Abstract
We have undertaken a detailed analysis of the biotransformation of five of the most therapeutically important benzimidazole anthelmintics - albendazole (ABZ), mebendazole (MBZ), thiabendazole (TBZ), oxfendazole (OxBZ) and fenbendazole (FBZ) - in Caenorhabditis elegans and the ruminant parasite Haemonchus contortus. Drug metabolites were detected by LC-MS/MS analysis in supernatants of C. elegans cultures with a hexose conjugate, most likely glucose, dominating for all five drugs. This work adds to a growing body of evidence that glucose conjugation is a major pathway of xenobiotic metabolism in nematodes and may be a target for enhancement of anthelmintic potency. Consistent with this, we found that biotransformation of albendazole by C. elegans reduced drug potency. Glucose metabolite production by C. elegans was reduced in the presence of the pharmacological inhibitor chrysin suggesting that UDP-glucuronosyl/glucosyl transferase (UGT) enzymes may catalyze benzimidazole glucosidation. Similar glucoside metabolites were detected following ex vivo culture of adult Haemonchus contortus. As a step towards identifying nematode enzymes potentially responsible for benzimidazole biotransformation, we characterised the transcriptomic response to each of the benzimidazole drugs using the C. elegans resistant strain CB3474 ben-1(e1880)III. In the case of albendazole, mebendazole, thiabendazole, and oxfendazole the shared transcriptomic response was dominated by the up-regulation of classical xenobiotic response genes including a shared group of UGT enzymes (ugt-14/25/33/34/37/41/8/9). In the case of fenbendazole, a much greater number of genes were up-regulated, as well as developmental and brood size effects suggesting the presence of secondary drug targets in addition to BEN-1. The transcriptional xenobiotic response of a multiply resistant H. contortus strain UGA/2004 was essentially undetectable in the adult stage but present in the L3 infective stage, albeit more muted than C. elegans. This suggests that xenobiotic responses may be less efficient in stages of parasitic nematodes that reside in the host compared with the free-living stages.
Keywords: Caenorhabditis elegans, Haemonchus contortus, Benzimidazoles, Xenobiotic metabolism, Transcriptomic, LC-MS/MS
Graphical abstract
Highlights
-
•
C. e. & H. c. display hexose conjugation (likely glucose) and excretion of 5 BZs.
-
•
C. elegans (C.e.) biotransformation of ABZ reduces drug potency.
-
•
UGT inhibitor chrysin reduces ABZ biotransformation by C. elegans.
-
•
Transcriptomic response of C. e. (ben-1) to 5 BZs dominated by xenobiotic response and additional targets for FBZ.
-
•
Minimal transcriptomic response of H. contortus to ABZ exposure.
1. Introduction
Metabolism of drugs can have a major effect on drug potency (Daborn et al., 2002; Li et al., 2007; Amenya et al., 2008), for example, in humans, the cytochrome P450s and UDP-glucuronsyltransferases together are responsible for clearing greater than 90% of all commercial drugs; CYP3A4, with its large active site, is responsible for metabolizing 30% of all commercial drugs tested (Rowland et al., 2013; Zanger and Schwab, 2013; Mazerska et al., 2016). Understanding anthelmintic metabolism and the enzyme pathways involved in gastrointestinal parasites should illuminate our understanding of factors that affect anthelmintic potency and may lead to the development of synergistic compounds that increase the potency of anthelmintics currently in use. Use of detoxifying enzyme inhibitors in gastrointestinal parasitic nematodes has been shown to increase anthelmintic drug potency, suggesting that xenobiotic metabolism occurs in these parasites. In the cattle parasites Cooperia oncophora and Ostertagia ostertagi, the use of the cytochrome P450 inhibitor, piperonyl butoxide made the larvae of both strains more sensitive to anthelmintics suggesting that cytochrome P450s play a role in metabolizing thiabendazole and macrocyclic lactones such as Ivermectin (AlGusbi et al., 2014). The P-glycoprotein inhibitor, verapamil, increased the susceptibility of benzimidazole resistant strains to thiabendazole, and increased the susceptibility of O. ostertagi strains to Ivermectin (AlGusbi et al., 2014). In the sheep parasite, H. contortus, the use of UGT inhibitor, 5-nitrouracil was able to make naphtalophos resistant worms more sensitive in a Larval Developmental Assay (LDA), and also increased the sensitivity of wild type worms to this anthelmintic (Kotze et al., 2014). Hence, there is an increasing amount of data to suggest that xenobiotic responses play an important role in modulating drug sensitivity and resistance in parasitic nematodes.
The benzimidazoles (BZ) are a family of broad-spectrum anthelmintics used in livestock, and are one of the few drugs available for the control of helminth parasites in humans (Albonico et al., 2003; Kaplan, 2004; Kaplan et al., 2004; Wolstenholme et al., 2004; von Samson-Himmelstjerna and Blackhall, 2005; Gilleard, 2006; Osei-Atweneboana et al., 2007). Although mutations in the β-tubulin drug target are clearly important (Kwa et al., 1995; Grant and Mascord, 1996; Silvestre and Humbert, 2000; Winterrowd et al., 2003; Alvarez-Sanchez et al., 2005; de Lourdes Mottier and Prichard, 2008; Hodgkinson et al., 2008), there is some evidence that other mechanisms of resistance may occur. For example, natural field populations of C. elegans exist in which there are several major QTL underlying benzimidazole resistance in addition to the ben-1 locus (Zamanian et al., 2017, 2018). Further, different field isolates of H. contortus, which have the same frequency of the isotype-1 β-tubulin resistance mutations have very different levels of drug resistance as measured by egg hatch assay LD50s (von Samson-Himmelstjerna et al., 2009). Also, benzimidazole resistant H. contortus strains have been reported to produce increased amounts of glucosidated metabolite products (Cvilink et al., 2008a, 2008b; Vokral et al., 2012, 2013).
Benzimidazoles are good substrates for biotransformation in a large number of organisms from mammals to nematodes (Velik et al., 2005; Laing et al., 2010; Aksit et al., 2015). There is also some evidence that xenobiotic biotransformation plays a role in sensitivity and resistance of parasitic helminths to the benzimidazole drugs. In an in vitro assay that examined oxidation of albendazole, the resistant strains of F. hepatica have been shown to produce more of the oxidized product than sensitive strains (Solana et al., 2001). The liver fluke D. dendriticum was found to biotransform albendazole to albendazole sulphoxide, and mebendazole and flubendazole were either reduced, or reduced and methylated (Cvilink et al., 2009a, 2009b). F. hepatica metabolizes triclabendazole (TCBZ) into sulfoxide (TCBZ-SO) and sulphone (TCBZ-SO2), these metabolized products were found at higher concentrations in resistance strains (Solana et al., 2001; Mottier et al., 2004; Robinson et al., 2004). The cestode, Moniezia expansa, biotransforms flubendazole and mebendazole by reduction and albendazole was oxidized to albendazole sulfone and sulfoxide (Prchal et al., 2015). The parasitic nematodes, Ascaris suum and Haemonchus contortus, have also been shown to biotransform several benzimidazoles. In an in vitro assay, A. suum produced a reduced metabolite (Solana et al., 2001). H. contortus has been shown to produce a variety of benzimidazole metabolites that are either reduced, hydrolyzed and/or glucosidated (Cvilink et al., 2008a, 2008b; Vokral et al., 2012, 2013; Stuchlikova et al., 2018) with resistant strains producing more of these metabolized product than the sensitive strains (Vokral et al., 2012, 2013; Stuchlikova et al., 2018).
Investigation of the molecular processes involved in metabolism requires a model more tractable than parasitic nematodes, which are notoriously difficult to perform functional analysis on (Burglin et al., 1998; Geary and Thompson, 2001; Gilleard, 2004; Gilleard et al., 2005). This raises the question – is the free living nematode model, Caenorhabditis elegans, a good model for the study of anthelmintic metabolism of parasitic species of nematode (Gilleard, 2004; Sommer and Streit, 2011; Burns et al., 2015; Ward, 2015)? We previously found that C. elegans exposed to the benzimidazole, albendazole, resulted in up-regulation of several Phase I and Phase II metabolizing genes, and that albendazole is metabolized to an albendazole-hexose derivative (Laing et al., 2010). It has also been reported that there was a glucose metabolite produced and up-regulation of UDP-glucosyltranserase enzyme activity in resistant strains of H. contortus (Cvilink et al., 2008b; Vokral et al., 2013).
In this paper, we undertake a detailed comparison of the biotransformation and xenobiotic responses to five of the most important benzimidazole drugs used therapeutically in humans and animals; albendazole (ABZ), mebendazole (MBZ), thiabendazole (TBZ), oxfendazole (OxBZ) and fenbendazole (FBZ) (Supplemental Figure 1). We show that they are metabolized to similar products in both C. elegans and H. contortus by glucosidation, and metabolites are released from the worms into culture supernatants. Transcriptomic analysis shows that many Phase I and Phase II enzymes are up-regulated in C. elegans but not H. contortus when exposed to these benzimidazoles consistent with the glucosidation results. This work suggests that although benzimidazole biotransformation is biochemically similar in C. elegans and H. contortus it is less efficient, perhaps due reduced xenobiotic response efficiency, in the latter species.
2. Material and methods
2.1. Strains used
Using nematode strains containing mutations in the gene encoding the benzimidazole drug target, allows for acute anthelmintic exposures at very high concentrations to be used to induce xenobiotic responses without the worms being visibly adversely affected - having apparently normal movement and development. The C. elegans benzimidazole resistant strain CB3474 ben-1(e1880)III(Driscoll et al., 1989), was used for all drug exposure experiments. This strain is benomyl resistant, and is dominant at 25 °C and recessive at 15 °C. The H. contortus UGA/2004(Williamson et al., 2011), which is highly resistant to benzimidazoles, levamisole, and ivermectin, was to determine the transcriptional response to the benzimidazoles. The H. contortus MHco3 (ISE) (Otsen et al., 2001; Roos et al., 2004) inbred genome reference strain, and MHco12 (von Samson-Himmelstjerna et al., 2009) a benzimidazole resistant strain were used to detect metabolites. Worms were vigorously motile after drug exposure times used.
2.2. C. elegans culture and benzimidazole exposure experiments for the detection and characterization of metabolites
C. elegans stock strains were maintained on NGM agar plates with OP50 E. coli as the food source as described (Brenner, 1974). The C. elegans benzimidazole resistant strain CB3474 ben-1(e1880)III was cultured in CeHR3 axenic culture (Nass and Hamza, 2007) to ensure that the metabolites produced were the product of worm metabolism and not the bacteria normally used as food source. Eggs were isolated and sterilized using sodium hypochlorite/sodium hydroxide, L1s were cultured in 175 cm2 tissue flasks (BD Falcon # 353112) with CeHR3 media as the sole food source, supplemented with Antibiotic-Antimycotic (Gibco # 15240062), and incubated at 20 °C with 50 tilts per minute rocking, until a density of >10 worms/μL was achieved. Triplicate axenic cultures, of greater than 2.5 × 105 C. elegans CB3474 ben-1(e1880)III (Driscoll et al., 1989), were exposed to either albendazole (Sigma Cat# A4673), mebendazole (Sigma Cat# M2523), thiabendazole (Sigma Cat#T8904), oxfendazole (Sigma Cat#34176), fenbendazole (Sigma Cat# F5396) dissolved in DMSO and a 0.1% DMSO solvent only control, then added to axenic cultures to achieve a final concentration of 56.5 μM. The final DMSO concentration was 0.1% in all cultures. The cultures were incubated for 72 h at 20 °C with 50 tilts per minute rocking. A time course experiment was first performed with albendazole to determine the appropriate time of incubation required to detect metabolites in culture supernatants. Metabolites could be detected at low levels after as little as 3 h of culture but continued to increase in abundance until 72 h (Supplemental Figure 2). After incubation the samples were centrifuged at 3000xg for 5 min to pellet the worms. The supernatant was removed and snap frozen at −80 °C. The worm pellet was washed with 40 mL of M9 solution and centrifuged at 3000xg for 15 min. The wash supernatant was removed from the worm pellet and both were snap frozen at −80 °C. Samples were kept at −80 °C until used to prepare samples for HPLC and LC-MS/MS.
2.3. Culturing of H. contortus and anthelmintic exposure for detection and characterization of metabolites
The H. contortus reference strain MHco3(ISE) was used to detect metabolites produced upon exposure to benzimidazole anthelmintics. Two sheep were orally infected with a dose of 5000 third stage infective larvae. Twenty-eight days post-infection, the animals were euthanized and sheep abomasa were surgically removed, all experimental procedures described here were approved by the Moredun Research Institute Experiments and Ethics Committee and were conducted under the legislation of a UK Home Office License (reference PPL 60/03899) in accordance with the Animals (Scientific Procedures) Act of 1986. During worm counting and manipulation the H. contortus adults were held in pre-incubation media of RPMI-1640 medium (pH6.8, 38 °C) containing 10X antibiotic-antimycotic (Gibco # 15240062). Triplicate samples of 150 adult worms were incubated in 7.5 mL of incubation media consisting of RPMI-1640 (pH6.8), supplemented with 0.8% (w/vol) glucose, 10 mM HEPES, 1X antibiotic-antimycotic (Gibco # 15240062) in 25 cm2 flat bottomed tissue flasks (BD Falcon Cat #353108). At time zero, the benzimidazole anthelmintics: albendazole (Sigma Cat# A4673), mebendazole (Sigma Cat# M2523), thiabendazole (Sigma Cat#T8904), oxfendazole (Sigma Fluka 34176), fenbendazole (Sigma Cat# F5396) were added to a final concentration of 10 μM, and the flasks then incubated at 38 °C in humid atmosphere with 10% CO2 for 16 h, DMSO was 0.1% in all treatments. After 16 h of culture, adult worms were manually removed from media, washed in PBS and snap frozen at −80 °C. The culture supernatants were snap frozen at −80 °C, and kept at −80 °C until sample preparation for HPLC and LC-MS/MS.
2.4. Metabolite extraction from C. elegans and H. contortus culture supernatants
The C. elegans CeHR-3 culture supernatant was centrifuged at 4000 rpm (3400xg) on a Sorvall Legend RT for 10 min; 20 mL of the 50 mL C. elegans culture supernatant, or the entire 7.5 mL of H. contortus culture supernatant was filtered through a 0.22 μm filter (Pall Life Sciences Acrodisc 25 mm syringe filter) and applied to an activated and equilibrated C18 solid phase extraction column (SPE; Sep-Pak Plus C18 cartridge (Waters Cat. No. WAT020515); column was activated with 5 mL 100% acetonitrile and equilibrated with 10 mL dH2O). The SPE column was washed with 5 mL dH20. Metabolites and parental drug were eluted in 5 mL 100% acetonitrile (OmniSolv grade), collected in three fractions. All flow-through steps were collected and analyzed to ensure nothing was missed. All samples were initially analyzed by HPLC prior to LC-MS/MS analysis.
2.5. Metabolite extraction from pelleted C. elegans and H. contortus cultures
The worm pellet of each culture, as well as the wash of each worm pellet, was thawed on ice. A fraction of the pellet wash was used to re-suspend the respective worm pellet to a final volume of 10 mL and a 1-mL aliquot was removed for protein quantification and stored at −80 °C. The worm pellet and pellet wash were separated by centrifugation. Metabolites and parental drug extraction from the pellet wash was carried out as described for metabolite extraction from worm culture supernatant (using the entire pellet wash). The worm pellet was ground under liquid nitrogen and metabolites and parental drug were extracted in a total of 3 mL of methanol over two extraction steps. Samples were vortexed, sonicated (1–2 min), and centrifuged to remove worm debris (2-step centrifugation: 1) 5 min at 4000 rpm in the Sorvall Legend RT centrifuge and 2) supernatant was transferred to a 2-mL microtube and centrifuged at full speed in a microcentrifuge for 5 min). The supernatant was filtered using a 0.22 μm filter, and analyzed by HPLC and LC-MS/MS.
2.6. HPLC and LC-MS/MS analysis
HPLC analysis of SPE flow-through collections and elutions, as well as worm pellet extractions, were performed on a Waters 2795 Separations Module coupled with a Waters 2996 Photodiode Array Detector (PDA). Compounds present in a 5 μL injection (10 μL injection of pellet extract) were separated by a Sunfire C18 column (Waters; 3.5 μm × 4.6 mm x 150 mm; coupled with a Waters Sunfire C18 guard column, 3.5 μm × 4.6 mm x 20 mm) and a linear gradient of 80:20% water+1% acetic acid:acetonitrile to 100% acetonitrile over 10 min with a flow rate of 1 mL min−1. Compounds were detected by UV within a 200–400 nm range. Analysis was conducted using Waters Empower software. The first two elution fractions from the SPE for both culture supernatant and pellet wash extractions were combined and diluted 5 fold in dH2O. Metabolite extracts of pellet were diluted 5 fold with dH2O. Five μL was injected on to an Eclipse Plus C18 column (Agilent; 1.8 μM × 21 mm x 50 mm) using an Agilent 1200 series LC system (G1379B degasser, G1312B binary pump SL, G1367D HiP-ALS SL+, and G1316B TCC SL) together with an Agilent 6410 Triple Quad LC-MS/MS. The mobile phase and gradient was identical to the HPLC with a flow rate of 0.4 mL min−1. Samples were initially run using MS2 scan (TIC) in positive mode with a fragmentor set to 80 V. Once metabolites were identified, product ion scan was performed using the following masses for each drug treatment: For product ion scan, the fragmentor was set to 150 V, with collision energy of 30eV. Analysis was conducted using MassHunter software (Agilent Technologies).
