Abstract
Accumulating evidence indicates that circular RNAs (circRNAs) are vital regulators of various biological functions involved in the progression of multiple cancers. Circular F-box and WD repeat domain containing 7 (circFBXW7) (hsa_circ_0001451) has been reported to act as a tumor suppressor by encoding a novel protein in glioma; however, its functions and mechanisms in triple-negative breast cancer (TNBC) remain elusive. In the current study, we validated by qRT-PCR that circFBXW7 was downregulated in TNBC cell lines and found that low expression of circFBXW7 was associated with poorer clinical outcomes. circFBXW7 expression was negatively correlated with tumor size and lymph node metastasis, and it was an independent prognostic factor for TNBC patients. We performed cell proliferation, colony formation, transwell, wound-healing, and mouse xenograft assays to confirm the functions of circFBXW7. Overexpression of circFBXW7 obviously inhibited cell proliferation, migration, and tumor growth in both in vitro and in vivo assays. Luciferase reporter assays and RNA immunoprecipitation assays revealed that circFBXW7 serves as a sponge of miR-197-3p and suppresses TNBC growth and metastasis by upregulating FBXW7 expression. In addition, the FBXW7-185aa protein encoded by circFBXW7 inhibited the proliferation and migration abilities of TNBC cells by increasing the abundance of FBXW7 and inducing c-Myc degradation. In summary, our research demonstrated that circFBXW7 sponges miR-197-3p and encodes the FBXW7-185aa protein to suppress TNBC progression through upregulating FBXW7 expression. Thus, circFBXW7 may act as a therapeutic target and prognostic biomarker for TNBC.
Keywords: circular RNAs, circFBXW7, FBXW7, competitive endogenous RNAs, triple negative breast cancer
Introduction
As the most pervasive malignancy and second leading cause of cancer-related death among women worldwide, breast cancer is regarded as a heterogeneous carcinoma with various molecular subtypes.1, 2 Triple-negative breast cancer (TNBC) is acknowledged as the subtype with the worst prognosis as a result of the lack of effective therapeutic targets.3, 4 Undoubtedly, it is a matter of great urgency for oncologists to develop more efficient molecular targets and novel biomarkers for TNBC therapy and monitoring.
Currently, circular RNAs (circRNAs) have attracted great research interest for their diversity of impacts on the progression and recurrence of cancers.5, 6, 7 Like many other kinds of noncoding RNAs (ncRNAs), circRNAs were once considered a waste of biological energy that is abundant in mammals.8 circRNAs are more stable and resistant than linear RNAs, with no 5′ cap or 3′ poly(A) tail.9 circRNAs are versatile regulators of cell biological activities and have various biological functions, including as microRNA (miRNA)-binding sponges, RNA-binding protein regulators, and protein translation templates.10, 11
On the basis of the competing endogenous RNA (ceRNA) hypothesis, RNAs can act as ceRNAs by competing for specific miRNAs.12, 13, 14 circRNAs are also ceRNAs because of their miRNA response elements (MREs) and predominant localization in the cytoplasm.15 For example, the notable circRNA ciRS-7 acts as a negative regulator of miR-7 and exerts influence on the development of tumors.16, 17, 18 In addition, circRAPGEF5 promotes proliferation and metastasis in papillary thyroid cancer by sponging miR-198 and upregulating FGFR1.19 Knockdown of hsa_circ_0061140 suppresses proliferation and migration in ovarian cancer through miR-370 sponge activity.20
In our previous study, circEPSTI1, circKIF4A, circGFRA1, and circRAD18 were identified and confirmed to have oncogenic roles in the progression of TNBC.21, 22, 23, 24 Conversely, an increasing number of circRNAs have been discovered to be tumor suppressors in different kinds of tumors. For instance, circZKSCAN1 can inhibit cell proliferation, migration, and invasion in hepatocellular carcinoma.25 circRNA derived from F-box and WD repeat domain containing 7 (circFBXW7) was reported as a tumor suppressor in the human brain by encoding a novel 21-kDa protein, FBXW7-185aa.26 However, the biological function of circFBXW7 in the progression of TNBC and whether it acts as a ceRNA in cells remain unclear.
In this study, we validated that circFBXW7 was downregulated in TNBC cell lines and that low expression of circFBXW7 was associated with poorer clinical outcomes in 473 breast cancer patients. circFBXW7 was negatively correlated with tumor size and lymph node metastasis, and it was an independent prognostic factor for TNBC patients. Both in vitro and in vivo assays revealed that circFBXW7 significantly suppressed cell proliferation and migration abilities. A further series of mechanistic experiments confirmed that circFBXW7 directly sponges miR-197-3p to relieve the silencing of FBXW7. In addition, FBXW7-185aa protein encoded by circFBXW7 inhibited the proliferation and migration ability of TNBC cells by increasing the abundance of FBXW7 and inducing c-Myc degradation. Taken together, our findings indicate that circFBXW7 sponges miR-197-3p and encodes the FBXW7-185aa protein to suppress TNBC progression though upregulating FBXW7 expression. Thus, circFBXW7 might be a novel prognostic biomarker and potential target for TNBC treatment.
Results
circFBXW7 Is Downregulated in TNBC and Correlates with Better Clinical Outcomes
circFBXW7 was reported as a tumor suppressor circRNA that is abundantly expressed in normal brain tissues and downregulated in glioma.26 According to the University of California, Santa Cruz (UCSC) and circBase databases, circFBXW7 (hsa_circ_0001451) is derived from exons 3 and 4 of FBXW7 (F-box and WD repeat domain containing 7) located on chromosome 4q31.3 (chr4:153332454–153333681), with backsplicing of exons 3 and 4.
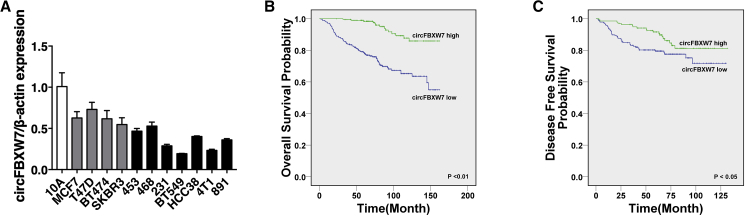
We first performed qRT-PCR to evaluate the expression level of circFBXW7 in cell lines, and we found low expression in breast cancer cell lines, especially in TNBC cell lines (Figure 1A). To explore the association between the circFBXW7 expression level and clinical significance, we next recruited a cohort of 473 breast cancer patients. We assigned the patients into two groups according to the expression level of circFBXW7. The average expression level was defined as the cutoff value. Kaplan-Meier survival analysis showed poorer overall survival (OS) and disease-free survival (DFS) of breast cancer patients with low circFBXW7 levels (Figures 1B and 1C). circFBXW7 was negatively correlated with tumor size and lymph node metastasis, and it was an independent prognostic factor for TNBC patients (Tables 1 and 2).
Figure 1.
circFBXW7 Is Downregulated in TNBC and Correlated with Better Clinical Outcomes
(A) The relative expression level of circFBXW7 in breast cancer cell lines. (B and C) Kaplan-Meier analysis of the (B) overall survival and (C) disease-free survival of 473 TNBC patients with high (green) or low (blue) circFBXW7 expression levels. SD is shown as error bars.
Table 1.
