Dear Editor,
FOXA1 (Forkhead Box Protein A1) is a pioneer transcription factor (TF) that functions to loosen the compact chromatin structure to facilitate binding of other TFs such as estrogen receptor and androgen receptor (AR).1,2 AR is a nuclear receptor functioning as a ligand-dependent TF that plays a pivotal role in driving the initiation of prostate cancer (PCa) and the development of castration-resistant PCa (CRPC).3 The chromatin binding of AR is dependent on FOXA1, which interacts with DNA through its Forkhead DNA binding domain (FKHD). Recent studies on PCa patient samples revealed genetic mutations of FOXA1 in primary PCa (~4%) and the mutation frequency is increased in the high-risk racial groups and in more aggressive metastatic CRPC (~10%),4–7 suggesting that PCa with FOXA1 mutations may be more aggressive and resistant to current therapies. The majority of FOXA1 mutations (~60%-80%) are found in a hot spot region located at the Wing2 region of FKHD. Due to a lack of functional studies, it remains unclear how these mutations affect PCa progression and patient outcome.
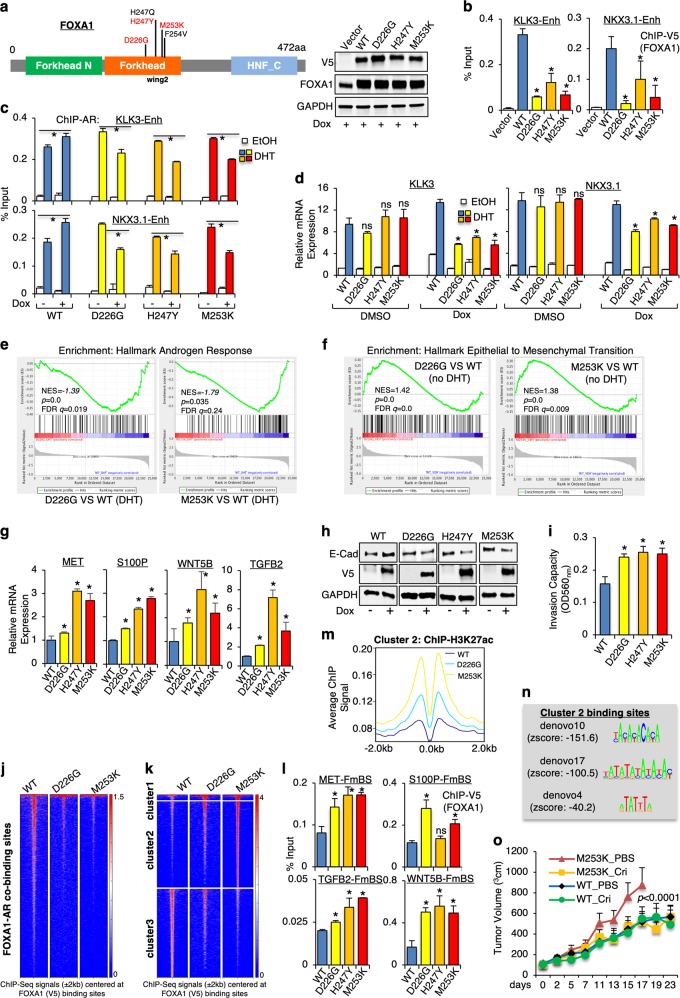
In this study, we focused on the missense mutations at the winged-helix FKHD (called FKHD-MSs) and selected three most frequent FKHD-MSs (D226G, H247Y and M253K) to generate LNCaP (an androgen-sensitive PCa cell line) stable cells with tetracycline-inducible expression of V5-tagged FKHD-MSs (Fig. 1a). Intriguingly, the chromatin binding of FOXA1 at previously identified AR-dependent enhancers8 was impaired by the expression of FKHD-MSs (Fig. 1b) although some mutants may have more stable protein expression and stronger interaction with AR (Supplementary information, Fig. S1a–e). As a result, AR recruitments at these enhancers were decreased (Fig. 1c) and the expression of androgen-regulated genes was repressed by FHKD-MSs (Fig. 1d). Expressing a FKHD-MS with simultaneous knockdown of endogenous FOXA1 resulted in a similar reduction of AR-regulated gene expression (Supplementary information, Fig. S1f, g). Using RNA-seq analyses, we found that AR activity was broadly suppressed by the expression of FKHD-MSs (Fig. 1e; Supplementary information, Fig. S1h, i). Overall, these results indicate that FKHD-MSs impair the chromatin binding of FOXA1 to AR-dependent enhancers and thus suppress AR transcriptional activity.
Fig. 1.