2.7. Incubation of C. elegans in the presence of UDP-glucuronosyltransferase inhibitor chrysin
Chrysin is an UGT inhibitor of human UGT2B7, UGT1A1, UGT1A6, and UGT1A9 (Walsky et al., 2012). Triplicate axenic cultures of greater than 1.5 × 106 C. elegans benzimidazole drug resistant strain, CB3474 ben-1(e1880)III (Driscoll et al., 1989), were exposed to 56.6 μM albendazole for 3 days either with or without the UGT inhibitor, chrysin (Sigma Cat# C80105) at 200 μM. The relative amounts of the albendazole-glucose metabolite, with and without chrysin, were determined by HPLC by measuring the area under the 7.4min peak at 306 nm wavelength (previously determined to be the albendazole-glucose metabolite peak). These data were made relative to the total protein in culture replicates using a BCA protein assay (Sigma Cat# 1001-491004).
2.8. Preconditioned supernatant containing metabolized albendazole used to test changes in potency
In 24 well plates containing 400 μL of liquid NGM, Gentamycin and OP50 E. coli (0.75%w/vol) as food source, the C. elegans benzimidazole drug resistant strain, CB3474 ben-1(e1880)III (Driscoll et al., 1989), were exposed for 10 days at 20 °C to a 2 fold serial dose range from 64 μM to 62.5 nM plus DMSO negative control (this is four times the final drug doses used in motility bioassay). 100 μL of this pre-conditioned supernatant along with 300 μL fresh NGM, containing Gentamycin, and OP50 E. coli (0.75% w/vol) and first larval stage (L1) of C. elegans N2 wild-type worms at final concentration of 1.5 L1/μL were aliquoted into each well, to achieve the standard drug curve range (16 μM–15.625 nM plus DMSO Negative control). To ensure that ten days incubation at 20 °C does not decrease the potency of albendazole, an additional control of four times the standard drug dose of albendazole in culture medium without worms for 10 days was used, which was then diluted to the standard drug curve range and used in the bioassay. To ensure that albendazole was not sticking to the cuticle of the worms during the pre-incubation step, we exposed dead worms to four times the standard drug dose overnight. A control of fresh albendazole exposed to C. elegans N2 was used as comparison.
2.9. Digital analysis of C. elegans motility bioassay in response to albendazole anthelmintic exposure
Worm motility was used as a measure of drug potency. First stage larvae at a density of 1.5 L1/μL were cultured in 400 μL in multiwell plate, in liquid NGM with OP50 E. coli as the food source along a standard drug curve range. A standard drug curve range for albendazole results in paralysed worm at the higher drug doses and fully motile worms at the lower drug doses, this was previously found to be a 2-fold serial dilution of albendazole from 16 μM to 15.625 nM plus a negative DMSO control. Worms were cultured at 20 °C, rocking for 60 h to the early adult stage. After 60 h these culture plates were then placed in the Larval Motility Digital Analyzer (LaMDA), which uses a scanning digital camera to scan the wells, pixel data from sequential picture frames are taken every 300 ms, for 3 s and are assessed for changes in greyscale for every pixel coordinate in the field of view and converted to Mean Squared Error (MSE) (Lee, 1980; Du, 2013) as a measure of total movement and degree of paralysis of the worms. MSE values are calculated according to the equation: MSE = ijΣ (aij–bij)2/(i · j), where i and j are the pixel coordinates and a and b are sequential picture frames. This gives a numerical value for total movement in a well, the MSE values were made relative to the DMSO negative control.
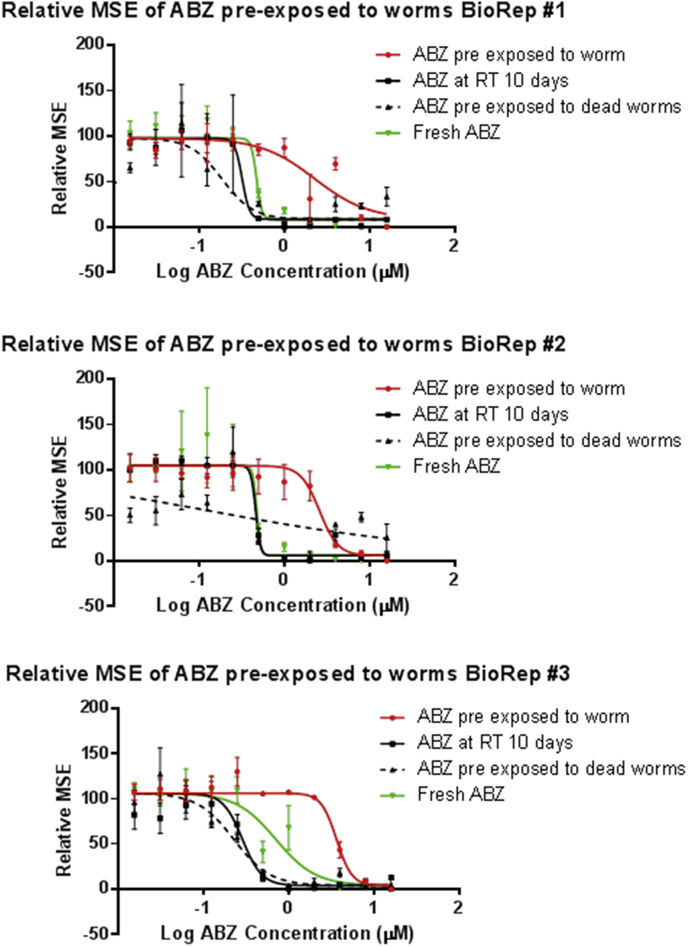
Three biological replicates in triplicate technical replicates were analyzed by non-linear regression analysis of the drug dose curves, using GraphPad Prism version 6.01, 4 parameter non-linear regression with top and base constrained. For one of the biological replicates, the 0.25 μM dose of ABZ exposed to dead worms did not receive the drug and this data point was removed from the analysis. Extra sum of square F test was used to analyse whether the logEC50 of fresh albendazole versus albendazole pre-exposed to live worms was significantly different at (p < 0.0001) in each of the biological replicates tested when the albendazole is pre-exposed to worms.
2.10. Transcriptional response to benzimidazole drug exposures
2.10.1. C. elegans culturing and drug exposure
C. elegans stock strains were maintained on NGM agar plates with OP50 E. coli as the food source as described (Brenner, 1974). Three biological replicates of approximately 20,000 L1 larvae C. elegans CB3474 ben-1(e1880)III were cultured in NGM media with E. coli OP50 at 1% (w/vol) at 20 °C for approximately 70 h until the adult stage, concentrations were kept consistent with micro-array experiment performed previously (Laing et al., 2010). Cultures were exposed to 1.13 mM of either albendazole (Sigma Cat# A4673), mebendazole (Sigma Cat# M2523), thiabendazole (Sigma Cat#T8904), oxfendazole (Sigma Cat#34176), fenbendazole (Sigma Cat# F5396) dissolved in DMSO as well as a DMSO negative control solution of for 4 h, care was taken to ensure that DMSO concentration were less than 1%. The worms were then harvested by centrifugation at 300xg for 3 min, no brake, and cleaned on a sucrose gradient, the supernatant was decanted and the pellet stored in TRIzol (Invitrogen # 15596-018) and immediately frozen at −80 °C until processing.
2.10.2. Adult stage of H. contortus culturing and drug exposure
The triple resistant H. contortus UGA/2004 strain (Williamson et al., 2011) was used to determine the transcriptional response to benzimidazole exposure. Two sheep per strain were orally infected with a dose of 5000 third stage infective larvae. Twenty-eight days post-infection, the animals were euthanized, sheep abomasa were surgically removed, and the adult H. contortus collected. All animal experimental procedures described here were carried out in accordance with Agriculture & Agrifood Canada (AAFC, Lethbridge, AB, Canada) and Animal Care Committee (ACC) protocol #1613.
During worm counting and manipulation the H. contortus adults were held in pre-incubation media of RPMI-1640 medium (pH6.8, 38 °C) containing 10X antibiotic-antimycotic (Gibco # 15240062). Triplicate samples of 50 adult worms were incubated in 7.5 mL of incubation media consisting of RPMI-1640 (pH6.8), supplemented with 0.8% (w/vol) glucose, 10mMol HEPES, 1X antibiotic-antimycotic (Gibco # 15240062) in 25 cm2 flat bottomed tissue flasks (BD Falcon Cat #353108). At time zero, worms were exposed to either DMSO solvent control or albendazole (Sigma Cat# A4673), mebendazole (Sigma Cat# M2523), thiabendazole (Sigma Cat#T8904), oxfendazole (Sigma Fluka 34176), fenbendazole (Sigma Cat# F5396) were added to a final concentration of 1.13 mM, and the flasks then incubated at 38 °C in humid atmosphere with 10% CO2 for 4 h. Worms were then picked from culture into Eppendorf tubes and stored in TRIzol (Invitrogen # 15596-018) and immediately frozen at −80 °C until processing.
2.10.3. H. contortus infective larvae (iL3) drug exposure
Quadruplicate samples of 1.75 × 105 H. contortus UGA/2004 strain were exsheathed with 0.2% sodium hypochlorite for 10–15 min and washed with PBS. Exsheathed larvae were cultured in Earles Buffered Saline Solution with either DMSO or benzimidazoles to final concentration 1.13 mM for 4 h to keep consistent with C. elegans exposures, care was taken to ensure that DMSO concentration were less than 1%. The worm strains were then harvested by centrifugation at 300xg for 3 min, no brake, washed and the supernatant was decanted and the pellet stored in TRIzol (Invitrogen # 15596-018) and immediately frozen at −80 °C until processing.
2.11. RNA extraction
RNA was extracted using TRIzol as described by the manufacturer protocol (Invitrogen # 15596-018); total RNA pellets were suspended in DEPC treated water. RNA was isolated and treated with DNaseI as per manufacturer protocol (Qiagen #74134). Only samples that had a minimum A260/A280 ratio of 1.7 and a RIN of greater than 8.0 were used. Total RNA extracted and sent to Genome Quebec [http://gqinnovationcenter.com] for template preparation and RNA-Seq Next Generations Sequencing on the Illumina HiSeq platform with PE100 to read depth of >50,000,000 aligned reads.
2.12. RNA-seq analysis
Transcripts were quantified using Kallisto 0.46.0 (Bray et al., 2016) (default settings) with the Haemonchus contortus (PRJEB506, WBPS13) or Caenorhabditis elegans (PRJNA13758, WS271) transcript coding sequences (CDS). All statistical analysis was done with R 3.6.0. Gene level analysis was done by first summarizing the transcripts to the gene level using tximport version 1.12.3 (Soneson et al., 2015) and then detecting differentially expressed genes with DESeq2 version 1.24.0 (Love et al., 2014), with an adjusted p-value cutoff of 0.05 for H. contortus samples and 0.01 for C. elegans samples. A stricter cutoff was used for C. elegans due to the large number of differentially expressed genes. Principal component analysis was done using the “plotPCA” function from the DESeq2 package. Functional analysis (over-representation test) for the H. contortus larva samples was done with the clusterProfiler (Yu et al., 2012) R package version 3.12.0 using the GO annotations from the PRJEB506 assembly.
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE130466 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE130466).
3. Results
3.1. Similar biotransformation products of benzimidazole drug family members are released into in vitro culture supernatants by C. elegans and H. contortus
We investigated the biotransformation and export of each of five major benzimidazole drug family members in C. elegans and H. contortus by assaying axenic in vitro culture supernatants of each species for the presence of both parental drug and metabolites using LC-MS/MS (Fig. 1, Fig. 2, Fig. 3, Fig. 4, Table 1). In each experiment, a control culture containing drug but no worms, and a control containing worms but no drug were included to ensure any benzimidazole metabolites detected were the product of nematode biotransformation. In the case of C. elegans, triplicate axenic cultures (Nass and Hamza, 2007) of approximately 1.5 × 106 C. elegans CB3474 ben-1(e1880)III were exposed to benzimidazoles: albendazole (ABZ: 225 m/z), mebendazole (MBZ; 295 m/z), fenbendazole (FBZ; 299 m/z), thiabendazole (TBZ; 201 m/z), and oxfendazole (OxBZ; 315 m/z) at 56.5 μM for 72 h; the time of culture being chosen on the basis of previous pilot time course experiments (Supplemental Figure 2) no additional compounds were found and the amount of compounds are comparable at time points, indicating the newly synthesized compounds are relatively stable, 72 h was chosen to better detect rare metabolites. In the case of H. contortus, triplicate cultures of 150 MHco3(ISE) adult worms were exposed to 10 μM of each benzimidazole for 16 h; the time of culture being limited by the viability of the adults worms in culture outside the host and the concentration chosen based on previous work (Cvilink et al., 2008b). As there was no evidence of a significant reduction of substrate in drug-exposure experiments using H. contortus, it is unlikely that any major biotransformed compounds other than those reported (Fig. 1, Fig. 2, Fig. 3, Fig. 4, Table 1) are produced by H. contortus.
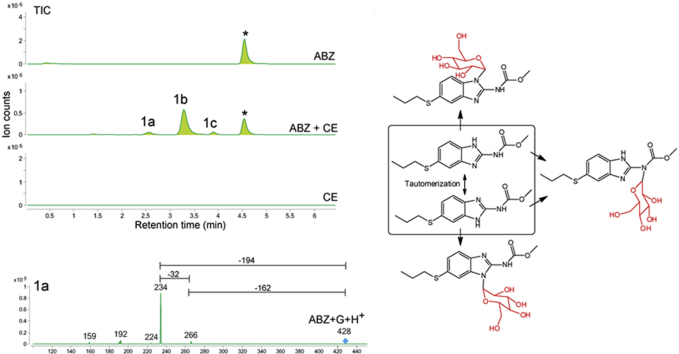
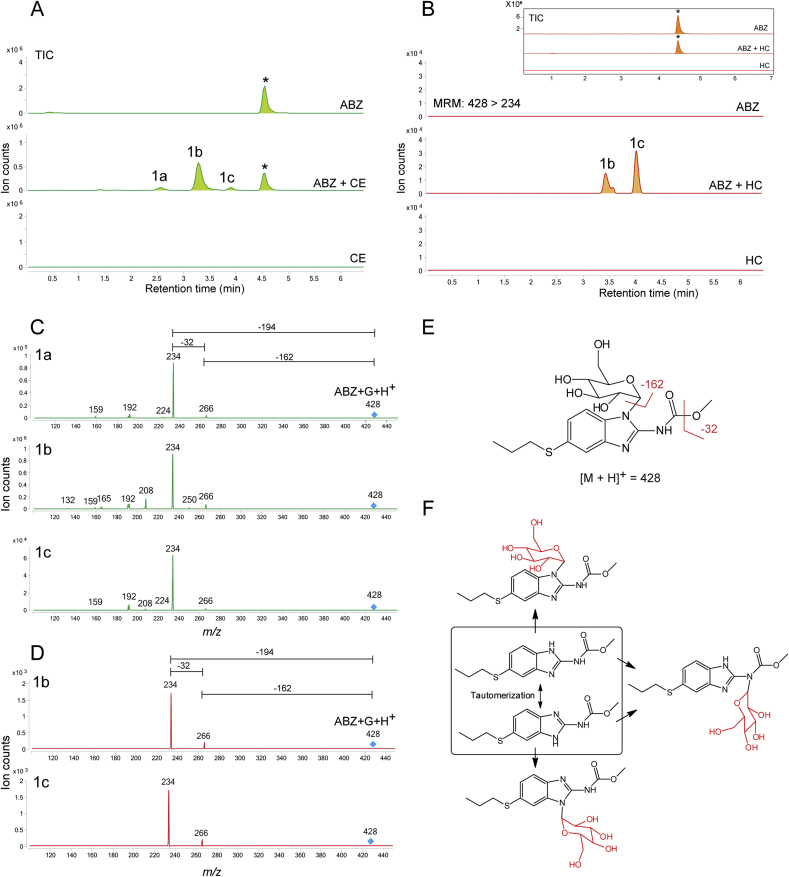
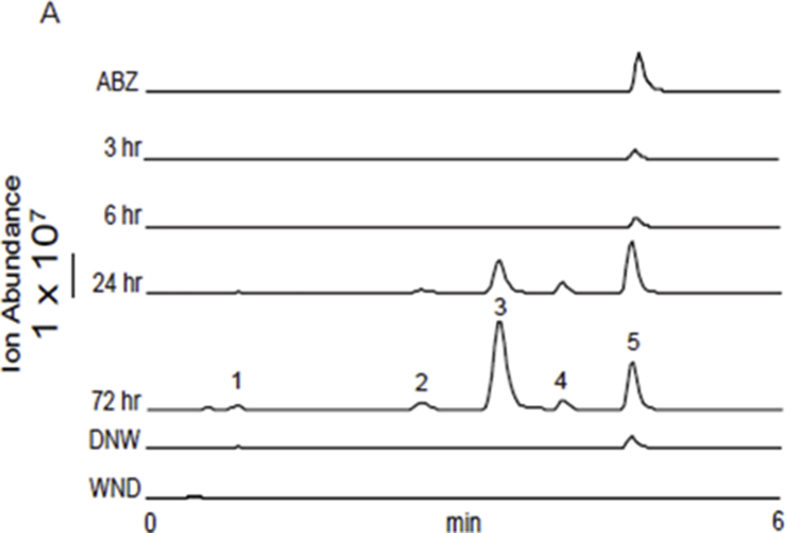
Fig. 1.
