Abstract
Noncoding RNAs play an important role in the pathogenesis of pulmonary arterial hypertension (PAH). In this study, we investigated the roles of hsa_circ_0016070, miR-942, and CCND1 in PAH. circRNA microarray was used to search circRNAs involved in PAH, whereas real-time PCR and western blot analysis were performed to detect miR-942 and CCND1 expression in different groups. In addition, the effect of miR-942 on CCND1 expression, as well as the effect of hsa_circ_0016070 on the expression of miR-942 and CCND1, was also studied using real-time PCR and western blot analysis. Moreover, MTT assay and flow cytometry were used to detect the effect of hsa _circ_0016070 on cell proliferation and cell cycle. According to the results of circRNA microarray analysis, hsa _circ_0016070 was identified to be associated with the risk of PAH in chronic obstructive pulmonary disease (COPD) patients. The miR-942 level in the COPD(+) PAH(+) group was much lower than that in the COPD(+) PAH(−) group, while the CCND1 level in the COPD(+) PAH(+) group was much higher. CCND1 was identified as a candidate target gene of miR-942, and the luciferase assay showed that the luciferase activity of wild-type CCND1 3′ UTR was inhibited by miR-942 mimics. In addition, hsa _circ_0016070 reduced miR-942 expression and enhanced CCND1 expression. Furthermore, hsa _circ_0016070 evidently increased cell viability and decreased the number of cells arrested in the G1/G0 phase. In summary, the results of this study suggested that hsa_circ_0016070 was associated with vascular remodeling in PAH by promoting the proliferation of pulmonary artery smooth muscle cells (PASMCs) via the miR-942/CCND1. Accordingly, has_circ_0016070 might be used as a novel biomarker in the diagnosis and treatment of PAH.
Keywords: hsa _circ_0016070, PAH, smooth muscle, miR-942, CCND1
Introduction
Hypoxia-induced pulmonary hypertension (HPH) is a devastating outcome of extended exposure to low oxygen tension in alveoli.1 Featured by hyperproliferative remodeling and vasoconstriction of pulmonary arteries, HPH can result in the failure of the right ventricle and death.2 However, no effective treatment is available for HPH. In fact, the vasodilators used in the treatment of pulmonary arterial hypertension (PAH) tend to aggravate the conditions of oxygenation and ventilation in HPH.1, 3 The pathology of PAH mainly involves aberrant vascular remodeling, which is caused by abnormal cellular proliferation as well as the tolerance to apoptotic stimuli in pulmonary artery smooth muscle cells (PASMCs).4 Thus, the suppression of cell proliferation and the promotion of cell apoptosis can become an efficient strategy for the treatment of PAH.5 In addition, PAH induced by chronic hypoxia is a frequently observed type of PAH, since hypoxia is a well-known stimulus to the pathogenesis of PAH by enhancing the proliferation of PASMCs.2
As a type of noncoding RNA, circular RNA (circRNA) is generated by the connection of a 3′ splice donor site with a 5′ splice acceptor site in a primary transcript.6 In fact, after two circRNAs—DCC and SRY—were discovered in the 1990s, various circRNAs have been discovered and validated.6, 7 Compared to other RNA transcripts, circRNAs are highly abundant, conserved, and stable, and they are predominantly located in the cytoplasm.8 In fact, circRNAs are synthesized through multiple unique mechanisms relying on sequence complementarity within lariat precursors, exon skippings, and flanking introns.8, 9, 10 Furthermore, circRNA expression is controlled by RNA-binding proteins and RNA-editing enzymes, including Quaking and ADAR.9, 11 Similar to linear RNAs, circRNAs are also synthesized from introns or exons at splice sites and, hence, require the spliceosomal machinery.12 circRNA is a novel type of endogenous noncoding RNA. It has been shown that circRNAs can act as the sponges of microRNAs (miRNAs) to regulate the expression of their parent genes and, hence, affect the onset and progression of diseases.13 It has been hypothesized that hsa_circ_0022342 and hsa_circ_0002062 can act as key circRNAs in the onset of chronic thromboembolic pulmonary hypertension (CTEPH). Therefore, the regulation of hsa_circ_0022342 and hsa_circ_0002062 may become an effective way for the treatment of CTEPH.14 Moreover, it has been demonstrated that, by inhibiting connective tissue growth factor (CTGF) expression, short hairpin RNAs (shRNAs) targeting CTGF suppressed the growth of PASMCs and inhibited the pulmonary vascular remodeling (PAR) induced by monocrotaline.15 It has also been shown that the knockdown of cyclin D1 (CCND1) by shRNA apparently suppressed the proliferation of PASMCs both in vitro and in vivo.16 In addition, it has been shown that the inhibition of miR-26b in PAR and the modulation in CCND1 and CTGF expression can exert a critical effect on the pathogenesis of PAH. Therefore, the regulation of these genes may lead to novel therapeutic treatments for PAH.17
Based on the previous data from circRNA microarrays, several circRNAs, are differentially expressed in PAH.14 In addition, we found that CCND1 was a virtual target of miR-942 by searching an online miRNA database (miRDB; http://www.mirdb.org). Meanwhile, CCND1 was considered as a gene involved in the pathogenesis of PAH by enhancing vascular remodeling.17 In this study, we investigated the roles of circRNA, miRNA, and CCND1 in PAH.
Results
Determination of circRNAs Involved in PAH
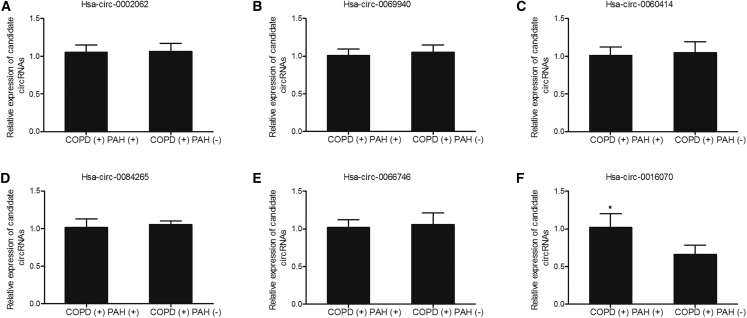
24 subjects diagnosed with chronic obstructive pulmonary disease (COPD) were recruited for our research and were further divided into two groups: 12 COPD patients with PAH, the COPD(+) PAH(+) group, and 12 COPD patients without PAH, the COPD(+) PAH(−) group. A circRNA microarray analysis was performed to search potential circRNAs involved in PAH, and the levels of hsa_circ_0002062, hsa_circ_0069940, hsa_circ_0060414, hsa_circ_0084265, hsa_circ_0066746, and hsa_circ_0016070 were measured in the aforementioned two groups. As shown in Figure 1, only the expression of hsa_circ_0016070 was increased in the COPD(+) PAH(+) group compared with that in the COPD(+) PAH(−) group, while the expression of hsa_circ_0002062, hsa_circ_0069940, hsa_circ_0060414, hsa_circ_0084265, and hsa-circ-0066746 in both groups was similar, suggesting that hsa_circ_0016070 was associated with risk of PAH in COPD patients.
