Role of RNA regulation in autophagy was rather unexplored until Horos et al. recently showed that vault RNA1-1 binds directly to p62/SQSTM1, a well-established autophagy receptor. This binding leads to inhibition of p62 oligomerization and negative regulation of selective autophagy pathways.
Autophagy is a conserved eukaryotic mechanism for the clearance of unwanted cellular components like protein aggregates (aggrephagy), damaged organelles (e.g., mitophagy) and pathogens (xenophagy).1 In this process, a membrane bilayer termed autophagosome encloses the disposable garbage. Autophagosomes fuse with lysosomes for the decomposition and recycling. There are two modes of autophagy: selective and nonselective. Whereas selective autophagy identifies ubiquitin chains on the cargo for the targeted delivery, nonselective autophagy randomly sequesters cytoplasmic components for lysosomal degradation.2
To harbor the cargo, selective autophagy makes use of several autophagy receptors, e.g., p62, OPTN, NDP52 and NBR1.3 p62/SQSTM1, one of the best-characterized autophagy receptor, contains LIR (LC3 interacting region) motif and UBA (ubiquitin associated) domain at its C-terminus that are required for the binding to Atg8 family members (LC3/GABARAPs) and ubiquitin chains, respectively.3 With the help of these interactions, p62 achieves binding to the ubiquitinated cargo on one hand and to the LC3-decorated phagosomes on the other hand. p62 plays a critical role in aggrephagy, mitophagy and xenophagy.4 p62 is itself degraded in autophagolysosomes together with the cargo and is also distinguished due to its ability to form higher-order oligomers via its N-terminal PB1 (Phox and Bem1p) domain, which help to attain high-avidity interactions with ubiquitin and LC3-coated surfaces.5 Oligomerization is dependent on ubiquitin chains (mainly K63- or M1-linked ubiquitin chains) and leads to phase separated bodies before subsequent clearance by autophagy.6,7
In a recent study, Horos et al., identified p62 as a RNA-binding protein using a proteome-wide method named RBDmap (RNA-binding domains map).8,9 In this method, UV-crosslinked protein-RNA complexes are purified using magnetic oligo(dT) beads in two steps and then subjected to mass spectrometric analysis. The peptide intensity ratios between the RNA-bound and -released fractions are then used to ascertain RNA-binding sites. Subsequently, p62-RNA interaction was confirmed using northern blotting, western blotting, PNK (polynucleotide kinase) assay and in vitro binding assays. Among the RNAs identified to bind p62, vault RNAs (vtRNAs) turned out to be the most prominent hit. vtRNAs are ~88–100 nt long non-coding RNAs transcribed by RNA polymerase III.10 All vtRNAs are predicted to fold into characteristic stem-loop structures. The role of stem loop in vtRNAs is still unknown, however, they potentially participate in interactions with RNAs and proteins. Interestingly, the analysis of iCLIP experiments suggested that the binding of vtRNAs to p62 is driven by structural signatures like loops rather than a consensus binding motif. The authors further identified vtRNA1-1 as a prime p62-interacting RNA. The vtRNA1-1 interacting region of p62 was mapped to the zinc cluster 1 of the Zinc Finger (ZZ) domain, which was further characterized by loss of binding of the R139A, K141A (RK/A) double mutant of p62. However, ZZ domain does not seem to be the only interaction surface as the oligomerization-deficient mutant of p62 (R21A, D69A, D73A triple mutant referred to as p62 PB1m) also showed diminished binding to vtRNA1-1.11
The authors also tested the functional consequences of vtRNA1-1 binding on p62 activity and autophagy. Knockdown of vtRNA1-1 by antisense LNAs (locked nucleic acids) induced the shift in LC3B-II/LC3B-I ratio, indicating elevation in autophagy flux. Under starvation conditions, the levels of vtRNA1-1 are diminished, which facilitates p62-mediated autophagy induction. This raised the questions: how does RNA binding affect autophagy via p62? What is the underlying mechanism? Oligomerization of p62 and the binding of p62 to ATG8-like proteins (LC3B and GABARAP used in this study) appears to be prevented in the presence of vtRNA1-1. In a so-called ‘laddering assay’ the authors showed that p62 oligomerization could be visualized in cellulo after UV treatment and this oligomerization was visualized by western blot in the form of p62 higher-order oligomers/ladders. In this assay the ZZ domain RK/A mutant appears to have the tendency to form more higher-order oligomers compared to the wild-type (WT) p62 at basal conditions, suggesting that vtRNA1-1 prevents p62 oligomerization.
To further address this issue, the authors utilized a chemical compound XIE62-1004-4 (XIE) that has been shown to bind ZZ domain of p62, inducing p62 oligomerization and further p62-dependent autophagy. Compared to the WT cells, vtRNA1-1 knockout cells showed increased levels of LC3B lipidation under XIE treatment, underlining the fact that vtRNA1-1 prevents p62-mediated autophagosome formation.
