | 5′-CACAAGCAGAGTGCTGAAGGTG-3′ |
| reverse | 5′-GATTCCTGAGAGTCCAAAGACAG-3′ | |
| N-cadherin | forward | 5′-CCTCCAGAGTTTACTGCCATGAC-3′ |
| reverse | 5′-GTAGGATCTCCGCCACTGATTC-3′ | |
| Vimentin | forward | 5′-GCTACAGCATGATGCAGGACCA-3′ |
| reverse | 5′-TCTGCGAGCTGGTCATGGAGTT-3′ | |
| REST | forward | 5′-CTTTGTCCTTACTCAAGTTCTCAG-3′ |
| reverse | 5′-ACCTGTCTTGCATGGCGGGTTA-3′ |
Western Blotting
The density of total proteins extracted from cells was examined by BCA Protein Assay kit (Takara). Afterward, proteins were segregated by 12% sodium lauryl sulfate-polyacrylamide gels and then transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, USA). After being blocked by 5% non-fat milk, the membranes were incubated with specific primary antibodies, followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibody (Santa Cruz, USA). Finally, proteins were visualized and determined by the use of enhanced chemiluminescence (Beyotime, China) following the manufacturer’s instructions. The primary antibodies used here were as below: anti-E-cadherin (#14472, 1:1,000; Cell Signaling Technology, Boston, MA, USA), anti-β-catenin (#8480, 1:1,000; Cell Signaling Technology), anti-N-cadherin (#13116, 1:1,000; Cell Signaling Technology), anti-STAT3 (#9139, 1:1,000; Cell Signaling Technology), anti-p-STAT3 (#9145, 1:2,000; Cell Signaling Technology), anti-Vimentin (ab193555, 1:1,000; Abcam, Cambridge, UK), anti-JAK1/2 (ab108596, 1:3,000; Abcam), anti-p-JAK1/2 (ab195055, 1:2,000; Abcam), anti-FUS (ab124923, 1:2,000; Abcam), and anti-GAPDH (ab8245, 1:5,000; Abcam).
Cell Viability and Proliferation Assays
Cell viability and proliferation were detected, respectively, by CCK-8 assay and colony formation assay. According to the manufacturer’s instructions, the viability of cells cultured for 24, 48, 72, and 96 h was examined using Cell Counting Kit-8 (CCK-8; Dojindo, Kumamoto, Japan). With respect to the colony formation assay, cells seeded in a six-well plate were cultured in complete culture medium for 2 weeks, with the medium changed every 3 days. Then colonies containing more than 50 cells were fixed, stained, and counted. The data collected from five random fields were finally applied for statistics.
Caspase-3 Activity Assay
To evaluate the impact of ITIH4-AS1 on CRC cell apoptosis, the caspase-3 activity was assayed by using colorimetric assay kit CASPASE-3 Cellular Activity Assay Kit PLUS (Biomol, Plymouth Meeting, PA, USA) based on the manufacturer’s instructions. The free pNA (p-nitroaniline) released from colorimetric substrate N-acetyl-Asp-Glu-Val-Asp-pNA was determined at 405 nm by the use of a multi-well microtiter plate reader (Spectra MAX 340pc; Molecular Devices, Sunnyvale, CA, USA). Caspase-3 activity was evaluated as the amount of released pNA (nmol) per minute per milligram of protein. The results were presented as a percentage of the control group.
Transwell Assay
The migratory and invasive capacities of CRC cells were respectively estimated by 24-well Transwell chambers without or with Matrigel (BD Pharmingen, San Jose, CA, USA). In brief, cells (2 × 105) cultured in 100 μL of serum-free RPMI 1640 were added into the upper inserts of the 8-μm Transwell chamber, whereas 500 μL of RPMI 1640 with 10% FBS was supplemented into the lower chamber. After 24 h of incubation, cells migrating or invading to the lower surface were fixed, stained, and counted.
Immunofluorescence (IF) Analysis
The PBS-rinsed cells were fixated by 4% paraformaldehyde and then permeabilized using 0.25% Triton X-100 in PBS, followed by blocking with 1% BSA. After being washed three times by PBS, cell incubation with primary antibodies was processed at 4°C overnight. Subsequently, cells were incubated with Cy3-conjugated secondary antibody for 1 h and the nuclei stained by DAPI. Images were captured under an immunofluorescence microscope (Olympus), with the analysis conducted by a Nikon and spot image acquisition system. The primary antibodies applied here were anti-E-cadherin (#14472, 1:50; Cell Signaling Technology), anti-N-cadherin (#13116, 1:200; Cell Signaling Technology), anti-FUS (ab124923, 1:100; Abcam), and anti-p-STAT3 (#9145, 1:100; Cell Signaling Technology).