Figure 1.
A circRNA Microarray Showed that hsa_circ_0016070 Was Involved in the Pathogenesis of PAH in COPD Patients
A circRNA microarray was used to identify the circRNAs involved in PAH and found that hsa_circ_0016070 was involved in the pathogenesis of PAH in COPD patients. *p < 0.05, versus the COPD(+) PAH(−) group. (A) hsa_circ_0002062. (B) hsa_circ_0069940. (C) hsa_circ_0060414. (D) hsa_circ_0084265. (E) hsa_circ_0066746. (F) hsa_circ_0016070.
Different Expression of miR-942 and CCND1 in Different Groups
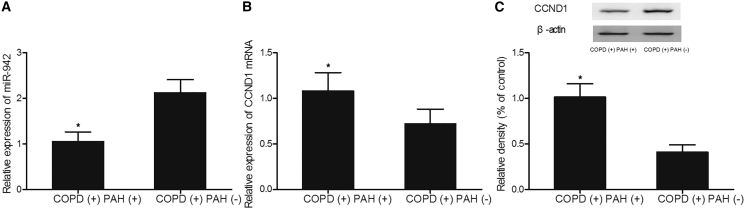
The underlying mechanism of hsa_circ_0016070 involvement in PAH was investigated using real-time PCR and western blot analysis. As shown in Figure 2, a lower level of miR-942 (Figure 2A) was observed in the COPD(+) PAH(+) group compared with that in the COPD(+) PAH(−) group, while higher levels of CCND1 mRNA (Figure 2B) and protein (Figure 2C) were observed in the COPD(+) PAH(+) group compared with those in the COPD(+) PAH(−) group.
Figure 2.
Differential Expression of miR-942 and CCND1 in Different Groups Was Detected Using Real-Time PCR and Western Blot Analysis
(A) The miR-942 level in the COPD(+) PAH(+) group was much lower than that in the COPD(+) PAH(−) group. *p < 0.05, versus the COPD(+) PAH(−) group. (B) The CCND1 mRNA level in the COPD(+) PAH(+) group was much higher than that in the COPD(+) PAH(−) group. *p < 0.05, versus the COPD(+) PAH(−) group. (C) The CCND1 protein level in the COPD(+) PAH(+) group was much higher than that in the COPD(+) PAH(−) group. *p < 0.05, versus the COPD(+) PAH(−) group.
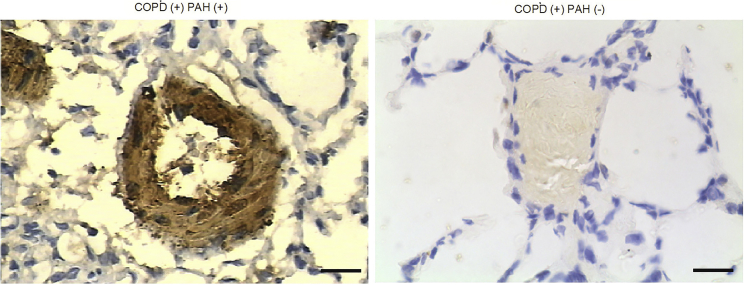
Meanwhile, immunohistochemistry (IHC) assay was also carried out to compare the CCND1 protein level between COPD(+) PAH(+) and COPD(+) PAH(−) groups, and the results showed that CCND1 protein level (Figure 3) in the COPD(+) PAH(+) group was much higher than that in the COPD(+) PAH(−) group. All results collectively indicated that miR-942 and CCND1 were associated with PAH.
Figure 3.
CCND1 Protein Level between COPD(+), PAH(+), and COPD(+) PAH(−) Groups
IHC assay was carried out to compare CCND1 protein levels between COPD(+) PAH(+) and COPD(+) PAH(−) groups. The CCND1 protein level in the COPD(+) PAH(+) group was evidently upregulated compared with that in the COPD(+) PAH(−) group.
CCND1 Was a Target Gene of miR-942
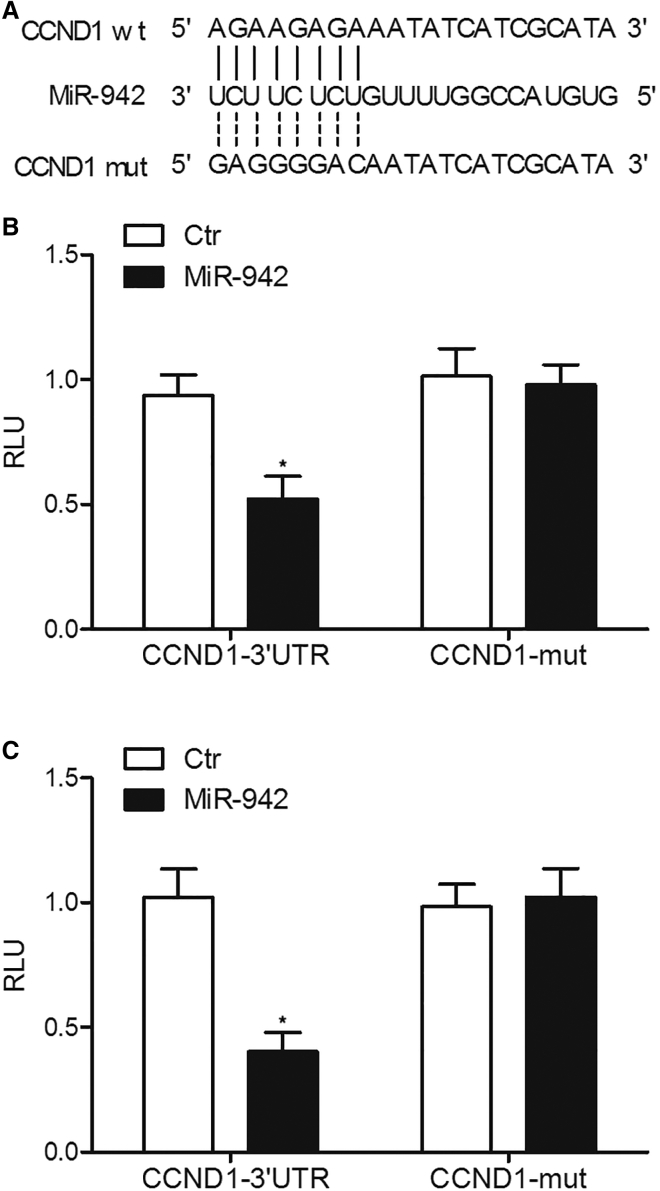
The results from computational screening (http://www.mirdb.org) and previous reports suggested that CCND1 was a virtual target gene of miR-942, with a complementary binding site located in the 3′ UTR of CCND1 (Figure 4). Subsequently, the QuikChange XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA) was used to generate a mutation in the 3′ UTR of CCND1, followed by luciferase assay analysis to validate the relationship between miR-942 and CCND1. As shown in Figure 4, HPASMCs or RPASMCs co-transfected with wild-type CCND1 and miR-942 mimics showed reduced luciferase activity, while the cells co-transfected with mutant CCND1 and miR-942 mimics showed no significant change in luciferase activity, suggesting that CCND1 was a virtual target of miR-942.