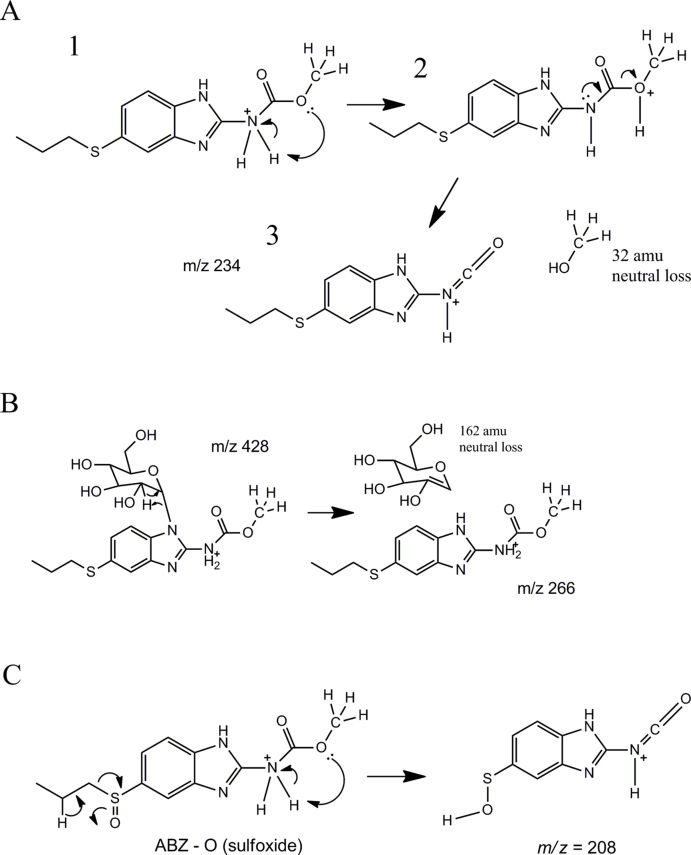
Chromatograms are the result of product total ion current (TIC), or a multiple reaction monitoring (MRM), of albendazole (ABZ) parental drug and its metabolites identified in the supernatant of cultures of either C. elegans benzimidazole resistant allele CB3474 ben-1(e1880)III (shown in green) or adult H. contortus drug sensitive MHco3(ISE) allele (shown in orange). The first (top) chromatograms corresponds to the culture containing benzimidazole but no nematodes, the second chromatograms corresponds to the culture containing either C. elegans or H. contortus, respectively, exposed to albendazole (ABZ), the third chromatogram corresponds to the control culture containing C. elegans or H. contortus but with no benzimidazole. C. elegans was exposed to 56.5 μM BZ for 3 days, and H. contortus was exposed to 10 μM BZ for 16 h respectively. A. TIC chromatogram of C. elegans exposed to albendazole (ABZ), B. MRM chromatogram of H. contortus exposed to albendazole (ABZ). TIC chromatogram is shown in an insert. C. Metabolites and their LC-MS/MS fragmentation patterns identified in supernatant of the ABZ-treated C. elegans cultures (as indicated by the numbered labels on the second trace of panels A and B). D. Metabolites and their LC-MS/MS fragmentation patterns identified in supernatant of the ABZ-treated adult H. contortus cultures. The fragmentation patterns were extracted from either a Total Ion Current (TIC) of ABZ-treated C. elegans culture supernatant, or multiple reaction monitoring (MRM) of H. contortus culture and resulted from a fragmentor setting of 150 V and a collision energy of 30eV. E. Two diagnostic daughter ions derived from the m/z 428 ion were m/z 234 and m/z 266 of the fragmentation data. F. Three proposed isoforms formed by N-linked sugar, the most likely structures of these three isomers. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article).
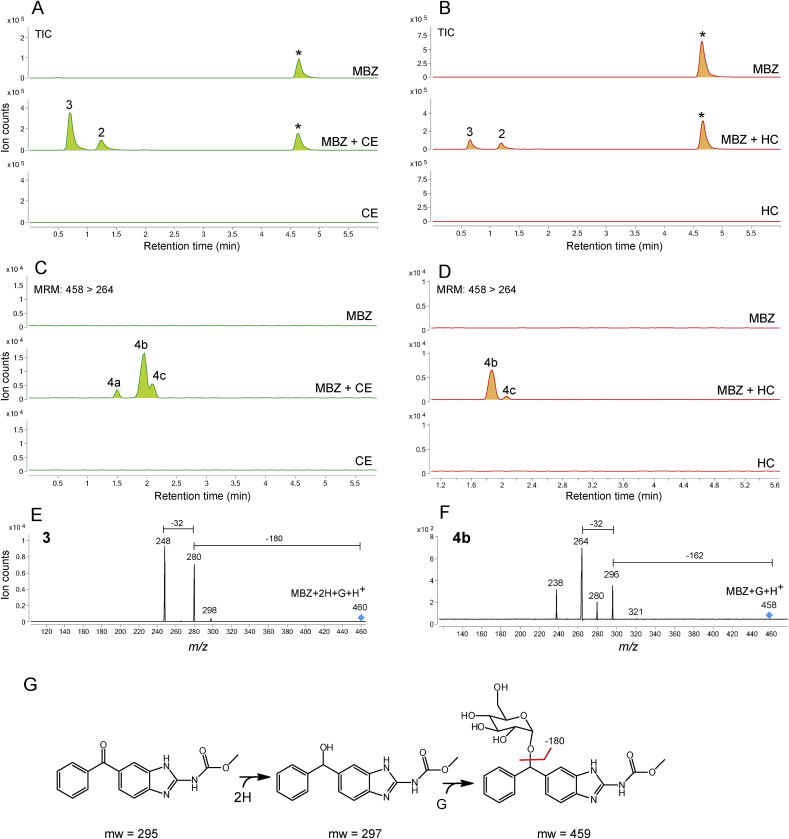
Fig. 2.
Chromatograms are the result of product Total Ion Current (TIC) or multiple reaction monitoring (MRM) of mebendazole (MBZ) parental drug and its metabolites identified in the supernatant of cultures of C. elegans benzimidazole resistant allele CB3474 ben-1(e1880)III (shown in green) or of adult H. contortus drug sensitive MHco3(ISE) allele (shown in orange). The first (top) chromatogram corresponds to the culture containing benzimidazole but no nematodes, the second chromatogram corresponds to the culture containing either C. elegans or H. contortus respectively, exposed to the benzimidazole, and the third chromatogram corresponds to the control culture containing C. elegans or H. contortus but with no benzimidazole. C. elegans was exposed to 56.5 μM BZ for 3 days, and H. contortus was exposed to 10 μM BZ for 16 h respectively. A. TIC chromatogram of C. elegans exposed to mebendazole (MBZ), B. TIC chromatogram of H. contortus exposed mebendazole (MBZ), C. Multiple reaction monitoring (MRM) chromatogram of C. elegans exposed to mebendazole (MBZ), D. Multiple reaction monitoring (MRM) chromatogram of H. contortus exposed to mebendazole (MBZ). Metabolites and their LC-MS/MS fragmentation patterns identified in supernatant of the MBZ-treated C. elegans or adult H. contortus cultures. These fragmentation patterns were extracted from a Total Ion Current (TIC) or multiple reaction monitoring (MRM) and resulted from a fragmentor setting of 150 V and a collision energy of 30eV. E. Metabolite 3 and its LC-MS/MS fragmentation pattern. F. Metabolite 4b and its LC-MS/MS fragmentation pattern. Two diagnostic daughter ions derived from the m/z 428 ion were m/z 234 and m/z 266 of the fragmentation data. G. Proposed sequential reaction of reduction and O-glycosylation to the phenyl ketone moiety in MBZ. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article).
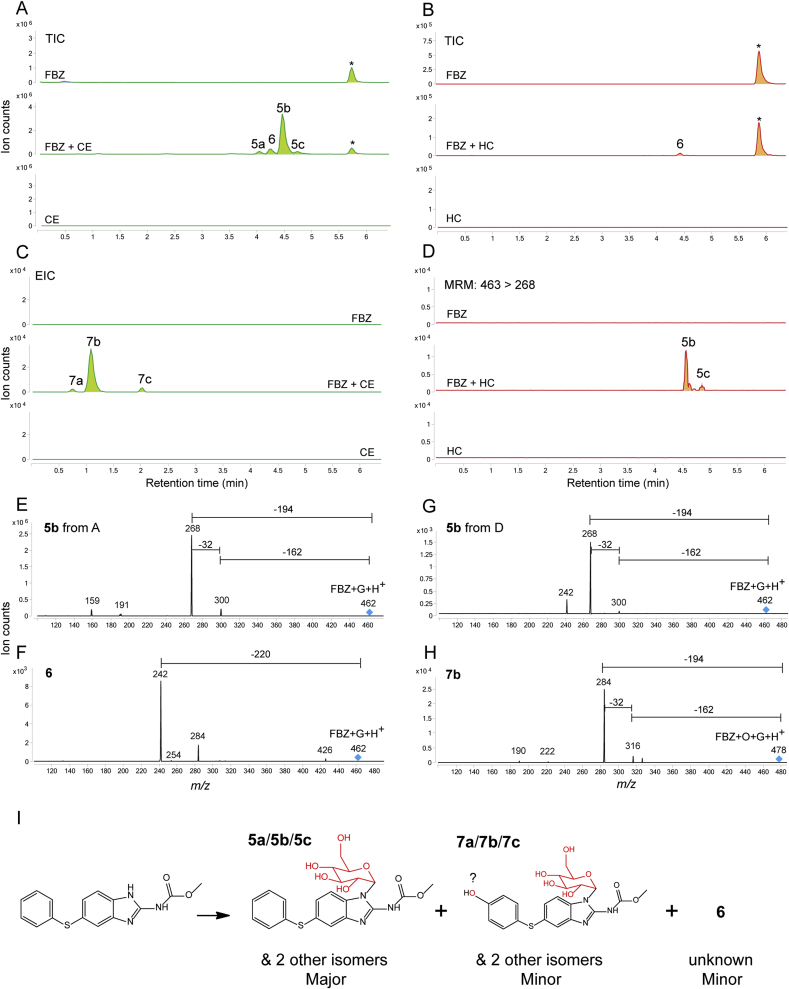
Fig. 3.
Chromatograms are the result of product Total Ion Current (TIC) or multiple reaction monitoring (MRM) of fenbendazole (FBZ) parental drug and its metabolites identified in the supernatant of cultures of C. elegans benzimidazole resistant allele CB3474 ben-1(e1880)III (shown in green) or of adult H. contortus drug sensitive MHco3(ISE) allele (shown in orange). The first (top) chromatogram corresponds to the culture containing benzimidazole but no nematodes, the second chromatogram corresponds to the culture containing either C. elegans or H. contortus respectively, exposed to the benzimidazole, and the third chromatogram corresponds to the control culture containing C. elegans or H. contortus but with no benzimidazole. C. elegans was exposed to 56.5 μM BZ for 3 days, and H. contortus was exposed to 10 μM BZ for 16 h respectively. A. TIC scan of C. elegans exposed to fenbendazole (FBZ), B. TIC scan of H. contortus exposed to fenbendazole (FBZ), C. An extracted-ion chromatogram (EIC) of C. elegans exposed to fenbendazole (FBZ), D. MRM scan of H. contortus exposed to fenbendazole (FBZ). Metabolites and their LC-MS/MS fragmentation patterns identified in supernatant of the FBZ-treated C. elegans or adult H. contortus cultures. These fragmentation patterns were extracted from a Total Ion Current (TIC) or multiple reaction monitoring (MRM) and resulted from a fragmentor setting of 150 V and a collision energy of 30eV. E. Metabolite 5b from C. elegans and its LC-MS/MS fragmentation pattern, F. Metabolite 6 and its LC-MS/MS fragmentation pattern, G. Metabolite 5b from H. contortus and its LC-MS/MS fragmentation pattern, H. Metabolite 7 and its LC-MS/MS fragmentation pattern. I. A schematic summary of FBZ oxidative biotransformation is shown. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article).
Fig. 4.
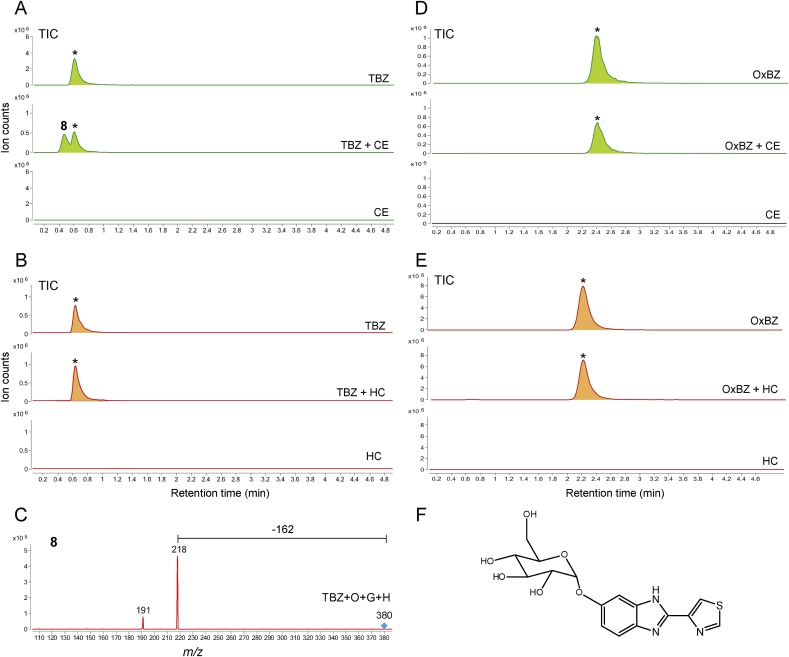
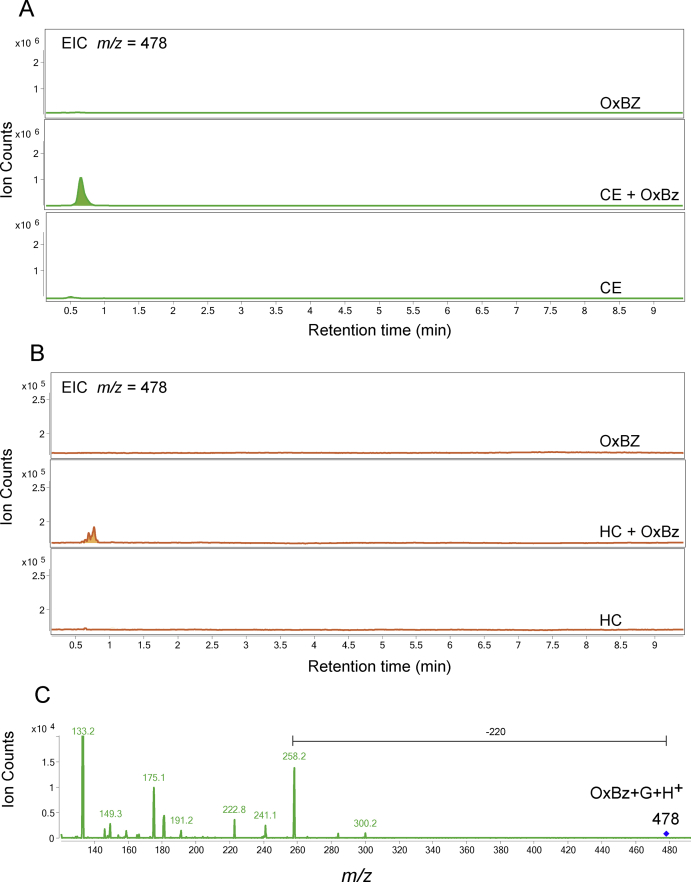
Chromatograms are the result of product Total Ion Current (TIC) of thiabendazole (TBZ) or oxfendazole (OxBZ) parental drug and its metabolites identified in the supernatant of cultures of C. elegans benzimidazole resistant allele CB3474 ben-1(e1880)III (shown in green) or of adult H. contortus drug sensitive MHco3(ISE) allele (shown in orange). The first (top) chromatogram corresponds to the culture containing benzimidazole but no nematodes. The second chromatogram corresponds to the culture containing either C. elegans or H. contortus respectively, exposed to the benzimidazole. The third chromatogram corresponds to the control culture containing C. elegans or H. contortus but with no benzimidazole. C. elegans was exposed to 56.5 μM BZ for 3 days, and H. contortus was exposed to 10 μM BZ for 16 h respectively. LC-MS/MS fragmentation patterns identified in supernatant of the TBZ-treated C. elegans or adult H. contortus cultures. These fragmentation patterns were extracted from a Total Ion Current (TIC) and resulted from a fragmentor setting of 150 V and a collision energy of 30eV. A. TIC scan of C. elegans exposed to thiabendazole (TBZ), B. TIC scan of H. contortus exposed to thiabendazole (TBZ), C. Metabolite 8 and its LC-MS/MS fragmentation pattern, D. TIC scan of C. elegans exposed to oxfendazole (OxBZ), E. TIC scan of H. contortus exposed to oxfendazole, F. Proposed structure of metabolite 8. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article).
Table 1.
Summary of metabolites produced by C. elegans CB3474 ben-1(e1880)III and H. contortus drug sensitive MHco3(ISE) as determined by LC-MS/MS analysis shown in Fig. 1, Fig. 2, Fig. 3, Fig. 4. C. elegans were cultured with each benzimidazole drug at 56.5 μM for 3 days in axenic culture. H. contortus adult worms were cultured in RPMI-1640 plus supplements with 10 μM of each benzimidazole for 16 h at 38 °C (10% CO2).
|
C. elegans metabolites |
H. contortus metabolites |
|||||
|---|---|---|---|---|---|---|
| Parental | Primary product | Secondary product | Tertiary product | Primary product | Secondary product | Tertiary product |
| ABZ | ABZ+hexose | ABZ+Oa | ABZ+O | ABZ+hexosea | ||
| MBZ | MBZ+hexose+2H | MBZ+2H | MBZ+hexosea | MBZ+hexose+2H | MBZ+2H | MBZ+hexosea |
| FBZ | FBZ+hexose | FBZ+O+hexose+Ha | FBZ+hexosea | Not found | ||
| TBZ | TBZ+O+hexose+Ha | Not found | ||||
| OxBZ | OxBZ+hexose+Ha | OxBZ+hexose+Ha | ||||
Trace amounts.
