Dear Editor,
Cancer cells often upregulate the expression of ligands for the inhibitory immune checkpoints receptor to escape anti-tumor immunity1. Targeting inhibitory immune checkpoints has become one of the most promising anticancer therapeutic inventions. Infiltration of myeloid cells, especially phagocytic cells, to tumors is often associated with chemotherapy resistance and progression of various cancer types. Phagocytosis of cancer cells is the major mechanism for these myeloid cells-mediated antitumor activity. However, many types of cancer cells express high level of ‘don’t eat me’ signals on the cell surface to escape the phagocytic cells-mediated antitumor immunity.
Transmembrane protein CD47 is highly expressed on many types of cancer cells and can directly bind to the receptor signal regulatory protein alpha (SIRPα), which is highly expressed on phagocytic cells. Binding of CD47 to SIRPα can protect cancer cells from phagocytosis by phagocytic cells and therefore functions as the major ‘don’t eat me’ signal2. Therapeutic blockade of CD47-SIRPα axis can efficiently promote the macrophage-mediated phagocytosis and elimination of cancer cells. Targeting CD47-SIRPα axis thus may function as an efficient immunotherapy treatment of various cancers. However, apart from being expressed on cancer cells, endogenous CD47 is also expressed on normal cells, such as erythrocytes, which may lead to side effects of CD47-targeting cancer therapies. Therefore, identification of novel CD47- SIRPα axis regulators that might be novel therapeutic targets for cancer treatments is needed.
Glutaminyl cyclase (QC, Glutaminyl-peptide cyclotransferase (QPCT)) and its isoenzyme isoQC (QPCTL) belong to a family of enzymes which catalyze the formation of pyroglutamate (pGlu, pE) at N-terminus of proteins by converting glutamate/glutamine to pGlu residue3. N-terminal pGlu modification of proteins may protect protein from degradation by proteases or promote protein aggregation. Unlike QC, a secreted enzyme, isoQC exclusively localizes within the Golgi complex via a transmembrane domain3. Although QC and isoQC exhibit nearly identical substrate specificity in vitro, data from knockout mice indicate that QC and isoQC have preference for different peptides and proteins4. Since important substrates, such as CCL2, MCP-1, and amyloid-β, have been identified for QC/isoQC, QC/isoQC are regarded as potential therapeutic targets for inflammatory disease, Huntington’s disease and Alzheimer disease5,6. Although studies suggest that QC/isoQC may be a risk factor for chronic lymphocytic leukemia and thyroid carcinomas7, it is largely unknown whether QC/isoQC, especially isoQC, play any role in cancer development, especially cancer immune surveillance.
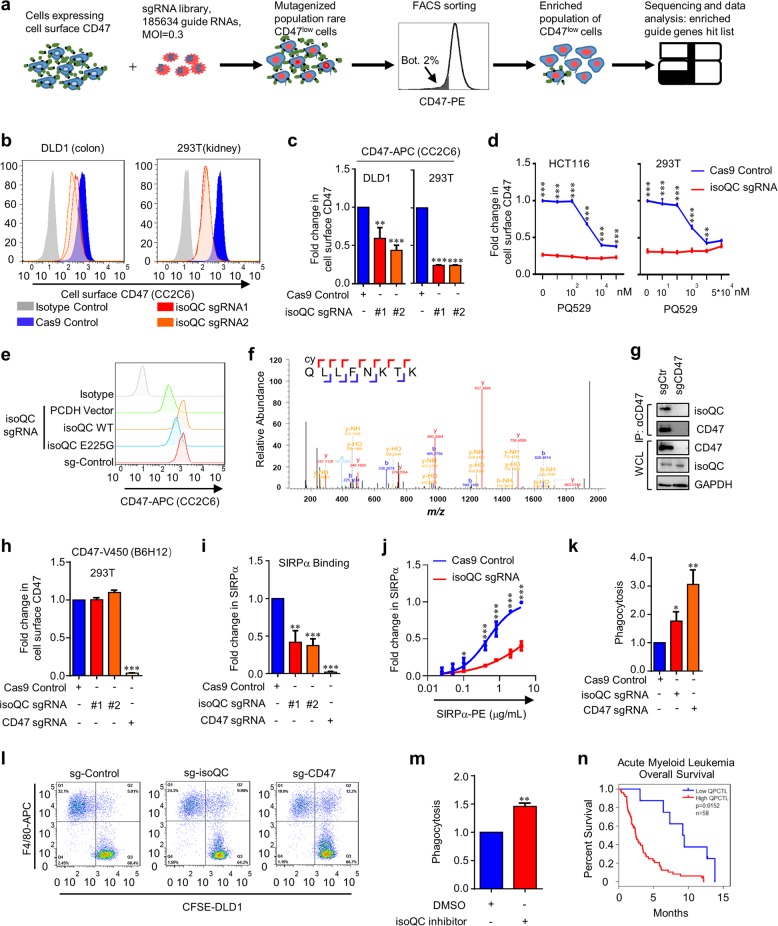
To identify novel regulators of CD47, we performed a FACS based whole-genome screen in colon cancer cell line HCT116 cells, which express relative high levels of CD47, using a CRISPR-Cas9 knockout library (Fig. 1a, Supplementary information, Data S1). We chose an anti-human CD47 monoclonal antibody CC2C6 from Biolegend (we then called CC2C6), which binds to the same site on CD47 as SIRPα and has been widely used for the detection of CD47 by FACS. We confirmed that this antibody could specifically recognize CD47 because knockout of CD47 by sgRNA almost completely blocked its binding to the cell surface (Supplementary information, Fig. S1a). Through analyzing our screening data using MAGeCK software, CD47 was identified as one of the most significant candidates in our screen (Supplementary information, Fig. S1b), indicating the successful screen of our system. Besides CD47, we also identified multiple potential candidates affecting the binding of CC2C6 to cell surface (Supplementary information, Fig. S1b).