FKHD-MSs impaired AR signaling while activated EMT in PCa cells. a Establishment of LNCaP stable cell lines expressing tetracycline-inducible V5-tagged FKHD mutants. b ChIP-qPCR of V5 at the indicated sites in FKDH-MS stable cells treated with doxycycline for 2 days (d). c ChIP-qPCR of AR at the indicated sites in FKDH-MS-expressing cells treated with doxycycline (2 d) and stimulated with 10 nM DHT (4 h). d The mRNA expression of AR-regulated genes was decreased by FKHD-MSs (by qRT-PCR). e RNA-seq analyses on FOXA1 WT-, D226G- or M253K-expressing cells treated with/without 10 nM DHT (24 h). FKHD-MS-downregulated genes were enriched in AR signaling (by GSEA). f FKHD-MS-upregulated genes were enriched in EMT (by GSEA). g Validation for a list of FKHD-MS-activated EMT genes (by qRT-PCR). h E-Cadherin expression was decreased by induced expression of FKHD-MSs (by immunoblotting). i LNCaP cells expressing FHKD-MSs had higher invasion capability (by Matrigel invasion assay). j ChIP-seq analyses of V5 in LNCaP cells expressing WT FOXA1 versus FKHD-MSs were performed under hormone-depleted condition. Heatmap view for V5 binding at the previously determined AR-FOXA1 overlapping sites was shown. k Heatmap view for V5 binding at clustered binding sites (top 10 k). l ChIP-qPCR of V5 at the indicated sites. m ChIP-seq of H3K27ac in LNCaP cells expressing WT FOXA1 versus FKHD-MSs were performed under hormone-depleted condition. Means of H3K27ac signals at cluster 2 binding sites were shown. n Motif enrichment analyses in the cluster 2 sites revealed previously unidentified motifs (three top ranked motifs were shown). o CWR22-RV1 cells expressing FOXA1 WT or M253K were injected into castrated male SCID mice. When tumors reached ~5 mm, mice were fed with doxycycline-supplemented food to induce FOXA1 expression and then were treated with PBS or crizotinib. Tumor volumes were measured by caliper. Student’s t-test was used to determine the statistical difference (*P < 0.05) for FKHD-MSs versus WT FOXA1 unless otherwise indicated. Two-way ANOVA test was performed in the animal study to compare tumor growth
The RNA-seq analyses also revealed that the FKHD-MSs upregulate a subset of genes independent of AR (in the absence of androgens), which were highly enriched in epithelial-mesenchymal transition (EMT), a key process for cancer cell metastasis (Fig. 1f; Supplementary information, Fig. S2a, b). Amongst these genes, MET, S100P, TGFB2, and WNT5B were selected for subsequent validation. While FKHD-MSs significantly increased levels of the EMT genes (Fig. 1g; Supplementary information, Fig. S2c), FOXA1 silencing had no such consistent effect (Supplementary information, Fig. S2d–f), indicating that this EMT-promoting function of FKHD-MSs is through a mechanism distinct from the loss of overall FOXA1 binding. Loss of E-cadherin is a critical step in EMT initiation and we next examined the effects of FKHD-MSs on E-cadherin expression. As shown in Fig. 1h, E-cadherin expression was decreased by FKHD-MSs while increased by wild-type (WT) FOXA1, indicating that FKHD-MSs may facilitate EMT of PCa cells. Consistently, increased levels of additional EMT markers (vimentin and phosphorylated MET) were also seen in FHKD-MS-expressing cells (Supplementary information, Fig. S2g, h). Using Matrigel invasion assays, we found that the FKHD-MSs increased PCa cell invasion in vitro (Fig. 1i; Supplementary information, Fig. S2i). In addition to the effects on EMT and invasion, FKHD-MSs also modestly increased cell cycle progression in the absence of androgens (Supplementary information, Fig. S2j).
Based on the above studies, we hypothesize that FKHD-MSs may activate a subset of enhancers regulating EMT-related genes independent of AR. To examine the chromatin binding of FKHD-MSs, we carried out ChIP-seq analyses of V5 (for exogenous FOXA1) in those stable cells under hormone-depleted condition (Supplementary information, Fig. S3a). Consistent with the repressive effect of FOXA1 binding at KLK3 and NKX3.1 enhancers, the chromatin binding intensity of the FKHD-MSs was substantially lower at the previously identified AR-FOXA1 overlapping sites8 (Fig. 1j). Intriguingly, the analyses also revealed a cluster of preferred binding sites for FKHD-MSs (Fig. 1k, cluster 2), indicating that FKHD-MSs may reprogram FOXA1 cistrome to occupy new chromatin sites independent of AR activity. FKHD-MS-preferred binding sites (called FmBS) were identified at those EMT gene loci (Supplementary information, Fig. S3b) and the increased bindings of FKHD-MSs at those sites were confirmed (Fig. 1l; Supplementary information, Fig. S3c, d). Activated enhancers are commonly associated with histone acetyltransferases (HATs) and increased histone acetylation.9 Therefore, we performed ChIP-seq of H3K27ac (acetylated lysine 27 of histone 3), a histone modification mediated by CBP/p300, to determine whether FmBSs are globally associated with enhancer activation. As shown in Fig. 1m and Supplementary information, Fig. S4a, the levels of H3K27ac were substantially increased at FmBSs, suggesting that many of these sites may be activated enhancers. Interestingly, the H3K27ac level was low in an identified FmBSs (Supplementary information, Fig. S4b, c), possibly due to the recruitment of distinct HATs other than CBP/p300 at those regions. Nonetheless, these results highly suggest that FKHD-MSs specifically activate a subset of enhancers.