Figure 4.
CCND1 Was Validated to Be a Target Gene of miR-942 via Schematic Comparison and Luciferase Assays
(A) Schematic comparison by utilization of an online miRNA database miRDB (http://www.mirdb.org) between miR-942 and the “seed sequence” in the 3′ UTR of CCND1. (B) miR-942 mimics decreased the luciferase activity of HPASMC cells co-transfected with wild-type CCND1 3′ UTR but not the luciferase activity of cells co-transfected with mutant CCND1 3′ UTR. *p < 0.05, versus cells co-transfected with mutant CCND1 3′ UTR. (C) miR-942 mimics decreased the luciferase activity of RPASMC cells co-transfected with wild-type CCND1 3′ UTR but not the luciferase activity of cells co-transfected with mutant CCND1 3′ UTR. *p < 0.05, versus cells co-transfected with mutant CCND1 3′ UTR.
Expression of hsa_circ_0016070, miR-942, and CCND1 in Different Groups
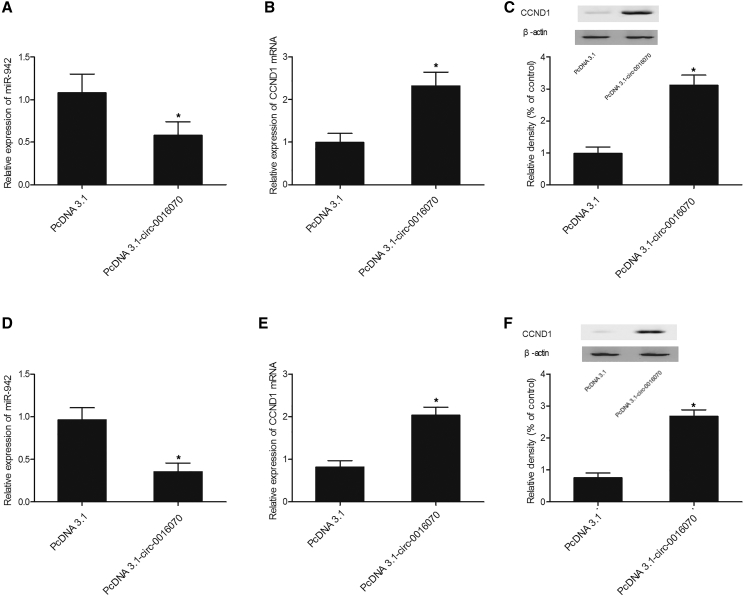
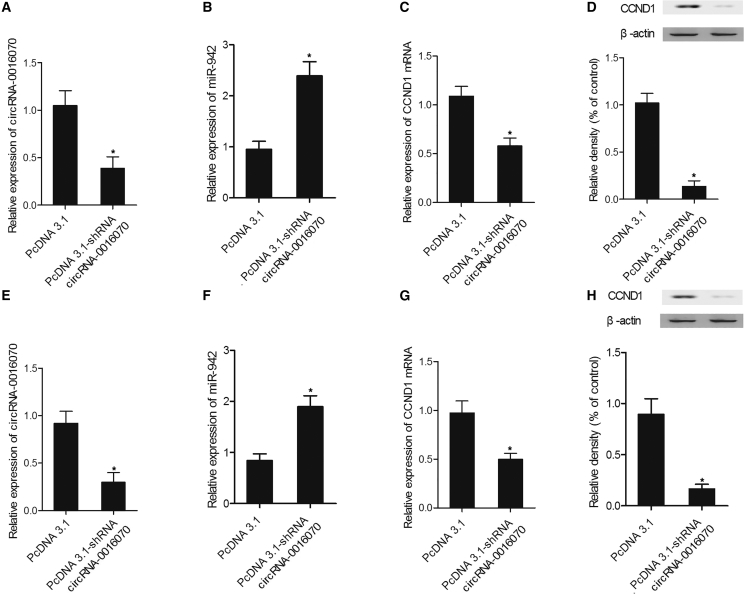
Real-time PCR and western blot analysis were used to measure the expression of hsa_circ_0016070, miR-942, and CCND1 in cells transfected with constructs carrying hsa_circ_0016070 and hsa_circ_0016070 shRNA. As shown in Figure 5, HPASMCs (Figure 5A) and RPASMCs (Figure 5D) transfected with hsa_circ_0016070 showed repressed miR-942 (Figures 5A and 5D) expression but evidently increased levels of CCND1 mRNA (Figures 5B and 5E) and protein (Figures 5C and 5F) compared with control cells. In addition, hsa_circ_0016070 shRNA downregulated hsa_circ_0016070 expression in HPASMCs (Figure 6A) and RPASMCs (Figure 6E). Furthermore, the downregulation of hsa_circ_0016070 increased miR-942 expression in HPASMCs (Figure 6B) and RPASMCs (Figure 6F). Finally, the levels of CCND1 mRNA (Figures 6C and 6G) and protein (Figures 6D and 6H) in HPASMCs (Figures 6C and 6D) and RPASMCs (Figures 6G and 6H) transfected with hsa_circ_0016070 shRNA were much lower than those in control cells.
Figure 5.
Expression of hsa_circ_0016070, miR-942, and CCND1 in Different Groups Was Detected Using Real-Time PCR and Western Blot Analysis
(A) hsa_circ_0016070 reduced the level of miR-942 in HPASMCs. *p < 0.05, versus control group. (B) hsa_circ_0016070 increased the level of CCND1 mRNA in HPASMCs. *p < 0.05, versus control group. (C) The CCND1 protein level in HPASMCs transfected with hsa_circ_0016070 was increased compared with that in negative control (NC). *p < 0.05, versus control group. (D) hsa_circ_0016070 reduced the level of miR-942 in RPASMCs. *p < 0.05, versus control group. (E) hsa_circ_0016070 increased the level of CCND1 mRNA in RPASMCs. *p < 0.05, versus control group. (F) The CCND1 protein level in RPASMCs transfected with hsa_circ_0016070 was increased compared with that in NC. *p < 0.05, versus control group.