Correlation of circFBXW7 Expression with Clinicopathologic Characteristics of Breast Cancer Patients
| Variable | Cases | circFBXW7 |
p Value | |
|---|---|---|---|---|
| Low | High | |||
| Age (Years) | ||||
| >50 | 218 | 140 (64.2%) | 78 (35.8%) | 0.061 |
| ≤50 | 255 | 141 (55.3%) | 114 (44.7%) | |
| Subtypes | ||||
| Non-TNBC | 191 | 111 (58.1%) | 80 (41.9%) | 0.684 |
| TNBC | 275 | 165 (60.0%) | 110 (40.0%) | |
| Tumor Size (cm) | ||||
| ≤2.0 | 120 | 60 (50.0%) | 60 (50.0%) | 0.014∗ |
| >2.0 | 352 | 221 (62.8%) | 131 (37.2%) | |
| Lymph Node Status | ||||
| Negative | 210 | 109 (51.9%) | 101 (48.1%) | 0.004∗ |
| Positive | 258 | 168 (65.1%) | 90 (34.9%) | |
| TNM Stage | ||||
| I-II | 292 | 168 (57.5%) | 124 (42.5%) | 0.302 |
| III-IV | 178 | 111 (62.4%) | 67 (37.6%) | |
| FBXW7 Expression | ||||
| Low | 261 | 166 (63.6%) | 95 (36.4%) | 0.039∗ |
| High | 212 | 115 (54.2%) | 97 (45.8%) | |
p < 0.05, statistically significant.
Table 2.
Univariate and Multivariate Cox Regression Analysis of circFBXW7 and Survival in Patients with Breast Cancer
| Parameter | Univariate Analysis |
Multivariate Analysis |
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p Value | HR | 95% CI | p Value | |
| Age (>50 versus ≤50 years) | 0.976 | 0.653–1.458 | 0.904 | NA | ||
| TNBC (yes versus no) | 1.214 | 0.796–1.851 | 0.368 | NA | ||
| Histological grade (G3 versus G1-2) | 1.722 | 1.097–2.702 | 0.018* | NA | ||
| Tumor size (>2.0 cm versus ≤2.0 cm) | 1.934 | 1.115–3.356 | 0.019* | 1.504 | 0.845–2.674 | 0.165 |
| Lymph node status (positive versus negative) | 1.978 | 1.269–3.081 | 0.003* | 1.468 | 0.913–2.361 | 0.113 |
| TNM stage (III-IV versus I-II) | 2.342 | 1.560–3.516 | 0.001* | 1.779 | 1.143–2.769 | 0.011* |
| circFBXW7 expression (high versus low) | 0.189 | 0.105–0.339 | 0.001* | 0.215 | 0.119–0.387 | 0.001* |
| FBXW7 expression (high versus low) | 0.314 | 0.193–0.509 | 0.001* | 0.349 | 0.212–0.574 | 0.001* |
NA, not analyzed; *p < 0.05, statistically significant.
Overexpression of circFBXW7 Inhibits the Proliferation and Metastasis of TNBC Cells
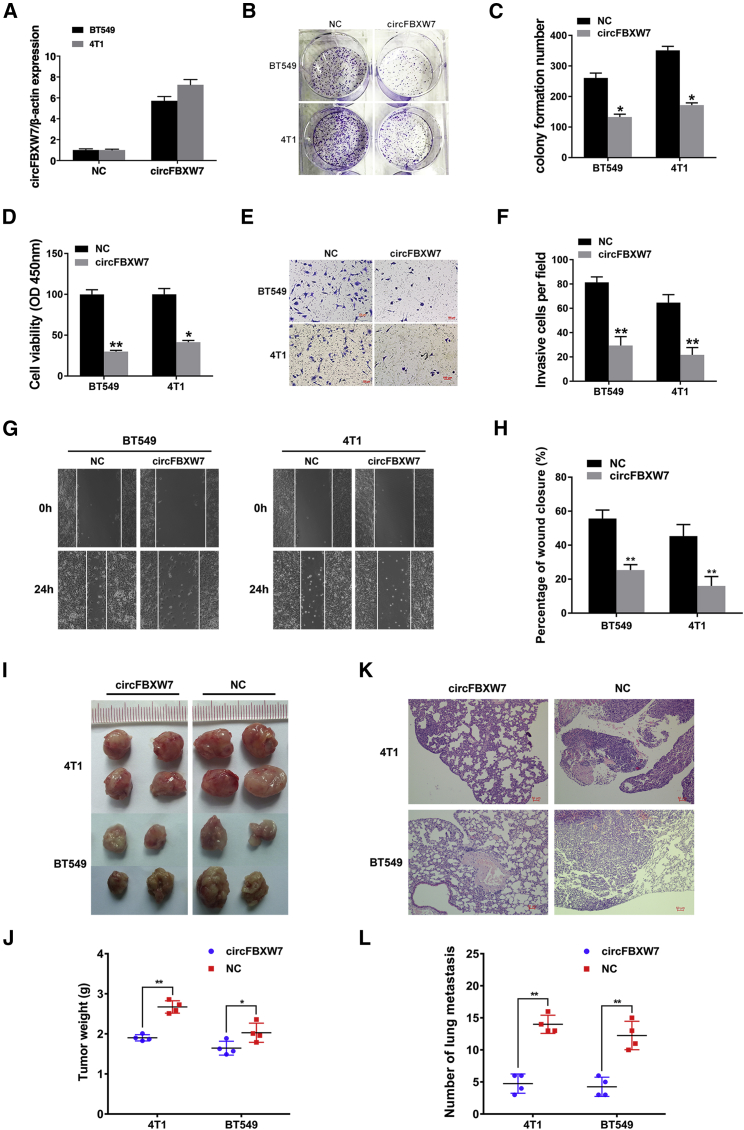
To investigate the biological function of circFBXW7 in TNBC cells, we constructed a circFBXW7 overexpression vector and verified its effectiveness in BT549 and 4T1 cell lines (Figure 2A). We conducted colony formation assays, and we found that the upregulation of circFBXW7 significantly inhibited the colony-forming ability of these two TNBC cell lines (BT549 and 4T1) (Figures 2B and 2C).
Figure 2.
Overexpression of circFBXW7 Inhibits the Proliferation and Metastasis of TNBC Cells
(A) Overexpression of circFBXW7, as assessed by qRT-PCR. (B) circFBXW7 inhibits the colony-forming ability of BT549 and 4T1 cells. (C) Statistical graph of the colony-forming assays. (D) CCK-8 assays were used to assess cell proliferation. (E) Transwell assays were used to evaluate the cell migration capability. (F) Statistical graph of the transwell assays. (G) Wound-healing assays were used to assess the impact of circFBXW7 on the cell migration ability. (H) Statistical graph of the wound-healing assays. (I) Xenograft models were established. (J) Tumor weights were measured and are shown. (K) H&E-stained sections of lung metastases. (L) The number of metastases was counted and recorded. *p < 0.05, **p < 0.01. SD is shown as error bars.
