Abstract
Macrophages have been suggested to contribute to constructing a cancer stem cell (CSC) niche. However, whether and how macrophages regulate the activity of CSCs through juxtacrine signaling are poorly understood. Here we report LSECtin, a transmembrane protein highly expressed on tumor-associated macrophages (TAMs), enhances stemness of breast cancer cells (BCCs). We identified BTN3A3, a B7 family member with previously unknown functions as the receptor for LSECtin on BCCs responsible for stemness-promoting effect of LSECtin. In mice bearing human tumor xenografts, either macrophage-specific ablation of LSECtin or silencing of BTN3A3 in BCCs decreased CSC frequency and tumor growth. Admixture of LSECtin-positive macrophages increased the tumorigenic activity of BCCs dependent on BTN3A3. Disruption of the LSECtin-BTN3A3 axis with BTN3A3-Fc or anti-BTN3A3 mAb has a therapeutic effect on breast cancer. These findings define a juxtacrine signaling mechanism by which TAMs promote cancer stemness. Targeting this axis in the CSC niche may provide potential therapies to breast cancer.
Subject terms: Cell biology, Cancer microenvironment
Introduction
As is the case for normal stem cells, CSCs exist in a cellular niche comprising numerous cell types that together provide a unique microenvironment to protect and promote CSC functions.1,2 Therefore, targeting CSC niche components may be a promising approach in cancer therapy. However, we are far from achieving this goal because our understanding of the complex communication network of the CSC niche is limited. An area of great interest is the role of immune cells in the CSC niche. Tumor-associated macrophages (TAMs) representing a major immune cell type in the tumor microenvironment, provide pivotal paracrine signals that promote CSC survival, self-renewal, maintenance, and migratory ability in various types of cancer.3–5 Some researchers have speculated that, like normal stem cells, CSCs utilize cell–cell contact to preserve their phenotype and exert their functions.1 However, whether and how macrophages regulate CSC activity through juxtacrine signaling is poorly understood.
Our previous studies have shown that LSECtin (coded by Clec4g), is a type II transmembrane protein that is constitutively expressed on Kupffer cells and liver sinusoidal endothelial cells and negatively regulates hepatic T cell immunity.6–8 We also demonstrated that melanoma cell-expressed LSECtin promotes tumor growth by dampening antitumor T cell responses.9 LSECtin is also detected in monocyte-derived macrophages at the RNA and protein level, which could be induced by IL-4.10 The transcriptome-based network analysis reveals that the immune-regulatory factors IL-4 and IL-13 could induce Lsectin expression on human macrophages.11 These findings encourage us to investigate whether LSECtin is expressed by macrophages in the tumor environment and how macrophage-derived LSECtin contributes to the tumor progression.
Butyrophilins (BTNs) contain one or two extracellular Ig domains that exhibit some structural features of the B7 family of co-receptors and are considered to be B7-related proteins.12–14 The BTN3A subfamily, which includes three members, BTN3A1, BTN3A2, and BTN3A3, has been shown to be expressed in various types of tumor cells.15,16 Specific single-nucleotide polymorphisms (SNPs) in BTN3A3 and BTN3A2 have recently been reported to be associated with increased susceptibility to ovarian and gastric cancer, respectively.17,18 However, little is known about the receptor-ligand interactions for BTN3A family or about their precise functions in the tumor microenvironment.
Macrophages constitute a major component of the leukocyte infiltrate in breast cancer. Here, we demonstrate that the breast cancer cell-intrinsic BTN3A3 receptor drives tumor stemness and that this effect requires interactions with its ligand, LSECtin, expressed on TAMs. Our study defines a juxtacrine signaling mechanism by which TAMs in CSC niche promote breast cancer stemness and growth. Targeting the LSECtin-BTN3A3 axis may be an exciting approach for breast cancer treatment.
Results
LSECtin is expressed on macrophages in the tumor microenvironment
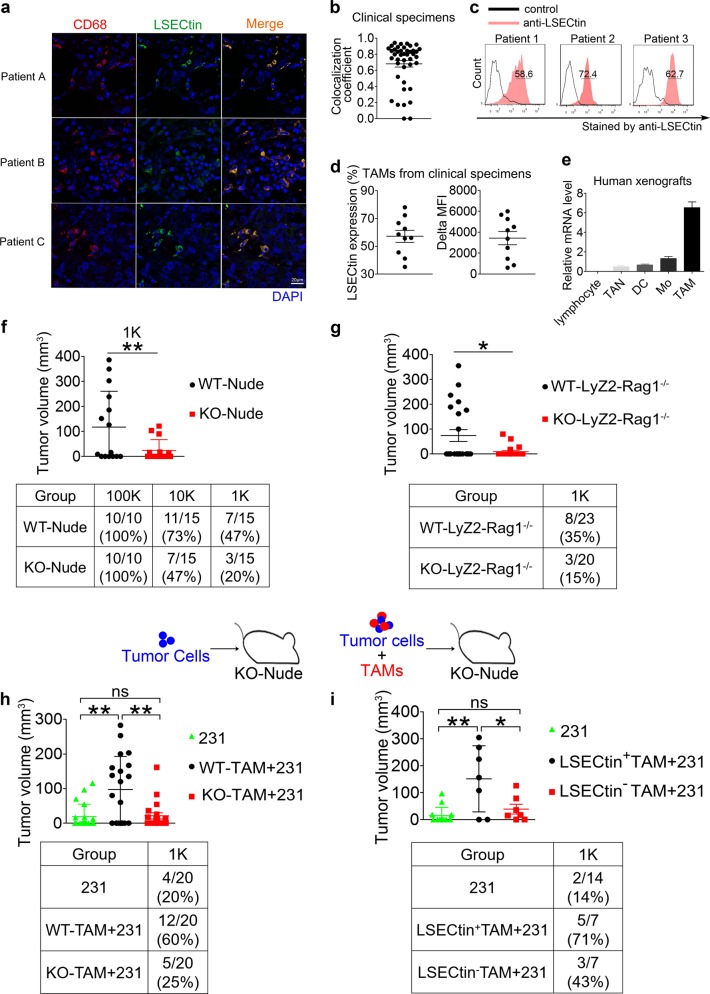
We first determined whether LSECtin is expressed on macrophages in human breast tumor tissues. We collected 42 samples expressing CD68, an indicator of macrophages and found LSECtin was expressed in 31/ 42 specimens (Supplementary information, Fig. S1a and Supplementary information, Table S1). The overlap coefficient of the 31/42 samples (>0.65) showed co-localization of LSECtin and CD68 in the clinical specimens (Fig. 1a, b and Supplementary information, Table S2), especially in the triple-negative breast cancer (TNBC) specimens (Supplementary information, Fig. S1b). Moreover, flow cytometric analysis revealed LSECtin surface expression by macrophages in 10/10 breast cancer specimens examined, with cell frequencies ranging from 35.2 to 78.1% (Fig. 1c, d, Supplementary information, Figs. S1c, d and Supplementary information, Table S3).19 Furthermore, RT-PCR revealed that Lsectin was highly expressed in macrophages compared with other immune subpopulations (Supplementary information, Fig. S1e).
Fig. 1.
LSECtin expressed on macrophages drives breast cancer growth. a Representative immunofluorescence double staining of clinical breast cancer biopsy for co-expression of LSECtin (green) and CD68 (red). Nuclei were counterstained with DAPI (blue) (scale bar = 20 µm). Details regarding the tumor pathology are presented in Supplementary information, Table S2 (See also Supplementary information, Fig. S1a). b Manders coefficient of LSECtin and CD68 in clinical tumor sections. Each symbol represents a specimen. c Representative histograms of macrophages stained by anti-LSECtin mAb (red) or isotype control mAb (black) isolated from the fresh clinical breast tumor tissue. The details of tumor pathology are outlined in Supplementary information, Table S3. d Percentage (left) and delta MFI (right) of LSECtin surface expression by clinical tumor biopsy-derived macrophages from 10 patients. The details of tumor pathology are outlined in Supplementary information, Table S3. e Real-time PCR analysis showing the expression of Lsectin relative to that of the housekeeping gene β-actin in lymphocyte, tumor-associated neutrophil (TAN), dendritic cell (DC), monocyte (Mo), or TAM from human xenograft tumors (n = 3). One of three experiments is shown. f MDA-MB-231 cells (hereinafter designated “231”) were injected into LSECtin knockout nude mice (KO-Nude in short) or littermate controls (WT-Nude in short) (n = 15 each). The tumor volume at 7 weeks after injection of 1 × 103 231 cells is shown (top). The table shows the number of tumors (defined by > 20 mm3) (bottom). One of two experiments is shown. g 1 × 103 231 cells were injected into Rag1-knockout immunocompromised mice with a macrophage-specific deletion of LSECtin (Rag1−/−; LyZ2-Cre; LSECtinfl/fl; KO-LyZ2-Rag1−/− in short, n = 20) or littermate controls (Rag1−/−; LSECtinfl/fl; WT-LyZ2- Rag1−/− in short, n = 23). The table shows the number of tumors. The tumor volume at 10 weeks after injection is shown. One of two experiments is shown. h, i 1 × 103 231 cells were injected with admixed 1 × 104 WT-TAMs or admixed KO-TAMs from mice (h), with admixed human primary LSECtin+TAMs or admixed LSECtin-TAMs from surgical breast cancer specimens (i) into KO-Nude mice. The tumor volume of KO-nude mice injected with 1 × 103 231 cells without admixed TAMs served as controls. The table shows the number of tumors. The tumor volume at 7 weeks after injection is shown. One of two experiments is shown. Data are presented as the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired Student’s t test for f, g, h, i). See also Supplementary information, Fig. S1
We next characterized LSECtin expression in mouse models, where LSECtin+ cells co-expressed the murine macrophage marker F4/80 in the human xenograft tumors (Supplementary information, Fig. S1f). With the gating strategy shown in Supplementary information, Fig. S1g,20 the major populations of myeloid cells including TAMs, monocytes (Mo) and tumor-associated neutrophils (TAN) were identified. RT-PCR revealed higher Lsectin expression in TAMs than in other subpopulations (Fig. 1e). The intratumoral factors may stimulate Lsectin expression on macrophages, because bone marrow-derived macrophages (BMDMs) were induced to express Lsectin by tumor extract solution (Supplementary information, Fig. S1h).21 Together, these results suggest that LSECtin is expressed by macrophages in the tumor microenvironment.
