Abstract
Artemisinin-based combination therapy (ACT) offers highly successful treatment of malaria. Emergence and spread of Plasmodium falciparum (Pf) parasites with decreased susceptibility to ACT in South-East Asia has caused concern worldwide. The current accepted criteria to assess artemisinin (ART) resistance relies upon data on treatment failure, delayed parasite clearance at day 3 (DPC3), parasite clearance half-life (PCHL) and in-vitro/ex-vivo ring stage survival assays (RSAs). Interestingly, some studies suggest that DPC3 does not provide a distinct separation between ART sensitive/resistant strains, and RSA differences may also be inconclusive. More recently, recrudescence of ART treated Pf, independent of the presence of Kelch 13 (K13) mutation (C580Y), has been reported in the monkey malaria model suggesting that genes other than K13 like coronin, dhps, dhfr, crt, mdr1 and plasmepsin1 may contribute towards ACT failure. Here we have collated the distribution of K13 mutants from Pf strains in South Asia. A total of fifty Pf-K13 mutations have been studied for ART resistance in South Asia of which nine have been validated while eleven are potentials for ART resistance. The remaining thirty K13 mutations have been reported from various locations in South Asia but lack corroborative clinical data on ART resistance/ACT failure. Of the fifty, fourteen K13 mutations have been identified in India including four novel mutations (S549Y, G625R, N657H, D702N). Structural mapping of these K13 mutations does not offer any coherent explanation for their contribution towards ART resistance as they are scattered in the K13 structure. Thus, K13 mutations likely provide only a partial synopsis, and we propose that all suspect cases of ACT failure be assessed by: 1) DPC3, 2) PCHL, 3) in-vitro/ex-vivo RSAs and 4) GWAS data in an effort to annotate the resistance status of the parasites. These efforts may help in surveillance and containment of ART resistance/ACT failure in South Asia.
Keywords: Artemisinin, Kelch, Malaria, Mutations, Plasmodium, Resistance
Graphical abstract
1. Background
Use of insecticide-treated nets, advances in vector control, timely malaria detection and administration of artemisinin-based combination therapy (ACT) have together contributed to reduction in the incidence of malaria from 72 to 59 cases per 1000 population between the years 2010 and 2017 worldwide (World Health Organisation (WHO), 2018) (https://www.who.int/malaria/publications/world-malaria-report-2018/en/). Yet malaria remains a major killer with ~219 million cases and ~435,000 deaths in 2017 alone (WHO, 2018) (https://www.who.int/malaria/publications/world-malaria-report-2018/en/). India has witnessed ~56% decline in malaria-related deaths between 2004 and 2016 but still ranks fourth in malaria incidence worldwide (www.malariasite.com/malaria-India) (National framework malaria elimination India, 2016-2030, 2015). Surveillance reports by WHO indicate that ~80% of cases in India occur in rural areas within 16 states covering the regions of East, Northeast (NE) and central India (http://www.searo.who.int/india/topics/malaria/en/) (National framework malaria elimination India: 2016-2030, 2015).
Despite numerous reports on the emergence of Plasmodium falciparum with reduced susceptibility to ACT, it remains the favored drug cocktail due to its effectiveness (Na and Efferth, 2019; WHO, 2015) (https://www.who.int/malaria/publications/atoz/9789241549127/en/). The isolation of P. falciparum parasites from Western Cambodia with delayed parasite clearance at day 3 (DPC3) that showed decreased susceptibility to ACT was alarming and marked the start of concern for ACT resistance (Ariey et al., 2014; Dondorp et al., 2009; Feng et al., 2015; Noedl et al., 2008; Thuy-Nhien et al., 2017; Tun et al., 2015; Wang et al., 2015). Mutations in K13 have been studied for their association with parasites’ decreased susceptibility to ACT (Amaratunga et al., 2016; Ariey et al., 2014; Phyo et al., 2016; Spring et al., 2015; Straimer et al., 2015). Kelch 13 (K13) is a member of Kelch-like superfamily (Ariey et al., 2014; Fairhurst, 2015; Tilley et al., 2016) and is dimeric as per its crystal structure (Protein Data Bank ID: 4YY8). The structure of K13 has previously been analyzed for the distribution of ART resistance mutations and its dimerization interfaces (Haldar et al., 2018; Singh et al., 2016). A previous structural exploration of K13 mutational landscape by our group revealed their partitioning into surface exposed and buried mutations (Singh et al., 2016). However, these analyses did not draw any structural link with the proposed ART failure phenotype.
In addition to the presence of K13 mutations, the current criteria for assessment of ART resistance rely upon DPC3, parasite clearance half-life (PCHL) as well as in-vitro and ex-vivo ring stage survival assays (RSAs) (described in later sections). Interestingly, a recent study on splenectomized monkey malaria model showed equal recrudescence of ART treated Pf infections with strains that either harbored or not the key K13 mutation C580Y (Sá et al., 2018). Further, the authors elegantly showed that parasite clearance time did not provide a distinct separation between the K13 C580Y mutant strain and its wild-type (Sá et al., 2018). This study therefore re-emphasized the importance of partner drugs in ACT, and supported the view from several other studies that mutations in other genes (dhps, dhfr, crt, mdr1, plasmepsin1) may also contribute towards ACT failure (Leang et al., 2013; Miotto et al., 2015; Mutabingwa et al., 2005; Pau et al., 2019; Saunders et al., 2014).
Aside from propelling basic research directed at identifying the molecular markers for ACT failure, the international community is also racing towards the identification of new drug targets and scaffolds to cover for the eventual emergency of complete ACT failure (Chhibber-Goel and Sharma, 2019; Dogovski et al., 2015; Jain et al., 2017; Jain, 2017; Khan et al., 2014; Yogavel et al., 2018a, Yogavel et al., 2018b). Over the past decade, numerous new drug targets have been identified, probed and then validated. Drug repurposing methodologies and structure-based conservation techniques that target invariant parasite house-keeping proteins may lead to novel foci for drug development (Dogovski et al., 2015; Jain et al., 2017; Jain V, 2017; Khan et al., 2014; Pazhayam et al., 2019; Yogavel et al., 2018a, Yogavel et al., 2018b). However, till then, surveillance and containment of mutant P. falciparum parasites that show decreased susceptibility to ART/ACT are of primary importance. The location and timing of efforts at containment likely need to be centered on India as well as the country nestles in the traditional path of anti-malarial drug resistance spread – i.e. from East Asia to Africa (Blasco et al., 2017; Dhingra et al., 2019; Mita et al., 2011).
Our analyses here provide a summation of K13 mutant data from South Asia with a focus on India. It is apparent that an integrated framework of surveillance and validation is required for tracking possible ART/ACT resistance in India. We emphasize the cataloging of single nucleotide polymorphisms (SNPs) on a whole genome level of parasites from malaria endemic regions that can then provide a genomic atlas. This will contribute towards both surveillance and containment of ART resistance/ACT failure.
2. Identification of ART/ACT failure
ART is highly active against all intra-erythrocytic stages of Plasmodium spp. and causes up to ~10,000 fold reduction in parasites within the first 48 h of administration (Dondorp et al., 2009; White, 2008; Nosten and White, 2007; Witkowski et al., 2013b). Since ART has a short plasma half-life it is administered in combination with long-acting partner drugs that clear the remaining parasite biomass (Dondorp et al., 2009; White, 2008; Nosten and White, 2007; Witkowski et al., 2013b). The standard assay for validating ART resistance relies on the identification of parasites with reduced susceptibility to ACT (via DPC3) and the presence of mutations in K13 protein (Dondorp et al., 2009; White, 2008; Nosten and White, 2007; Witkowski et al., 2013b). Resistance to partner drugs is assessed by tracking any recrudescence of infection at day 28 or 42, depending on the half-life of the partner drug (De Lucia et al., 2018; Krishna and Kremsner, 2013; WHO, 2017) (https://www.who.int/malaria/publications/atoz/artemisinin-resistance-april2017/en/). Delayed parasite clearance (DPC) is expressed as PCHL, which is determined by plotting log-linear parasite clearance curve (Dondorp et al., 2011). The curve is drawn between log parasitemia (calculated every 6 h from the beginning of medication till no parasites are detected) against time interval (White, 2011). PCHL is then calculated via slope of the linear part of parasite clearance curve using the parasitemia clearance estimator (PCE) software (https://www.wwarn.org/pce/) (Flegg et al., 2011; White, 2011). PCHL of ≥5 h after ACT administration is defined as ART resistance relative to the PCHL values determined for ART sensitive isolates (Ashley et al., 2014; Fairhurst, 2015; White et al., 2015). In addition, parasite survival rate of ≥1% via in-vitro RSAs is also used as marker for possible ART resistance (Witkowski et al., 2013a). For in-vitro RSAs (0-3h), cultured synchronized ring stage parasites are subjected to a dose of 700 nM of dihydroartemisinin (DHA) for 6 h and parasite numbers are estimated after 66 h of growth (Ariey et al., 2014; Witkowski et al., 2013b, 2013a). The ratio of number of parasites surviving DHA to parasites surviving DMSO (control) is used to calculate percent survival (Ariey et al., 2014; Witkowski et al., 2013b, 2013a). In an alternative version of RSA, called ex-vivo RSA, P. falciparum isolated directly from patients can be used to distinguish between fast and slow clearing parasites after ACT administration (Witkowski et al., 2013a). A malaria endemic area is considered for ART resistance if either 10% or more of its patients harbor parasites with PCHL ≥5 h or 10% or more of the patients show DPC3 after ACT administration (WHO, 2017) (https://www.who.int/malaria/publications/atoz/artemisinin-resistance-april2017/en/). Further, a malaria endemic region is confirmed for ART resistance if both of the above parameters are met along with detection of K13 P. falciparum mutants (WHO, 2017) (https://www.who.int/malaria/publications/atoz/artemisinin-resistance-april2017/en/). These are the current WHO standards for annotation of ART resistance. However, the landscape of K13 mutants reported from South Asia, and particularly India, presents an interesting scenario. For instance, there is discordance in the annotation of ART resistance (using DPC3, PCHL and/or RSA) for the same K13 mutants (E252Q, E270K, P441L, G538V and A675V) (Boullé et al., 2016; Mukherjee et al., 2017; Witkowski et al., 2013a). This situation therefore necessitates new integrated efforts to annotate whether a given clinical isolate is indeed resistance to ART or not. It is also important to dissect whether the same K13 mutants behave differently (in context of ART resistance) depending on their clinical/geographical/genomic background.
