Abstract
Oriented cell divisions are controlled by a conserved molecular cascade involving Gαi, LGN, and NuMA. Here, we show that NDP52 regulates spindle orientation via remodeling the polar cortical actin cytoskeleton. siRNA-mediated NDP52 suppression surprisingly revealed a ring-like compact subcortical F-actin architecture surrounding the spindle in prophase/prometaphase cells, which resulted in severe defects of astral microtubule growth and an aberrant spindle orientation. Remarkably, NDP52 recruited the actin assembly factor N-WASP and regulated the dynamics of the subcortical F-actin ring in mitotic cells. Mechanistically, NDP52 was found to bind to phosphatidic acid-containing vesicles, which absorbed cytoplasmic N-WASP to regulate local filamentous actin growth at the polar cortex. Our TIRFM analyses revealed that NDP52-containing vesicles anchored N-WASP and shortened the length of actin filaments in vitro. Based on these results we propose that NDP52-containing vesicles regulate cortical actin dynamics through N-WASP to accomplish a spatiotemporal regulation between astral microtubules and the actin network for proper spindle orientation and precise chromosome segregation. In this way, intracellular vesicles cooperate with microtubules and actin filaments to regulate proper mitotic progression. Since NDP52 is absent from yeast, we reason that metazoans have evolved an elaborate spindle positioning machinery to ensure accurate chromosome segregation in mitosis.
Subject terms: Mitosis, Mitotic spindle
Introduction
Mitotic spindle orientation is essential for proper cell division and precise chromosome segregation during organismal development and tissue homeostasis. Emerging evidence shows that errors in spindle orientation have been correlated with developmental diseases1,2 and tumorigenesis3–11 in multiple tissues and organisms, underscoring the importance of understanding the mechanisms regulating these processes. Spindle orientation control has been extensively studied from three different perspectives: from the view of the cell cortex level; from that of force generators linking cortex and microtubule and from the view of the astral microtubule network. A core model in which the evolutionarily conserved Gαi/LGN/NuMA complex regulates spindle orientation and positioning through polarizing cortical force generators has been well established.12–16 In this model, Gαi subunits (GDP-bound inhibitory α-subunits of heterotrimeric G protein) anchored to the plasma membrane by myristoylation recruit the complex to the cell cortex through interacting with the C terminus of LGN (G protein regulator leucine-glycine-asparagine repeat protein).17 The N terminus of LGN binds to NuMA (nuclear and mitotic apparatus protein),18 which interacts with microtubules and the dynein-dynactin motor complex.19,20 Both the dynein-dynactin motor complex and NuMA contribute to pulling force generation on astral microtubules.
Recent studies revealed the involvement of cortical actin-related pathways in the control of spindle orientation.21–23 One of these findings is that myosin 10 acts as the link between polarized subcortical cluster of actin (actin cloud) and astral microtubules to regulate spindle orientation in a LGN/dynein-independent manner in cells cultured on micropatterns.23 In dorsal epiblast cell divisions during zebrafish gastrulation, anthrax toxin receptor 2a (Antxr2a) accumulates in a polarized cortical actin filament cap, which is aligned with the embryonic animal-vegetal axis and forecasts the division plane.21 This suggests that anthrax toxin receptor may function as a key Wnt determinant to control the orientation of cell division. In Drosophila cells, the RhoA/diaphanous/actin pathway regulates spindle orientation in a Dishevelled-mediated fashion.22
In mitosis, astral microtubules are thought to aid in spindle orientation through interactions between various microtubule plus-end binding proteins (e.g., Dynein, Khc-73, EB1-APC, and CLASPs) and the cell cortex.14 However, how astral microtubules coordinate with cortical/subcortical actin to promote proper spindle orientation remains elusive. Although recent studies have shown that myosin 10 regulates spindle orientation by modulating MT plus-end dynamics,23 little is known about the specific coordination between actin and astral MTs in this process and the underlying mechanisms.
NDP52 is a key regulator for selective autophagy that recognizes invading pathogens by binding the poly-ubiquitin coat of bacteria and the poly-ubiquitin coat and Galectin 8 (danger adaptor) of damaged bacteria-containing vacuoles through its C terminus.24,25 It then targets these structures for autophagosomes by interacting with LC3C via its N terminus,24–26 and promotes the fusion of autophagosomes with lysosomes through myosin VI.27 Furthermore, NDP52 is also directed to autophagy targets by Rab35 GTPase.28 Recent study has showed that NDP52 activates nuclear myosin VI to drive transcription.29 Up till now, whether NDP52 plays any roles beyond autophagic and transcriptional regulation and how this might be regulated is largely unexplored.
In this study, we found that NDP52 localizes at phosphatidic acid (PA)-positive vesicles by direct binding to PA and plays an essential role in mitotic spindle orientation. Suppression of NDP52 by siRNA induced the formation of a premature subcortical F-actin ring and subsequently impaired astral microtubules. This resulted in aberrant spindle orientation, chronic mitotic arrest and abnormal segregation of chromosomes. Treatment of NDP52-depleted HeLa cells with latrunculin B largely restored astral MT and proper spindle orientation. This suggested that there was some type of coordination between membrane vesicles decorated with actin and microtubule networks in the regulation of spindle orientation. Further results revealed that NDP52 both binds and recruits the actin assembly factor N-WASP to inhibit the premature formation of a subcortical F-actin ring in prophase/prometaphase. Supporting this notion, our in vitro TIRF imaging analyses showed that NDP52-positive liposomes recruit N-WASP and restrain the formation of long actin filaments. Taken together, these results suggest a novel vesicle-based regulatory mechanism that coordinates dynamic F-actin and microtubule networks to modulate mitotic spindle orientation.
Results
NDP52 is required for proper mitotic progression and spindle orientation
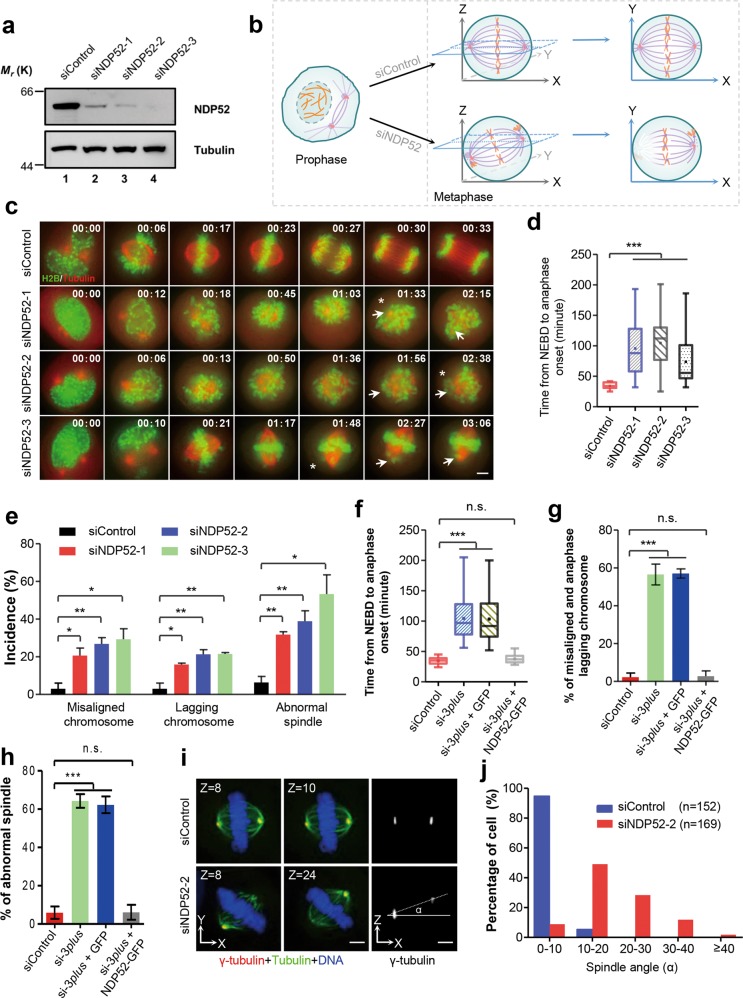
To elucidate the function of NDP52 in mitosis, we employed three independent sets of siRNA oligonucleotides to suppress endogenous NDP52 protein level in HeLa cells. Our trial experiments were employed to determine the dose and time to achieve an optimal suppression of NDP52 protein judged by Western blotting analyses, which indicated that treatment of 40 nM siRNA oligonucleotides for 48 h resulted in maximal suppression of NDP52 protein (Supplementary information, Fig. S1a; lanes 5 and 7, respectively). Interestingly, depletion of NDP52 resulted in an apparent increase in the number of mitotic cells with condensed chromosomes judged by fluorescence microscopic examination of nuclear dye DAPI staining and quantitative analyses (Supplementary information, Fig. S1b, c), suggesting that NDP52 exhibits non-canonical function in cell division control.
