Abstract
Pancreatic ductal adenocarcinoma (PDAC) is one of the most aggressive malignancies; it preferentially metastasizes to the liver and is the main cause of death from this disease. In previous studies, small activating RNA against CCAAT/enhancer-binding protein-α (C/EBPα-saRNA) demonstrated efficacy of PDAC in a local subcutaneous tumor model. In this study, we focused on the efficacy of C/EBPα-saRNA in advanced stage PDAC. For targeted delivery, we selected a new anti-transferrin receptor aptamer (TR14), which demonstrated a high binding affinity to target proteins. The TR14 aptamer was internalized with clathrin-mediated endocytosis, distributed in early endosome, late endosome, and lysosome subcellularly. To investigate its anti-tumor effects to advanced PDAC, we conjugated C/EBPα-saRNA to TR14. Treatment of pancreatic cancer cells with the conjugates upregulated expression of C/EBPα and its downstream target p21, and inhibited cell proliferation. For in vivo assays, we established an advanced PDAC mouse model by engrafting luciferase reporter-PANC-1 cells directly into the livers of non-obese diabetic/severe combined immunodeficiency (NOD/SCID) mice. After treatment of aptamer-C/EBPα conjugates, we observed significant reduction of tumor growth in this advanced PDAC mouse model. Combinational treatment of the conjugates with gemcitabine also demonstrated enhanced anti-tumor effects in advanced PDAC. This suggests that aptamer-C/EBPα conjugates could be used as an adjuvant, along with other conventional anti-cancer drugs in advanced PDAC. In conclusion, targeted delivery of C/EBPα-saRNAs by aptamers might have potential therapeutic effects in advanced PDAC.
Keywords: C/EBPα-saRNA, targeted delivery, transferrin receptor aptamer, clathrin-mediated endocytosis, advanced PDAC, anti-tumor effects, adjuvant, truncation, albumin affinity tag
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is one of the most aggressive malignant tumors, with few effective therapeutic options and high mortality rates.1, 2 Although a cure can be achieved through surgical resection, the majority of PDAC patients are diagnosed at metastatic stages in which the tumor is surgically unresectable. This type of advanced disease is the main cause of death for patients with PDAC. PDAC preferentially metastasizes to the liver, which is explained by the theory of the hepatic pre-metastatic niche.3 The current standard of care for advanced PDAC is limited to mono- (gemcitabine) or combinational chemotherapy (gemcitabine combined with other chemotherapeutic agents such as fluorouracil [5-FU], erlotinib, cisplatin, capecitabine, docetaxel, and oxaliplatin).4 Induction chemotherapy prior to treatment with radiation and dose escalation shows slightly improved survival rates in locally advanced pancreatic cancer patients.5 However, overall, these combination chemotherapies do not show statistically significant survival benefits for PDAC.6
The development of cancer therapeutics has focused on targeted delivery in the past few decades to increase therapeutic efficacy. Therapeutics are packaged in carriers to achieve targeted delivery of cancer therapeutics with systemic administration. This is achieved via two main approaches: passive targeting or active targeting. Passive targeting depends solely on enhanced permeability and retention effects; for this reason, less than 1% accumulates in xenografted tumors.7 In contrast, active targeting uses affinity ligands such as antibodies or aptamers for target-specific homing, resulting in increased target efficiency with improved tumor localization and retention. Aptamers are structured nucleic acid ligands that hold unique three-dimensional structures based on defined nucleic acid sequences that are selected using the systematic evolution of ligands by exponential enrichment (SELEX) strategy in vitro.8, 9 Because aptamers can be selected against numerous varieties of targets, including small molecules, proteins, cultured cells, and even ex vivo organ cultures,10, 11, 12, 13, 14, 15 they have been popularized as ligands for active targeting. Compared with antibodies, aptamers hold significant advantages as delivery vehicles, including structural stability and flexibility, ease of synthesis, as well as very limited toxicity and immunogenicity.16 For these reasons, multiple groups have isolated aptamers recognizing specific epitopes of plasma membrane receptors on cancers for internalization into target cells. These cancer-specific aptamers have been successfully utilized to deliver various therapeutic payloads such as antibodies, peptides, small inhibiting RNAs (siRNAs), small activating RNAs (saRNAs), and toxins.17 For targeted delivery of therapeutic payload, we developed aptamer-drug conjugates (ApDCs) that intrinsically incorporated active metabolites of the nucleoside analogs gemcitabine or 5-FU.18 In that study, we showed that gemcitabine was more potent for induction of DNA damage than 5-FU in PDAC as an anti-cancer drug.18
To develop an active targeting ligand, most of the strategies adopted to date typically target receptors that are selectively overexpressed on diseased tissues or cells. This approach dramatically increases the therapeutic index and reduces unwanted effects on non-targeted cells.18, 19, 20 For example, human transferrin receptor 1 (hTfR1), which is involved in cellular iron uptake to maintain intracellular homeostasis, is overexpressed on and internalized into multiple cancer cell types through the clathrin-mediated endocytosis pathway.21 Thus, hTfR1 is considered an attractive target for the targeted delivery of therapeutic agents against various cancers.22 Recently, hTfR2, another receptor for transferrin, was cloned.23 The main difference between hTfR1 and hTfR2 is in their expression patterns: hTfR1 is expressed on most cell types, except mature erythrocytes and terminally differentiated cells, whereas hTfR2 is highly expressed in the liver, erythroid cells, and peripheral mononuclear cells.24 More recently, transferrin was shown to pass through blood-brain barrier endothelial cells into the brain via receptor-mediated transcytosis.25 In turn, hTfRs have attracted attention as candidates for targeted drug delivery to multiple cancers and the CNS. In pancreatic cancer, overexpressed hTfR is a specific malignant marker: 82% positive in primary tumor and 75% in metastatic tumors.26 Therefore, hTfR is a good cell surface target for targeted delivery in pancreatic cancer.
saRNAs offer an emerging therapeutic strategy for transcriptional gene activation in mammalian cells, in the form of short 21-mer nucleotide duplexes that target the promoter regions of genes.27, 28 The therapeutic potential of saRNAs has been explored in multiple cancers.29 The most successful therapeutic saRNA is CCAAT/enhancer-binding protein-α (C/EBPα)-saRNA that shows potent anti-tumor effects through the inhibition of cell proliferation in hepatocellular carcinoma by upregulation of C/EBPα and its downstream targets, cyclin-dependent kinase inhibitor 1 (p21) in vivo.30 C/EBPα-saRNA encapsulated by liposomal carrier molecules is currently under a phase I clinical trial in patients with advanced liver cancer. Given the success of C/EBPα-saRNA in hepatocellular carcinoma, it indicates that C/EBPα-saRNA might have anti-tumor effects in other type of cancers through inhibition of cancer cell proliferation. Consistent with this, in a previous study, we showed anti-tumor effects following targeted delivery of C/EBPα-saRNA in a local subcutaneous xenografted PDAC mouse model.31 Based on our previous results, it suggests that C/EBPα might be a novel druggable target in patients with advanced PDAC.
We herein determine the anti-tumor effects of C/EBPα-saRNA by using TfR aptamers for targeted delivery in an advanced PDAC mouse model. Also, the feasibility of aptamer-C/EBPα-saRNA for adjuvant in combination with gemcitabine is determined in an advanced PDAC mouse model. We observed the significant reduction of PDAC tumor burden in the treatment of TfR aptamer-C/EBPα-saRNA conjugates in an advanced PDAC mouse model. Our results suggest that our approach could be translated into the clinic to cure advanced PDAC cancer patients.
Results
Anti-hTfR Aptamer Is Selected through Protein SELEX
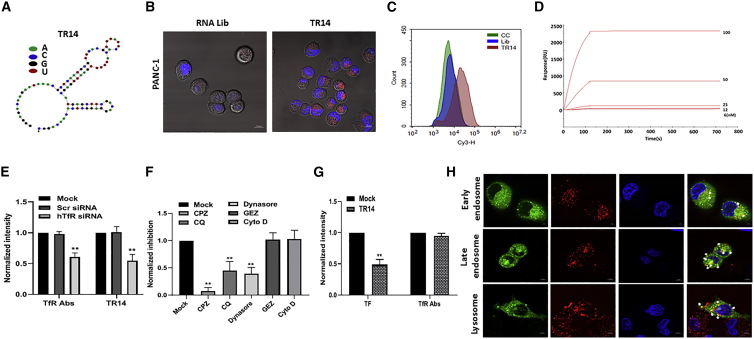
We used protein SELEX to select RNA aptamers against hTfR. As a target for SELEX, the extracellular domain of hTfR with a six-histidine (His6) tag to immobilize to beads was expressed in HEK293 cells (Figure S1A). We first incubated an RNA aptamer library pool with agarose beads to remove non-specific binders. Subsequently, we incubated the supernatant with the His6-hTfR target protein for positive selection, then amplified the aptamers bound to hTfR using PCR and in vitro transcription, as depicted in Figure S1B. After nine rounds of SELEX, we identified the 87-nt anti-hTfR aptamer TR14 (Table 1). The frequency of TR14 was depicted in Table S1. We predicted the structure of TR14 using Mfold, which showed multiple stem-loop structures (Figure 1A).
Table 1.