3. On K13 as a marker of artemisinin resistance
Analysis of P. falciparum mutants with reduced susceptibility to ACT via genome-wide association studies (GWAS), along with gene manipulation studies, have indicated an association between mutations in K13 and altered PCHL/parasite survival in RSAs (Ariey et al., 2014; Cheeseman et al., 2012; Ghorbal et al., 2014; Straimer et al., 2015; Takala-Harrison et al., 2015, 2013; WHO, 2017) (https://www.who.int/malaria/publications/atoz/artemisinin-resistance-april2017/en/). K13 protein is a 726 amino-acid member of Kelch-like superfamily with a C-terminal six-blade β-propeller domain, a Plasmodium-specific N-terminal domain and a BTB/POZ domain (Ariey et al., 2014; Fairhurst, 2015; Singh et al., 2016; Tilley et al., 2016). K13 exists as a dimer as per its deposited crystal structure (PDB ID: 4YY8). K13 has been analyzed for distribution of resistance mutations and for its dimerization interfaces (Haldar et al., 2018; Singh et al., 2016). To date, no cohesive or testable hypothesis links K13 mutations to ART failure. A recent study has suggested the use of protein structural modelling to generate testable predictions to determine the impact of Pf K13 mutations on in vivo (and potentially in vitro) ART susceptibility (He et al., 2019). Independently, P. falciparum K13 down-regulation has been linked with increased parasite clearance times thereby suggesting that reduced Pf K13 transcriptional response may be a first step towards ART resistance (Silva et al., 2019).
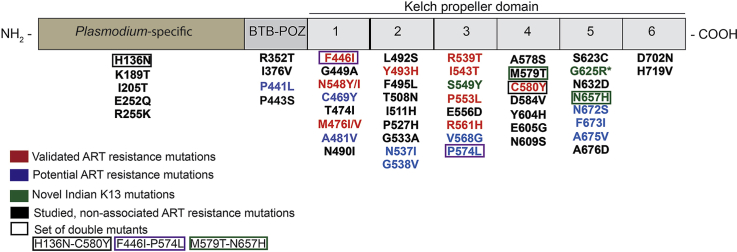
More than 200 non-synonymous mutations have been identified in K13 protein from P. falciparum strains (Fairhurst, 2015; WHO, 2017) (https://www.who.int/malaria/publications/atoz/artemisinin-resistance-april2017/en/). While most mutations occur at low prevalence and have uncertain functional significance, fifty of these were assessed for reduced susceptibility to ACT/ART resistance using DPC3, PCHL and/or RSA (Cooper et al., 2015; Fairhurst and Dondorp, 2016; Ménard et al., 2016; Nyunt et al., 2017; Sá et al., 2018; Tun et al., 2016; Ye et al., 2016) (Fig. 1a and b). Of these, only nine mutations have been validated for ART resistance ex-vivo (F446I, N458Y/I, M476I/V, Y493H, R539T, I543T, P553L, R561H, C580Y) (Fig. 1a (in red)) (Boullé et al., 2016; Ghorbal et al., 2014; Straimer et al., 2015; WHO, 2017) (https://www.who.int/malaria/publications/atoz/artemisinin-resistance-april2017/en/). A further eleven mutations have either been marked as possible candidates or remain associated but have not been validated for ART resistance (P441L, G449A, C469Y, A481V, N537I, G538V, V568G, P574L, N672S, F673I, A675V) (Fig. 1a). Although C580Y mutation remains a dominant marker for ART/ACT resistance across SEA, a recent study has suggested that parasite clearance time does not distinguish between the ART sensitive and ART resistant C580Y strains (Sá et al., 2018). In this work, the authors showed that IC50 values of DHA were at similar nM levels for both C580 (wildtype) and C580Y mutant strain (a validated K13 mutant for ART resistance) (Sá et al., 2018). However, it is noteworthy that the splenectomized Aotus monkey, which is a nonhuman primate model for malaria, is semi-immune and therefore an artificial system. This work and other studies have nonetheless reinforced the need for a more holistic approach to study ART resistance in the field. Such efforts may minimally include validation of clinical data, assessment of reduced drug sensitivity via DPC3 and PCHL, study of corresponding RSAs and possibly genome wide association studies (GWAS) to determine SNPs across the parasite genome.
Fig. 1.
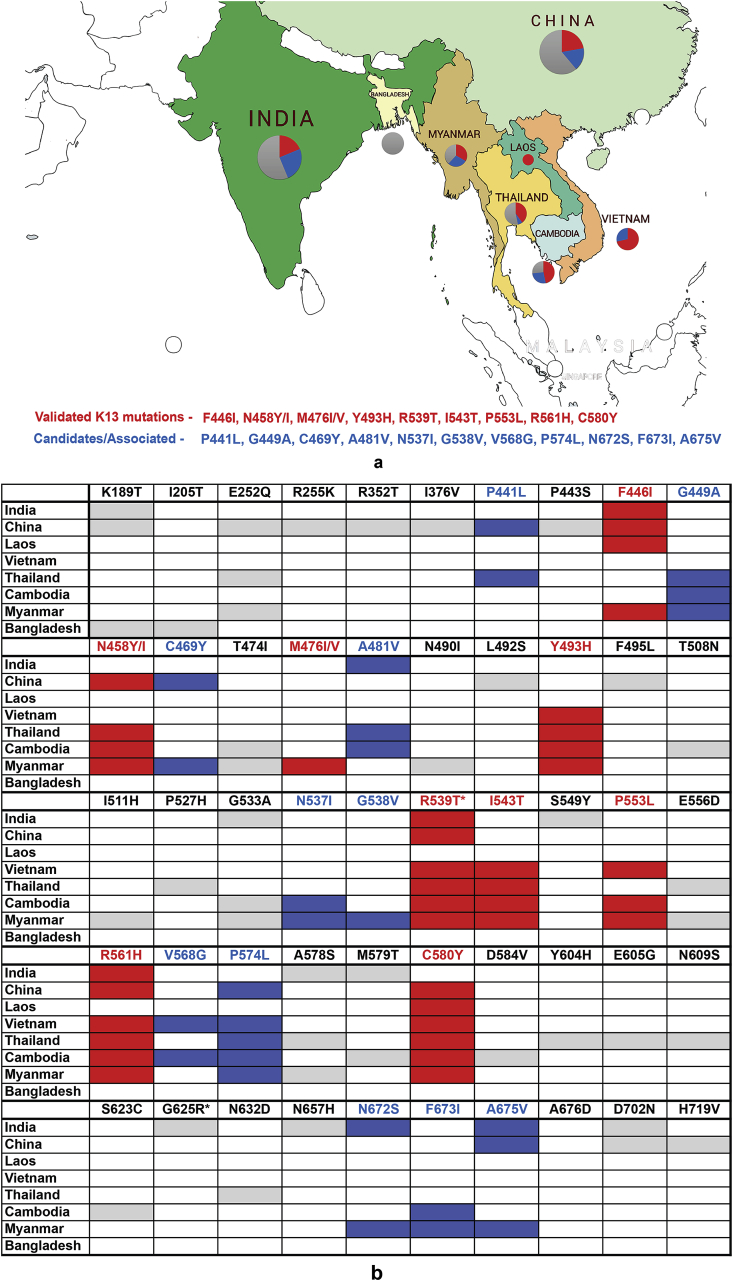
Geographical distribution of K13 domain mutations in South Asia. a) K13 mutations that are either validated (red) or are associated/candidates (blue) for ART resistance are shown. Pie chart depicts K13 mutant distribution based on their status i.e. validated (red), candidates (blue) or uncategorized/un-associated (grey) as defined by the World Health Organization (WHO) in context with ART resistance. b) Tabular representation of the K13 mutations identified from different countries of South Asia. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Landscape of K13 mutations
SEA is considered an epicenter for emergence and spread of malaria drug resistance (Mita et al., 2011; Mita and Tanabe, 2012; Roper C, Pearce R, Nair S, Sharp B, Nosten F, 2004; Wernsdorfer WH, 1991) (Fig. 1a). Mutations in K13 were first reported from West Cambodia in 2008 and C580Y, Y493H and R539T were the most prevalent then (>5% frequency) (Ariey et al., 2014). These along with other K13 protein mutants either spread slowly or emerged independently in various other SEA countries - Vietnam, Thailand and Myanmar – and eventually C580Y became the most dominant (75–90%) mutant in a span of ten years (Ariey et al., 2014; Kyaw et al., 2013; Leang et al., 2015; Phyo et al., 2016; Putaporntip et al., 2016; Takala-Harrison et al., 2015; Talundzic et al., 2015; Wang et al., 2015). However, reports from China and China-Myanmar border showed the presence of a different set of K13 mutations with F446I being the most dominant followed by R539T, P574L, N458Y, R561H and A676D (Feng et al., 2015; Huang et al., 2015; Wang et al., 2015; Ye et al., 2016). It has been suggested that the presence of K13 mutants at China-Myanmar border may enable spread of above strains to bordering NE states of India (Tun et al., 2015). This situation therefore calls for stringent monitoring of K13 mutant parasites isolated from malaria-infected patients in NE (and mainland) India. Recent studies from India have identified fourteen K13 mutations in parasites isolated from malaria infected Indian patients. These mutations are: K189T, F446I, A481V, G533 A/S, R539T, S549Y, R561H, A578S, M579T, G625R, N657H, N672S, A675V and D702N (Fig. 1b & Table 1) (Das et al., 2017, 2018; Mishra et al., 2016, 2015; 2017). These mutations were isolated from the states of Assam, Tripura, Mizoram, Arunachal Pradesh, West Bengal, Madhya Pradesh and Chhattisgarh (Das et al., 2017, 2018; Mishra et al., 2016, 2015; 2017). A number of studies have failed to show a significant co-relation between these K13 mutants and their propensity for ACT failure in Indian population (Bharti et al., 2016; Das et al., 2017; Miraclin et al., 2016; Mishra et al., 2016, 2015, 2017). However, a more recent study from India identified a novel Pf mutation – G625R - and a previously validated mutation – R539T - from malaria-infected patients in Eastern India; each is associated with ART resistance based on DPC3 (presence of parasite at day 3), PCHL> 5 h and RSA0-3hr (parasite survival rate, >1%) criteria (Das et al., 2018). This report on the identification of G625R and R539T and their association with ART resistance, along with detection of early treatment failure, has raised concerns in the malaria research community in India (Rasmussen et al., 2019). While Das et al. emphasize that the criteria required to define ART resistance are met, the points raised by the malaria research community in India are also valid (Rasmussen et al., 2019). Intriguingly, another study from India by Chakrabarti et al. in 2019 showed reduced sensitivity to DHA (Chakrabarti et al., 2019). Further to this, the authors reported A675V mutation within the Kelch propeller domain (RSA of 2%). These cases from India therefore present an opportunity for adoption of a more robust and thorough approach for annotating field strains as ART/ACT resistant or otherwise.