Chromatin Immunoprecipitation (ChIP)
The ChIP assay was performed to validate the binding of REST to ITIH4-AS1 promoter as described previously.35 In short, the cell lysates were incubated with REST antibody (17-641, 1:100; Millipore, MA, USA) or IgG antibody (negative control; #3900, 1:100; Cell Signaling Technology). The DNA fragments in the immunoprecipitated complex eluted from Dynabeads G (Thermo Fisher Scientific, Waltham, MA, USA) were then reverse transcribed, amplified, and determined by quantitative real-time PCR.
Luciferase Reporter Assay
The effect of REST on ITIH4-AS1 transcription was assessed by performing luciferase reporter assays in HEK293T cells. In brief, the recombinant plasmids (pGL3 vector with ITIH4-AS1 promoter) were transfected into REST-overexpressed or -inhibited HEK293T cells using Lipofectamine 2000 (Invitrogen). Forty-eight hours after transfection, cells were collected and the luciferase activity was measured by the Dual-Luciferase Reporter Assay System (Promega, Wisconsin, WI, USA), and the pGL3 vectors were purchased from Promega (Madison, WI, USA).
RNA-Binding Protein Immunoprecipitation (RIP) Assay
The RIP experiments were conducted by using a Magna RNA-binding protein immunoprecipitation kit (Millipore, Billerica, MA, USA), following the manufacturer’s instructions. In brief, cell extracts were mixed with agarose beads with antibodies against FUS or IgG (negative control). Then the precipitated RNAs were analyzed by quantitative real-time PCR.
RNA Pull-Down Assay
First, the full length of ITIH4-AS1 and its antisense sequences were biotin labeled and transcribed by the Biotin RNA Labeling Mix (Roche) and AmpliScripe T7/SP6-flash Transcription Kit (Epicenter), respectively. Thereafter, cell lysates were mixed with the above biotin-labeled RNAs and then further incubated with Streptavidin Dynabeads (Dyna beads M-280 Streptavidin, #11205D; Invitrogen). At last, the proteins pulled down by ITIH4-AS1 or its antisense sequence were purified and examined by western blotting.
Co-immunoprecipitation (coIP) Assay
The coIP assays were carried out to detect the interactions among FUS, STAT3, and ITIH4-AS1 in HT-29 cells in light of protocols provided by a previous report.20
Animal Experiments
The animal experiments were all carried out in line with the guidelines of China Association of Laboratory Animal Care. All of the BALB/c nude mice the age of 6 weeks were purchased from Vital River (China) and kept in germ-free conditions. For the xenograft assay, LoVo cells transfected with shCtrl or shITIH4-AS1 were then inoculated subcutaneously into the left flanks of mice (5 × 106 cells/mouse; n = 6 for each group). The tumor volume was examined every 4 days and calculated according to the equation length × (width)2 × 0.5. Four weeks later, the mice were sacrificed, and the formed tumors were excised and weighted. Finally, the paraffin-embedded tumors were sliced and used to evaluate Ki67 expression by conducting immunohistochemical (IHC) staining as previously described.36 For in vivo metastatic assay, the indicated LoVo cells were injected into the tail vein of mice. After being injected for 5–6 weeks, the mice were euthanatized with the liver dissected out and paraffin embedded. The animal experiments were approved by the Ethics Committee of First Hospital/First Clinical College of Shanxi Medical University, and all the procedures were conducted in line with the national guidelines for the care and use of laboratory animals (GB14925–2010).
Statistical Analysis
All data collected from three independent experiments were statistically analyzed by using the SPSS 18.0 statistical software package (SPSS, Chicago, IL, USA), with results displayed as means ± SD. Differences between two groups were determined by Student’s t test, whereas differences among at least three groups were assessed using one-way ANOVA. p < 0.05 was thought to be statistically significant.
Author Contributions
C.L. and T.Z. obtained several data through bioinformatics analyses, subsequently designed the experiments in this study, and also performed cell transfection, western blot, and all of the mechanism-related assays, and wrote the Results in the manuscript as well. Quantitative real-time PCR and all of the in vitro functional assays were conducted by H.L. and F.H., and the Abstract and Discussion in the manuscript were also done by them. The in vivo experiments were carried out by X.Z. and Y.Z., whereas the schedule model in Figure 7 was accomplished by X.C. and C.H., and the rest of this manuscript was finished by Y.Q. and Y.D. L.M. and J.G. are the corresponding authors.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We thank all of the supporters of our work for their assistance. Funding:This study was funding by Shanxi Provincial Key Research and Development Program(201703D32111143), Shanxi Medical University Doctor’s Startup Fund Project (XD1802).
Contributor Information
Liang Ming, Email: liang8184107300@163.com.
Jiansheng Guo, Email: huajiang29974939@163.com.