Fig. 1.
a Genome-wide CRISPR-Cas9 screen identified genes essential for cell surface CD47 expression. HCT116 colon cancer cells were mutagenized with a pooled lentiviral sgRNA library and CD47low cells were enriched by FACS sorting. b, c Fluorescence-activated cell sorting (FACS) analysis of surface CD47 in DLD1 and HEK293T cells stably expressing the indicated sgRNAs. Shown is a representative experiment (b). Mean fluorescent intensity (MFI) values are shown in c. The data were normalized using Cas9 control. Shown is the mean ± SD of three independent experiments. t test, **P < 0.01, ***P < 0.001. d FACS analysis of cell surface CD47 using CC2C6 antibody in HCT116 (left) and HEK293T (right) cells stably expressing the indicated sgRNAs after treatment with the increasing concentrations of isoQC inhibitor (PQ529) for 48 h. The data were normalized using DMSO control. Shown is the mean ± SD of three independent experiments. two-way ANOVA, **P < 0.01, ***P < 0.001. e FACS analysis of cell surface CD47 expression in isoQC knockout HEK293T cells with the indicated overexpressed-plasmids. Shown is a representative experiment. Data represent three independent experiments with similar results. f pGlu modification of human CD47 N-terminal peptide by purified isoQC was monitored by nanoESI-MS mass spectrometry. g Co-immunoprecipitation of endogenous CD47 and isoQC in HEK293T cells (IP, CD47 antibody: sc-59079; IB, CD47 antibody: CST 63000). Shown is a representative experiment. Data represent three independent experiments with similar results. h FACS analysis of cell surface CD47 using anti-CD47 B6H12 monoclonal antibody (clone B6H12) in the indicated knockout HEK293T cells. The data were normalized using Cas9 control. Shown is the mean ± SD of three independent experiments. t test, ***P < 0.001. i FACS analysis of the binding of SIRPα to cell surface in the indicated knockout HEK293T cells. The data were normalized using Cas9 control. Shown is the mean ± SD of three independent experiments. t test, **P < 0.01, ***P < 0.001. j FACS analysis of the interaction between SIRPα and CD47 in indicated knockout HEK293T cells. Cells were treated with increasing concentrations of SIRPα. The data were normalized using Cas9 control. Shown is a mean ± SD of three independent experiments. two-way ANOVA, *P < 0.05, ***P < 0.001. k, l Phagocytosis of knockout DLD1 cells by macrophages was analyzed via FACS. The data were normalized using Cas9 control. Shown is the mean ± SD of three independent experiments (k). t test, *p < 0.5, **p < 0.01. Phagocytosis was evaluated as a sum of the CFSE+ macrophages, expressed as a percentage of the total macrophages, as depicted in the representative FACS shown in (l). m FACS analysis of the phagocytosis of the indicated DLD1 cells treated with isoQC inhibitor (PQ529, 10 μM) or vehicle control for 48 h before co-culture. The data were normalized using vehicle control. Shown is the mean ± SD of five independent experiments. t test, **P < 0.01. n Correlation of isoQC expression with prognosis in Acute Myeloid Leukemia patients (n = 58)