Table 1.
K13 mutations from Indian patients.
| Mutation | City (State) | Reference |
|---|---|---|
| K189T | Lunglei (Mizoram) Gomati (Tripura) |
Mishra et al. (2016) |
| F446I | Changlang (Arunanchal Pradesh) | Mishra et al. (2016) |
| A481V | Changlang (Arunanchal Pradesh) | Das et al. (2017) |
| G533 A/S | Gomati (Tripura) | Mishra et al. (2015) |
| R539T | - (West Bengal) | Das et al. (2018) |
| S549Y | Jalpaiguri (West Bengal) | Mishra et al. (2015) |
| R561 H/C | Changlang (Arunanchal Pradesh) | Mishra et al. (2015) |
| A578S | Lunglei (Mizoram) | (Mishra et al., 2016, 2015) |
| M579 T/H | Bastar (Chattisgarh) Balaghat, Annupur, Jhabua (Madhya Pradesh) |
Mishra et al. (2017) |
| G625R | (West Bengal) | Das et al. (2018) |
| N657H | Balaghat, Annupur (Madhya Pradesh) | Mishra et al. (2017) |
| N672S | - (West Bengal) | Das et al. (2018) |
| A675V | - (Arunanchal Pradesh and Assam) | Das et al. (2017) |
| D702N | - (Arunanchal Pradesh) | Das et al. (2017) |
5. Structural analysis of K13 mutations
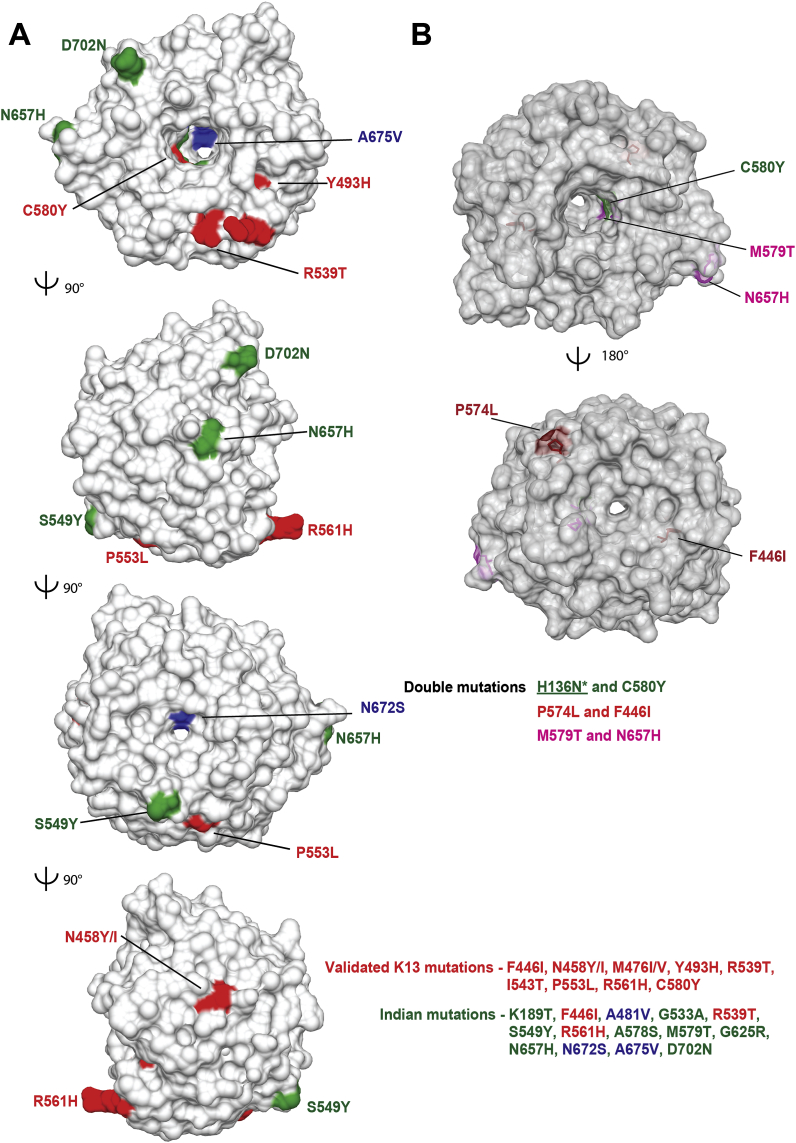
We mapped fourteen mutations observed in Indian isolates on the K13 crystal structure (PDB ID: 4YY8) (Fig. 2a). Of these, eleven mutations - G533A, R539T, S549Y, R561H, A578S, M579H, G625R, N657H, N672S, A675V and D702N - are exposed while two - F446I and A481V – are in the central protein cavity (Fig. 2a). Of the fourteen, six (G533A, A578S, M579H, G625R, N672S and A675V) are located on the small channel that runs through the center of K13 (Fig. 2a), and as K189T/I205T are located outside the propeller domain their significance is especially unclear. Furthermore, three K13 mutations – F446I, R539T and R561H – have been categorized as validated for ART resistance in SEA countries by WHO (Fig. 1) (WHO, 2017) (https://www.who.int/malaria/publications/atoz/artemisinin-resistance-april2017/en/). Of these three, a recent report has suggested that R539T from (two) Indian isolates showed signs of ART resistance and early treatment failure as assessed by DPC3, PCHL and RSAs) (Das et al., 2018). In yet another contrary finding, the WHO-validated F446I and R561H do not seem to affect treatment when isolated from Indian patients (as per DPC data alone) (Mishra et al., 2016; Das et al., 2018).
Fig. 2.
Structural mapping of K13 mutations found in India. a) Orthogonal views of single nucleotide polymorphisms (SNPs) where validated K13 mutations are in red, associated/candidate mutations are in blue and those from India are in green (PDB ID: 4YY8). b) Mapping of three sets of double mutations found in K13 along with validated C580Y mutation (in green) . As H136N does not map on K13 propeller domain it is not shown. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Strikingly, our structural mapping of the fourteen Indian mutations does not offer any coherent explanation for their contribution towards ART resistance (Fig. 2a). The mutations are spatially spread over K13 propeller domain and do not congregate to any particular surface or groove (Fig. 2a). There seem to be no clear links between the sets of K13 mutations and their association with ART failure. This suggests that the ART-K13 linkage for drug resistance is not evidentially and structurally robust. More intriguingly, a few Indian K13 mutations (R561H, F446I, A481V, A675V) display contradictory phenotypes in context of ACT failure when compared to data from SEA. For instance, the mutation R561H was previously reported in SEA and sub-Saharan Africa and displays PCHL ≥5 h (Fairhurst and Dondorp, 2016). The same mutation was isolated from a malaria patient in Changlang district of Arunachal Pradesh, India but was not associated with ACT failure as estimated by monitoring treatment failure at day 28 (Mishra et al., 2015). The F446I mutation (buried) has been isolated from a patient in Changlang district of Arunachal Pradesh (close to Myanmar, Fig. 3) (Mishra et al., 2016) and it is also highly prevalent at the China-Myanmar border (in Northeast Myanmar and in Yunnan province, China) (Wang et al., 2015). However, F446I shows no association with ACT failure in India as estimated by lack of treatment failure at day 28 (Mishra et al., 2016) as in the case for samples from China-Myanmar border (as per RSAs) (Ye et al., 2016). However, the F446I mutation has been associated with DPC3 in Central and Northern Myanmar (Tun et al., 2016). The above two examples of R561H and F446I highlight lack of coherence in association between K13 SNPs and treatment failure when isolates from different geographical regions are compared. These contradictory phenotypes also suggest that reliance on K13 protein as a marker for ACT failure may be reconsidered.
Fig. 3.