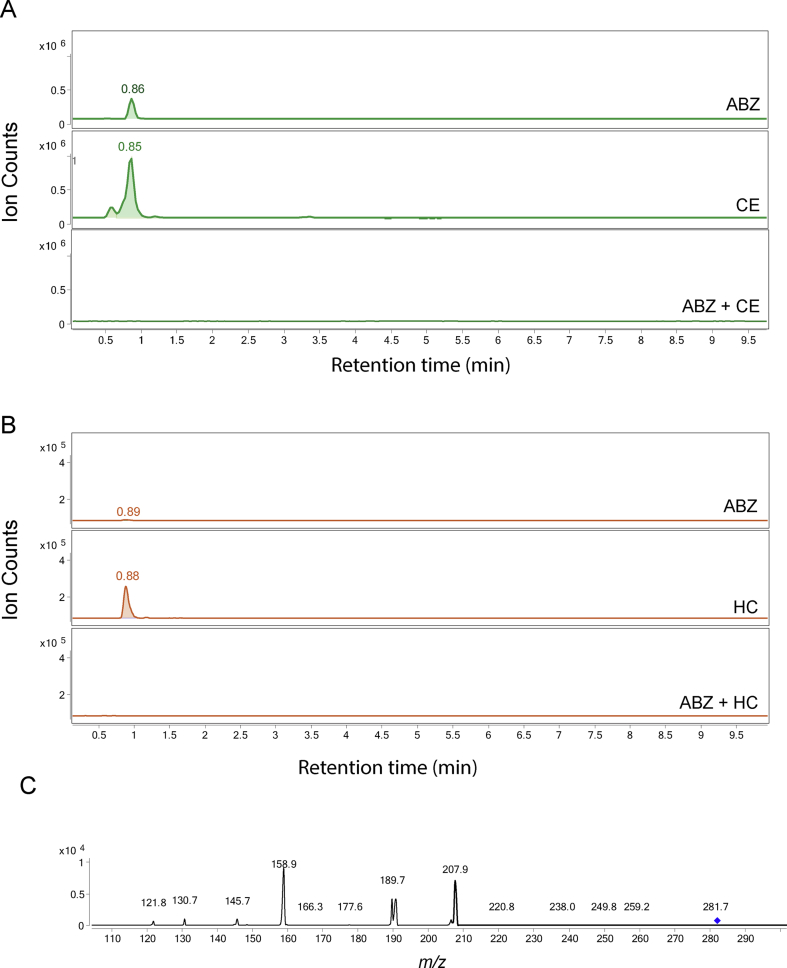
Three major new compounds of [M+H]+ ion at m/z 428 were detected when ABZ was added to C. elegans cultures (Fig. 1A, peaks 1a-1c), and the mass increase (+162) from ABZ indicated an addition of a hexose sugar. The MS/MS fragmenting pattern of these compounds were identical (Fig. 1C), implying they are structural isomers. Two diagnostic daughter ions derived from the m/z 428 ion were m/z 234 and m/z 266, which can be explained by neutral loss of the methoxide and the sugar linked to one of three nitrogen atoms, respectively (Fig. 1E). These neutral losses are common and predictable, and the mechanism of methoxide and N-linked sugar loss is shown in (Supplemental Figure 3A/B). Based on these observations and possible three isoforms formed by N-linked sugar, the most likely structures of these three isomers are proposed in (Fig. 1F). The same bio-transformation of ABZ was also previously reported (Laing et al., 2010). When the similar ABZ exposure experiments were performed in the parasite H. contortus, three sugar-conjugated ABZs were barely detectable in total ion current (TIC) scan analysis. However, using a sensitive multiple reaction monitoring (MRM) that traces a specific parent and a daughter ion (428 → 234), two new compounds could be identified (Fig. 1B peaks 1b and 1c), of which the MS/MS patterns were the same as those from C. elegans (Fig. 1D). In addition to the sugar-conjugated ABZ, an oxygen-added ABZ, [ABZ+O], was identified in trace amounts in both worms by a targeted ion search using m/z 282. Although the oxygenated ABZ was also found in the ABZ-control without worms, co-incubation of worms with ABZ increased the production of oxygenated ABZ by 4.5-fold in C. elegans and by 34-fold in H. contortus (Supplemental Figure 4A/B). It appears that H. contortus produces relatively more oxygenated ABZ than C. elegans because ABZ is not efficiently glycosylated in H. contortus, thus providing more substrate for oxygenation. Similar H. contortus metabolites have previously been reported (Cvilink et al., 2008b; Laing et al., 2010) (ABZ with S O) by a characteristic daughter ion m/z 208 (Supplemental Figure 3C; Supplemental Figure 4C). Recent work from Stuchlikova et al. used UHPLC/MS/MS and identified additional novel ABZ metabolites in H. contortus: ABZ-sulphone, two N-glycosides with a hydrolyzed carbamate side chain, and two ABZ.SO-N-glycosides (Stuchlikova et al., 2018).
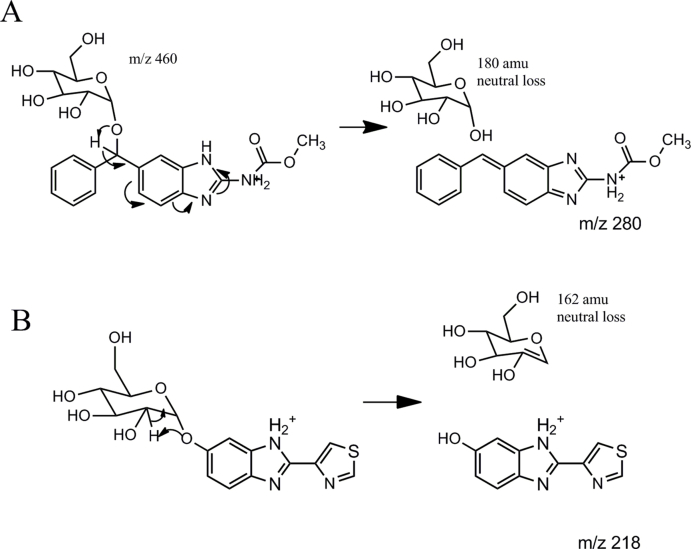
In MBZ exposure assays, two new major compounds were produced from both C. elegans and H. contortus (Fig. 2A/2B). The compound 2 showed a [M+H] ion of m/z 298 while the compound 3 showed a [M+H] ion at m/z 460, indicating a 2 mass increase for 2 and an additional 162 mass increase for 3. The MS/MS analysis of 3 showed a 32 neutral loss as shown in ABZ but also showed a 180 mass neutral loss. The latter is a typical MS/MS fragmenting pattern for O-linked glycosylation on non-aromatic carbon (Fig. 2E) and the mechanism is shown in Supplemental Figure 5A. To explain the observed data, we propose the reaction scheme shown in Fig. 2G after considering structural differences between ABZ and MBZ. As the phenyl ketone group distinguishes MBZ from ABZ, the reduction and glycosylation should occur in the phenyl ketone group of MBZ. In this transformation, a reduction of the MBZ phenyl ketone group firstly occurred, followed by a hexose sugar conjugation to the newly formed hydroxyl group. Additional to these compounds, the new compounds with the MS/MS pattern (32 and 162 neutral losses) identical to that of the sugar-conjugated ABZ were also detected by MRM (Fig. 2C/2D/2F peaks 4a-4c), but these compounds (MBZ+hexose) were present significantly lower (~1%) than the compound 2 and 3 (MBZ+2H and MBZ+2H+hexose). These results demonstrated that a sequential reaction of reduction and O-glycosylation is a dominant metabolic route for MBZ in both C. elegans and H. contortus. This reaction is due to the unique phenyl ketone moiety in MBZ.
In FBZ exposure assays, the hexose sugar-conjugated products with 32 and 162 neutral losses in MS/MS were found in C. elegans (Fig. 3A/3E peaks 5a-5c) and were also detected in H. contortus only by MRM (Fig. 3D/3G peaks 5b and 5c). These are the compounds analogous to 1a-1c when ABZ was exposed to C. elegans. A second species of sugar-conjugated FBZ (Fig. 3A/3B peak 6) was detected in both C. elegans and H. contortus, but their MS/MS patterns do not match to the known MS/MS patterns for sugar-conjugated compound (i.e., 162 or 180 mass loss; Fig. 3F). Another type of sugar-conjugated FBZ was detected with typical 32 and 162 mass losses (Fig. 3C/3H peaks 7a-7c) only in C. elegans, but they showed a 16 mass increase in addition to a sugar conjugation. This mass increase suggested an oxygen addition as well as a hexose sugar, indicating that the detected molecule is a [FBZ+O+hexose+H]+ion. As the phenyl sulfide group is unique in FBZ, we considered the formation of phenyl sulfoxide (phenyl S O) to produce OxBZ from FBZ, but OxBZ could not be detected in FBZ exposed C. elegans, indicating that an oxygen atom was not added to sulfur but likely to be added to the phenyl ring. These data suggested that the major metabolized species for FBZ in C. elegans are N-linked glycosylated products (5a-5c) similar to those from ABZ. The compounds 6 and 7a-7c are minor products, and their chemical identities remain unknown.
In TBZ exposure assays, a new compound 8 with [M+H]+ ion at m/z 380 was detected in C. elegans (Fig. 4A), whereas no biotransformation of TBZ was detected in H. contortus (Fig. 4B). The mass increase of 178 from TBZ, together with the MS/MS pattern (Fig. 4C), suggested that an oxygen (16 mass) and a hexose sugar (162 mass) were added to TBZ to form [TBZ+O+hexose+H]+ ion. TBZ derivatives attached with a hexose sugar alone (i.e., [M+sugar+H]+ with m/z 364 ion) could not be detected. Therefore, different from ABZ, MBZ, and FBZ, nitrogen atoms in TBZ are not glycosylated, and an oxygen addition to TBZ is required for the next hexose conjugation. The neutral loss of 162 mass indicated that the sugar is conjugated to the oxygen attached to the aromatic ring, and it mechanism is shown in (Supplemental Figure 5B). Taken together, a possible structure for 8 is shown in Fig. 4F. The same TBZ bio-transformation pattern was recently reported (Jones et al., 2015).
When OxBZ was exposed to either C. elegans or H. contortus, no major compound metabolized from OxBz were initially detected in both worms (Fig. 4D/4E), indicating OxBZ is not efficiently metabolized in worms. Further targeted ion searches for glycosylated OxBZ (M+hexose+H; m/z = 478) identified trace amounts of glycosylated OxBz in both worms (Supplemental Figure 6A/B). However, as was the case shown in FBZ (Fig. 3F), the MS/MS pattern of this compound does not match to typical MS/MS patterns of sugar-conjugated compound (i.e., 162 or 180 mass loss; Supplemental Figure 6C) and it structure remains unknown.
We examined whether the glycosylated drugs accumulate inside worms after grinding C. elegans in methanol. LC-MS analysis showed that the worm extracts accumulate the glycosylated drugs from five drugs less than 5% of those found in medium. We concluded that glycosylated drugs are efficiently secreted to medium.
A summary of the described metabolites of C. elegans and H. contortus are presented in Table 1 and highlights several things of interest. Firstly, similar metabolites are produced by these two nematodes, with two exceptions (i.e.: FBZ+hexose+O and TBZ+O+hexose+H), where these metabolites were found in C. elegans in trace amounts only. Secondly, as illustrated from Table 1, biotransformation of the BZs with a hexose or a hexose plus other moiety is a common reaction for all the drugs tested. A BZ drug resistant strain of H. contortus (MHco12) (von Samson-Himmelstjerna et al., 2009) was compared to the drug sensitive ISE strain to determine whether there were any differences in the amount of any of the metabolites produced. No significant differences were found and so this was not investigated further (data not shown).
3.2. Exposure of albendazole to live C. elegans worms decreases the potency of the anthelmintic
We tested whether the anthelmintic albendazole is modified to a less potent product by pre-exposing the anthelmintic to the benzimidazole resistant strain C. elegans CB3474 ben-1(e1880)III for 10 days and then testing the pre-conditioned supernatant which contains the metabolized albendazole, alongside several controls, in a motility bio-assay on wild type N2 C. elegans. To ensure that albendazole does not lose potency after ten days incubation at 20 °C, we included a control which contained albendazole in culture medium without worms for ten days. Another control, to ensure that albendazole was modified by the worms rather than removed from the experiment due to absorption to the worm's cuticle during the pre-incubation step, involved albendazole exposed to dead worms. Finally, fresh albendazole exposed to C. elegans N2 was used as a negative control and as comparison. In all three bioreplicates the dose response curve was shifted to the right for the medium that had been pre-exposed to the C. elegans cultures relative to the various controls, suggesting that C. elegans modifies the albendazole to a less potent product (Fig. 5).
Fig. 5.
The plots show the total movement of N2 Bristol C. elegans (relative MSE) versus log concentration ABZ. Three biological replicates of C. elegans N2 were exposed to supernatants containing different concentrations of ABZ drug (16 μM–0 μM) for 60 h. Red Line, ABZ pre-exposed to C. elegans CB3474 ben-1(e1880)III for 10 days; Green line, fresh ABZ; Dashed line, ABZ pre-exposed to dead worms for 10 days; black line, ABZ at room temperature (RT) for 10 days. Extra sum of square F test was used to analyse whether the logEC50 of fresh albendazole versus albendazole pre-exposed to live worms was significantly different at (p < 0.0001) Bio#1, F = 31.63 (1,62); Bio#2 F = 19.43 (1,62); Bio#3 F = 59.98, (1,62). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article).
3.3. The UDP-glucuronosyltransferase inhibitor chrysin reduces albendazole biotransformation by C. elegans in culture
A toxicity assay was performed with several pharmacological inhibitors- piperonyl butoxide (a CYP inhibitor), T-cinnaminic acid (a GST inhibitor), verapamil (a PGP inhibitor), and chrysin (a UGT inhibitor) - to determine the highest dose that did not affect C. elegans development or brood size. Doses of 100 μM, 200 μM, 100 μM, 200 μM respectively, did not produce a toxic effect on worms (data not shown). The effect on the abundance of metabolite produced on single biological replicates with each of these inhibitors was then performed using LC LC-MS/MS; these results suggested that only chrysin (UGT inhibitor) caused a decrease in the amount of metabolite produced (data not shown). Therefore, the effect of chrysin on metabolite production was chosen for further analysis.
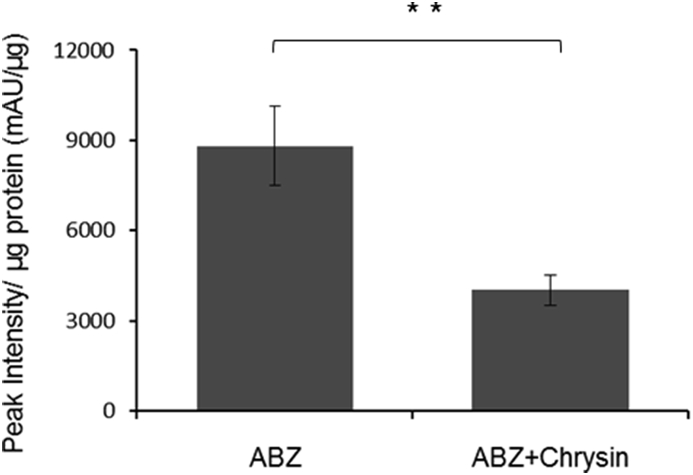
Approximately 1.5 × 106 C. elegans CB3474 ben-1(e1880)III were axenically cultured, in triplicate, with 56.5 μM of albendazole either with or without 200 μM chrysin for 72 h. A 20 mL of culture supernatant was extracted and examined by HPLC and the area under the 7.4min peak at 306 nm corresponding to the ABZ+hexose metabolite was measured for each sample. The absorbance values were then divided by the total amount of worm protein extracted from the worm pellet (BCA protein assay) in each culture to normalize for the total number of worms. The mean peak intensity of the 306 nm absorbance peak corresponding to the ABZ +hexose metabolite was significantly less, for the chrysin containing cultures compared to the control cultures (2.19 fold reduction, P = 0.0042) (Fig. 6).
Fig. 6.
Reduction of albendazole-glucose metabolite production in the presence of the UDP-glucuronosyltransferase inhibitor, chrysin. Triplicate samples of C. elegans CB3474 ben-1(e1880)III in axenic culture exposed to albendazole at 56.5 μM with and without the UGT inhibitor chrysin (200 μM) for 3 days, HPLC was performed on 20 mL worm supernatant, area under the metabolite peak (at 7.4min, 306 nm) was measured and normalized to total protein in sample using BCA assay. The error bars represent ± SEM of three biological reps. ** represents a significant difference at, p > 0.005, Student T test.
3.4. Comparison of the transcriptional response of C. elegans to different members of the benzimidazole drug family
We compared the transcriptional response of C. elegans to five different benzimidazole drug family members, at drug exposure concentrations as used previously (Laing et al., 2010), in order to identify both shared and distinct features of the transcriptional response. Three biological replicates of approximately 20,000 L1 larvae C. elegans CB3474 ben-1(e1880)III were cultured in liquid media, with E. coli OP50 at 1% (w/vol) at 20 °C for approximately 70 h until worms reached the adult stage. Worms were then exposed to 1.13 mM of either: ABZ, MBZ, TBZ, FBZ or OxBZ as well as a negative control solution of DMSO for 4 h.