We next chose several candidates we are interested in and synthesized two specific sgRNAs for each gene to examine their effect on CD47 by FACS using CC2C6 antibody. Interestingly, we found that knockout of isoQC, but not other candidates, including ERBB2IP, JAKMIP2, RPL31, USP7, REEP6 and AHCTF1, significantly reduced the binding of CC2C6 to cell surface in HCT116 cells (Supplementary information, Fig. S1c). Moreover, knockout of isoQC significantly reduced the surface binding of CC2C6 in multiple cell lines, including 293T, DLD1 (Fig. 1b, c), HCT116 and ovarian cancer cell (SKOV3) cells (Supplementary information, Fig. S1d, e). The efficient knockout of isoQC by sgRNA was confirmed using Western Blot (Supplementary information, Fig. S1f). Because N-terminal pGlu modification of protein can be catalyzed by both QC and isoQC, we examined whether QC can affect the binding of CC2C6 to cell surface. To this end, we generated two QC knockout HCT116 cell lines and found that knockout of QC had minor effect on the binding of CC2C6 to HCT116 cells (Supplementary information, Fig. S2a, b, c). These data suggest that isoQC, but not QC, regulates the binding of CC2C6 to cancer cells.
Because isoQC is mainly localized within the Golgi complex, we asked whether isoQC is a general regulator of cell surface protein. To this end, we examined the effect of isoQC depletion on surface expression of another membrane protein PDL1. Our data indicated that knockout of isoQC had minor effect on the surface expression of PDL1 as detected by FACS assay (Supplementary information, Fig. S2d, e). These data indicate that isoQC specifically regulates the surface expression of CD47 in a broad range of cancer types.
To examine whether the enzymatic activity of isoQC is required for its regulation of CD47, we treated 293T cells with isoQC inhibitor, PQ529, which can efficiently inhibit the activity of both QC and isoQC. Our data showed that treatment with PQ529 for 48 h significantly blocked the binding of CC2C6 to the cell surface in a dosage-dependent manner in wild-type, but not in isoQC-deficient cells (Fig. 1d). Similar results were obtained by using different cell lines, including DLD1, 293T as well as leukemia cell lines ARP1, NU-DUL-1, Jurkat and H929 (Supplementary information, Fig. S3). To further confirm that the enzymatic activity of isoQC is required for its effect on CD47, we re-introduced either wild-type (WT) isoQC or E225G mutant isoQC that has partial enzymatic activity into isoQC-deficient 293T cells and detected the binding of CC2C6 to cell surface. Our data showed that WT, but not E225G, significantly promoted the binding of CC2C6 to the cell surface (Fig. 1e). Together, these data indicate that the enzymatic activity of isoQC is required for binding of CD47-specific antibody CC2C6 to cell surface.
Biochemical analysis of the native CD47 protein has predicted that the N-terminal residue of CD47 is pyroglutamate8. However, no direct evidence shows that the N-terminal residue of CD47 is modified to pGlu by isoQC. Through analyzing the protein sequence of CD47, we found a conserved glutamine at the N-terminus of CD47 exactly after the signal peptide, as similar to other pGlu modified substrates (Supplementary information, Fig. S4). Thus, we examined whether CD47 is a substrate of isoQC. To this end, we performed an in vitro cyclization assay by examining the cyclization of CD47 N-terminal peptide catalyzed by purified isoQC and the cyclization was analyzed by a nanoESI-MS. Our data confirmed that isoQC could promote the N-terminal pGlu formation of CD47 (Fig. 1f). Moreover, our co-IP data showed that isoQC was a binding partner of CD47 in cells (Fig. 1g). These data suggest that CD47 is a potential substrate of isoQC.