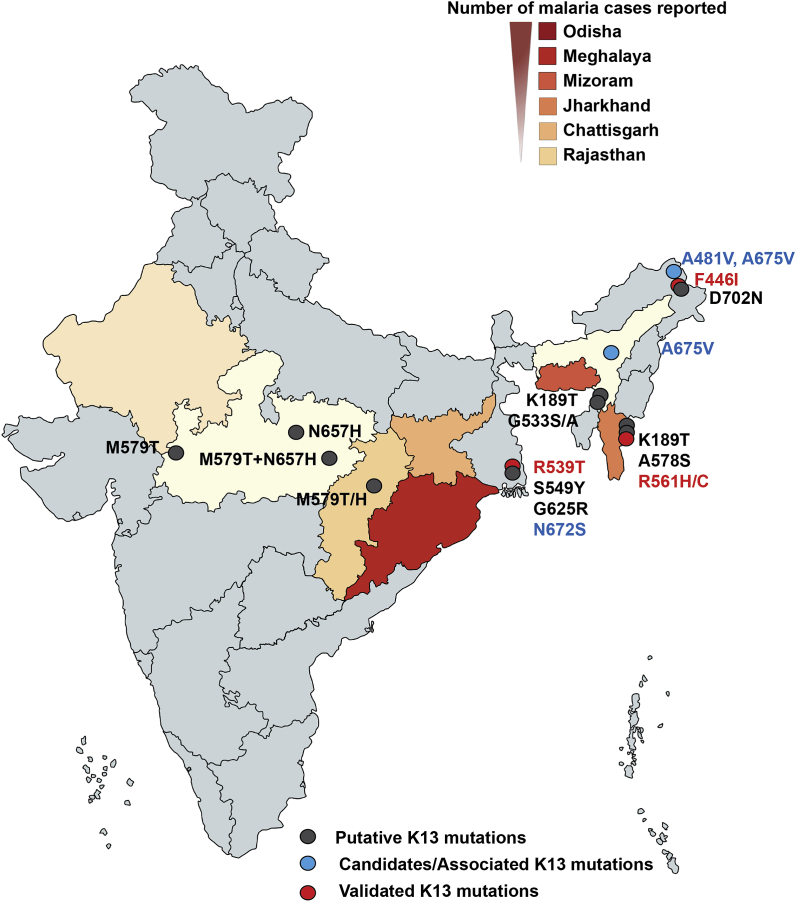
Kelch 13 (K13) mutations in India. Geographical distribution of Kelch 13 (K13) mutations in Plasmodium sp. clinical isolates. The states with highest number of malaria cases reported are shown based on their ranking (i.e., Odisha followed by Meghalaya, Mizoram, Jharkhand, Chattisgarh and Rajasthan) along with the K13 mutations categorized based on validated (in red), candidates/associated (in blue) and putative (in grey) for ART resistance. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
In context of India, only the G533A mutation, identified from one isolate in Gomati district of Tripura (bordering Bangladesh), has been claimed for treatment failure at day 28 (no DPC3, PCHL or RSA data are available for this sample) (Mishra et al., 2015). However, as noted earlier, treatment failure cannot be attributed to ART resistance alone and could be due to resistance towards the partner drug or due to several other factors (Leang et al., 2013; Saunders et al., 2014). The most frequent mutations observed amongst Indian isolates include K189T (Tripura and Mizoram - bordering Bangladesh) (Mishra et al., 2016), A481V (Arunachal Pradesh) (Das et al., 2017) and A675V (from Assam and Arunachal Pradesh) (Das et al., 2017). Although the role of K189T in ACT resistance is yet to be established, A481V and A675V have been categorized as potential candidates for ART resistance (WHO, 2017) (https://www.who.int/malaria/publications/atoz/artemisinin-resistance-april2017/en/). Once again, there is lack of agreement between WHO declared annotation of A481V and A675V mutants (Das et al., 2017). A novel mutation - D702N – has been observed amongst isolates from Arunachal Pradesh (Fig. 2, Fig. 3) (Das et al., 2017). It has been postulated that D702N mutation emerged independently and therefore validation assays are required to unlock its role in ACT resistance, if any (Das et al., 2017). Two additional novel mutations - M579T and N657H - have been identified from different districts of Madhya Pradesh in India (Fig. 3). Though M579T was found to occur alone among 1.5% of isolates, N657H was observed along with M579T in 37% of isolates; however, none of these isolates were found to be associated with DPC3 (Bharti et al., 2016; Mishra et al., 2017). The above double mutant (M579T/N657H) parasites could be due to infection of multiple parasite clones or due to the presence of double mutation in a single parasite line (discussed later). The structural mapping of M579T and N657H reveals that these residues reside on the blades IV and VI respectively. Further, these two mutations are located on the small channel that runs through the center of K13 (Fig. 2b). M579T is proximal to the well-studied C580Y while N657H resides on blade V within the central channel (Fig. 2b).
Identification of S549Y mutation from Jalpaiguri district of West Bengal (bordering Bhutan and Bangladesh, near Assam) (Fig. 3) could be indicative of slow migration of mutant parasites from NE to the mainland, or of another independent origin (Mishra et al., 2015). S549Y remains to be studied for ART resistance in SEA and is uncategorized as yet. Further, the mutation A578S has been identified in Bangladesh with no reported cases of ACT failure or ART resistance (Mohon et al., 2014; Starzengruber et al., 2012). A retrospective study conducted on 68 malaria patients showed day three parasitemia/DPC3 amongst 57% of the patients (as monitored by microscopic examination) post ACT (Artesunate + Sulfadoxine-Pyrimethamine administration (Mishra et al., 2016). However, this study lacks corroborative markers on K13 mutations and/or PCHL/RSA data (Miraclin et al., 2016).
Studies on K13 mutations isolated from patients reveal dominance of single mutations and double ones are rare (Table 2 & Fig. 2b) (Ariey et al., 2014; Ashley et al., 2014; Mishra et al., 2017; Mukherjee et al., 2017; Ouattara et al., 2015; Talundzic et al., 2017; Thuy-Nhien et al., 2017; Tun et al., 2016; Wang et al., 2015). Nonetheless, double mutations of H136N-C580Y and P574L-F446I have been identified from single parasite clones isolated in China (1/191) and Cambodia (2/157) respectively (Mukherjee et al., 2017; Wang et al., 2015). K13 double mutants were also seen in one clinical isolate from Mali and four from Senegal but these were attributed to infection by two parasite clones (Ouattara et al., 2015; Talundzic et al., 2017) (Table 2 & Fig. 2b).
Table 2.
Clinical isolates with K13 double mutants.
| Mutations | Location | Isolates | Reference |
|---|---|---|---|
| P574L and F446I | China-Myanmar border | 1/191 | Wang et al. (2015) |
| M579T and N657H | Madhya Pradesh | 50/186 | Mishra et al. (2017) |
| H136N and C580Y | Cambodia | 2/36 | Mukherjee et al. (2017) |
6. K13-independent artemisinin resistance
ART resistance in the absence of K13 mutations has been reported (Table 3) (Ariey et al., 2014; Cerqueira et al., 2017; Davis et al., 2017; Demas et al., 2018; Lee and Fidock, 2016; Miotto et al., 2015; Mukherjee et al., 2017; Sá et al., 2018; Takala-Harrison et al., 2013, 2015; Wang et al., 2016). Identification of four parasite isolates from Cambodia with wild type K13 protein but RSA ≥0.8% had suggested K13-independent ART resistance (Mukherjee et al., 2017). Indeed, genomic analysis performed on both sensitive and resistant isolates have identified 37 novel mutations spread over nineteen non K 13 genes, with eleven present in the four isolates without K13 mutations with RSA ≥0.8% (Mukherjee et al., 2017). Maximum mutations were identified in phosphoinositide 3-kinase (PI3K) and in multidrug resistance protein 1/2 (mdr1/2) (Mukherjee et al., 2017). In another study, three cases of treatment failure at days 21 and 28 were reported from Bastar district of Chhattisgarh in India but all three-harbored wild-type K13 protein (Bharti et al., 2016). However, recrudescence was confirmed using only two of the three recommended markers (Bharti et al., 2016). Further, SNPs outside chromosome 13 have also been weakly associated with ART resistance (Ariey et al., 2014; Cerqueira et al., 2017; Lee and Fidock, 2016; Miotto et al., 2015; Takala-Harrison et al., 2013; Wang et al., 2016). Thus, it is imperative to understand if mutations in genes other than K13 contribute towards emergence and spread of ACT failure (Cerqueira et al., 2017; Miotto et al., 2015; Winzeler, 2017).
Table 3.
Wild type K13 with postulated Artemisinin (ART) resistance.
| Location | Isolates | Assessment criteria | Reference |
|---|---|---|---|
| Odisha, India | 3 | DPC3 +ve | Bharti et al. (2016) |
| Uganda | 3 | Ex-vivo RSA >10% | (Ikeda et al., 2018) |
| Asia wide study | 21 | PCHL > 5 h | Ashley et al. (2014) |
| China-Myanmar border | 1 | Ex-vivo RSA >14% | Ye et al. (2016) |
| Cambodia | 4 | In-vitro RSA > 0.8% | Mukherjee et al. (2017) |
A GWAS performed by Miotto et. al has earlier suggested that mutations in multidrug resistance protein 2 (mdr2, T484I), ferredoxin (fd, D193Y), apicoplast ribosomal protein S10 (arpS10, V127M) and chloroquine resistance transporter (crt, I356T and N326S) may form a genetic background on which mutations in K13 emerge (Miotto et al., 2015). Mutations in fd, arsp10 and mdr2 genes from ART resistant K13 mutant parasites have also been isolated from Myanmar (Dondorp et al., 2017; Nyunt et al., 2017). Surprisingly, D193Y and V127M mutations (in ferredoxin and ribosomal protein respectively) were observed in only three ‘R539T’ K13 mutant isolates from China-Myanmar border whereas both D193Y and V127M showed high association with the ‘R539T’ K13 mutant isolated from Thai-Cambodia border (Ye et al., 2016). Thus it was suggested that SNP analysis of fd and arpS10 genes may shed additional light on the lineage of mutant haplotypes (Ye et al., 2016). Studies have also indicated weak association of SNPs in nuclear LIM-interactor interacting factor (NIF4)-like phosphatase (Miotto et al., 2015; Wang et al., 2016), and ‘T38N’ change in pfatg18 to be associated with ART resistance (Wang et al., 2016). Pfatg18 has also been proposed as a potential target for ART resistance (Dondorp et al., 2017). Various DNA repair genes including mlh1, pms1 and exo1 are overexpressed in ART resistant parasites and thus considered to be associated with ART resistance (Lee and Fidock, 2016). The genome analysis of parasites with K13 mutations has revealed yet another Kelch-domain containing protein Kelch10 (K10) that may epistatically modulate ART resistance phenotype (Cerqueira et al., 2017; Winzeler, 2017). The mutation P623T in K10 was shown to increase parasite clearance rates in C580Y and E252Q K13 mutant strains (Cerqueira et al., 2017). Further, mutations in actin-binding protein coronin have been shown to reduce parasite susceptibility to ART in-vitro (Demas et al., 2018). The above studies therefore present a varied landscape of alternations in the genomes of resistant parasites isolated from field, and offer no clear causal relationship for ART resistance with a one-gene locus.
7. Surveillance strategies for tracking ACT failure
Isolation of K13 mutant parasites from various Indian states highlights the need to understand their co-relation with ART resistance/ACT failure. This requires robust surveillance and quick detection of K13 mutant parasites, especially amongst the populations living in NE of India. The identification of K13-independent isolates which may show treatment failure is also equally vital. Hence, to cover these issues holistically, we propose that new surveillance strategies be devised based on state-of-the-art molecular technologies. A few possibilities are:
-
1.
GWAS: It will provide a complete readout of the genomic content of field parasites and will contribute to the growing worldwide genome database (Cerqueira et al., 2017; Miotto et al., 2015; Takala-Harrison et al., 2015). Study of microsatellite loci along with SNPs in genes such as fd, crt, mdr2 and arps10 will contribute towards dissecting the lineages of these mutations in field isolates. Such studies will also shed light on the phylogenetic relationships between field strains (Imwong et al., 2017; Miotto et al., 2015; Talundzic et al., 2015; Thuy-Nhien et al., 2017).
-
2.
Implementation of filter paper based blood sample collection methodology for genomic surveillance (Oyola et al., 2016).
-
3.
Establishment of biobanks for maintaining stocks of parasites isolated from various Indian regions spanning the country so that geographical linkages can be drawn for migration of resistant parasite strains.
-
4.