Figure 6.
Effect of hsa_circ_0016070 shRNA on the Expression of hsa_circ_0016070, miR-942, and CCND1 Was Detected Using Real-Time PCR and Western Blot Analysis
(A) hsa_circ_0016070 shRNA reduced hsa_circ_0016070 expression in HPASMCs. *p < 0.05, versus control group. (B) Inhibition of hsa_circ_0016070 increased the expression of miR-942 in HPASMCs. *p < 0.05, versus control group. (C) The level of CCND1 mRNA in HPASMCs transfected with hsa_circ_0016070 shRNA was downregulated compared with that in NC. *p < 0.05, versus control group. (D) The level of CCND1 protein in HPASMC cells transfected with hsa_circ_0016070 shRNA was downregulated compared with that in NC. *p < 0.05, versus control group. (E) hsa_circ_0016070 shRNA reduced hsa_circ_0016070 expression in RPASMCs. *p < 0.05, versus control group. (F) Inhibition of hsa_circ_0016070 increased the expression of miR-942 in RPASMCs. *p < 0.05, versus control group. (G) The level of CCND1 mRNA in RPASMCs transfected with hsa_circ_0016070 shRNA was downregulated compared with that in NC. *p < 0.05, versus control group. (H) The level of CCND1 protein in RPASMCs transfected with hsa_circ_0016070 shRNA was downregulated compared with that in NC. *p < 0.05, versus control group.
Effect of hsa_circ_0016070 on Cell Proliferation and Cell Cycle
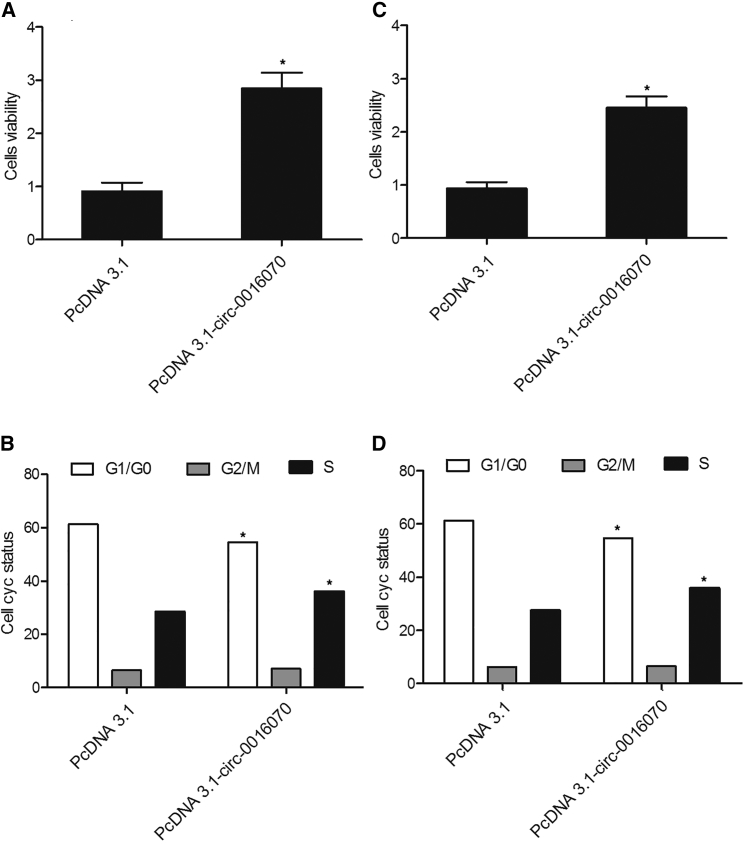
MTT assay and flow cytometry (FCM) analysis were performed to assess the effect of hsa_circ_0016070 on cell proliferation and cell cycle. As shown in Figure 7, HPASMCs (Figure 7A) and RPASMCs (Figure 7C) transfected with hsa_circ_0016070 showed increased cell viability. In addition, fewer HPASMCs (Figure 7B) and RPASMCs (Figure 7D) in the has_circ_0016070 group were arrested in the G1/G0 phase compared with those in the control group, accompanied by an increased number of cells in the S phase, suggesting that hsa_circ_0016070 increased cell viability.
Figure 7.
Effect of hsa_circ_0016070 on Cell Proliferation and Cell Cycle Was Detected via MTT Assay and Flow Cytometry
(A) HPASMCs transfected with hsa_circ_0016070 showed a higher growth rate than NC cells. *p < 0.05, versus control group. (B) Fewer HPASMCs were in the G1/G0 phase after transfection with hsa_circ_0016070, but the number of cells in the S phase was much higher after transfection. *p < 0.05, versus control group. (C) RPASMCs transfected with hsa_circ_0016070 showed a higher growth rate than NC cells. *p < 0.05, versus control group. (D) Fewer RPASMCs were in the G1/G0 phase after transfection with hsa_circ_0016070, but the number of cells in the S phase was much higher after transfection. *p < 0.05, versus control group.
Effect of hsa_circ_0016070 shRNA on Cell Proliferation and Cell Cycle
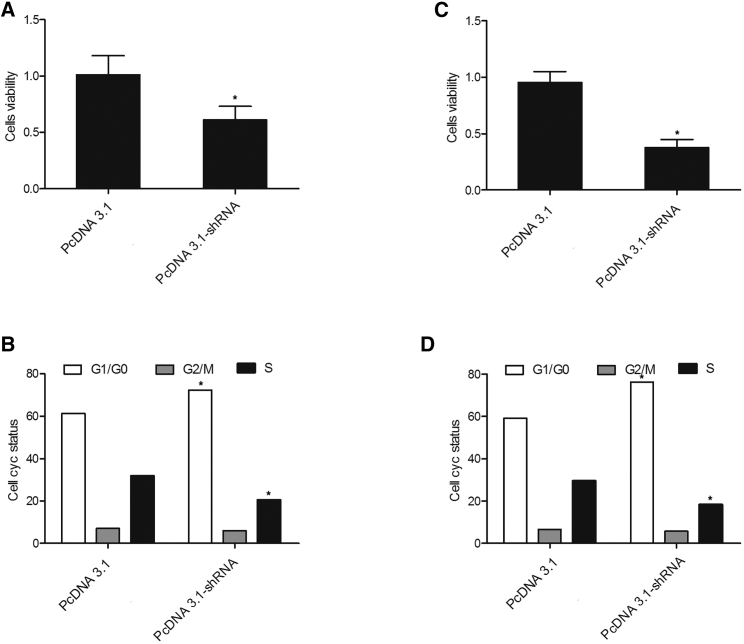
MTT assay and FCM analysis were performed to assess the effect of hsa_circ_0016070 shRNA on cell proliferation and cell cycle. As shown in Figure 8, HPASMCs (Figure 8A) and RPASMCs (Figure 8C) transfected with hsa_circ_0016070 shRNA showed decreased cell viability. In addition, more HPASMCs (Figure 8B) and RPASMCs (Figure 8D) in the hsa_circ_0016070 shRNA group were arrested in the G1/G0 phase compared with those in the control group, accompanied by an decreased number of cells in the S phase, suggesting that hsa_circ_0016070 downregulation decreased cell viability by arresting cell-cycle progression.