pGlu formation of protein usually protects targeting protein from degradation by proteases. Moreover, exclusive localization of isoQC within the Golgi complex suggests isoQC may also be involved in the regulation of protein maturation. We therefore examined whether isoQC affected the protein levels of CD47. First, our data from qPCR data showed isoQC knockout had no significant effect on CD47 mRNA level (Supplementary information, Fig. S5a). Surprisingly, we did not detect any significant effect on CD47 protein levels examined by Western Blot as well using a commercial rabbit anti-CD47 antibody from CST, which is generated with a synthetic peptide corresponding to residues surrounding Leu72 of human CD47 protein (Supplementary information, Fig. S5b). These inconsistent results from FACS and WB led us to propose that the CC2C6 antibody we used in FACS analysis may recognize CD47 in a pGlu formation-dependent manner. To this end, we obtained another commercial anti-CD47 antibody B6H12 from BD which could be used for FACS analysis. Compared to CC2C6, we did not detect any significant effect of isoQC on the cell surface expression of CD47 using B6H12 antibody (Fig. 1h; Supplementary information, Fig. S5c, d). These data suggest that CC2C6 is a pGlu-dependent CD47 antibody.
To further prove this hypothesis, we obtained a construct containing a GFP tag at the N-terminus9, which thus may block the exposure of N-terminal glutamine and should block the pGlu residue formation. We expressed GFP-CD47 in 293T cells and analyzed its expression on cell surface using CC2C6 and B6H12 antibodies. The efficient expression of GFP-CD47 was measured by FACS (Supplementary information, Fig. S6a). Moreover, although both B6H12 and CC2C6 could recognize the endogenous CD47, B6H12, but not CC2C6, could detect the surface expression of GFP-CD47 (Supplementary information, Fig. S6b). To further confirm the conclusion that the N-terminal residue of CD47 is modified to pGlu, we generated an antibody specifically recognizing the Q19 pGlu-modified CD47. The specificity of this antibody for recognition of the pGlu-modified CD47 was confirmed using Dot-blot (Supplementary information, Fig. S6c). Our data from both Western blot and FACS showed that depletion of isoQC significantly reduced the levels of pGlu-containing CD47 (Supplementary information, Fig. S6d, e). Based on the above findings, we concluded that the N-terminus of CD47 can be modified to form a pGlu residue by isoQC and CC2C6 antibody recognizes CD47 in a pGlu modification-dependent manner.
The crystal structure of CD47-SIRPα complex indicates that pGlu of CD47 is located in the CD47-SIRPα binding interface10. We therefore examined whether isoQC depletion affects the binding of CD47 to SIRPα using a cell-based SIRPα binding assay. Our data indicated knockout of isoQC dramatically reduced the binding of SIRPα to cell surface (Fig. 1i, j; Supplementary information, Fig. S7a). Moreover, treatment with isoQC inhibitor (PQ529) also reduced the binding of SRIPα (Supplementary information, Fig. S7b). These data together indicate that the enzymatic activity of isoQC is required for the sufficient binding of SIRPα to CD47.
Because CD47-SIRPα axis is the major ‘don’t eat me’ signal to block the phagocytosis of cancer cells by macrophage, we asked whether isoQC is required for the inhibition of phagocytosis of cancer cells by macrophage. To this end, we performed a widely used phagocytotic assay by co-culture of DLD1 cells with mouse bone marrow derived macrophages. Our data showed that depletion of isoQC dramatically promoted the macrophage-mediated phagocytosis of DLD1 cells (Fig. 1k, l). Similar results were obtained by treatment of DLD1 cells with isoQC inhibitors (PQ529) (Fig. 1m; Supplementary information, Fig. S8a). Interestingly, our data showed that the monoclonal antibody CC2C6 could also promote the phagocytosis of DLD1 by macrophages (Supplementary information, Fig. S8b), confirming that pGlu modification is required for the sufficient function of CD47. We also examined whether isoQC-mediated inhibition of the phagocytosis is dependent on CD47. To this end, we generated an isoQC and CD47 double knockout DLD1 cell line and performed phagocytosis assay. Our data showed that there was no significant difference in the phagocytosis of DLD1 cells by macrophages between isoQC/CD47 double knockout and CD47 knockout DLD1 cells (Supplementary information, Fig. S9). Together, these data indicate that isoQC catalyzed pGlu formation is required for the efficient phagocytosis of cancer cells by macrophages in a CD47-dependent manner.