4. Health care worker training on the use of new web tools for analysis of parasites including the PCE developed by World Wide Antimalarial Resistance Network and ‘Shiny web’ (available at ‘bit.ly/id_artemisinin_resistance’) which estimates PCHL (Flegg et al., 2011; Tun et al., 2017). Another software called Genome-wide mixed-model association (GEMMA) and/or PLINK could be used for establishing co-relations between mutations in K13 and DPC3 data (Purcell et al., 2007; Wang et al., 2016; Zhou and Stephens, 2012).
8. Concluding remarks
Artemisinin-based combination therapy (ACT) is a highly efficacious treatment for malaria but emergence and spread of P. falciparum parasites with reduced susceptibility to ACTs is threatening malaria control. Stringent monitoring of ART resistance/ACT failure, possibly via development of rapid and economical surveillance tools, as well as systematic cataloguing of epidemiological, clinical (DPC3, PCHL, RSA) and genomic data (GWAS) is hence of utmost importance. We propose emphasis on whole genome sequencing of field strains so that data obtained can be integrated with genomic data from South Asia and African isolates. Further, in light of emerging reports on K13 mutants from various regions in India, it is imperative that India employ the latest technologies for annotation of any treatment failure cases. There is thus an urgent need to track ART resistance/ACT failure via clinical, epidemiological and genomic screenings. Such assessments may include 1) DPC3, 2) PCHL, 3) in-vitro and ex-vivo RSAs and 4) GWAS to identify mutations. These efforts may help in surveillance and containment of ART resistance/ACT failure in India, and may contribute towards integration with worldwide databanks that are monitoring the growth of drug resistance in malaria.
Funding
AS is supported by JC Bose fellowship (JCB-41) from the Government of India. Department of Biotechnology (DBT) supports JCG under the BioCARe scheme. DBT, Department of Science and Technology (DST), Global Health Initiative (GHIT) and Medicines for Malaria Venture (MMV) fund the laboratory of AS.
Declaration of competing interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgements
JCG and AS researched the literature, analyzed the data and wrote the manuscript. We thank Dr Rai for inspiration and Dr DLS Armstrong for disturbing insights.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijpddr.2019.10.001.
Contributor Information
Jyoti Chhibber-Goel, Email: jyoti.chhibber@gmail.com.
Amit Sharma, Email: amitpsharma68@gmail.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Amaratunga C., Lim P., Suon S., Sreng S., Mao S., Sopha C., Sam B., Dek D., Try V., Amato R., Blessborn D., Song L., Tullo G.S., Fay M.P., Anderson J.M., Tarning J., Fairhurst R.M. Dihydroartemisinin-piperaquine resistance in Plasmodium falciparum malaria in Cambodia: a multisite prospective cohort study. Lancet Infect. Dis. 2016;16:357–365. doi: 10.1016/S1473-3099(15)00487-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariey F., Witkowski B., Amaratunga C., Beghain J., Langlois A.C., Khim N., Kim S., Duru V., Bouchier C., Ma L., Lim P., Leang R., Duong S., Sreng S., Suon S., Chuor C.M., Bout D.M., Ménard S., Rogers W.O., Genton B., Fandeur T., Miotto O., Ringwald P., Le Bras J., Berry A., Barale J.C., Fairhurst R.M., Benoit-Vical F., Mercereau-Puijalon O., Ménard D. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley E.A., Dhorda M., Fairhurst R.M., Amaratunga C., Lim P., Suon S., Sreng S., Anderson J.M., Mao S., Sam B., Sopha C., Chuor C.M., Nguon C., Sovannaroth S., Pukrittayakamee S., Jittamala P., Chotivanich K., Chutasmit K., Suchatsoonthorn C., Runcharoen R., Hien T.T., Thuy-Nhien N.T., Thanh N.V., Phu N.H., Htut Y., Han K.-T., Aye K.H., Mokuolu O.A., Olaosebikan R.R., Folaranmi O.O., Mayxay M., Khanthavong M., Hongvanthong B., Newton P.N., Onyamboko M.A., Fanello C.I., Tshefu A.K., Mishra N., Valecha N., Phyo A.P., Nosten F., Yi P., Tripura R., Borrmann S., Bashraheil M., Peshu J., Faiz M.A., Ghose A., Hossain M.A., Samad R., Rahman M.R., Hasan M.M., Islam A., Miotto O., Amato R., MacInnis B., Stalker J., Kwiatkowski D.P., Bozdech Z., Jeeyapant A., Cheah P.Y., Sakulthaew T., Chalk J., Intharabut B., Silamut K., Lee S.J., Vihokhern B., Kunasol C., Imwong M., Tarning J., Taylor W.J., Yeung S., Woodrow C.J., Flegg J.A., Das D., Smith J., Venkatesan M., Plowe C.V., Stepniewska K., Guerin P.J., Dondorp A.M., Day N.P., White N.J. Spread of artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2014;371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharti P.K., Shukla M.M., Ringwald P., Krishna S., Singh P.P., Yadav A., Mishra S., Gahlot U., Malaiya J.P., Kumar A., Prasad S., Baghel P., Singh M., Vadadi J., Singh M.P., Bustos M.D.G., Ortega L.I., Christophel E.M., Kashyotia S.S., Sonal G.S., Singh N. Therapeutic efficacy of artemether-lumefantrine for the treatment of uncomplicated Plasmodium falciparum malaria from three highly malarious states in India. Malar. J. 2016;15:498. doi: 10.1186/s12936-016-1555-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco B., Leroy Di, Fidock D.A. Antimalarial drug resistance: linking Plasmodium falciparum parasite biology to the clinic. Nat. Med. 2017;23:917–928. doi: 10.1038/nm.4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boullé M., Witkowski B., Duru V., Sriprawat K., Nair S.K., McDew-White M., Anderson T.J.C., Phyo A.P., Menard D., Nosten F. Artemisinin-resistant Plasmodium falciparum K13 mutant alleles, Thailand-Myanmar border. Emerg. Infect. Dis. 2016;22:1503–1505. doi: 10.3201/eid2208.160004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerqueira G.C., Cheeseman I.H., Schaffner S.F., Nair S., McDew-White M., Phyo A.P., Ashley E.A., Melnikov A., Rogov P., Birren B.W., Nosten F., Anderson T.J.C., Neafsey D.E. Longitudinal genomic surveillance of Plasmodium falciparum malaria parasites reveals complex genomic architecture of emerging artemisinin resistance. Genome Biol. 2017;18:78. doi: 10.1186/s13059-017-1204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti R., White J., Babar P.H., Kumar S., Mudeppa D.G., Mascarenhas A., Pereira L., Dash R., Maki J.N., Sharma A., Gogoi K., Sarma D.K., Bhowmick I.P., Manoharana S.K., Gomes E., Mahanta J., Mohapatra R.K., Chery L., Rathod P.K. Decreased in vitro artemisinin sensitivity of Plasmodium falciparum across India. Antimicrob. Agents Chemother. 2019;63(10) doi: 10.1128/AAC.00101-19. [Epub ahead of print]), pii: e00101-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.H., Miller B.A., Nair S., Nkhoma S., Tan A., Tan J.C., Al Saai S., Phyo A.P., Ler Moo C., Lwin K.M., McGready R., Ashley E., Imwong M., Stepniewska K., Yi P., Dondorp A.M., Mayxay M., Newton P.N., White N.J., Nosten F., Ferdig M.T., Anderson T.J.C. A major genome region underlying artemisinin resistance in malaria. Science. 2012;336:79–82. doi: 10.1126/science.1215966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhibber-Goel J., Sharma A. 2019. Side Chain Rotameric Changes and Backbone Dynamics Enable Specific Cladosporin Binding in Plasmodium Falciparum Lysyl‐tRNA Synthetase. Proteins. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Cooper R.A., Conrad M.D., Watson Q.D., Huezo S.J., Ninsiima H., Tumwebaze P., Nsobya S.L., Rosenthal P.J. Lack of artemisinin resistance in Plasmodium falciparum in Uganda based on parasitological and molecular assays. Antimicrob. Agents Chemother. 2015;59:5061–5064. doi: 10.1128/AAC.00921-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das M.K., Kalita M.C., Chetry S D.P. PfK13 kelch propeller domain and Pfmdr1 sequence polymorphism in Plasmodium falciparum field isolates from Northeast region, India. Hum. Parasit. Dis. 2017;9:1–9. [Google Scholar]
- Das S., Saha B., Haiti A.K., Roy S. Evidence of artemisinin-resistant Plasmodium falciparum malaria in Eastern India. N. Engl. J. Med. 2018;379:1962–1964. doi: 10.1056/NEJMc1713777. [DOI] [PubMed] [Google Scholar]
- Davis S.Z., Checkley L., Pinapati R.S., Vaughan A., Fishbaugher M., Camargo N., McDew-White M., Nair S., Nosten F.H., Kappe S., Cheeseman I., Anderson T.J.C., Ferdig M.T. A malaria genetic cross generated in a humanized mouse indicate multi-gene control of resistances to artemisinin and piperaquine. Am. J. Trop. Med. Hyg. 2017;97:403. [Google Scholar]
