Dear Editor,
Current approaches to analyze cellular structures in the bone tissues have depended mainly on tissue sections. For instance, a recent study visualized different cellular populations in the 300-μm sections of the mouse femurs1. Similarly, the distribution of hematopoietic stem cells could be examined in the half-bone sections2. Notably, such conventional methods obscure specific cellular structures, e.g., neural innervations, that would be better observed with the intact, unsectioned bones. Moreover, histological sectioning could be challenging for many bone tissues, e.g., vertebral column, skull and limbs.
Development of the optical clearing techniques, e.g., CLARITY3, iDISCO+4, uDISCO5, and CUBIC6, has enabled the 3D imaging of diverse cellular structures in intact tissues. In particular, 3D fluorescence imaging of the mouse bones has been recently achieved. For example, uDISCO visualized the Cx3cr1-positive cells genetically labeled by EGFP in the femur5; Bone CLARITY imaged the tdTomato-labeled osteoprogenitor cells in Sox9CreER; Rosa26-LSL-tdTomato mice7. However, these methods relied exclusively on the fluorescence markers, and their general implementation could be limited by several factors: (1) lack of an available genetic fluorescence labeling approach; (2) research need to simultaneously image two or more different cellular structures; (3) time and cost of breeding a mouse line to integrate multiple genetic alleles. Therefore, a whole-tissue immunolabeling procedure to detect endogenous epitopes in the intact bones, and at the same time, to be compatible with optical clearing for 3D fluorescence imaging, has been highly desired but not yet been successfully attempted.
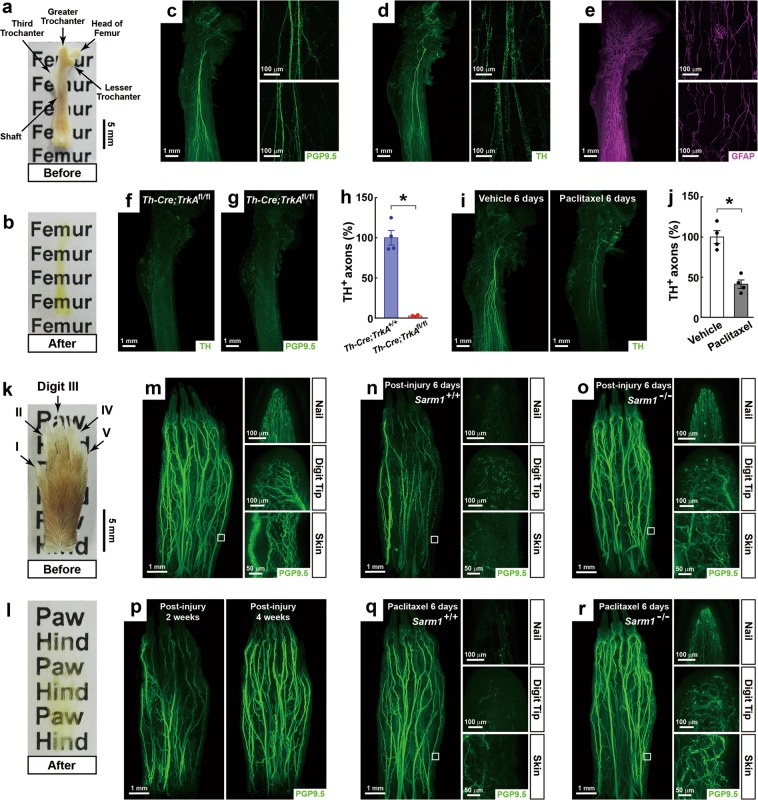
We reported here a new BoneClear method for the robust immunolabeling of different cellular structures in the intact, unsectioned bone tissues of adult mice (Supplementary information, Data S1). First, we compared BoneClear with iDISCO+4, uDISCO5, and Bone CLARITY7 for the optical clearing of the mouse bones. The intact femurs became nearly transparent after being processed by BoneClear (Fig. 1a, b), which produced the highest quality of optical clearing among the examined methods (Supplementary information, Fig. S1a). Of note, uDISCO resulted in tissue shrinkage (Supplementary information, Fig. S1d), while Bone CLARITY led to tissue expansion (Supplementary information, Fig. S1e). BoneClear (and also iDISCO+) did not cause such changes in tissue size of the femurs (Supplementary information, Fig. S1b, c). Next, we compared BoneClear with the published methods for the whole-tissue immunolabeling and 3D imaging of intended cellular structures. The intact, unsectioned mouse femurs were processed by BoneClear for the immunolabeling of PGP9.5, a specific pan-neural marker, which revealed a neural architecture within the bone marrow that had been previously undetected (Fig. 1c; Supplementary information, Movie S1). The XZ- and YZ-sections showed the imaging depth of > 3 mm through the femur (Supplementary information, Fig. S2a). In addition, the femurs were processed by BoneClear for the immunolabeling of tyrosine hydroxylase (TH), a specific marker for sympathetic axons, which exhibited the intraosseous sympathetic innervations (Fig. 1d). In contrast, the TH immunolabeling by iDISCO+, which lacks the decalcification step as been optimized for BoneClear that