Macrophage-expressed LSECtin promotes breast cancer growth
To further examine the role of macrophage-expressed LSECtin in tumor progression, we detected xenograft tumor formation by MDA-MB-231 (231) cells using limiting-dilution assays. As shown in Fig. 1f, no significant difference of tumor incidence at 100,000 cells per injection was detected. Meanwhile, 231 cells resulted in markedly reduced tumor incidence and tumor volume at 1000 cells per injection in LSECtin KO-Nude mice compared with littermate controls (WT-Nude) (Supplementary information, Fig. S1i). To further verify that macrophage-expressed LSECtin promoted tumor growth at 1000 cells per injection, we generated Rag1−/−; LyZ2-Cre; LSECtinfl/fl mice (KO-LyZ2-Rag1−/−), where Lsectin expression is reduced by 95% in macrophages (Supplementary information, Fig. S1j). Compared to littermate controls, macrophage-specific deletion of LSECtin led to reduction in the tumor incidence and tumor growth (Fig. 1g and Supplementary information, Fig. S1k). Similar observations were made when the sorted tumor cell populations from MMTV-PyMT mice were implanted orthotopically into mice with macrophage-specific deletion of LSECtin (KO-LyZ2) or littermate controls (WT-LyZ2) (Supplementary information, Fig. S1l). To further examine the opposite effect, we co-injected MDA-MB-231 cells at 1000 cells per injection with either mouse TAMs or human TAMs from patients. The admixture of either mouse TAMs from WT mice (WT-TAMs) or human LSECtin+ macrophages resulted in increased tumor growth; in contrast, TAMs from LSECtin knockout mice (KO-TAMs) or human LSECtin- macrophages did not have the effect (Fig. 1h, i and Supplementary information, Figs. S1m, n). Taken together, these findings suggest that macrophage-expressed LSECtin promotes breast cancer growth.
Macrophage-expressed LSECtin promotes breast cancer stemness in a contact-dependent manner
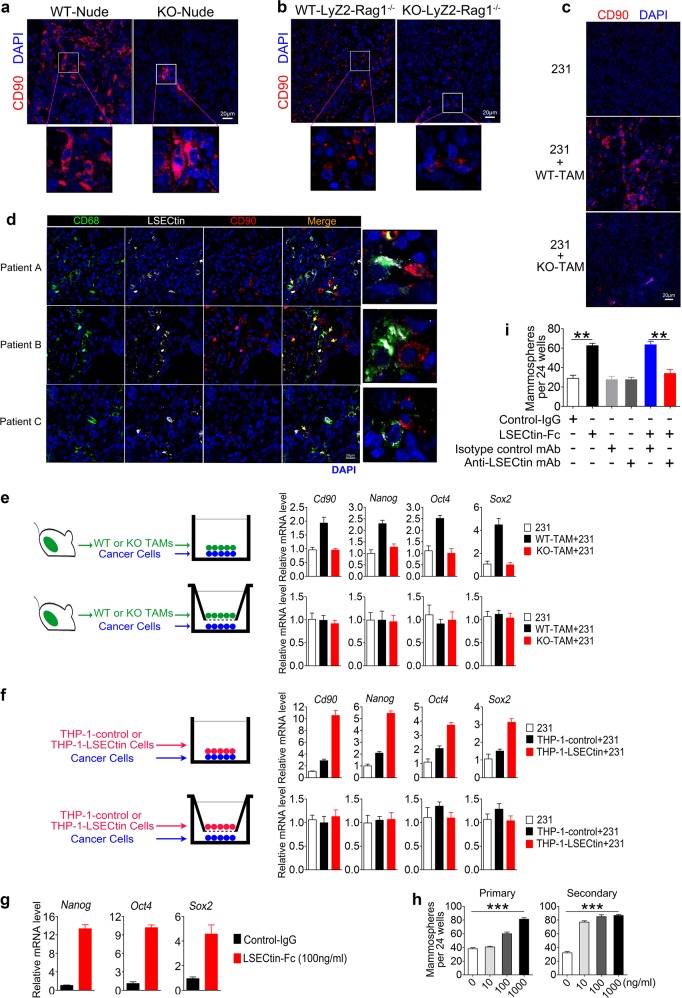
We next examined the mechanism by which macrophage-expressed LSECtin promotes tumor growth. There was no difference in the mRNA levels of polarization-related genes between primary WT-TAMs and KO-TAMs from human xenograft tumors (Supplementary information, Fig. S2a). Furthermore, LSECtin systemic knockout or LSECtin conditional knockout in macrophages did not lead to significant change in the frequency of intratumoral TAMs from human xenograft tumors (Supplementary information, Figs. S2b, c). The role of the cross-talk between macrophages and tumor cells in promoting tumor stemness and growth has been investigated.3–5 Our previous data showed that LSECtin knockout in host reduced tumor incidence and tumor volume in limiting-dilution assays, implying that macrophage-expressed LSECtin regulates stemness in vivo. CD90 expressed by BCCs has been reported to be a CSC marker, which mediates the physical interactions of CSCs with TAMs.22 We found that the CD90+ CSCs were significantly reduced in human xenograft tumors in hosts with systemic LSECtin knockout or macrophage-specific LSECtin knockout (Fig. 2a, b and Supplementary information, Figs. S2d, e). Additionally, co-injection of WT-TAMs but not KO-TAMs with MDA-MB-231 cells at 1000 cells per injection increased the frequency of CD90+ CSCs (Fig. 2c and Supplementary information, Fig. S2f). These data above suggest that macrophage-expressed LSECtin promotes BCCs stemness in vivo.
Fig. 2.
Macrophage-expressed LSECtin promotes breast cancer stemness in a contact-dependent manner. a–c Immunofluorescent staining of tumor tissue for human CD90 (red) in human xenograft tumors in WT-Nude or KO-Nude mice (a); in immunocompromised macrophage-specific LSECtin knockout mice (KO-LyZ2-Rag1−/− in short) and littermate controls (WT-LyZ2-Rag1−/− in short) (b); in the KO-Nude mice with co-injection of 231 cells and WT-TAMs or KO-TAMs (c). The anti-human CD90 antibody without reactivity with mouse CD90 was used in the immunofluorescence analysis. Nuclei were counterstained with DAPI (blue). One typical region is shown (scale bar = 20 µm). d Confocal images of primary patient-derived breast tumor sections co-stained for LSECtin+TAMs (white), CD68+ TAMs (green) and CD90+ CSCs (red) showed LSECtin+TAMs mediated juxtaposed TAM-CSC pairs in the patient (yellow arrow). Nuclei were counterstained with DAPI (blue) (scale bar = 20 µm). The details of tumor pathology are outlined in Supplementary information, Table S2. e, f Real-time analysis of the expression of Cd90, Nanog, Oct4 and Sox2 in MDA-MB-231 mammosphere cultures with or without primary WT-TAMs or KO-TAMs (e), THP-1-control or THP-1-LSECtin cells (f), in a contact fashion (top) or in a chamber with a transwell plate (bottom) (n = 3 each). One of two experiments is shown. g Real-time analysis of the expression of Cd90, Nanog, Oct4 and Sox2 in MDA-MB-231 cells stimulated with LSECtin-Fc recombinant protein or control-IgG (100 ng/ml) (n = 3 each). One of three experiments is shown. h Sphere-forming assay showing the representative mean number of spheres stimulated with LSECtin-Fc recombinant protein in a concentration gradient (n = 3 each). One of three experiments is shown. i Sphere-forming assay showing the increased sphere formation stimulated with LSECtin-Fc recombinant protein (100 ng/ml) was blocked by anti-LSECtin mAb (2 μg/ml) but not by isotype control mAb at the same concentration (n = 3 each). One of three experiments is shown. *P < 0.05, **P < 0.01, ***P < 0.001 in one way ANOVA (h) or unpaired Student’s t test (i). See also Supplementary information, Fig. S2
Some previous studies have examined direct cell–cell adhesion between macrophages and tumor cells in situ in both human and mouse specimens.22,23 Both the CD68+ macrophages in human and the F4/80+ macrophages in mouse can contact with CD90+ CSCs (Supplementary information, Figs. S2g, h and Supplementary information, Table S2). Moreover, a high density of infiltrating LSECtin+ macrophages was observed in contact with CD90+ tumor cells in the clinical specimens, suggesting that LSECtin-mediated TAM-CSC interaction occurs in vivo (Fig. 2d and Supplementary information, Table. S2). We then examined whether mouse or human macrophage-expressed LSECtin drives stemness via interaction with tumor cells. TAMs from WT mice (WT-TAMs) or LSECtin-overexpressed THP-1 cells (THP-1-LSECtin), but not TAMs from LSECtin knockout mice (KO-TAMs) or THP-1-control cells induced increased expression of Cd90 and stemness-related TFs (Nanog, Oct4, Sox2) in a co-culture system (Fig. 2e, f, top).24 Direct cell–cell interaction appeared to be critical for this function because WT-TAMs or THP-1-LSECtin cells in separate compartments in the Boyden chambers had no apparent effect (Fig. 2e, f, bottom). These data suggest that both mouse and human macrophage-expressed LSECtin promotes breast cancer stemness in a contact-dependent manner.