To assess the precise defects of mitotic cells resulting from NDP52 deficiency, we sought to carry out real-time analyses of chromosome movements in HeLa cells transfected with three independent siRNAs. To accurately relate the phenotypic changes seen in real-time mitosis to the siRNA-elicited knock-down efficiency, we performed Western blotting analyses to assess the NDP52 protein levels from the same populations of transfected cells in which dual-color real-time imaging of chromosome and microtubules was conducted. As shown in Fig. 1a, transfection of three independent siRNAs for 48 h resulted in maximal suppression of NDP52 proteins in HeLa cells. In the same population of NDP52-suppressed cells, real-time imaging of GFP-H2B and mCherry-tubulin was carried out to assess the phenotypes associated with NDP52 deficiency. Figure 1b illustrates a characteristic phenotype seen in NDP52-depleted cells in which two spindle poles are often not seen in the same focal plane (Fig. 1b; siNDP52; x–y axis projection). Our real-time imaging analyses using three independent siRNAs revealed that NDP52 deficiency resulted in chromosome segregation defects, including chromosome misalignment and anaphase lagging chromosomes (Fig. 1c, e). Although these NDP52-suppressed cells finally completed mitosis, the duration of mitotic process was dramatically extended judged by the time from nuclear envelope breakdown (NEBD) to anaphase onset (Fig. 1c, d). Surprisingly, almost all the cells undergoing abnormal mitosis showed perturbation of accurate spindle positioning (Fig. 1b, c and e). To ensure that the above phenotypes are not due to off-target effects, we performed rescue experiments by expressing exogenous NDP52-GFP or GFP in HeLa cells that were deprived of NDP52 with siRNA-3 and measured their ability to restore accurate mitosis using live-cell imaging, respectively. The expression of exogenous NDP52-GFP restored normal spindle morphology and chromosome segregation in HeLa cells deficient in endogenous NDP52 (Fig. 1f–h and Supplementary information, Fig. S1d–f; Supplementary information, Movies S1–8). Thus, NDP52 is essential for accurate mitotic progression and spindle formation during cell division.
Fig. 1.
NDP52 is essential for proper mitotic progression and spindle orientation. a Western blotting analyses of HeLa cells treated with control siRNA, NDP52 siRNA-1, NDP52 siRNA-2 or NDP52 siRNA-3 at 40 nM for 48 h paralleling to the live-cell imaging experiments shown in c. b Scheme of prophase and metaphase indicating spindle formation and chromosome alignment in mitotic HeLa cells treated with control siRNA or NDP52 siRNA. Note that loss of NDP52 causes slope of spindle in the z direction, which means that, when one spindle pole is just right on the focus plane, the second pole usually stays out of sight. c Representative mitotic phenotypes in NDP52-depleted HeLa cells expressing mCherry-tubulin and GFP-H2B shown by live-cell imaging (arrows, misalignment; asterisks, abnormal spindle; numbers at top left of images indicate elapsed time in the form of hour:minute). HeLa cells were treated with three different siRNAs for approximately 46 h prior to real-time imaging analyses. Scale bar, 5 μm. d Statistics of the time from nuclear envelope breakdown to anaphase onset in live HeLa cells treated with control siRNA (n = 23) or NDP52 siRNAs (n = 19, siNDP52–1; n = 19, siNDP52–2; n = 20, siNDP52–3). Values (whiskers: min to max) are from three independent experiments. Mean were shown as “+”. ***P < 0.001, Student’s t-test. e Quantification of chromosome segregation and spindle orientation defects in above-mentioned cells (n ≥ 19). Chromosomes failing to align at the metaphase plate within 60 min after nuclear envelope breakdown were considered to be misaligned. Spindle rotating all the time before anaphase onset was thought to be abnormal. Data represent mean ± SEM from three independent experiments. *P < 0.05, **P < 0.01, Student’s t-test. f Statistic analysis of the time from nuclear envelope breakdown to anaphase onset in live NDP52-depleted HeLa cells (si-3plus) expressing GFP or NDP52-GFP (n = 46, control siRNA; n = 41, si-3plus; n = 41, si-3plus + GFP; n = 40, si-3plus + NDP52-GFP). Values (whiskers: min to max) are from three independent experiments. Mean were shown as “+”. ***P < 0.001, Student’s t-test; n.s., no significant difference. g Quantification of chromosome segregation defects in above-mentioned cells (n ≥ 40). Data represent mean ± SEM from three independent experiments. ***P < 0.001, Student’s t-test; n.s., no significant difference. h Quantification of spindle defects in above-mentioned cells (n ≥ 40). Data represent mean ± SEM from three independent experiments. ***P < 0.001, Student’s t-test; n.s., no significant difference. i Representative immunofluorescence images of z-sections (0.2 μm per stack) with maximum spindle pole intensity in HeLa cells treated with control siRNA and NDP52 siRNA-2. Orthogonal views (x-z) of the depicted cells with the spindle angle α were obtained using SoftWorx software. Fixed Cells were co-stained for γ-tubulin (red), α-tubulin (green), and DNA (blue). Scale bar, 5 μm. See also Supplementary Fig. S1. j A histogram of measured spindle angle profiles of HeLa cells treated with control siRNA (n = 152) and NDP52 siRNA-2 (n = 169)
To further examine the abnormal spindle in NDP52-suppressed cells, we labeled the mitotic spindle with anti-α-tubulin antibody and anti-γ-tubulin (the centrosomal marker) antibody, respectively. Two spindle poles were found to be located on different z planes in NDP52-depleted cells, whereas in control transfected cells they were almost on the same focal plane of Z-axis (Fig. 1i and Supplementary information, Fig. S1g). Consistently, statistical analyses of spindle angles revealed that most NDP52-depleted cells exhibited a spindle angle α greater than 10°, whereas most control cells had a spindle angle of less than 10° (Fig. 1j). Thus, we conclude that NDP52 plays an essential role in accurate mitosis by determining accurate spindle orientation.
NDP52 localizes at PA vesicles in mitotic cells
To determine the subcellular distribution of NDP52 in mitotic cells, we carried out immunofluoresence microscope by which NDP52 exists as cytoplasmic puncta in mitotic cells (Supplementary information, Fig. S2a). To characterize the nature of subcellular localization of NDP52, we selected several markers for membranes such as early endosome, Golgi and endoplasmic reticulum (ER) for immunofluorescence analysis. No major overlap was observed between NDP52 and Rab5/EEA-1 or GM130 (Supplementary information, Fig. S2b), suggesting that NDP52 is not associated with early endosome nor Golgi apparatus. Only a fraction of NDP52-GFP overlapped with the endoplasmic reticulum marker calnexin, the recycling endosome marker Rab11 and the lysosomal marker LAMP-1 (Supplementary information, Fig. S2b). To test if the subcellular distribution of NDP52 was associated with membrane containing phospholipid, we characterized the phospholipid binding property of NDP52 using protein-lipid overlay assay and found that NDP52 prefers binding phosphatidic acid (PA) (Supplementary information, Fig. S2c). Further liposome co-sedimentation assays showed that purified NDP52 protein only bound to the synthetic liposomes with PA (Supplementary information, Fig. S2d), suggesting that NDP52 is likely to associate with the PA-containing membrane structures. To further ascertain the precise NDP52 localization, we fused the endogenous NDP52 protein with a C-terminal 3 × Flag and GFP tag using the CRISPR/Cas9 technique in HeLa cells (Fig. 2a and Supplementary information, Fig. S2e). As expected, NDP52-GFP co-localized with PA marker mCherry-PABD-Spo20p (mCh-PABD)30,31 from prophase to anaphase A in mitosis (Fig. 2b, c). To understand the biochemical property of NDP52-positive PA vesicles, we carried out magnetic bead-based immuno-isolation assays followed by mass spectrometric analyses. Our mass spectrometric analyses identified a wide range of cytoplasmic proteins bound to NDP52 vesicles (Supplementary information, Fig. S2f and Table S1). Then we analyzed these potential binding candidates using the PANTHER classification system and found that they were not exclusive to markers for a specific type of membrane structure like ER, Golgi or endosome (Supplementary information, Fig. S2g–i), which was in accordance with previous results (Fig. 2c and Supplementary information, Fig. S2b) and suggested that NDP52-rich vesicles represent a distinctly different membrane population from all known defined structures.
Fig. 2.
NDP52 localizes at the vesicles containing phosphatidic-acid (PA) a Schematic representation of fusing the endogenous NDP52 gene with a 3 × Flag and GFP tag via CRISPR/Cas9-meidated gene editing. Gray, red, green and black boxes indicate last coding exon, stop codon, insertion sequence and 3’-UTR region at NDP52 gene locus, respectively. b NDP52 co-localizes with mCherry-PABD-Spo20p (mCh-PABD, PA marker) in NDP52-GFP knock-in HeLa cells from prophase to anaphase A in mitosis. The NDP52-GFP knock-in HeLa cells expressing mCherry-PABD-Spo20p were fixed and stained for DNA (DAPI). Scale bar, 5 μm. c Co-localization analyses of NDP52 with mCherry-PABD-Spo20p, Golgi marker GM130, early endosome markers Rab5 and EEA-1, recycling endosome marker Rab11, endoplasmic reticulum marker Calnexin as well as lysosomal marker LAMP-1 during metaphase. Shown is the mean percentage of NDP52 co-localized with each marker (n = 5–8). Error bars represent SD. See also Supplementary Fig. S2 for images. d Schematic representation of NDP52 and deletion mutants. Abbreviations: CLIR, LC3C-specific binding region; ZnF, zinc finger domain; SKICH, skeletal muscle and kidney enriched inositol phosphatase carboxyl homology. “ + + ” represents strong interaction; “+” indicates weak interaction; “–” means no interaction. e Representative immunofluorescence images of z-sections (0.2 μm per stack) with maximum spindle pole intensity in control or NDP52 3’-UTR shRNA-treated HeLa cells with and without expression of HA-NDP52 or HA-NDP52Δ319–355. Scale bar, 5 μm. See also Supplementary Fig. S2. f Scatter plots showing the spindle angles in indicated HeLa cells (n = 37, shcontrol; n = 31, shNDP52; n = 40, shNDP52 + HA; n = 40, shNDP52 + HA-NDP52; n = 27, shNDP52 + HA-NDP52Δ319–355). Data represent mean ± SEM from three independent experiments. Student’s t-test, ***P < 0.001
To assess if PA binding activity of NDP52 is necessary for its regulation of mitotic spindle orientation, we first conducted deletion mapping experiments and found that the amino acid sequence from Ile319 to Ser355 in the coiled-coil domain of NDP52 is responsible for its binding to PA (Fig. 2d and Supplementary information, Fig. S2j, k). Then we performed rescue experiments by expressing HA-NDP52 or mutant HA-NDP52Δ319–355 deficient in PA-binding in NDP52-depleted HeLa cells. Careful examination of the spindle poles revealed that HA-NDP52Δ319–355 failed to restore the normal spindle angle in shNDP52-expressing HeLa cells whereas HA-NDP52 restored the normal spindle angle (Fig. 2e, f and Supplementary information, Fig. S2l, m), demonstrating the critical role of PA-binding activity of NDP52 in regulating spindle orientation.
