Abstract
Background
Oral squamous cell carcinoma (OSCC) is one of the most common malignant tumors worldwide. Patients with poorly differentiated OSCC often exhibit a poor prognosis. AUNIP (Aurora Kinase A and Ninein Interacting Protein), also known as AIBp, plays a key role in cell cycle and DNA damage repair. However, the function of AUNIP in OSCC remains elusive.
Methods
The differentially expressed genes (DEGs) were obtained using R language. Receiver operating characteristic curve analysis was performed to identify diagnostic markers for OSCC. The effectiveness of AUNIP in diagnosing OSCC was evaluated by machine learning. AUNIP expression was analyzed in publicly available databases and clinical specimens. Bioinformatics analysis and in vitro experiments were conducted to explore biological functions and prognostic value of AUNIP in OSCC.
Findings
The gene integration analysis revealed 90 upregulated DEGs. One candidate biomarker, AUNIP, for the diagnosis of OSCC was detected, and its expression gradually increased along with malignant differentiation of OSCC. Bioinformatics analysis demonstrated that AUNIP could be associated with tumor microenvironment, human papillomavirus infection, and cell cycle in OSCC. The suppression of AUNIP inhibited OSCC cell proliferation and resulted in G0/G1 phase arrest in OSCC cells. The survival analysis showed that AUNIP overexpression predicted poor prognosis of OSCC patients.
Interpretation: AUNIP could serve as a candidate diagnostic and prognostic biomarker for OSCC and suppression of AUNIP may be a potential approach to preventing and treating OSCC.
Fund
Taishan Scholars Project in Shandong Province (ts201511106) and the National Natural Science Foundation of China (Nos. 61603218).
Keywords: Oral squamous cell carcinoma, Receiver operating characteristic curve, Biomarker, Weighted gene co-expression network analysis, Survival analysis, AUNIP
Research in context.
Evidence before this study
Patients with poorly differentiated oral squamous cell carcinoma (OSCC) often have a poor prognosis. Tumor biomarkers can highlight biological differences among cancers and help predict treatment outcomes. However, tumor biomarkers with 100% sensitivity and specificity have not been discovered so far. Although a variety of potential biomarkers of OSCC have been discovered using the bioinformatics method, their verifications failed in large samples. AUNIP, also known as AIBp, is demonstrated to regulate mitotic entry and mitotic spindle assembly during cell cycle. However, the role of AUNIP in solid tumors such as OSCC remains elusive.
Added value of this study
Our research creatively integrated microarray data of OSCC with relative large samples from publicly available databases and clinical specimens. Bioinformatics analysis and in vitro experiments were carried out to speculate the potential biological functions and clinical significance of AUNIP in OSCC. We found that AUNIP was overexpressed in OSCC tissues and increased AUNIP expression predicted poor clinical outcomes in OSCC patients. The decreased expression of AUNIP caused an abnormity of OSCC cell cycle transition. In addition, AUNIP was associated with tumor microenvironment and HPV infection in OSCC.
Implications of all the available evidence
The findings of our research suggest that AUNIP could serve as a potential biomarker and the inhibition of AUNIP may be a new strategy for the treatment of OSCC. Integrating AUNIP with well-known biomarkers to detect OSCC and predict treatment outcomes could promote clinical practice.
Alt-text: Unlabelled Box
1. Introduction
Cancer morbidity and mortality have experienced a gradual increase all over the world. There were approximate 18·1 million cancer cases and 9·6 million cancer deaths worldwide in 2018, with oral cancer accounting for 2% and 1·9% respectively [1,2]. Oral squamous cell carcinoma (OSCC), exhibiting high morbidity and malignancy, is the most common type of oral cancer [3,4]. It normally involves the front two thirds of the tongue, gingiva (gums), floor of mouth, lips, alveolar ridge, buccal mucosa, hard palate, and retromolar trigone (https://www.cancer.gov/). The excision of primary tumor with or without dissection of lymph nodes is the routine treatment of OSCC [4,5]. Most patients with OSCC are in advanced clinical stage at the time of diagnosis [6]. Although the clinical multidisciplinary collaboration and sequential therapy can improve the prognosis [7], the 5-year survival rate of OSCC patients remains about 50% and 25%–50% of them will suffer from local relapse and distant metastasis after treatment [8,9]. It is thus necessary to search for effective diagnostic and prognostic biomarkers for OSCC.
A cancer biomarker is defined as a substance or a process that is indicative of the presence of cancer in the body (https://en.wikipedia.org). There have been many papers reporting the discovery of OSCC biomarkers, but only few biomarkers have been validated and successfully applied in routine clinical practice [10]. Moreover, most biomarkers possess limitations for the early detection of OSCC, and the prognostic value of them is plagued by inaccuracies [11]. By taking advantage of bioinformatics analysis, it is more likely to overcome deficiencies in the known biomarkers. Bioinformatics analysis allows researchers to delve into integrated data of numerous clinical samples from different independent studies, which provides data infrastructure for discovering promising biomarkers and updating our understanding about cancers [6,12,13].
The aim of this study was to explore potential diagnostic and prognostic biomarkers and their biological functions in OSCC by bioinformatics analysis. We finally determined Aurora Kinase A and Ninein Interacting Protein (AUNIP) as our gene of interest and confirmed its overexpression in OSCC tissues. AUNIP, also known as AIBp, was demonstrated to interact with Aurora-A at the C-terminal and with hNinein at the N-terminal to promote centrosomal structure maintenance and spindle formation during cell cycle [14]. By controlling activation of both Aurora-A and Plk1, AUNIP could also regulate mitotic entry and mitotic spindle assembly [15]. Apart from these, AUNIP and Aurora-A were co-overexpressed in various brain tumors [14] and researches suggested that AUNIP might have an oncogenic role in human cancers and regulate cell cycle progression. Recently, AUNIP was discovered to be connected with CtIP and be required for DNA double-strand breaks repair [16]. Our present study shed light on the critical role of AUNIP in regulating cell cycle in OSCC cells and promoting OSCC cell proliferation and proved its diagnostic and prognostic value for OSCC patients. Therefore, AUNIP inhibition could serve as a potential strategy to treat OSCC.
2. Materials and methods
2.1. Ethics statement
The study protocol was approved by Taizhou Hospital Ethics Committee (Zhejiang, China) and Shandong University Research Ethics Committee (Shandong, China). All experiments were performed complying with the relevant regulations, and written informed consent was obtained from patients.
2.2. Clinical specimens
Tissue microarray chips containing 89 samples of OSCC and 16 samples of normal control (nine samples of adjacent normal oral tissue) were purchased from Shanghai Qutdo Biotech Company (Shanghai, China). These samples were collected from Taizhou Hospital (Zhejiang, China). Clinicopathological characteristics of these patients were shown in (Table S1). Four pairs of OSCC and matched adjacent normal oral tissues, obtained from Stomatological Hospital of Shandong University (Shandong, China) and Qilu Hospital of Shandong University (Shandong, China), were frozen in liquid nitrogen until further analysis. Clinicopathological characteristics of these four patients were shown in (Table S2).
2.3. Analysis of upregulated differentially expressed genes (DEGs)
In the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/), we selected datasets of GSE30784 [17], GSE3524 [18], GSE78060 [19], GSE41613 [20], and downloaded the original files (.CEL files) and platform files. Background correction, quantile normalization, pro summarization, log2 conversion, and missing values supplement for the matrix data of each GEO dataset were performed using Robust Multi-array Average algorithm with the “affy” package and the “impute” package in R/Bioconductor software (version 3·5·3). Upregulated DEGs of GSE30784, GSE3524, and GSE78060 displaying an overlap region in Venn diagram, were screened and constructed matrices by means of the “limma” package in R/Bioconductor when log2FoldChange (FC) > 1 and adjusted P-value <0·01.
2.4. Receiver operating characteristic (ROC) curve and logistic regression analysis
By screening out the relevant documents (.count files) and clinical information related to OSCC in The Cancer Genome Atlas (TCGA) database (https://www.cancer.gov) and FireBrowse database (http://www.firebrowse.org) [21], we gained RNA-sequence data and clinicopathological characteristics over two groups of samples, the OSCC tumor category and the adjacent normal one. The “edgeR” package in R/Bioconductor software was used to normalize and process data before log2 conversion. It was reported that infiltrating stromal and immune cells had important roles in tumor development. ESTIMATE score is the estimation of stromal and immune cells in malignant tumor tissues using expression data of TCGA samples. We calculated ESTIMATE, stromal, and immune scores in OSCC by adopting the ESTIMATE algorithm from (https://bioinformatics.mdanderson.org/estimate/) [22]. A total of 306 OSCC patients with detailed survival information and their clinical samples of oral cavity which included the front two thirds of the tongue, gingiva, floor of mouth, lips, alveolar ridge, buccal mucosa, hard palate, and retromolar trigone were involved in subsequent analysis. Then ROC curve analysis was performed to evaluate the sensitivity (true positive rate) and specificity (true negative rate) of the upregulated DEGs for OSCC diagnosis and we investigated how large the area under the curve (AUC) was by using the statistical package IBM SPSS Statistics software (SPSS, version 23).