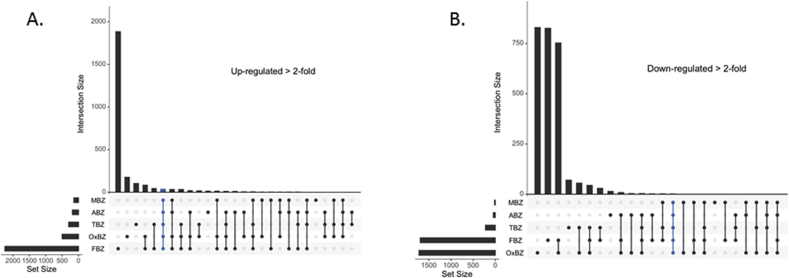
Relatively few genes are up regulated > 2-fold (adjusted p value < 0.01) in response to exposure to each of oxfendazole (506 genes), thiabendazole (304 genes), albendazole (193 genes) and mebendazole (148 genes) (Fig. 7A). There was a common subset of 41 genes that were up regulated (>2-fold, adjusted p value < 0.01) in response to exposure to all of the five benzimidazoles (listed in Table 2). Over half of these were genes encoding enzymes known to have a role in the biotransformation of small xenobiotic molecules including: 10 cytochrome P450s; 8 UDP-glycosyltransferases; and 2 glutathione-S- transferases; as well as oxidoreductases, dehydrogenases and reductases (Table 2). In addition, one nuclear hormone receptor (nhr-142), and several other genes (cdr-1, irg-2, and vem-1) known to be induced by other xenobiotics or bacterial infections were up regulated by each of the five benzimidazoles in all cases (Liao et al., 2002; Runko and Kaprielian, 2004; Wong et al., 2007; Estes et al., 2010). Hence, the core transcriptomic response of the C. elegans CB3474 ben-1(e1880)III strain, in terms of genes up-regulated to four of the benzimidazole family members, is overwhelming dominated by classical xenobiotic response genes. In contrast, fenbendazole exposure resulted in the transcriptional up-regulation of the greatest number of genes by far; a total of 2262 were up regulated >2-fold relative to the DMSO control (adjusted p value < 0.01) with 1963 of these genes being specific to fenbendazole and not up regulated in response to the other benzimidazoles (Fig. 7A).
Fig. 7.
Transcriptomic analysis of differentially expressed genes visualized in Interaction plots using the UpSet method. Panel A.C. elegans CB3474 ben-1(e1880)III genes significantly (adjusted p value of less than 0.01) up-regulated greater than 2 fold to each of the benzimidazoles tested. Mebendazole (MBZ), albendazole (ABZ), thiabendazole (TBZ), oxfendazole (OxBZ) and fenbendazole (FBZ). Horizontal histogram to the left of the drug labels represent the total number of genes significantly up-regulated greater than 2 fold to each drug, vertical histogram to the right represents each sub-set of significantly up-regulated genes specific to each drug combination represented by filled circles. Panel B. Interaction Plot of C. elegans CB3474 ben-1(e1880)III genes significantly (adjusted p value < 0.01) down-regulated greater than 2 fold to each of the benzimidazoles tested. Mebendazole (MBZ), albendazole (ABZ), thiabendazole (TBZ), oxfendazole (OxBZ) and fenbendazole (FBZ). Transcriptomic analysis of differentially expressed genes visualized with the UpSet method.
Table 2.
Sub-set of genes significantly (adjusted p values < 0.01) up-regulated greater than 2 fold, followed by their log2 fold change for C. elegans CB3474 ben-1(e1880)III exposed to 1.13 mM BZ for 4 h in response to all benzimidazoles tested.
| Gene ID | Putavie enzyme | Gene description | ABZ Δ log2 fold | FBZ Δ log2 fold | MBZ Δ log2 fold | OxBZ Δ log2 fold | TBZ Δ log2 fold |
|---|---|---|---|---|---|---|---|
| alh-5 | Phase I | Aldehyde dehydrogenase | 2.25 | 1.64 | 1.25 | 1.28 | 1.74 |
| dhs-23 | Phase I | DeHydrogenases, Short chain | 3.28 | 3.76 | 2.82 | 8.20 | 3.90 |
| E02C12.10 | Phase I | Uncharacterized oxidoreductase Dhs-27 | 7.20 | 12.83 | 6.38 | 15.47 | 8.33 |
| C52A10.2 | Phase I | Carboxylic ester hydrolase | 3.00 | 5.43 | 4.27 | 6.96 | 2.19 |
| F13H6.3 | Phase I | Esterase CM06B1 | 2.35 | 2.15 | 2.74 | 4.07 | 1.27 |
| gba-2 | Phase I | Putative glucosylceramidase 2 | 4.28 | 6.95 | 3.34 | 7.60 | 5.53 |
| Y73C8C.10 | Phase I | FMN binding activity and oxidoreductase activity | 5.73 | 5.54 | 4.48 | 7.40 | 5.83 |
| T25G12.13 | Phase I | ortholoug of dhrs7b (dehydrogenase/reductase(SDR family) | 5.91 | 7.07 | 5.09 | 9.62 | 7.20 |
| T10B5.8 | Phase I | predicted FMN binding and oxidoreductase activity | 2.96 | 1.95 | 1.62 | 5.33 | 4.16 |
| cyp-13A6 | Phase I | CYtochrome P450 family | 2.16 | 4.93 | 8.83 | 6.92 | 2.02 |
| cyp-14A4 | Phase I | CYtochrome P450 family | 2.26 | 3.03 | 3.47 | 8.14 | 1.78 |
| cyp-34A9 | Phase I | CYtochrome P450 family | 1.60 | 3.05 | 3.94 | 3.81 | 1.36 |
| cyp-35A1 | Phase I | CYtochrome P450 family | 8.61 | 7.53 | 6.79 | 9.63 | 6.06 |
| cyp-35A2 | Phase I | CYtochrome P450 family | 5.00 | 3.72 | 3.93 | 2.97 | 1.54 |
| cyp-35A3 | Phase I | CYtochrome P450 family | 6.06 | 7.44 | 4.97 | 8.25 | 7.61 |
| cyp-35A4 | Phase I | CYtochrome P450 family | 5.26 | 5.63 | 5.46 | 6.52 | 5.88 |
| cyp-35A5 | Phase I | CYtochrome P450 family | 7.11 | 6.40 | 5.88 | 7.10 | 7.67 |
| cyp-35B2 | Phase I | CYtochrome P450 family | 4.37 | 4.96 | 2.91 | 7.04 | 3.80 |
| cyp-35C1 | Phase I | CYtochrome P450 family | 7.18 | 5.45 | 5.61 | 5.55 | 3.24 |
| ugt-8 | Phase II | UDP-GlucuronosylTransferase | 5.26 | 3.91 | 3.42 | 5.69 | 4.73 |
| ugt-9 | Phase II | UDP-GlucuronosylTransferase | 4.32 | 4.53 | 3.56 | 6.90 | 4.54 |
| ugt-14 | Phase II | UDP-GlucuronosylTransferase | 3.41 | 2.52 | 2.56 | 4.29 | 2.26 |
| ugt-25 | Phase II | UDP-glucuronosylTransferase | 3.02 | 1.27 | 2.08 | 3.73 | 1.94 |
| ugt-33 | Phase II | UDP-GlucuronosylTransferase | 3.50 | 5.49 | 2.44 | 9.24 | 2.76 |
| ugt-34 | Phase II | UDP-GlucuronosylTransferase | 5.41 | 5.76 | 3.06 | 6.99 | 4.62 |
| ugt-37 | Phase II | UDP-GlucuronosylTransferase | 1.68 | 4.09 | 2.09 | 3.39 | 3.32 |
| ugt-41 | Phase II | UDP-GlucuronosylTransferase | 1.89 | 1.61 | 1.54 | 3.38 | 1.30 |
| gst-5 | Phase II | Probable glutathione S-transferase 5 | 2.29 | 3.18 | 2.24 | 4.42 | 1.93 |
| gst-21 | Phase II | Glutathione S-Transferase | 1.73 | 2.08 | 1.35 | 5.89 | 2.15 |
| C29F7.2 | Phase II | Predicted transferase, transfers phophorous-cantaining groups | 4.10 | 4.09 | 2.87 | 4.07 | 2.25 |
| H23N18.4 | Phase II | Predicted transferase, transfers hexosyl groups | 5.83 | 7.02 | 4.19 | 7.33 | 7.16 |
| nhr-142 | Nuclear Hormone Receptor family | 1.29 | 1.65 | 1.23 | 2.79 | 1.44 | |
| apy-1 | Apyrase, guanosine and uridine-diphosphatase | 1.17 | 1.47 | 1.85 | 2.67 | 1.19 | |
| cdr-1 | CaDmium Responsive; Cadmium-inducible lysosomal protein CDR-1 | 4.26 | 5.24 | 8.76 | 8.19 | 10.37 | |
| irg-2 | Infection Response protein | 2.88 | 3.69 | 3.03 | 6.23 | 1.50 | |
| clec-206 | C-type LECtin | 6.47 | 5.70 | 5.91 | 7.44 | 6.43 | |
| ptr-22 | PaTched Related family | 5.16 | 4.37 | 3.66 | 4.75 | 3.99 | |
| F54B8.4 | death domain binding activity; involved in innate immune response | 1.62 | 1.33 | 1.52 | 2.56 | 1.53 | |
| vem-1 | ortholog of PGRMC1; is involved in innate immune response | 2.52 | 2.86 | 2.06 | 3.73 | 1.59 | |
| Y102A11A.9 | Uncharacterized, up-regulated in response to chemicals | 1.68 | 2.82 | 1.98 | 3.19 | 1.39 | |
| R09E12.9 | Uncharacterized, up-regulated in response to chemicals | 5.29 | 5.56 | 3.37 | 7.50 | 6.73 | |
Complete Gene Ontology (GO) analysis was performed on the genes up-regulated >2-fold in response to exposure by each of the benzimidazole drug using Panther (Mi et al., 2013). A summary of top hits (based upon a greater than 4.5 fold enrichment and p values less than 1.0e-4) of the Panther Over-representation Test (release 20190711) (Mi et al., 2017) of GO-Slim biological processes are summarized in Table 3. The Panther Over-representation Test found an over representation of the response to xenobiotic stimulus (GO:0009410) for all anthelmintics tested, and response to drug (GO:0042493), organic acid metabolic process (GO:0006082), cellular catabolic process (GO:0044248) and drug metabolic process (GO:0017144) for albendazole, mebendazole, thiabendazole, and oxfenbendazole, again consistent with a classical xenobiotic response (Table 3).
Table 3.
Summary of Complete Gene Ontology (GO Slim) biological process analysis of the C. elegans CB3474 ben-1(e1880)III for differentially expressed genes, significantly up-regulated greater than 2-fold using Panther Overrepresentation Test (release 20190711) with FDR correction. Cut off for GO terms selected for this table: fold enrichment of > 4.5 fold.
| PANTHER GO-Slim Biological Process | C. elegans REFLIST (19921) | Input | Expected | over/ under | Fold Enrichment | P-value |
|---|---|---|---|---|---|---|
| ABZ | ||||||
| response to xenobiotic stimulus (GO:0009410) | 40 | 12 | 0.39 | + | 31.13 | 6.0E-14 |
| response to drug (GO:0042493) | 58 | 12 | 0.56 | + | 21.47 | 2.7E-12 |
| organic acid metabolic process (GO:0006082) | 76 | 12 | 0.73 | + | 16.38 | 4.4E-11 |
| cellular catabolic process (GO:0044248) | 91 | 12 | 0.88 | + | 13.68 | 2.9E-10 |
| drug metabolic process (GO:0017144) | 91 | 12 | 0.88 | + | 13.68 | 2.9E-10 |
| innate immune response (GO:0045087) | 120 | 13 | 1.16 | + | 11.24 | 4.8E-10 |
| cellular response to chemical stimulus (GO:0070887) | 112 | 12 | 1.08 | + | 11.12 | 2.5E-09 |
| defense response (GO:0006952) | 123 | 13 | 1.19 | + | 10.97 | 6.3E-10 |
| response to stress (GO:0006950) | 212 | 14 | 2.04 | + | 6.85 | 3.8E-08 |
| response to chemical (GO:0042221) | 187 | 12 | 1.8 | + | 6.66 | 4.9E-07 |
| MBZ | ||||||
| response to xenobiotic stimulus (GO:0009410) | 40 | 17 | 0.3 | + | 57.21 | 2.4E-23 |
| response to drug (GO:0042493) | 58 | 17 | 0.43 | + | 39.45 | 4.5E-21 |
| organic acid metabolic process (GO:0006082) | 76 | 17 | 0.56 | + | 30.11 | 2.4E-19 |
| cellular catabolic process (GO:0044248) | 91 | 17 | 0.68 | + | 25.15 | 3.4E-18 |
| drug metabolic process (GO:0017144) | 91 | 17 | 0.68 | + | 25.15 | 3.4E-18 |
| cellular response to chemical stimulus (GO:0070887) | 112 | 19 | 0.83 | + | 22.83 | 1.6E-19 |
| response to chemical (GO:0042221) | 187 | 20 | 1.39 | + | 14.4 | 6.3E-17 |
| response to stimulus (GO:0050896) | 627 | 23 | 4.66 | + | 4.94 | 4.4E-10 |
| TBZ | ||||||
| response to xenobiotic stimulus (GO:0009410) | 40 | 12 | 0.6 | + | 19.92 | 1.1E-11 |
| response to drug (GO:0042493) | 58 | 12 | 0.87 | + | 13.74 | 4.5E-10 |
| organic acid metabolic process (GO:0006082) | 76 | 12 | 1.14 | + | 10.48 | 6.9E-09 |
| cellular response to chemical stimulus (GO:0070887) | 112 | 16 | 1.69 | + | 9.49 | 7.8E-11 |
| cellular catabolic process (GO:0044248) | 91 | 12 | 1.37 | + | 8.76 | 4.2E-08 |
| drug metabolic process (GO:0017144) | 91 | 12 | 1.37 | + | 8.76 | 4.2E-08 |
| innate immune response (GO:0045087) | 120 | 13 | 1.81 | + | 7.19 | 9.5E-08 |
| defense response (GO:0006952) | 123 | 13 | 1.85 | + | 7.02 | 1.2E-07 |
| response to chemical (GO:0042221) | 187 | 19 | 2.82 | + | 6.75 | 2.7E-10 |
| OxBZ | ||||||
| response to xenobiotic stimulus (GO:0009410) | 40 | 24 | 1 | + | 24.1 | 1.1E-22 |
| response to drug (GO:0042493) | 58 | 24 | 1.44 | + | 16.62 | 9.1E-20 |
| organic acid metabolic process (GO:0006082) | 76 | 24 | 1.89 | + | 12.68 | 1.5E-17 |
| drug metabolic process (GO:0017144) | 91 | 25 | 2.27 | + | 11.03 | 5.0E-17 |
| cellular catabolic process (GO:0044248) | 91 | 24 | 2.27 | + | 10.59 | 4.6E-16 |
| cellular response to chemical stimulus (GO:0070887) | 112 | 28 | 2.79 | + | 10.04 | 5.3E-18 |
| response to chemical (GO:0042221) | 187 | 29 | 4.66 | + | 6.23 | 8.0E-14 |
| FBZ | ||||||
| intraciliary transport (GO:0042073) | 11 | 11 | 1.23 | + | 8.94 | 2.6E-06 |
| regulation of trans-synaptic signaling (GO:0099177) | 14 | 11 | 1.57 | + | 7.02 | 1.2E-05 |
| modulation chemical synaptic transmission (GO:0050804) | 14 | 11 | 1.57 | + | 7.02 | 1.2E-05 |
| cell surface receptor signaling pathway (GO:1905114) | 9 | 7 | 1.01 | + | 6.95 | 5.2E-04 |
| glutamate receptor signaling pathway (GO:0007215) | 8 | 6 | 0.9 | + | 6.7 | 1.5E-03 |
| regulation of signaling (GO:0023051) | 15 | 11 | 1.68 | + | 6.55 | 1.9E-05 |
| synaptic transmission, glutamatergic (GO:0035249) | 15 | 11 | 1.68 | + | 6.55 | 1.9E-05 |
| cilium organization (GO:0044782) | 30 | 20 | 3.36 | + | 5.96 | 2.5E-08 |
| protein localization to cilium (GO:0061512) | 28 | 16 | 3.13 | + | 5.11 | 2.8E-06 |
| response to xenobiotic stimulus (GO:0009410) | 40 | 22 | 4.48 | + | 4.92 | 6.6E-08 |
| cAMP-mediated signaling (GO:0019933) | 25 | 13 | 2.8 | + | 4.65 | 5.1E-05 |
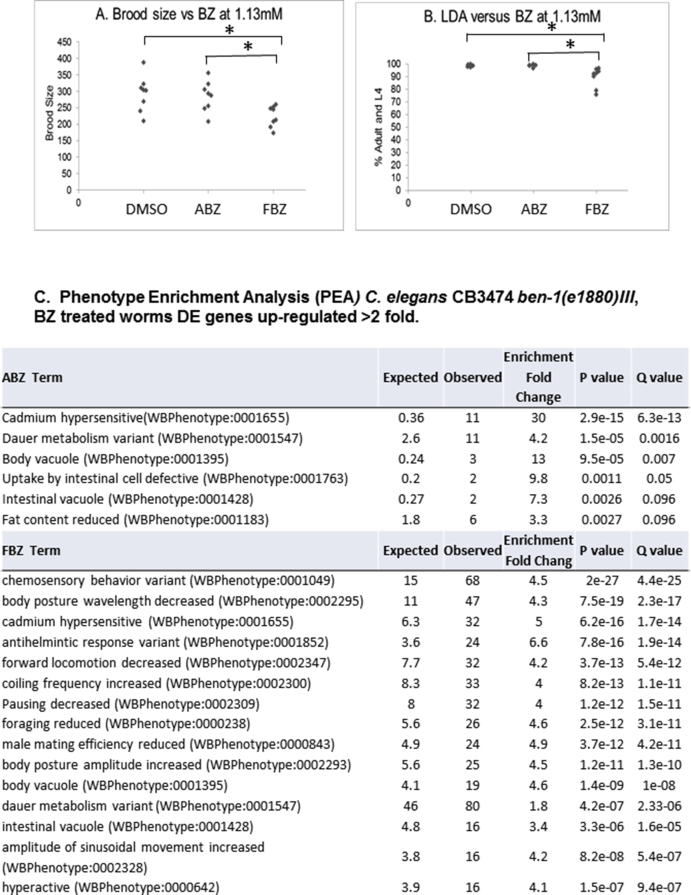
In the case of fenbendazole, for which a much greater number of unique genes were up-regulated, there was an enrichment of interciliary transport (GO:0042073), chemical and trans-synaptic transmission (GO:0099177, GO:0050804), cell surface, glutamate receptor and cAMP-mediated signaling pathways (GO:1905114, GO:0007215, GO:0019933, GO:0023051), and cilium organization (GO:0044782) (Table 3) suggesting secondary targets. This was investigated further, exposure of C. elegans CB3474 ben-1(e1880)III to 1.13 mM FBZ throughout development and resulted in a reduction in brood size and larval development that was not observed in ABZ exposed worms (Supplemental Figs. 8A and B); phenotype enrichment analysis (Angeles-Albores et al., 2016) showed chemosensory behavior variant (WBPhenotype:0001049) and various movement variants (WBPhenotype:0002295, WBPhenotype:0002347, WBPhenotype:0002300, WBPhenotype:0000238), as well as male mating efficiency reduced phenotype (WBPhenotype:0000843) and foraging reduced phenotype (WBPhenotype:0000238) (Supplemental Fig. 8C).