Loss of NDP52 leads to premature subcortical F-actin ring formation and subsequently impaired astral microtubules
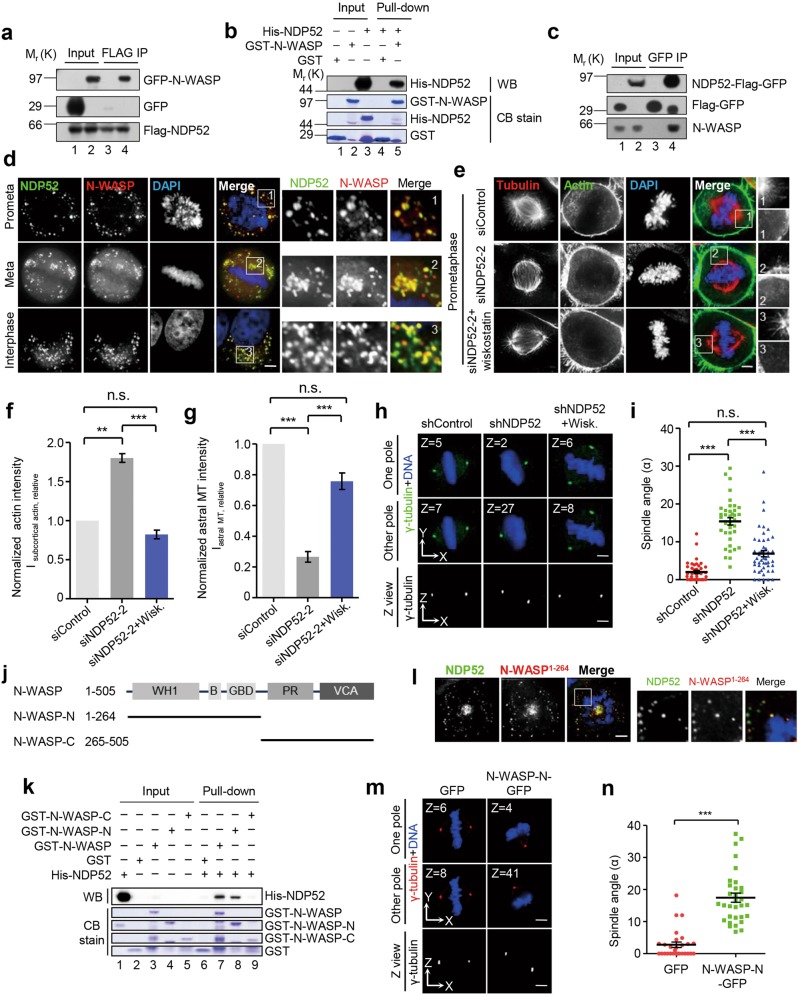
To delineate the role of NDP52 in spindle orientation during mitosis, we searched for binding partners of NDP52 using mass spectrometric identification and identified microtubule-associated proteins and actin-related candidates such as TIP150, CKAP2, Myosin family proteins, N-WASP, Actinin, CapZ etc. (Supplementary information, Fig. S3a). Given this discovery, we tested whether loss of NDP52 impacts the organization of microtubule and/or actin networks. In order to project precisely the molecular dynamics of NDP52 in spindle positioning during mitosis, we sought to use lattice light sheet microscopy,32 which enables us to visualize three-dimensional molecular dynamics at subsecond intervals and diffraction limit. Using microtubule plus-ends marked EB1-GFP cells, we found that NDP52 depletion led to a significant loss of astral microtubules and resulted in perturbation of accurate spindle positioning (Supplementary information, Fig. S3b). Besides, live-cell experiments showed that suppression of NDP52 resulted in premature formation of ring-like subcortical compact filamentous actin network in prophase/prometaphase, while in control cells it formed just a few minutes before anaphase onset (Fig. 3a, b), consistent with our previous study.33 We further performed rescue experiments by expressing GFP-NDP52 or GFP in NDP52-suppressed HeLa cells, and found that the expression of exogenous GFP-NDP52 restored accurate astral microtubule and actin networks (Fig. 3c–e).
Fig. 3.
Loss of NDP52 led to premature cortical actin filaments formation and subsequently impaired astral microtubules. a Representative mitotic phenotypes in NDP52-depleted HeLa cells (siNDP52–2) expressing mCherry-UtrCH and GFP-tubulin shown by live-cell imaging (numbers at top right of images indicate elapsed time in the form of hour: minute). The red ring-like region depicted, in the cartoon, by dashed line represents subcortical actin in prometaphase/metaphase HeLa cells. Based on dual color real-time mitosis, we defined prometaphase and metaphase in control and NDP52 siRNA-treated cells. Typically, the prometaphase cells selected from control group were taken from the 20 min time point while metaphase cells were taken from 50 min time point. For siRNA-treated cells, the typical prometaphase cells selected were taken from the 35 min time point while metaphase-like cells were taken from 130 min time point. The quantification strategy of this real-time imaging experiment was shown on the right, in which Isubcortical actin, prometaphase indicates the intensity of subcortical actin in early prometaphase; Isubcortical actin, just before anaphase indicates the intensity of subcortical actin just before anaphase onset. Note that the subcortical actin intensity was increased in the NDP52 siRNA-treated cells compared to that of control group. Scale bar, 5 μm. b Quantitative analyses of subcortical actin intensity from real-time imaging. The ratio of Isubcortical actin, prometaphase to Isubcortical actin, just before anaphase was calculated based on the subcortical intensity of mCherry-UtrCH from real-time imaging experiments as shown in a. The statistic analyses were taken from three independent experiments while data were expressed as mean ± SEM. Student’s t-test, ***P < 0.001. Note that the suppression of NDP52 protein by siRNA treatment increased the actin filament assembly judged by an increase in mCherry-UtrCH intensity. c Immunofluorescence staining for α-tubulin and F-actin in NDP52-depleted HeLa cells (si-3plus) with and without expression of GFP or GFP-NDP52. Scale bar, 5 μm. d Quantification of subcortical actin fluorescence intensity (Isubcortical actin, relative) in above-mentioned cells (n = 6 for each group). Intensities of intracellular actin (Iintra = Igreen + Iblue), spindle-associated actin (Ispindle = Iblue) and the total actin (Itotal = Igrey + Igreen + Iblue) were determined with ImageJ software. Relative subcortical actin intensity (Isubcortical, rel) was calculated by [(Iintra – Ispindle)/Itotal] that is also indicated as [Igreen/(Igrey + Igreen + Iblue)]. Normalization was carried out by setting control value to 1. Data represent mean ± SEM from three independent experiments. Student’s t-test, **P < 0.01; n.s., no significant difference. e Quantification of astral microtubule fluorescence intensity (Iastral MT, relative) in above-mentioned cells (n = 6 for each group). Intensities of spindle microtubules (Ispindle) and the total microtubules (Itotal) were determined with ImageJ. Relative Astral microtubule intensity (Iastral, rel) was calculated by [(Itotal – Ispindle)/Itotal]. Normalization was carried out by setting control value to 1. Data represent mean ± SEM from three independent experiments. Student’s t-test, ***P < 0.001; n.s., no significant difference. f Representative immunofluorescence images of z-sections (0.2 μm per stack) with maximum spindle pole intensity in NDP52-depleted HeLa cells with and without the treatment of latrunculin B. Scale bar, 5 μm. See also Supplementary Fig. S3. g Scatter plots showing the spindle angles in indicated HeLa cells (n = 59, shcontrol; n = 65, shcontrol + latrunculin B; n = 76, shNDP52; n = 79, shNDP52 + latrunculin B). Data represent mean ± SEM from three independent experiments. Student’s t-test, ***P < 0.001; n.s., no significant difference. h Immunofluorescence staining for α-tubulin and F-actin in NDP52-depleted HeLa cells (siNDP52–2) with and without the treatment of latrunculin B. Scale bar, 5 μm. i Quantification of subcortical actin fluorescence intensity (Isubcortical actin, relative) in above-mentioned cells (n = 12 for each group). Data represent mean ± SEM from three independent experiments. Student’s t-test, ***P < 0.001. j Quantification of astral microtubule fluorescence intensity (Iastral MT, relative) in above-mentioned cells (n = 12 for each group). Data represent mean ± SEM from three independent experiments. Student’s t-test, ***P < 0.001; n.s., no significant difference. k Model indicating the effects of NDP52 depletion on coordination between subcortical actin and microtubule networks in mitosis
We then examined whether there was a relationship between the abnormal actin network and the aberrant spindle when NDP52 was suppressed. To this end, we treated NDP52-depleted mitotic cells with latrunculin B, an actin polymerization inhibitor, and measured the spindle angle. To our surprise, latrunculin B treatment minimized the spindle misorientation phenotype seen in NDP52-suppressed HeLa cells (Fig. 3f, g and Supplementary information, Fig. S3c). This suggested that the cortical actin growth or hyper-stabilization in NDP52-suppressed cells caused spindle misorientation. One possibility is that the abnormal F-actin growth under NDP52 depletion affects spindle orientation by inhibiting astral microtubule growth. To test this hypothesis, we treated the NDP52-depleted mitotic cells with latrunculin B and stained microtubules and F-actin with anti-α-tubulin antibody and Alexa594-conjugated phalloidin, respectively. Consistent with above results, the compact subcortical F-actin occurred in early prophase/prometaphase and a large number of astral microtubules disappeared in NDP52-depleted cells (Fig. 3h–j). When we treated these cells with latrunculin B to depolymerize the abnormal compact subcortical F-actin, the astral microtubules reappeared in these cells (Fig. 3h–j). These results support the hypothesis that loss of NDP52 causes premature formation of compact subcortical actin filaments, perturbation of astral microtubules, and finally spindle misorientation during mitotic progression (Fig. 3k).