To further assess the efficacy of the gene in diagnosing OSCC, we performed five-fold cross-validation using logistic regression model in TCGA database [23]. The randomly selected 60 tumor samples and 29 control ones were formed a new dataset, which avoided the size imbalance of two groups. The logistic regression model was built by using “scikit-learn” package in Python software (version 3.6, https://www.python.org) [24]. The confusion matrix of two classifications (the positive and negative) was displayed in (Table S3). Precision, recall, accuracy and F1-score were introduced to evaluate the performance of this classification model. The precision of a classifier was the number of true positives divided by the number that we predicted as positive. High precision meant high accuracy of predicting OSCC by AUNIP. Recall was defined as the number of true positives divided by the number of actual positives. Accuracy referred to the proportion of correctly predicted results among all samples. Because precision and recall affected each other and the values of them could not be ideally simultaneously large, we considered computing the F1-score as a comprehensive evaluation index. F1-score was the harmonic average of precision and recall.
2.5. Immunohistochemistry (IHC) staining
The tissue microarray chips through deparaffinization and dehydration were incubated with monoclonal rabbit anti-human AUNIP (dilution 1:500, Bioss, bs-15019R) overnight at 4 °C after epitope retrieval, H2O2 treatment and non-specific antigens blocking. Chips were next incubated with secondary antibody, followed by signal detection with DAB staining kit (Vector Laboratories, USA), and Histochemistry Score (H-Score = ∑ (PI × I) = (percentage of cells of weak intensity × 1) + (percentage of cells of moderate intensity × 2) + percentage of cells of strong intensity × 3) [25] was obtained with Quant Center Analysis tool.
2.6. Construction of weighted gene co-expression network analysis (WGCNA)
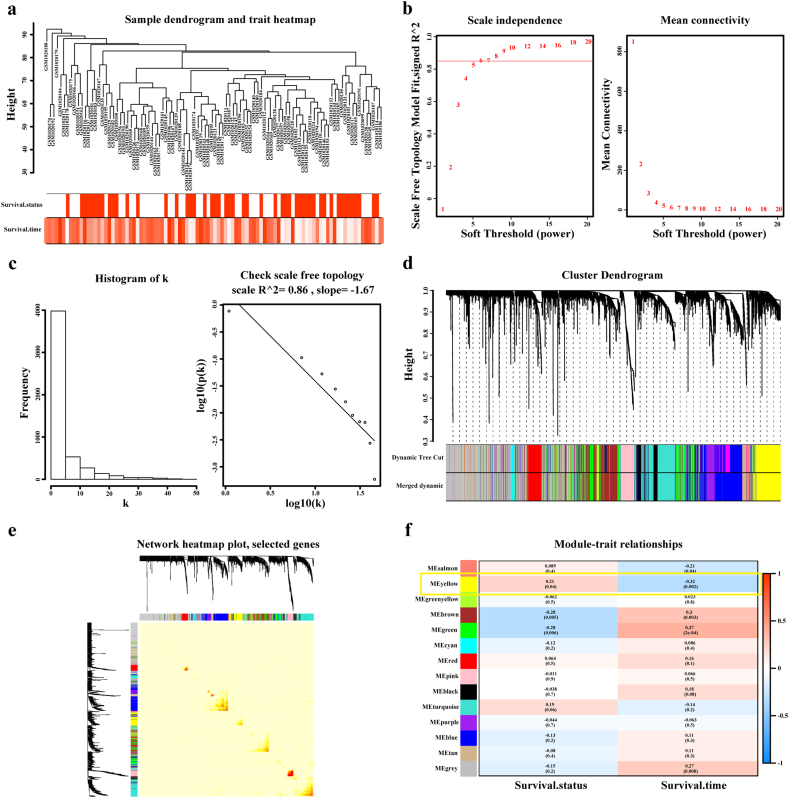
GSE41613 dataset including 97 OSCC patients with detailed overall survival information was appropriate for the construction of WGCNA. We could link module eigengenes with survival data of OSCC patients. The data matrix of gene expression in GSE41613 was constructed and the top 25% of genes in tumor samples with the largest variance were selected as input data set for the subsequent WGCNA using “WGCNA” package in R/Bioconductor software. Outlier samples were detected and excluded with sample hierarchically clustering method, before selecting an appropriate soft threshold power in order to achieve standard scale-free networks. In the next stage, through the construction of adjacency and topological overlap matrix (TOM) and the calculation of corresponding dissimilarity (1-TOM), gene dendrogram and module identification were accomplished with dynamic tree cut and the minimum module size was 30. Then clustering of module eigengenes was implemented to merge highly similar modules with the dissimilarity of <0·25 and we calculated the correlation between module eigengene and clinical phenotypes of OSCC. The module including AUNIP was chosen for further analysis and gene significance (GS) and module membership (MM) were obtained to identify modules related to clinical traits of OSCC.
2.7. Biological function and pathway enrichment analysis
In gene networks conforming to scale-free distribution, genes with similar expression patterns could be co-regulated, functionally related or pathway shared. Through calculating the corresponding topological overlap, genes in the above selected module positively associated with AUNIP were found out (displayed by Cytoscape 3·6·1) and they were next executed gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis via the “clusterprofiler” package in R/Bioconductor software to acquire the enriched biological process and KEGG pathway for next analysis. In addition, samples from TCGA were divided into two groups based on the expression of AUNIP (median value) and gene set enrichment analysis (GSEA) software (http://software.broadinstitute.org/gsea/index.jsp) was applied in the two groups to verify the results of GO and KEGG analysis. The cut-off criteria for GSEA were nominal P-value <0·05 and false discovery rate (FDR) < 0·25.
2.8. Cell culture and transient transfection
Human OSCC cell lines SCC-15 and SCC-9 were obtained from ATCC (Beijing Beina Chuanglian Biotechnology Institute) and they were cultured in F12 and DMEM respectively containing 10% fetal bovine serum (Gibco, Carlsbad, CA, USA). Both cell lines were maintained in a humidified incubator with 37 °C, 5% CO2. Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was used to transfect Negative Control (NC) and AUNIP siRNAs (Sigma-Aldrich, USA) into OSCC cells as the manufacturer's protocol suggested. Target sequences for siRNAs were UUCUCCGAACGUGUCACGUdTdT ACGUGACACGUUCGGAGAAdTdT (NC si), CCAUUUGAUCCCAGGCUUAdTdT UAAGCCUGGGAUCAAAUGGdTdT (AUNIP si1), and GAAACAAAGCCGUAGUCCAdTdT UGGACUACGGCUUUGUUUCdTdT (AUNIP si2).
2.9. RNA extraction, RT-PCR and qRT-PCR
Total RNA extracted from SCC-15 and SCC-9 with Trizol Reagent (Invitrogen, Carlsbad, CA, USA) were reverse transcribed with RT reagent Kit gDNA Eraser (TaKaRa). Next, SYBR-Green (TaKaRa) and qRT-PCR analysis were used for detecting cDNA expression levels and β-ACTIN was used as internal reference. Primers were shown as follows: hβ-ACTIN, Forward (F): 5′-AGTTGCGTTACACCCTTTCTTG-3′, Reverse (R): 5′-CACCTTCACCGTTCCAGTTTT-3′; hAUNIP, Forward (F): 5′- GCGGAAAGTGCAGACACATTT -3′, Reverse (R): 5′- TCTCTGGTGAATGCCTGTAGAT -3′.
2.10. Western blotting
Total proteins were extracted with lysis buffer including protease inhibitors, and the concentration was measured. PVDF membranes (Millipore, Bedford, MA) containing proteins separated on SDS-PAGE gels were blocked with 5% nonfat dry milk and incubated with primary antibodies for overnight at 4 °C. Immunoblots were detected using ECL detection reagent (Millipore) according to manufacturer's protocol. Antibodies used were: β-ACTIN (A1978, Sigma-Aldrich, RRID: AB_476692), GAPDH (Santa Cruz Biotechnology, sc-47724), AUNIP (Bioss, bs-15019R).
2.11. Colony formation and cell cycle analysis
Cells (500–800 cells/well) were seeded in 6-well plate and supported for 7–14 days in a humidified incubator with 37 °C, 5% CO2 until colonies of cells appeared. The colonies were fixed with methanol and stained with Giemsa in order to be counted. The colonies containing >50 cells were counted for analysis. For cell cycle analyses, cells were fixed with 70% ethanol at 4 °C overnight and stained with RNase A containing Propidium Iodide (Sigma-Aldrich, USA). Cell cycle distribution was determined using flow cytometry with ModFit software. Blank control, si-NC, and si-AUNIP groups were established.