This study nicely shows that p62 is a bona fide RNA-binding protein and its physical interaction with vtRNA1-1 blocks oligomerization of p62 and disables effective clearance of the accumulated ubiquitinated cargo by selective autophagy (Fig. 1). Yet, several intriguing questions follow this study. First, how does starvation downregulate the levels of vtRNA1-1? The authors tested the effect of increased expression of vtRNA1-1 by transfection on autophagy and found that increased vtRNA1-1 led to prevention of p62-dependent autophagy and p62 accumulation itself. Since p62 accumulation has been implicated in a variety of cancer types, the question is whether the cancer cells also use vtRNA1-1 as a tool to preserve p62 levels that in turn promotes tumor growth by mTORC1, Nrf2 and NF-κB signaling.4 Malfunctions in autophagic machinery or p62 mutations have been shown to result in p62 aggregates, which form inclusion bodies characteristic of several pathological conditions in brain and liver.12 Thus, it is interesting to determine the structural basis of the interactions of vtRNA1-1 with the PB1 and ZZ domains of p62, which may help design novel therapeutic interventions to inhibit p62 multimerization in such diseases. Other than vtRNAs, tRNAs also appeared as a hit for p62 binding. The physiological consequences of the binding of other vtRNA paralogs and tRNA to p62 need further investigation. Particularly, the study raises the question of whether binding of other RNA molecules can regulate autophagy at several levels of this multi-step process or it is very unique to only one autophagy receptor as shown for p62. Perhaps this is only the tip of the iceberg and we have a lot to learn about riboregulation in cellular catabolism.
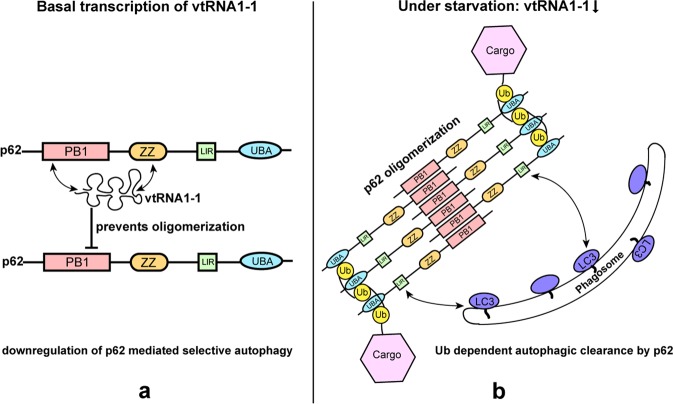
Fig. 1.
RNA binding negatively regulates p62-mediated autophagy. a p62 contains an N-terminal PB1 domain that is responsible for its oligomeric character followed by a zinc finger domain. C-terminal LIR motif and UBA domain are responsible for binding to Atg8 family members and ubiquitin (Ub) chains, respectively. Vault RNA1-1, a small non-coding RNA, blocks p62 oligomerization via interaction with the PB1 and ZZ domains. This results in the inhibition of p62 activity as autophagy receptor. b Starved cells show low levels of vtRNA1-1, which enables p62 multimerization. Thus, Ub-dependent selective autophagy is favored under starvation in a p62-mediated manner. The members of ATG8 family (LC3s/GABARAPs) are colored in purple
References
- 1.Mizushima N, Komatsu M. Cell. 2011;147:728–41. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 2.Dikic I, Elazar Z. Nat. Rev. Mol. Cell Biol. 2018;19:349–64. doi: 10.1038/s41580-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 3.Johansen T, Lamark T. Autophagy. 2011;7:279–96. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanchez-Martin P, Saito T, Komatsu M. FEBS J. 2019;286:8–23. doi: 10.1111/febs.14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciuffa R, et al. Cell Rep. 2015;11:748–58. doi: 10.1016/j.celrep.2015.03.062. [DOI] [PubMed] [Google Scholar]
- 6.Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. Mol. Cell. 2011;44:279–89. doi: 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- 7.Sun D, Wu R, Zheng J, Li P, Yu L. Cell Res. 2018;28:405–15. doi: 10.1038/s41422-018-0017-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horos R, et al. Cell. 2019;176:1054–67. doi: 10.1016/j.cell.2019.01.030. [DOI] [PubMed] [Google Scholar]
- 9.Castello A, et al. Nat. Protoc. 2017;12:2447–64. doi: 10.1038/nprot.2017.106. [DOI] [PubMed] [Google Scholar]
- 10.Stadler PF, et al. Mol. Biol. Evol. 2009;26:1975–91. doi: 10.1093/molbev/msp112. [DOI] [PubMed] [Google Scholar]
- 11.Wurzer B, et al. Elife. 2015;4:e08941. doi: 10.7554/eLife.08941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bjorkoy G, et al. J. Cell Biol. 2005;171:603–14. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]