- De Lucia S., Tsamesidis I., Pau M.C., Kesely K.R., Pantaleo A., Turrini F. Induction of high tolerance to artemisinin by sub-lethal administration: a new in vitro model of P. falciparum. PLoS One. 2018;13:e0191084. doi: 10.1371/journal.pone.0191084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demas A.R., Sharma A.I., Wong W., Early A.M., Redmond S., Bopp S., Neafsey D.E., Volkman S.K., Hartl D.L., Wirth D.F. Mutations in Plasmodium falciparum actin-binding protein coronin confer reduced artemisinin susceptibility. Proc. Natl. Acad. Sci. 2018;115:12799–12804. doi: 10.1073/pnas.1812317115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhingra S.K., Gabryszewski S.J., Small-Saunders J.L., Yeo T., Henrich P.P., Mok S., Fidock D.A. Global spread of mutant PfCRT and its pleiotropic impact on Plasmodium falciparum multidrug resistance and fitness. mBio. 2019;10 doi: 10.1128/mBio.02731-18. e02731-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogovski C., Xie S.C., Burgio G., Bridgford J., Mok S., McCaw J.M., Chotivanich K., Kenny S., Gnädig N., Straimer J., Bozdech Z., Fidock D.A., Simpson J.A., Dondorp A.M., Foote S., Klonis N., Tilley L. Targeting the cell stress response of Plasmodium falciparum to overcome artemisinin resistance. PLoS Biol. 2015;13:e1002132. doi: 10.1371/journal.pbio.1002132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondorp A.M., Nosten F., Yi P., Das D., Phyo A.P., Tarning J., Lwin K.M., Ariey F., Hanpithakpong W., Lee S.J., Ringwald P., Silamut K., Imwong M., Chotivanich K., Lim P., Herdman T., An S.S., Yeung S., Singhasivanon P., Day N.P., Lindegardh N., Socheat D., White N.J. Artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2009;365:1073–1075. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondorp A.M., Fairhurst R.M., Slutsker L., MacArthur J.R., J G.B., Guerin P.J., Wellems T.E., Ringwald P., Newman R.D., Plowe C.V. The threat of artemisinin-resistant malaria. N. Engl. J. Med. 2011;365:1073–1075. doi: 10.1056/NEJMp1108322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondorp A.M., Smithuis F.M., Woodrow C., Seidlein L. von. How to contain artemisinin- and multidrug-resistant falciparum malaria. Trends Parasitol. 2017;33:353–363. doi: 10.1016/j.pt.2017.01.004. [DOI] [PubMed] [Google Scholar]
- Fairhurst R.M. Understanding artemisinin-resistant malaria: what a difference a year makes. Curr. Opin. Infect. Dis. 2015;28:417–425. doi: 10.1097/QCO.0000000000000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairhurst R.M., Dondorp A.M. Artemisinin-resistant Plasmodium falciparum malaria. Microbiol. Spectr. 2016;4 doi: 10.1128/microbiolspec.EI10-0013-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Zhou D., Lin Y., Xiao H., Yan H., Xia Z. Amplification of pfmdr1, pfcrt, pvmdr1, and K13 propeller polymorphisms associated with Plasmodium falciparum and Plasmodium vivax isolates from the China-Myanmar border. Antimicrob. Agents Chemother. 2015;59:2554–2559. doi: 10.1128/AAC.04843-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flegg J.A., Guerin P.J., White N.J., Stepniewska K. Standardizing the measurement of parasite clearance in falciparum malaria: the parasite clearance estimator. Malar. J. 2011;10:339. doi: 10.1186/1475-2875-10-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghorbal M., Gorman M., MacPherson C.R., Martins R.M., Scherf A., Lopez-Rubio J.J. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat. Biotechnol. 2014;32:819–821. doi: 10.1038/nbt.2925. [DOI] [PubMed] [Google Scholar]
- Haldar K., Bhattacharjee S., Safeukui I. Drug resistance in plasmodium. Nat. Rev. Microbiol. 2018;16:156–170. doi: 10.1038/nrmicro.2017.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Campino S., Benavente E.D., Warhurst D.C., Beshir K.B., Lubis I., Gomes A.R., Feng J., Jiazhi W., Sun X., Huang F., Tang L. hua, Sutherland C.J., Clark T.G. Artemisinin resistance-associated markers in Plasmodium falciparum parasites from the China-Myanmar border: predicted structural stability of K13 propeller variants detected in a low-prevalence area. PLoS One. 2019;14:e0213686. doi: 10.1371/journal.pone.0213686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Deng C., Yang T., Xue L., Wang Q., Huang S., Su X.Z., Liu Y., Zheng S., Guan Y., Xu Q., Zhou J., Yuan J., Bacar A., Abdallah K.S., Attoumane R., Mliva A.M.S.A., Zhong Y., Lu F., Song J. Polymorphisms of the artemisinin resistant marker (K13) in Plasmodium falciparum parasite populations of Grande Comore Island 10 years after artemisinin combination therapy. Parasites Vectors. 2015;8:634. doi: 10.1186/s13071-015-1253-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M., Kaneko M., Tachibana S.I., Balikagala B., Sakurai-Yatsushiro M., Yatsushiro S., Takahashi N., Yamauchi M., Sekihara M., Hashimoto M., Katuro O.T., Olia A., Obwoya P.S., Auma M.A., Anywar D.A., Odongo-Aginya E.I., Okello-Onen J., Hirai M., Ohashi J., Palacpac N.M.Q., Kataoka M., Tsuboi T., Kimura E., Horii T., Mita T. Artemisinin-Resistant plasmodium falciparum with high survival rates, Uganda, 2014-2016. Emerg Infect Dis. 2018;24:718–726. doi: 10.3201/eid2404.170141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imwong M., Suwannasin K., Kunasol C., Sutawong K., Mayxay M., Rekol H., Smithuis F.M., Hlaing T.M., Tun K.M., van der Pluijm R.W., Tripura R., Miotto O., Menard D., Dhorda M., Day N.P.J., White N.J., Dondorp A.M. The spread of artemisinin-resistant Plasmodium falciparum in the Greater Mekong subregion: a molecular epidemiology observational study. Lancet Infect. Dis. 2017;17:491–497. doi: 10.1016/S1473-3099(17)30048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain V.,S.A. Repurposing of potent drug candidates for multiparasite targeting. Trends Parasitol. 2017;33:158–161. doi: 10.1016/j.pt.2016.12.007. [DOI] [PubMed] [Google Scholar]
- Jain V., Sharma Arvind, Singh G., Yogavel M., Sharma Amit. Structure-based targeting of orthologous pathogen proteins accelerates antiparasitic drug discovery. ACS Infect. Dis. 2017;3:281–292. doi: 10.1021/acsinfecdis.6b00181. [DOI] [PubMed] [Google Scholar]
- Khan S., Sharma A.A., Belrhali H., Yogavel M., Sharma A.A. Structural basis of malaria parasite lysyl-tRNA synthetase inhibition by cladosporin. J. Struct. Funct. Genom. 2014;15:63–71. doi: 10.1007/s10969-014-9182-1. [DOI] [PubMed] [Google Scholar]
- Krishna S., Kremsner P.G. Antidogmatic approaches to artemisinin resistance: reappraisal as treatment failure with artemisinin combination therapy. Trends Parasitol. 2013;29:313–317. doi: 10.1016/j.pt.2013.04.001. [DOI] [PubMed] [Google Scholar]
- Kyaw M.P., Nyunt M.H., Chit K., Aye M.M., Aye K.H., Aye M.M., Lindegardh N., Tarning J., Imwong M., Jacob C.G., Rasmussen C., Perin J., Ringwald P., Nyunt M.M. Reduced susceptibility of Plasmodium falciparum to artesunate in Southern Myanmar. PLoS One. 2013;8:e57689. doi: 10.1371/journal.pone.0057689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leang R., Barrette A., Bouth D.M., Menard D., Abdur R., Duong S., Ringwald P. Efficacy of dihydroartemisinin-piperaquine for treatment of uncomplicated Plasmodium falciparum and plasmodium vivax in Cambodia, 2008 to 2010. Antimicrob. Agents Chemother. 2013;57:818–826. doi: 10.1128/AAC.00686-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leang R., Taylor W.R.J., Bouth D.M., Song L., Tarning J., Char M.C., Kim S., Witkowski B., Duru V., Domergue A., Khim N., Ringwald P., Menard D. Evidence of Plasmodium falciparum malaria multidrug resistance to artemisinin and piperaquine in Western Cambodia: dihydroartemisinin-piperaquine open-label multicenter clinical assessment. Antimicrob. Agents Chemother. 2015;59:4719–4726. doi: 10.1128/AAC.00835-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.H., Fidock D.A. Evidence of a mild mutator phenotype in cambodian Plasmodium falciparum malaria parasites. PLoS One. 2016;11:e0154166. doi: 10.1371/journal.pone.0154166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménard D., Khim N., Beghain J., Adegnika A.A., Shafiul-Alam M., Amodu O., Rahim-Awab G., Barnadas C., Berry A., Boum Y., Bustos M.D., Cao J., Chen J.-H., Collet L., Cui L., Thakur G.-D., Dieye A., Djallé D., Dorkenoo M.A., Eboumbou-Moukoko C.E., Espino F.-E.-C.J., Fandeur T., Ferreira-da-Cruz M.-F., Fola A.A., Fuehrer H.-P., Hassan A.M., Herrera S., Hongvanthong B., Houzé S., Ibrahim M.L., Jahirul-Karim M., Jiang L., Kano S., Ali-Khan W., Khanthavong M., Kremsner P.G., Lacerda M., Leang R., Leelawong M., Li M., Lin K., Mazarati J.-B., Ménard S., Morlais I., Muhindo-Mavoko H., Musset L., Na-Bangchang K., Nambozi M., Niaré K., Noedl H., Ouédraogo J.-B., Pillai D.R., Pradines B., Quang-Phuc B., Ramharter M., Randrianarivelojosia M., Sattabongkot J., Sheikh-Omar A., Silué K.D., Sirima S.B., Sutherland C., Syafruddin D., Tahar R., Tang L.-H., Touré O.A., Tshibangu-wa-Tshibangu P., Vigan-Womas I., Warsame M., Wini L., Zakeri S., Kim S., Eam R., Berne L., Khean C., Chy S., Ken M., Loch K., Canier L., Duru V., Legrand E., Barale J.-C., Stokes B., Straimer J., Witkowski B., Fidock D.A., Rogier C., Ringwald P., Ariey F., Mercereau-Puijalon O. A worldwide map of Plasmodium falciparum K13-propeller polymorphisms. N. Engl. J. Med. 2016;374:2453–2464. doi: 10.1056/NEJMoa1513137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto O., Amato R., Ashley E.A., Macinnis B., Almagro-Garcia J., Amaratunga C., Lim P., Mead D., Oyola S.O., Dhorda M., Imwong M., Woodrow C., Manske M., Stalker J., Drury E., Campino S., Amenga-Etego L., Thanh T.N.N., Tran H.T., Ringwald P., Bethell D., Nosten F., Phyo A.P., Pukrittayakamee S., Chotivanich K., Chuor C.M., Nguon C., Suon S., Sreng S., Newton P.N., Mayxay M., Khanthavong M., Hongvanthong B., Htut Y., Han K.T., Kyaw M.P., Faiz M.A., Fanello C.I., Onyamboko M., Mokuolu O.A., Jacob C.G., Takala-Harrison S., Plowe C.V., Day N.P., Dondorp A.M., Spencer C.C.A., Mcvean G., Fairhurst R.M., White N.J., Kwiatkowski D.P. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat. Genet. 2015;47:226–234. doi: 10.1038/ng.