To further clarify whether macrophage-expressed LSECtin plays a direct role in regulating tumor cell stemness, direct interaction between human LSECtin recombinant protein and BCCs was verified (Supplementary information, Fig. S2i). Additionally, the LSECtin recombinant protein induced the increase in stemness-related TFs expression and sphere formation in a dose-dependent manner (Fig. 2g, h), which could be blocked by anti-LSECtin mAb (Fig. 2i). Collectively, these results show that macrophage-expressed LSECtin promotes stemness through physical interactions between macrophages and BCCs.
LSECtin can interact with BTN3A3 expressed on human breast cancer cells
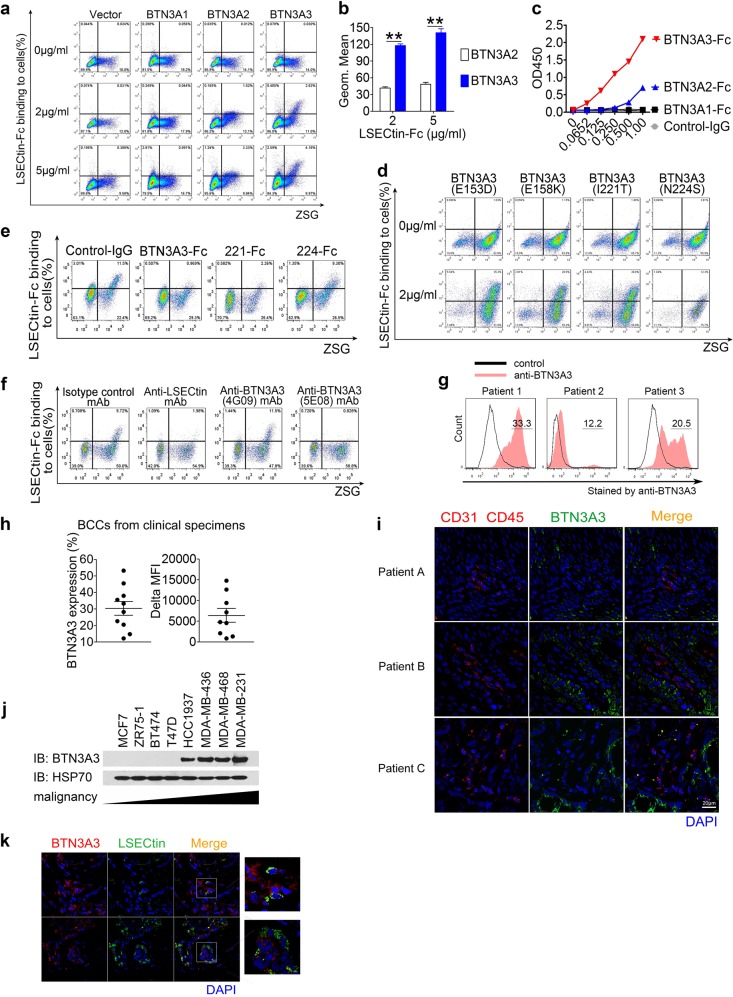
We speculated that protein(s) that interact with LSECtin are present on the surfaces of BCCs. To identify the membrane interaction proteins through expression cloning, we transduced a commercial human cDNA library into HEK293 cells that were negative for LSECtin binding. After five successive rounds of sorting, the predominant plasmids from the LSECtin-binding cells were identified as BTN3A3, a B7-related butyrophilin family member with unclear functions. The BTN3A subfamily comprises three members, BTN3A1, BTN3A2, and BTN3A3, whose extracellular protein sequences show more than 92% amino acid identity.12–14 BTN3A3 rather than other BTNs, when ectopically expressed on HEK293 cells, enabled the cells to bind recombinant human LSECtin with high affinity, while BTN3A2 enabled the cells to bind recombinant human LSECtin with low affinity (Fig. 3a, b and Supplementary information, Fig. S3a). BTN3As expression levels were determined by an anti-CD277 mAb that has been shown to react with BTN3A1, BTN3A2 and BTN3A3 (Supplementary information, Fig. S3b). Reciprocally, recombinant BTN3A3-Fc bound to HEK293 cells overexpressing LSECtin with higher affinity than BTN3A1-Fc or BTN3A2-Fc (Supplementary information, Fig. S3c). Furthermore, immobilized LSECtin protein interacted directly with BTN3A3 but not with the control BTN3A1 as shown by Elisa (Fig. 3c). Structurally, BTNs consist of an extracellular IgV-like domain followed by an IgC-like domain and a heptad repeated sequence. Both the IgV-like domain and the IgC-like domain were required for interaction with LSECtin as measured in an adhesion assay (Supplementary information, Fig. S3d). Comparison of the extracellular sequences of the BTN3As shows strong homology in the IgV domains. The most significant differences are found within the IgC domain (the differences are indicated by gray shades in the figure). Of note, BTN3A1 contains some unique amino acids in this domain (unique amino acids are indicated by red shades in the figure) (Supplementary information, Fig. S3e). The four BTN3A3 mutants (E153D, E158K, I221T, and N224S) were overexpressed and their expression levels were determined by anti-CD277 mAb (Supplementary information, Fig. S3f). Recombinant LSECtin bound to the N224S mutant-overexpressing cells with lower affinity than to other cells (Fig. 3d). Immobilized LSECtin protein interacted strongly with wild-type BTN3A3 protein, but its interaction with the N224S mutant was much weaker (Supplementary information, Fig. S3g).
Fig. 3.
LSECtin can interact with BTN3A3 expressed on human breast cancer cells. a, b Binding of human LSECtin-Fc recombinant protein to HEK293 cells transfected with BTN3As at different concentrations. The adhesion rate (a) and geometric mean fluorescence intensity (MFI) (b) are shown (n = 3 each). One of three experiments is shown. (c) LSECtin-his recombinant protein bound to the BTN3A3-Fc recombinant protein but did not bind to BTN3A1-Fc. One of three experiments is shown. d Binding of LSECtin-Fc recombinant protein at 2 μg/ml to HEK293 cells transfected with BTN3A3 mutations. One of three experiments is shown. e The interaction between LSECtin-Fc recombinant protein at 2 μg/ml and BTN3A3 expressed in the HEK293 cells was blocked by 20 μg/ml BTN3A3-Fc or BTN3A3(I221T)-Fc (“221-Fc”) but not by BTN3A3(N224S)-Fc (“224-Fc”). One of three experiments is shown. f The interaction between LSECtin-Fc recombinant protein at 1 μg/ml and BTN3A3 expressed in the HEK293 cells was blocked by anti-LSECtin mAb or anti-BTN3A3 (5E08) mAb at 5 μg/ml but not by anti-BTN3A3(4G09) mAb or by isotype control mAb at the same concentration. One of three experiments is shown. g Representative histograms of BCCs stained by anti-BTN3A3 (5E08) mAb (red) or isotype control mAb (black) isolated from the clinical breast tumor tissues. The details of tumor pathology are outlined in Supplementary information, Table S3. h Percentage (left) and delta MFI (right) of BTN3A3 surface expression by clinical tumor biopsy-derived BCCs from 10 patients. The details of tumor pathology are outlined in Supplementary information, Table S3. i Immunofluorescent staining for CD45 (red), CD31 (red) or BTN3A3 (green) of clinical breast cancer biopsy. Nuclei were counterstained with DAPI (blue). A high proportion of CD31-CD45- cells are BTN3A3-positive (scale bar = 20 µm). Details regarding the tumor pathology are presented in Supplementary information, Table S2. (See also Supplementary information, Fig. S3m). j Western blotting (WB) showing the expression of BTN3A3 relative to that of the housekeeping gene HSP70 in breast cancer cell lines. One of three experiments is shown. k Immunofluorescent staining of clinical breast cancer biopsy for the direct contact between LSECtin-positive cells (green) and BTN3A3-positive cells (red). Nuclei were counterstained with DAPI (blue) (scale bar = 20 µm). One typical region is shown. Data are presented as the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired Student’s t test for b). See also Supplementary information, Fig. S3
To validate the interaction between LSECtin and BTN3A3, we used recombinant BTN3A3-Fc to antagonize LSECtin-BTN3A3 interactions. BTN3A3-Fc with high-affinity for LSECtin, rather than BTN3A1-Fc or BTN3A3 mutant-Fc, antagonized LSECtin-BTN3A3 interactions (Supplementary information, Fig. S3h and Fig. 3e). We also generated mAbs against human BTN3A3 by immunizing BALB/c mice with the BTN3A3-Fc fusion protein. One anti-BTN3A3 mAb, clone 5E08, specifically bound BTN3A3-transfected cells but not control cells (Supplementary information, Fig. S3i). Importantly, the anti-BTN3A3 (5E08) mAb or anti-LSECtin mAb were able to block the LSECtin-BTN3A3 interaction completely (Fig. 3f). We also verified that recombinant mouse LSECtin bound BTN3A3-overexpressing cells but not control cells (Supplementary information, Fig. S3j).