Figure 8.
Effect of hsa_circ_0016070 shRNA on Cell Proliferation and Cell Cycle Was Detected via MTT Assay and Flow Cytometry
(A) HPASMCs transfected with hsa_circ_0016070 shRNA showed a slower growth rate than NC cells. *p < 0.05, versus control group. (B) More HPASMCs were in the G1/G0 phase after transfection with hsa_circ_0016070 shRNA, but the number of cells in the S phase was much lower after transfection. *p < 0.05, versus control group. (C) RPASMCs transfected with hsa_circ_0016070 shRNA showed a slower growth rate than NC cells. *p < 0.05, versus control group. (D) More RPASMCs were in the G1/G0 phase after transfection with hsa_circ_0016070 shRNA, but the number of cells in the S phase was much lower after transfection. *p < 0.05, versus control group.
Discussion
As a progressive disease, PAH is caused by a wide range of cardiac or pulmonary disorders and is associated with high mortality and morbidity. PAH is featured by a progressive elevation in vascular remodeling, right heart dysfunction, and pulmonary arterial pressure. Consequently, PAH results in the failure of the right ventricle.18 Specific characteristics of vascular remodeling associated with PAH include the muscularization of distal arterioles in the lungs, the proliferation and apoptosis of PASMCs, perivascular inflammation, and the sedimentation of extracellular matrix protein.19
Previously, circRNAs have been shown to function as sponges of miRNAs and are believed to regulate the expression of their miRNA targets, thus contributing to the competing endogenous RNA network.20 It has been shown that circRNAs synthesized from the Sry gene (circSry) functions as a miR-138 sponge.21 In addition, circRNAs play a key role in diseases, including tumor, and thus may become novel therapeutic and diagnostic targets in disease treatment.22 Some other studies have indicated that circRNAs are implicated in the pathogenesis of disorders of the nervous system and atherosclerosis.23 It has been shown that the hsa_circ_0002062-hsa-miR-942-5p-CDK6 signaling pathway and the hsa_circ_0022342-hsa-miR-940-CRKL-ErbB pathway can play important roles in CTEPH development.14 In this study, we measured the expression of candidate circRNAs to identify the circRNAs involved in PAH and found that hsa_circ_0016070 was associated with the risk of HPH. In addition, using real-time PCR and western blot analysis, we investigated the potential mechanism underlying the role of hsa_circ_0016070 in PAH and found that miR-942 and CCND1 were also associated with PAH.
Target genes of hsa-miR-942–5p, such as CDK6, are primarily enriched in tumor pathway.22 Auger et al. have reported that the presence of an organized thrombus in major pulmonary arteries is typically associated with diseases including lung tumor.24 Therefore, it has been inferred that such pathways and axon guidance might be related to CTEPH.24 Furthermore, in pulmonary hypertension, CDK6 was shown to be involved in cell cycle and proliferation.25 It has been shown that miR-942 can activate the Wnt/β-catenin pathway by directly inhibiting the expression of TLE1, GSK3β, and sFRP4.26
CCND1 is a critical regulator of cell cycle in many types of cells. In addition, CCND1 can bind and activate cyclin-dependent kinase (CDK) 4, which then inactivates retinoblastoma (Rb) protein and results in the progression from G1 to S phase.27 CCND1 was also found to accumulate in the nuclei in the G1 phase and then translocate to the cytoplasm, where CCND1 is degraded by the ubiquitin-proteasome system.28 During the proteolysis of CCND1, the necessary phosphorylation of threonine-286 (T286) residue is mainly mediated by glycogen synthase kinase 3 beta (GSK3β).29 In addition, the phosphorylation of T286 can also be induced by other kinases such as p38 mitogen-activated protein kinase (MAPK), which can also result in CCND1 degradation.30 It has also been shown that CCND1 and CTGF played important roles in cigarette-smoke-induced PAH by accelerating the proliferation of PASMCs.31, 32 Furthermore, as an important member of the cyclin family, CCND1 can regulate cell-cycle progression, thus controlling the transition from G1 to S phase.33 Previously, a growing amount of evidence suggested that CTGF is implicated in the control of cell-cycle progression.34 It was also shown that CCND1 works as a “mitogenic sensor” to mediate PASMC proliferation by pushing the cells beyond the restriction point of the G1 phase.35, 36 It has also been demonstrated that the intratracheal administration of CCND1 or CTGF shRNA could apparently reverse the PAH induced by monocrotaline.15, 16 In addition, it was shown that CCND1 is involved in the function of CTGF by accelerating the G1/S transition, thus promoting the proliferation of PASMCs and facilitating the onset of PAH.37
In summary, we were the first to suggest that hsa_circ_0016070 was associated with vascular remodeling in PAH by promoting the proliferation of SMCs via miR-942-5p/CCND1. It has been found that hsa_circ_0016070, miR-942, and CCND1 were differentially expressed in PAH patients. In this study, we found that CCND1 was a virtual target of miR-942. Meanwhile, CCND1 was involved in the pathogenesis of PAH by enhancing vascular remodeling. Our research also suggested that hsa_circ_0016070 might be used as a novel biomarker in the diagnosis and treatment of PAH.
Materials and Methods
Human Subject Sample Collection
In this study, lung tissues from 24 COPD patients were collected. Among the subjects, 12 patients also suffered from PAH—the COPD(+) PAH(+) group—and the remaining 12 patients were PAH free—the COPD(+) PAH(−) group. Collected tissue samples were immediately frozen in liquid nitrogen and subsequently stored in a −80°C freezer. All patients had complete clinical data. This study was approved by the Ethical Committee of Anhui Medical University and conducted according to the Declaration of Helsinki. All subjects have signed informed consent.