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miraclin T.A., Matthew A., Rupali P. Decreased response to artemisinin combination therapy in falciparum malaria: a preliminary report from South India. Tropenmed. Parasitol. 2016;6:85–86. doi: 10.4103/2229-5070.175125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra N., Prajapati S.K., Kaitholia K., Bharti R.S., Srivastava B., Phookan S., Anvikar A.R., Dev V., Sonal G.S., Dhariwal A.C., White N.J., Valecha N. Surveillance of artemisinin resistance in Plasmodium falciparum in India using the kelch13 molecular marker. Antimicrob. Agents Chemother. 2015;59:2548–2553. doi: 10.1128/AAC.04632-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra N., Bharti R.S., Mallick P., Singh O.P., Srivastava B., Rana R., Phookan S., Gupta H.P., Ringwald P., Valecha N. Emerging polymorphisms in falciparum Kelch 13 gene in Northeastern region of India. Malar. J. 2016;15:583. doi: 10.1186/s12936-016-1636-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S., Bharti P.K., Shukla M.M., Ali N.A., Kashyotia S.S., Kumar A., Dhariwal A.C., Singh N. Clinical and molecular monitoring of Plasmodium falciparum resistance to antimalarial drug (artesunate+sulphadoxine-pyrimethamine) in two highly malarious district of Madhya Pradesh, Central India from 2012–2014. Pathog. Glob. Health. 2017;111:186–194. doi: 10.1080/20477724.2017.1331875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita T., Tanabe K. Evolution of Plasmodium falciparum drug resistance: implications for the development and containment of artemisinin resistance. Jpn. J. Infect. Dis. 2012;65:465–475. doi: 10.7883/yoken.65.465. [DOI] [PubMed] [Google Scholar]
- Mita T., Venkatesan M., Ohashi J., Culleton R., Takahashi N., Tsukahara T., Ndounga M., Dysoley L., Endo H., Hombhanje F., Ferreira M.U., Plowe C.V., Tanabe K. Limited geographical origin and global spread of sulfadoxine-resistant dhps alleles in Plasmodium falciparum populations. J. Infect. Dis. 2011;204:1980–1988. doi: 10.1093/infdis/jir664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohon A.N., Alam M.S., Bayih A.G., Folefoc A., Shahinas D., Haque R., Pillai D.R. Mutations in Plasmodium falciparum K13 propeller gene from Bangladesh (2009-2013) Malar. J. 2014;13:431. doi: 10.1186/1475-2875-13-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A., Bopp S., Magistrado P., Wong W., Daniels R., Demas A., Schaffner S., Amaratunga C., Lim P., Dhorda M., Miotto O., Woodrow C., Ashley E.A., Dondorp A.M., White N.J., Wirth D., Fairhurst R., Volkman S.K. Artemisinin resistance without pfkelch13 mutations in Plasmodium falciparum isolates from Cambodia. Malar. J. 2017;16:195. doi: 10.1186/s12936-017-1845-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutabingwa T.K., Anthony D., Heller A., Hallett R., Ahmed J., Drakeley C., Greenwood B.M., Whitty C.J.M. Amodiaquine alone, amodiaquine+sulfadoxine-pyrimethamine, amodiaquine+artesunate, and artemether-lumefantrine for outpatient treatment of malaria in Tanzanian children: a four-arm randomised effectiveness trial. Lancet. 2005;365:1474–1480. doi: 10.1016/S0140-6736(05)66417-3. [DOI] [PubMed] [Google Scholar]
- Na J., Efferth T. Development of artemisinin resistance in malaria therapy. Pharmacol. Res. 2019;146:10427. doi: 10.1016/j.phrs.2019.104275. [DOI] [PubMed] [Google Scholar]
- National Framework Malaria Elimination India: 2016-2030. National Vector Borne Disease Control Programme; 2015. [Google Scholar]
- Noedl H., Se Y., Schaecher K., Smith B.L., Socheat D., Fukuda M.M. Evidence of artemisinin-resistant malaria in western Cambodia. N. Engl. J. Med. 2008;359:2619–2620. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- Nosten F., White N.J. Artemisinin-based combination treatment of falciparum malaria. Am. J. Trop. Med. Hyg. 2007;77:181–192. [PubMed] [Google Scholar]
- Nyunt M.H., Shein T., Zaw N.N., Han S.S., Muh F., Lee S.K., Han J.H., Thant K.Z., Han E.T., Kyaw M.P. Molecular evidence of drug resistance in asymptomatic malaria infections, Myanmar, 2015. Emerg. Infect. Dis. 2017;23:517–520. doi: 10.3201/eid2303.161363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouattara A., Kone A., Adams M., Fofana B., Maiga A.W., Hampton S., Coulibaly D., Thera M.A., Diallo N., Dara A., Sagara I., Gil J.P., Bjorkman A., Takala-Harrison S., Doumbo O.K., Plowe C.V., Djimde A.A. Polymorphisms in the K13-propeller gene in artemisinin-susceptible Plasmodium falciparum parasites from Bougoula-Hameau and Bandiagara, Mali. Am. J. Trop. Med. Hyg. 2015;92:1202–1206. doi: 10.4269/ajtmh.14-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyola S.O., Ariani C.V., Hamilton W.L., Kekre M., Amenga-Etego L.N., Ghansah A., Rutledge G.G., Redmond S., Manske M., Jyothi D., Jacob C.G., Otto T.D., Rockett K., Newbold C.I., Berriman M., Kwiatkowski D.P. Whole genome sequencing of Plasmodium falciparum from dried blood spots using selective whole genome amplification. Malar. J. 2016;15:597. doi: 10.1186/s12936-016-1641-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pau M.C., Pantaleo A., Tsamesidis I., Hoang H., Tran A.T., Nguyen T.L.H., Phan T.H.G., Nu P.A.T., Ngo T.M.C., Marchetti G., Schwarzer E., Fiori P.L., Low P.S., Huynh C.D., Turrini F.M. Clinical impact of the two ART resistance markers, K13 gene mutations and DPC3 in Vietnam. PLoS One. 2019;14:e0214667. doi: 10.1371/journal.pone.0214667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazhayam N.M., Chhibber-Goel J., Sharma A. New leads for drug repurposing against malaria. Drug Discov. Today. 2019;24:263–271. doi: 10.1016/j.drudis.2018.08.006. [DOI] [PubMed] [Google Scholar]
- Phyo A., Ashley E., Anderson T., Bozdech Z., Carrara V., Sriprawat K., Nair S., White M., Dziekan J., Ling C., Proux S., Konghahong K., Jeeyapant A., Woodrow C., Imwong M., McGready R., Lwin K., Day N., White N., Nosten F. Declining efficacy of artemisinin combination therapy against P. Falciparum malaria on the Thai-Myanmar border (2003-2013): the role of parasite genetic factors. Clin. Infect. Dis. 2016;63:784–791. doi: 10.1093/cid/ciw388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putaporntip C., Kuamsab N., Kosuwin R., Tantiwattanasub W., Vejakama P., Sueblinvong T., Seethamchai S., Jongwutiwes S., Hughes A.L. Natural selection of K13 mutants of Plasmodium falciparum in response to artemisinin combination therapies in Thailand. Clin. Microbiol. Infect. 2016;22:285e1–285e8. doi: 10.1016/j.cmi.2015.10.027. [DOI] [PubMed] [Google Scholar]
- Rasmussen C., Valecha N., Ringwald P. Lack of convincing evidence of artemisinin resistance in India. Clin. Infect. Dis. 2019;69:1461–1462. doi: 10.1093/cid/ciz166. pii: ciz166. [DOI] [PubMed] [Google Scholar]
- Roper C., Pearce R., Nair S., Sharp B., Nosten F., A.T. Intercontinental spread of pyrimethamine-resistant malaria. Science. 2004;305:1124. doi: 10.1126/science.1098876. [DOI] [PubMed] [Google Scholar]
- Sá J.M., Kaslow S.R., Krause M.A., Melendez-Muniz V.A., Salzman R.E., Kite W.A., Zhang M., Moraes Barros R.R., Mu J., Han P.K., Mershon J.P., Figan C.E., Caleon R.L., Rahman R.S., Gibson T.J., Amaratunga C., Nishiguchi E.P., Breglio K.F., Engels T.M., Velmurugan S., Ricklefs S., Straimer J., Gnädig N.F., Deng B., Liu A., Diouf A., Miura K., Tullo G.S., Eastman R.T., Chakravarty S., James E.R., Udenze K., Li S., Sturdevant D.E., Gwadz R.W., Porcella S.F., Long C.A., Fidock D.A., Thomas M.L., Fay M.P., Sim B.K.L., Hoffman S.L., Adams J.H., Fairhurst R.M., Su X., Wellems T.E. Artemisinin resistance phenotypes and K13 inheritance in a Plasmodium falciparum cross and Aotus model. Proc. Natl. Acad. Sci. 2018;115:12513–12518. doi: 10.1073/pnas.1813386115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders D.L., Vanachayangkul P., Lon C. Dihydroartemisinin–Piperaquine failure in Cambodia. N. Engl. J. Med. 2014;371:484–485. doi: 10.1056/NEJMc1403007. [DOI] [PubMed] [Google Scholar]
- Silva M., Ferreira P., Otienoburu S., Calçada C., Ngasala B., Björkman A., Mårtensson A., Gil J., Veiga M. Plasmodium falciparum K13 expression associated with parasite clearance during artemisinin-based combination therapy. J. Antimicrob. Chemother. 2019;74:1890–1893. doi: 10.1093/jac/dkz098. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Singh G.P., Goel P., Sharma A. Structural mapping of Kelch13 mutations associated with artemisinin resistance in malaria. J. Struct. Funct. Genom. 2016;17:51–56. doi: 10.1007/s10969-016-9205-1. [DOI] [PubMed] [Google Scholar]
- Spring M.D., Lin J.T., Manning J.E., Vanachayangkul P., Somethy S., Bun R., Se Y., Chann S., Ittiverakul M., Sia-ngam P., Kuntawunginn W., Arsanok M., Buathong N., Chaorattanakawee S., Gosi P., Ta-aksorn W., Chanarat N., Sundrakes S., Kong N., Heng T.K., Nou S., Teja-isavadharm P., Pichyangkul S., Phann S.T., Balasubramanian S., Juliano J.J., Meshnick S.R., Chour C.M., Prom S., Lanteri C.A., Lon C., Saunders D.L. Dihydroartemisinin-piperaquine failure associated with a triple mutant including kelch13 C580Y in Cambodia: an observational cohort study. Lancet Infect. Dis. 2015;15:683–691. doi: 10.1016/S1473-3099(15)70049-6. [DOI] [PubMed] [Google Scholar]