2.12. Statistical analysis
The Kaplan-Meier analysis for overall survival was proceeded based on the gene's expression level whose cut-off level was set at the median value with the aid of GraphPad Prism 8 software and the Log-Rank was utilized to test. SPSS was employed to perform univariate and multivariable analysis so as to establish a Cox proportional hazard regression model. Patients were divided into groups in the light of the target gene's expression whose correlation with clinicopathological characteristics in TCGA was studied by SPSS with the application of two-tailed χ2 test. In addition, Pearson correlation analysis was adopted to determine the linear relationship between two groups. One-way ANOVA test and two-tailed Student's t-tests were used for expression data comparisons by using GraphPad Prism 8 software. Each experiment was repeated three times or more and all data were presented as mean ± standard deviation (SD). Statistical significance was described as follows: n.s, not significant; *P ≤ 0·05; **P ≤ 0·01; ***P ≤ 0·001; ****P ≤ 0·0001.
3. Results
3.1. Identification of upregulated DEGs in OSCC after data integration
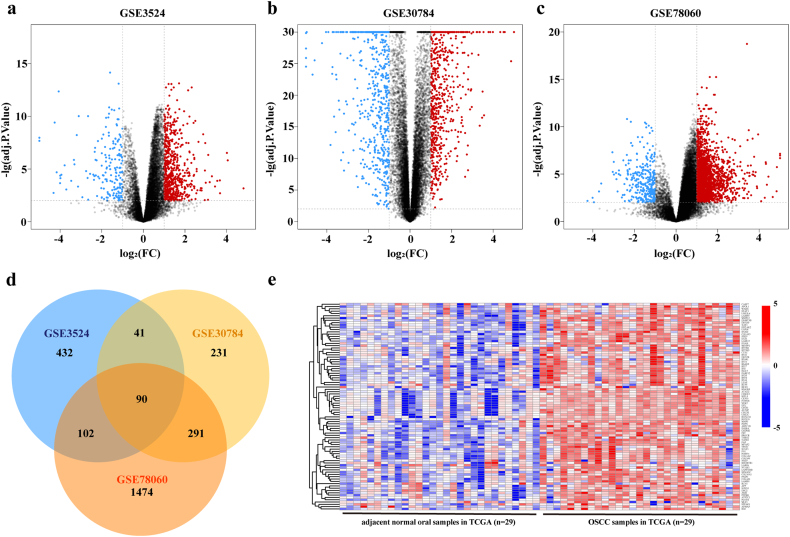
As the volcano plots illustrated, gene expression profiles from GSE3524 identified 843 differentially expressed genes with 665 genes upregulated and 178 genes downregulated in OSCC samples compared with the expression in normal control tissues (Fig. 1a). From GSE30784 data, we recognized 1266 differentially expressed genes, of which 653 genes were upregulated and 613 genes were downregulated in OSCC (Fig. 1b). 2264 differentially expressed genes were ascertained in GSE78060, comprising 1957 genes upregulated and 307 genes downregulated in OSCC (Fig. 1c). Grounded on the cut-off criteria, DEGs were selected for integrated analysis and we gained 90 overlapping upregulated DEGs from the above mentioned three GSE datasets (Fig. 1d, Table S4). Gene expression profiles from paired adjacent and tumor samples in TCGA database confirmed the overexpression of these 90 genes in OSCC samples (Fig. 1e).
Fig. 1.
Identification of differentially upregulated expressed genes.
a–c. Volcano plots of gene expression profiles in GSE3524, GSE30784, and GSE78060. Red/blue symbols classify the upregulated/downregulated genes according to the criteria: log2FC > 1 and adjusted P-value <0·01. d. Common upregulated DEGs among GSE3524, GSE30784, and GSE78060. e. The expression matrix of 90 common upregulated DEGs in 29 pairs of OSCC and adjacent normal tissues followed by unsupervised hierarchical clustering in TCGA database. Blue, red and white respectively represents a lower expression level, a higher expression level and no expression difference among the genes. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Identification of key genes for diagnosis of OSCC
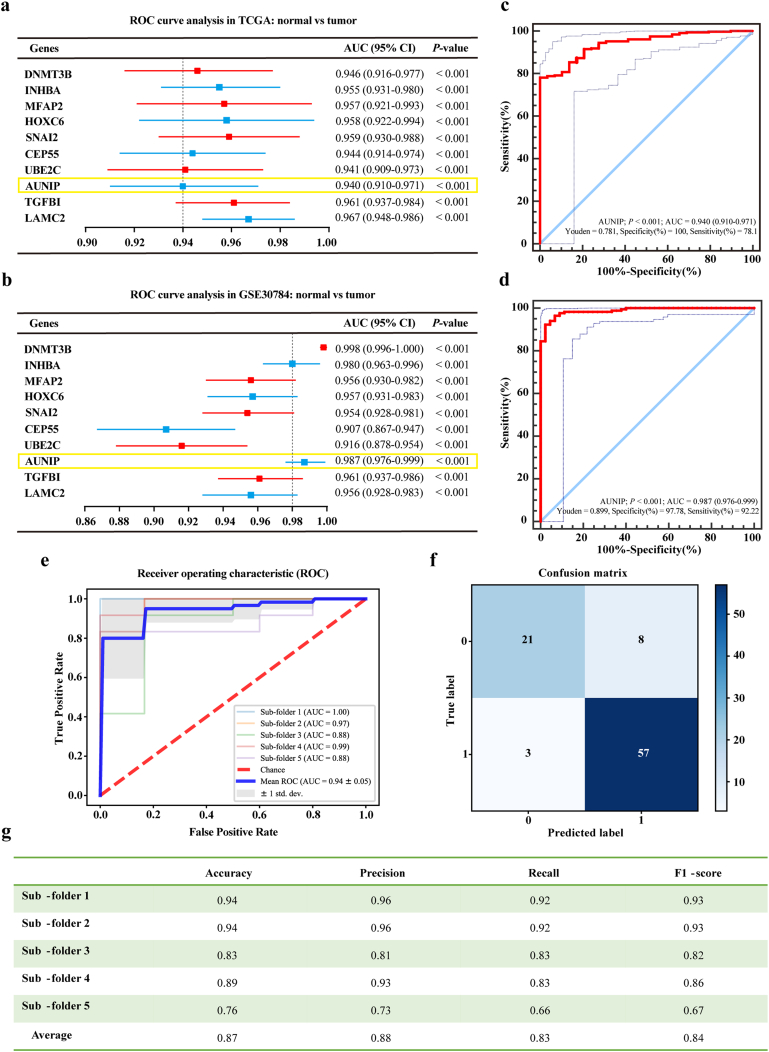
We conducted ROC curve analysis and compared the AUC value to evaluate the sensitivity and specificity of the above-mentioned 90 selected genes for the diagnosis of OSCC in TCGA database. The top ten genes were listed according to their AUC values which were no <0·940, demonstrating that these genes had the highest diagnostic significance for OSCC (Fig. 2a). To verify the diagnostic significance of these ten genes for OSCC, ROC curve analysis was performed in GSE30784 dataset again (Fig. 2b). Judging from the previous studies on OSCC or even all other cancers, we found one barely reported gene among the list, namely AUNIP. ROC curves of AUNIP in TCGA database and GSE30784 dataset were displayed (Fig. 2c–d), showing good sensitivity and specificity with average AUC of 0·940 and 0·987 respectively. AUNIP bore 78·10% prediction accuracy and 100% specificity at the Youden Index of 0·781 in TCGA database, and 92·22% prediction accuracy and 97·78% specificity at the Youden Index of 0·899 in GSE30784 dataset. AUNIP has been confirmed to play crucial roles in cell cycle control and DNA damage repair. Therefore, its aberrant expression in pathological conditions, such as tumor development, may contribute to the occurrence of diseases. Of note, the expression and function of AUNIP in cancers remain largely unknown. Therefore, we proposed AUNIP as the target gene of this study and tried to discover its role in OSCC development. Logistic regression was next modeled to further appraise the effectiveness of AUNIP in diagnosing OSCC. In ROC curve of AUNIP, the mean of AUC (0·940 ± 0·050) (Fig. 2e) was obtained after the construction of confusion matrix (Fig. 2f). The average values of accuracy, precision, recall, and F1-score were 0·870, 0·880, 0·830, and 0·840 respectively (Fig. 2g). These results meant that AUNIP showed a positive performance to classify the OSCC samples from the normal controls.
Fig. 2.
Identification of key genes for diagnosis of OSCC.
a. ROC curve analysis of the top ten DEGs in TCGA database. Genes with an AUC value are shown. b. ROC curve analysis of the selected ten genes in GSE30784. Genes with an AUC value are shown. c, d. ROC curve to assess sensitivity and specificity of AUNIP expression as a diagnostic biomarker for OSCC in TCGA database and GSE30784 dataset. e. ROC curve of AUNIP in the five-fold cross-validation. f. Construction of confusion matrix. g. Evaluation metrics of each fold. All data are represented by mean ± SD.