Although the major focus of this work was to determine which biotransformation enzymes may be transcriptionally up-regulated to the benzimidazole drugs, we also mined the data for those genes down-regulated >2 fold in response to drug exposure (Fig. 7B). Of the five drugs, oxfendazole exposure resulted in the greatest number of down regulated genes (total of 1728 genes), closely followed by fenbendazole exposure (total of 1695 genes). A high proportion of these genes were down-regulated in response to both drugs (a total of 759 shared down regulated genes). There were fewer genes down regulated in response to exposure to the other drugs; thiabendazole (225 genes), albendazole (50 genes), and mebendazole (20 genes). There were only four genes down-regulated in response to all the drugs (pud-3, bath-26, cest-1 and C30G12.2), bath-26 has been shown to be regulated by several xenobiotics (Rudgalvyte et al., 2013). As fenbendazole and oxfendazole had the greatest number of genes down-regulated Complete Gene Ontology (GO) analysis was performed on the genes down-regulated >2-fold in response to these two benzimidazoles using Panther Overrepresentation Test of biological processes (Mi et al., 2017) (Supplementary Table 1). Transmembrane tyrosine kinase signaling pathway (GO:0007169), very long-chain fatty acid metabolic process (GO:0000038), protein phosphorylation and autophosphorylation (GO:0006468, GO:0046777), had the highest fold enrichment, again suggesting secondary targets.
3.5. Comparison of the H. contortus transcriptional response following exposure to different members of the benzimidazole drug family
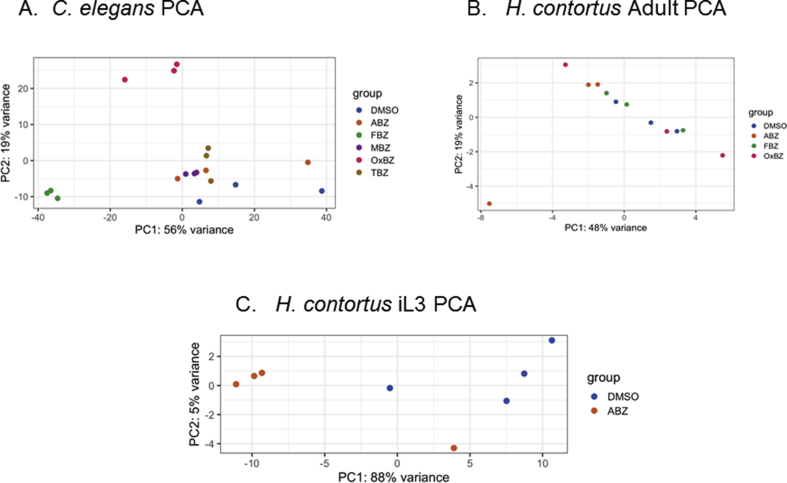
Triplicate ex-vivo cultures of fifty H. contortus UGA/2004 (Williamson et al., 2011) adult worms, collected fresh from the sheep abomasum, were exposed to 1.13 mM of ABZ, MBZ, TBZ, FBZ, OxBZ or DMSO solvent control for 4 h. RNA was extracted and RNAseq analysis performed using the same approaches as the C. elegans experiments. No genes were identified as expressed >2-fold with an adjusted p-value of >0.05 between any of the drug exposed populations and the DMSO control. Consequently, the experiment was repeated for three of these drugs - albendazole, fenbendazole and oxfendazole –but at a higher dose (3 mM). Principal component analysis (PCA plot) to visualize the variation in the samples found little clustering of treatment replicates (Supplemental Figure 7B) signifying the majority of variance was not due to drug treatment. Few genes were significantly differentially expressed at any fold change (at adjusted p < 0.05) with albendazole having the greatest number of differentially expressed genes (357 genes). However, none of these were differentially expressed at >2 fold. Fenbendazole had no significantly differentially regulated genes, and oxfendazole had five genes which were differentially regulated, only 1 of these HCON 00141910 was regulated >2 fold (results not shown).
Quadruplicate samples of exsheathed infective larval stage of the triple-resistant field isolate UGA/2004 H. contortus were exposed to either 1.13 mM albendazole or DMSO solvent control for 4 h. The PCA plot for the infective larval RNA-Seq did indicate that the majority (up to 88%) of the variance in these samples is due to the differences between drug treatments with the exception of single outliers (Supplemental Figure 7C). A total of 2920 transcripts were significantly differentially up-regulated (adj P < 0.05), and of these 226 genes were up regulated >2-fold (Supplemental Table 2). The genes up-regulated >2-fold did not include any classical xenobiotic response genes; however examination of all differentially expressed genes indicated that several classical xenobiotic response genes were up-regulated, albeit less than 2-fold, in contrast no xenobiotic response genes were found to be down-regulated (Supplemental Table 3). These included 2 genes with protein motif homology to short chain dehydrogenases, 8 genes with protein motif homology to cytochrome P450s, 2 genes with glutathione S-transferase domain containing protein motifs, and 6 genes with UDP-glucuronosyl UDP-glucosyltransferase domain containing protein motifs (Supplemental Table 3). Functional analysis of the differentially expressed genes performed with the clusterProfiler R package version 3.12.0 using the GO annotations from the PRJEB506 assembly showed a top hit of oxidation-reduction processes (GO:0055114). A total of 2537 H. contortus infective larvae genes were significantly (at adj P < 0.05) down regulated when exposed to albendazole, of which 509 genes were differentially down regulated greater than 2 fold (Supplemental Table 2).
4. Discussion
4.1. Glucose conjugation is the predominant biotransformation pathway for multiple benzimidazole drugs in both C. elegans and H. contortus
We, and others, have previously shown that the predominant biotransformation reaction of albendazole in C. elegans and H. contortus is the conjugation of glucose to the parental molecule (Cvilink et al., 2008b; Laing et al., 2010; Vokral et al., 2013; Stuchlikova et al., 2018). In this work we demonstrate that hexose conjugation (mostly likely glucose) is also the predominant biotransformation reaction for four of the most therapeutically important benzimidazole drugs but with some differences (Table 1). We found that some of the metabolites were also reduced or oxidized (ABZ+O, MBZ+hexose+2H, FBZ+O+hexose+H, TBZ+O+hexose+H, or OxBZ+hexose+H) and several less abundant non-glucosidated products were also produced to varying degrees for each compound. In future experiments, different time courses or advancements in MS/MS technologies may elucidate more of the trace metabolites and provide further insight into metabolism of the benzimidazoles.
The observation that glucose conjugation is the predominant pathway for benzimidazole metabolism in these two nematode species is different to mammals where, to our knowledge, hexose conjugation of a benzimidazole drug has not been reported to date. In liver microsomes from deer, cattle, pigs, and sheep (Velik et al., 2005), albendazole is quickly metabolized to a sulfoxide derivative (ABZ-SO), the active form of the drug, and then slowly oxidized to albendazole sulfone. In the kidney, it was found that cattle, sheep, rats and mice metabolized albendazole to various albendazole sulfoxides and sulfones found in the urine (Gyurik et al., 1981). Our results for the benzimidazoles add to the growing evidence that hexose conjugation (particularly glucose) is a common biotransformation reaction of small xenobiotic compounds in nematodes. This contrasts with the situation in mammals where the major conjugation reaction of small xenobiotic compounds is either oxidation/reduction (Ding and Kaminsky, 2003; Gueguen et al., 2006), or the addition of glucuronic acid (Meech et al., 2012; Rowland et al., 2013). There is a growing list of examples where sugar conjugation of small xenobiotic molecules occurs in C. elegans. For example, C. elegans metabolizes genistein to various sugar conjugates, predominately genistein-O-glucosides (Soukup et al., 2012). An analysis of 12 metabolites produced in a drug screen using C. elegans, showed that 8 of the 12 were modified by conjugation of sugars, either: O-hexosidation, O-glucosidation or N-glucosidation, some of these were also modified by demethylation or sulfoxidation (Burns et al., 2010). C. elegans metabolized 1-hydroxyphenazine, and indole by both O- and N-glucosylation and additional 3′-O-phosphorylation, it was found that these biotransformation products had decreased toxicity to the worms (Stupp et al., 2013). These fundamental differences in small molecular biotransformation pathways between mammals and nematodes may provide a potential for synergistic compounds that enhance drug potency in parasitic nematodes but not their mammalian hosts.
4.2. Albendazole biotransformation by C. elegans reduces drug potency and is inhibited by the UGT inhibitor chrysin
As the majority of metabolites produced in C. elegans were glucosidated, we tested the effect of inhibiting the UDP-glucuronosyltransferase enzymes with chrysin to determine the effects on metabolite production. There was a 2-fold reduction in the ABZ+hexose metabolite when cultured in the presence of chrysin (at 200 μM). Since, chrysin has a relatively high degree of specificity as a UGT inhibitor (Walsky et al., 2012), this result suggests that UDP-glucuronosyltransferase enzymes are likely to play a role in the production of this metabolite. We also found that the biotransformation of albendazole in culture supernatants by C. elegans reduces the potency of the drug (Fig. 5). This suggests that upregulation of the relevant biotransformation enzymes, likely the UGTs, could be protective and so play a role in benzimidazole resistance. There is growing evidence to support this hypothesis; two benzimidazole resistant H. contortus strains (WR and BR) were shown to have greater UDP-glucosyl transferases enzyme activity compared to a sensitive strain (ISE) (Vokral et al., 2013). Also, there was an up-regulation of UGT368B2 in two BZ resistant strains (IRE and WR) compared to the sensitive ISE strain (Matouskova et al., 2018) Further, three flubendazole resistant strains (WR, BR and ISE-S) produced more of the metabolites compared to the ISE sensitive strain (Vokral et al., 2012). In contrast, we did not detect a significant increase in the production of benzimidazole glucoside metabolites between the MHco12 benzimidazole resistant strain (von Samson-Himmelstjerna et al., 2009) compared to MHco3(ISE) (Redman et al., 2008) strain for any of the BZs tested in our experiments (data not shown). It is not clear whether this is due the parasite strains, drugs used, or other methodological differences but warrants further investigation.
4.3. The dominant xenobiotic transcriptional response to benzimidazole in C. elegans is more muted in H. contortus with stage-specific differences
We examined the transcriptomic response in C. elegans to five different benzimidazoles to determine whether each benzimidazole induces its own specific xenobiotic response, or, whether there was a common benzimidazole xenobiotic response. The C. elegans benzimidazole resistant strain CB3474 ben-1(e1880)III, which carries a null mutation in the gene encoding the major β-tubulin drug target, was used to minimize stress responses and so enable us to focus on the xenobiotic response itself. Analysis of the Gene Ontology (GO) of complete biological processes (Mi et al., 2017) show that up-regulated genes to each of the benzimidazole drugs were dominated by xenobiotic metabolic response terms. A core set of 41 genes were up-regulated to all the benzimidazoles tested and over 50% (31 genes) of this core group were associated with classic Phase I and Phase II xenobiotic response genes based on their gene descriptions (Table 2). Given the nature of the benzimidazole biotransformation products in C. elegans the UDP-glucuronosyltransferase enzymes and the cytochrome P450 genes are of particular interest. Of the UDP-glucuronosyltransferase enzymes UGT – 8, 9, 14, 25, 33, 34, 37, 41 would be priorities for investigation of enzymes that might be involved in benzimidazole glucosidation. Similarly, CYP–13A6, 14A4, 34A9, 35A1-5, 35B2, and 35C1 may be involved in the oxidation/reduction biotransformation reactions. Interestingly, the CYP-35 group has been shown in several other studies to be a core xenobiotic response group in C. elegans references (Menzel et al., 2001, 2005; Reichert and Menzel, 2005; Roh et al., 2007). Several members of the core group of BZ xenobiotic metabolizing genes which we identified (cyp-35a5, ugt-8, ugt-25 and ugt-37), were also found to be up-regulated in response to several xenobiotics, including thiabendazole (Jones et al., 2013), which further validates our results, and suggests that these 43 genes are the key xenobiotic response group to benzimidazoles in C. elegans.
The transcriptional response of C. elegans CB3474 ben-1(e1880)III when exposed to fenbendazole showed many more uniquely differentially expressed genes, as well as reduced brood size and larval development, fenbendazole inhibition of ubiquitin-proteasome function and induction of endoplasmic reticulum stress in human cancer cells has been reported (Dogra and Mukhopadhyay, 2012), suggesting fenbendazole has secondary targets in addition to the well characterised β-tubulin target.
Albendazole, mebendazole and fenbendazole, and oxfendazole metabolites similar to those found in C. elegans were also present in adult H. contortus cultures suggesting that similar xenobiotic biotransformation may occur in these nematodes. There were inevitable differences in the culture conditions and tissue mass between C. elegans and H. contortus, however, the results suggest the biotransformation processes are less efficient in adult H. contortus than in C. elegans. Consistent, with this hypothesis, the global transcriptomic response to benzimidazole drugs of adult H. contortus worms was less marked than that of C. elegans with no genes being significantly up-regulated >2-fold (adjusted p-value >0.05) and with no apparent enrichment of xenobiotic response genes in response to any of the five drugs. These experiments were conducted at the same (and also higher) drug concentrations, and same exposure times as for C. elegans, H. contortus was viable for the length of the in vitro culture as demonstrated by vigorous movement after the 4 h drug exposure. Nevertheless, one caveat is that the adult parasites are in a highly abnormal environment when in vitro culture and may not behave in a physiologically relevant manner. The investigation of transcriptional responses of H. contortus to benzimidazoles in vivo in the host would be of interest to test this possibility.
We also investigated the transcriptional response to albendazole exposure of H. contortus infective L3 to test for stage-specific differences in xenobiotic responses. Although H. contortus infective L3 had a less marked transcriptomic response than C. elegans, they did show some evidence of a muted xenobiotic response to albendazole exposure. It has been hypothesised that parasites in the host environment have less need for reactive xenobiotic responses than organisms living in a more dynamic free-living environment (Zarowiecki and Berriman, 2015). Recent genomic analysis (review see (Zarowiecki and Berriman, 2015; Matouskova et al., 2016)) of several helminths has shown that the free living nematode C. elegans have many xenobiotic metabolizing genes (86 CYPs, 68 SDR, 48 GSTs, 72 UGTs and 60 ABC transporters) (Lindblom and Dodd, 2006). In contrast, parasitic platyhelminths, with life cycles less exposed to the environment, have very small numbers of drug metabolizing gene families (e.g. 1 or 2 CYPs each and no UGTs detected) (Berriman et al., 2009; Olson et al., 2012; Young et al., 2012; Cwiklinski et al., 2015). The parasitic nematodes are more variable; however, there is a trend that those parasites that do not have free-living stages (e.g. Trichinella and filarial parasites) have fewer drug metabolizing gene family members. In contrast, the strongylid nematodes (which have free-living stages as part of their life cycle) tend to have larger xenobiotic metabolizing enzyme gene families (Zarowiecki and Berriman, 2015); H. contortus has, 42 CPYs, 70 SDR, 28 GSTs, 34 UGTs and 46 ABC transporters in the current published draft genome assembly (Laing et al., 2013). The transcriptional profile of a panel of H. contortus cyp genes showed that 15 out of 19 tested were significantly down-regulated in the adult stage compared to the free-living L3 stage (Laing et al., 2015). Hence, an interesting hypothesis to test will be whether the regulatory elements of genes encoding xenobiotic response elements are less responsive to induction in the parasitic stages compared to the free-living stages of parasites such as H. contortus.