Sequences of Parent and Truncated TR14 Transferrin Receptor Aptamers
| Name | Sequence |
|---|---|
| TR14 (parent, 87 nt) | 5′-GGGAGACAAGAAUAAACGCUCAAUGCGUUCACGUUUAUUCACAUUUUUGAAUUGAGCAUGAGCUUCGACAGGAGGCUCACAACAGGC-3′ |
| TR14 S1 (46 nt) | 5′-GGGGCUCAAUGCGUUCACGUUUAUUCACAUUUUGAAUUGAGCAUG-3′ |
| TR14 S2 (43 nt) | 5′-GGGGCUCAAUGCGUUCACGUUUAUUCACAUUUUUGAAUUGAGC-3′ |
| TR14 ST1-1 (40 nt) | 5′-GCUCAAUGCGUUCACGUUUAUUCACAUUUUUGAAUUGAGC-3′ |
| TR14 ST1-2 (32 nt) | 5′-AAUGCGUUCACGUUUAUUCACAUUUUUGAAUU-3′ |
| TR14 ST1-3 (22 nt) | 5′-UUUAUUCACAUUUUUGAAUUGA-3′ |
Figure 1.
Identification of an RNA Aptamer against the Human Transferrin Receptor (hTfR)
(A) The expected secondary structure of anti-hTfR aptamer, TR14, was predicted using Mfold. (B) To confirm the ability of the anti-hTfR TR14 aptamer to enter target cells, we performed cell internalization assays in PANC-1. 200 nM Cy3-labeled RNA aptamer library or Cy3-labeled TR14 was incubated on live cells and visualized using confocal microscopy. Red: Cy3-labeled RNAs; blue: Hoechst 33342 for nuclei. Scale bar: 10 μm. (C) The binding of TR14 to target cells, PANC-1, was assessed by flow cytometry. (D) TR14-hTfR binding affinity and kinetics were determined using label-free biosensor assays and a Biacore T100 instrument. Positive response units (RUs) were observed following injection of hTfR proteins. (E) PANC-1 cells transfected with anti-hTfR siRNA or scramble siRNA were incubated with anti-hTfR Abs or TR14 labeled with Cy3. The intensity of fluorescence was normalized with mock. Student’s t test was used to determine statistical significance: *p ≤ 0.05, **p ≤ 0.01. (F) The uptake mechanism of TR14 was determined by small-molecule inhibitors (clathrin-mediated endocytosis [CME]: chlorpromazine [CPZ], chloroquine [CQ], and dynasore; clathrin-independent endocytosis [CIE]: genistein [GEZ; caveolae- and lipid-mediated endocytosis inhibitor], and cytochalasin D [Cyto D]). (G) The competition assay of TR14 was assessed with transferrin and anti-TfR antibodies. (H) Subcellular co-localization of TR14 with early endosome, late endosome, and lysosomes was determined on live cells by confocal microscopy with Airyscan. The co-localized areas of TR14 with subcellular organs are presented in yellow where indicated by white arrows. Red: Cy3-labeled RNAs; green: GFP fused to Rab5a (early endosome marker), Rab7a (late endosome marker), or Lamp1 (lysosomal marker); blue: Hoechst 33342 for nuclei. Scale bar: 5 μm.
The Anti-hTfR Aptamer (TR14) Is Efficiently Internalized into Cancer Cells and Shows High Binding Affinity
Because we isolated the anti-hTfR aptamer for use in therapeutic delivery, we performed cell internalization assays using confocal microscopy of live cells for targeted delivery. To determine intracellular uptake, we incubated Cy3-labeled TR14 aptamers at 200 nM on PANC-1 cells. For the control, the Cy3-labeled initial RNA library was treated on both cell lines as for flow cytometry. After a 2-h incubation, we observed a typical punctate fluorescence pattern in the cytoplasm of PANC-1 cells, suggesting that TR14 was successfully internalized into cancer cells (Figure 1B). To confirm the binding of TR14 to cancer cells, we incubated the Cy3-labeled initial non-selected aptamer library as negative control or TR14 aptamers on PANC-1 cells. We observed enriched cell surface binding to the cells, compared with the initial RNA library pool by flow cytometry (Figure 1C).
To characterize TR14-hTfR binding affinity and kinetics, we used a label-free biosensor assay in real time. We measured the dissociation constant (KD) of TR14 for hTfR as 3.17 × 10−11 mol/L, with 6.78 × 103 M−1s−1 and 1.68 × 10−6 s−1 as the corresponding association (kon) and dissociation (koff) rate constants, respectively (Figure 1D; Table 2).
Table 2.
Measured Association Rate (kon), Dissociation Rate (koff), and Dissociation Constant (KD)
| Name | kon (M–1s–1) | koff (s–1) | KD (M) | Remarks |
|---|---|---|---|---|
| Parent TR14 (87 nt) | 3.70E+7 | 1.17E−3 | 3.17E−11 | |
| TR14 S1 (46 nt) | 1.30E+4 | 1.89E−5 | 1.46E−9 | kon and KD: decreased; koff increased |
| TR14 S2 (43 nt) | 8.34E+8 | 5.77E−4 | 6.92E−13 | kon, koff, and KD: increased |
| TR14 ST1-1 (40 nt) | N/A | N/A | N/A | N/A |
| TR14 ST1-2 (32 nt) | N/A | N/A | N/A | N/A |
| TR14 ST1-3 (22 nt) | 6.30E+6 | 1.38E−4 | 2.20E−11 | kon, koff, and KD: stayed in similar range |
The TR14 aptamer Is Internalized into Cells via Clathrin-Mediated Endocytosis and Competes with Transferrin, but Not Anti-TfR Antibodies
To verify the specificity of TR14 for hTfR, we performed siRNA knockdown experiments. After PANC-1 cells were transfected with anti-hTfR siRNA, control siRNA, or no siRNA, the uptake of TR14 was assessed. It showed diminished uptake upto 50% in treatment of anti-hTfR siRNA (Figure 1E).
To determine the mechanism of endocytosis, we utilized small molecules. In the pre-treatment of clathrin-mediated endocytosis (CME) inhibitors such as chlorpromazine (CPZ), chloroquine (CQ), or dynasore, the uptake of TR14 was inhibited significantly (Figure 1F). But in the pre-treatment of clathrin-independent endocytosis (CIE) inhibitors such as genistein (GEZ; caveolae- and lipid-mediated endocytosis inhibitor) or cytochalasin D (Cyto D; phagocytosis or micropinocytosis), the uptake was not interfered with (Figure 1F). With pre-incubation of TR14 on PANC-1 cells, internalized transferrin was significantly inhibited (Figure 1G, left). But anti-TfR aptamer did not compete with TfR antibodies, suggesting that the binding site of TR14 and TfR antibodies might be different (Figure 1G, right).
The TR14 aptamer Is Distributed into Endosome and Lysosome Subcellularly
To confirm subcellular co-localization of TR14, we incubated chemically synthesized Cy3-labeled TR14 on live cells, where it expressed GFP fused to Rab5a (early endosome marker), Rab7a (late endosome marker), or Lamp1 (lysosomal marker). The co-localization was assessed on live cells with confocal microscopy with Airyscan. After 2-h incubation of TR14, we observed the multiple co-localizations of TR14 with early endosome, late endosome, and lysosome (Figure 1H).
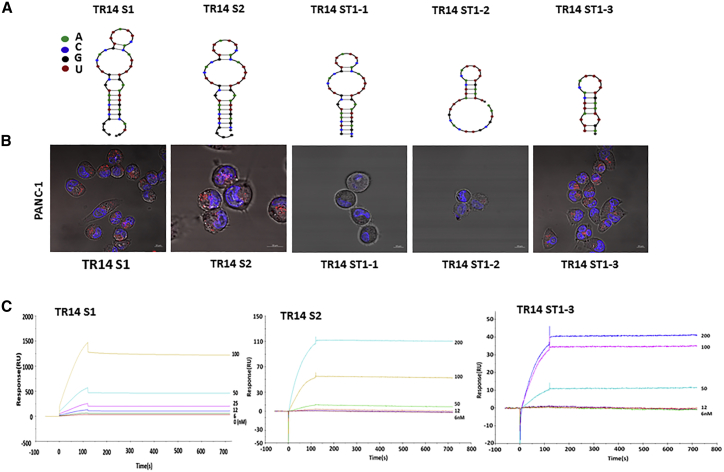
Truncated TR14 Aptamers Show Efficient Cell Internalization in Cancer Cells
Based on structural analysis using computational prediction, we truncated the TR14 aptamer into the smallest functional unit that was expected to maintain binding to hTfR. We generated five truncated TR14 aptamers: S1 (46 nt), S2 (43 nt), ST1-1 (40 nt), ST1-2 (32 nt), and ST1-3 (22 nt) (Table 1). The series of truncated TR14 aptamers was chemically synthesized, and the expected structures of the truncated aptamers were depicted using NUPACK (Figure 2A). To confirm the efficacy of the truncated aptamers for targeted delivery, we performed cell internalization assays using confocal microscopy on live cells with chemically synthesized Cy3-labeled truncates. The two truncated aptamers, TR14 S1 and TR14 S2, were internalized into PANC-1 cells (Figure 2B). In contrast, two truncated aptamers, TR14 ST1-1 and TR14 ST1-2, were not internalized into the cells. Interestingly, another truncated aptamer, TR14 ST1-3, was internalized into PANC-1 cells (Figure 2B).
Figure 2.
Truncation of TR14 Anti-hTfR RNA Aptamers
(A) The expected secondary structures of truncated TR14 aptamers (S1 [46 nt], S2 [43 nt], ST1-1 [40 nt], ST1-2 [32 nt], and ST1-3 [22 nt]) were predicted using NUPACK. The indicated color code represents RNA nucleotides. (B) Internalization of each truncated TR14 aptamer was confirmed using live-cell imaging on PANC-1 (pancreatic cancer cells). 200 nM of each Cy3-labeled truncated aptamer was added to cancer cells and visualized using confocal microscopy. Red: Cy3-labeled RNAs; blue: Hoechst 33342 for nuclei. Scale bars: 10 μm. (C) The binding affinity and kinetics of truncated TR14 aptamers (TR14 S1, TR14 S2, and TR14 ST1-3) against hTfR were determined using label-free biosensor assays and a Biacore T100 instrument.