3.3. High expression of AUNIP is associated with the progression and tumor purity of OSCC
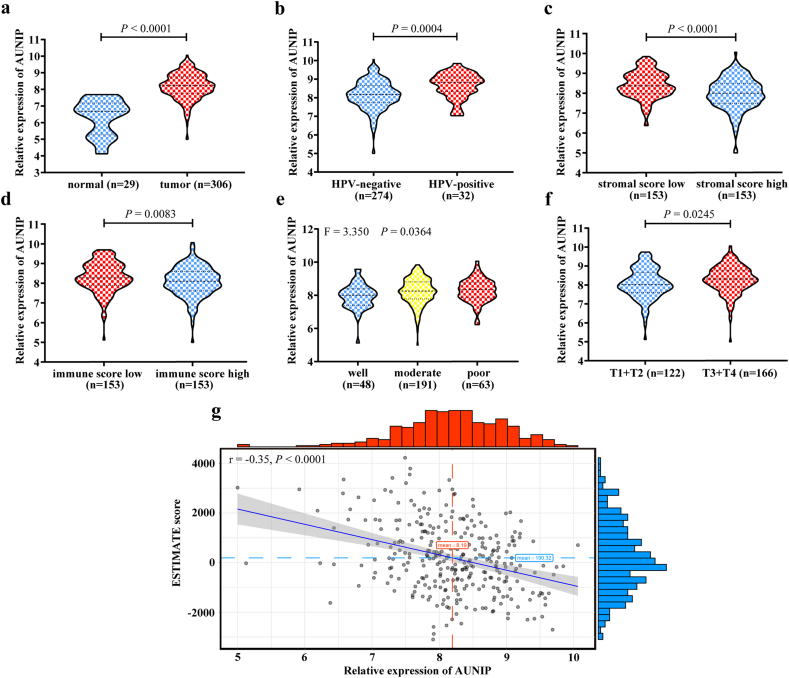
To assess the role of AUNIP in the progression of OSCC, we exploited TCGA database to analyze its expression. The mRNA level of AUNIP was significantly increased in OSCC tissues compared to normal oral tissues (Fig. 3a), and the similar results were observed in human papillomavirus (HPV)-positive, low stromal score (median value), and low immune score (median value) group (Fig. 3b–d). Further studies showed that the expression level of AUNIP gradually increased along with malignant progression of the tumor (indicated by histologic grades and tumor T stages) (Fig. 3e–f). In accordance with the different expression levels of AUNIP, the correlation among several clinicopathological characteristics of OSCC patients was explored. High expression of AUNIP (median value) was statistically associated with HPV infection (positive; P < 0·001), stromal score (low; P < 0·001), immune score (low; P = 0·016), lymphovascular invasion (yes; P = 0·049), histologic grade (poor; P = 0·036) and tumor T stage (T3 + T4; P = 0·006; Table 1). The expression of AUNIP negatively correlated with ESTIMATE score (r = −0·348; P < 0·0001; Fig. 3g), suggesting that AUNIP was positively connected with tumor purity of OSCC.
Fig. 3.
AUNIP expression is elevated in OSCC samples from TCGA database.
a. TCGA database analysis of the relatively differential expression level (log2) of AUNIP in OSCC (n = 306) and adjacent normal oral (n = 29) tissues. b. Quantification of AUNIP mRNA levels (log2) in HPV-negative (n = 274) and HPV-positive (n = 32) OSCC tissues from TCGA database. c, d. Analysis of AUNIP mRNA levels (log2) in different stromal score and immune score (low, high, n = 153) of OSCC tissues from TCGA database. e, f. Analysis of AUNIP mRNA levels (log2) in different histologic grades (well, n = 48; moderate, n = 191; poor, n = 63) and tumor T stages (T1 + T2, n = 122; T3 + T4, n = 166) of OSCC tissues from TCGA database. g. The negative correlation between AUNIP mRNA expression level (log2) and ESTIMATE score in TCGA database. P-values were obtained by Student's t-test, One-way ANOVA test, and Pearson correlation analysis. All data are represented by mean ± SD.
Table 1.
Correlations between AUNIP and clinicopathological characteristics.
| AUNIP expression |
|||
|---|---|---|---|
| Characteristics | Low, no. cases |
High, no. cases |
P-value |
| Age (years) | |||
| < 60 | 62 | 69 | 0.419 |
| ≥ 60 | 91 | 84 | |
| Gender | |||
| Female | 55 | 44 | 0.179 |
| Male | 98 | 109 | |
| Alcohol history | |||
| No | 57 | 46 | 0.143 |
| Yes | 91 | 105 | |
| HPV infection | |||
| Negative | 146 | 128 | < 0.001 |
| Positive | 7 | 25 | |
| Stromal score | |||
| Low | 58 | 95 | < 0.001 |
| High | 95 | 58 | |
| Immune score | |||
| Low | 66 | 87 | 0.016 |
| High | 87 | 66 | |
| Radiation therapy | |||
| No | 62 | 51 | 0.089 |
| Yes | 72 | 90 | |
| Lymphovascular invasion | |||
| No | 82 | 76 | 0.049 |
| Yes | 26 | 43 | |
| Perineural invasion | |||
| No | 52 | 55 | 0.802 |
| Yes | 62 | 70 | |
| Histologic grade | |||
| Well | 32 | 16 | 0.036 |
| Moderate | 88 | 103 | |
| Poor | 30 | 33 | |
| Tumor stage | |||
| I–II | 42 | 29 | 0.075 |
| III–IV | 100 | 113 | |
| T stage | |||
| T1–2 | 73 | 49 | 0.006 |
| T3–4 | 72 | 94 | |
| N stage | |||
| N0 | 61 | 54 | 0.327 |
| Non N0 | 69 | 78 | |
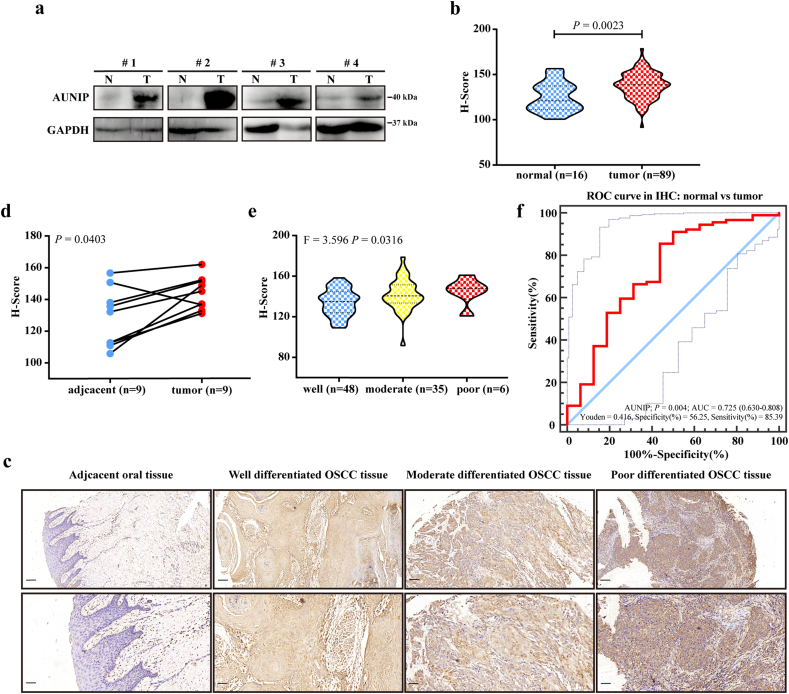
Then the protein expression of AUNIP was detected by western blotting and IHC staining, and the results of which were similar to the mRNA expression in OSCC tissues. Western blotting revealed the overexpression of AUNIP in OSCC patients (Fig. 4a). In IHC staining, AUNIP was overexpressed in OSCC tissues compared with normal controls (Fig. 4b–c) and with paired adjacent normal oral tissues (Fig. 4d). And AUNIP protein expression grew along with the malignant progression of tumors (indicated by change of histologic grades) (Fig. 4c; Fig. 4e). The AUC value obtained by ROC curve analysis from IHC staining was 0·725, which held the statistical significance to support the diagnostic value of AUNIP for OSCC (85·39% prediction accuracy and 56·25% specificity at the Youden Index of 0·416; Fig. 4f). Altogether, these data implied the potential oncogenic role of AUNIP in OSCC.
Fig. 4.
AUNIP expression is upregulated in clinical OSCC samples.
a. The protein levels of AUNIP in four pairs of OSCC tissues (T) and adjacent non-tumor tissues (N) measured by western blot. b. Quantification of AUNIP IHC staining in OSCC (n = 16) and normal oral (n = 89) tissues. c. Representative images of IHC staining for AUNIP in normal oral tissues and different histologic grades of OSCC tissues. Scale bars: 100 μm (insets 50 μm). d. Quantification of AUNIP IHC staining in OSCC and paired adjacent normal tissues (n = 9). e. Quantification of AUNIP IHC staining in different histologic grades of OSCC tissues (well, n = 48; moderate, n = 35; poor, n = 6). f ROC curve from IHC staining shows AUNIP is a marker to distinguish OSCC tissues from normal oral tissues. P-values were obtained by Student's t-test and One-way ANOVA test. All data are represented by mean ± SD.