References
- 1.Jemal A., Bray F., Center M.M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA Cancer J. Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Brenner H., Kloor M., Pox C.P. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 3.Böckelman C., Engelmann B.E., Kaprio T., Hansen T.F., Glimelius B. Risk of recurrence in patients with colon cancer stage II and III: a systematic review and meta-analysis of recent literature. Acta Oncol. 2015;54:5–16. doi: 10.3109/0284186X.2014.975839. [DOI] [PubMed] [Google Scholar]
- 4.Wolpin B.M., Meyerhardt J.A., Mamon H.J., Mayer R.J. Adjuvant treatment of colorectal cancer. CA Cancer J. Clin. 2007;57:168–185. doi: 10.3322/canjclin.57.3.168. [DOI] [PubMed] [Google Scholar]
- 5.Mayer R.J. Targeted therapy for advanced colorectal cancer—more is not always better. N. Engl. J. Med. 2009;360:623–625. doi: 10.1056/NEJMe0809343. [DOI] [PubMed] [Google Scholar]
- 6.Corley D.A., Jensen C.D., Quinn V.P., Doubeni C.A., Zauber A.G., Lee J.K., Schottinger J.E., Marks A.R., Zhao W.K., Ghai N.R. Association Between Time to Colonoscopy After a Positive Fecal Test Result and Risk of Colorectal Cancer and Cancer Stage at Diagnosis. JAMA. 2017;317:1631–1641. doi: 10.1001/jama.2017.3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolf A.M.D., Fontham E.T.H., Church T.R., Flowers C.R., Guerra C.E., LaMonte S.J., Etzioni R., McKenna M.T., Oeffinger K.C., Shih Y.T. Colorectal cancer screening for average-risk adults: 2018 guideline update from the American Cancer Society. CA Cancer J. Clin. 2018;68:250–281. doi: 10.3322/caac.21457. [DOI] [PubMed] [Google Scholar]
- 8.Kung J.T., Colognori D., Lee J.T. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibb E.A., Brown C.J., Lam W.L. The functional role of long non-coding RNA in human carcinomas. Mol. Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 11.Han D., Wang M., Ma N., Xu Y., Jiang Y., Gao X. Long noncoding RNAs: novel players in colorectal cancer. Cancer Lett. 2015;361:13–21. doi: 10.1016/j.canlet.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 12.Luo J., Qu J., Wu D.-K., Lu Z.-L., Sun Y.-S., Qu Q. Long non-coding RNAs: a rising biotarget in colorectal cancer. Oncotarget. 2017;8:22187–22202. doi: 10.18632/oncotarget.14728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Damas N.D., Marcatti M., Côme C., Christensen L.L., Nielsen M.M., Baumgartner R., Gylling H.M., Maglieri G., Rundsten C.F., Seemann S.E. SNHG5 promotes colorectal cancer cell survival by counteracting STAU1-mediated mRNA destabilization. Nat. Commun. 2016;7:13875. doi: 10.1038/ncomms13875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taniue K., Kurimoto A., Sugimasa H., Nasu E., Takeda Y., Iwasaki K., Nagashima T., Okada-Hatakeyama M., Oyama M., Kozuka-Hata H. Long noncoding RNA UPAT promotes colon tumorigenesis by inhibiting degradation of UHRF1. Proc. Natl. Acad. Sci. USA. 2016;113:1273–1278. doi: 10.1073/pnas.1500992113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He F., Song Z., Chen H., Chen Z., Yang P., Li W., Yang Z., Zhang T., Wang F., Wei J. Long noncoding RNA PVT1-214 promotes proliferation and invasion of colorectal cancer by stabilizing Lin28 and interacting with miR-128. Oncogene. 2019;38:164–179. doi: 10.1038/s41388-018-0432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chong J.A., Tapia-Ramírez J., Kim S., Toledo-Aral J.J., Zheng Y., Boutros M.C., Altshuller Y.M., Frohman M.A., Kraner S.D., Mandel G. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- 17.Negrini S., Prada I., D’Alessandro R., Meldolesi J. REST: an oncogene or a tumor suppressor? Trends Cell Biol. 2013;23:289–295. doi: 10.1016/j.tcb.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 18.Slattery M.L., Lundgreen A., Kadlubar S.A., Bondurant K.L., Wolff R.K. JAK/STAT/SOCS-signaling pathway and colon and rectal cancer. Mol. Carcinog. 2013;52:155–166. doi: 10.1002/mc.21841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiong H., Zhang Z.G., Tian X.Q., Sun D.F., Liang Q.C., Zhang Y.J., Lu R., Chen Y.X., Fang J.Y. Inhibition of JAK1, 2/STAT3 signaling induces apoptosis, cell cycle arrest, and reduces tumor cell invasion in colorectal cancer cells. Neoplasia. 