Dear Editor,
FOXA1, a forkhead (FKHD) family transcription factor, is highly expressed in the epithelium of endoderm-derived organs, including the prostate gland.1 Transgenic mouse studies have shown that FOXA1 expression is required for prostate epithelial cell differentiation and ductal morphogenesis during development and for the maintenance of this differentiated epithelial phenotype in the adult. Mechanistically, FOXA1 binds FKHD motifs in the DNA to open chromatin and increase local accessibility, thereby recruiting androgen receptor (AR) to prostate lineage-specific enhancers.2,3 AR mediates prostatic transcriptional program and normal prostate development and function. However, AR also plays a pivotal role in prostate cancer (PCa). Equilibrium between nuclear FOXA1 and AR levels is essential for defining a prostatic, rather than an oncogenic, AR program and for balancing cell differentiation and growth.4,5 In addition, FOXA1 has been shown to play androgen-independent roles in regulating epithelial-to-mesenchymal transition (EMT), cell invasion, and tumor metastasis.6 Recent studies have found FOXA1 among the most frequently mutated genes in PCa, with ~4% and ~12% mutation rates in localized tumors and metastatic castration-resistant prostate cancer (CRPC), respectively.7,8 A majority of these mutations cluster at the FKHD domain, especially around the Wing2 region.
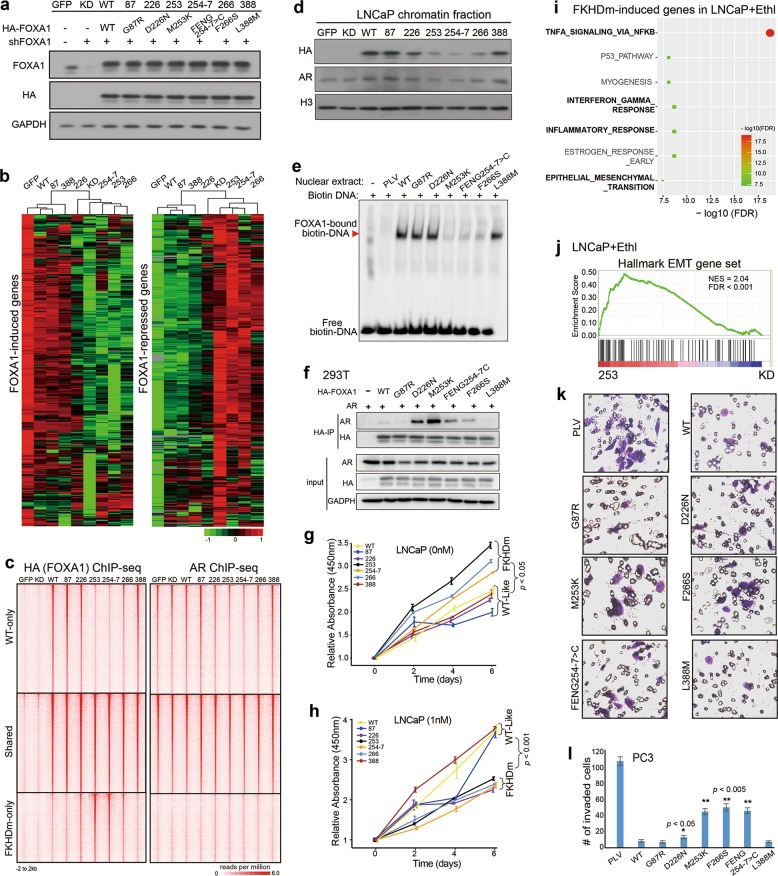
To examine the roles of FOXA1 mutations in PCa, we cloned four frequent mutations at the FKHD domain (D226N, M253K, FENG254-7>C (del), F266S), and one mutation (G87R) in the N-terminal and one (L388M) in the C-terminal region of the protein, all with 2× HA tag. To avoid overexpressing non-physiological levels of FOXA1, we performed FOXA1 knockdown and re-expression to generate nine stable LNCaP cell lines including control (GFP), FOXA1 knockdown (KD), and KD with wild type (WT), G87R, D226N, M253K, FENG254-7>C, F266S, and L388M re-expression, denoted as GFP, KD, WT, 87, 226, 253, 254-7, 266, and 388, respectively (Fig. 1a). Gene expression analysis revealed that re-introduction of FKHD domain mutants (FKHDm), namely 226, 253, 254-7, and 266, failed to rescue FOXA1-mediated gene expression under both androgen-stimulated (Fig. 1b) and -deprived conditions (Supplementary information, Fig. S1), and showed expression patterns similar to that of the KD cells. In contrast, 87 and 388 lines clustered together with the GFP and WT cells, exhibiting WT-like expression patterns.
Fig. 1.