Truncated TR14 Aptamers Show Improved or Equivalent Binding Affinity Compared with the Parent TR14 Aptamer
To measure binding affinity and kinetics of the truncated TR14 aptamers for hTfR1, we performed a label-free biosensor assay in real time using a Biacore T100 instrument again. The resulting Biacore sensorgrams for TR14 S1, TR14 S2, and TR14 ST1-3 are presented in Figure 2C; neither TR14 ST1-1 nor TR14 ST1-2 was measurable. Based on these results, we calculated KD and kon and koff rate constants. As a result, TR14 S2 showed improved kinetic constants. The TR14 ST1-3 remained equivalent kinetic constants compared with parent ones (Table 2).
The parent TR14 and Truncated TR14 ST1-3 (22 nt) aptamers Show Cross-Reactivity to hTfR2
Reportedly, two human glioblastoma cell lines (U87MG and TB10) express hTfR1 or hTfR2 selectively.32 Therefore, to determine the potential cross-reactivity of the parent TR14 and the truncated TR14 S2 and TR14 ST1-3 aptamers for hTfR1 and hTfR2, we incubated chemically synthesized Cy3-labeled aptamers on two live glioblastoma cells. We showed that the parent TR14 bound to both hTfR1 and hTfR2, TR14 S2 bound selectively to hTfR1, and TR14 ST1-3 bound both hTfR1 and hTfR2 (Figure S1C).
The parent TR14 and Truncated TR14 aptamers without or with Albumin Affinity Tag Conjugated to C/EBPα-saRNA Demonstrate Upregulation of C/EBPα and p21, and Show Anti-proliferative Effects In Vitro
To achieve targeted delivery of C/EBPα-saRNA into pancreatic cancer cells, we constructed multiple conjugates that linked the TR14, truncates of TR14, or albumin affinity tagged truncates of TR14 with C/EBPα-saRNA. To maintain the functional integrity of the molecule, we placed a “sticky” sequence between TR14 and the C/EBPα-saRNA oligonucleotide to prevent structural hindrance (Table 3), as we described for the construction of P19-CEBPA in our previous study.31
Table 3.
Sequences of TR14 ST1-3 with CEBPA Sense and/or Albumin Affinity Tag and CEBPA Anti-sense
| Name | Sequence |
|---|---|
| TR14-STICK | 5′-GGGAGACAAGAAUAAACGCUCAA UGCGUUCACGUUUAUUCACAUUUUUG AAUUGAGCAUGAGC UUCGACAGGAGGCUCACAACAGGCoooooGUACAUUCUAGAUACGC-3′ |
| CEBPA sense-STICK | 5′-GCGGUCAUUGUCACUGGUCUUoooooGCGUAUCUAGAAUGUAC-3′ |
| IRRE-TR14-STICK | 5′-GGGAGACAAGAAUAAACGCUCAA UUAUCACAUGCGUACCUUACCGUUCC CAUUUCUUCCCUGUUUCGACAGGAGGCUCACAACAGGCoooooGUACAUUCUAGAUACGC-3′ |
| IRRE-tTR14 | 5′-GCGGUCAUUGUCACUGGUCUUoooooGCUCAAUGCGUUCACGUUUAUUCA CAUUUUUGAAUUGAGC-3′ |
| TR14 ST1-3 with CEBPA sense (CT) | 5′-UUUAUUCACAUUUUUGAAUUGAoooooGCGGUCAUUGUCACUGGUCUUX-3′ |
| TR14 ST1-3 with CEBPA sense-affinity TAG (TCT) | 5′-UUUAUUCACAUUUUUGAAUUGAoooooGCGGUCAUUGUCACUGGUCUUX-affinity TAG-3′ |
| TR14 ST1-3 with CEBPA sense-spacer-affinity-TAG (TCUT) | 5′-UUUAUUCACAUUUUUGAAUUGAoooooGCGGUCAUUGUCACUGGUCUUUUUUUUUUUUX-affinity-TAG-3′ |
| CEBPA anti-sense | 5′-GACCAGUGACAAUGACCGCUU-3′ |
Underscore indicates sticky sequences. ooooo, five C3 -carbon linkers.
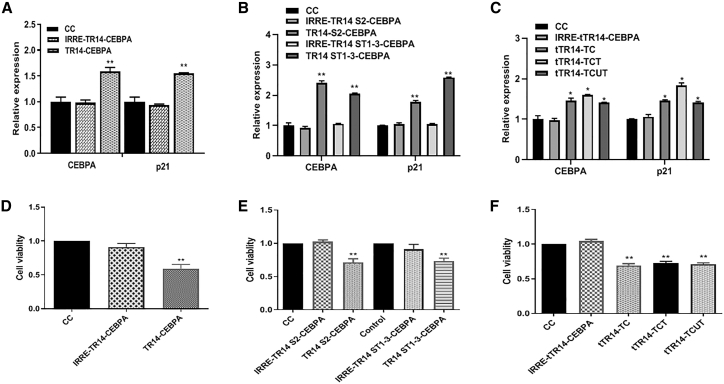
To assess gene activation in vitro, we added TR14-CEBPA or IRRE-TR14-CEBPA (irrelevant or non-targeting aptamer control) to cultured PANC-1 cells, then used qPCR to measure mRNA expression of C/EBPα and its downstream target, p21. Cells treated with TR14-CEBPA showed significantly higher mRNA expression of C/EBPα and p21 (Figure 3A) compared with the IRRE control group. To measure the inhibition of cell proliferation by TR14-CEBPA, we performed 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) cell proliferation assays on PANC-1 cells treated with TR14-CEBPA or IRRE-TR14-CEBPA for 72 h. We also observed a significant reduction in cell proliferation following treatment with TR14-CEBPA compared with the IRRE control group at the time point of 72 h (Figure 3D). In treatment of conjugates of truncated TR14 (TR14-S3 or TR14 ST1-3) with C/EBPα-saRNA, we observed upregulated mRNA expression of C/EBPα and its downstream target, p21, and inhibition of cancer cell proliferation by MTS assay (Figures 3B and 3E).
Figure 3.
Upregulation of CEBPA and p21 In Vitro and Inhibition of Cancer Cell Proliferation
(A) PANC-1 cells were treated with cell control (CC), IRRE-TR14-CEBPA (irrelevant aptamer control), or TR14-CEBPA for 72 h. mRNA expression of C/EBPα and its downstream target p21 were measured using qPCR. (B) PANC-1 cells were treated with cell control (CC), IRRE-TR14 S2-CEBPA (irrelevant aptamer control), IRRE-TR14 ST1-3-CEBPA (irrelevant aptamer control), TR14 S2-CEBPA, or TR14 ST1-3-CEBPA for 72 h. mRNA expression of C/EBPα and its downstream target p21 were measured using qPCR. (C) PANC-1 cells were treated with cell control (CC), IRRE-tTR14-CEBPA (irrelevant aptamer control), tTR14-TC, tTR14-TCT, or tTR14-TCUT for 72 h. mRNA expression of C/EBPα and its downstream target p21 were measured using qPCR. One-way ANOVA test was used to determine statistical significance: *p ≤ 0.05, **p ≤ 0.01. (D) Inhibition of cell proliferation by IRRE-TR14-CEBPA or TR14-CEBPA was determined using MTS assay. (E) Inhibition of cell proliferation by cell control (CC), IRRE-TR14 S2-CEBPA (irrelevant aptamer control), IRRE-TR14 ST1-3-CEBPA (irrelevant aptamer control), TR14 S2-CEBPA, or TR14 ST1-3-CEBPA was determined using MTS assay. (F) Inhibition of cell proliferation by cell control (CC), IRRE-tTR14-CEBPA (irrelevant aptamer control), tTR14-TC, tTR14-TCT, or tTR14-TCUT for 72 h was determined using MTS assay. One-way ANOVA test was used to determine statistical significance: *p ≤ 0.05, **p ≤ 0.01.
To increase pharmacokinetics of the conjugates in vivo, we chemically attached an albumin affinity tag to TR14 ST1-3 (termed tTR14). The tTR14 aptamers without or with an albumin affinity tag were named TC or TCT, respectively. We also made a tTR14 with an affinity tag and a 10-uracil spacer (TCUT). A schematic illustration of these conjugates is depicted in Figure S1D; sequences are shown in Table 3. After treatment of PANC-1 cells with these conjugates, we also observed upregulation of C/EBPα and p21 (Figure 3C) and inhibition of tumor cell proliferation (Figure 3F), compared with the IRRE control group.