2008;10:287–297. doi: 10.1593/neo.07971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu C., Zhang Y., She X., Fan L., Li P., Feng J., Fu H., Liu Q., Liu Q., Zhao C. A cytoplasmic long noncoding RNA LINC00470 as a new AKT activator to mediate glioblastoma cell autophagy. J. Hematol. Oncol. 2018;11:77. doi: 10.1186/s13045-018-0619-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huarte M. The emerging role of lncRNAs in cancer. Nat. Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 22.Schmitt A.M., Chang H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma Y., Yang Y., Wang F., Moyer M.-P., Wei Q., Zhang P., Yang Z., Liu W., Zhang H., Chen N. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/β-catenin signalling pathway via suppression of activator protein 2α. Gut. 2016;65:1494–1504. doi: 10.1136/gutjnl-2014-308392. [DOI] [PubMed] [Google Scholar]
- 24.Taniue K., Kurimoto A., Takeda Y., Nagashima T., Okada-Hatakeyama M., Katou Y., Shirahige K., Akiyama T. ASBEL-TCF3 complex is required for the tumorigenicity of colorectal cancer cells. Proc. Natl. Acad. Sci. USA. 2016;113:12739–12744. doi: 10.1073/pnas.1605938113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lan Y., Xiao X., He Z., Luo Y., Wu C., Li L., Song X. Long noncoding RNA OCC-1 suppresses cell growth through destabilizing HuR protein in colorectal cancer. Nucleic Acids Res. 2018;46:5809–5821. doi: 10.1093/nar/gky214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ooi L., Wood I.C. Chromatin crosstalk in development and disease: lessons from REST. Nat. Rev. Genet. 2007;8:544–554. doi: 10.1038/nrg2100. [DOI] [PubMed] [Google Scholar]
- 27.Griffith E.C., Cowan C.W., Greenberg M.E. REST acts through multiple deacetylase complexes. Neuron. 2001;31:339–340. doi: 10.1016/s0896-6273(01)00386-5. [DOI] [PubMed] [Google Scholar]
- 28.Westbrook T.F., Martin E.S., Schlabach M.R., Leng Y., Liang A.C., Feng B., Zhao J.J., Roberts T.M., Mandel G., Hannon G.J. A genetic screen for candidate tumor suppressors identifies REST. Cell. 2005;121:837–848. doi: 10.1016/j.cell.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 29.Yu H., Lee H., Herrmann A., Buettner R., Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat. Rev. Cancer. 2014;14:736–746. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 30.Liang Q., Ma D., Zhu X., Wang Z., Sun T.-T., Shen C., Yan T., Tian X., Yu T., Guo F. RING-Finger Protein 6 Amplification Activates JAK/STAT3 Pathway by Modifying SHP-1 Ubiquitylation and Associates with Poor Outcome in Colorectal Cancer. Clin. Cancer Res. 2018;24:1473–1485. doi: 10.1158/1078-0432.CCR-17-2133. [DOI] [PubMed] [Google Scholar]
- 31.Xue X., Ramakrishnan S.K., Weisz K., Triner D., Xie L., Attili D., Pant A., Győrffy B., Zhan M., Carter-Su C. Iron Uptake via DMT1 Integrates Cell Cycle with JAK-STAT3 Signaling to Promote Colorectal Tumorigenesis. Cell Metab. 2016;24:447–461. doi: 10.1016/j.cmet.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan A.Y., Manley J.L. TLS/FUS: a protein in cancer and ALS. Cell Cycle. 2012;11:3349–3350. doi: 10.4161/cc.21875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan A.Y., Manley J.L. The TET family of proteins: functions and roles in disease. J. Mol. Cell Biol. 2009;1:82–92. doi: 10.1093/jmcb/mjp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo H., Liu J., Ben Q., Qu Y., Li M., Wang Y., Chen W., Zhang J. The aspirin-induced long non-coding RNA OLA1P2 blocks phosphorylated STAT3 homodimer formation. Genome Biol. 2016;17:24. doi: 10.1186/s13059-016-0892-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiang J.-F., Yin Q.-F., Chen T., Zhang Y., Zhang X.-O., Wu Z., Zhang S., Wang H.B., Ge J., Lu X. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014;24:513–531. doi: 10.1038/cr.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ling J., Wang F., Liu C., Dong X., Xue Y., Jia X., Song W., Li Q. FOXO1-regulated lncRNA LINC01197 inhibits pancreatic adenocarcinoma cell proliferation by restraining Wnt/β-catenin signaling. J. Exp. Clin. Cancer Res. 2019;38:179. doi: 10.1186/s13046-019-1174-3. [DOI] [PMC free article] [PubMed] [Google Scholar]