Conflicts of interest
The authors have no competing interests to declare.
Acknowledgements
The authors thank Ray Kaplan for the use of the H. contortus UGA/2004 strain and the CGC (C. elegans Genetics Center, Minnesota, Minneapolis, MN USA) for the C. elegans strains. We also thank Dr Andreas Wissman for advice on C. elegans motility assays. This work was supported by Canadian Institutes of Health Research (CIHR) grant 230927 and the (NSERC-CREATE Host-Pathogens Interactions HPI graduate training program at the University of Calgary, Canada).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijpddr.2019.09.001.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
List of differentially expressed genes of exsheathed infective larvae H. contortus (UGA/2004) exposed to 1.13 mM albendazole for 4 hours, which were significantly differentially expressed (adjusted P value 0.05).
Fig. 1.
Structure of benzimidazole drugs: albendazole (265 Mw), mebendazole (295 Mw), fenbendazole (299 Mw), thiabendazole (201 Mw), and oxfendazole (315 Mw).
Fig. 2.
Metabolite accumulation over time in C. elegans CB3474 ben-1(e1880)III cultures treated with 56.5 μM ABZ and incubated for 3, 6, 24, and 72 h, compared to the controls, drug-no-worm (DNW) culture and worm-no-drug (WND) culture, and an ABZ standard. Metabolites produced include ABZ+SO (1), ABZ+G (2), ABZ+G (3), and ABZ+G (4), from the ABZ parental drug (5). Chromatograms are result of product ion scan from LS-MS/MS
Note: The 3 h and 6 h cultures were performed together, beginning with the same concentration of worms. The 24 h and 72 h cultures were performed together, beginning with the same concentration of worms, but not at the same time as the 3 h and 6 h cultures, therefore worm concentration between these 2 different groups were not the same and thus metabolite accumulation is not directly comparable
Fig. 3.
Proposed mechanism of methoxide loss from albendazole to produce m/z 234 fragmentation product, B. N-linked sugar loss from albendazole + hexose metabolite to produce the m/z 266 product. C. An oxygen addition to the aromatic ring is required for hexose conjugation, a proposed mechanism for the m/z 208 fragmentation product
Fig. 4.
Chromatograms are the result of product Total Ion Current (TIC), or a multiple reaction monitoring (MRM), of albendazole (ABZ) parental drug and its metabolites identified in the supernatant of cultures of either C. elegans benzimidazole resistant allele CB3474 ben-1(e1880)III (shown in green) or adult H. contortus drug sensitive MHco3(ISE) allele (shown in orange). The first (top) chromatograms contains benzimidazole but no nematodes, the second chromatograms corresponds to the culture containing either C. elegans or H. contortus, respectively, exposed to albendazole (ABZ), the third chromatogram corresponds to the control culture containing C. elegans or H. contortus but with no benzimidazole. C. elegans was exposed to 56.5 μM BZ for 3 days, and H. contortus was exposed to 10 μM BZ for 16 h respectively. [ABZ+O], was identified in trace amounts in both worms by a targeted ion search using m/z 282. Metabolites and their LC-MS/MS fragmentation patterns identified in supernatant of the ABZ-treated nematode cultures. The fragmentation patterns were extracted from either a Total Ion Current (TIC) of ABZ-treated C. elegans culture supernatant, or multiple reaction monitoring (MRM) of H. contortus culture and resulted from a fragmentor setting of 150V and a collision energy of 30eV
Fig. 5.
A. Proposed mechanism of O-linked glycosylation on non-aromatic carbon of mebendazole and 180amu neutral to produce m/z 280 fragmentation product, B. Proposed mechanism of neutral loss of 162 mass indicated that the sugar is conjugated to the oxygen attached to the aromatic ring of thiabendazole to produce m/z 218 fragmentation product
Fig. 6.
Chromatograms of oxfendazole parental drug and its metabolites identified in the supernatant of cultures of either C. elegans benzimidazole resistant allele CB3474 ben-1(e1880)III (shown in green) or adult H. contortus drug sensitive MHco3(ISE) allele (shown in orange). The first (top) chromatograms contains benzimidazole but no nematodes, the second chromatograms corresponds to the culture containing either C. elegans or H. contortus, respectively, exposed to oxfendazole, the third chromatogram corresponds to the control culture containing C. elegans or H. contortus but with no benzimidazole. C. elegans was exposed to 56.5 μM BZ for 3 days, and H. contortus was exposed to 10 μM BZ for 16 h respectively. [OxBZ+hexose], was identified in trace amounts in both worms by a targeted ion search using m/z 478. Metabolites and their LC-MS/MS fragmentation patterns identified in supernatant of the OxBZ-treated worm cultures. The fragmentation patterns were extracted from either a Total Ion Current (TIC) of ABZ-treated C. elegans culture supernatant, or multiple reaction monitoring (MRM) of H. contortus culture and resulted from a fragmentor setting of 150V and a collision energy of 30eV
Fig. 7.
Principal component analysis (PCA) plots using plot PCA from DESeq2 package of the top 500 most variable genes after normalization and log transformation. Samples are colored by treatment to show the grouping. The amount of variation attributable to each principal component is indicated in the axis titles. Panel A.C. elegans CB3474 ben-1(e1880)III exposed to 1.13 mM BZ for 4 h. Panel B. H. contortus (UGA/2004), exposed to 3 mM BZ for 4 h. Panel C.H. contortus (UGA/2004) exsheathed infective larvae exposed to 1.13 mM ABZ for 4 h
Fig. 8.
Brood size, Larval Developmental Assay and Phenotype Enrichment Assay (PEA) of C. elegans CB3474 ben-1(e1880)III worms exposed to 1.13 mM albendazole or fenbendazole versus DMSO control. Eight C. elegans CB3474 ben-1(e1880)III worms exposed to 1.13 mM albendazole or fenbendazole versus DMSO control throughout development and resulting Panel A. Brood size and Panel B. Larval development of those broods. T.test shows a significant reduction in brood size of FBZ versus DMSO (p, 00753) and FBZ versus ABZ (p, 0.00861) and a significant reduction in larval development of FBZ versus DMSO (p, 0.00523) and FBZ versus ABZ (p, 0.00546). Panel C. Phenotype Enrichment Analysis of RNA-Seq data set for albendazole and fenbendazole exposed C. elegans CB3474 ben-1(e1880)III analyzed on https://wormbase.org/tools/enrichment/tea/tea.cgi
References
- Aksit D., Yalinkilinc H.S., Sekkin S., Boyacioglu M., Cirak V.Y., Ayaz E., Gokbulut C. Comparative pharmacokinetics and bioavailability of albendazole sulfoxide in sheep and goats, and dose-dependent plasma disposition in goats. BMC Vet. Res. 2015;11:124. doi: 10.1186/s12917-015-0442-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albonico M., Bickle Q., Ramsan M., Montresor A., Savioli L., Taylor M. Efficacy of mebendazole and levamisole alone or in combination against intestinal nematode infections after repeated targeted mebendazole treatment in Zanzibar. Bull. World Health Organ. 2003;81:343–352. [PMC free article] [PubMed] [Google Scholar]
- AlGusbi S., Krucken J., Ramunke S., von Samson-Himmelstjerna G., Demeler J. Analysis of putative inhibitors of anthelmintic resistance mechanisms in cattle gastrointestinal nematodes. Int. J. Parasitol. 2014;44:647–658. doi: 10.1016/j.ijpara.2014.04.007. [DOI] [PubMed] [Google Scholar]
- Alvarez-Sanchez M.A., Perez-Garcia J., Cruz-Rojo M.A., Rojo-Vazquez F.A. Real time PCR for the diagnosis of benzimidazole resistance in trichostrongylids of sheep. Vet. Parasitol. 2005;129:291–298. doi: 10.1016/j.vetpar.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Amenya D.A., Naguran R., Lo T.C., Ranson H., Spillings B.L., Wood O.R., Brooke B.D., Coetzee M., Koekemoer L.L. Over expression of a cytochrome P450 (CYP6P9) in a major African malaria vector, Anopheles Funestus, resistant to pyrethroids. Insect Mol. Biol. 2008;17:19–25. doi: 10.1111/j.1365-2583.2008.00776.x. [DOI] [PubMed] [Google Scholar]
- Angeles-Albores D., RY N.L., Chan J., Sternberg P.W. Tissue enrichment analysis for C. elegans genomics. BMC Bioinf. 2016;17:366. doi: 10.1186/s12859-016-1229-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriman M., Haas B.J., LoVerde P.T., Wilson R.A., Dillon G.P., Cerqueira G.C., Mashiyama S.T., Al-Lazikani B., Andrade L.F., Ashton P.D., Aslett M.A., Bartholomeu D.C., Blandin G., Caffrey C.R., Coghlan A., Coulson R., Day T.A., Delcher A., DeMarco R., Djikeng A., Eyre T., Gamble J.A., Ghedin E., Gu Y., Hertz-Fowler C., Hirai H., Hirai Y., Houston R., Ivens A., Johnston D.A., Lacerda D., Macedo C.D., McVeigh P., Ning Z., Oliveira G., Overington J.P., Parkhill J., Pertea M., Pierce R.J., Protasio A.V., Quail M.A., Rajandream M.A., Rogers J., Sajid M., Salzberg S.L., Stanke M., Tivey A.R., White O., Williams D.L., Wortman J., Wu W., Zamanian M., Zerlotini A., Fraser-Liggett C.M., Barrell B.G., El-Sayed N.M. The genome of the blood fluke Schistosoma mansoni. Nature. 2009;460:352–358. doi: 10.1038/nature08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burglin T.R., Lobos E., Blaxter M.L. Caenorhabditis elegans as a model for parasitic nematodes. Int. J. Parasitol. 1998;28:395–411. doi: 10.1016/s0020-7519(97)00208-7. [DOI] [PubMed] [Google Scholar]
- Burns A.R., Luciani G.M., Musso G., Bagg R., Yeo M., Zhang Y., Rajendran L., Glavin J., Hunter R., Redman E., Stasiuk S., Schertzberg M., Angus McQuibban G., Caffrey C.R., Cutler S.R., Tyers M., Giaever G., Nislow C., Fraser A.G., MacRae C.A., Gilleard J., Roy P.J. Caenorhabditis elegans is a useful model for anthelmintic discovery. Nat. Commun. 2015;6:7485. doi: 10.1038/ncomms8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns A.R., Wallace I.M., Wildenhain J., Tyers M., Giaever G., Bader G.D., Nislow C., Cutler S.R., Roy P.J. A predictive model for drug bioaccumulation and bioactivity in Caenorhabditis elegans. Nat. Chem. Biol. 2010;6:549–557. doi: 10.1038/nchembio.380. [DOI] [PubMed] [Google Scholar]
- Cvilink V., Kubicek V., Nobilis M., Krizova V., Szotakova B., Lamka J., Varady M., Kubenova M., Novotna R., Gavelova M., Skalova L. Biotransformation of flubendazole and selected model xenobiotics in Haemonchus contortus. Vet. Parasitol. 2008;151:242–248. doi: 10.1016/j.vetpar.2007.10.010. [DOI] [PubMed] [Google Scholar]
- Cvilink V., Skalova L., Szotakova B., Lamka J., Kostiainen R., Ketola R.A. LC-MS-MS identification of albendazole and flubendazole metabolites formed ex vivo by Haemonchus contortus. Anal. Bioanal. Chem. 2008;391:337–343. doi: 10.1007/s00216-008-1863-9. [DOI] [PubMed] [Google Scholar]
- Cvilink V., Szotakova B., Krizova V., Lamka J., Skalova L. Phase I biotransformation of albendazole in lancet fluke (Dicrocoelium dendriticum) Res. Vet. Sci. 2009;86:49–55. doi: 10.1016/j.rvsc.2008.05.006. [DOI] [PubMed] [Google Scholar]
- Cvilink V., Szotakova B., Vokral I., Bartikova H., Lamka J., Skalova L. Liquid chromatography/mass spectrometric identification of benzimidazole anthelminthics metabolites formed ex vivo by Dicrocoelium dendriticum. Rapid Commun. Mass Spectrom. : RCM (Rapid Commun. Mass Spectrom.) 2009;23:2679–2684. doi: 10.1002/rcm.4170. [DOI] [PubMed] [Google Scholar]
- Cwiklinski K., Dalton J.P., Dufresne P.J., La Course J., Williams D.J., Hodgkinson J., Paterson S. The Fasciola hepatica genome: gene duplication and polymorphism reveals adaptation to the host environment and the capacity for rapid evolution. Genome Biol. 2015;16:71. doi: 10.1186/s13059-015-0632-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daborn P.J., Yen J.L., Bogwitz M.R., Le Goff G., Feil E., Jeffers S., Tijet N., Perry T., Heckel D., Batterham P., Feyereisen R., Wilson T.G., ffrench-Constant R.H. A single p450 allele associated with insecticide resistance in Drosophila. Science (New York, N.Y. 2002;297:2253–2256. doi: 10.1126/science.1074170. [DOI] [PubMed] [Google Scholar]
- de Lourdes Mottier M., Prichard R.K. Genetic analysis of a relationship between macrocyclic lactone and benzimidazole anthelmintic selection on Haemonchus contortus. Pharmacogenetics Genom. 2008;18:129–140. doi: 10.1097/FPC.0b013e3282f4711d. [DOI] [PubMed] [Google Scholar]
- Ding X., Kaminsky L.S. Human extrahepatic cytochromes P450: function in xenobiotic metabolism and tissue-selective chemical toxicity in the respiratory and gastrointestinal tracts. Annu. Rev. Pharmacol. Toxicol. 2003;43:149–173. doi: 10.1146/annurev.pharmtox.43.100901.140251. [DOI] [PubMed] [Google Scholar]
- Dogra N., Mukhopadhyay T. Impairment of the ubiquitin-proteasome pathway by methyl N-(6-phenylsulfanyl-1H-benzimidazol-2-yl)carbamate leads to a potent cytotoxic effect in tumor cells: a novel antiproliferative agent with a potential therapeutic implication. J. Biol. Chem. 2012;287:30625–30640. doi: 10.1074/jbc.M111.324228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll M., Dean E., Reilly E., Bergholz E., Chalfie M. Genetic and molecular analysis of a Caenorhabditis elegans beta-tubulin that conveys benzimidazole sensitivity. J. Cell Biol. 1989;109:2993–3003. doi: 10.1083/jcb.109.6.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du S., Rahman C.A., Badawy W. 2013 IEEE 17TH INTERNATIONAL SYMPOSIUM ON CONSUMER ELECTRONICS (ISCE) Natl Chiao Tung Univ, Microelectron & Informat Syst Res Ctr; Hsinchu, TAIWAN: 2013. A novel automated microscopy system for the study of population statistics of parasitic nematode; pp. 43–44. [Google Scholar]
- Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes K.A., Dunbar T.L., Powell J.R., Ausubel F.M., Troemel E.R. bZIP transcription factor zip-2 mediates an early response to Pseudomonas aeruginosa infection in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 2010;107:2153–2158. doi: 10.1073/pnas.0914643107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geary T.G., Thompson D.P. Caenorhabditis elegans: how good a model for veterinary parasites? Vet. Parasitol. 2001;101:371–386. doi: 10.1016/s0304-4017(01)00562-3. [DOI] [PubMed] [Google Scholar]
- Gilleard J.S. The use of Caenorhabditis elegans in parasitic nematode research. Parasitology. 2004;128(Suppl. 1):S49–S70. doi: 10.1017/S003118200400647X. [DOI] [PubMed] [Google Scholar]
- Gilleard J.S. Understanding anthelmintic resistance: the need for genomics and genetics. Int. J. Parasitol. 2006;36:1227–1239. doi: 10.1016/j.ijpara.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Gilleard J.S., Woods D.J., Dow J.A. Model-organism genomics in veterinary parasite drug-discovery. Trends Parasitol. 2005;21:302–305. doi: 10.1016/j.pt.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Grant W.N., Mascord L.J. Beta-tubulin gene polymorphism and benzimidazole resistance in trichostrongylus colubriformis. Int. J. Parasitol. 1996;26:71–77. doi: 10.1016/0020-7519(95)00092-5. [DOI] [PubMed] [Google Scholar]
- Gueguen Y., Mouzat K., Ferrari L., Tissandie E., Lobaccaro J.M., Batt A.M., Paquet F., Voisin P., Aigueperse J., Gourmelon P., Souidi M. [Cytochromes P450: xenobiotic metabolism, regulation and clinical importance] Ann. Biol. Clin. 2006;64:535–548. [PubMed] [Google Scholar]
- Gyurik R.J., Chow A.W., Zaber B., Brunner E.L., Miller J.A., Villani A.J., Petka L.A., Parish R.C. Metabolism of albendazole in cattle, sheep, rats and mice. Drug Metab. Dispos. 1981;9:503–508. [PubMed] [Google Scholar]
- Hodgkinson J.E., Clark H.J., Kaplan R.M., Lake S.L., Matthews J.B. The role of polymorphisms at beta tubulin isotype 1 codons 167 and 200 in benzimidazole resistance in cyathostomins. Int. J. Parasitol. 2008;38:1149–1160. doi: 10.1016/j.ijpara.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Jones L.M., Flemming A.J., Urwin P.E. NHR-176 regulates cyp-35d1 to control hydroxylation-dependent metabolism of thiabendazole in Caenorhabditis elegans. Biochem. J. 2015;466:37–44. doi: 10.1042/BJ20141296. [DOI] [PubMed] [Google Scholar]