Altered chromatin recruitment by FKHDm promotes androgen independence and cell invasion in PCa. a LNCaP cells were infected with GFP or 2× HA-tagged WT or mutant FOXA1 (named 87, 226, 253, 254-7, 266, 388) and with control or FOXA1-targeting shRNA (shFOXA1, KD), followed by puromycin selection and immunoblotting. Ectopic and total FOXA1 proteins were measured using HA and FOXA1 antibodies, respectively. GAPDH was used as a loading control. b FOXA1-induced or -repressed genes were derived by comparing GFP with KD cells and genes/samples were grouped by hierarchical clustering. c Heatmaps of ChIP-seq data showing the occupancy of ectopic FOXA1 and endogenous AR in various LNCaP cells. All rows were centered on FOXA1 binding sites (± 2 kb) identified in WT and/or 254-7 cells. d Immunoblot showing AR and ectopic HA-FOXA1 levels in the chromatin-bound fraction of LNCaP cells. H3 was used as a chromatin control. e Cell lysates were harvested from 293T cells transfected with control (PLV), WT, or mutant FOXA1 and incubated with a biotin-labeled DNA probe containing a FKHD motif for EMSA assays. FOXA1-DNA complex is indicated by a red arrowhead. f The 293T cells were transfected by AR and WT or mutant FOXA1 plasmids. Whole cell lysates (input) were subjected to immunoprecipitation using anti-HA antibody followed by immunoblotting to detect HA-FOXA1 and AR. g, h LNCaP cells with FOXA1 KD and re-introduction of WT or mutant FOXA1 were hormone-starved for 3 days, grown in 0 (g) or 1 nM (h) of synthetic androgen R1881, and then subjected to cell growth assays. i GSEA analysis of the genes that were induced by FKHDm 253 as compared to KD cells. Y-axis shows the enriched hallmark gene sets. X-axis indicates FDR values of the enrichment. Molecular concepts in bold are related to inflammatory response and EMT. j GSEA of hallmark EMT genes in 253 vs KD LNCaP cells grown in hormone-deprived medium. PC3 cells with WT or mutant FOXA1 overexpression were subjected to cell invasion assays (k) and data were quantified (l). *P < 0.05 and **P < 0.005 compared to WT
To identify binding sites of ectopic FOXA1 WT or mutant proteins, we performed ChIP-seq using an anti-HA antibody. Despite the presence of some gained binding sites, FKHDm overall showed dramatically reduced chromatin binding, with 253, 254-7, and 266 being the most greatly impaired, while 87 and 388 demonstrated similar binding profiles to WT (Fig. 1c and Supplementary information, Fig. S2a–c). FKHDm reduced FOXA1 binding to chromatin under both androgen-stimulated and -deprived conditions (Supplementary information, Fig. S2d, e). Western blot analysis confirmed less chromatin-bound FKHDm proteins as compared to WT, 87, and 388 in LNCaP and 293T cells (Fig. 1d and Supplementary information, Fig. S2f–h). To investigate whether this is due to impaired DNA-binding ability of FKHDm, we performed electrophoretic mobility shift assay (EMSA) using a biotin-labeled DNA probe containing a FKHD motif and found that FKHDm, especially M253K, FENG254-7>C, and F266S, markedly reduced the ability of FOXA1 to interact with DNA (Fig. 1e).
To determine how FOXA1 mutations alter AR cistrome, we performed AR ChIP-seq and observed concordant changes of AR binding at lost, shared, and gained FOXA1-binding sites in FKHDm cells as compared to WT (Fig. 1c and Supplementary information, Fig. S3a, b). Surprisingly, FKHDm decreased not only FOXA1-dependent, but also FOXA1-independent (i.e., AR-only) sites, indicating a global inhibition (Supplementary information, Fig. S3d–f). FKHDm expression further decreased AR binding in FOXA1 KD cells, suggesting a gain of function of FKHDm in reducing AR cistrome. Immunoblotting confirmed an overall reduction in chromatin-bound AR in the FKHDm cells, which was independently validated by immunofluorescent staining of cells with or without pre-extraction and removal of chromatin-free proteins (Fig. 1d and Supplementary information, Fig. S3g, h). Importantly, co-immunoprecipitation experiments revealed that FKHDm proteins, which have impaired DNA-binding ability, formed much stronger interaction with the AR proteins, thus preventing them from binding to the chromatin (Fig. 1f). Concordantly, re-introduction of FKHDm to FOXA1 KD cells failed to rescue AR signaling and resulted in androgen irresponsiveness (Supplementary information, Fig. S3i). On the other hand, FOXA1 KD in hormone-deprived cells induced basal AR signaling as previously reported5 (Supplementary information, Fig. S3j). Importantly, FKHDm likewise failed to affect AR signaling and exhibited expression patterns similar to that of KD cells, i.e., with enhanced AR activities under hormone-deprived conditions. Concordantly, FKHDm cells showed reduced growth as compared with WT-like cells in the presence of androgen, but induced growth under hormone-deprived conditions, favoring CRPC (Fig. 1g, h). Interestingly, 226 demonstrated intermediate effects. It showed a decrease in androgen-stimulated cell growth and gene expression (Supplementary information, Fig. S3i), possibly because of its increased ability to interact with AR in a way similar to other FKHDm (Fig. 1f). On the other hand, 226 clustered with WT-like cells in androgen-independent cell growth and gene expression (Supplementary information, Fig. S3j), likely due to its largely remained ability to bind DNA (Fig. 1c, e).