conceivably facilitates the tissue penetration of antibodies, revealed only a few sympathetic axons (Supplementary information, Fig. S2b). Because uDISCO and Bone CLARITY only visualized fluorescence markers5,7, we utilized Cx3cr1EYFP/+ mice, in which the Cx3cr1-positive immune cells are EYFP labeled. Indeed, some EYFP-positive cells could be imaged in the femurs processed by uDISCO or Bone CLARITY. However, the EYFP immunolabeling by BoneClear offered a significantly stronger signal than the endogenous EYFP fluorescence (Supplementary information, Fig. S2c). These results have demonstrated the advantages of BoneClear over the reported methods for the whole-tissue immunolabeling and 3D imaging of the intact mouse bones.
Fig. 1.
Whole-tissue immunolabeling of the intact mouse femurs and hindpaws to image neural anatomy and pathology. a, b The intact, unsectioned femurs of adult mice before (a) and after (b) BoneClear. c–e The femurs were processed for the whole-tissue immunolabeling of PGP9.5 (c), TH (d) or GFAP (e). Representative 3D-projection images at 1.26× magnification on the lightsheet microscope (left panels), and representative 3D-projection images of 300 μm-depth bone marrow in the proximal (upper-right panels) and middle (lower-right panels) shafts at 20× magnification on the confocal microscope were shown. f–h Ablation of the intraosseous sympathetic innervations in the femurs of Th-Cre; TrkAfl/fl mice. The femurs were processed for the immunolabeling of TH (f) or PGP9.5 (g). Representative 3D-projection images at 1.26× magnification on the lightsheet microscope were shown. The length of the TH-positive sympathetic axons in the femurs was quantified (h). n = 4, mean ± SEM, *P < 0.01 (Student’s t-test). i, j Chemotherapy-induced neuropathy of the intraosseous sympathetic innervations. The femurs of the vehicle-treated (left panel) or Paclitaxel-treated (right panel) mice were processed for the TH immunolabeling. Representative 3D-projection images at 1.26× magnification on the lightsheet microscope were shown (i). The length of the TH-positive sympathetic axons in the femurs was quantified (j). n = 4, mean ± SEM, *P < 0.01 (Student’s t-test). k, l The intact, unsectioned hindpaws of adult mice before (k) and after (l) BoneClear. m The hindpaw was processed for the whole-tissue PGP9.5 immunolabeling. n, o Traumatic injury-induced neuropathy in the hindpaw. Sarm1+/+ (n) or Sarm1−/− (o) mice were subjected to the sciatic nerve injury. The hindpaws were processed for the PGP9.5 immunolabeling at 6 days post injury. p Post-injury neural regeneration in the hindpaw. The hindpaws of wild-type mice were processed for the PGP9.5 immunolabeling at 2 weeks (left panel) or 4 weeks (right panel) after the sciatic nerve injury. q, r Chemotherapy-induced neuropathy in the hindpaw. Sarm1+/+ (q) or Sarm1−/− (r) mice were treated with Paclitaxel for 6 days, and the hindpaws were processed for the PGP9.5 immunolabeling. Representative 3D-projection images at 1.26× magnification on the lightsheet microscope (left panels, the squares denote the skin regions imaged at 12.6× magnification), and representative 3D-projection images of the nail (upper-right panels), digit tip (middle-right panels) and 300 μm-depth skin (lower-right panels) at 12.6× magnification were shown (m, n, o, q, r)