- Jones L.M., Rayson S.J., Flemming A.J., Urwin P.E. Adaptive and specialised transcriptional responses to xenobiotic stress in Caenorhabditis elegans are regulated by nuclear hormone receptors. PLoS One. 2013;8 doi: 10.1371/journal.pone.0069956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan R.M. Drug resistance in nematodes of veterinary importance: a status report. Trends Parasitol. 2004;20:477–481. doi: 10.1016/j.pt.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Kaplan R.M., Klei T.R., Lyons E.T., Lester G., Courtney C.H., French D.D., Tolliver S.C., Vidyashankar A.N., Zhao Y. Prevalence of anthelmintic resistant cyathostomes on horse farms. J. Am. Vet. Med. Assoc. 2004;225:903–910. doi: 10.2460/javma.2004.225.903. [DOI] [PubMed] [Google Scholar]
- Kotze A.C., Ruffell A.P., Ingham A.B. Phenobarbital induction and chemical synergism demonstrate the role of UDP-glucuronosyltransferases in detoxification of naphthalophos by Haemonchus contortus larvae. Antimicrob. Agents Chemother. 2014;58:7475–7483. doi: 10.1128/AAC.03333-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwa M.S., Veenstra J.G., Van Dijk M., Roos M.H. Beta-tubulin genes from the parasitic nematode Haemonchus contortus modulate drug resistance in Caenorhabditis elegans. J. Mol. Biol. 1995;246:500–510. doi: 10.1006/jmbi.1994.0102. [DOI] [PubMed] [Google Scholar]
- Laing R., Bartley D.J., Morrison A.A., Rezansoff A., Martinelli A., Laing S.T., Gilleard J.S. The cytochrome P450 family in the parasitic nematode Haemonchus contortus. Int. J. Parasitol. 2015;45:243–251. doi: 10.1016/j.ijpara.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing R., Kikuchi T., Martinelli A., Tsai I.J., Beech R.N., Redman E., Holroyd N., Bartley D.J., Beasley H., Britton C., Curran D., Devaney E., Gilabert A., Hunt M., Jackson F., Johnston S.L., Kryukov I., Li K., Morrison A.A., Reid A.J., Sargison N., Saunders G.I., Wasmuth J.D., Wolstenholme A., Berriman M., Gilleard J.S., Cotton J.A. The genome and transcriptome of Haemonchus contortus, a key model parasite for drug and vaccine discovery. Genome Biol. 2013;14:R88. doi: 10.1186/gb-2013-14-8-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing S.T., Ivens A., Laing R., Ravikumar S., Butler V., Woods D.J., Gilleard J.S. Characterization of the xenobiotic response of Caenorhabditis elegans to the anthelmintic drug albendazole and the identification of novel drug glucoside metabolites. Biochem. J. 2010;432:505–514. doi: 10.1042/BJ20101346. [DOI] [PubMed] [Google Scholar]
- Lee J.S. Digital image enhancement and noise filtering by use of local statistics. IEEE Trans. Pattern Anal. Mach. Intell. 1980;2:165–168. doi: 10.1109/tpami.1980.4766994. [DOI] [PubMed] [Google Scholar]
- Li X., Schuler M.A., Berenbaum M.R. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu. Rev. Entomol. 2007;52:231–253. doi: 10.1146/annurev.ento.51.110104.151104. [DOI] [PubMed] [Google Scholar]
- Liao V.H., Dong J., Freedman J.H. Molecular characterization of a novel, cadmium-inducible gene from the nematode Caenorhabditis elegans. A new gene that contributes to the resistance to cadmium toxicity. J. Biol. Chem. 2002;277:42049–42059. doi: 10.1074/jbc.M206740200. [DOI] [PubMed] [Google Scholar]
- Lindblom T.H., Dodd A.K. Xenobiotic detoxification in the nematode Caenorhabditis elegans. J. Exp. Zool. A Comp. Exp. Biol. 2006;305:720–730. doi: 10.1002/jez.a.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matouskova P., Lecova L., Laing R., Dimunova D., Vogel H., Raisova Stuchlikova L., Nguyen L.T., Kellerova P., Vokral I., Lamka J., Szotakova B., Varady M., Skalova L. UDP-glycosyltransferase family in Haemonchus contortus: phylogenetic analysis, constitutive expression, sex-differences and resistance-related differences. Int. J. Parasitol. Drugs Drug Resist. 2018;8:420–429. doi: 10.1016/j.ijpddr.2018.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matouskova P., Vokral I., Lamka J., Skalova L. The role of xenobiotic-metabolizing enzymes in anthelmintic deactivation and resistance in helminths. Trends Parasitol. 2016;32:481–491. doi: 10.1016/j.pt.2016.02.004. [DOI] [PubMed] [Google Scholar]
- Mazerska Z., Mroz A., Pawlowska M., Augustin E. The role of glucuronidation in drug resistance. Pharmacol. Ther. 2016;159:35–55. doi: 10.1016/j.pharmthera.2016.01.009. [DOI] [PubMed] [Google Scholar]
- Meech R., Miners J.O., Lewis B.C., Mackenzie P.I. The glycosidation of xenobiotics and endogenous compounds: versatility and redundancy in the UDP glycosyltransferase superfamily. Pharmacol. Ther. 2012;134:200–218. doi: 10.1016/j.pharmthera.2012.01.009. [DOI] [PubMed] [Google Scholar]
- Menzel R., Bogaert T., Achazi R. A systematic gene expression screen of Caenorhabditis elegans cytochrome P450 genes reveals CYP35 as strongly xenobiotic inducible. Arch. Biochem. Biophys. 2001;395:158–168. doi: 10.1006/abbi.2001.2568. [DOI] [PubMed] [Google Scholar]
- Menzel R., Rodel M., Kulas J., Steinberg C.E. CYP35: xenobiotically induced gene expression in the nematode Caenorhabditis elegans. Arch. Biochem. Biophys. 2005;438:93–102. doi: 10.1016/j.abb.2005.03.020. [DOI] [PubMed] [Google Scholar]
- Mi H., Huang X., Muruganujan A., Tang H., Mills C., Kang D., Thomas P.D. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., Casagrande J.T., Thomas P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottier L., Virkel G., Solana H., Alvarez L., Salles J., Lanusse C. Triclabendazole biotransformation and comparative diffusion of the parent drug and its oxidized metabolites into Fasciola hepatica. Xenobiotica. 2004;34:1043–1057. doi: 10.1080/00498250400015285. [DOI] [PubMed] [Google Scholar]
- Nass R., Hamza I. 2007. The Nematode C. elegans as an Animal Model to Explore Toxicology in Vivo: Solid and Axenic Growth Culture Conditions and Compound Exposure Parameters. Current Protocols in Toxicology/ Editorial Board. Mahin D. Maines (Chapter 1), Unit1 9. [DOI] [PubMed] [Google Scholar]
- Olson P.D., Zarowiecki M., Kiss F., Brehm K. Cestode genomics - progress and prospects for advancing basic and applied aspects of flatworm biology. Parasite Immunol. 2012;34:130–150. doi: 10.1111/j.1365-3024.2011.01319.x. [DOI] [PubMed] [Google Scholar]
- Osei-Atweneboana M.Y., Eng J.K., Boakye D.A., Gyapong J.O., Prichard R.K. Prevalence and intensity of Onchocerca volvulus infection and efficacy of ivermectin in endemic communities in Ghana: a two-phase epidemiological study. Lancet. 2007;369:2021–2029. doi: 10.1016/S0140-6736(07)60942-8. [DOI] [PubMed] [Google Scholar]
- Otsen M., Hoekstra R., Plas M.E., Buntjer J.B., Lenstra J.A., Roos M.H. Amplified fragment length polymorphism analysis of genetic diversity of Haemonchus contortus during selection for drug resistance. Int. J. Parasitol. 2001;31:1138–1143. doi: 10.1016/s0020-7519(01)00225-9. [DOI] [PubMed] [Google Scholar]
- Prchal L., Bartikova H., Becanova A., Jirasko R., Vokral I., Stuchlikova L., Skalova L., Kubicek V., Lamka J., Trejtnar F., Szotakova B. Biotransformation of anthelmintics and the activity of drug-metabolizing enzymes in the tapeworm Moniezia expansa. Parasitology. 2015;142:648–659. doi: 10.1017/S0031182014001711. [DOI] [PubMed] [Google Scholar]
- Redman E., Packard E., Grillo V., Smith J., Jackson F., Gilleard J.S. Microsatellite analysis reveals marked genetic differentiation between Haemonchus contortus laboratory isolates and provides a rapid system of genetic fingerprinting. Int. J. Parasitol. 2008;38:111–122. doi: 10.1016/j.ijpara.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Reichert K., Menzel R. Expression profiling of five different xenobiotics using a Caenorhabditis elegans whole genome microarray. Chemosphere. 2005;61:229–237. doi: 10.1016/j.chemosphere.2005.01.077. [DOI] [PubMed] [Google Scholar]
- Robinson M.W., Lawson J., Trudgett A., Hoey E.M., Fairweather I. The comparative metabolism of triclabendazole sulphoxide by triclabendazole-susceptible and triclabendazole-resistant Fasciola hepatica. Parasitol. Res. 2004;92:205–210. doi: 10.1007/s00436-003-1003-6. [DOI] [PubMed] [Google Scholar]
- Roh J.Y., Jung I.H., Lee J.Y., Choi J. Toxic effects of di(2-ethylhexyl)phthalate on mortality, growth, reproduction and stress-related gene expression in the soil nematode Caenorhabditis elegans. Toxicology. 2007;237:126–133. doi: 10.1016/j.tox.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Roos M.H., Otsen M., Hoekstra R., Veenstra J.G., Lenstra J.A. Genetic analysis of inbreeding of two strains of the parasitic nematode Haemonchus contortus. Int. J. Parasitol. 2004;34:109–115. doi: 10.1016/j.ijpara.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Rowland A., Miners J.O., Mackenzie P.I. The UDP-glucuronosyltransferases: their role in drug metabolism and detoxification. Int. J. Biochem. Cell Biol. 2013;45:1121–1132. doi: 10.1016/j.biocel.2013.02.019. [DOI] [PubMed] [Google Scholar]
- Rudgalvyte M., VanDuyn N., Aarnio V., Heikkinen L., Peltonen J., Lakso M., Nass R., Wong G. Methylmercury exposure increases lipocalin related (lpr) and decreases activated in blocked unfolded protein response (abu) genes and specific miRNAs in Caenorhabditis elegans. Toxicol. Lett. 2013;222:189–196. doi: 10.1016/j.toxlet.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runko E., Kaprielian Z. Caenorhabditis elegans VEM-1, a novel membrane protein, regulates the guidance of ventral nerve cord-associated axons. J. Neurosci. 2004;24:9015–9026. doi: 10.1523/JNEUROSCI.2385-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestre A., Humbert J.F. A molecular tool for species identification and benzimidazole resistance diagnosis in larval communities of small ruminant parasites. Exp. Parasitol. 2000;95:271–276. doi: 10.1006/expr.2000.4542. [DOI] [PubMed] [Google Scholar]
- Solana H.D., Rodriguez J.A., Lanusse C.E. Comparative metabolism of albendazole and albendazole sulphoxide by different helminth parasites. Parasitol. Res. 2001;87:275–280. doi: 10.1007/pl00008578. [DOI] [PubMed] [Google Scholar]
- Sommer R.J., Streit A. Comparative genetics and genomics of nematodes: genome structure, development, and lifestyle. Annu. Rev. Genet. 2011;45:1–20. doi: 10.1146/annurev-genet-110410-132417. [DOI] [PubMed] [Google Scholar]
- Soneson C., Love M.I., Robinson M.D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2015;4:1521. doi: 10.12688/f1000research.7563.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup S.T., Spanier B., Grunz G., Bunzel D., Daniel H., Kulling S.E. Formation of phosphoglycosides in Caenorhabditis elegans: a novel biotransformation pathway. PLoS One. 2012;7 doi: 10.1371/journal.pone.0046914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuchlikova L.R., Matouskova P., Vokral I., Lamka J., Szotakova B., Seckarova A., Dimunova D., Nguyen L.T., Varady M., Skalova L. Metabolism of albendazole, ricobendazole and flubendazole in Haemonchus contortus adults: sex differences, resistance-related differences and the identification of new metabolites. Int. J. Parasitol. Drugs Drug Resist. 2018;8:50–58. doi: 10.1016/j.ijpddr.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupp G.S., von Reuss S.H., Izrayelit Y., Ajredini R., Schroeder F.C., Edison A.S. Chemical detoxification of small molecules by Caenorhabditis elegans. ACS Chem. Biol. 2013;8:309–313. doi: 10.1021/cb300520u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velik J., Baliharova V., Skalova L., Szotakova B., Wsol V., Lamka J. Liver microsomal biotransformation of albendazole in deer, cattle, sheep and pig and some related wild breeds. J. Vet. Pharmacol. Ther. 2005;28:377–384. doi: 10.1111/j.1365-2885.2005.00671.x. [DOI] [PubMed] [Google Scholar]
- Vokral I., Bartikova H., Prchal L., Stuchlikova L., Skalova L., Szotakova B., Lamka J., Varady M., Kubicek V. The metabolism of flubendazole and the activities of selected biotransformation enzymes in Haemonchus contortus strains susceptible and resistant to anthelmintics. Parasitology. 2012;139:1309–1316. doi: 10.1017/S0031182012000595. [DOI] [PubMed] [Google Scholar]
- Vokral I., Jirasko R., Stuchlikova L., Bartikova H., Szotakova B., Lamka J., Varady M., Skalova L. Biotransformation of albendazole and activities of selected detoxification enzymes in Haemonchus contortus strains susceptible and resistant to anthelmintics. Vet. Parasitol. 2013;196:373–381. doi: 10.1016/j.vetpar.2013.03.018. [DOI] [PubMed] [Google Scholar]
- von Samson-Himmelstjerna G., Blackhall W. Will technology provide solutions for drug resistance in veterinary helminths? Vet. Parasitol. 2005;132:223–239. doi: 10.1016/j.vetpar.2005.07.014. [DOI] [PubMed] [Google Scholar]
- von Samson-Himmelstjerna G., Walsh T.K., Donnan A.A., Carriere S., Jackson F., Skuce P.J., Rohn K., Wolstenholme A.J. Molecular detection of benzimidazole resistance in Haemonchus contortus using real-time PCR and pyrosequencing. Parasitology. 2009;136:349–358. doi: 10.1017/S003118200800543X. [DOI] [PubMed] [Google Scholar]
- Walsky R.L., Bauman J.N., Bourcier K., Giddens G., Lapham K., Negahban A., Ryder T.F., Obach R.S., Hyland R., Goosen T.C. Optimized assays for human UDP-glucuronosyltransferase (UGT) activities: altered alamethicin concentration and utility to screen for UGT inhibitors. Drug Metab. Dispos. 2012;40:1051–1065. doi: 10.1124/dmd.111.043117. [DOI] [PubMed] [Google Scholar]
- Ward J.D. Rendering the intractable more tractable: tools from Caenorhabditis elegans ripe for import into parasitic nematodes. Genetics. 2015;201:1279–1294. doi: 10.1534/genetics.115.182717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson S.M., Storey B., Howell S., Harper K.M., Kaplan R.M., Wolstenholme A.J. Candidate anthelmintic resistance-associated gene expression and sequence polymorphisms in a triple-resistant field isolate of Haemonchus contortus. Mol. Biochem. Parasitol. 2011;180:99–105. doi: 10.1016/j.molbiopara.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Winterrowd C.A., Pomroy W.E., Sangster N.C., Johnson S.S., Geary T.G. Benzimidazole-resistant beta-tubulin alleles in a population of parasitic nematodes (Cooperia oncophora) of cattle. Vet. Parasitol. 2003;117:161–172. doi: 10.1016/j.vetpar.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Wolstenholme A.J., Fairweather I., Prichard R., von Samson-Himmelstjerna G., Sangster N.C. Drug resistance in veterinary helminths. Trends Parasitol. 2004;20:469–476. doi: 10.1016/j.pt.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Wong D., Bazopoulou D., Pujol N., Tavernarakis N., Ewbank J.J. Genome-wide investigation reveals pathogen-specific and shared signatures in the response of Caenorhabditis elegans to infection. Genome Biol. 2007;8:R194. doi: 10.1186/gb-2007-8-9-r194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young N.D., Jex A.R., Li B., Liu S., Yang L., Xiong Z., Li Y., Cantacessi C., Hall R.S., Xu X., Chen F., Wu X., Zerlotini A., Oliveira G., Hofmann A., Zhang G., Fang X., Kang Y., Campbell B.E., Loukas A., Ranganathan S., Rollinson D., Rinaldi G., Brindley P.J., Yang H., Wang J., Wang J., Gasser R.B. Whole-genome sequence of Schistosoma haematobium. Nat. Genet. 2012;44:221–225. doi: 10.1038/ng.1065. [DOI] [PubMed] [Google Scholar]
- Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanian M., Cook D.E., Lee D., Lee J., Andersen E. bioRxiv; 2017. Heritable Small RNAs Regulate Nematode Benzimidazole Resistance. [Google Scholar]
- Zamanian M., Cook D.E., Zdraljevic S., Brady S.C., Lee D., Lee J., Andersen E.C. Discovery of genomic intervals that underlie nematode responses to benzimidazoles. PLoS Neglected Trop. Dis. 2018;12 doi: 10.1371/journal.pntd.0006368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanger U.M., Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 2013;138:103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- Zarowiecki M., Berriman M. What helminth genomes have taught us about parasite evolution. Parasitology. 2015;142(Suppl. 1):S85–S97. doi: 10.1017/S0031182014001449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of differentially expressed genes of exsheathed infective larvae H. contortus (UGA/2004) exposed to 1.13 mM albendazole for 4 hours, which were significantly differentially expressed (adjusted P value 0.05).