Consistent with these findings, CCK-8 assays revealed that circFBXW7 obviously suppressed cell proliferation (Figure 2D). To further examine the influence of circFBXW7 on the metastatic capacity of TNBC cells, transwell assays and wound-healing assays were also conducted. Our results revealed that the overexpression of circFBXW7 markedly inhibited the migration ability of these two TNBC cell lines, as demonstrated by transwell assays (Figures 2E and 2F) and wound-healing assays (Figures 2G and 2H). Mouse xenograft models were established to assess the function of circFBXW7 in vivo. circFBXW7 obviously inhibited tumor growth (Figures 2I and 2J) and decreased the number of lung metastases (Figures 2K and 2L), which indicated the significant role of circFBXW7 in tumor suppression.
circFBXW7 Acts as a Sponge of miR-197-3p
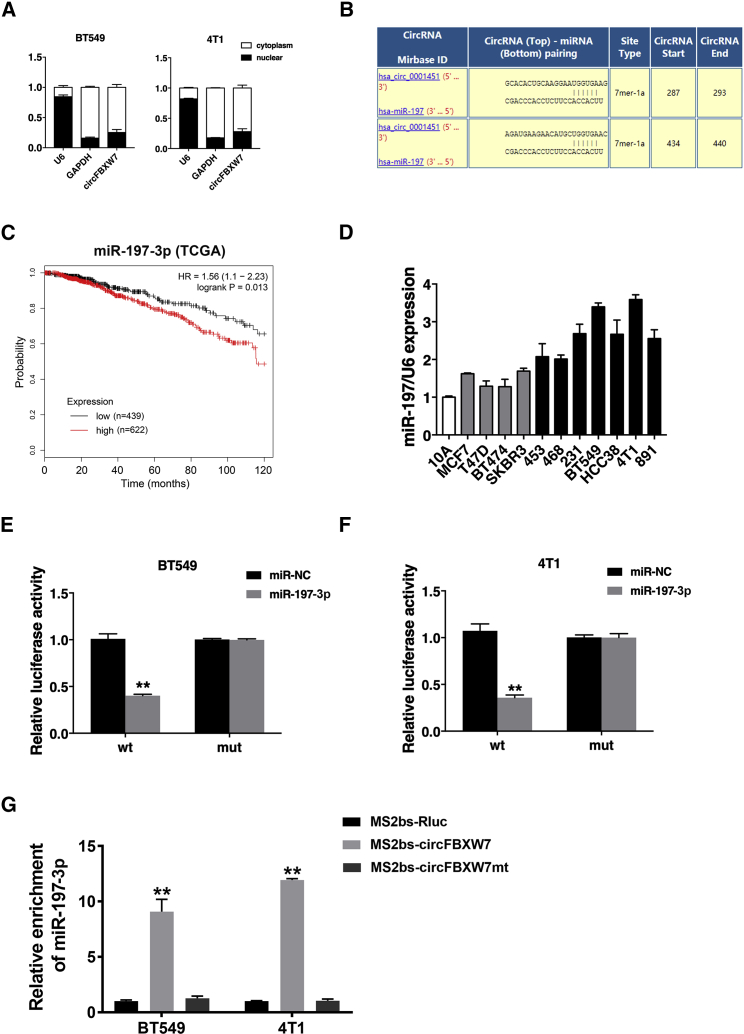
Next, we examined the intracellular location of circFBXW7 in TNBC cell lines. qRT-PCR analysis of nuclear and cytoplasmic RNA isolated from cells revealed that circFBXW7 was abundantly distributed in the cytoplasm of cells (Figure 3A). Considering that circFBXW7 was mainly localized and stable in the cytoplasm, we speculated that it might function as a sponge of miRNA. According to the circRNA Interactome website (https://circinteractome.nia.nih.gov/index.html), binding sites for miR-197-3p were discovered within the circFBXW7 sequence (Figure 3B). As reported in several studies, miR-197-3p acts as a tumor-promoting miRNA in many cancers, including breast cancer.27, 28, 29, 30, 31 miR-197-3p was positively associated with worse overall survival in breast cancer patients, according to The Cancer Genome Atlas (TCGA) database (Figure 3C).
Figure 3.
circFBXW7 Acts as a Sponge for miR-197-3p
(A) U6 (nuclear control transcript), GAPDH (cytoplasmic control transcript), and circFBXW7 transcription in the nuclear and cytoplasmic fractions, as analyzed by qRT-PCR. (B) Predicted binding sites of miR-197-3p in circFBXW7. (C) Kaplan-Meier analysis of the association between miR-197-3p and overall survival in patients with breast cancer from TCGA database. (D) The relative expression level of miR-197-3p in breast cancer cell lines. (E and F) Luciferase reporter assay of BT549 (E) and 4T1 (F) cells cotransfected with miR-197-3p mimics and the circFBXW7 wild-type or mutant luciferase reporter. (G) MS2-based RIP assay of cells transfected with MS2bs-circFBXW7, MS2bs-circFBXW7-mt, or control. *p < 0.05, **p < 0.01. SD is shown as error bars.
We measured the expression level of miR-197 in cell lines and found its upregulation in TNBC (Figure 3D). Subsequently, we performed dual luciferase reporter assays to confirm the interaction between circFBXW7 and miR-197-3p. We cotransfected a luciferase (luc)-circFBXW7-wild-type (WT) or a luc-circFBXW7-mutant (mut) reporter vector with miR-197-3p mimics or negative control (NC) mimics into BT549 and 4T1 cells. Overexpression of miR-197-3p decreased the relative luciferase activity of the wild-type reporter by over 50%, whereas it had no influence on the mutant reporter (Figures 3E and 3F). To further verify the direct binding of circFBXW7 and miR-197-3p, we conducted a RNA immunoprecipitation (RIP) assay. The results showed that miR-197-3p was predominantly enriched in the MS2bs-circFBXW7 group, which indicates that circFBXW7 directly interacts with miR-197-3p and acts as a sponge for it (Figure 3G).
miR-197-3p Decreases the Expression of the Tumor Suppressor Gene FBXW7
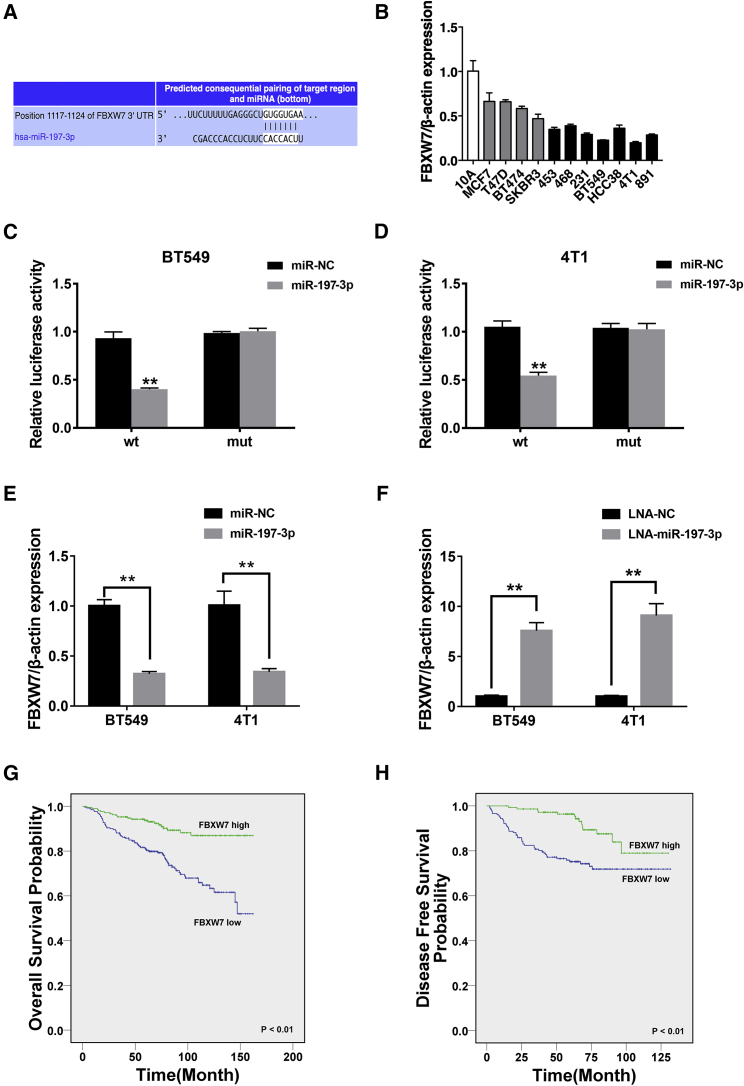
To determine the downstream targets of miR-197-3p, we used TargetScan32 to identify putative genes, and FBXW7 was predicted (Figure 4A). FBXW7 has been confirmed as a tumor suppressor gene in multiple cancers, including breast cancer, by directly binding to cyclin E and degrading it via a ubiquitin-mediated process.33, 34, 35, 36 FBXW7 expression was downregulated in TNBC cell lines compared to that in mammary epithelial cell lines, as evidenced by qRT-PCR analysis (Figure 4B).