3.4. Weighted co-expression network construction and identification of key modules, especially the yellow module containing AUNIP
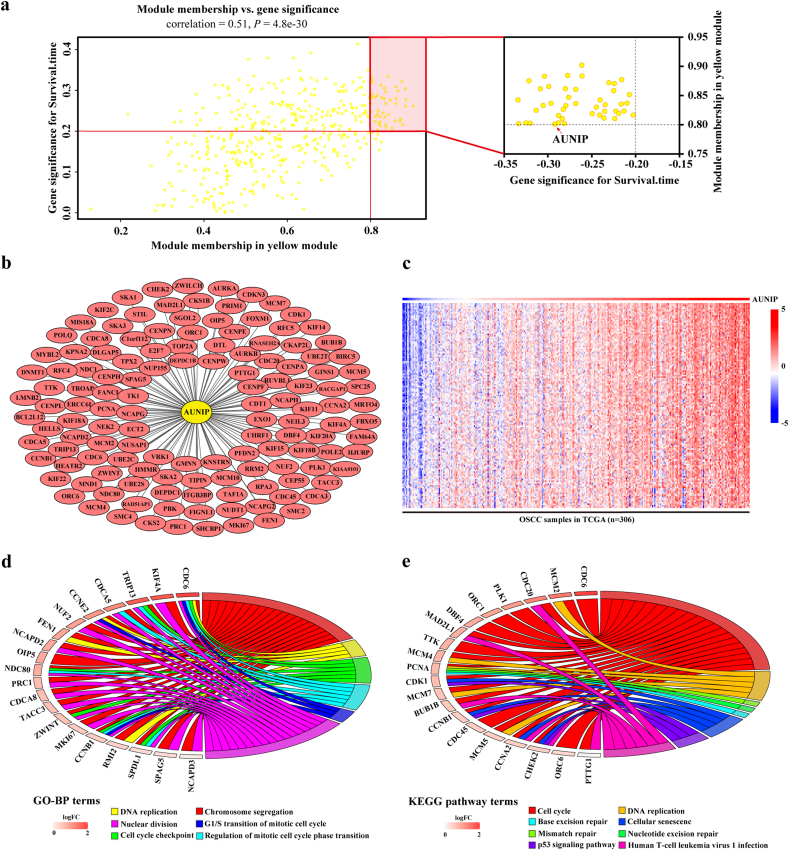
WGCNA is a systematic biological method to analyze the expression patterns of multiple genes in different samples and it can form clusters or modules containing genes with the same expression pattern [26]. If certain genes are located in one module, they are likely to possess the same biological functions and the association between modules and sample characteristics such as clinical traits can be studied [27,28]. GSM1020115 was excluded from subsequent analysis in GSE41613 as an outlier sample, so a total of 96 samples with survival data were included in WGCNA (Fig. 5a). In this study, we selected the power of β = 8 (scale free R2 = 0·850) as the soft-thresholding to achieve a scale-free network (Fig. 5b–c). As a result, 13 gene co-expression modules were identified after excluding the grey module using merged dynamic tree cut (Fig. 5d). The heat map plotted the TOM among 1000 selected genes in the analysis, which revealed that every module was an independent validation to each other (Fig. 5e). Afterwards, we found the module containing AUNIP, i.e. yellow module, as a key module for accounting for the highest correlation with OSCC prognosis (R2 = −0·320, P = 0·002 with survival time; R2 = 0·21, P = 0·04 with survival status; Fig. 5f). We drew scatter plots of GS vs. MM in the yellow module with survival time of OSCC patients (correlation = 0·51, P = 4·80e−30), elaborating that AUNIP was one of the genes under the condition of MM > 0·80 and GS > 0·20 (Fig. 6a). AUNIP was thus deemed as one of the hub genes in the yellow module.
Fig. 5.
Weighted co-expression network construction and identification of the key module containing AUNIP.
a. Hierarchical clustering dendogram of samples from GSE41613. The clinical traits of survival status and survival time are displayed at the bottom. b, c. Analysis of scale-free fit index and the mean connectivity for various soft-thresholding powers. Testing the scale free topology when β = 8. d. Hierarchical clustering dendogram of genes with dissimilarity based on topological overlap. Modules are the branches of the clustering tree. e. The heat map describes the TOM among selected 1000 genes in WGCNA, and darker colour represents higher overlap and lighter colour corresponds to lower overlap. The gene dendrogram and module assignment are shown along the left side and the top. f. Correlation between module eigengenes and clinical traits. Each row corresponds to a module eigengene and columns represent clinical traits. Each cell contains the correlation and P-value, and yellow module containing AUNIP is selected. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 6.
Genes positively associated with AUNIP are enriched in cell cycle.
a. Scatter plot of genes in yellow module. The vertical line represents cutoff of module membership = 0·8, and the horizontal line represents cutoff of gene significances for survival time = 0·2. Genes on upper right contains AUNIP. b. Network of genes positively associated with AUNIP expression according to the topological overlap. c. Heat map describes expression profiles of 134 genes positively associated with AUNIP in 306 OSCC samples in TCGA database. d. Gene ontology analysis of genes positively associated with AUNIP. e. KEGG pathway enrichment analysis of genes positively associated with AUNIP. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.5. Genes positively associated with AUNIP are enriched in cell cycle related processes and HPV infection
To further explore the potential biological function of AUNIP in OSCC, we constructed the network of 134 genes positively associated with AUNIP on the basis of the topological overlap in WGCNA (Fig. 6b) and found that expression profiles of these genes in 306 OSCC samples from TCGA database were similar (Fig. 6c). Moreover, we uploaded the selected genes to R/Bioconductor software to conduct GO and KEGG pathway analysis. GO analysis showed that the genes were enriched in DNA replication, chromosome segregation, nuclear division, G1/S transition of mitotic cell cycle, cell cycle checkpoint, regulation of mitotic cell cycle phase transition, etc. (Fig. 6d). The signaling network resulted from KEGG pathway analysis established the association between AUNIP and cell cycle such as cell cycle pathway, DNA replication, mismatch repair, etc. (Fig. 6e).
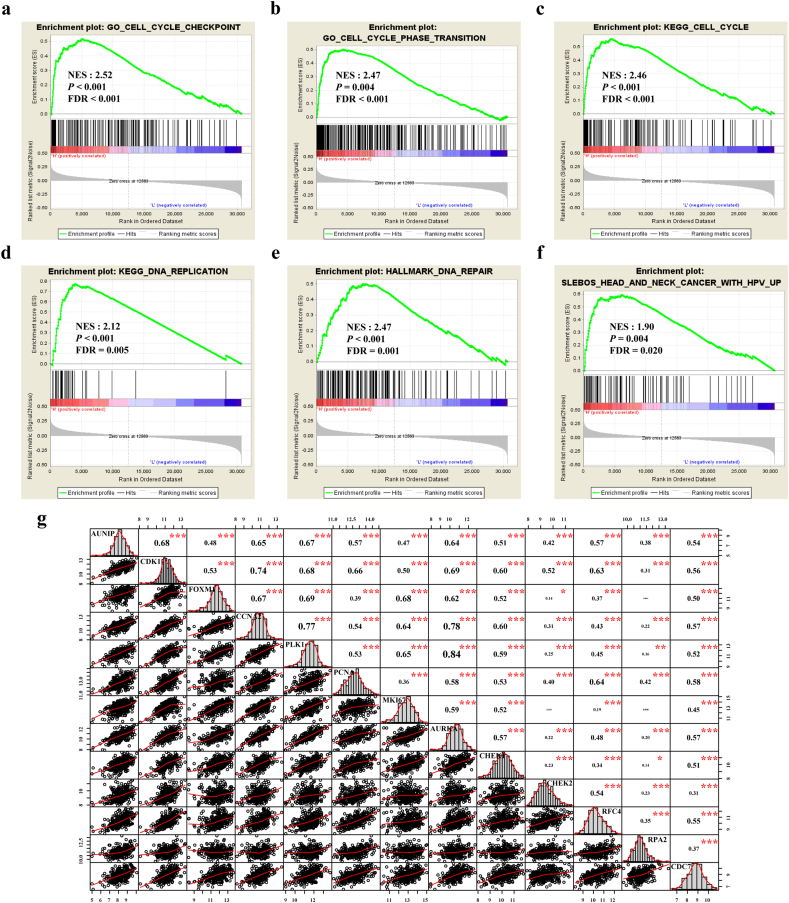
The relation between AUNIP expression and Hallmark, GO, KEGG pathway gene sets were analyzed with GSEA software. Cell cycle checkpoint, cell cycle phase transition, cell cycle, DNA replication, DNA repair, head and neck cancer with HPV up were significantly enriched (Fig. 7a–f). Given the vital functions of key molecules in the regulation of cell cycle, DNA repair and HPV infection in OSCC, we made correlation analysis to assess the relationship between AUNIP and several well-known genes in OSCC samples from TCGA database. AUNIP was highly correlated with CDK1 (Cyclin Dependent Kinase 1), FOXM1 (Forkhead Box M1), CCNA2 (Cyclin A2), PLK1 (Polo Like Kinase 1), PCNA (Proliferating Cell Nuclear Antigen), MKI67 (Marker of Proliferation Ki-67), AURKA (Aurora Kinase A), CHEK1 (Checkpoint Kinase 1), CHEK2 (Checkpoint Kinase 2), RFC4 (Replication Factor C Subunit 4), RPA2 (Replication Protein A2), and CDC7 (Cell Division Cycle 7) in 306 OSCC samples from TCGA database (Fig. 7g). CDK1, FOXM1, CCNA2, PLK1, PCNA, MKI67, AURKA, CHEK2 and RFC4 were included in the network of genes positively associated with AUNIP constructed by WGCNA (Fig. 6b). Based on these results, we deduced that AUNIP could regulate mitotic cell cycle progression, connect with HPV infection in OSCC and play an important role during tumorigenesis.