We then examined BTN3A3 protein expression using the Human Protein Atlas database.25 The total 11 breast cancer samples contained BTN3A3-positive tumor cells (Supplementary information, Fig. S3k). Flow cytometric analysis of single-cell suspensions derived from clinical tumor specimens revealed BTN3A3 surface expression by BCC subpopulations negative for the pan-lymphocyte marker, CD45, and the endothelial marker, CD31, in 10/10 breast cancer specimens examined, with BTN3A3+ cell frequencies ranging from 12.5% to 53.2% (Fig. 3g, h, Supplementary information, Fig. S3l and Supplementary information, Table S3). Furthermore, immunofluorescence analysis of tissue sections showed strong BTN3A3 expression by CD31-CD45- cells (Fig. 3i, Supplementary information, Fig. S3m and Supplementary information, Table S4). Immunofluorescence analysis of tissue sections also showed strong BTN3A3 expression by EPCAM+ cells (Supplementary information, Fig. S3n and Supplementary information, Table S2). Besides, several human breast cancer cell lines of high-grade malignancy (MDA-MB-436, MDA-MB-468 and MDA-MB-231) expressed high levels of BTN3A3, whereas cell lines of low-grade malignancy (MCF7, ZR-75-1, BT474, and T-47D) expressed low levels of BTN3A3 (Fig. 3j and Supplementary information, Figs. S3o, p). Moreover, the immunofluorescence analysis of clinical specimens also showed the physical contact between LSECtin+ cells and BTN3A3+ cells (Fig. 3k). These data demonstrate that BTN3A3 expressed on the human BCCs is the potential receptor for LSECtin.
The LSECtin-BTN3A3 axis promotes breast cancer stemness in vitro
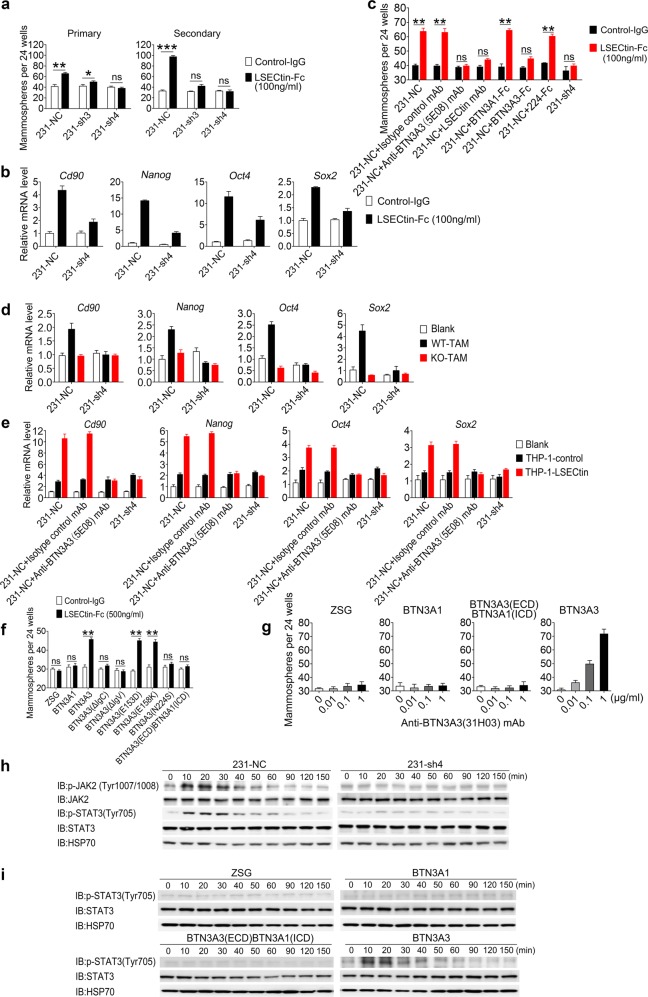
To determine whether LSECtin drives stemness through interaction with BTN3A3 in vitro, we constructed BTN3A3 knockdown (KD) MDA-MB-231 cells (231-sh3 or 231-sh4 cells) or control cells (231-NC cells) (Supplementary information, Fig. S4a, b). Recombinant LSECtin promoted mammosphere growth and mRNA expression of Cd90 and stemness-related TFs by 231-NC cells but not by 231-sh3 or 231-sh4 cells (Fig. 4a, b). To further verify the importance of the activation of LSECtin-BTN3A3 axis, we also generated BTN3A3 agonists. One anti-BTN3A3 mAb, clone 31H03, could act as an agonist that specifically promoted mammosphere formation by 231-NC cells but not by 231-sh4 cells (Supplementary information, Fig. S4c). Consistently, recombinant LSECtin promoted sphere formation by BTN3A3-OE BT474 cells, by BTN3A2-OE cells partly, but not by BTN3A1-OE cells (Supplementary information, Figs. S4d, e). We overexpressed BTN3A3 in 231-sh4 cells and found that re-overexpressing BTN3A3 could rescue the promotion of sphere formation by recombinant LSECtin (Supplementary information, Figs. S4f, g). We also found BTN3A3-Fc and anti-BTN3A3 (5E08) mAb exhibited great ability to block sphere formation induced by recombinant LSECtin (Fig. 4c). We then verified whether LSECtin-BTN3A3 axis medicates breast cancer stemness in a macrophage-BCC contact-dependent fashion. Co-cultured WT-TAMs or THP-1-LSECtin cells increased mRNA expression of Cd90 and stemness-related TFs in 231-NC cells but not in 231-sh4 cells (Fig. 4d, e). Furthermore, the anti-BTN3A3 (5E08) mAb interrupted expression of Cd90 and the stemness-related TFs induced by human macrophage-expressed LSECtin (Fig. 4e). These findings suggest that the LSECtin-BTN3A3 axis drives stemness in vitro.
Fig. 4.
The LSECtin-BTN3A3 axis promotes breast cancer stemness in vitro. a Sphere-forming assay showing the representative mean number of spheres of BTN3A3-KD cells (231-sh3 and 231-sh4) or control cells (231-NC) in primary and secondary passaged cultures stimulated with LSECtin-Fc or control-IgG (n = 3 each). One of three experiments is shown. b, d Real-time analysis of the expression of Cd90, Nanog, Oct4 and Sox2 in 231-NC and 231-sh4 mammosphere cultures with LSECtin-Fc or control-IgG (b); with or without primary WT-TAMs or KO-TAMs (d) (n = 3 each). The gene expression shown in Fig. 2e of 231 cells co-cultured with TAMs in a contact fashion is shown for reference. One of three experiments is shown. c Sphere-forming assay showing the representative mean number of spheres. 231-NC cells, 231-sh4 were stimulated with LSECtin-Fc or control-IgG (100 ng/ml). Isotype control mAb, anti-BTN3A3 (5E08) mAb, Anti-LSECtin mAb, BTN3A1-Fc, BTN3A3-Fc or 224-Fc at the same concentration (10 μg/ml) were added to the culture medium (n = 3 each). One of three experiments is shown. e Real-time analysis of the expression of Cd90, Nanog, Oct4 and Sox2 in 231-NC and 231-sh4 mammosphere cultures with or without THP-1-control or THP-1-LSECtin cells (n = 3 each). Anti-BTN3A3 (5E08) mAb or isotype control mAb at the same concentration (10 μg/ml) were added to culture medium to interrupt the LSECtin-BTN3A3 axis. The gene expression shown in Fig. 2f of 231 cells co-cultured with THP-1 cells in a contact fashion is shown for reference. One of three experiments is shown. f, g Sphere-forming assay showing the representative mean number of spheres formed by BT474 cells overexpressing BTN3A3, BTN3A3 deletions, mutants, chimeric proteins or vector in primary passaged cultures stimulated with LSECtin-Fc recombinant protein or control-IgG (f); with anti-BTN3A3 (31H03) mAb in a concentration gradient (g) (n = 4 each). One of three experiments is shown. h 231-NC or 231-sh4 cells stimulated with anti-BTN3A3 (31H03) agonist mAb at 10 μg/ml were lysed and fractionated to obtain cell extracts for WB. JAK, p-JAK, STAT3 and p-STAT3 were immunoblotted to demonstrate signal transduction pathway activation. HSP70 served as a control. i BT474 cells overexpressing vector, BTN3A1, BTN3A3 or BTN3A3 chimeric proteins stimulated with anti-BTN3A3 (31H03) mAb at 10 μg/ml were lysed and fractionated to obtain cell extracts for WB. STAT3 and p-STAT3 were immunoblotted to demonstrate signal transduction pathway activation. HSP70 served as a control. Data are presented as the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired Student’s t test for a, c, f). See also Supplementary information, Fig. S4
We previously observed that both the IgC and the IgV domains of BTN3A3 are required for its interaction with LSECtin (Supplementary information, Fig. S3d). Consistently, recombinant LSECtin failed to promote sphere formation by cells harboring BTN3A3 deletions and the residue N224 of BTN3A3 is important for the function of LSECtin-BTN3A3 axis (Fig. 4f). We also constructed BTN3A3(ECD)BTN3A1(ICD), a chimeric protein that consists of the extracellular domain (ECD) and transmembrane domains of BTN3A3 as well as the intracellular domain (ICD) of BTN3A1 (Supplementary information, Fig. S4h). LSECtin recombinant protein or anti-BTN3A3 (31H03) agonist mAb was shown to promote sphere formation by BTN3A3-OE cells rather than BTN3A3(ECD)BTN3A1(ICD)-OE cells, indicating that the ICD of BTN3A3 is required for its role in promoting stemness (Fig. 4f, g).