BoneClear enables the immunolabeling of diverse cellular structures in the mouse bones. For instance, the immunolabeling of PECAM1, a specific marker for vascular endothelial cells, revealed the vascular network inside the femur (Supplementary information, Fig. S3a). Notably, 82.4% ± 4.7% (mean ± SEM, n = 4) of the intraosseous neural innervations appeared spatially engaged with the vasculatures as shown by the co-immunolabeling of PGP9.5 and PECAM1 (Supplementary information, Fig. S5f). Chondrocytes in the femur could also be visualized by the immunolabeling of SOX9 (Supplementary information, Fig. S3b), a transcription factor involved in chondrocyte differentiation. Interestingly, the immunolabeling of GFAP, a specific glial marker, exhibited a striking glial network in the femur that had not been realized before (Fig. 1e; Supplementary information, Movie S2). The co-immunolabeling of GFAP and TH showed that 38.8% ± 5.2% (mean ± SEM, n = 4) of the glial cells were entangled with the sympathetic axons (Supplementary information, Fig. S3c). In addition, the co-immunolabeling of GFAP and PECAM1 revealed that 74.6% ± 6.9% (mean ± SEM, n = 4) of the glial cells were in apposition to the vasculatures (Supplementary information, Fig. S3d), elucidating the potential axonal-glial-vascular interaction within the bone marrow.
Moreover, BoneClear could process the unsectioned mouse vertebral column. The dorsal root ganglion was imaged through the vertebrae by the PGP9.5 immunolabeling (Supplementary information, Fig. S4a, b). In addition, BoneClear was applied to the intact mouse skull. The immunolabeling of VEGFR3, a specific marker for lymphatic endothelial cells, visualized the lymphatic vessels in the meninges underneath the skull (Supplementary information, Fig. S4c, d), as been recently discovered8.
To exhibit the utilization of this new BoneClear method, we assessed the intraosseous sympathetic innervations in detail. Th-Cre; TrkAfl/fl mice were bred to delete TrkA, the high-affinity receptor for nerve growth factor, specifically in sympathetic neurons. Gross appearance, length, and tissue weight of the Th-Cre; TrkAfl/fl femurs appeared comparable with those of control femurs (Supplementary information, Fig. S5a-c). However, the sympathetic innervations were ablated entirely in the Th-Cre; TrkAfl/fl femurs as evidenced by the TH immunolabeling (Fig. 1f, h). Intriguingly, the immunolabeling of PGP9.5 or Synaptophysin, two specific pan-neural markers, showed a similar loss of the total neural innervations in the Th-Cre; TrkAfl/fl femurs (Fig. 1g; Supplementary information, Fig. S5d). This finding implicated that the neural innervations in the femur are predominantly of sympathetic origin, though an indirect effect of the TrkA deletion on other neural types cannot be excluded. We noted that such ablation of the neural innervations did not affect the vasculatures in the Th-Cre; TrkAfl/fl femurs (Supplementary information, Fig. S5e, f). Next, we looked into chemotherapy-induced neuropathy of the intraosseous sympathetic innervations. Paclitaxel is a common chemotherapy drug for cancer treatment. As one of its adverse effects, Paclitaxel causes peripheral neuropathy in patients. There was a profound loss of the sympathetic innervations in the femurs of the Paclitaxel-treated mice (Fig. 1i, j), consistent with the prior report by the conventional histochemistry method9. These results have demonstrated the strength of BoneClear for the 3D imaging of neural anatomy and pathology in the mouse bones.