We next conducted a motif enrichment analysis on the cluster 2 versus cluster 3 sites to characterize the binding motifs for FKHD-MSs at FmBSs. While the cluster 3 sites were enriched for classic FOXA1 binding motifs, the cluster 2 sites contained a number of structurally altered FKHD binding motifs (Fig. 1n and Supplementary information, Fig. S4d, e). Using gel shift assay, we further show that D226G and M253K mutants but not WT FOXA1 can directly bind to the DNA probe containing a top ranked motif (denovo10) (Supplementary information, Fig. S4f). These results suggest that FKHD-MSs may have increased binding affinity to the new DNA motifs that were not previously identified.
To further determine the role of FKHD-MSs in CRPC progression, we used CWR22-RV1 cells (an AR-positive CRPC cell line) to generate stable lines with tetracycline-inducible expression of V5-tagged FKHD-MSs (Supplementary information, Fig. S5a). Similar to LNCaP-derived models, FKHD-MSs significantly decreased FOXA1 binding at the KLK3 enhancer and the expression of AR-regulated genes while increased the expression of EMT genes (Supplementary information, Fig. S5b–d). Amongst those FKHD-MS-activated EMT genes, MET has been intensively studied and was shown to associate with PCa progression.10 Importantly, CRPC cells expressing FKHD-MSs were more sensitive to treatment of a MET inhibitor, crizotinib (Supplementary information, Fig. S6a). We next established CRPC xenograft models using cells expressing FOXA1 WT or M253K and the increased expression of MET was confirmed in the tumor biopsies carrying M253K mutation (Supplementary information, Fig. S6b). Interestingly, M253K-expressing tumors grew faster than WT tumors; however, they were sensitive to the crizotinib treatment (Fig. 1o), whereas the development of WT tumors was not affected by crizotinib. These results suggest that MET inhibition may be a novel strategy to treat CRPC with FKHD-MSs.
In summary, our data demonstrate that FKHD-MSs reduce the PCa cell dependency on AR signaling through impairing FOXA1 chromatin binding to the AR-dependent enhancers and thus suppressing AR activity, and promote PCa progression through increasing FOXA1 binding to a subset of AR-independent enhancers that regulates transcription of genes mediating EMT and metastasis. The GEO accession for RNA-seq and ChIP-seq data is GSE133455.
Supplementary information
Acknowledgements
We thank Drs. Franklin Huang (University of California, San Francisco) and Alexandra E. Shields (Massachusetts General Hospital) for the advice and support of this project. This work was supported by grants from NIH (R01CA211350 to C.C. and U54CA156734 to C.C. and J.A.M.), DOD (W81XWH-15-1-0554 to S.G. and W81XWH-16-1-0445 to C.C.), CIHR (142246, 152863, 152864, and 159567 to H.H.H.), Prostate Cancer Canada (RS2016-1022 and TAG2018-2061 to H.H.H.), NSERC (498706 to H.H.H.), Terry Fox New Investigator Award (1069 to H.H.H.), and Princess Margaret Cancer Foundation (to H.H.H.).
Author contributions
S.G., S.C., H.H.H. and C.C. designed the study. S.G., S.C., D.H., D.B., W.H. and M.A. performed the experiments and analyzed the results. S.P. and J.A.M performed deep sequencing analyses. S.G., S.C., H.H.H. and C.C. wrote and revised the manuscript. All authors discussed the results and commented on the manuscript.
Competing interests
The authors declare no competing interests.
Contributor Information
Housheng Hansen He, Email: hansenhe@uhnresearch.ca.
Changmeng Cai, Email: changmeng.cai@umb.edu.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-019-0203-2.
References
- 1.Carroll JS, et al. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 2.Lupien M, et al. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuan X, et al. Oncogene. 2014;33:2815–2825. doi: 10.1038/onc.2013.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbieri CE, et al. Nat. Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research Network. Cell. 2015;163:1011–1125. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson D, et al. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang FW, et al. Cancer Discov. 2017;7:973–983. doi: 10.1158/2159-8290.CD-16-0960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai C, et al. Cell Rep. 2014;9:1618–1627. doi: 10.1016/j.celrep.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calo E, Wysocka J. Mol. Cell. 2013;49:825–837. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varkaris A, et al. Expert Opin. Investig. Drugs. 2011;20:1677–1684. doi: 10.1517/13543784.2011.631523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.