We next explored the signal transduction mechanisms that might be responsible for the LSECtin-BTN3A3 axis. Recent studies highlight that the JAK2/STAT3 signaling pathway regulates breast cancer stemness.26,27 We found that LSECtin recombinant protein promoted phosphorylation of STAT3 but not other STATs in control cells but not in BTN3A3-KD cells (Supplementary information, Fig. S4i). We next found that anti-BTN3A3 (31H03) agonist mAb promoted phosphorylation of JAK2 and STAT3 in 231-NC cells but not in 231-sh4 cells (Fig. 4h). Besides, anti-BTN3A3 (31H03) agonist mAb promoted mRNA expression of stemness-related TFs by 231-NC cells but not by 231-sh3 or 231-sh4 cells (Supplementary information, Fig. S4j). Moreover, 31H03 also promoted phosphorylation of STAT3 in BTN3A3-OE cells but not in BTN3A3(ECD)BTN3A1(ICD)-OE cells (Fig. 4i), suggesting the ICD of BTN3A3 is necessary for the STAT3 phosphorylation.
The LSECtin-BTN3A3 axis promotes breast cancer stemness in vivo
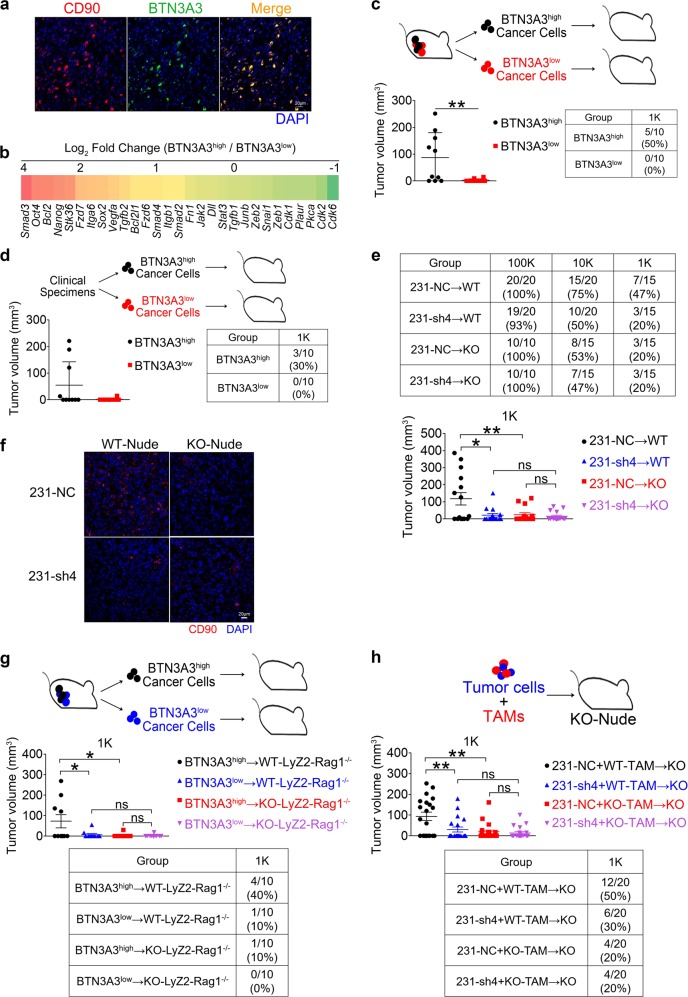
To verify whether the LSECtin-BTN3A3 axis promotes tumor stemness in vivo, we first verified the association of BTN3A3 expression and stemness in vivo. Histological analysis of tissue sections from human xenograft tumors showed strong BTN3A3 expression by CD90+ CSCs (Fig. 5a). We then analyzed the differences between BTN3A3high and BTN3A3low tumor cells from human xenograft tumors (Supplementary information, Fig. S5a). RT-PCR was used to verify BTN3A3 expression in BTN3A3high and BTN3A3low cells (Supplementary information, Fig. S5b). Notably, BTN3A3high cells from human xenograft tumors exhibited higher expression levels of stemness-related TFs (Nanog, Sox2, Oct4) than BTN3A3low cells (Fig. 5b). Besides, the BTN3A3high cells at 1000 cells per injection formed tumors but the BTN3A3low populations rarely formed tumors (Fig. 5c and Supplementary information, Fig. S5c). Similar observations were made in the patient-derived xenograft (PDX) models (Fig. 5d and Supplementary information, Figs. S5d,e). These data indicate that BTN3A3high cells exhibit higher tumorigenesis ability. In contrast, BTN3A3-KD (231-sh4) cells displayed markedly reduced tumorigenesis capabilities in nude mice (Fig. 5e and Supplementary information, Fig. S5f). Besides, the CD90+ CSC subpopulations were significantly reduced in mice injected with 231-sh4 cells compared to control mice (Fig. 5f and Supplementary information, Fig. S5g). Although we found that knockdown of BTN3A3 in tumor cells markedly reduced tumor growth in WT mice, there was no similar effect in the mice with systemic knockout of LSECtin (Fig. 5e and Supplementary information, Fig. S5f), suggesting that tumor growth driven by BTN3A3 is LSECtin-dependent.
Fig. 5.
The LSECtin-BTN3A3 axis promotes breast cancer stemness in vivo. a Immunofluorescent staining of human xenograft tumors for human CD90 (red) or BTN3A3 (green). The anti-human CD90 antibody without reactivity with mouse CD90 was used in the immunofluorescence analysis. A high proportion of CD90-positive cells are BTN3A3-positive (scale bar = 20 µm). b Heatmap showing the expression of stemness TFs (Nanog, Sox2, Oct4), cell cycle checkpoints (Cdk1, Cdk2, Cdk6), and EMT genes (Zeb1, Zeb2, Snail1) in BTN3A3low cells or BTN3A3high cells from human xenograft tumors determined by real-time PCR analysis. c, d BTN3A3high cells or BTN3A3low cells from human xenograft tumors or clinical specimens were injected into WT-Nude mice (n = 10 each). The tables indicate the number of tumors. The tumor volume at 7 weeks (c) or at 4 months (d) after injection are shown. One of two experiments is shown. e 231-NC or 231-sh4 cells were injected into KO-nude mice or littermate controls. The table indicates the number of tumors (top). The tumor volume at 7 weeks after injection of 1 × 103 cells is shown (n = 15 each) (bottom). One of two experiments is shown. f Immunofluorescent staining of tumor tissue for human CD90 (red) in human xenograft tumors formed by 231-NC and 231-sh4 cells. Nuclei were counterstained with DAPI (blue) (scale bar = 20 µm). Typical images are shown. g 1 × 103 BTN3A3high cells or BTN3A3low cells from human xenograft tumors were injected into Rag1-knockout immunocompromised mice with a macrophage-specific deletion of LSECtin (KO-LyZ2-Rag1−/−) or littermate controls (WT-LyZ2- Rag1−/−) (n = 10 each). The tumor volume at 7 weeks after injection is shown. One of two experiments is shown. h An admixture of 1 × 103 231-NC or 231-sh4 cells and 1 × 104 WT-TAMs or KO-TAMs were injected into KO-Nude mice (n = 20 each). The tumor volume at 7 weeks after injection is shown. One of two experiments is shown. Data are presented as the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired Student’s t test for c, d, e, g, h). See also Supplementary information, Fig. S5
Our previous data demonstrated that macrophage-expressed LSECtin promotes tumor though BCC-expressed BTN3A3. We next verified whether the role of LSECtin-BTN3A3 axis is macrophage-dependent. Mice with macrophage-specific deletion of LSECtin (KO-LyZ2-Rag1−/−) showed markedly reduced tumor incidence and tumor volume as compared with the littermate controls (WT-LyZ2-Rag1−/−) after injection of 1000 BTN3A3high cells (Fig. 5g and Supplementary information, Fig. S5h). Moreover, injections with BTN3A3high cells or BTN3A3low cells caused no obvious difference in tumor growth in mice with macrophage-specific deletion of LSECtin (Fig. 5g and Supplementary information, Fig. S5h). Furthermore, we co-injected 231-NC or 231-sh4 cells at 1000 cells per injection with WT or LSECtin KO-TAMs into LSECtin KO-Nude mice. Although we found that knockdown of BTN3A3 in tumor cells markedly reduced tumor incidence and growth in mice with admixture of WT-TAMs, there was no similar effect in mice with admixture of KO-TAMs (Fig. 5h and Supplementary information, Fig. S5i). Taken together, our data clearly demonstrate that the LSECtin-BTN3A3 axis promotes breast cancer stemness in vivo.