Figure 4.
miR-197-3p Decreases the Expression of the Tumor Suppressor Gene FBXW7
(A) Predicted binding sites of miR-197-3p in the 3′ UTR of FBXW7 mRNA according to TargetScan. (B) The relative expression level of FBXW7 in breast cancer cell lines. (C and D) Luciferase reporter assay of BT549 (C) and 4T1 (D) cells cotransfected with miR-197-3p mimics and the FBXW7 3′ UTR wild-type or mutant luciferase reporter. (E) The expression of FBXW7 was decreased after transfection with miR-197-3p mimics. (F) The expression of FBXW7 was increased after transfection with miR-197-3p inhibitors. (G and H) Kaplan-Meier analysis of the (G) overall survival and (H) disease-free survival of 473 TNBC patients with high (green) or low (blue) FBXW7 expression levels. *p < 0.05, **p < 0.01. SD is shown as error bars.
Subsequently, we conducted dual luciferase reporter assays, and we found that the relative luciferase activity was significantly decreased in BT549 and 4T1 cell lines after cotransfection with miR-197-3p mimics and the WT-3′ UTR FBXW7 vector compared with that produced by cotransfection with the mutant FBXW7 vector (Figures 4C and 4D). Moreover, overexpression of miR-197-3p in TNBC cell lines significantly reduced the expression level of FBXW7 mRNA (Figure 4E). Consistent with these results, the FBXW7 expression level was 7-fold higher in the miR-197-3p inhibition group than in the control group (Figure 4F).
Then, we analyzed the clinical significance of FBXW7 in TNBC patients, and we found that low expression of FBXW7 was associated with poorer OS and DFS in breast cancer patients (Figures 4G and 4H). The expression level of FBXW7 was positively correlated with that of circFBXW7, as validated in 473 breast cancer patients (Table 1).
circFBXW7 and FBXW7 Act as ceRNAs in TNBC through the Regulation of miR-197-3p
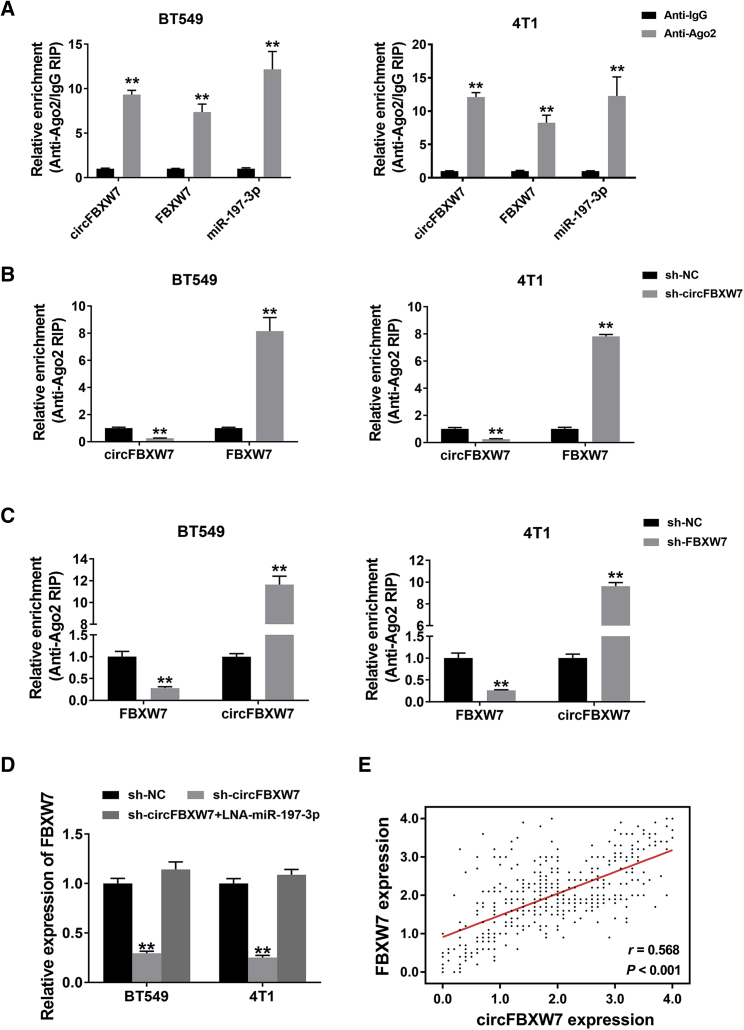
To validate whether circFBXW7 acts as a ceRNA to sponge miR-197-3p and rescue the expression of potential downstream targets, RIP assays were performed. circFBXW7, FBXW7, and miR-197-3p were predominantly enriched on Ago2, demonstrating the recruitment of both circFBXW7 and FBXW7 to an Ago2-related RNA-induced silencing complex (RISC), where they bind with miR-197-3p (Figure 5A). Additionally, the relative enrichment of Ago2 on circFBXW7 was decreased, while the expression of FBXW7 was increased after knockdown of circFBXW7 (Figure 5B). Similarly, silencing FBXW7 reduced the relative enrichment of Ago2 on FBXW7 and increased the abundance of circFBXW7 (Figure 5C). Furthermore, knockdown of circFBXW7 resulted in a reduction in FBXW7 expression, while this impact could be reversed by transfection with miR-197-3p inhibitors (Figure 5D). Spearman correlation analysis showed that circFBXW7 expression was positively correlated with FBXW7 expression in the 473 TNBC samples (r = 0.568, p < 0.001) (Figure 5E). These results illustrate that circFBXW7 and FBXW7 act as ceRNAs in TNBC through the regulation of miR-197-3p.
Figure 5.
circFBXW7 and FBXW7 Act as ceRNAs in TNBC through the Regulation of miR-197-3p
(A) Enrichment of circFBXW7, FBXW7, and miR-197-3p on Ago2, as assessed by a RIP assay. (B) The enrichment of Ago2 on circFBXW7 was decreased, while FBXW7 expression was increased after knockdown of circFBXW7. (C) Silencing FBXW7 reduced the relative enrichment of Ago2 on FBXW7 and increased the abundance of circFBXW7. (D) Knockdown of circFBXW7 resulted in a reduction in FBXW7 expression, which was reversed by miR-197-3p inhibitors. (E) Spearman correlation analysis showed that circFBXW7 expression was positively correlated with FBXW7 expression in 473 TNBC samples (r = 0.568, p < 0.001). *p < 0.05, **p < 0.01. SD is shown as error bars.