TR14-CEBPA Shows Potent Anti-tumor Effects in a Mouse Model of Advanced PDAC
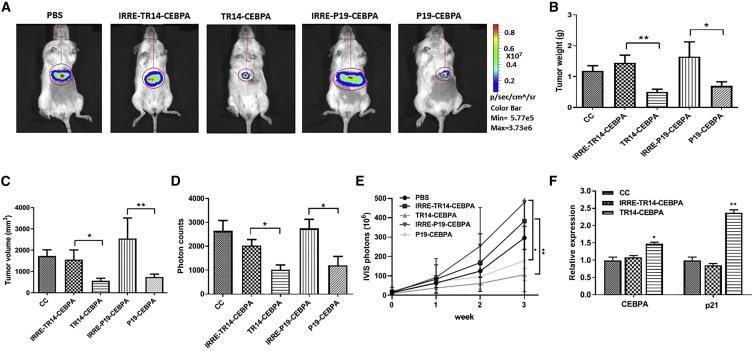
To determine the anti-tumor effects of the aptamer conjugates in advanced PDAC, we established a traceable animal model by implanting firefly luciferase-engineered PANC-1 cells (PANC-Luc) into the livers of NOD/SCID mice (i.e., intrahepatic pancreatic cancer cell implantation). In our previous studies, we demonstrated that P19-CEBPA showed potent anti-tumor effects in a local subcutaneous pancreatic cancer mouse model.31 Therefore, in this study, we included P19-CEBPA in parallel with TR14-CEBPA to determine the anti-tumor efficacy in an advanced PDAC model engrafted in the liver of PANC-Luc. We randomly divided mice into five groups (n = 4–6/group) and injected them with PBS, IRRE-TR14-CEBPA (1 nmol), TR14-CEBPA (1 nmol), IRRE-P19-CEBPA (1 nmol), or P19-CEBPA (1 nmol) via the tail vein 3 times/week for 3 weeks. We monitored tumor growth by quantifying bioluminescence using an IVIS 200 in vivo imaging platform (Figure 4A).
Figure 4.
Comparison of Anti-tumor Effects with TR14-CEBPA and P19-CEBPA in an Advanced PDAC Mouse Model
PANC-Luc xenografted mice were injected with PBS (CC), IRRE-TR14-CEBPA, TR14-CEBPA, IRRE-P19-CEBPA, or P19-CEBPA (1 nM) via tail vein. (A) Representative traceable tumor images show bioluminescence in the liver. (B–E) Liver tumor weight (B) and volume (C) were measured from liver biopsies. Data are presented as mean ± SD. (D) Tumor growth was monitored by evaluating the difference in bioluminescence before the first injection and 1 day after the last injection. Data are presented as the mean ± SD. (E) Tumor growth was monitored by evaluating bioluminescence before the first injection and weekly throughout the 3-week treatment. Data are presented as the mean ± SD. Student’s t test was used to determine statistical significance: *p ≤ 0.05, **p ≤ 0.01. (F) The expression of C/EBPα and p21 in tumor was determined using real-time qPCR. Data are presented as mean ± SD; n = 4–6. Unpaired t test with Welch’s correction was used to determine statistical significance: *p ≤ 0.05, **p ≤ 0.01.
After treatment of mice with the aptamer-conjugates as described above, we assessed the burden of pancreatic cancer tumors on each implanted liver by measuring tumor weight and volume for 3 weeks. Both TR14-CEBPA and P19-CEBPA treatment groups showed significant reduction of tumor burden compared with the control groups (IRRE conjugates) (Figures 4B and 4C). We subsequently quantified the bioluminescent signal, indicative of PANC-Luc tumor growth, which showed a significant inhibition of tumor growth following treatment with TR14-CEBPA and P19-CEBPA compared with controls (Figure 4D). By tracing tumor growth using bioluminescence over time (0–3 weeks), we observed persistent anti-tumor effects following treatment with TR14-CEBPA and P19-CEBPA compared with controls in our model of advanced PDAC (Figure 4E). The expression of C/EBPα in engrafted tumor was analyzed. We observed significant upregulation of C/EBPα and p21 in the treatment group of TR14-CEBPA compared with control (Figure 4F). In summary, our results showed significant reduction of advanced PDAC tumor burden in two groups: TR14-CEBPA and P19-CEBPA.
The Truncated TR14-CEBPA Attached to an Albumin Affinity Tag Shows Potent Anti-tumor Effects in a Mouse Model of Advanced PDAC in Combination with Gemcitabine
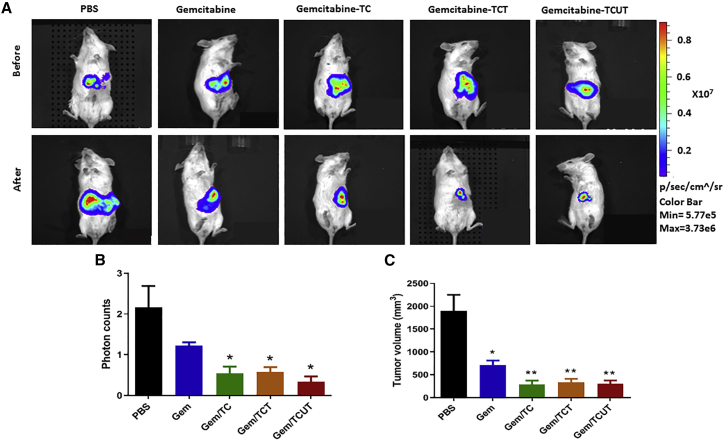
The current standard care for advanced PDAC is gemcitabine-based monotherapy or combinational therapies. Therefore, we tested the efficacy of albumin affinity-tagged tTR14-C/EBPα-saRNA conjugates (TC, TCT, TCUT; 1 nmol) in combination therapy with gemcitabine (50 mg/kg) in a mouse model of advanced PDAC. Representative bioluminescence images taken before and after treatment are shown in Figure 5A. We quantitated anti-tumor effects by measuring tumor volume and change in photons. Compared with untreated control (PBS), all three conjugates (TC, TCT, TCUT) combined with gemcitabine caused a significant reduction of tumor burden in the advanced PDAC by measuring photon increase (Figure 5B). Compared with vehicle-treated control, our studies showed that gemcitabine alone reduced tumor volume ∼70%; the three conjugates (TC, TCT, TCUT) combined with gemcitabine reduced tumor growth up to ∼85% (Figure 5C), suggesting that aptamer-C/EBPα-saRNAs might be used as an adjuvant in combination with chemotherapy.
Figure 5.
Anti-tumor Effects of Three Conjugates of Truncated TR14 (tTR14) with Albumin Affinity Tag in Combination with Gemcitabine in an Advanced PDAC Mouse Model
PANC-Luc xenografted mice were injected with PBS or gemcitabine (50 mg/kg) via tail vein. The TC, TCT, and TCUT (1 nmol) were injected via i.p. (A) Representative traceable tumor images before and after treatment show bioluminescence in the liver. (B) Tumor growth was monitored by evaluating the difference in bioluminescence before the first injection and 1 day after the last injection. Data are presented as the mean ± SD. (C) Liver tumor volume was measured from liver biopsies. Data are presented as mean ± SD. Student’s t test was used to determine statistical significance: *p ≤ 0.05, **p ≤ 0.01.
Discussion
More than 75% of pancreatic cancer patients are diagnosed with metastatic advanced PDAC, which shows a dismal prognosis of only 3% overall survival rate at 5 years.33 Although surgical resection of the primary tumor has been considered to improve the survival rate of PDAC patients, 85%–90% are ineligible.34 Moreover, more than 80% of patients have suffered systemic spread following surgery.35 Therefore, metastatic advanced PDAC remains the main cause of pancreatic cancer mortality. The currently approved standard of care for metastatic advanced PDAC is systemic gemcitabine-based therapy. However, because the anti-tumor effects of gemcitabine monotherapy are disappointing, the current trend for therapeutic options in advanced PDAC is combinational therapeutics to improve survival of patients such as a combination of gemcitabine and nab-paclitaxel.36 However, combinational therapeutics with multiple chemo-drugs does not show statistically significant survival benefits for PDAC, leaving severe side effects in patients.6 Therefore, more effective treatments for metastatic advanced PDAC had been explored in this study.
Molecularly targeted therapies have been explored in unresectable pancreatic cancer patients, targeting epidermal growth factor receptor (EGFR), vascular endothelial growth factor (VEGF), platelet-derived growth factor (PDGF), HER2, tyrosine kinase (TK), insulin-like growth factor 1 receptor (IGF1R), or hedgehog signaling elements.37 Despite these intensive studies, the effect is too small to adopt for therapeutic options. The transcription factor C/EBPα is known to suppress tumor growth in PDAC, because its expression is typically reduced during disease progression.31, 38, 39, 40 We previously reported using the pancreatic cancer-specific P19 and P1 aptamer to deliver C/EBPα-saRNAs, and it showed the significant anti-tumor effects in a locally engrafted subcutaneous PDAC mouse model,31 suggesting that C/EBPα could be an effective target in aggressive advanced pancreatic cancer. Therefore, herein, we determined the therapeutic efficacy of molecularly targeted therapy targeting C/EBPα in an advanced PDAC mouse model.
To determine the therapeutic efficacy in vivo, herein, we established an advanced PDAC mouse model by implanting pancreatic cancer cells in the lobe of the liver intrahepatically. Typically, the use of orthotopic mouse models with intrasplenically injected pancreatic cancer cells to study the molecular mechanism of metastasis and to evaluate therapeutic regimens is well established.41 However, the progression of metastasis of pancreatic cancer cells to the liver takes up to 145 days in this model. The aim of this study was focused on determining the anti-tumor effects of aptamer-based conjugates targeting already metastasized advanced PDAC, rather than investigating inhibition of the metastasis process to other body organs. Direct implanting of tumor cells in the liver is a more clinically relevant approach to test anti-tumor effects in an advanced PDAC mouse model.