Targeting LSECtin-BTN3A3 axis inhibits breast cancer stemness and tumor growth
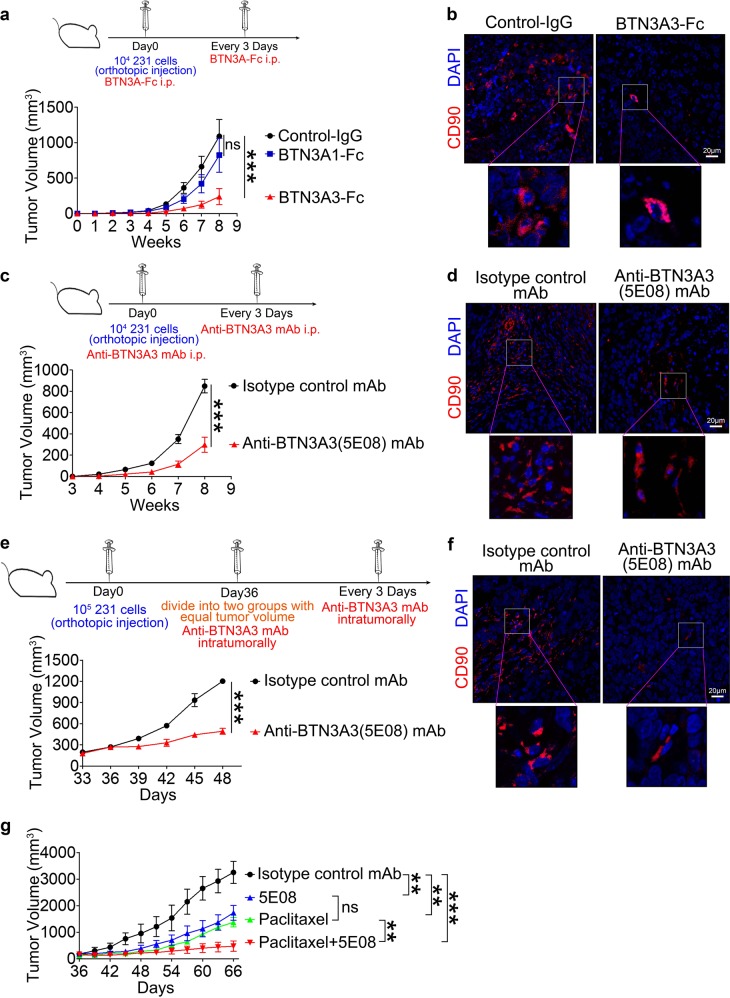
Because the LSECtin-BTN3A3 axis promotes breast cancer stemness and tumor growth, we speculated that targeting this axis could inhibit human breast cancer progression. To evaluate whether treatment with BTN3A3-Fc or anti-BTN3A3 (5E08) mAb, which were verified to functionally inhibit LSECtin-BTN3A3 interactions, could reduce human breast cancer progression, we orthotopically inoculated WT-Nude mice with MDA-MB-231 cells and administered BTN3A3-Fc or anti-BTN3A3 (5E08) mAb intraperitoneally (i.p.). Compared to BTN3A1-Fc, control-IgG or isotype control mAb treatment, systemic BTN3A3-Fc (Fig. 6a, b and Supplementary information, Figs. S6a, b) or anti-BTN3A3 (5E08) mAb treatment (Fig. 6c, d and Supplementary information, Fig. S6c,d) decreased tumor progression and CD90+ subpopulations without damage of other tissues.
Fig. 6.
Targeting LSECtin-BTN3A3 axis inhibits breast cancer stemness and tumor growth. a WT-Nude mice were injected with 1 × 104 231-NC cells and treated with 10 µg of BTN3A1-Fc, BTN3A3-Fc or control-IgG intraperitoneally (i.p.) every three days (n = 10 each). The tumor volume was monitored weekly. One of two experiments is shown. b, d, f Immunofluorescent staining of tumor tissues for human CD90 (red) in human xenograft tumors after administration of BTN3A3-Fc or control-IgG i.p. (b), anti-BTN3A3 (5E08) mAb or isotype control mAb i.p. (d) or anti-BTN3A3 (5E08) mAb or isotype control mAb intratumorally (f). One typical region is shown (scale bar = 20 µm). (c) WT-Nude mice were injected with 231-NC cells and treated with 50 µg of anti-BTN3A3 (5E08) mAb or isotype control mAb i.p. every three days (n = 10 each). The tumor volume was monitored weekly. One of two experiments is shown. e WT-Nude mice were injected with 231-NC cells. Mice were divided into two groups with equal tumor volume and treated with 50 µg of anti-BTN3A3 (5E08) mAb or isotype control mAb intratumorally five weeks after injection (n = 5 each). One of three experiments is shown. g WT-Nude mice were injected with MDA-MB-231 cells. Mice were divided into four groups with equal tumor volumes and intratumorally treated with 50 µg of paclitaxel, anti-BTN3A3 (5E08) mAb, isotype control mAb or a combination of paclitaxel and anti-BTN3A3 (5E08) mAb (n = 5 each). One of three experiments is shown. Data are presented as the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 (two-way ANOVA test for a, c, e, g). See also Supplementary information, Fig. S6
In addition, we examined whether administering anti-BTN3A3 (5E08) mAb intratumorally reduced tumor progression. Five weeks after inoculation, anti-BTN3A3 (5E08) mAb or isotype control mAb was administered intratumorally into human xenograft tumors every three days. Compared to isotype control mAb treatment, anti-BTN3A3 (5E08) mAb treatment delayed tumor progression without damage of other tissues (Fig. 6e and Supplementary information, Fig. S6e). We then found that CD90+ subpopulations were significantly reduced in mice injected with anti-BTN3A3 (5E08) mAb (Fig. 6f and Supplementary information, Fig. S6f). Furthermore, we administered anti-BTN3A3 (5E08) mAb and paclitaxel intratumorally into human xenograft tumors. Compared with the single-agent treatments, the combination of anti-BTN3A3 (5E08) mAb and paclitaxel significantly decreased tumor growth (Fig. 6g). These data indicate that BTN3A3-Fc and anti-BTN3A3 (5E08) mAb possess potential therapeutic activity for the treatment of breast cancer.
Signatures of BTN3A3 and LSECtin abundance in breast cancer
We further assessed the translational relevance of the LSECtin-BTN3A3 axis. Btn3a3 expression was significantly higher in ER− samples than in ER+ samples based on our analysis using the Cancer Genome Atlas (TCGA) data set28 (Supplementary information, Fig. S7a) or real-time PCR results using commercially available human breast cancer tissue cDNAs (Supplementary information, Fig. S7b). Moreover, ER- patients with high Btn3a3 expression (>40% of breast tumor tissue) showed decreased overall survival (Supplementary information, Fig. S7c). Meanwhile, Btn3a3 expression was not associated with PR or HER2 status (Supplementary information, Figs. S7d,e).