To further prove its imaging power, BoneClear was applied to the intact, unsectioned hindpaws of adult mice (Fig. 1k, l). The whole-tissue PGP9.5 immunolabeling revealed the striking entirety of the neural innervations in the hindpaw (Fig. 1m; Supplementary information, Movie S3). The high-magnification images showed the fine details of the neural innervations at different anatomical regions. For example, the neural fibers projecting along the luminal surface inside the nail was visualized (Fig. 1m; Supplementary information, Movie S4). We then examined traumatic injury-induced neuropathy in the hindpaw. Following the sciatic nerve injury, the widespread destruction of the neural innervations occurred (Fig. 1n). Interestingly, the neural fibers in the digits I and II remained intact because the sciatic nerve projects to the center and lateral side of the hindpaw but the saphenous nerve innervates the medial side10. Previous studies reported that traumatic injury-induced neuropathy depends on Sarm1 and its downstream MAPK signal11,12. Indeed, such a protective effect could be visualized in the Sarm1−/− hindpaw (Fig. 1o). We also followed the process of neural regeneration. The regenerating axons began to re-innervate the proximal and middle portions of the hindpaw at 2 weeks post injury, and the overall pattern of the neural innervations was re-established at 4 weeks (Fig. 1p). Finally, we examined chemotherapy-induced neuropathy in the hindpaw. While the major neural bundles were preserved under the Paclitaxel-treated condition, destruction of the local neural fibers was evident in several anatomical regions, i.e., nail, digit tip and skin (Fig. 1q). Consistent with the recent reports that Sarm1 also regulates the chemotherapy-induced neuropathy12–14, we observed the significant protection in the Sarm1−/− hindpaws under the Paclitaxel-treated condition (Fig. 1r; Supplementary information, Fig. S6a-c). These results have exhibited the application of BoneClear for the 3D imaging of neural anatomy and pathology in the mouse limbs.
In summary, we have developed a new BoneClear method to achieve the robust whole-tissue immunolabeling and 3D imaging of diverse cellular structures in the intact mouse bone tissues. This technical advance is poised to serve the research field in future investigations.
Supplementary information
Supplementary information, Methods and Figures
Acknowledgements
This study was supported by the State Key Laboratory of Membrane Biology, the IDG/McGovern Institute for Brain Research, the Center for Life Sciences and the School of Life Sciences at Peking University, and by the National Natural Science Foundation of China (31522024 and 31771111).
Author Contributions
J.Y. conceived and designed the project as the senior author. Q.W., K.L., L.Y. and H.W. performed the experiments and analyzed the results under the supervision of J.Y. J.Y. wrote the manuscript with the assistance of Q.W., K.L., L.Y. and H.W.
Competing interests
The authors declare no competing interests.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-019-0217-9.
References
- 1.Coutu DL, Kokkaliaris KD, Kunz L, Schroeder T. Nat. Biotechnol. 2017;35:1202–1210. doi: 10.1038/nbt.4006. [DOI] [PubMed] [Google Scholar]
- 2.Acar M, et al. Nature. 2015;526:126–130. doi: 10.1038/nature15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chung K, et al. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renier N, et al. Cell. 2016;165:1789–1802. doi: 10.1016/j.cell.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan C, et al. Nat. Methods. 2016;13:859–867. doi: 10.1038/nmeth.3964. [DOI] [PubMed] [Google Scholar]
- 6.Susaki EA, et al. Cell. 2014;157:726–739. doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- 7.Greenbaum A, et al. Sci. Transl. Med. 2017;9:eaah6518. doi: 10.1126/scitranslmed.aah6518. [DOI] [PubMed] [Google Scholar]
- 8.Louveau A, et al. Nature. 2015;523:337–341. doi: 10.1038/nature14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lucas D, et al. Nat. Med. 2013;19:695–703. doi: 10.1038/nm.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Decosterd I, Woolf CJ. Pain. 2000;87:149–158. doi: 10.1016/S0304-3959(00)00276-1. [DOI] [PubMed] [Google Scholar]
- 11.Osterloh JM, et al. Science. 2012;337:481–484. doi: 10.1126/science.1223899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang J, et al. Cell. 2015;160:161–176. doi: 10.1016/j.cell.2014.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geisler S, et al. Brain. 2016;139:3092–3108. doi: 10.1093/brain/aww251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turkiew E, Falconer D, Reed N, Hoke A. J. Peripher. Nerv. Syst. 2017;22:162–171. doi: 10.1111/jns.12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information, Methods and Figures