FBXW7-185aa, Encoded by circFBXW7, Inhibits the Proliferation and Metastasis of TNBC Cells by Downregulating c-Myc Expression
In a previous study, circFBXW7 was found to be downregulated in glioma, where it is translated into a 185-amino acid (aa) protein (FBXW7-185aa) that competitively interacts with the deubiquitinating enzyme USP28, preventing USP28 from binding to FBXW7 and antagonizing USP28-induced c-Myc stabilization.26 Thus, we next investigated the potential function of the FBXW7-185aa protein encoded by circFBXW7 in TNBC.
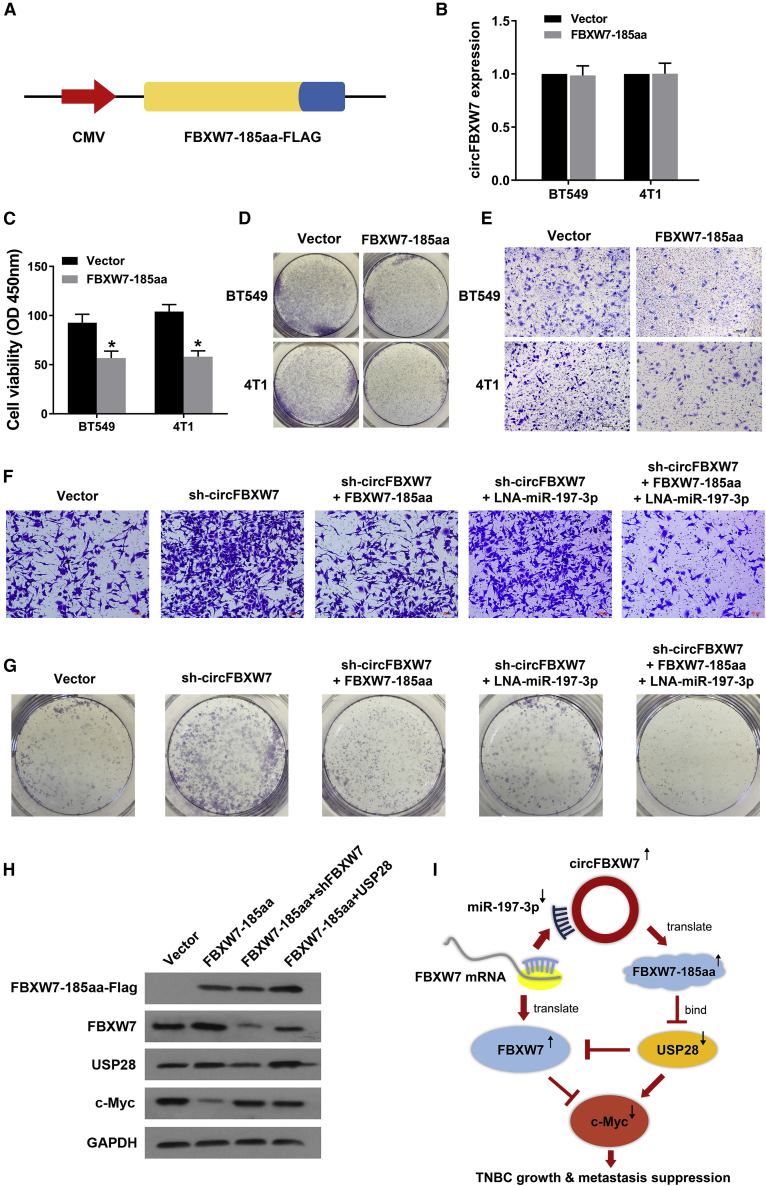
As shown in Figure 6A, the linearized FBXW7-185aa open reading frame (ORF) and a FLAG tag were cloned into a plasmid (FBXW7-185aa-FLAG). Overexpression of FBXW7-185aa did not affect the expression level of the circFBXW7 transcript, as measured by qRT-PCR (Figure 6B). Subsequently, we conducted CCK-8, colony formation, and transwell assays to assess the influence of FBXW7-185aa on TNBC cell growth and proliferation. FBXW7-185aa significantly inhibited the growth, colony-forming, and migration abilities of BT549 cells (Figures 6C–6E). Additionally, the prooncogenic effect enhanced by sh-circFBXW7 was reversed after cotransfection with miR-197-3p inhibitors and the FBXW7-185aa protein expression plasmid (Figures 6F and 6G). Western blot analysis revealed that overexpression of FBXW7-185aa increased the abundance of FBXW7 and induced c-Myc degradation. Overexpressing USP28 reduced the expression of FBXW7 and suppressed FBXW7-185aa-induced c-Myc destabilization (Figure 6H). In summary, circFBXW7 sponges miR-197-3p and encodes the FBXW7-185aa protein to suppress TNBC progression though upregulating FBXW7 expression (Figure 6I).
Figure 6.
FBXW7-185aa, Encoded by circFBXW7, Inhibited the Proliferation and Metastasis of TNBC Cells through Downregulating c-Myc Expression
(A) Schematic diagram of plasmid construction. The linearized FBXW7-185aa ORF and a FLAG tag were cloned downstream of a cytomegalovirus (CMV) promoter (FBXW7-185aa-FLAG). (B) The expression level of circFBXW7 was measured in BT549 cells after transfection with the FBXW7-185aa-FLAG vector. (C–E) FBXW7-185aa inhibits (C) cell proliferation, as assessed by CCK-8 assays; (D) the colony-forming ability and (E) migration ability were assessed via transwell assays. (F and G) The prooncogenic effect enhanced by sh-circFBXW7 was reversed after cotransfection with miR-197-3p inhibitors and the FBXW7-185aa protein expression plasmid, as evaluated by (F) colony formation assays and (G) transwell assays. (H) FBXW7-185aa-FLAG-overexpressing BT549 cells were cotransfected with sh-FBXW7 or USP28 vectors. The FBXW7-185aa-FLAG, FBXW7, USP28, and c-Myc expression levels were quantified by western blot analysis. (I) The schematic illustrates the biological mechanism of circFBXW7, which sponges miR-197-3p and encodes FBXW7-185aa to suppress TNBC progression through upregulating FBXW7 expression. SD is shown as error bars.
Discussion
Due to the rapid development of high-throughput sequencing technology and bioinformatics, numerous circRNAs have been discovered as critical modulators of various biological processes in recent years. circRNAs are a type of noncoding RNA that is well conserved and mainly localized in the cytoplasm.9 Compared to linear RNAs, circRNAs are abundantly and stably expressed, with covalently linked ends.37 The presence of circRNAs in exosomes, which can be secreted into the blood and saliva, has been evidenced.38 These characteristics will make circRNAs potential diagnostic and prognostic biomarkers for many diseases, including cancers.
Emerging evidence shows that circRNAs act as oncogenic stimuli or tumor suppressors by regulating protein expression in multiple cancers.39 For example, the circRNA ciRS-7 acts as a negative regulator of miR-7 and affects the carcinogenesis and progression of many tumors.15, 16, 17 Additionally, circHIPK3 and circMTO1 inhibit cell proliferation and migration by activating different signaling pathways.40, 41 circFBXW7 (hsa_circ_0001451), which can encode the novel 21-kDa protein FBXW7-185aa, has been reported as one of the tumor suppressor circRNAs in glioma.26 In renal cell carcinoma, the silencing of circFBXW7 promoted cell growth and migration.42 However, the role of circFBXW7 and its underlying mechanisms remain unknown in breast cancer. In the current study, we validated that circFBXW7 was significantly downregulated in TNBC cell lines and that low expression of circFBXW7 was associated with worse clinical outcome in breast cancer patients. Subsequently, functional experiments revealed that overexpression of circFBXW7 inhibited TNBC cell proliferation and migration. These results indicated the biological significance of circFBXW7 and its prognostic value for TNBC patients.