Fig. 7.
AUNIP is associated with cell cycle and HPV status in OSCC.
a–f. GSEA of Hallmarks, GO, KEGG pathway gene sets in AUNIP high versus low samples from TCGA database. Normalized enrichment score (NES), nominal P-value and FDR are shown in each plot. g. Correlation between AUNIP and cell cycle, HPV infection related molecules in OSCC samples from TCGA database. *P ≤ 0·05, **P ≤ 0·01, and ***P ≤ 0·001 were obtained by Pearson correlation analysis.
3.6. AUNIP promotes OSCC cell proliferation and controls cell cycle progression in vitro
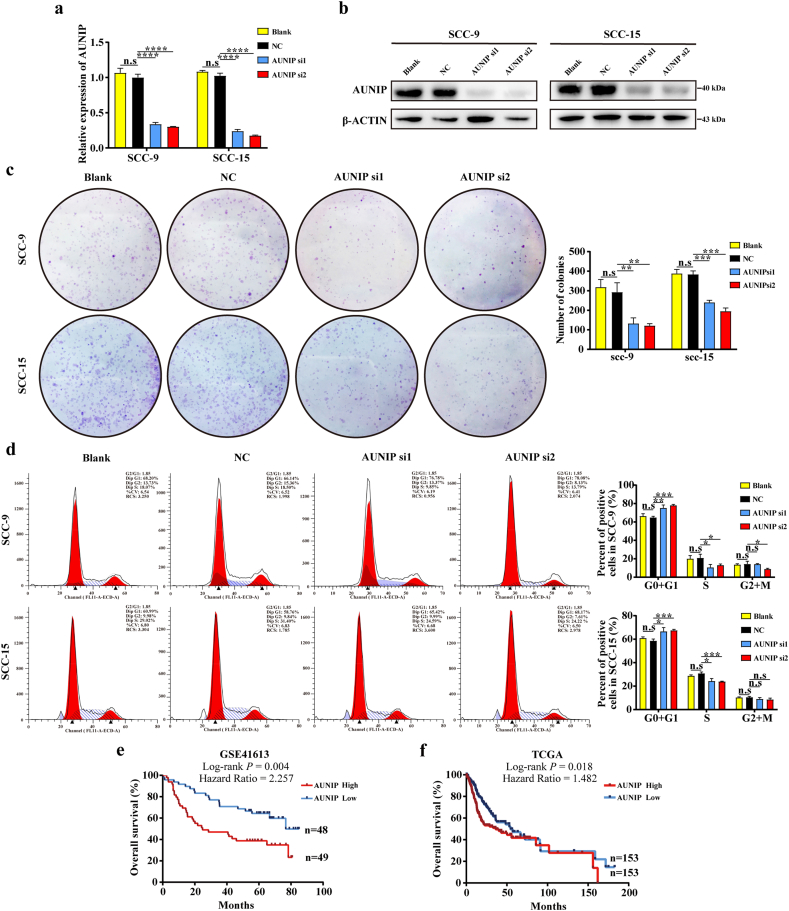
In order to verify the results of above bioinformatics analysis, AUNIP was knocked down in SCC-9 and SCC-15 OSCC cell lines (Fig. 8a–b). It turned out that colony formation in OSCC cells was significantly reduced after AUNIP depletion (Fig. 8c). Flow cytometry was performed to analyze cell cycle, as shown in (Fig. 8d). The OSCC cells with AUNIP suppression were observed to arrest in G0/G1 phase, confirming the role of AUNIP in controlling cell cycle progression.
Fig. 8.
AUNIP controls cell cycle progression in vitro and its expression is associated with poor survival in OSCC patients.
a. AUNIP mRNA levels in indicated cells transfected with AUNIP siRNA. b. AUNIP protein levels in indicated cells transfected with AUNIP siRNA. c. Representative images (left) and quantification (right) of Blank, Negative Control (NC) siRNA-, and AUNIP siRNA- transfected SCC-9 and SCC-15 cells were analyzed in a colony formation assay. d. Representative images (left) and quantification (right) of Blank, Negative Control (NC) siRNA-, and AUNIP siRNA- transfected SCC-9 and SCC-15 cells were analyzed in cell cycle assay. e, f. Kaplan-Meier analysis of overall survival was performed to indicate higher expression of AUNIP was correlated with poor survival of OSCC patients in GSE41613 and TCGA datasets. P-values were obtained from the log-rank test. n.s, not significant, *P ≤ 0·05, **P ≤ 0·01, ***P ≤ 0·001, and ****P ≤ 0·0001 were obtained by Student's t-test. All data are represented by mean ± SD.
3.7. High expression of AUNIP predicts poor prognosis in OSCC
AUNIP's prognostic value in OSCC was worth exploring, considering its oncogenic potential implied by the above results. The expression level of AUNIP was introduced into GSE41613 and TCGA data for Kaplan-Meier survival analysis and Log-Rank test. As a result, OSCC patients with high expression of AUNIP (median value) had a markedly lower overall survival rate in two datasets (Fig. 8e–f). The median survival lengths of low AUNIP expression groups were 76·75 and 54·70 months, whereas in the AUNIP high expression groups were 24·87 and 36·33 months respectively. For the COX univariate regression analysis, seven elements with statistical significance were AUNIP expression, radiation therapy, lymphovascular invasion, perineural invasion, tumor stage, tumor T stage, and tumor N stage. Finally, COX multivariate regression analysis manifested that AUNIP could be deemed to be an independent prognostic factor for OSCC (hazard ratio (HR) = 1·421, 95% confidence interval (CI) = 1·012 to 2·459, P = 0·043; Table 2).
Table 2.
Univariate and multivariate analysis of various factors associated with overall survival in patients with OSCC.
| Characteristics | Univariate analysis |
Multivariate analysis |
||
|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
| AUNIP expression High expression |
1.491 (1.067–2.082) | 0.019 | 1.421 (1.012–2.459) | 0.043 |
| Age (years) ≥ 60 |
1.181 (0.843–1.654) | 0.333 | ||
| Gender Male |
0.917 (0.649–1.296) | 0.623 | ||
| Alcohol history Yes |
1.074 (0.760–1.518) | 0.687 | ||
| HPV infection Positive |
1.213 (0.719–2.046) | 0.470 | ||
| Histologic grade Moderate and Poor |
1.533 (0.955–2.461) | 0.077 | ||
| Stromal score High |
0.878 (0.630–1.224) | 0.444 | ||
| Immune score High |
0.874 (0.627–1.218) | 0.427 | ||
| Radiation therapy Yes |
0.568 (0.406–0.795) | 0.001 | 0.359 (0.248–0.518) | < 0.001 |
| Lymphovascular invasion Yes |
1.617 (1.112–2.351) | 0.012 | 1.622 (1.070–2.459) | 0.023 |
| Perineural invasion Yes |
1.538 (1.105–2.143) | 0.011 | 1.295 (0.902–1.859) | 0.162 |
| Tumor stage III–IV |
1.926 (1.298–2.858) | 0.001 | 1.008 (0.478–2.126) | 0.984 |
| T stage T3–4 |
2.217 (1.543–3.186) | < 0.001 | 2.253 (1.241–4.093) | 0.008 |
| N stage Non N0 |
1.570 (1.125–2.189) | 0.008 | 1.301 (0.839–2.016) | 0.240 |
4. Discussion
OSCC, one of the most common epithelial malignancies in head and neck squamous cell carcinoma (HNSCC) [29], exerts an adverse influence on the appearance and life quality of patients [30,31]. It often involves the tongue, lips and the floor of mouth and easily spreads to regional lymph nodes and/or distant organs [9,32,33]. At present, the comprehensive clinical examination and histological analysis of suspicious areas are implemented to diagnose OSCC, but early diagnosis is still difficult due to the limited sensitivity and specificity [34,35]. Also the unaffordable and disfiguring-prone therapeutic alternatives sometimes do not work because of the poor prognosis [11,36]. The search for more effective diagnostic and prognostic markers thus represents a continuing challenge for the biomedical research. Thanks to advances in bioinformatics techniques, researchers are able to seek promising biomarkers for OSCC and some relevant studies have been published. For instance, a set of prognostic signatures including PLAU (Plasminogen Activator, Urokinase), CLDN8 (Claudin 8), and CDKN2A (Cyclin Dependent Kinase Inhibitor 2A) were identified from the database to predict overall survival in OSCC patients [6]. PSMA7 (Proteasome Subunit Alpha 7), ITGA6 (Integrin Subunit Alpha 6), ITGB4 (Integrin Subunit Beta 4), and APP (Amyloid Beta Precursor Protein) were overexpressed in HNSCC and significantly correlated with poor overall survival in patients [37]. Additionally, LGALS1 (Galectin 1) was found to be linked to oral cancer progression and metastasis, and potentially served as a prognostic biomarker for oral cancer therapy [9].