Isolation of PASMCs
Human PASMCs (HPASMCs) were obtained from ATCC. Rat PASMCs (RPASMCs) were isolated as previously described.32, 37 The tissues collected were cut into pieces of 1-mm3 blocks under aseptic conditions, washed with D-Hank’s solution, and transferred into culture dishes. After adding 1 mg/mL collagenase I (Invitrogen, Carlsbad, CA, USA) about twice the size of the tissue samples and digested for 2 h at 37°C, PASMCs were collected by filtration using nylon nets with a 70-μm diameter. After centrifugation and washing with PBS, the cells were resuspended in DMEM containing 10% fetal bovine serum (FBS). When the cells reached a confluence rate of 80% to 90%, they were trypsinized with a 0.25% EDTA-trypsin solution. The cells were then passaged at a 1:3 ratio. The cells in the logarithmic growth phase were collected for subsequent experiments.
RNA Isolation and Real-Time PCR
Using a previously illustrated method,38, 39, 40 an RNeasy Mini Kit (QIAGEN, Hilden, Germany) was used according to the manufacturer’s protocol to extract total RNA from the tissue samples. The ratio of A260/A280, as well as RNA concentration, was measured by a UV spectrophotometer. Subsequently, isolated RNA was converted into cDNA using a Promega reverse transcription kit (Madison, WI, USA). The qPCR reaction was carried out using an ABI 7500 Real-Time qPCR machine (Applied Biosystems, Foster City, CA, USA). The reaction system was 20 μL, and the reaction condition included 40 cycles of 3 min at 95°C, 12 s at 95°C, and 50 s at 62°C. The quantification of circRNAs (has_circ_0002062, hsa_circ_0069940, hsa_circ_0060414, hsa_circ_0084265, hsa_circ_0066746, and hsa_circ_0016070), as well as of miR-942 and CCND1 mRNA, was carried out using U6 as an internal control. The 2−ΔΔCT method was used in the calculation of relative expression of the aforementioned transcripts.
Cell Culture and Transfection
Using a previously illustrated method,41, 42 HPASMCs and RPASMCs were cultured in DMEM containing 10% imported FBS, 100 mg/L streptomycin, and 100 U/mL penicillin. The cells were incubated at 37°C under 5% CO2 and saturated humidity. Subsequently, the cells were seeded into 96-well plates and transfected with different vectors (see the following text for more details) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. At 48 h after transfection, the cells were collected for subsequent assays.
Vector Construction and Mutagenesis
The full length of hsa_circ_0016070 was amplified by PCR. The PCR product was cloned into a pcDNA3.1 vector (Promega, Madison, WI, USA) downstream of the luciferase reporter gene. At the same time, an shRNA against hsa_circ_0016070 was also designed and inserted into the pcDNA3.1 vector to create hsa_circ_0016070 shRNA, which was later transfected into HPASMCs and RPASMCs to suppress the expression of endogenous hsa_circ_0016070. Similarly, the 3′ UTR region of the CCND1 gene was amplified and cloned into the pcDNA3.1 vector (wild-type CCND1 3′ UTR) downstream of the luciferase reporter gene. Subsequently, the binding site of miR-942 in the 3′ UTR of CCND1 was mutated by site-directed mutagenesis to create a vector carrying the mutant CCND1 3′ UTR and used in parallel with the wild-type CCND1 3′ UTR.
Luciferase Assay
At 48 h after transfection, the cells were collected, and the luciferase activity in each well of 96-well plates was measured on a Luminometer (TD-20/20; Turner Designs, Sunnyvale, CA, USA) using a Stop & Glo kit (Promega, Madison, WI, USA). The expression of each target gene was calculated from the ratio between the firefly and renilla (internal control) luciferase activities following the instruction of the kit.
Cell Proliferation Assay
The experiments were performed as previously described.43 The MTT (Sigma-Aldrich, St. Louis, MO, USA) assay was used according to the manufacturer’s protocol. The third generation of HPASMCs and RPASMCs was used for transfection, and the cell viability was measured using the MTT assay at 48 h after transfection. The proliferation rates of HPASMCs and RPASMCs were calculated by measuring their optical density (OD) value at 570 nm.
Western Blot Analysis
Both transfected cells and tissue samples were rinsed with PBS and subsequently lysed with 100 μL of a cell lysis solution at 4°C for 30 min. In the next step, the lysate was centrifuged at 12,000 × g for 10 min, while the concentration of protein in the lysate was measured by a BCA assay kit. Subsequently, 12% SDS-PAGE was used to dissolve sample proteins, which were then blotted onto a polyvinylidene fluoride (PVDF) membrane. After being blocked with 5% skim milk for 1 h at room temperature, the membrane was incubated with a 1:200 dilution of monoclonal anti-human CCND1 and anti-GAPDH (internal control) primary antibodies overnight at 4°C. In the next step, a 1:2,000 dilution of anti-mouse immunoglobulin G (IgG) secondary antibody was added onto the membrane and incubated for 1 h in the dark at room temperature. All antibodies were purchased from Abcam (Cambridge, MA, USA). Subsequently, the membrane was visualized in a dual-color infrared laser imaging system. The OD of each protein band was measured, and the expression of CCND1 was calculated as the ratio of the total OD of the CCND1 band to that of the GAPDH band.
Cell-Cycle Analysis
Cell-cycle analysis was done with the following procedures.44 At 48 h after transfection, cells were collected and washed with pre-cooled PBS solution and then centrifuged to collect cell pellets. After adjusting the cell concentration to 1 × 105/mL, the cells were immobilized by 1 mL 75% ethyl alcohol and incubated overnight at 4°C. Afterward, cells were incubated with 400 μL propidium iodide (PI) (Sigma-Aldrich, St. Louis, MO, USA) for 30 min at 37°C, and the cell-cycle profile was detected by a FACSCanto II flow cytometer (BD Biosciences, San Jose, CA, USA) at the wavelength of 488 nm.
Immunohistochemistry
The experiments were performed as previously described.45 The specimens were fixed in 10% formalin, embedded in paraffin, dried in a 60°C oven for 1 h, dewaxed by xylene, and dehydrated in graded alcohol. Subsequently, the slides were incubated in 3% H2O2 (Sigma-Aldrich, St. Louis, MO, USA) at 37°C for 30 min, boiled in 0.01 M citrate buffer at 95°C for 20 min, and then incubated with a normal sheep serum at 37°C for 10 min prior to incubation with anti-CCND1 primary antibody and horseradish-peroxidase-labeled secondary antibody (Bioss, Beijing, China) according to the manufacturer’s protocol. After being exposed to diaminobenzidine (Sigma-Aldrich, St. Louis, MO, USA) and hematoxylin staining, the expression of CCND1 in each sample slide was scored independently by two operators.
Statistical Analysis
SPSS 19.0 statistical software was used in the statistical analysis. The measurement data were displayed as , and the differences among different groups were evaluated by ANOVA, unpaired two-tailed t test. A p value of <0.05 was considered statistically significant.