To compare genes regulated by FOXA1 WT and FKHDm, we performed Venn diagram analysis and identified a large number of genes that were uniquely regulated by FKHDm (Supplementary information, Fig. S4a–d). Gene Set Enrichment Analysis (GSEA) revealed that FKHDm-induced genes were strongly enriched for hallmark gene sets involved in EMT, interferon-gamma response, and inflammatory responses, especially under androgen-deprived conditions (Fig. 1i, j; Supplementary information, Fig. S4e–g). Hallmark EMT signature was markedly upregulated in FKHDm cells and the expression of epithelial marker E-cadherin was dramatically decreased. This is in striking contrast to WT FOXA1-regulated genes, which have been previously shown to suppress these pathways and inhibit cell invasion.6 Mechanistically, this could be caused by loss of the ability of FKHDm to inhibit EMT genes, such as TGFB3 (Supplementary information, Fig. S1d), or by a gain of function of FKHDm to directly induce these genes. The latter was supported by our observation of some gained FOXA1-binding sites in FKHDm cells (Fig. 1c). De novo motif analysis revealed a second to FKHD motif in the FKHDm binding sites that is similar to the FKHD motif, but is more AT-rich and is associated with the ARID5A protein (Supplementary information, Fig. S5a). Interestingly, ARID5A has been previously implicated in the regulation of inflammation and interferon-gamma responses.9 Indeed, GSEA analysis of target genes associated with the gained FKHDm binding sites revealed a significant (q = 8.22E−04) enrichment of EMT gene set. Concordantly, FKHDm cells showed increased cell invasion as compared to WT in androgen-dependent LNCaP cells, androgen-independent but AR-dependent C4-2B cells (Supplementary information, Fig. S5b–e), and, in particular, in AR-negative PC3 cells (Fig. 1k, l). As this function is independent of AR signaling, 226 demonstrated effects similar to WT, rather than other FKHDm cells.
In summary, we show that PCa-associated FOXA1 mutations around residue 253 in the FKHD domain were defective in DNA binding but enhanced in AR interaction, thereby hampering both FOXA1 and AR cistromes. These mutations, albeit caused a decrease in AR signaling and cell growth in the presence of androgen, substantially increased cell growth under hormone-deprived conditions, promoting CRPC. In addition, FKHDm FOXA1 regulated a unique transcriptome enriched in EMT and inflammatory responses, and concordantly induced cell invasion. Moreover, these mutations were shown to promote CRPC tumor metastasis in xenograft models (personal communications).10 The clinical relevance of our findings was validated in the TCGA dataset wherein we observed decreased AR signaling but enhanced EMT in PCa with FKHDm FOXA1 (Supplementary information, Fig. S6). In aggregates, our study provides the mechanistic details by which FKHDm FOXA1 promotes PCa progression.
Supplementary information
Acknowledgements
We thank Dr Daniel Foltz (Northwestern University) for helpful discussions. This work was supported in part by the NIH R50CA211271 (to J.C.Z.), NIH prostate SPORE P50CA180995 (to J.Y.), and Prostate Cancer Foundation 2017CHAL2008 (to J.Y., J.C.Z.).
Author contributions
J.Y. and J.C.Z. conceived the project. B.S., X.L., B.X., and J.K. performed the experiments. B.X., J.C.Z., and J.Y. designed and conducted bioinformatics analysis. M.H. provided guidance on statistical inference of the data. B.X., J.C.Z., and J.Y. wrote the manuscript. All authors read and approved the manuscript.
Competing interests
The authors declare no competing interests.
Contributor Information
Jonathan C. Zhao, Email: jonathan-zhao@northwestern.edu
Jindan Yu, Email: jindan-yu@northwestern.edu.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-019-0204-1.
References
- 1.Lee CS, Friedman JR, Fulmer JT, Kaestner KH. Nature. 2005;435:944–947. doi: 10.1038/nature03649. [DOI] [PubMed] [Google Scholar]
- 2.Lupien M, et al. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Q, et al. Cell. 2009;138:245–256. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang D, et al. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin HJ, Zhao JC, Wu L, Kim J, Yu J. Nat. Commun. 2014;5:3972. doi: 10.1038/ncomms4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song B, et al. J. Clin. Investig. 2019;129:569–582. doi: 10.1172/JCI122367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbieri CE, et al. Nat. Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iyer MK, et al. Nat. Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higa M, et al. Proc. Natl Acad. Sci. USA. 2018;115:E1214–E1220. doi: 10.1073/pnas.1719921115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao, S. et al. Cell Res. 2019. 10.1038/s41422-019-0203-2.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.