What we focused on in this study were potential oncogenes with diagnostic and prognostic values. We found a total of 90 genes were upregulated in OSCC samples from three GSE datasets. Ten genes were confirmed as potential markers for the diagnosis of OSCC with high sensitivity and specificity using ROC curve analysis. Among these, we discovered a candidate AUNIP and evaluated its efficacy in diagnosing OSCC with machine learning. The expression of this gene was further validated in TCGA database and clinical samples, which proved that AUNIP was overexpressed in tumorous tissues by comparison with that in normal oral tissues, especially in patients with HPV infection, low stromal score, low immune score, poor histologic grade, and advanced tumor T stage. To further investigate the role of AUNIP in OSCC, gene co-expression network was constructed and yellow module containing AUNIP was selected. Before conducting function and pathway enrichment analysis, we stumbled upon a high correlation between the yellow module and the survival rate of OSCC patients. The biological functions of AUNIP in OSCC cells were researched and we verified that increased expression of AUNIP can predict a poor prognosis in patients with OSCC in independent cohorts.
Cancer is characterized by uncontrolled cell proliferation caused by the aberrant activity of cell cycle controlling genes [38,39]. Several cell cycle related inhibitors have shown antitumor activities in cancer researches, such as Dinaciclib for CDK1 [40], Rigosertib and Volasertib for PLK1 [39]. In our study, the oncogenic role of AUNIP in OSCC development was initially revealed. G0/G1 phase arrest and inhibition of OSCC cell proliferation were observed resulting from depletion of AUNIP expression. The interaction between AUNIP and cell cycle related molecules was foundation of cell cycle blockade, so we analyzed their relations and found out that AUNIP was highly correlated with CDK1, FOXM1, CCNA2, PLK1, PCNA, MKI67, AURKA, CHEK1 and CHEK2 in OSCC.
The tumor microenvironment of HNSCC critically functions as both positive and negative regulators of hallmarks of cancer. Its comprehensive understanding could provide important insights into tumor progression and prognosis. For example, immunohistochemical analysis reveals that tumors heavily infiltrated by tumor-infiltrating lymphocytes are associated with a better outcome of patients with HNSCC [41]. In our work, the negative correlation between AUNIP and stromal and immune score suggested that AUNIP could be one of the tumor microenvironment related genes that affected the recruitment of infiltrating stromal and immune cells into the tumor microenvironment of OSCC. Patients with relatively high expression level of AUNIP may poorly response to immunotherapy, which could be one of the explanations that AUNIP has predictive value for OSCC prognosis. And the relationship between tumor microenvironment and AUNIP could impact on the curative effect of radiotherapy and HPV status in OSCC. However, further studies are needed to elucidate the interaction network between AUNIP and infiltrating stromal and immune cells.
DNA double-strand breaks are the most deleterious form of DNA damage, and they are mainly repaired through two pathways: non-homologous end joining and homologous recombination. A number of determinants influence which pathway should be chosen. One of these is cell cycle. Studies have shown that efficient DNA end processing is restricted to the S and G2 phases, and is regulated by cell cycle-dependent CDK activity [42,43]. Furthermore, cells have checkpoints to halt cell cycle progression in response to DNA damage, allowing time for DNA repair. Some DNA damage checkpoints have influences on the activity of specific CDK complexes. PLK1 is crucial for mitotic entry following recovery from DNA damage-induced cell cycle arrest [39], and PCNA is associated with DNA damage tolerance pathways [44]. AUNIP is regarded as a sensor of DNA damage [16]. This research went further to detect G0/G1 phase arrest after depletion of AUNIP expression and the positive correlation between AUNIP and CDK1, CHEK1, CHEK2, PLK1, PCNA demonstrated that AUNIP could be involved in DNA damage repair processes and the role in DNA damage repair processes could affect cell cycle progression in OSCC. However, AUNIP might change the radiotherapy effect and promote cellular resistance to drugs through DNA damage repair processes as a potential target for the treatment of OSCC, which needs to be confirmed by more work.
HPV infection was associated with a subset of HNSCC and it was estimated to cause 5% of OSCC worldwide [45]. We found AUNIP was overexpressed in HPV-positive OSCC samples compared with HPV-negative ones, and it was highly correlated with RPA2, RFC4, and CDC7 which were associated with HPV status in HNSCC as reported [46]. These findings uncovered that AUNIP could have a biological role in HPV infection in OSCC. Further studies should be warranted in the future to clarify the relationship between AUNIP and HPV status in OSCC and to illustrate detailed molecular mechanism concerning the effects of AUNIP in carcinogenesis.
Our research creatively integrated microarray data of OSCC with relative large samples from GEO and TCGA database. WGCNA and GSEA were carried out to speculate the potential biological functions and clinicopathological significance of AUNIP in OSCC. AUNIP was found to be overexpressed in OSCC, and its decreased expression caused an abnormity of OSCC cell cycle transition. Therefore, suppressing AUNIP may be a strategy for OSCC prevention and treatments.
Funding sources
This study was supported by the Taishan Scholars Project in Shandong Province (ts201511106) and the National Natural Science Foundation of China (Nos. 61603218).
Author contributions
XX and XQ designed the study. ZY, XL, LZ, FL, TL and XY performed the experiments. ZY, YL and YF analyzed the data. XX and XQ obtained the funding. ZY, XX and XQ prepared the figs. ZY, XL, YL, LZ, XX and XQ wrote the manuscript. XX and XQ supervised the study. All authors read and approved the final manuscript.
Declaration of Competing Interest
The authors declare no conflicts of interest.
Acknowledgements
We thank Professor Jihui Jia and Professor Chunyan Chen for offering expert advice concerning this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.08.013.
Contributor Information
Xu Qiao, Email: qiaoxu@sdu.edu.cn.
Xin Xu, Email: xinxu@sdu.edu.cn.