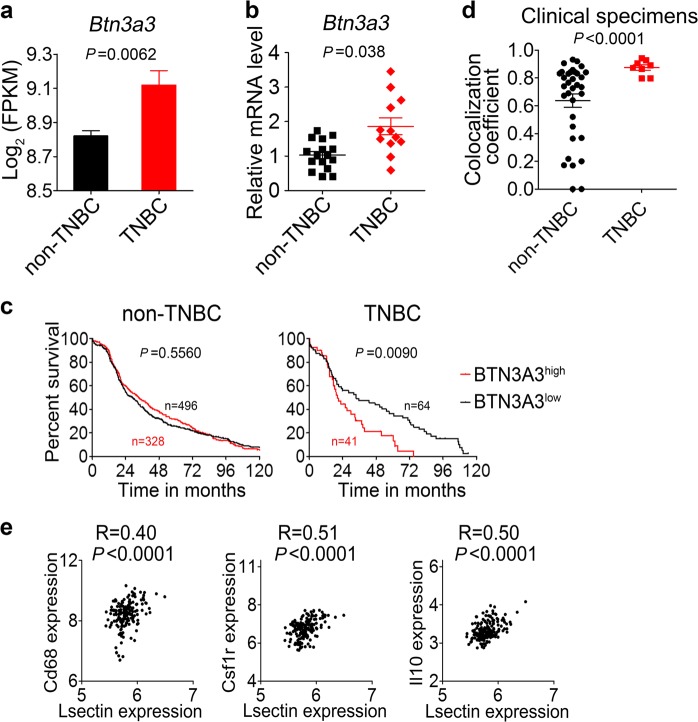
In breast cancer cells of triple-negative (ER−, PR−, HER2−) subtype, several genes have been identified that amplify the output of cell survival and stemness pathways.29 Compared with non-TNBC samples, Btn3a3 expression was significantly higher in TNBC samples, analyzed using either the TCGA data set (Fig. 7a) or commercially available cDNAs from patients (Fig. 7b). Moreover, TNBC patients with high Btn3a3 expression (>40% of breast tumor tissue) showed a significant decrease in overall survival (Fig. 7c). Besides, the expression of Btn3a1 or Btn3a2 were not associated with overall survival time in patients with TNBC (Supplementary information, Figs. S7f, g).
Fig. 7.
Signatures of BTN3A3 and LSECtin abundance in the breast cancer. a Btn3a3 mRNA expression in non-TNBC and TNBC samples from the TCGA data set (non-TNBC n = 824; TNBC n = 105). b Real-time PCR analysis showing Btn3a3 expression with cDNAs from non-TNBC patients (n = 16, black) and TNBC patients (n = 12, red) as templates. c Kaplan–Meier survival curve of breast cancer patients from the TCGA data set (For non-TNBC samples, BTN3A3low: 60%, n = 496, BTN3A3high: 40%, n = 328; for TNBC samples, BTN3A3low: 60%, n = 64, BTN3A3high: 40%, n = 41). d Manders coefficient of LSECtin and CD68 in non-TNBC and TNBC clinical tumor sections. Each symbol represents a specimen. Details regarding the tumor pathology are presented in Supplementary information, Table S1. e Correlation between Lsectin expression and TAM-associated gene expression in human TNBC samples from GSE76250 data set (n = 165). Data are presented as the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired Student’s t test for a, b, d). See also Supplementary information, Fig. S7
We previously demonstrated that TAM-expressed LSECtin was required for BTN3A3-driven tumor stemness and LSECtin co-expressed with CD68 in the clinical specimens (Fig. 1a, Supplementary information, Fig. S1a and Supplementary information, Table S1). We further verified that the co-localization coefficient was higher in the TNBC patients in situ (Fig. 7d). Importantly, the association between the expression of Lsectin and that of Cd68, Csf1r (another indicator of TAMs) and Il10 (an indicator of M2-type macrophage polarization) was relatively stronger in the TNBC patients (R > 0.3) than in the non-TNBC patients, analyzed using multiple databases (Fig. 7e and Supplementary information, Fig. S7h).30,31
Discussion
Our study identifies a cellular, molecular, and clinical network involving the LSECtin-BTN3A3 axis that extrinsically controls cancer stemness and tumor growth and impacts patient outcome.
TAMs act on CSCs at each stage of tumor progression. In breast cancer, macrophage-mediated paracrine signaling enhances cancer stem cell-like properties by promoting the epithelial-mesenchymal transition (EMT), inducing chemotherapy resistance of CSCs and driving CSC expansion.3–5 A recent study showed that macrophages physically interact with mouse breast CSCs and thereby maintain the stem-like state of CSCs, suggesting that juxtacrine signaling by macrophages represents an alternative means by which macrophages can support the physical CSC niche.22 In our study, we specifically investigated the role of cell–cell contact-dependent signaling in the CSC niche, in which macrophages promote BCCs stemness and tumor growth through an axis involving the transmembrane proteins LSECtin and BTN3A3. Our study provides a novel molecular mechanism for understanding how environmental cues regulate the fate of CSCs in a cell–cell contact-dependent manner. Nevertheless, whether the LSECtin-BTN3A3 axis exhibits similar functions in tumor types other than breast cancer awaits further investigations.
LSECtin, which belongs to the C-type lectin receptor family, was originally cloned by our group and was shown to be expressed in the liver and melanoma cells. Our previous studies have revealed that LSECtin has emerged as a novel tissue-resident immune-regulatory molecule. LSECtin, when expressed in the liver, acted as a co-inhibitory molecule and limited T-cell immune responses to promote HBV tolerance.7,8 Moreover, expression of LSECtin in melanoma cells engenders immune escape through blunting tumor-specific T-cell responses.9 Beyond its constitutive expression on both human and mouse kupffer cells, LSECtin is also induced on monocyte-derived macrophages by Th2 cytokines, such as IL-4 and IL-13.11 However, whether LSECtin is expressed by TAMs and how TAM-derived LSECtin functions in the tumor microenvironment is unknown. Here we characterized a specific expression pattern of LSECtin on TAMs and found that the association between the expression of LSECtin and CD68 was relatively strong in triple-negative breast cancer patients by analyzing multiple databases. TAM-expressed LSECtin directly advances breast cancer stemness and growth via its functional receptor BTN3A3, even in the absence of adaptive immunity. Thus, our study reveals a novel mechanism of LSECtin in cancer cell biology and suggests that LSECtin plays key roles in tumor microenvironment, and influences tumour progression. Therefore, we speculate that targeting LSECtin has promising antitumor activity.
Beyond elucidating a crucial role of LSECtin on macrophages that contributes to breast cancer stemness, one of the most important discoveries in this study is that BTN3A3, a B7-related butyrophilin family member with unclear functions, is a functional receptor for LSECtin. The BTN3A subfamily has been shown to be expressed on tumor cells15,16 and has recently been reported to be associated with increased susceptibility to ovarian and gastric cancers.17,18 However, our understanding of the role of BTN3A in the tumor microenvironment is still at a very preliminary stage. Here, for the first time, we reveal the function of BTN3A3 in promoting cancer cell stemness. Particularly, unlike the typical B7 family members such as B7-H1 that shows tumor cell-extrinsic functions through inhibiting tumor-specific T cell immunity, BTN3A3 exhibits tumor cell-intrinsic receptor function through interaction with its ligand LSECtin, leading to phosphorylation of STAT3, an important kinase that affects the expression of genes driving stemness.26,32 We also noticed that another pathway including microRNAs/CTBP2 regulated stemness of cancer cells.33 Whether LSECtin-BTN3A3 axis promoted stemness by other mechanism remains to be further investigated. Besides, our result demonstrated that LSECtin-BTN3A3 axis promotes stemness of TNBC cells. A recent study demonstrates that aerobic glycolysis affects G-CSF and GM-CSF expression and regulates myeloid-derived suppressor cells.34 Whether cancer metabolism regulates BTN3A3 expression in TNBC cells or LSECtin expression in TAMs needs further study. Taken together, our findings provide unique insights into the function of Butyrophilin pathway in tumor, and provide an extra dimension to the understanding of B7 family members in the tumor microenvironment.
Since the tumor microenvironment has the potential to support and initiate stem- cell-like programs in cancer cells, developing CSC-targeting therapy through blocking communication between CSCs and the microenvironment appears promising.1,35 Our results define a novel type of TAM-BCC cross-talk in a cell–cell contact manner via the LSECtin-BTN3A3 axis, and show that both exogenous BTN3A3-Fc-mediated blockade of LSECtin and antibody-mediated blockade of BTN3A3 slowed tumor growth in human xenograft models. Therefore, we speculate that targeting the LSECtin-BTN3A3 axis in the CSC niche could serve as an exciting and distinctive approach for breast cancer therapy. Future studies directed at expanding the therapeutic potential of the LSECtin-BTN3A3 axis will have important clinical implications.
Methods
For a detailed description of all methods, see the detailed methods in Supplementary information.
Animals and clinical samples
All animals were handled in strict accordance with the ‘Guide for the Care and Use of Laboratory Animals’ and the ‘Principles for the Utilization and Care of Vertebrate Animals’, and all animal work was approved by the Institutional Animal Care and Use Committee (IACUC) at the Beijing Institute of Lifeomics. Specific pathogen-free (SPF) female Rag1-knockout mice (C57BL/6), macrophage-specific LSECtin knockout mice (C57BL/6), female nude mice (BALB/c) were bred in our SPF facilities. The use of clinical breast cancer specimens was performed with the approval of the Medical Ethical Committee of Beijing Institute of Lifeomics. Informed consent was obtained from all subjects.
Isolation of single cells
Isolation of single cells from tumor tissue was performed as described previously.19,20 Briefly, tumor tissues were dissected, minced into small pieces and digested for 1 h at 37 °C in RPMI medium containing 2% FBS supplemented with 300 U/ml type IV collagenase (Sigma-Aldrich) and 100 U/ml DNase I (Sigma-Aldrich). The pieces and fluid were passed through a sieve. The cells were suspended by vigorous pipetting, washed twice in RPMI medium, and collected by centrifugation.