Currently, a challenge of the oligonucleotide therapeutic field is delivery. Since the first US Food and Drug Administration (FDA) approval in 2004 for an aptamer-based treatment for neovascular age-related macular degeneration,42 RNA aptamers have become very attractive therapeutic modalities.43 TfR is a transmembrane protein to deliver ferric iron to cells.44 TfR is overexpressed, almost 100-fold, in high-proliferative cancer cells.45 As a consequence, TfR is a very attractive target for targeted therapy development. In previous studies, murine-specific DNA and RNA aptamers against TfR have been isolated.46 But, unfortunately, these murine TfR aptamers showed high susceptibility to nucleases, which limit their utility. For nuclease-stabilized aptamers, two RNA aptamers, named c247 or Waz,48 employed with 2′-fluoro (2′F)-modified pyrimidines were isolated against hTfRs in the same group. The binding affinity of c2 and minimized c2 was 17 and 102 nmol/L, respectively. The binding affinity of Waz was 390 nmol/L. To isolate c2 and Waz aptamers, they employed a two-stage selection method; recombinant proteins expressed insect cells followed by target cell-SELEX; HeLa or Jurkat cells. This two-stage selection approach allows for solving the issue that aptamers isolated against recombinant proteins sometimes do not bind to mammalian cells because of glycosylation. Even though this two-stage selection is quite a smart approach, it is a time-consuming SELEX process. Therefore, in this study, to reduce the time of the SELEX procedure, we used the extracellular domain of hTfR recombinant proteins that were expressed in mammalian cells to isolate RNA aptamers, and intracellular uptake was tested in pancreatic cancer cells. In turn, we successfully isolated new anti-hTfR RNA aptamers with high binding affinity that showed better binding affinity than c2 and Waz. The newly isolated anti-hTfR aptamers were internalized into cells via CME.
With the aim to develop a new therapeutic regimen of molecularly targeted therapy and to test the suitability of targeting C/EBPα for anti-cancer effects in aggressive advanced PDAC, we employed two aptamers (TR14 and P19) that were conjugated with C/EBPα-saRNAs using sticky bridge sequences for targeted delivery. We observed that both conjugates significantly inhibited tumor growth in a mouse model of advanced PDAC. However, no significant difference in anti-tumor effects was observed between TR14-CEBPA or P19-CEBPA treatment. For cost-effective chemical synthesis for clinical trials, the TR14 was truncated to reduce the size. The truncation of aptamer is a process of trial and error. The binding affinity of aptamers has been often completely lost or compromised during the tedious truncation process. Cowperthwaite and Ellington49 suggest that the fixed regions do not contribute to or constrain the binding properties of aptamers, and they are minimally involved in the overall structure by bioinformatics analysis. Currently, computer simulation programs, such as Mfold,50 RNAstructure,51 and NUPACK,52 have been used to predict the secondary structure of aptamers. Thus, using this secondary structure information, conserved stem-loop regions are truncated, which is predicted to be necessary for binding to hTfR. However, in previous studies, the influence of nucleotide bases, which are incorporated into aptamers from the promoter region during the in vitro transcription (IVT) to truncate the aptamers, has not been investigated. To investigate the influence of the nucleotide bases incorporated from the promoter region throughout IVT for the function of aptamers, we generated truncates of TR14 that hold the “5′-GGG transcription start codon”: TR14 S1 and TR14 S2. The truncates of TR14 that do not hold the “5′-GGG transcription start codon” are TR14 ST1-1 and TR14 ST1-2. Based on our data, TR14 S2 containing the “5′-GGG in vitro transcription start codon” keeps the functionality and increased binding affinity, compared with the parent TR14 aptamer. However, TR14 ST1-1 without the “5′-GGG transcription start codon” completely lost the functionality and binding affinity, even though the structure of TR14 S2 and TR14 ST1-1 remains the same by NUPACK. In this study, we observed that three extra Gs at the 5′ end are indispensable in some truncates of aptamers. Therefore, “5′-GGG transcription start codon” incorporated from the promoter region will be considered to truncate the aptamers after selection. However, we cannot conclude that our observation can be generally applied to all of the truncations of aptamers. Surprisingly, TR14 ST1-2 completely lost its function, but TR14 ST1-3 kept similar functionality of parent TR14.
Finally, TR14 ST1-3 aptamer (termed tTR14, 22-mer) showing the similar range of binding kinetics to target, compared with parent TR14, was employed further in an in vivo experiment. To improve efficacy and pharmacokinetic properties,53 we chemically attached an albumin affinity tag to the 3′ end of the selected TR14 ST1-3 aptamer (termed tTR14): TC (without albumin tag), TCT (with albumin affinity tag), and TCUT (extra Us with albumin affinity tag). Reportedly, molecularly targeted therapy of EGFR inhibitors in combination with gemcitabine showed the evident therapeutic efficacy: improved overall survival rate in advanced PDAC patients.37 Therefore, the efficacy of conjugates in combination with gemcitabine was assessed in an advanced PDAC mouse model, which can be used as for adjuvant. The results showed significant reduction of tumor burden in combination with gemcitabine in an advanced PDAC mouse model, suggesting that aptamer-C/EBPα-saRNAs might have potential as an adjuvant in combination with small drug-based anti-cancer drugs.
We expected that the attachment of albumin affinity tag would improve the tissue biodistribution (PD) and pharmacokinetics (PK) profile of the aptamers. Based on our current study, we cannot conclude how the albumin-tag affects delivery of total payload to target tissues (i.e., % of injected saRNA dose delivered per gram of tissue [%ID/g]); we will complement our current findings with direct quantification of saRNA biodistribution in tissues in further studies.
In summary, our study provides strong evidence that targeted delivery of CEBPα-saRNA by anti-hTfR or pancreatic cancer-specific aptamers leads to potent anti-tumor effects in a mouse model of advanced PDAC. We also showed that truncated anti-hTfR1 TR14 aptamers delivered C/EBPα-saRNA into cancer cells, and that combined with gemcitabine they showed an anti-tumor effect. Our results suggest that C/EBPα-saRNAs could be used as an adjuvant in combination with gemcitabine in advanced PDAC. Finally, given that the hTfR is overexpressed in multiple cancer cells, we expect that hTfR targeting will allow delivery of therapeutic payloads to other types of cancers.
Materials and Methods
Chemicals and Reagents
CPZ (C8138), CQ (C6658), dynasore (D7693), GEZ (D6649), Cyto D (C8273), and Hank’s balanced salt solution (HBSS; 6648) were purchased from Sigma. CellLight Early Endosome-GFP (C10588), Late Endosome-GFP (C10586), and lysosome-GFP (C10507) were purchased from Thermo Fisher Scientific. Anti-TfR antibodies (ab47095) were purchased from Abcam. Human transferrin-Alexa 488 was purchased from Invitrogen (1780257).
Recombinant Target Protein
hTfR was purchased from Sino Biological (11020-H07H; Beijing, P.R. China). The extracellular domain of hTfR (NP_003225.2) (Cys 89-Phe 760) was expressed with a His6-tag at the N terminus in human cells (HEK293).
Cell Line
PANC-1 (pancreatic epithelioid carcinoma, CRL-1469) and U-87 MG (glioblastoma, HTB-14) were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). The cells were cultured according to the suppliers’ instructions. The TB10 human glioma cell line was obtained from Vittorio de Franciscis lab in Italy.
Protein SELEX
In vitro selection was carried out essentially as described previously,54 with a few modifications. The 2′F-RNA aptamers were selected from 40-nt randomized sequences constructed by IVT of synthetic DNA templates with nucleoside triphosphates (NTPs) (2′F dUTP [2'-Fluoro-2'-deoxyuridine-5'-triphosphate], 2′F dCTP [2'-Fluoro-2'-deoxycytidine-5'-triphosphate], guanosine triphosphate [GTP], adenosine triphosphate [ATP]; Epicenter Biotechnologies, Madison, WI, USA) and T7 RNA polymerase. To remove RNAs that bound nonspecifically to agarose beads, 1.44 μM of the RNA library was pre-incubated with 20 μL of Ni-NTA agarose beads in 100 μL of binding buffer (30 mM Tris-HCl [pH 7.5]; 150 mM NaCl; 5 mM MgCl2; 2 mM dithiothreitol; 1% BSA; 100 μg/mL yeast tRNA) for 30 min at room temperature with shaking, precipitated by centrifugation, and discarded. The precleared supernatant was transferred to a new tube and incubated with 300 nM His6-tagged hTfR for 30 min at room temperature. RNAs that bound to hTfR were recovered, amplified by RT-PCR and IVT, and used in subsequent selection rounds. In subsequent rounds, hTfR concentration was reduced by 2-fold at every three rounds for more stringent conditions. After nine rounds of SELEX, the resulting cDNA was amplified. The amplified DNA was cloned, and individual clones were identified by DNA sequencing. Aptamer structures were predicted using Mfold50 (available at http://www.bioinfo.rpi.edu/applications/mfold/), using a salt correction algorithm and temperature correction for 25°C, or were predicted using NUPACK52 (available at http://www.nupack.org/).
Surface Plasmon Resonance-Based Biosensor Assay
The Biacore T100 (GE Healthcare, Uppsala, Sweden) was used to monitor label-free interactions of TR14-hTfR in real time. The biotinylated aptamer was coupled to a streptavidin-coated Biacore chip (SensorChip SA, BR-1003-98; General Electric Company) by an injection in binding buffer at a concentration of 25 μg/mL (30 mM Tris-HCl [pH 7.5]; 150 mM NaCl; 5 mM MgCl2) at 10 μL/min. The RNA was refolded by heating to 65°C, followed by cooling to 37°C, before immobilization. To measure binding kinetics, we injected five concentrations of purified hTfR protein at a flow rate of 10 μL/min. After binding, the surface was regenerated by injecting 50 mM NaOH at a flow rate of 15 μL/min for 20 s. Data from the control surface were subtracted. BIAevaluation software (GE Healthcare) was used for analysis. The binding data were fit to a 1:1 binding with a mass transfer model to calculate kinetics parameters as previously described.55, 56
Live-Cell Confocal Imaging for Aptamer Internalization
A total of 1 × 105 PANC-1 cells were seeded in 35-mm glass-bottom dishes (MatTek, Ashland, MA, USA) and grown in appropriate media for 24 h. Aptamer RNAs were labeled with Cy3 fluorescent dye using the Cy3 Silencer siRNA labeling kit (Thermo Fisher Scientific, Waltham, MA, USA). Truncated aptamers or full-length aptamers were chemically conjugated with Cy3. Cy3-labeled aptamers in binding buffer (30 mM Tris-HCl [pH 7.5]; 150 mM NaCl; 5 mM MgCl2; 100 μg/mL yeast tRNA) were added to the cells at 200 nM and incubated at 37°C for 2 h. Before imaging, cells were washed with Dulbecco’s PBS (DPBS) twice. Live-cell confocal imaging was performed with a Zeiss LSM 510 Meta inverted two-photon confocal microscope system using a C-Apo 40×/1.2 NA water immersion objective and AIM 4.2 software (Carl Zeiss, Jena, Germany).