As one type of noncoding RNA, circRNAs are crucial posttranscriptional regulators. The well-known ceRNA hypothesis illustrates the communication and regulation among mRNAs, pseudogenes, and lncRNAs as well as circRNAs by MREs. circRNAs contain multiple miRNA-binding sites and can sponge various miRNAs, as predicted by different algorithms. For instance, ciRS-7 contains multiple miR-7-binding sites and regulates several oncogenes via miR-7 in diverse cancers.16, 17, 18 In TNBC, miR-34a is obviously downregulated and promotes tumor growth by targeting GFRA1.23
To gain a better understanding of the molecular mechanism of circFBXW7, we used MRE analysis to predict the miRNA-binding sites. Among the candidates, miR-197-3p was selected as the downstream target because of solid evidence of its role in tumor promotion.27, 28, 29, 30, 31 In addition, we found that miR-197-3p was overexpressed in breast cancer cell lines and was positively associated with poorer overall survival, according to TCGA database. Further luciferase reporter assays and RIP assays confirmed the direct binding of circFBXW7 and miR-197-3p. Our study reverified the cancer-promoting function of miR-197-3p, indicating its therapeutic value in breast cancer. Additionally, we found that the FBXW7-185aa protein encoded by circFBXW7 suppressed the proliferation and migration abilities of TNBC cells by increasing the abundance of FBXW7 and inducing c-Myc degradation.
FBXW7 was reported as a tumor suppressor in diverse malignancies.33, 34, 35, 36, 43 FBXW7 can directly bind to cyclin E and degrade it via a ubiquitin-mediated process. Low expression of FBXW7 indicates an increased risk for tumor progression and recurrence. FBXW7 is the target of several miRNAs, such as the miR-497∼195 cluster.44 In this study, we showed that FBXW7 was the target of miR-197-3p and confirmed this finding by luciferase reporter assays and RIP assays. Further survival analysis revealed the crucial role of FBXW7 in tumor suppression in breast cancer.
Due to the absence of druggable molecular targets, TNBC treatment is very limited compared with the treatments for luminal or HER2+ subtypes. Patients with TNBC have the worst prognosis among patients with breast cancer because chemotherapy is the only choice. Therefore, it is of great importance to develop an efficient molecular target and novel biomarker for TNBC therapy. In recent years, many novel molecules have been found to have the potential to become therapeutic targets. AKR1B1 and UGT8 promote basal-like breast cancer metastasis by activating the epithelial-mesenchymal transition (EMT) pathway and sulfatide-αVβ5 axis, respectively.45, 46 Tinagl1 inhibits TNBC growth by downregulating focal adhesion kinase (FAK) and epidermal growth factor receptor (EGFR) signaling.47 Targeting KDM4B in combination with PI3K inhibition induces further activation of the unfolded protein response (UPR), leading to robust synergy in apoptosis in TNBC.48 By regulating the tumor microenvironment, stromal cells facilitate breast cancer progression and may have important implications for patient precision therapeutics.49, 50 In the current study, circFBXW7 suppressed TNBC growth and metastasis, which could become a novel therapeutic method in the future.
In summary, we showed the significant regulatory mechanisms of circFBXW7 in TNBC, and we found that circFBXW7 regulated the expression of FBXW7 through blocking miR-197-3p and encoding the FBXW7-185aa protein to carry out its functions in TNBC. Therefore, circFBXW7 might be a novel prognostic biomarker and potential treatment strategy for TNBC.
Materials and Methods
Clinical Data and Patient Samples
Fresh tumor samples were collected from patients with breast cancer at Sun Yat-sen University Cancer Center (SYSUCC, Guangzhou, Guangdong, China). All resected tissues were instantaneously infiltrated in RNAlater (Ambion, TX). All patients were followed up periodically, and the clinical data were recorded. This study was approved by the Ethics Committee of Sun Yat-sen University Cancer Center Health Authority and conducted in accordance with the Declaration of Helsinki. Written informed consent was obtained from all patients before participation in this study.
Cell Culture
All cell lines (MCF-10A, MCF-7, T47D, BT474, SKBR-3, MDA-MB-453, MDA-MB-468, MDA-MB-231, BT549, HCC38, 4T1, and MA-891) used in this study were obtained from the American Type Culture Collection, and they were cultured appropriately and passaged for no more than 6 months. All of the above cell lines were free of mycoplasma infection and verified occasionally by DNA fingerprinting.
Quantitative Real-Time PCR
TRIzol reagent (Invitrogen) was used to extract total RNA. NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific) were utilized to isolate the nuclear and cytoplasmic portions of cellular RNA. qRT-PCR was performed with SYBR Premix Ex Taq (Takara). Primer information is listed in Table S1.
Vector Construction and Transfection
The full-length cDNA of human circFBXW7 was amplified and cloned into the pCDNA3.0 vector to construct the overexpression plasmid, and the efficiency was then evaluated by qRT-PCR. We mutated circFBXW7 and the FBXW7 3′ UTR by changing the conserved binding sites of miR-197-3p using a Gene Mutation Kit (Takara). Transfection was conducted with Lipofectamine 2000 (Invitrogen). The miRNA inhibitors and mimics were synthesized by GeneCopoeia (Rockville).
CCK-8 Assay
Briefly, 1 × 103 cells were plated into each well of a 96-well plate. 10 μL CCK-8 solution (Dojindo, Japan) was added to each well on a certain day. The absorbance at a wavelength of 450 nM was assessed with a microtiter plate reader after incubation for 2 h at 37°C.
Colony Formation Assay
A total of 1 × 103 cells was resuspended and plated into each well of a 6-well plate. After incubation at 37°C for 14 days, cell colonies were fixed with methanol and stained with 0.3% crystal violet for 30 min. Images were acquired soon after staining, and colonies were counted by ImageJ software.
Transwell Assay and Wound-Healing Assay
Generally, transwell assays were conducted using migration chambers (BD Biosciences) to which 2 × 104 cells in suspension were added (serum-free medium). Medium containing 20% fetal bovine serum (FBS) was added to the lower chambers and incubated for 24 h. Subsequently, cells in the upper chambers were removed, and methanol was used to fix the remaining cells. After staining with crystal violet, the migrated cells were imaged and counted. For the wound-healing assay, cells were plated in 6-well plates, and an artificial linear wound was made by scratching with a sterile 200 μL pipette tip. Wounds were imaged with an inverted microscope at 0 and 24 h.
Dual Luciferase Reporter Assay
HEK293 cells at a density of 5 × 103 cells/well were added to 96-well plates. Constructed plasmids and miRNA mimics were cotransfected into BT549 and 4T1 cells for 48 h before luciferase activity was detected by the dual luciferase reporter assay system (Promega), according to its instructions. Renilla luciferase activity was defined as the internal control. We conducted independent experiments in triplicate.
RIP
Cells were cotransfected with MS2bs-circFBXW7, MS2bs-circFBXW7mt, and MS2bs-Rluc. After 48 h, RIP was conducted with a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore). The level of miR-197-3p was quantified after the RNA complexes were purified. The RIP assay for Ago2 was conducted with an anti-Ago2 antibody (Millipore). The abundances of circFBXW7, FBXW7, and miR-197-3p were determined after purification.