NDP52 restrains the early formation of subcortical F-actin ring via actin nucleation factor N-WASP
Next, we asked how NDP52 inhibits the early formation of the subcortical F-actin ring in prophase/prometaphase. Co-sedimentation assays in vitro showed that NDP52 did not bind F-actin or microtubules directly (Supplementary information, Fig. S4a, b). We further screened the actin-associated candidates from NDP52-interacting proteins obtained from immunoprecipitation and found that N-WASP is an NDP52-binding partner (Fig. 4a and Supplementary information, Fig. S4c). GST pull-down assay in vitro also validated that NDP52 interacted with N-WASP directly (Fig. 4b and Supplementary information, Fig. S4d). To examine if NDP52 and N-WASP form a cognate complex in mitotic cells, we performed an immunoprecipitation using NDP52-Flag-GFP knock-in cell lines. As shown in Fig. 4c, N-WASP bound NDP52-Flag-GFP (lane 3), but not the negative control (lane 4), indicating that N-WASP and NDP52 are cognate binding partners. Immunofluorescence analyses demonstrated that NDP52 co-localized with N-WASP in mitosis and interphase (Fig. 4d and Supplementary information, Fig. S4e). Furthermore, depletion of NDP52 resulted in a significant decrease of N-WASP dots (Supplementary information, Fig. S4f). Since N-WASP is a key promoter for actin nucleation and polymerization, we tested whether NDP52 restrains the formation of subcortical F-actin ring by clustering N-WASP during prophase/prometaphase. We treated NDP52-depleted HeLa cells with wiskostatin, a chemical inhibitor of N-WASP, and found that wiskostatin rescued the defects of F-actin and astral microtubules caused by loss of NDP52 (Fig. 4e–g). To confirm that N-WASP is crucial to NDP52-mediated regulation of spindle orientation, we treated shNDP52-expressing HeLa cells with wiskostatin and measured the spindle angle. Wiskostatin treatment also rescued the abnormal spindle angle caused by NDP52 depletion (Fig. 4h, i and Supplementary information, Fig. S4g).
Fig. 4.
NDP52 inhibits premature formation of subcortical F-actin ring via N-WASP a HEK293T cells co-expressing 3 × Flag-NDP52 and GFP-N-WASP were lysed, immunoprecipitated with anti-FLAG beads and analyzed by Western blotting. Lanes 1–2 annotate the input while lanes 3 and 4 mark immunoprecipitates of Flag-NDP52. See also Supplementary Fig. S4. b Pull-down assay with GST-N-WASP and His-NDP52. GST-N-WASP affinity matrix was used to bind His-NDP52. The binding fractions were subjected to SDS-PAGE and stained with Coomassie Brilliant Blue or analyzed by Western blotting. Lanes 1–3 represent inputs which contain GST, GST-N-WASP and His-NDP52 protein, respectively, while lanes 4 and 5 mark pull-down fractions which contain proteins bound to GST or GST-N-WASP. Note that His-NDP52 binds to GST-N-WASP (lane 5) but not GST (lane 4). See also Supplementary Fig. S4. c The NDP52–3 × Flag-GFP knock-in HeLa cells synchronized by nocodazole treatment were lysed, and the clarified cell lysates were immunoprecipitated with GFP trap beads followed by Western blotting analyses. In negative control group, HeLa cells expressing 3 × Flag-GFP were used. Lanes 1–2 annotate the input while lanes 3 and 4 represent immunoprecipitates of NDP52-Flag-GFP and Flag-GFP, respectively. d Co-localization of NDP52-GFP and N-WASP-mCherry in mitosis and interphase. HeLa cells expressing NDP52-GFP and N-WASP-mCherry were synchronized with monastrol (prometaphase) or MG132 (metaphase) after release from the thymidine treatment. Scale bar, 5 μm. e Immunofluorescence staining for α-tubulin and F-actin in NDP52-depleted HeLa cells (siNDP52–2) with and without the treatment of wiskostatin. Scale bar, 5 μm. f Quantification of subcortical actin fluorescence intensity (Isubcortical actin, relative) in above-mentioned cells (n = 6 for each group). Data represent mean ± SEM from three independent experiments. Student’s t-test, **P < 0.01, ***P < 0.001; n.s., no significant difference. g Quantification of astral microtubule fluorescence intensity (Iastral MT, relative) in above-mentioned cells (n = 6 for each group). Data represent mean ± SEM from three independent experiments. Student’s t-test, ***P < 0.001; n.s., no significant difference. h Representative immunofluorescence images of z-sections (0.2 μm per stack) with maximum spindle pole intensity in NDP52-depleted HeLa cells with and without the treatment of wiskostatin. Scale bar, 5 μm. See also Supplementary Fig. S4. i Scatter plots showing the spindle angles in indicated HeLa cells (n = 40, shcontrol; n = 38, shNDP52; n = 48, shNDP52 + wiskostatin). Data represent mean ± SEM from three independent experiments. Student’s t-test, ***P < 0.001; n.s., no significant difference. j Schematic representation of N-WASP and deletion mutants. Abbreviations are as follows: WH1, WASP homology 1 domain; B, basic domain; GBD, GTPase-binding domain; PR, proline-rich domain; VCA, verprolin-cofilin-acidic domain. Note that VCA domain binds actin and promotes actin polymerization. k Identification of NDP52-binding domain within the N-WASP protein by pull down assay. Lanes 1–5 represent inputs which contain GST, GST-N-WASP (full-length and truncations) and His-NDP52 protein, respectively, while lanes 6–9 mark the pull-down fractions which contain proteins bound to GST (negative control) or GST-N-WASP. Note that N-terminal N-WASP protein interacts with NDP52 directly (lane 8). l N-WASP-N-mCherry co-localizes with NDP52-GFP in mitosis. HeLa cells expressing NDP52-GFP and N-WASP-N-mCherry were synchronized with monastrol after release from the thymidine treatment. Scale bar, 5 μm. m Representative immunofluorescence images of z-sections (0.2 μm per stack) with maximum spindle pole intensity in HeLa cells overexpressing GFP or N-WASP-N-GFP. Scale bar, 5 μm. See also Supplementary Fig. S4. n Scatter plots showing the spindle angles in indicated HeLa cells (n = 26, GFP; n = 32, N-WASP-N-GFP). Data represent mean ± SEM from three independent experiments. Student’s t-test, ***P < 0.001
To further characterize the role of N-WASP in NDP52-dependent regulation of spindle orientation, we performed deletion analyses and found that the N-terminal portion of N-WASP which loses the function of promoting actin nucleation and polymerization interacts with NDP52 directly (Fig. 4j, k and Supplementary information, Fig. S4h). Consistently, fluorescence analysis showed that N-terminal N-WASP co-localized with NDP52 in mitosis (Fig. 4l). Next, we overexpressed GFP-tagged N-terminal portion of N-WASP in cells to disturb the interaction of endogenous N-WASP with NDP52 and measured the spindle angle. Most cells expressing N-terminal N-WASP showed an abnormal spindle angle α of higher than 10°, whereas most control cells had a normal spindle angle of lower than 10° (Fig. 4m, n and Supplementary information, Fig. S4i), indicating that N-WASP is a crucial downstream factor of NDP52 in regulating spindle orientation. Overall, these findings suggest that NDP52 restrains the premature formation of subcortical ring-like F-actin in prophase/prometaphase via N-WASP, and thereby promotes astral microtubule growth to ensure accurate spindle orientation.
NDP52 inhibits long F-actin formation by clustering N-WASP
To further clarify the mechanism underlying NDP52’s role in modulating subcortical F-actin formation, we employed total internal reflection fluorescence microscopy (TIRFM) to analyze the status of F-actin formation in the presence or absence of NDP52. First, we assessed the liposome binding activity of NDP52-GFP and RFP-N-WASP proteins (Supplementary information, Fig. S5a). Consistent with the PIP strip assay results (Supplementary information, Fig. S2c and S5b), the fluorescence images revealed that purified NDP52-GFP protein associated selectively with the liposomes containing PA, and that RFP-N-WASP protein bound to the liposomes in an NDP52-dependent manner (Fig. 5a). Based on these preliminary results, we designed a strategy to explore NDP52’s effects on F-actin formation using TIRFM (Fig. 5b). We polymerized actin filaments from F-actin seeds immobilized on a microscope coverslip. In the presence of purified NDP52-GFP and RFP-N-WASP, as well as PA-containing liposomes, the F-actin structures were imaged by TIRFM after F-actin growth for about 30 min (Fig. 5b). F-actin showed a strong preference to NDP52- and N-WASP-positive liposomes and rarely formed long filaments (≥6 μm) (Fig. 5c–e). In contrast, in the absence of NDP52, N-WASP and F-actin failed to associate with the liposomes and formed long actin filaments with an average length of near 9 μm (Fig. 5c–e). These results demonstrated that NDP52 inhibits F-actin formation by clustering N-WASP in vitro.