Flow Cytometry-Based Binding Assays
Aptamer binding was assessed by flow cytometry. For the assay, the PANC-1 cells were detached using Accutase, washed with PBS, and suspended in binding buffer. Next, chemically synthesized aptamers labeled with Cy3 at 500 nM were added to target cells for 20 min in ice. Cells were washed with binding buffer and immediately analyzed by NovoCyte (ACEA Biosciences). For the exclusion of dead cells, DAPI (1 μg/mL) was used. The data were analyzed with NovoExpress software.
Cellular Uptake Inhibition Assay
A total of 5 × 103 PANC-1 cells/well were seeded in 96-well plates 1 day before assay. PANC-1 cells were left either pretreated or untreated with CPZ (clathrin endocytosis inhibitor, 10 μg/mL), CQ (clathrin endocytosis inhibitor, 20 μg/mL), dynasore (dynamin inhibitor, 80 μM), GEZ (caveolae- and lipid-mediated endocytosis inhibitor, 50 μg/mL), or Cyto D (5 μM) at 37°C for 30 min. Subsequently, cells were washed with DPBS and incubated with Cy3-labeled aptamers at 200 nM at 37°C for 1 h. Following incubation, cells were washed with HBSS to remove surface-bound aptamers. Cellular uptake was quantified by fluorescence using a plate reader (SpectraMax iD3; Molecular device). The inhibition of uptake was normalized with inhibitor non-treated groups (mock).
Competition Assays
For competition assays with human transferrin or hTfR antibodies, 5 × 103 PANC-1 cells/well were seeded in 96-well plates and grown in 1 day. Cells were pre-incubated with TR14 at 200 nM for 20 min on ice with PBS. After washing with PBS, human transferrin conjugated with Alexa 488 at 25 μg/mL or hTfR antibodies (2 μL/1 × 104 cells) conjugated with fluorescein isothiocyanate (FITC) were added to cells, and the cells were incubated at 37°C for 40 min. Afterward, the cells were washed with HBSS twice to remove cell surface binders. The intensity of fluorescence was quantified using a plate reader (SpectraMax iD5; Molecular Devices). The intensity was normalized with aptamer non-treated control cells (mock).
Receptor Specificity by siRNA Knockdown
Receptor knockdown was performed using a pre-designed anti-hTfR siRNA (Ambion Catalog 16708, assay ID: 12925) or control siRNA (AM4611; Ambion). siRNA transfection was performed using RNAiMax (Invitrogen) in 48-well plates according to the manufacturer’s protocol. Forty-eight hours after transfection, Cy3-labeled TR14 aptamers at 100 nM or anti-hTfR antibodies (1 μL/1 × 104 cells) were incubated at 37°C for 1 h. The intensity of fluorescence was quantified using a plate reader (SpectraMax iD3; Molecular Devices).
Co-localization Assay
A total of 1 × 105 PANC-1 cells were seeded in 35-mm glass-bottom dishes (MatTek, Ashland, MA, USA) and grown in appropriate media for 24 h. A total of 20 μL of CellLight Early endosome-GFP BacMam 2.0 (C10586; Thermo Fisher), CellLight Late endosome-GFP BacMam 2.0 (C10588; Thermo Fisher), or CellLight Lysosome-GFP BacMam 2.0 lysosome-GFP (C10596; Thermo Fisher) was added and incubated at 37°C for 16 h. After confirmation of GFP expression, chemically synthesized TR14 labeled with Cy3 at 200 nM was incubated at 37°C for 2 h. The co-localization was assessed on live cells by confocal microscopy.
Aptamer Conjugation to saRNA Using a “Sticky” Sequence (STICK)
A “sticky” sequence (a 16-nt sequence that prevents structural hindrance) was placed between TR14 and the C/EBPα-saRNA oligonucleotide, as we described for the construction of P19-CEBPA in our previous study.31 TR14-STICK-sense, P19-STICK, control-STICK, Sense-STICK, and antisense RNAs were chemically synthesized. The TR14-STICK, P19-STICK, or control-STICK RNAs were refolded in binding buffer, heated to 95°C for 3 min, slowly cooled to 37°C, and then incubated at 37°C for 10 min. To form the STICK-C/EBPα RNAs, we annealed the sense-STICK and antisense strand to the complementary strand using the same molar amounts. The same amount of refolded TR14-, P19-, or control-STICK was added and incubated at 37°C for 10 min in binding buffer to make the chimeric conjugates. For truncated TR14, the same molar amounts of aptamer-STICK-sense and CEBPA anti-sense were annealed in binding buffer by heating to 95°C for 3 min and slowly cooled to 37°C.
Relative Gene Expression Analysis by qPCR In Vitro
For analyzing gene activation and protein expression, PANC-1 cells were seeded in duplicate into 24-well plates at a density of 1 × 105 cells/well. TR14 or control aptamer (IRRE) conjugated to C/EBPα-saRNAs was added directly to the cells, at a final concentration of 100 nM. The treatment was repeated 24 h later, and cells were harvested at final incubation times of 72 h. Total RNA was extracted for reverse transcription using an RNeasy kit (QIAGEN). Target cDNA amplification and real-time PCR were performed using a Bio-Rad kit (SsoAdvanced Universal SYBR Green Supermix). For normalization, the reference gene 18S was used.
MTS Assay
To determine the inhibition of cell proliferation, 5 × 103 PANC-1 cells/well were seeded in 96-well plates and grown in appropriate media 1 day before the treatment. Cells were treated with TR14-CEBPA and IRRE-CEBPA at 100 nM twice at 24-h intervals. Inhibition of cell proliferation was measured using MTS assay (Promega, Madison, WI, USA) at a final incubation time of 72 h.
Chemical Synthesis of Albumin Tag to Aptamers
4-(p-Iodophenyl) butyric acid (Sigma, St. Louis, MO, USA) was converted into N-hydroxysuccinimide (NHS) ester using standard procedures. 4-(p-Iodophenyl) butyric acid NHS ester was used for the conjugation with 3′-amino or 5′-amino-labeled oligonucleotides. One micromole of oligonucleotide was dissolved in 750 μL of water, and 400 μL of DMSO was added, followed by 150 μL of 1.0 M phosphate buffer (pH 7.5). A solution of 5 equiv of 4-(p-Iodophenyl) butyric acid NHS ester in 100 μL of DMSO was added. The reaction mixture was vigorously stirred at room temperature. Progress of the reaction was monitored using high-pressure liquid chromatography (HPLC) on a PRP1 column (Hamilton, Reno, NV, USA) in triethylammonium acetate (TEAA) buffer. Buffer A was 50 mM TEAA in water; buffer B was 50 mM TEAA in acetonitrile-water, 9:1. After 1 h, the reaction was completed. To remove the DMSO, the reaction mixture was precipitated into isopropanol, kept at −20°C overnight, and centrifuged at 4°C for 20 min. The supernatant was discarded, and the resulting pellet was washed with an 8:2 solution of cold ethanol/water.
Aptamer Conjugation to C/EBPα-saRNA
The tTR14-C/EBPα-sense strand with (TCT) or without (TC) an albumin affinity tag and antisense CEBPA RNA were chemically synthesized at City of Hope. The RNAs were refolded in binding buffer, heated to 95°C for 3 min, slowly cooled to 37°C, and then incubated at 37°C for 10 min. To form the tTR14-C/EBPα RNA, the antisense strand of C/EBPα was annealed to the complementary strand using the same molar amounts. The same amount of refolded tTR14 was added and incubated at 37°C for 10 min in binding buffer to make the chimeric conjugates.
Intrahepatic Pancreatic Cancer Liver-Metastatic Mouse Model
To create an animal model harboring traceable tumors, we inserted a firefly luciferase fragment into the pLKO.1-AS3 backbone encoding the neo gene (National RNAi Core; Academia Sinica, Taiwan). One day before transduction, 293T cells were plated onto a six-well plate. On the day of transduction, the medium was replaced with DMEM containing serial dilutions of the transfer plasmids and incubated for 5 h; then the medium was replaced. After 2 days, the culture medium containing recombinant lentiviral particles was obtained. PANC-1 cells were incubated with recombinant lentiviral particles for 24 h. The following day, culture medium was replaced with standard medium containing 1.2 mg/mL G418 (Merck, Germany) for stable clone selection. Two weeks after selection, a single stable cell line was picked and maintained in medium containing G418. Luciferase expression was assessed using the Luciferase Assay System.