Author Contributions
R.W. designed the project. S.Z. carried out most of the experiments, analyzed the data, and wrote the manuscript. H.J. acquired, analyzed, and interpreted data and drafted the manuscript. M.L., P.W., L.S., Y.L., K.Z., and B.Z. were responsible for the concept, analyzed and interpreted data, and performed critical revision of the manuscript. G.S. and C.C. helped to design and coordinate the experiment.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This research was supported by the funding from the Natural Science Foundation of China (81300041, 81970051), the fund for the academic backbone of the excellent young and middle-age people of Anhui Medical University (2013), the fund from The First Affiliated Hospital of Anhui Medical University for reserve talents (2014), and the fund for Excellent Top Talent Cultivation Project of Anhui Higher Education Institutions (gxyqZD2017030).
Contributor Information
Gengyun Sun, Email: sungengyun@hotmail.com.
Chao Cao, Email: caochaoningbo@126.com.
Ran Wang, Email: ranwangtjmu@hotmail.com.
References
- 1.Hoeper M.M., Barberà J.A., Channick R.N., Hassoun P.M., Lang I.M., Manes A., Martinez F.J., Naeije R., Olschewski H., Pepke-Zaba J. Diagnosis, assessment, and treatment of non-pulmonary arterial hypertension pulmonary hypertension. J. Am. Coll. Cardiol. 2009;54(1, Suppl):S85–S96. doi: 10.1016/j.jacc.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 2.Stenmark K.R., Fagan K.A., Frid M.G. Hypoxia-induced pulmonary vascular remodeling: cellular and molecular mechanisms. Circ. Res. 2006;99:675–691. doi: 10.1161/01.RES.0000243584.45145.3f. [DOI] [PubMed] [Google Scholar]
- 3.Ghofrani H.A., Wiedemann R., Rose F., Schermuly R.T., Olschewski H., Weissmann N., Gunther A., Walmrath D., Seeger W., Grimminger F. Sildenafil for treatment of lung fibrosis and pulmonary hypertension: a randomised controlled trial. Lancet. 2002;360:895–900. doi: 10.1016/S0140-6736(02)11024-5. [DOI] [PubMed] [Google Scholar]
- 4.Tuder R.M., Abman S.H., Braun T., Capron F., Stevens T., Thistlethwaite P.A., Haworth S.G. Development and pathology of pulmonary hypertension. J. Am. Coll. Cardiol. 2009;54(1, Suppl):S3–S9. doi: 10.1016/j.jacc.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 5.McMurtry M.S., Archer S.L., Altieri D.C., Bonnet S., Haromy A., Harry G., Bonnet S., Puttagunta L., Michelakis E.D. Gene therapy targeting survivin selectively induces pulmonary vascular apoptosis and reverses pulmonary arterial hypertension. J. Clin. Invest. 2005;115:1479–1491. doi: 10.1172/JCI23203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salzman J., Gawad C., Wang P.L., Lacayo N., Brown P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nigro J.M., Cho K.R., Fearon E.R., Kern S.E., Ruppert J.M., Oliner J.D., Kinzler K.W., Vogelstein B. Scrambled exons. Cell. 1991;64:607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- 8.Li J., Yang J., Zhou P., Le Y., Zhou C., Wang S., Xu D., Lin H.K., Gong Z. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am. J. Cancer Res. 2015;5:472–480. [PMC free article] [PubMed] [Google Scholar]
- 9.Ivanov A., Memczak S., Wyler E., Torti F., Porath H.T., Orejuela M.R., Piechotta M., Levanon E.Y., Landthaler M., Dieterich C., Rajewsky N. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170–177. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 10.Barrett S.P., Wang P.L., Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. eLife. 2015;4:e07540. doi: 10.7554/eLife.07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A., Roslan S., Schreiber A.W., Gregory P.A., Goodall G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 12.Chen L.L., Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qu S., Yang X., Li X., Wang J., Gao Y., Shang R., Sun W., Dou K., Li H. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Miao R., Wang Y., Wan J., Leng D., Gong J., Li J., Liang Y., Zhai Z., Yang Y. Microarray expression profile of circular RNAs in chronic thromboembolic pulmonary hypertension. Medicine (Baltimore) 2017;96:e7354. doi: 10.1097/MD.0000000000007354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang R., Zhou S.J., Zeng D.S., Xu R., Fei L.M., Zhu Q.Q., Zhang Y., Sun G.Y. Plasmid-based short hairpin RNA against connective tissue growth factor attenuated monocrotaline-induced pulmonary vascular remodeling in rats. Gene Ther. 2014;21:931–937. doi: 10.1038/gt.2014.62. [DOI] [PubMed] [Google Scholar]
- 16.Zeng D.X., Xu G.P., Lei W., Wang R., Wang C.G., Huang J.A. Suppression of cyclin D1 by plasmid-based short hairpin RNA ameliorated experimental pulmonary vascular remodeling. Microvasc. Res. 2013;90:144–149. doi: 10.1016/j.mvr.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 17.Wang R., Ding X., Zhou S., Li M., Sun L., Xu X., Fei G. Microrna-26b attenuates monocrotaline-induced pulmonary vascular remodeling via targeting connective tissue growth factor (CTGF) and cyclin D1 (CCND1) Oncotarget. 2016;7:72746–72757. doi: 10.18632/oncotarget.10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai Y.C., Potoka K.C., Champion H.C., Mora A.L., Gladwin M.T. Pulmonary arterial hypertension: the clinical syndrome. Circ. Res. 2014;115:115–130. doi: 10.1161/CIRCRESAHA.115.301146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schermuly R.T., Ghofrani H.A., Wilkins M.R., Grimminger F. Mechanisms of disease: pulmonary arterial hypertension. Nat. Rev. Cardiol. 2011;8:443–455. doi: 10.1038/nrcardio.2011.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu H.J., Zhang C.Y., Zhang S., Chang M., Wang H.Y. Microarray expression profile of circular RNAs in heart tissue of mice with myocardial infarction-induced heart failure. Cell. Physiol. Biochem. 2016;39:205–216. doi: 10.1159/000445617. [DOI] [PubMed] [Google Scholar]