As CD47 is highly expressed in various cancers, we analyzed whether expression of isoQC also correlated with cancer development in patients. Data from TCGA indicate that isoQC is significantly upregulated in multiple types of tumors and high expression of isoQC is reversely correlated with the overall survival rate of several types of cancer patients (Fig.1n; Supplementary information, Fig. S10a, b). These data suggest that isoQC is a potential regulator of CD47 in cancer development.
Collectively, we demonstrated that isoQC is an essential regulator of CD47-SIRPα axis and required for efficient phagocytic cells-mediated clearance of cancer cells. Moreover, we provided evidence showing that the N-terminal Q19 of CD47 was modified to a pGlu residue by isoQC. pGlu formation at the N-terminus is required for binding of CD47 to SIRPα. Our data suggest that the antibody recognizing pGlu may be used for anti-cancer immunotherapy. Moreover, we also found that the commercial monoclonal antibody CC2C6 is a pGlu- dependent antibody for CD47. Thus, our study suggests that isoQC is a potential therapeutic target for treatments of various cancers.
Supplementary information
Acknowledgements
We thank Dr. Weirui Ma and Yufeng Shi for kindly providing CD47 plasmids, Yingcong Wang and Prof. Jumei Shi for providing cell lines. We also thank the members of the Wang lab for their assistance. This work was supported by the National Natural Science Foundation of China (81625019, 31830053, 31871398, 31801177, 31500735, 31801178, 31800757), the Science Technology Commission of Shanghai Municipality (16JC1404500, 18410722000), the Commission of Health and Family Planning of Shanghai Municipality (2017BR014 to P.W.; 2017YQ068 to L.F.), the Young Elite Scientist Sponsorship Program by CAST(YESS)(2018QNRC001), the Shanghai Sailing Program (18YF1419500, 18YF1419300), and the Fundamental Research Funds For the Central Universities (22120170259, 22120180043, 22120180045, 22120180049).
Author contributions
Z.W., L.W., T.Z., and P.W. conceived the project and designed experiments. Z.W., L.W., T.Z., H.T., L.F., H.T., W.Z., J.G., Y.H., Y.L., and H.Z. performed experiments. Z.W., L.W., T.Z., and P.W. analysis the data. Z.W., L.W., and P.W. wrote the manuscript.
Competing interests
The authors declare no competing interests.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-019-0177-0.
References
- 1.Pardoll DM. Nat. Rev. Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barclay AN, Van den Berg TK. Ann. Rev. Immunol. 2014;32:25–50. doi: 10.1146/annurev-immunol-032713-120142. [DOI] [PubMed] [Google Scholar]
- 3.Cynis H, et al. J. Mol. Biol. 2008;379:966–980. doi: 10.1016/j.jmb.2008.03.078. [DOI] [PubMed] [Google Scholar]
- 4.Becker A, et al. Biol. Chem. 2016;397:45–55. doi: 10.1515/hsz-2015-0192. [DOI] [PubMed] [Google Scholar]
- 5.Chen YL, et al. Biochem. J. 2012;442:403–412. doi: 10.1042/BJ20110535. [DOI] [PubMed] [Google Scholar]
- 6.Cynis H, Scheel E, Saido TC, Schilling S, Demuth HU. Biochemistry. 2008;47:7405–7413. doi: 10.1021/bi800250p. [DOI] [PubMed] [Google Scholar]
- 7.Speedy HE, et al. Nat .Genet. 2014;46:56–60. doi: 10.1038/ng.2843. [DOI] [PubMed] [Google Scholar]
- 8.Mawby WJ, Holmes CH, Anstee DJ, Spring FA, Tanner MJ. Biochem. J. 1994;304:525–530. doi: 10.1042/bj3040525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berkovits BD, Mayr C. Nature. 2015;522:363–367. doi: 10.1038/nature14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hatherley D, et al. Mol. Cell. 2008;31:266–277. doi: 10.1016/j.molcel.2008.05.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.