Fig. 5.
NDP52 inhibits long F-actin formation by clustering N-WASP a Fluorescence images showing NDP52-GFP-mediated recruitment of RFP-N-WASP to liposome consisting of DOPC, DOPE and DOPA as aforementioned. Scale bar, 5 μm. b Schematic representation of in vitro TIRF system studying how NDP52 vesicles influence actin polymerization via N-WASP. c TIRF images showing situation of F-actin formation with or without NDP52. Scale bar, 5 μm. d Quantification of F-actin length in the presence or absence of NDP52 (n = 94, −NDP52; n = 97, + NDP52). Data represent mean ± SEM from three independent experiments. Student’s t-test, ***P < 0.001. e Quantification of amount of long F-actin (≥6 μm) per 51.2 × 51.2 μm2. Data represent mean ± SD from three independent experiments. f Working model illustrating NDP52 vesicles-mediated coordination of subcortical actin and microtubule networks through N-WASP during prophase/prometaphase
Discussion
Actin networks are well known to play important roles in mitotic spindle orientation. For example, an intact actin cortex is not only required for correct LGN cortical localization in cultured cells and the mouse embryonic skin,34–36 but is also responsible for balancing the forces that pull the spindle in C. elegans zygote.37 In addition, an actin cap that recruits Antxr2a to activate zDia2 for allowing spindle rotation in the direction of the cap has been observed in epiblast cell divisions during zebrafish gastrulation.21 Interestingly, besides cortical actin, a subcortical cluster of actin, also called the actin cloud, has been reported to regulate the rotation of the mitotic spindle along with the unconventional microtubule binding of myosin 10 in cells cultured on micropatterns.23,38 In this study, we found a novel periodic ring-like compact subcortical F-actin structure emerging just before anaphase onset in HeLa cells (Fig. 3a, b). Premature formation of this compact F-actin ring prior to prophase/prometaphase in response to NDP52 depletion resulted in dramatic defects of astral microtubules and spindle orientation (Fig. 3c–k). This suggests that a precise spatiotemporal coordination between actin filament and microtubule dynamics is essential for proper spindle orientation. It would be of great interest to delineate the molecular architect of this NDP52-containing subcortical F-actin network.
An important aspect of this work is that intracellular vesicles may regulate actin and microtubule networks during mitosis by their sequestration of actin. We found that NDP52 vesicles act as a novel suppressor for preventing premature formation of the subcortical compact F-actin ring in prophase/prometaphase via the actin nucleator N-WASP (Fig. 5f). Consistent with this observation, our in vitro TIRFM results showed that NDP52-positive liposomes inhibit the formation of long F-actin by clustering N-WASP (Fig. 5c–e). Similarly, in mouse oocytes, Rab11a-positive vesicles regulate the dynamics and density of the actin network that consists of long, unbranched actin filaments through sequestering and clustering the nucleators of the network.39 Furthermore, similar working models have been proposed based on the studies conducted in a range of experimental systems such as Xenopus egg extracts40,41 and zebrafish embryos.41
Two studies have reported that autophagy activity is inhibited during normal mitosis.42,43 This inhibition has been thought to defend cell components undergoing drastic structural rearrangements from unintended loss.44 In this case, it is tempting to speculate that autophagy-related factors are primarily involved in the regulation of mitotic events rather than autophagy during mitosis. Indeed, growing evidence has suggested the non-autophagic functions of autophagy regulatory proteins.45 For instance, Beclin-1, a well-known autophagy protein, has been reported to associate with the KMN (KNL-1/Mis12/Ndc80) complex which is crucial for kinetochore-microtubule attachments to promote the recruitment of outer kinetochore proteins and accurate kinetochore anchoring to the spindle in mitosis.46 Interestingly, our work has uncovered a novel and non-autophagic function of NDP52 in mitotic spindle orientation. In mitotic cells, NDP52 interacts with N-WASP to ensure the formation of the subcortical compact F-actin ring in a precisely regulated temporal manner, which is essential for proper astral microtubule network formation and spindle orientation. The excitement and challenge ahead are to delineate the molecular mechanism underlying context-dependent function of NDP52 in cell fate decision control.47 Furthermore, another actin nucleation factor Arp2/3 that cooperates with N-WASP to promote actin polymerization may also be involved in the NDP52-mediated coordination between actin and microtubule networks. Future studies are needed to explore the potential role of Arp2/3 in this pathway.
It is worth noting that the loss of NDP52 led to chromosome segregation errors in addition to spindle misorientation (Fig. 1). This observation raised the question of whether chromosome segregation errors were independent of spindle misorientation in NDP52-depleted cells. Our rescue experiments using wild type NDP52 and NDP52 mutant deficient in PA-binding (NDP52Δ319–355) showed that NDP52Δ319–355 failed to restore accurate chromosome segregation in NDP52-depleted cells (Supplementary information, Fig. S6a–d), demonstrating the importance of PA-binding in NDP52 function in accurate mitosis. It remains to be examined whether NDP52-dependent regulation of chromosome segregation is parallel to its control of spindle orientation via spatial interaction with PA-containing vesicles or through other binding partners, just like it restrains the premature formation of subcortical ring-like F-actin in prophase/prometaphase via N-WASP (Fig. 5f). If the two pathways are not parallel events, one probably acts upstream of the other. Compared with the possibility that spindle checkpoint-dependent mitotic delay causes dramatic defects of astral microtubules and spindle orientation, it seems more logical and reasonable that perturbation of astral microtubules and spindle orientation leads to defects in kinetochore-microtubule attachment and chromosome segregation. Thus, further exploration is necessary to delineate the molecular mechanisms underlying NDP52-dependent regulation of accurate chromosome segregation.
In conclusion, our study suggests that a vesicle-based regulatory mechanism, distinctly different from previously reported actin/dynein and NuMA/dynein pathways, orchestrates the dynamic interaction between the actin and microtubule networks that cooperate to guide mitotic spindle orientation. This cooperation between intracellular vesicles and the cytoskeleton in fine regulation of proper spindle orientation in mitosis offers new perspectives on how NDP52 may serve as a novel sensor of cortical actin interaction with astral microtubules for accurate mitosis. As NDP52 is absent from yeast, we reasoned that metazoans evolved an elaborate spindle positioning machinery to ensure faithful chromosome segregation in mitosis.
Materials and methods
Plasmids, RNA interference and transfection
Various NDP52 constructs were generated by inserting human NDP52 isoform 3 cDNA (NCBI reference sequence: NM_005831.4) into different kinds of vectors via EcoRI and SalI sites. To construct NDP52-GFP-His, gene encoding NDP52 was cloned into a modified pET-22b plasmid containing a GFP open reading frame. His-RFP-N-WASP was generated by cloning gene encoding N-WASP into a modified pET-28a plasmid containing a RFP open reading frame. Other N-WASP plasmids were constructed with infusion kit provided by Xiaoming Tu’s lab in USTC. PLKO-mCherry-shNDP52 was obtained by inserting mCherry cDNA behind hPGK promoter of PLKO.1 vector and NDP52 shRNA sequence behind U6 promoter via EcoRI and AgeI sites. The nucleotide sequence for shRNA against NDP52 was 5′-GACAGGUGAGAGGAUGAUACAGCUAGAGU-3′ (3-UTR). The target sequences of NDP52 siRNAs were: siNDP52–1, 5′-GGAGGAGCUAGAAACCCUA-3′ (Ambion); siNDP52–2, 5′-UUCAGUUGAAGCAGCUCUGUCUCCC-3′ (Qiagen)25; siNDP52–3, 5′-GAGGAUGAUACAGCUAGAG-3′ (3′-UTR; Qiagen). siRNAs containing same but scramble sequences were used for controls. For immunofluorescence and live-cell imaging, trial experiments were used to determine an optimal condition for suppressing NDP52 by assessing concentrations and time of siRNA transfection. The optimal concentrations of various NDP52 siRNAs used in this study were: siNDP52–1, 40 nM; siNDP52–2, 40 nM; siNDP52–3, 40 nM. To precisely relate the mitotic phenotypes to NDP52 levels in siRNA-treated cells, same populations of transfected cells with three independent siRNAs were first used for real-time imaging followed by Western blotting analyses of NDP52 protein levels. Some rescue experiments were performed with NDP52 siRNA-3 plus an additional siRNA 5′-CAGUGGAGUUAUGUCACUG-3′ for maximal suppression of endogenous NDP52 protein. Specifically, we annotated those experiments as si-3plus which are shown in Fig. 1f–h, Fig. 3c–e and Fig. S1d–f. All plasmid and siRNA transfections were performed with Lipofectamine 2000 or 3000 (Invitrogen) according to user’s manual.