Western Blot Analysis
Total protein was extracted, separated by 12% SDS-PAGE, and subsequently transferred to polyvinylidene fluoride (PVDF) membranes (Millipore). Membranes were blocked with 5% skim milk at room temperature for 1 h and subsequently incubated with the primary antibody anti-FBXW7 (1:1,000, Abcam, USA), anti-FLAG antibody (1:1,000, Affinity, USA), anti-USP28 antibody (1:1,000, Affinity, USA), anti-c-Myc antibody (1:1,000, Cell Signaling Technology, USA), or anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibody (1:1,000, Affinity, USA). A secondary antibody (Cell Signaling Technology) was used and detected by chemiluminescence.
Mouse Xenograft Model
All animal procedures and care were performed in accordance with the guidelines of the institutes and the approval of the Institute Research Ethics Committee of SYSUCC. BT549 and 4T1 cells stably overexpressing circFBXW7 and mock vector BT549 and 4T1 cells (1 × 107) were subcutaneously inoculated into the dorsal flanks of female BALB/c nude mice (four in each group). We estimated the volume of tumors every 4 days by the following formula: 0.5 × length × width2. After 4 weeks, mice were euthanized and tumors were weighed.
For the lung metastasis experiments, 1 × 105 cells were intravenously injected into the tail vein of mice (four mice per group). After 8 weeks, the lungs were excised after the mice were euthanized, and the numbers of lung metastases were counted visually and subsequently confirmed via microscopy of H&E-stained sections.
Statistical Analysis
All statistical analyses were conducted with SPSS 25.0 software (SPSS, Chicago, IL, USA). Quantitative data are presented as the mean ± SD. Groups were compared using a two-tailed Student’s t test. Kaplan-Meier analysis and the log rank test were implemented to generate the overall survival curves and compare differences between the two cohorts respectively. p < 0.05 was considered statistically significant.
Author Contributions
H.T. and X.X. designed the experiments. F.Y., G.G., and Y.Z. performed the experiments. S.Z., L.Z., and X.O. analyzed and interpreted the data. F.Y. and Y.Z. were the major contributors to writing the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by funds from the National Natural Science Foundation of China (81772961 to H.T. and 81872152 to X.X.), the National Natural Science Foundation of Guangdong (2018A0303130285 to F.Y.), Science and Technology Planning Projects of Guangdong (2017A020215197 to F.Y.), and Science and Technology Planning Projects of Guangzhou (201704020188 to X.X.).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.07.023.
Contributor Information
Xiaoming Xie, Email: xiexm@sysucc.org.cn.
Hailin Tang, Email: tanghl@sysucc.org.cn.
Supplemental Information
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Harbeck N., Gnant M. Breast cancer. Lancet. 2017;389:1134–1150. doi: 10.1016/S0140-6736(16)31891-8. [DOI] [PubMed] [Google Scholar]
- 3.Carey L., Winer E., Viale G., Cameron D., Gianni L. Triple-negative breast cancer: disease entity or title of convenience? Nat. Rev. Clin. Oncol. 2010;7:683–692. doi: 10.1038/nrclinonc.2010.154. [DOI] [PubMed] [Google Scholar]
- 4.Xiao W., Zheng S., Yang A., Zhang X., Zou Y., Tang H., Xie X. Breast cancer subtypes and the risk of distant metastasis at initial diagnosis: a population-based study. Cancer Manag. Res. 2018;10:5329–5338. doi: 10.2147/CMAR.S176763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kristensen L.S., Hansen T.B., Venø M.T., Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arnaiz E., Sole C., Manterola L., Iparraguirre L., Otaegui D., Lawrie C.H. CircRNAs and cancer: Biomarkers and master regulators. Semin. Cancer Biol. 2018 doi: 10.1016/j.semcancer.2018.12.002. S1044-579X(18)30099-3. [DOI] [PubMed] [Google Scholar]
- 7.Bach D.H., Lee S.K., Sood A.K. Circular RNAs in Cancer. Mol. Ther. Nucleic Acids. 2019;16:118–129. doi: 10.1016/j.omtn.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X., Yang L., Chen L.L. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol. Cell. 2018;71:428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 11.Shang Q., Yang Z., Jia R., Ge S. The novel roles of circRNAs in human cancer. Mol. Cancer. 2019;18:6. doi: 10.1186/s12943-018-0934-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen H., Xu Z., Liu D. Small non-coding RNA and colorectal cancer. J. Cell. Mol. Med. 2019;23:3050–3057. doi: 10.1111/jcmm.14209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 16.Su C., Han Y., Zhang H., Li Y., Yi L., Wang X., Zhou S., Yu D., Song X., Xiao N. CiRS-7 targeting miR-7 modulates the progression of non-small cell lung cancer in a manner dependent on NF-κB signalling. J. Cell. Mol. Med. 2018;22:3097–3107. doi: 10.1111/jcmm.13587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li R.C., Ke S., Meng F.K., Lu J., Zou X.J., He Z.G., Wang W.F., Fang M.H. CiRS-7 promotes growth and metastasis of esophageal squamous cell carcinoma via regulation of miR-7/HOXB13. Cell Death Dis. 2018;9:838. doi: 10.1038/s41419-018-0852-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weng W., Wei Q., Toden S., Yoshida K., Nagasaka T., Fujiwara T., Cai S., Qin H., Ma Y., Goel A. Circular RNA ciRS-7-A Promising Prognostic Biomarker and a Potential Therapeutic Target in Colorectal Cancer. Clin. Cancer Res. 2017;23:3918–3928. doi: 10.1158/1078-0432.CCR-16-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu W., Zhao J., Jin M., Zhou M. circRAPGEF5 Contributes to Papillary Thyroid Proliferation and Metastatis by Regulation miR-198/FGFR1. Mol. Ther. Nucleic Acids. 2019;14:609–616. doi: 10.1016/j.omtn.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Chen Q., Zhang J., He Y., Wang Y. hsa_circ_0061140 Knockdown Reverses FOXM1-Mediated Cell Growth and Metastasis in Ovarian Cancer through miR-370 Sponge Activity. Mol. Ther. Nucleic Acids. 2018;13:55–63. doi: 10.1016/j.omtn.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Chen B., Wei W., Huang X., Xie X., Kong Y., Dai D., Yang L., Wang J., Tang H., Xie X. circEPSTI1 as a Prognostic Marker and Mediator of Triple-Negative Breast Cancer Progression. Theranostics. 2018;8:4003–4015. doi: 10.7150/thno.24106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang H., Huang X., Wang J., Yang L., Kong Y., Gao G., Zhang L., Chen Z.S., Xie X. circKIF4A acts as a prognostic factor and mediator to regulate the progression of triple-negative breast cancer. Mol. Cancer. 2019;18:23. doi: 10.1186/s12943-019-0946-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He R., Liu P., Xie X., Zhou Y., Liao Q., Xiong W., Li X., Li G., Zeng Z., Tang H. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J. Exp. Clin. Cancer Res. 2017;36:145. doi: 10.1186/s13046-017-0614-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zou Y., Zheng S., Xiao W., Xie X., Yang A., Gao G., Xiong Z., Xue Z., Tang H., Xie X. circRAD18 sponges miR-208a/3164 to promote triple-negative breast cancer progression through regulating IGF1 and FGF2 expression. Carcinogenesis. 2019:bgz071. doi: 10.1093/carcin/bgz071. [DOI] [PubMed] [Google Scholar]