Appendix A. Supplementary data
Supplementary material
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.Shi J., Bao X., Liu Z., Zhang Z., Chen W., Xu Q. Serum miR-626 and miR-5100 are promising prognosis predictors for Oral squamous cell carcinoma. Theranostics. 2019;9(4):920–931. doi: 10.7150/thno.30339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang T.H., Li K.Y., Choi W.S. Lymph node ratio as prognostic variable in oral squamous cell carcinomas: systematic review and meta-analysis. Oral Oncol. 2019;89:133–143. doi: 10.1016/j.oraloncology.2018.12.032. [DOI] [PubMed] [Google Scholar]
- 5.Gharat S.A., Momin M., Bhavsar C. Oral squamous cell carcinoma: current treatment strategies and nanotechnology-based approaches for prevention and therapy. Crit Rev Ther Drug Carrier Syst. 2016;33(4):363–400. doi: 10.1615/CritRevTherDrugCarrierSyst.2016016272. [DOI] [PubMed] [Google Scholar]
- 6.Zhao X., Sun S., Zeng X., Cui L. Expression profiles analysis identifies a novel three-mRNA signature to predict overall survival in oral squamous cell carcinoma. Am J Cancer Res. 2018;8(3):450–461. [PMC free article] [PubMed] [Google Scholar]
- 7.Yen C.J., Tsou H.H., Hsieh C.Y. Sequential therapy of neoadjuvant biochemotherapy with cetuximab, paclitaxel, and cisplatin followed by cetuximab-based concurrent bioradiotherapy in high-risk locally advanced oral squamous cell carcinoma: final analysis of a phase 2 clinical trial. Head Neck. 2019;41(6):1703–1712. doi: 10.1002/hed.25640. [DOI] [PubMed] [Google Scholar]
- 8.Gan C.P., Sam K.K., Yee P.S. IFITM3 knockdown reduces the expression of CCND1 and CDK4 and suppresses the growth of oral squamous cell carcinoma cells. Cell Oncol (Dordr) 2019;42(4):477–490. doi: 10.1007/s13402-019-00437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J.M., Tseng C.W., Lin C.C. Upregulation of LGALS1 is associated with oral cancer metastasis. Ther Adv Med Oncol. 2018;10 doi: 10.1177/1758835918794622. 1758835918794622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hussein A.A., Forouzanfar T., Bloemena E. A review of the most promising biomarkers for early diagnosis and prognosis prediction of tongue squamous cell carcinoma. Br J Cancer. 2018;119(6):724–736. doi: 10.1038/s41416-018-0233-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rivera C., Oliveira A.K., Costa R.A.P., De Rossi T., Paes Leme A.F. Prognostic biomarkers in oral squamous cell carcinoma: a systematic review. Oral Oncol. 2017;72:38–47. doi: 10.1016/j.oraloncology.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 12.Liu X., Wu J., Zhang D. Identification of potential key genes associated with the pathogenesis and prognosis of gastric cancer based on integrated bioinformatics analysis. Front Genet. 2018;9:265. doi: 10.3389/fgene.2018.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ni M., Liu X., Wu J. Identification of candidate biomarkers correlated with the pathogenesis and prognosis of non-small cell lung cancer via integrated bioinformatics analysis. Front Genet. 2018;9:469. doi: 10.3389/fgene.2018.00469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lieu A.S., Cheng T.S., Chou C.H. Functional characterization of AIBp, a novel Aurora-a binding protein in centrosome structure and spindle formation. Int J Oncol. 2010;37(2):429–436. doi: 10.3892/ijo_00000691. [DOI] [PubMed] [Google Scholar]
- 15.Chou C.H., Loh J.K., Yang M.C. AIBp regulates mitotic entry and mitotic spindle assembly by controlling activation of both Aurora-A and Plk1. Cell Cycle. 2015;14(17):2764–2776. doi: 10.1080/15384101.2015.1066536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lou J., Chen H., Han J. AUNIP/C1orf135 directs DNA double-strand breaks towards the homologous recombination repair pathway. Nat Commun. 2017;8(1):985. doi: 10.1038/s41467-017-01151-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foy J.P., Bertolus C., Ortiz-Cuaran S. Immunological and classical subtypes of oral premalignant lesions. Oncoimmunology. 2018;7(12) doi: 10.1080/2162402X.2018.1496880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y., Fan H., Zheng L. Biological information analysis of differentially expressed genes in oral squamous cell carcinoma tissues in GEO database. J BUON. 2018;23(6):1662–1670. [PubMed] [Google Scholar]
- 19.Wang R., Zhou X., Wang H. Integrative analysis of gene expression profiles reveals distinct molecular characteristics in oral tongue squamous cell carcinoma. Oncol Lett. 2019;17(2):2377–2387. doi: 10.3892/ol.2018.9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bai S., Zhang P., Zhang J.C. A gene signature associated with prognosis and immune processes in head and neck squamous cell carcinoma. Head Neck. 2019;41(8):2581–2590. doi: 10.1002/hed.25731. [DOI] [PubMed] [Google Scholar]
- 21.Chabanais J., Labrousse F., Chaunavel A., Germot A., Maftah A. POFUT1 as a promising novel biomarker of colorectal cancer. Cancers (Basel) 2018;10(11) doi: 10.3390/cancers10110411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshihara K., Shahmoradgoli M., Martinez E. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hosmer D.W., Lemeshow S., Sturdivant R.X. 3rd ed. Wiley; Hoboken, New Jersey: 2013. Applied logistic regression. [Google Scholar]
- 24.Müller A.C., Guido S. 1st ed. O'Reilly Media, Inc; Sebastopol, CA: 2016. Introduction to machine learning with Python: A guide for data scientists. [Google Scholar]
- 25.Azim H.A., Jr., Peccatori F.A., Brohee S. RANK-ligand (RANKL) expression in young breast cancer patients and during pregnancy. Breast Cancer Res. 2015;17:24. doi: 10.1186/s13058-015-0538-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beckerman P., Qiu C., Park J. Human kidney tubule-specific gene expression based dissection of chronic kidney disease traits. EBioMedicine. 2017;24:267–276. doi: 10.1016/j.ebiom.2017.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang J., Kong D., Cui Q. Prognostic genes of breast cancer identified by gene co-expression network analysis. Front Oncol. 2018;8:374. doi: 10.3389/fonc.2018.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosen E.Y., Wexler E.M., Versano R. Functional genomic analyses identify pathways dysregulated by progranulin deficiency, implicating Wnt signaling. Neuron. 2011;71(6):1030–1042. doi: 10.1016/j.neuron.2011.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carnielli C.M., Macedo C.C.S., De Rossi T. Combining discovery and targeted proteomics reveals a prognostic signature in oral cancer. Nat Commun. 2018;9(1):3598. doi: 10.1038/s41467-018-05696-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang S., Yin S., Zhang Z.L., Su X., Xu Z.F. Quality of life after oral cancer resection and free flap reconstruction. J Oral Maxillofac Surg. 2019;77(8):1724–1732. doi: 10.1016/j.joms.2019.02.029. [DOI] [PubMed] [Google Scholar]
- 31.Kansy K., Hoffmann J., Alhalabi O. Subjective and objective appearance of head and neck cancer patients following microsurgical reconstruction and associated quality of life horizontal line a cross-sectional study. J Craniomaxillofac Surg. 2018;46(8):1275–1284. doi: 10.1016/j.jcms.2018.05.024. [DOI] [PubMed] [Google Scholar]
- 32.Sequeira I., Neves J.F., Carrero D. Immunomodulatory role of keratin 76 in oral and gastric cancer. Nat Commun. 2018;9(1):3437. doi: 10.1038/s41467-018-05872-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y., Guo W., Li Z. Role of the EZH2/miR-200 axis in STAT3-mediated OSCC invasion. Int J Oncol. 2018;52(4):1149–1164. doi: 10.3892/ijo.2018.4293. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Keshavarzi M., Darijani M., Momeni F. Molecular imaging and oral cancer diagnosis and therapy. J Cell Biochem. 2017;118(10):3055–3060. doi: 10.1002/jcb.26042. [DOI] [PubMed] [Google Scholar]
- 35.Baykul T., Yilmaz H.H., Aydin U., Aydin M.A., Aksoy M., Yildirim D. Early diagnosis of oral cancer. J Int Med Res. 2010;38(3):737–749. doi: 10.1177/147323001003800302. [DOI] [PubMed] [Google Scholar]
- 36.Murphy J., Isaiah A., Wolf J.S., Lubek J.E. Quality of life factors and survival after total or extended maxillectomy for sinonasal malignancies. J Oral Maxillofac Surg. 2015;73(4):759–763. doi: 10.1016/j.joms.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 37.Yang B., Chen Z., Huang Y., Han G., Li W. Identification of potential biomarkers and analysis of prognostic values in head and neck squamous cell carcinoma by bioinformatics analysis. Onco Targets Ther. 2017;10:2315–2321. doi: 10.2147/OTT.S135514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 39.Otto T., Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer. 2017;17(2):93–115. doi: 10.1038/nrc.2016.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang P., Kawakami H., Liu W. Targeting CDK1 and MEK/ERK overcomes apoptotic resistance in BRAF-mutant human colorectal cancer. Mol Cancer Res. 2018;16(3):378–389. doi: 10.1158/1541-7786.MCR-17-0404. [DOI] [PubMed] [Google Scholar]
- 41.Peltanova B., Raudenska M., Masarik M. Effect of tumor microenvironment on pathogenesis of the head and neck squamous cell carcinoma: a systematic review. Mol Cancer. 2019;18(1):63. doi: 10.1186/s12943-019-0983-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huertas P., Cortes-Ledesma F., Sartori A.A., Aguilera A., Jackson S.P. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455(7213):689–692. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chapman J.R., Taylor M.R., Boulton S.J. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47(4):497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 44.Kanao R., Masutani C. Regulation of DNA damage tolerance in mammalian cells by post-translational modifications of PCNA. Mutat Res. 2017;803–805:82–88. doi: 10.1016/j.mrfmmm.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 45.Fakhry C., Blackford A.L., Neuner G. Association of oral human papillomavirus DNA persistence with cancer progression after primary treatment for oral cavity and oropharyngeal squamous cell carcinoma. JAMA Oncol. 2019;5(7):985–992. doi: 10.1001/jamaoncol.2019.0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Slebos R.J., Yi Y., Ely K. Gene expression differences associated with human papillomavirus status in head and neck squamous cell carcinoma. Clin Cancer Res. 2006;12(3):701–709. doi: 10.1158/1078-0432.CCR-05-2017. Pt 1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material