Cell culture, synchronization, and drug treatments
HEK293T and HeLa cells were purchased from the American Type Culture Collection and routinely maintained in advanced DMEM (Invitrogen) with 10% (vol/vol) FBS (HyClone) and 100 units/mL of penicillin plus 100 μg/ml streptomycin (GIBCO). To enrich mitotic cells, HeLa cells were treated with 2.5 mM thymidine (Invitrogen) for 14 h and then released in fresh DMEM medium. For Monastrol (50 μM, Sigma) and MG132 (10 μM, Sigma) treatment, drug was added into culture medium 8 h after release and maintained for 2 h; for latrunculin B (10 μM, Sigma) or wiskostatin (10 μM, Calbiochem) treatment in Figs. 3h and 4e, drug was added into culture medium 8.5 h after release and maintained for 20 min. Metaphase HeLa cells for quantification of spindle angle and chromosome misalignment were enriched with MG132 treatment. For latrunculin B (10 μM, Sigma) or wiskostatin (10 μM, Calbiochem) treatment in Figs. 3f and 4h, drug was added into culture medium and maintained for 20 min after cells had been treated with MG132 (10 μM, Sigma) for 100 min. To identify endogenous proteins bound to NDP52 in mitosis, HeLa cells expressing knock-in NDP52-GFP were treated with nocodazole (0.1 ng/μl) for 14 h followed by GFP immunoprecipitation.
Generation of NDP52–3 × FLAG-GFP stable cell line
NDP52–3 × Flag-GFP stable cell was generated by knocking in a 3 × Flag and a following GFP tag using CRISPR-Cas9 technology.48 The target sequence was: 5′-GCAGATAGGGCATTTCTTGATGG-3′. The sequences of 5′ and 3′ homology arms were listed in Supplementary Table S2. Cas9 and single guide RNA were expressed in PX330-puromycin, a kind gift from Dr. Ge Shan (University of Science and Technology of China).
Immunofluorescence and live-cell imaging
For visualization and quantitative analysis of spindle angle with γ-tubulin staining, HeLa cells were fixed in methanol at −20 °C for 3 min and then in PBS containing 3.7% paraformaldehyde for 2 min. For other immunofluorescence experiments, HeLa cells were fixed with PBS containing 3.7% paraformaldehyde for 5–10 min. Then the fixed cells were blocked with PBST (PBS with 0.05% Tween-20) buffer containing 1% BSA (Sigma) for 30 min at room temperature and sequentially incubated with the primary and secondary antibodies followed by DNA staining with DAPI (Sigma). Deconvolution images were collected using a Deltavision wide field deconvolution microscope system built on an Olympus IX-71 inverted microscope base. For imaging, a 60×, NA = 1.42 lens was used, and optical sections were taken at intervals of 0.2 μm. Images for display were generated by projecting single optical sections as described previously. Besides the images with γ-tubulin staining for quantitative analysis of spindle angle, the others were processed with Adobe Photoshop after deconvolution and z-stack projection using SoftWorx software (Applied Precision). The processing methods for the images with γ-tubulin staining were almost the same as the others with the exception of z-stack projection. All co-localization analyses and fluorescence intensity measurements were performed with ImageJ software.
For Live-cell imaging, HeLa cells were maintained in glass-bottomed culture dishes (MatTek) and cultured at 37 °C in CO2-independent medium (Gibco) supplemented with 10% FBS (vol/vol) and 2 mM glutamine. Images were taken with the DeltaVision microscope (Applied Precision).
Quantification of spindle angle
Orthogonal views (x–z) of metaphase HeLa cells stained for γ-tubulin were obtained with SoftWorx software from raw data. Spindle angle was calculated with tangent function by measuring the distances between two spindle poles in x and z direction using ImageJ software.
Fluorescence intensity quantification and spatial distribution measurements
Deconvolution images were collected using a Deltavision wide field deconvolution microscope system built on an Olympus IX-71 inverted microscope base. For imaging, a 60×, NA = 1.42 lens was used, and optical sections were taken at intervals of 0.2 μm. Quantification of the level of plasma membrane-associated polar cortical F-actin or astral microtubules was conducted as described by Lu et al.33 In brief, the average pixel intensities from at least 20 mitotic spindles were measured. For quantitative analysis of subcortical actin, fluorescence intensities of intracellular actin (Iintra), spindle-associated actin (Ispindle) and the total actin (Itotal) were determined with ImageJ software (NIH). Subcortical actin intensity (Isubcortical) was calculated by (Iintra ‒ Ispindle). Relative subcortical actin intensity (Isubcortical, rel) was calculated by [(Iintra ‒ Ispindle)/Itotal]. For quantitative analysis of astral MT, intensities of spindle microtubules (Ispindle) and the total microtubules (Itotal) were determined with ImageJ. Relative astral microtubule intensity (Iastral, rel) was calculated by [(Itotal – Ispindle)/Itotal]. Normalization was carried out by setting control value to 1.
Antibodies
Rabbit Anti-NDP52 (ab68588, 1:1000 for WB; 1:400 for IF) and biotinylated anti-GFP (ab6658) antibodies were purchased from Abcam. Anti-α-tubulin (DM1A, 1:10,000), anti-γ-tubulin (1:400 for IF) and anti-FLAG antibodies (M2, 1:2000 for WB; 1:400 for IF) antibodies were from Sigma. Anti-His-tag (12698, 1:2000 for WB), anti-HA-tag (3724, 1:400 for IF), and anti-N-WASP (4848, 1:400 for IF) antibodies were obtained from Cell Signaling Technology. For all Western blotting, signals were detected using HRP-conjugated anti-mouse or anti-rabbit antibodies (Pierce).
Lattice light sheet microscopy
In order to image mitotic NDP52-depleted HeLa cells expressing EB1-GFP and mCherry-H2B, lattice light sheet (LLS) scope was aligned and the imaging chamber with heating block was preheated to 37 °C.32 NDP52-depleted HeLa cells were seeded in 5 mm coverslips (Warner Instruments), synchronized and placed into the imaging chamber. The samples were then imaged in LLS scope using Bessel beams arranged in a square lattice configuration in dithered mode. Live cells immersed into imaging medium were imaged by exciting each plane alternatively with a 488 nm and 560 nm laser at ~10 µW (at the rear aperture of the excitation objective) intensity. The fluorescence signal generated within the specimen was collected by a detection objective (CFI Apo LWD 25XW, 1.1 NA, Nikon), filtered through a 440/521/607/700 nm BrightLine quad-band bandpass filter (Semrock), and eventually recorded by an ORCA-Flash 4.0 sCMOS camera (Hamamatsu).
The image volumes were taken by sampling scanning mode and the dithered light sheet at 300 nm step size, thereby capturing a volume of ~25 µm × 51 µm × 30 µm (corresponding to 500 × 512 × 100 pixels in deskewed data). Each image volume includes about 100 image frames, which were acquired at 20 ms exposure time with minimal photobleaching. The collected raw images have spatial resolution of 230 nm in X/Y and 370 nm in Z axes. The generated raw images were deskewed and then deconvoluted by running 10 Richardson-Lucy iterations. 3D visualization of deskewed and deconvoluted image stacks was performed by using Imaris 8.0 software (BITPLANE).
Recombinant protein preparation and purification
His-NDP52, NDP52-GFP-His, His-RFP-N-WASP, GST-NDP52 and GST-N-WASP plasmids were transformed into E. coli strain Rosetta (DE3), and protein expression was induced with 0.2 mM IPTG at 16 °C for 20 h. Bacteria expressing His-NDP52, NDP52-GFP-His, His-RFP-N-WASP were lysed by sonication in Ni-NTA binding buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 10 mM imidazole) and incubated with Ni-NTA agarose (Qiagen) for 1 h at 4 °C. The agarose was washed three times in Ni-NTA binding buffer supplemented with 30 mM imidazole and eluted with Ni-NTA binding buffer supplemented with 250 mM imidazole.
Bacterially expressed recombinant proteins of GST-NDP52 and GST-N-WASP were lysed by sonication in PBS buffer supplemented with 0.5% Triton X-100 and incubated with glutathione-Sepharose 4B (GE Healthcare Life Science) for 1 h at 4 °C. GST-NDP52 and GST-N-WASP protein was eluted with 20 mM glutathione. All purification procedures were performed at 4 °C, and protease inhibitor cocktail (Sigma) was added to prevent protein degradation.
Immunoprecipitaion and pull-down assay
For immunoprecipitation, HEK293T cells expressing 3 × Flag-tagged NDP52 and GFP-N-WASP were lysed in IP buffer (50 mM HEPES, 150 mM NaCl, 2 mM EDTA, pH 7.4) supplemented with 0.1% Triton X-100 plus protease inhibitor cocktail and incubated with anti-FLAG M2 resin for 2 h followed by washing three times with IP buffer. The FLAG-M2 beads were then resolved by SDS-PAGE and immunoblotted with anti-FLAG-tag and anti-GFP-tag antibodies. For the anti-GFP immunoprecipitation, the NDP52–3 × Flag-GFP knock-in HeLa cells synchronized by nocodazole treatment were lysed in IP buffer (50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH 7.4) supplemented with 0.1% Triton X-100 plus protease inhibitor cocktail. Clarified cell lysates were subjected to immunoprecipitation using GFP Trap A beads (ChromoTek). The binding fractions were analyzed by Western blotting with anti-GFP-tag and anti-N-WASP antibodies. In control group, HeLa cells expressing 3 × Flag-GFP were used for parallel immunoprecipitation. The pull-down assay with purified GST-N-WASP and His-NDP52 was performed as previously described.49
Flag bead-based immunoprecipitation assay for LC-MS
HEK293T cells expressing 3 × Flag or 3 × Flag-NDP52 were lysed in IP buffer (50 mM HEPES, 150 mM NaCl, 2 mM EDTA, pH 7.4) supplemented with 0.1% Triton X-100 plus protease inhibitor cocktail and incubated with anti-FLAG M2 resin for 2 h followed by washing three times with IP buffer. The binding fractions were subjected to SDS-PAGE and stained with Coomassie Brilliant Blue. The gel samples were harvested and sent for LC-MS. HEK293T cells expressing 3 × Flag was used for negative control to eliminate non-specific binders.