- 25.Yao Z., Luo J., Hu K., Lin J., Huang H., Wang Q., Zhang P., Xiong Z., He C., Huang Z. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol. Oncol. 2017;11:422–437. doi: 10.1002/1878-0261.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Y., Gao X., Zhang M., Yan S., Sun C., Xiao F., Huang N., Yang X., Zhao K., Zhou H. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J. Natl. Cancer Inst. 2018;110:304–315. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang T., Li H., Chen T., Ren H., Shi P., Chen M. LncRNA MALAT1 Depressed Chemo-Sensitivity of NSCLC Cells through Directly Functioning on miR-197-3p/p120 Catenin Axis. Mol. Cells. 2019;42:270–283. doi: 10.14348/molcells.2019.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaker O., Maher M., Nassar Y., Morcos G., Gad Z. Role of microRNAs -29b-2, -155, -197 and -205 as diagnostic biomarkers in serum of breast cancer females. Gene. 2015;560:77–82. doi: 10.1016/j.gene.2015.01.062. [DOI] [PubMed] [Google Scholar]
- 29.Hu Z., Wang P., Lin J., Zheng X., Yang F., Zhang G., Chen D., Xie J., Gao Z., Peng L., Xie C. MicroRNA-197 Promotes Metastasis of Hepatocellular Carcinoma by Activating Wnt/β-Catenin Signaling. Cell. Physiol. Biochem. 2018;51:470–486. doi: 10.1159/000495242. [DOI] [PubMed] [Google Scholar]
- 30.Lehmann U., Streichert T., Otto B., Albat C., Hasemeier B., Christgen H., Schipper E., Hille U., Kreipe H.H., Länger F. Identification of differentially expressed microRNAs in human male breast cancer. BMC Cancer. 2010;10:109. doi: 10.1186/1471-2407-10-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang T., Cheng Y., She Q., Jiang Y., Chen Y., Yang W., Li Y. Long non-coding RNA TUG1 sponges miR-197 to enhance cisplatin sensitivity in triple negative breast cancer. Biomed. Pharmacother. 2018;107:338–346. doi: 10.1016/j.biopha.2018.07.076. [DOI] [PubMed] [Google Scholar]
- 32.Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwatsuki M., Mimori K., Ishii H., Yokobori T., Takatsuno Y., Sato T., Toh H., Onoyama I., Nakayama K.I., Baba H., Mori M. Loss of FBXW7, a cell cycle regulating gene, in colorectal cancer: clinical significance. Int. J. Cancer. 2010;126:1828–1837. doi: 10.1002/ijc.24879. [DOI] [PubMed] [Google Scholar]
- 34.Mao J.H., Kim I.J., Wu D., Climent J., Kang H.C., DelRosario R., Balmain A. FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science. 2008;321:1499–1502. doi: 10.1126/science.1162981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao J., Wang Y., Mu C., Xu Y., Sang J. MAGEA1 interacts with FBXW7 and regulates ubiquitin ligase-mediated turnover of NICD1 in breast and ovarian cancer cells. Oncogene. 2017;36:5023–5034. doi: 10.1038/onc.2017.131. [DOI] [PubMed] [Google Scholar]
- 36.Xia W., Zhou J., Luo H., Liu Y., Peng C., Zheng W., Ma W. MicroRNA-32 promotes cell proliferation, migration and suppresses apoptosis in breast cancer cells by targeting FBXW7. Cancer Cell Int. 2017;17:14. doi: 10.1186/s12935-017-0383-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang F., Nazarali A.J., Ji S. Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am. J. Cancer Res. 2016;6:1167–1176. [PMC free article] [PubMed] [Google Scholar]
- 38.Li Z., Yanfang W., Li J., Jiang P., Peng T., Chen K., Zhao X., Zhang Y., Zhen P., Zhu J., Li X. Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett. 2018;432:237–250. doi: 10.1016/j.canlet.2018.04.035. [DOI] [PubMed] [Google Scholar]
- 39.Verduci L., Strano S., Yarden Y., Blandino G. The circRNA-microRNA code: emerging implications for cancer diagnosis and treatment. Mol. Oncol. 2019;13:669–680. doi: 10.1002/1878-0261.12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han D., Li J., Wang H., Su X., Hou J., Gu Y., Qian C., Lin Y., Liu X., Huang M. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–1164. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 41.Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B., Luo Y., Lyu D., Li Y., Shi G. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang G., Xue W., Jian W., Liu P., Wang Z., Wang C., Li H., Yu Y., Zhang D., Zhang C. The effect of Hsa_circ_0001451 in clear cell renal cell carcinoma cells and its relationship with clinicopathological features. J. Cancer. 2018;9:3269–3277. doi: 10.7150/jca.25902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao Y., Yin C., Wang Y., Lv H., Wang W., Huang Y., Perez-Losada J., Snijders A.M., Mao J.H., Zhang P. FBXW7 deletion contributes to lung tumor development and confers resistance to gefitinib therapy. Mol. Oncol. 2018;12:883–895. doi: 10.1002/1878-0261.12200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang M., Li C.J., Sun X., Guo Q., Xiao Y., Su T., Tu M.L., Peng H., Lu Q., Liu Q. MiR-497∼195 cluster regulates angiogenesis during coupling with osteogenesis by maintaining endothelial Notch and HIF-1α activity. Nat. Commun. 2017;8:16003. doi: 10.1038/ncomms16003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu X., Li X., Fu Q., Cao Q., Chen X., Wang M., Yu J., Long J., Yao J., Liu H. AKR1B1 promotes basal-like breast cancer progression by a positive feedback loop that activates the EMT program. J. Exp. Med. 2017;214:1065–1079. doi: 10.1084/jem.20160903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cao Q., Chen X., Wu X., Liao R., Huang P., Tan Y., Wang L., Ren G., Huang J., Dong C. Inhibition of UGT8 suppresses basal-like breast cancer progression by attenuating sulfatide-αVβ5 axis. J. Exp. Med. 2018;215:1679–1692. doi: 10.1084/jem.20172048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen M., Jiang Y.Z., Wei Y., Ell B., Sheng X., Esposito M., Kang J., Hang X., Zheng H., Rowicki M. Tinagl1 Suppresses Triple-Negative Breast Cancer Progression and Metastasis by Simultaneously Inhibiting Integrin/FAK and EGFR Signaling. Cancer Cell. 2019;35:64–80.e7. doi: 10.1016/j.ccell.2018.11.016. [DOI] [PubMed] [Google Scholar]
- 48.Wang W., Oguz G., Lee P.L., Bao Y., Wang P., Terp M.G., Ditzel H.J., Yu Q. KDM4B-regulated unfolded protein response as a therapeutic vulnerability in PTEN-deficient breast cancer. J. Exp. Med. 2018;215:2833–2849. doi: 10.1084/jem.20180439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raz Y., Cohen N., Shani O., Bell R.E., Novitskiy S.V., Abramovitz L., Levy C., Milyavsky M., Leider-Trejo L., Moses H.L. Bone marrow-derived fibroblasts are a functionally distinct stromal cell population in breast cancer. J. Exp. Med. 2018;215:3075–3093. doi: 10.1084/jem.20180818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shee K., Yang W., Hinds J.W., Hampsch R.A., Varn F.S., Traphagen N.A., Patel K., Cheng C., Jenkins N.P., Kettenbach A.N. Therapeutically targeting tumor microenvironment-mediated drug resistance in estrogen receptor-positive breast cancer. J. Exp. Med. 2018;215:895–910. doi: 10.1084/jem.20171818. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.