Six-week-old female NOD/SCID mice (BioLasco, Taiwan) were used in these experiments. Animal studies were performed in compliance with approval from the Institutional Animal Care and Use Committee of College of Medicine, National Taiwan University. Mice were kept in a conventional, specific pathogen-free facility. To establish a liver-metastatic pancreatic cancer model, we performed intrahepatic tumor implantation by injecting 30 μL of a monocellular suspension (in PBS) containing 1 × 106 PANC-Luc cells into a region in the middle lobe of the livers of 6-week-old female NOD/SCID mice (BioLasco, Taiwan). Tumors were allowed to grow for 1 week after inoculation; then the mice were randomly divided into five groups and injected with PBS, IRRE-TR14-CEBPA (1 nmol), TR14-CEBPA (1 nmol), IRRE-P19-CEBPA (1 nmol), or P19-CEBPA (1 nmol) via the tail vein 3 times/week for 3 weeks.
For the combinational treatment with gemcitabine, tumors were allowed to grow for 2 weeks after inoculation until they could be detected using an IVIS system; then mice were randomly divided into five groups of 10 animals/group. Each treatment group followed a specific schedule. The Gem group was treated with gemcitabine (50 mg/kg, 2 times/week, intravenously [i.v.]). The Gem/TC group was treated with gemcitabine (50 mg/kg, 2 times/week, i.v.) and TC (1 nmol, 3 times/week, intraperitoneally [i.p.]). The Gem/TCT group was treated with gemcitabine (50 mg/kg, 2 times/week, i.v.) and TCT (1 nmol, 3 times/week, i.p.). The Gem/TCUT group was treated with gemcitabine (50 mg/kg, 2 times/week, i.v.) and TCUT (1 nmol, 3 times/week, i.p.). The control group was treated with PBS only.
Tumor growth was monitored by evaluating bioluminescence using an IVIS 200 in vivo imaging platform (Caliper Life Sciences, Alameda, CA, USA) and measuring the difference from before the first injection and 1 day after the last injection. To do this, prior to in vivo imaging, the mice were anesthetized using isoflurane. A solution of 150 μg/kg D-luciferin (Biosynth, USA) was then injected i.p. The mice were imaged using the IVIS 200, and bioluminescent signals were analyzed using Living Image Software (Caliper Life Sciences, Alameda, CA, USA). The mice were euthanized 2 days after the last injection. Tumors were removed from mice, and tumor size was measured by caliper and further analysis of gene expression.
Statistical Analysis
Data were analyzed using Student’s t test, ANOVA, or unpaired t test with Welch’s correction to assess statistical significance (*p ≤ 0.05, **p ≤ 0.01).
Author Contributions
S.Y., N.H., and J.J.R. developed the concept. S.Y. designed and performed SELEX to isolate RNA aptamers and in vitro experiments, including cell internalization, measurements of cell binding affinity by SPR, qPCR, data organization, and statistical analyses. K.-W.H. performed in vivo assays. P.A., V.R., and D.V. performed qPCR and data organization. S.Y., N.H., J.J.R., K.-W.H., P.A., V.R., M.S., and D.V. prepared the manuscript.
Conflicts of Interest
J.J.R. and N.H. are co-founders of privately owned Apterna Ltd. UK.
Acknowledgments
We would like to thank the Light Microscopy Digital Imaging (Loren Quintanar and Michael Nelson) and Drug Discovery & Structural Biology Cores at City of Hope for technical assistance. Research reported in this publication included work performed in the Drug Discovery & Structural Biology Core supported by the National Cancer Institute of the NIH under award number P30 CA033572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The authors would like to acknowledge support from Apterna Ltd. UK and NIH grants R01 AI029329 and R01 AI042552 to J.J.R. We thank Sarah T. Wilkinson, scientific writer, at City of Hope for language editing.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.08.017.
Supplemental Information
References
- 1.Huguet F., Girard N., Guerche C.S., Hennequin C., Mornex F., Azria D. Chemoradiotherapy in the management of locally advanced pancreatic carcinoma: a qualitative systematic review. J. Clin. Oncol. 2009;27:2269–2277. doi: 10.1200/JCO.2008.19.7921. [DOI] [PubMed] [Google Scholar]
- 2.Shaib Y.H., Davila J.A., El-Serag H.B. The epidemiology of pancreatic cancer in the United States: changes below the surface. Aliment. Pharmacol. Ther. 2006;24:87–94. doi: 10.1111/j.1365-2036.2006.02961.x. [DOI] [PubMed] [Google Scholar]
- 3.Houg D.S., Bijlsma M.F. The hepatic pre-metastatic niche in pancreatic ductal adenocarcinoma. Mol. Cancer. 2018;17:95. doi: 10.1186/s12943-018-0842-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mohammad A.A. Advanced pancreatic cancer: The standard of care and new opportunities. Oncol. Rev. 2018;12:370. doi: 10.4081/oncol.2018.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma S.J., Prezzano K.M., Hermann G.M., Singh A.K. Dose escalation of radiation therapy with or without induction chemotherapy for unresectable locally advanced pancreatic cancer. Radiat. Oncol. 2018;13:214. doi: 10.1186/s13014-018-1158-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paulson A.S., Tran Cao H.S., Tempero M.A., Lowy A.M. Therapeutic advances in pancreatic cancer. Gastroenterology. 2013;144:1316–1326. doi: 10.1053/j.gastro.2013.01.078. [DOI] [PubMed] [Google Scholar]
- 7.Rosenblum D., Joshi N., Tao W., Karp J.M., Peer D. Progress and challenges towards targeted delivery of cancer therapeutics. Nat. Commun. 2018;9:1410. doi: 10.1038/s41467-018-03705-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 9.Tuerk C. Using the SELEX combinatorial chemistry process to find high affinity nucleic acid ligands to target molecules. Methods Mol. Biol. 1997;67:219–230. doi: 10.1385/0-89603-483-6:219. [DOI] [PubMed] [Google Scholar]
- 10.Ulrich H., Magdesian M.H., Alves M.J., Colli W. In vitro selection of RNA aptamers that bind to cell adhesion receptors of Trypanosoma cruzi and inhibit cell invasion. J. Biol. Chem. 2002;277:20756–20762. doi: 10.1074/jbc.M111859200. [DOI] [PubMed] [Google Scholar]
- 11.Wang J., Jiang H., Liu F. In vitro selection of novel RNA ligands that bind human cytomegalovirus and block viral infection. RNA. 2000;6:571–583. doi: 10.1017/s1355838200992215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blank M., Weinschenk T., Priemer M., Schluesener H. Systematic evolution of a DNA aptamer binding to rat brain tumor microvessels. selective targeting of endothelial regulatory protein pigpen. J. Biol. Chem. 2001;276:16464–16468. doi: 10.1074/jbc.M100347200. [DOI] [PubMed] [Google Scholar]
- 13.Daniels D.A., Chen H., Hicke B.J., Swiderek K.M., Gold L. A tenascin-C aptamer identified by tumor cell SELEX: systematic evolution of ligands by exponential enrichment. Proc. Natl. Acad. Sci. USA. 2003;100:15416–15421. doi: 10.1073/pnas.2136683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hicke B.J., Marion C., Chang Y.F., Gould T., Lynott C.K., Parma D., Schmidt P.G., Warren S. Tenascin-C aptamers are generated using tumor cells and purified protein. J. Biol. Chem. 2001;276:48644–48654. doi: 10.1074/jbc.M104651200. [DOI] [PubMed] [Google Scholar]
- 15.Wilson D.S., Szostak J.W. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 16.Que-Gewirth N.S., Sullenger B.A. Gene therapy progress and prospects: RNA aptamers. Gene Ther. 2007;14:283–291. doi: 10.1038/sj.gt.3302900. [DOI] [PubMed] [Google Scholar]
- 17.Yoon S., Rossi J.J. Emerging cancer-specific therapeutic aptamers. Curr. Opin. Oncol. 2017;29:366–374. doi: 10.1097/CCO.0000000000000389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoon S., Huang K.W., Reebye V., Spalding D., Przytycka T.M., Wang Y., Swiderski P., Li L., Armstrong B., Reccia I. Aptamer-Drug Conjugates of Active Metabolites of Nucleoside Analogs and Cytotoxic Agents Inhibit Pancreatic Tumor Cell Growth. Mol. Ther. Nucleic Acids. 2017;6:80–88. doi: 10.1016/j.omtn.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kratschmer C., Levy M. Targeted Delivery of Auristatin-Modified Toxins to Pancreatic Cancer Using Aptamers. Mol. Ther. Nucleic Acids. 2018;10:227–236. doi: 10.1016/j.omtn.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Powell Gray B., Kelly L., Ahrens D.P., Barry A.P., Kratschmer C., Levy M., Sullenger B.A. Tunable cytotoxic aptamer-drug conjugates for the treatment of prostate cancer. Proc. Natl. Acad. Sci. USA. 2018;115:4761–4766. doi: 10.1073/pnas.1717705115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayle K.M., Le A.M., Kamei D.T. The intracellular trafficking pathway of transferrin. Biochim. Biophys. Acta. 2012;1820:264–281. doi: 10.1016/j.bbagen.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daniels T.R., Bernabeu E., Rodríguez J.A., Patel S., Kozman M., Chiappetta D.A., Holler E., Ljubimova J.Y., Helguera G., Penichet M.L. The transferrin receptor and the targeted delivery of therapeutic agents against cancer. Biochim. Biophys. Acta. 2012;1820:291–317. doi: 10.1016/j.bbagen.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawabata H., Yang R., Hirama T., Vuong P.T., Kawano S., Gombart A.F., Koeffler H.P. Molecular cloning of transferrin receptor 2. A new member of the transferrin receptor-like family. J. Biol. Chem. 1999;274:20826–20832. doi: 10.1074/jbc.274.30.20826. [DOI] [PubMed] [Google Scholar]