Magnetic bead-based immunoprecipitation assay for LC-MS
Nocodazole-synchronized (0.1 ng/μl) HeLa cells expressing Protein A or Protein A-NDP52 fusion protein were collected and lysed in IP buffer (20 mM Tris-HCl, 200 mM NaCl, pH 7.6, protease inhibitor cocktail, without detergent) by hundreds of passages through a 25 G needle. The lysates were centrifuged at 1000 × g for 15 min to remove cell debris and nuclei, incubated with IgG magnetic beads for 2 h and then immuno-isolated using a magnetic separator. The Binding fractions were eluted with 0.1% SDS, subjected to SDS-PAGE and stained with Coomassie Brilliant Blue. The gel samples were harvested and sent for LC-MS.
In-gel trypsin digestion, LC-MS and data analysis
For in-gel digestion, gel pieces were sequentially destained in 50% acetonitrile (50 mM NH4HCO3, v/v) until clear, dehydrated with 100 μl of 100% acetonitrile for 5 min, rehydrated in 10 mM dithiothreitol (in 50 mM NH4HCO3) and incubated at 56 °C for 60 min. Gel pieces were then again dehydrated in 100% acetonitrile and rehydrated with 55 mM iodoacetamide. After being incubated in the dark for 45 min at room temperature, gel pieces were washed with 50 mM NH4HCO3 and dehydrated with 100% acetonitrile. Next, gel pieces were incubated with 10 ng/μl trypsin (in 50 mM NH4HCO3) on ice for 1 h. Excess enzyme solution was removed and proteins in gel pieces were digested with trypsin at 37 °C overnight. Peptides were extracted with 60% acetonitrile/5% trifluoroacetic acid, followed by 100% acetonitrile. The extraction solution was then dried down. The pellet was resuspended in 0.1% formic acid and ready for MS.
The trypsin digest was analyzed with a QE-Plus mass spectrometer (ThermoFisher Scientific) coupled to an Easy-nLC nanoflow HPLC (Proxeon Biosciences) with a self-packed 75 μm id x 15 cm C18 column in the data-dependent acquisition and positive ion mode at a flow rate of 300 nl/min. Spectra were manually inspected to confirm the fragment ions aligned with the assigned peptide sequences. Raw files were processed using Proteome Discoverer 1.3. The database search was performed using Mascot (Matrix Science) against the Uniprot human protein database. Trypsin was specified as cleavage enzyme allowing up to 2 missing cleavages. Carbamidomethylation on cysteine was specified as fixed modification and oxidation on methionine was specified as variable modification. Peptide confidence was set at high, and peptide ion score was set >20.
PIP strip assay
The PIP strip membranes were blocked with 1% BSA in Buffer A (20 mM HEPES, 50 mM KCl, pH 7.4) for 30 min, and His-NDP52 was added at 1 μg/ml and incubated for 1 h at room temperature. Then the bound protein was detected using anti-His-tag mouse antibody (cell signaling) and HRP conjugated anti-mouse antibody. HRP was visualized by using ECL reagent (Pierce).
Microtubule and F-actin co-sedimentation assay
Microtubule assembly and co-sedimentation were performed as described previously.50–52 Briefly, purified recombinant GST-NDP52 protein was incubated with taxol-stabilized microtubules at 4 °C for 30 min followed by sedimentation for 30 min at 70,000 rpm in TLA100 rotor at 4 °C. Supernatants and pellets were collected for SDS-PAGE and Coomassie Brilliant Blue staining. For F-actin co-sedimentation assay, stabilized actin filaments were polymerized at 37 °C for 30 min with a 1 μg/μl G-actin using unlabeled actin (Cytoskeleton) with 4 μM phalloidin, 4 mM MgCl2 and 5 mM ATP, all diluted in Buffer A. Purified recombinant NDP52-GFP-His protein with gradually increasing concentrations was incubated with phalloidin-stabilized F-actin at 4 °C for 30 min followed by sedimentation for 30 min at 70,000 rpm in TLA100 rotor at 4 °C. Supernatants and pellets were subjected to SDS-PAGE and Coomassie Brilliant Blue staining for visualization and quantitative analyses.
Liposome co-sedimentation experiment
Liposomes containing PA were prepared at a 1 μg/μl lipids concentration using DOPC, DOPE and DOPA at a mole ratio of 6:2:2; Liposomes without PA were prepared at a 1 μg/μl lipids final concentration using DOPC and DOPE at a mole ratio of 6:4. Liposomes with or without PA were incubated with His-NDP52 in Buffer A (20 mM HEPES, 50 mM KCl, pH 7.4) supplemented with 1 mM Ca2+ for 20 min at 4 °C and centrifuged at 70,000 rpm in TLA100 rotor at 4 °C. Supernatants and pellets were collected for SDS-PAGE and Coomassie Brilliant Blue staining. All lipids were purchased from Avanti Polar Lipids.
Preparation of stabilized F-actin seeds
Stabilized F-actin seeds were prepared at 37 °C for 1 h at a 3 μg/μl final G-actin concentration using unlabeled actin (Cytoskeleton), Hilyte 647 labeled G-actin (Cytoskeleton) and biotinylated actin (Cytoskeleton) at a ratio of 24:4:1 with 4 μM phalloidin, 4 mM MgCl2 and 5 mM ATP, all diluted in Buffer A. Before use, Stabilized filamentous actin seeds were diluted at 1:50 ratio in Buffer A and sheared twice with 25G needle to generate even-length filamentous fluorescence actin.
In Vitro actin filaments formation assay with TIRF microscopy
Microscope slides and coverslips were passivated with PEG and doped with biotin-PEG as previously described.53 Biotinylated anti-GFP antibody was immobilized using NeutrAvidin.53 The flow chamber were blocked with 5% (wt/vol) Pluronic F-127 for 10 min, diluted in Buffer A, and then incubated with prepared F-actin seeds for 10 min. After unbound actin seeds were flush out, the chamber were sequentially incubated with 0.2 mg/ml NeutrAvidin and 20 nM Biotinylated anti-GFP antibody to allow NDP52-GFP-His-bounded liposomes binding. Formation of actin filaments was initiated in the presence of 1.5 μg/μl actin (18% of Alexa647-labeled and 72% unlabelled actin), 0.04 μg/μl His-RFP-N-WASP, 0.01 μg/μl NDP52-GFP-His and 0.5 μg/μl liposomes containing PA in freshly prepared TIRF buffer (20 mM HEPES, 50 mM KCl, pH 7.4, 5 mM DTT, 0.1% methylcellulose, 5 mM ATP, 4 mM MgCl2, 4 μM phalloidin, 0.25 μg/μl K-casein, 4.5 mg/ml glucose, 35 μg/ml catalase and 200 μg/ml glucose oxidase). The images were acquired from 30 min after initiation with a TIRF microscopy system and analyzed using the ZEN Light Edition software.
Supplementary information
Supplementary information, Movies Legend
Acknowledgements
We are grateful to Drs. Xiaoming Tu, Ge Shan and Kai Jiang for reagents. This work was supported in part by National Key Research and Development Program of China (2017YFA0503600, 2016YFA0100500, 2014CB964803, 2002CB713703), National Natural Science Foundation of China (31621002, 31320103904, 31430054, 91854203, 91313303, 31501093, 31500601, 91753000, 91853115, 31671405, 31671407, 31871359, 31601097, 31601098), ‘Strategic Priority Research Program’ of the Chinese Academy of Sciences (XDB19000000), MOE Innovative team (IRT_17R102), the Fundamental Research Funds for the Central Universities (KB2070000023) and China Postdoctoral Science Foundation Grant 2015M580547.
Author contributions
X.Y. and H.Y. conceived the project. H.Y. performed most cell biology experiments, in vitro reconstitution experiments and data analyses. F.Y. optimized siRNA treatments and performed correlated real-time imaging and Western blotting analyses. P.D., Z.L., W.L., J.Z., X.L. and J.L-S. contributed reagents. All authors contributed to the writing or editing of the manuscript.
Competing interests
The authors declare no competing interests.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-019-0189-9.