- 21.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 22.Hansen T.B., Kjems J., Damgaard C.K. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 23.Burd C.E., Jeck W.R., Liu Y., Sanoff H.K., Wang Z., Sharpless N.E. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6:e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fedullo P., Kerr K.M., Kim N.H., Auger W.R. Chronic thromboembolic pulmonary hypertension. Am. J. Respir. Crit. Care Med. 2011;183:1605–1613. doi: 10.1164/rccm.201011-1854CI. [DOI] [PubMed] [Google Scholar]
- 25.White K., Loscalzo J., Chan S.Y. Holding our breath: The emerging and anticipated roles of microRNA in pulmonary hypertension. Pulm. Circ. 2012;2:278–290. doi: 10.4103/2045-8932.101395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ge C., Wu S., Wang W., Liu Z., Zhang J., Wang Z., Li R., Zhang Z., Li Z., Dong S. miR-942 promotes cancer stem cell-like traits in esophageal squamous cell carcinoma through activation of Wnt/β-catenin signalling pathway. Oncotarget. 2015;6:10964–10977. doi: 10.18632/oncotarget.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsushime H., Roussel M.F., Ashmun R.A., Sherr C.J. Colony-stimulating factor 1 regulates novel cyclins during the G1 phase of the cell cycle. Cell. 1991;65:701–713. doi: 10.1016/0092-8674(91)90101-4. [DOI] [PubMed] [Google Scholar]
- 28.Lukas J., Pagano M., Staskova Z., Draetta G., Bartek J. Cyclin D1 protein oscillates and is essential for cell cycle progression in human tumour cell lines. Oncogene. 1994;9:707–718. [PubMed] [Google Scholar]
- 29.Diehl J.A., Zindy F., Sherr C.J. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 1997;11:957–972. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- 30.Zou Y., Ewton D.Z., Deng X., Mercer S.E., Friedman E. Mirk/dyrk1B kinase destabilizes cyclin D1 by phosphorylation at threonine 288. J. Biol. Chem. 2004;279:27790–27798. doi: 10.1074/jbc.M403042200. [DOI] [PubMed] [Google Scholar]
- 31.Zeng D.X., Liu X.S., Xu Y.J., Wang R., Xiang M., Xiong W.N., Ni W., Chen S.X. Plasmid-based short hairpin RNA against cyclin D1 attenuated pulmonary vascular remodeling in smoking rats. Microvasc. Res. 2010;80:116–122. doi: 10.1016/j.mvr.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 32.Wang R., Xu Y.J., Liu X.S., Zeng D.X., Xiang M. Knockdown of connective tissue growth factor by plasmid-based short hairpin RNA prevented pulmonary vascular remodeling in cigarette smoke-exposed rats. Arch. Biochem. Biophys. 2011;508:93–100. doi: 10.1016/j.abb.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 33.Sherr C.J. G1 phase progression: cycling on cue. Cell. 1994;79:551–555. doi: 10.1016/0092-8674(94)90540-1. [DOI] [PubMed] [Google Scholar]
- 34.Kothapalli D., Grotendorst G.R. CTGF modulates cell cycle progression in cAMP-arrested NRK fibroblasts. J. Cell. Physiol. 2000;182:119–126. doi: 10.1002/(SICI)1097-4652(200001)182:1<119::AID-JCP13>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 35.Wu S.H., Wu X.H., Lu C., Dong L., Chen Z.Q. Lipoxin A4 inhibits proliferation of human lung fibroblasts induced by connective tissue growth factor. Am. J. Respir. Cell Mol. Biol. 2006;34:65–72. doi: 10.1165/rcmb.2005-0184OC. [DOI] [PubMed] [Google Scholar]
- 36.Qin X.Q., Livingston D.M., Kaelin W.G., Jr., Adams P.D. Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc. Natl. Acad. Sci. USA. 1994;91:10918–10922. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang R., Xu Y.J., Liu X.S., Zeng D.X., Xiang M. CCN2 promotes cigarette smoke-induced proliferation of rat pulmonary artery smooth muscle cells through upregulating cyclin D1 expression. J. Cell. Biochem. 2012;113:349–359. doi: 10.1002/jcb.23361. [DOI] [PubMed] [Google Scholar]
- 38.Wang R., Li M., Zhou S., Zeng D., Xu X., Xu R., Sun G. Effect of a single nucleotide polymorphism in miR-146a on COX-2 protein expression and lung function in smokers with chronic obstructive pulmonary disease. Int. J. Chron. Obstruct. Pulmon. Dis. 2015;10:463–473. doi: 10.2147/COPD.S74345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou S., Li M., Zeng D., Sun G., Zhou J., Wang R. Effects of basic fibroblast growth factor and cyclin D1 on cigarette smoke-induced pulmonary vascular remodeling in rats. Exp. Ther. Med. 2015;9:33–38. doi: 10.3892/etm.2014.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou S., Li M., Zeng D., Xu X., Fei L., Zhu Q., Zhang Y., Wang R. A single nucleotide polymorphism in 3′ untranslated region of epithelial growth factor receptor confers risk for pulmonary hypertension in chronic obstructive pulmonary disease. Cell. Physiol. Biochem. 2015;36:166–178. doi: 10.1159/000374061. [DOI] [PubMed] [Google Scholar]
- 41.Zhou S.J., Li M., Zeng D.X., Zhu Z.M., Hu X.W., Li Y.H., Wang R., Sun G.Y. Expression variations of connective tissue growth factor in pulmonary arteries from smokers with and without chronic obstructive pulmonary disease. Sci. Rep. 2015;5:8564. doi: 10.1038/srep08564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ding X., Zhou S., Li M., Cao C., Wu P., Sun L., Fei G., Wang R. Upregulation of SRF is associated with hypoxic pulmonary hypertension by promoting viability of smooth muscle cells via increasing expression of Bcl-2. J. Cell. Biochem. 2017;118:2731–2738. doi: 10.1002/jcb.25922. [DOI] [PubMed] [Google Scholar]
- 43.Wang R., Zhou S., Wu P., Li M., Ding X., Sun L., Xu X., Zhou X., Zhou L., Cao C., Fei G. Identifying involvement of H19-miR-675-3p-IGF1R and H19-miR-200a-PDCD4 in treating pulmonary hypertension with melatonin. Mol. Ther. Nucleic Acids. 2018;13:44–54. doi: 10.1016/j.omtn.2018.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou S., Sun L., Cao C., Wu P., Li M., Sun G., Fei G., Ding X., Wang R. Hypoxia-induced microRNA-26b inhibition contributes to hypoxic pulmonary hypertension via CTGF. J. Cell. Biochem. 2018;119:1942–1952. doi: 10.1002/jcb.26355. [DOI] [PubMed] [Google Scholar]
- 45.Zhou S., Liu Y., Li M., Wu P., Sun G., Fei G., Xu X., Zhou X., Zhou L., Wang R. Combined effects of PVT1 and miR-146a single nucleotide polymorphism on the lung function of smokers with chronic obstructive pulmonary disease. Int. J. Biol. Sci. 2018;14:1153–1162. doi: 10.7150/ijbs.25420. [DOI] [PMC free article] [PubMed] [Google Scholar]