- 24.Kawabata H., Nakamaki T., Ikonomi P., Smith R.D., Germain R.S., Koeffler H.P. Expression of transferrin receptor 2 in normal and neoplastic hematopoietic cells. Blood. 2001;98:2714–2719. doi: 10.1182/blood.v98.9.2714. [DOI] [PubMed] [Google Scholar]
- 25.Paterson J., Webster C.I. Exploiting transferrin receptor for delivering drugs across the blood-brain barrier. Drug Discov. Today. Technol. 2016;20:49–52. doi: 10.1016/j.ddtec.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 26.Ryschich E., Huszty G., Knaebel H.P., Hartel M., Büchler M.W., Schmidt J. Transferrin receptor is a marker of malignant phenotype in human pancreatic cancer and in neuroendocrine carcinoma of the pancreas. Eur. J. Cancer. 2004;40:1418–1422. doi: 10.1016/j.ejca.2004.01.036. [DOI] [PubMed] [Google Scholar]
- 27.Janowski B.A., Younger S.T., Hardy D.B., Ram R., Huffman K.E., Corey D.R. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat. Chem. Biol. 2007;3:166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 28.Li L.C., Okino S.T., Zhao H., Pookot D., Place R.F., Urakami S., Enokida H., Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proc. Natl. Acad. Sci. USA. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoon S., Rossi J.J. Therapeutic Potential of Small Activating RNAs (saRNAs) in Human Cancers. Curr. Pharm. Biotechnol. 2018;19:604–610. doi: 10.2174/1389201019666180528084059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reebye V., Sætrom P., Mintz P.J., Huang K.W., Swiderski P., Peng L., Liu C., Liu X., Lindkaer-Jensen S., Zacharoulis D. Novel RNA oligonucleotide improves liver function and inhibits liver carcinogenesis in vivo. Hepatology. 2014;59:216–227. doi: 10.1002/hep.26669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoon S., Huang K.W., Reebye V., Mintz P., Tien Y.W., Lai H.S., Sætrom P., Reccia I., Swiderski P., Armstrong B. Vivo. Mol. Ther. Vol. 24. 2016. Targeted Delivery of C/EBPalpha -saRNA by Pancreatic Ductal Adenocarcinoma-specific RNA Aptamers Inhibits Tumor Growth; pp. 1106–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calzolari A., Oliviero I., Deaglio S., Mariani G., Biffoni M., Sposi N.M., Malavasi F., Peschle C., Testa U. Transferrin receptor 2 is frequently expressed in human cancer cell lines. Blood Cells Mol. Dis. 2007;39:82–91. doi: 10.1016/j.bcmd.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 33.Loehrer P.J., Sr., Feng Y., Cardenes H., Wagner L., Brell J.M., Cella D., Flynn P., Ramanathan R.K., Crane C.H., Alberts S.R., Benson A.B., 3rd Gemcitabine alone versus gemcitabine plus radiotherapy in patients with locally advanced pancreatic cancer: an Eastern Cooperative Oncology Group trial. J. Clin. Oncol. 2011;29:4105–4112. doi: 10.1200/JCO.2011.34.8904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lillemoe K.D., Barnes S.A. Surgical palliation of unresectable pancreatic carcinoma. Surg. Clin. North Am. 1995;75:953–968. doi: 10.1016/s0039-6109(16)46739-3. [DOI] [PubMed] [Google Scholar]
- 35.Garrido-Laguna I., Hidalgo M. Pancreatic cancer: from state-of-the-art treatments to promising novel therapies. Nat. Rev. Clin. Oncol. 2015;12:319–334. doi: 10.1038/nrclinonc.2015.53. [DOI] [PubMed] [Google Scholar]
- 36.Blomstrand H., Scheibling U., Bratthäll C., Green H., Elander N.O. Real world evidence on gemcitabine and nab-paclitaxel combination chemotherapy in advanced pancreatic cancer. BMC Cancer. 2019;19:40. doi: 10.1186/s12885-018-5244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheahan A.V., Biankin A.V., Parish C.R., Khachigian L.M. Targeted therapies in the management of locally advanced and metastatic pancreatic cancer: a systematic review. Oncotarget. 2018;9:21613–21627. doi: 10.18632/oncotarget.25085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamamoto K., Tateishi K., Kudo Y., Sato T., Yamamoto S., Miyabayashi K., Matsusaka K., Asaoka Y., Ijichi H., Hirata Y. Loss of histone demethylase KDM6B enhances aggressiveness of pancreatic cancer through downregulation of C/EBPα. Carcinogenesis. 2014;35:2404–2414. doi: 10.1093/carcin/bgu136. [DOI] [PubMed] [Google Scholar]
- 39.Kumagai T., Akagi T., Desmond J.C., Kawamata N., Gery S., Imai Y., Song J.H., Gui D., Said J., Koeffler H.P. Epigenetic regulation and molecular characterization of C/EBPalpha in pancreatic cancer cells. Int. J. Cancer. 2009;124:827–833. doi: 10.1002/ijc.23994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Timchenko N.A., Wilde M., Nakanishi M., Smith J.R., Darlington G.J. CCAAT/enhancer-binding protein alpha (C/EBP alpha) inhibits cell proliferation through the p21 (WAF-1/CIP-1/SDI-1) protein. Genes Dev. 1996;10:804–815. doi: 10.1101/gad.10.7.804. [DOI] [PubMed] [Google Scholar]
- 41.Zhang G., Du Y.N. Orthotopic Pancreatic Tumor Mouse Models of Liver Metastasis. Methods Mol. Biol. 2019;1882:309–320. doi: 10.1007/978-1-4939-8879-2_27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gragoudas E.S., Adamis A.P., Cunningham E.T., Jr., Feinsod M., Guyer D.R., VEGF Inhibition Study in Ocular Neovascularization Clinical Trial Group Pegaptanib for neovascular age-related macular degeneration. N. Engl. J. Med. 2004;351:2805–2816. doi: 10.1056/NEJMoa042760. [DOI] [PubMed] [Google Scholar]
- 43.Zhou J., Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov. 2017;16:181–202. doi: 10.1038/nrd.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daniels T.R., Delgado T., Rodriguez J.A., Helguera G., Penichet M.L. The transferrin receptor part I: Biology and targeting with cytotoxic antibodies for the treatment of cancer. Clin. Immunol. 2006;121:144–158. doi: 10.1016/j.clim.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 45.Daniels T.R., Delgado T., Helguera G., Penichet M.L. The transferrin receptor part II: targeted delivery of therapeutic agents into cancer cells. Clin. Immunol. 2006;121:159–176. doi: 10.1016/j.clim.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 46.Chen C.H., Dellamaggiore K.R., Ouellette C.P., Sedano C.D., Lizadjohry M., Chernis G.A., Gonzales M., Baltasar F.E., Fan A.L., Myerowitz R., Neufeld E.F. Aptamer-based endocytosis of a lysosomal enzyme. Proc. Natl. Acad. Sci. USA. 2008;105:15908–15913. doi: 10.1073/pnas.0808360105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilner S.E., Wengerter B., Maier K., de Lourdes Borba Magalhães M., Del Amo D.S., Pai S., Opazo F., Rizzoli S.O., Yan A., Levy M. An RNA alternative to human transferrin: a new tool for targeting human cells. Mol. Ther. Nucleic Acids. 2012;1:e21. doi: 10.1038/mtna.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maier K.E., Jangra R.K., Shieh K.R., Cureton D.K., Xiao H., Snapp E.L., Whelan S.P., Chandran K., Levy M. A New Transferrin Receptor Aptamer Inhibits New World Hemorrhagic Fever Mammarenavirus Entry. Mol. Ther. Nucleic Acids. 2016;5:e321. doi: 10.1038/mtna.2016.32. [DOI] [PubMed] [Google Scholar]
- 49.Cowperthwaite M.C., Ellington A.D. Bioinformatic analysis of the contribution of primer sequences to aptamer structures. J. Mol. Evol. 2008;67:95–102. doi: 10.1007/s00239-008-9130-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mathews D.H. RNA secondary structure analysis using RNAstructure. Curr. Protoc. Bioinformatics. 2014;46:12.6.1–12.6.25. doi: 10.1002/0471250953.bi1206s46. [DOI] [PubMed] [Google Scholar]
- 52.Zadeh J.N., Steenberg C.D., Bois J.S., Wolfe B.R., Pierce M.B., Khan A.R., Dirks R.M., Pierce N.A. NUPACK: Analysis and design of nucleic acid systems. J. Comput. Chem. 2011;32:170–173. doi: 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
- 53.Huang Y., Reis E.S., Knerr P.J., van der Donk W.A., Ricklin D., Lambris J.D. Conjugation to albumin-binding molecule tags as a strategy to improve both efficacy and pharmacokinetic properties of the complement inhibitor compstatin. ChemMedChem. 2014;9:2223–2226. doi: 10.1002/cmdc.201402212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoon S., Lee G., Han D., Song J.Y., Kang K.S., Lee Y.S. Neutralization of infectivity of porcine circovirus type 2 (PCV2) by capsid-binding 2'F-RNA aptamers. Antiviral Res. 2010;88:19–24. doi: 10.1016/j.antiviral.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 55.Hernandez F.J., Kalra N., Wengel J., Vester B. Aptamers as a model for functional evaluation of LNA and 2′-amino LNA. Bioorg. Med. Chem. Lett. 2009;19:6585–6587. doi: 10.1016/j.bmcl.2009.10.039. [DOI] [PubMed] [Google Scholar]
- 56.Soontornworajit B., Zhou J., Snipes M.P., Battig M.R., Wang Y. Affinity hydrogels for controlled protein release using nucleic acid aptamers and complementary oligonucleotides. Biomaterials. 2011;32:6839–6849. doi: 10.1016/j.biomaterials.2011.05.074. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.