References
- 1.Fish JL, Kosodo Y, Enard W, Paabo S, Huttner WB. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc. Natl Acad. Sci. USA. 2006;103:10438–10443. doi: 10.1073/pnas.0604066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morin X, Bellaiche Y. Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Dev. Cell. 2011;21:102–119. doi: 10.1016/j.devcel.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 3.Caussinus E, Gonzalez C. Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nat. Genet. 2005;37:1125–1129. doi: 10.1038/ng1632. [DOI] [PubMed] [Google Scholar]
- 4.Fleming ES, Temchin M, Wu Q, Maggio-Price L, Tirnauer JS. Spindle misorientation in tumors from APC(min/+) mice. Mol. Carcinog. 2009;48:592–598. doi: 10.1002/mc.20506. [DOI] [PubMed] [Google Scholar]
- 5.Hehnly H, et al. A mitotic kinase scaffold depleted in testicular seminomas impacts spindle orientation in germ line stem cells. Elife. 2015;4:e09384. doi: 10.7554/eLife.09384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iden S, et al. Tumor type-dependent function of the par3 polarity protein in skin tumorigenesis. Cancer Cell. 2012;22:389–403. doi: 10.1016/j.ccr.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 7.McCaffrey LM, Macara IG. Epithelial organization, cell polarity and tumorigenesis. Trends Cell Biol. 2011;21:727–735. doi: 10.1016/j.tcb.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 8.McCaffrey LM, Montalbano J, Mihai C, Macara IG. Loss of the Par3 polarity protein promotes breast tumorigenesis and metastasis. Cancer Cell. 2012;22:601–614. doi: 10.1016/j.ccr.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pease JC, Tirnauer JS. Mitotic spindle misorientation in cancer--out of alignment and into the fire. J. Cell. Sci. 2011;124:1007–1016. doi: 10.1242/jcs.081406. [DOI] [PubMed] [Google Scholar]
- 10.Quyn AJ, et al. Spindle orientation bias in gut epithelial stem cell compartments is lost in precancerous tissue. Cell Stem Cell. 2010;6:175–181. doi: 10.1016/j.stem.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 11.Thoma CR, et al. VHL loss causes spindle misorientation and chromosome instability. Nat. Cell Biol. 2009;11:994–1001. doi: 10.1038/ncb1912. [DOI] [PubMed] [Google Scholar]
- 12.di Pietro F, Echard A, Morin X. Regulation of mitotic spindle orientation: an integrated view. EMBO Rep. 2016;17:1106–1130. doi: 10.15252/embr.201642292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kardon JR, Vale RD. Regulators of the cytoplasmic dynein motor. Nat. Rev. Mol. Cell Biol. 2009;10:854–865. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu MS, Johnston CA. Molecular pathways regulating mitotic spindle orientation in animal cells. Development. 2013;140:1843–1856. doi: 10.1242/dev.087627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seldin L, Macara I. Epithelial spindle orientation diversities and uncertainties: recent developments and lingering questions. F1000Res. 2017;6:984. doi: 10.12688/f1000research.11370.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siller KH, Doe CQ. Spindle orientation during asymmetric cell division. Nat. Cell Biol. 2009;11:365–374. doi: 10.1038/ncb0409-365. [DOI] [PubMed] [Google Scholar]
- 17.Siderovski DP, Diverse-Pierluissi M, De Vries L. The GoLoco motif: a Galphai/o binding motif and potential guanine-nucleotide exchange factor. Trends Biochem. Sci. 1999;24:340–341. doi: 10.1016/S0968-0004(99)01441-3. [DOI] [PubMed] [Google Scholar]
- 18.Du Q, Stukenberg PT, Macara IG. A mammalian partner of inscuteable binds NuMA and regulates mitotic spindle organization. Nat. Cell Biol. 2001;3:1069–1075. doi: 10.1038/ncb1201-1069. [DOI] [PubMed] [Google Scholar]
- 19.Merdes A, Heald R, Samejima K, Earnshaw WC, Cleveland DW. Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J. Cell Biol. 2000;149:851–862. doi: 10.1083/jcb.149.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merdes A, Ramyar K, Vechio JD, Cleveland DW. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 1996;87:447–458. doi: 10.1016/S0092-8674(00)81365-3. [DOI] [PubMed] [Google Scholar]
- 21.Castanon I, et al. Anthrax toxin receptor 2a controls mitotic spindle positioning. Nat. Cell Biol. 2013;15:28–39. doi: 10.1038/ncb2632. [DOI] [PubMed] [Google Scholar]
- 22.Johnston CA, et al. Formin-mediated actin polymerization cooperates with Mushroom body defect (Mud)-Dynein during Frizzled-Dishevelled spindle orientation. J. Cell Sci. 2013;126:4436–4444. doi: 10.1242/jcs.129544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwon M, Bagonis M, Danuser G, Pellman D. Direct microtubule-binding by Myosin-10 orients centrosomes toward retraction fibers and subcortical actin clouds. Dev. Cell. 2015;34:323–337. doi: 10.1016/j.devcel.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat. Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 25.Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–418. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boyle KB, Randow F. The role of ‘eat-me’ signals and autophagy cargo receptors in innate immunity. Curr. Opin. Microbiol. 2013;16:339–348. doi: 10.1016/j.mib.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 27.Tumbarello DA, et al. Autophagy receptors link myosin VI to autophagosomes to mediate Tom1-dependent autophagosome maturation and fusion with the lysosome. Nat. Cell Biol. 2012;14:1024–1035. doi: 10.1038/ncb2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minowa-Nozawa A, Nozawa T, Okamoto-Furuta K, Kohda H, Nakagawa I. Rab35 GTPase recruits NDP52 to autophagy targets. EMBO J. 2017;36:2790–2807. doi: 10.15252/embj.201796463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fili N, et al. NDP52 activates nuclear myosin VI to enhance RNA polymerase II transcription. Nat. Commun. 2017;8:1871. doi: 10.1038/s41467-017-02050-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakanishi H, de los Santos P, Neiman AM. Positive and negative regulation of a SNARE protein by control of intracellular localization. Mol. Biol. Cell. 2004;15:1802–1815. doi: 10.1091/mbc.e03-11-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Young BP, et al. Phosphatidic acid is a pH biosensor that links membrane biogenesis to metabolism. Science. 2010;329:1085–1088. doi: 10.1126/science.1191026. [DOI] [PubMed] [Google Scholar]
- 32.Chen BC, et al. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science. 2014;346:1257998. doi: 10.1126/science.1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu H, et al. Characterization of ring-like F-actin structure as a mechanical partner for spindle positioning in mitosis. PLoS ONE. 2014;9:e102547. doi: 10.1371/journal.pone.0102547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luxenburg C, Pasolli HA, Williams SE, Fuchs E. Developmental roles for Srf, cortical cytoskeleton and cell shape in epidermal spindle orientation. Nat. Cell Biol. 2011;13:203–214. doi: 10.1038/ncb2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Machicoane M, et al. SLK-dependent activation of ERMs controls LGN-NuMA localization and spindle orientation. J. Cell Biol. 2014;205:791–799. doi: 10.1083/jcb.201401049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng Z, et al. Evidence for dynein and astral microtubule-mediated cortical release and transport of Galphai/LGN/NuMA complex in mitotic cells. Mol. Biol. Cell. 2013;24:901–913. doi: 10.1091/mbc.e12-06-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Redemann S, et al. Membrane invaginations reveal cortical sites that pull on mitotic spindles in one-cell C. elegans embryos. PLoS ONE. 2010;5:e12301. doi: 10.1371/journal.pone.0012301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitsushima M, et al. Revolving movement of a dynamic cluster of actin filaments during mitosis. J. Cell Biol. 2010;191:453–462. doi: 10.1083/jcb.201007136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holubcova Z, Howard G, Schuh M. Vesicles modulate an actin network for asymmetric spindle positioning. Nat. Cell Biol. 2013;15:937–947. doi: 10.1038/ncb2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taunton J, et al. Actin-dependent propulsion of endosomes and lysosomes by recruitment of N-WASP. J. Cell Biol. 2000;148:519–530. doi: 10.1083/jcb.148.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Field CM, et al. Actin behavior in bulk cytoplasm is cell cycle regulated in early vertebrate embryos. J. Cell Sci. 2011;124:2086–2095. doi: 10.1242/jcs.082263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eskelinen EL, et al. Inhibition of autophagy in mitotic animal cells. Traffic. 2002;3:878–893. doi: 10.1034/j.1600-0854.2002.31204.x. [DOI] [PubMed] [Google Scholar]
- 43.Furuya T, et al. Negative regulation of Vps34 by Cdk mediated phosphorylation. Mol. Cell. 2010;38:500–511. doi: 10.1016/j.molcel.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mathiassen SG, De Zio D, Cecconi F. Autophagy and the cell cycle: a complex landscape. Front. Oncol. 2017;7:51. doi: 10.3389/fonc.2017.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Subramani S, Malhotra V. Non-autophagic roles of autophagy-related proteins. EMBO Rep. 2013;14:143–151. doi: 10.1038/embor.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fremont S, et al. Beclin-1 is required for chromosome congression and proper outer kinetochore assembly. EMBO Rep. 2013;14:364–372. doi: 10.1038/embor.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yao, X & Smolka, A. J. Gastric Parietal Cell Physiology and Helicobacter pylori–Induced Disease. Gastroenterology156, 2158–2173 (2019). [DOI] [PMC free article] [PubMed]
- 48.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang K, et al. TIP150 interacts with and targets MCAK at the microtubule plus ends. EMBO Rep. 2009;10:857–865. doi: 10.1038/embor.2009.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fu C, et al. Mitotic phosphorylation of PRC1 at Thr470 is required for PRC1 oligomerization and proper central spindle organization. Cell Res. 2007;17:449–457. doi: 10.1038/cr.2007.32. [DOI] [PubMed] [Google Scholar]
- 51.Ward T, et al. Regulation of a dynamic interaction between two microtubule-binding proteins, EB1 and TIP150, by the mitotic p300/CBP-associated factor (PCAF) orchestrates kinetochore microtubule plasticity and chromosome stability during mitosis. J. Biol. Chem. 2013;288:15771–15785. doi: 10.1074/jbc.M112.448886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xia P, et al. EB1 acetylation by P300/CBP-associated factor (PCAF) ensures accurate kinetochore-microtubule interactions in mitosis. Proc. Natl Acad. Sci. USA. 2012;109:16564–16569. doi: 10.1073/pnas.1202639109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jain A, Liu R, Xiang YK, Ha T. Single-molecule pull-down for studying protein interactions. Nat. Protoc. 2012;7:445–452. doi: 10.1038/nprot.2011.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information, Movies Legend