- Starzengruber P., Swoboda P., Fuehrer H.P., Khan W.A., Hofecker V., Siedl A., Fally M., Graf O., Teja-Isavadharm P., Haque R., Ringwald P., Noedl H. Current status of artemisinin-resistant falciparum malaria in South Asia: a randomized controlled artesunate monotherapy trial in Bangladesh. PLoS One. 2012;7:e52236. doi: 10.1371/journal.pone.0052236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straimer J., Gnädig N.F., Witkowski B., Amaratunga C., Duru V., Ramadani A.P., Dacheux M., Khim N., Zhang L., Lam S., Gregory P.D., Urnov F.D., Mercereau-Puijalon O., Benoit-Vical F., Fairhurst R.M., Ménard D., Fidock D.A. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science. 2015;347:428–431. doi: 10.1126/science.1260867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takala-Harrison S., Clark T.G., Jacob C.G., Cummings M.P., Miotto O., Dondorp A.M., Fukuda M.M., Nosten F., Noedl H., Imwong M., Bethell D., Se Y., Lon C., Tyner S.D., Saunders D.L., Socheat D., Ariey F., Phyo A.P., Starzengruber P., Fuehrer H.-P., Swoboda P., Stepniewska K., Flegg J., Arze C., Cerqueira G.C., Silva J.C., Ricklefs S.M., Porcella S.F., Stephens R.M., Adams M., Kenefic L.J., Campino S., Auburn S., MacInnis B., Kwiatkowski D.P., Su X. -z., White N.J., Ringwald P., Plowe C.V. Genetic loci associated with delayed clearance of Plasmodium falciparum following artemisinin treatment in Southeast Asia. Proc. Natl. Acad. Sci. 2013;110:240–245. doi: 10.1073/pnas.1211205110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takala-Harrison S., Jacob C.G., Arze C., Cummings M.P., Silva J.C., Dondorp A.M., Fukuda M.M., Hien T.T., Mayxay M., Noedl H., Nosten F., Kyaw M.P., Nhien N.T.T., Imwong M., Bethell D., Se Y., Lon C., Tyner S.D., Saunders D.L., Ariey F., Mercereau-Puijalon O., Menard D., Newton P.N., Khanthavong M., Hongvanthong B., Starzengruber P., Fuehrer H.P., Swoboda P., Khan W.A., Phyo A.P., Nyunt M.M., Nyunt M.H., Brown T.S., Adams M., Pepin C.S., Bailey J., Tan J.C., Ferdig M.T., Clark T.G., Miotto O., MacInnis B., Kwiatkowski D.P., White N.J., Ringwald P., Plowe C.V. Independent emergence of artemisinin resistance mutations among Plasmodium falciparum in Southeast Asia. J. Infect. Dis. 2015;211:670–679. doi: 10.1093/infdis/jiu491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talundzic E., Okoth S.A., Congpuong K., Plucinski M.M., Morton L., Goldman I.F., Kachur P.S., Wongsrichanalai C., Satimai W., Barnwell J.W., Udhayakumar V. Selection and spread of artemisinin-resistant alleles in Thailand prior to the global artemisinin resistance containment campaign. PLoS Pathog. 2015;11:e1004789. doi: 10.1371/journal.ppat.1004789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talundzic E., Ndiaye Y.D., Deme A.B., Olsen C., Patel D.S., Biliya S., Daniels R., Vannberg F.O., Volkman S.K., Udhayakumar V., Ndiaye D. Molecular epidemiology of Plasmodium falciparum kelch13 mutations in Senegal determined by using targeted amplicon deep sequencing. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.02116-16. e02116-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuy-Nhien N., Tuyen N.K., Tong N.T., Vy N.T., Thanh N.V., Van H.T., Huong-Thu P., Quang H.H., Boni M.F., Dolecek C., Farrar J., Thwaites G.E., Miotto O., White N.J., Hien T.T. K13 propeller mutations in Plasmodium falciparum populations in regions of malaria endemicity in Vietnam from 2009 to 2016. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.01578-16. e01578-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilley L., Straimer J., Gnädig N.F., Ralph S.A., Fidock D.A. Artemisinin action and resistance in Plasmodium falciparum. Trends Parasitol. 2016;32:682–696. doi: 10.1016/j.pt.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tun K.M., Imwong M., Lwin K.M., Win A.A., Hlaing T.M., Hlaing T., Lin K., Kyaw M.P., Plewes K., Faiz M.A., Dhorda M., Cheah P.Y., Pukrittayakamee S., Ashley E.A., Anderson T.J.C., Nair S., McDew-White M., Flegg J.A., Grist E.P.M., Guerin P., Maude R.J., Smithuis F., Dondorp A.M., Day N.P.J., Nosten F., White N.J., Woodrow C.J. Spread of artemisinin-resistant Plasmodium falciparum in Myanmar: a cross-sectional survey of the K13 molecular marker. Lancet Infect. Dis. 2015;15:415–421. doi: 10.1016/S1473-3099(15)70032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tun K.M., Jeeyapant A., Imwong M., Thein M., Aung S.S.M., Hlaing T.M., Yuentrakul P., Promnarate C., Dhorda M., Woodrow C.J., Dondorp A.M., Ashley E.A., Smithuis F.M., White N.J., Day N.P.J. Parasite clearance rates in Upper Myanmar indicate a distinctive artemisinin resistance phenotype: a therapeutic efficacy study. Malar. J. 2016;15:185. doi: 10.1186/s12936-016-1240-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tun S.T.T., Lubell Y., Dondorp A.M., Fieldman T., Tun K.M., Celhay O., Chan X.H., Saralamba S., White L.J. Identifying artemisinin resistance from parasite clearance half-life data with a simple Shiny web application. PLoS One. 2017;12:e0177840. doi: 10.1371/journal.pone.0177840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Shrestha S., Li X., Miao J., Yuan L., Cabrera M., Grube C., Yang Z., Cui L. Prevalence of K13-propeller polymorphisms in Plasmodium falciparum from China-Myanmar border in 2007-2012. Malar. J. 2015;14:168. doi: 10.1186/s12936-015-0672-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Cabrera M., Yang J., Yuan L., Gupta B., Liang X., Kemirembe K., Shrestha S., Brashear A., Li X., Porcella S.F., Miao J., Yang Z., Su X.Z., Cui L. Genome-wide association analysis identifies genetic loci associated with resistance to multiple antimalarials in Plasmodium falciparum from China-Myanmar border. Sci. Rep. 2016;6:33891. doi: 10.1038/srep33891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernsdorfer W.H., Payne D. The dynamics of drug resistance in Plasmodium falciparum. Pharmacol. Ther. 1991;50:95–121. doi: 10.1016/0163-7258(91)90074-v. [DOI] [PubMed] [Google Scholar]
- White N.J. Qinghaosu (artemisinin): the price of success. Science. 2008;320:330–334. doi: 10.1126/science.1155165. [DOI] [PubMed] [Google Scholar]
- White N.J. The parasite clearance curve. Malar. J. 2011;10:278. doi: 10.1186/1475-2875-10-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White L.J., Flegg J.A., Phyo A.P., Wiladpai-ngern J.H., Bethell D., Plowe C., Anderson T., Nkhoma S., Nair S., Tripura R., Stepniewska K., Pan-Ngum W., Silamut K., Cooper B.S., Lubell Y., Ashley E.A., Nguon C., Nosten F., White N.J., Dondorp A.M. Defining the in vivo phenotype of artemisinin-resistant falciparum malaria: a modelling approach. PLoS Med. 2015;12:e1001823. doi: 10.1371/journal.pmed.1001823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler E.A. Longitudinal study of Plasmodium pathogens identifies new loci associated with artemisinin resistance. Genome Biol. 2017;18:79. doi: 10.1186/s13059-017-1219-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkowski B., Amaratunga C., Khim N., Sreng S., Chim P., Kim S., Lim P., Mao S., Sopha C., Sam B., Anderson J.M., Duong S., Chuor C.M., Taylor W.R.J., Suon S., Mercereau-Puijalon O., Fairhurst R.M., Menard D. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in-vitro and ex-vivo drug-response studies. Lancet Infect. Dis. 2013;13:1043–1049. doi: 10.1016/S1473-3099(13)70252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkowski B., Khim N., Chim P., Kim S., Ke S., Kloeung N., Chy S., Duong S., Leang R., Ringwald P., Dondorp A.M., Tripura R., Benoit-Vical F., Berry A., Gorgette O., Ariey F., Barale J.C., Mercereau-Puijalon O., Menard D. Reduced artemisinin susceptibility of Plasmodium falciparum ring stages in western Cambodia. Antimicrob. Agents Chemother. 2013;57:914–923. doi: 10.1128/AAC.01868-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organisation - WHO . 2015. WHO - Guidelines for the Treatment of Malaria. [Google Scholar]
- World Health Organisation (WHO) World Health Organization; 2018. World Malaria Report 2018. [Google Scholar]
- World Health Organization . Global Malaria Programme; 2017. Artemisinin and Artemisinin-Based Combination Therapy Resistance. [Google Scholar]
- Ye R., Hu D., Zhang Y., Huang Y., Sun X., Wang J., Chen X., Zhou H., Zhang D., Mungthin M., Pan W. Distinctive origin of artemisinin-resistant Plasmodium falciparum on the China-Myanmar border. Sci. Rep. 2016;6:20100. doi: 10.1038/srep20100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogavel M., Chaturvedi R., Babbar P., Malhotra N., Jain V., Sharma A., Manickam Y., Chaturvedi R., Babbar P., Malhotra N., Jain V., Sharma A. Drug targeting of one or more aminoacyl-tRNA synthetase in the malaria parasite Plasmodium falciparum. Drug Discov. Today. 2018;23:1233–1240. doi: 10.1016/j.drudis.2018.01.050. [DOI] [PubMed] [Google Scholar]
- Yogavel M., Nettleship J.E., Sharma A., Harlos K., Jamwal A., Chaturvedi R., Sharma M., Jain V., Chhibber-Goel J., Sharma A. Structure of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase–dihydropteroate synthase from Plasmodium vivax sheds light on drug resistance. JBC. 2018;293(39):14962–14972. doi: 10.1074/jbc.RA118.004558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012;44:821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.