Sphere-forming assay
Mammosphere cultures were prepared as described.36 Single cells were plated in 24-well ultralow attachment plates (Corning) at a density of 500 cells/ml for BTN3A3-KD cells, 103 cells/ml for BT474-OE cells. Cells were grown in DMEM:F12 HAM (1:1) and supplemented with 5 μg/ml insulin, 20 ng/ml EGF, 4 μg/ml heparin and 0.5 μg/ml hydrocortisone. After 7 days mammospheres larger than 75 μm in diameter were photographed and counted.
Statistics
For adhesion rate comparison, statistical significance was determined with an unpaired Student’s t test. Statistical differences in the sphere-forming assay were determined with an unpaired Student’s t test. Statistics analysis of tumor growth was performed by two-way ANOVA and unpaired Student’s t test to compare the means. P values were considered significant as follows: *P < 0.05; **P < 0.01 and ***P < 0.0001. More details can be found in the supplemental information.
Supplementary information
Acknowledgements
We thank Imaging Facility of National Center for Protein Sciences· Beijing (NCPSB) (Mrs. Ping Wu) for Microscopy Imaging, Animal Facility of NCPSB (Mr. Chen Qiu), Flow Cytometry Facility of NCPSB (Mr. Yunxiang Sun and Ms. Mingxin Zhao) for FACS. We thank Mr. Pumin Zhang and Mrs. Jin Peng for cell culturing, Mr. Lichun Tang for gene editing (National Center for Protein Sciences·Beijing), Mr Feng Xu for WB (National Center for Protein Sciences·Beijing). This work was supported by the National Natural Science Foundation (31570901), Beijing Science and Technology Program Foundation (Z141100000214015), National Key R&D Program of China (2018YFA0507500), State Key Laboratory of Proteomics Foundation (SKLP-K201504, SKLP-K201701) and National Basic Research Program of China (973 Program) (2014CBA02000). Project was also funded by China Postdoctoral Science Foundation (2018M633740).
Author contributions
LT and FH planned the project. DL, QL, XW, JW, NL, ZJ, XH, JL, JL and DZ carried out experimental work. PC, GP, and YT analyzed data. DL and LT wrote the paper. All authors discussed the results and commented on the paper.
Competing interests
The authors declare no competing interests.
Contributor Information
Fuchu He, Phone: +86-10-61777004, Email: hefc@nic.bmi.ac.cn.
Li Tang, Phone: +86-10-61777091, Email: tangli731@163.com.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-019-0155-6.
References
- 1.Plaks V, Kong N, Werb Z. The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell. Stem. Cell. 2015;16:225–238. doi: 10.1016/j.stem.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oskarsson T, Batlle E, Massague J. Metastatic stem cells: sources, niches, and vital pathways. Cell. Stem. Cell. 2014;14:306–321. doi: 10.1016/j.stem.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P. Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 2017;14:399–416. doi: 10.1038/nrclinonc.2016.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J, et al. CCL18 from tumor-associated macrophages promotes breast cancer metastasis via PITPNM3. Cancer Cell. 2011;19:541–555. doi: 10.1016/j.ccr.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu W, et al. Characterization of a novel C-type lectin-like gene, LSECtin: demonstration of carbohydrate binding and expression in sinusoidal endothelial cells of liver and lymph node. J. Biol. Chem. 2004;279:18748–18758. doi: 10.1074/jbc.M311227200. [DOI] [PubMed] [Google Scholar]
- 7.Liu B, et al. Liver sinusoidal endothelial cell lectin inhibits CTL-dependent virus clearance in mouse models of viral hepatitis. J. Immunol. 2013;190:4185–4195. doi: 10.4049/jimmunol.1203091. [DOI] [PubMed] [Google Scholar]
- 8.Tang L, et al. Liver sinusoidal endothelial cell lectin, LSECtin, negatively regulates hepatic T-cell immune response. Gastroenterology. 2009;137:1498–1508. doi: 10.1053/j.gastro.2009.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu F, et al. LSECtin expressed on melanoma cells promotes tumor progression by inhibiting antitumor T-cell responses. Cancer Res. 2014;74:3418–3428. doi: 10.1158/0008-5472.CAN-13-2690. [DOI] [PubMed] [Google Scholar]
- 10.Dominguez-Soto A, et al. The DC-SIGN-related lectin LSECtin mediates antigen capture and pathogen binding by human myeloid cells. Blood. 2007;109:5337–5345. doi: 10.1182/blood-2006-09-048058. [DOI] [PubMed] [Google Scholar]
- 11.Xue J, et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 2014;40:274–288. doi: 10.1016/j.immuni.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abeler-Dorner L, Swamy M, Williams G, Hayday AC, Bas A. Butyrophilins: an emerging family of immune regulators. Trends Immunol. 2012;33:34–41. doi: 10.1016/j.it.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 13.Rhodes DA, Reith W, Trowsdale J. Regulation of Immunity by Butyrophilins. Annu. Rev. Immunol. 2016;34:151–172. doi: 10.1146/annurev-immunol-041015-055435. [DOI] [PubMed] [Google Scholar]
- 14.Arnett HA, Viney JL. Immune modulation by butyrophilins. Nat. Rev. Immunol. 2014;14:559–569. doi: 10.1038/nri3715. [DOI] [PubMed] [Google Scholar]
- 15.Cubillos-Ruiz JR, et al. CD277 is a negative co-stimulatory molecule universally expressed by ovarian cancer microenvironmental cells. Oncotarget. 2010;1:329–338. doi: 10.18632/oncotarget.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Compte E, Pontarotti P, Collette Y, Lopez M, Olive D. Frontline: characterization of BT3 molecules belonging to the B7 family expressed on immune cells. Eur. J. Immunol. 2004;34:2089–2099. doi: 10.1002/eji.200425227. [DOI] [PubMed] [Google Scholar]
- 17.Zhu M, et al. Exome array analysis identifies variants in SPOCD1 and BTN3A2 that affect risk for gastric cancer. Gastroenterology. 2017;152:2011–2021. doi: 10.1053/j.gastro.2017.02.017. [DOI] [PubMed] [Google Scholar]
- 18.Peedicayil A, et al. Risk of ovarian cancer and inherited variants in relapse-associated genes. PLoS One. 2010;5:e8884. doi: 10.1371/journal.pone.0008884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruffell B, et al. Leukocyte composition of human breast cancer. Proc. Natl. Acad. Sci. USA. 2012;109:2796–2801. doi: 10.1073/pnas.1104303108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franklin RA, et al. The cellular and molecular origin of tumor-associated macrophages. Science. 2014;344:921–925. doi: 10.1126/science.1252510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu D, et al. Comprehensive proteomics analysis reveals metabolic reprogramming of tumor-associated macrophages stimulated by the tumor microenvironment. J. Proteome Res. 2017;16:288–297. doi: 10.1021/acs.jproteome.6b00604. [DOI] [PubMed] [Google Scholar]
- 22.Lu H, et al. A breast cancer stem cell niche supported by juxtacrine signalling from monocytes and macrophages. Nat. Cell Biol. 2014;16:1105–1117. doi: 10.1038/ncb3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Q, Zhang XH, Massague J. Macrophage binding to receptor VCAM-1 transmits survival signals in breast cancer cells that invade the lungs. Cancer Cell. 2011;20:538–549. doi: 10.1016/j.ccr.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ben-Porath I, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uhlen M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 26.Marotta LL, et al. The JAK2/STAT3 signaling pathway is required for growth of CD44(+)CD24(-) stem cell-like breast cancer cells in human tumors. J. Clin. Invest. 2011;121:2723–2735. doi: 10.1172/JCI44745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou B, et al. Erythropoietin promotes breast tumorigenesis through tumor-initiating cell self-renewal. J. Clin. Invest. 2014;124:553–563. doi: 10.1172/JCI69804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vanharanta S, Massague J. Origins of metastatic traits. Cancer Cell. 2013;24:410–421. doi: 10.1016/j.ccr.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu YR, et al. Comprehensive transcriptome profiling reveals multigene signatures in triple-negative breast cancer. Clin. Cancer Res. 2016;22:1653–1662. doi: 10.1158/1078-0432.CCR-15-1555. [DOI] [PubMed] [Google Scholar]
- 31.Burstein MD, et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin. Cancer Res. 2015;21:1688–1698. doi: 10.1158/1078-0432.CCR-14-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng D, et al. Myeloid-derived suppressor cells endow stem-like qualities to breast cancer cells through IL6/STAT3 and NO/NOTCH cross-talk signaling. Cancer Res. 2016;76:3156–3165. doi: 10.1158/0008-5472.CAN-15-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui TX, et al. Myeloid-derived suppressor cells enhance stemness of cancer cells by inducing microRNA101 and suppressing the corepressor CtBP2. Immunity. 2013;39:611–621. doi: 10.1016/j.immuni.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li W, et al. Aerobic glycolysis controls myeloid-derived suppressor cells and tumor immunity via a specific CEBPB isoform in triple-negative breast cancer. Cell. Metab. 2018;28:87–103 e106. doi: 10.1016/j.cmet.2018.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pattabiraman DR, Weinberg RA. Tackling the cancer stem cells - what challenges do they pose? Nat. Rev. Drug. Discov. 2014;13:497–512. doi: 10.1038/nrd4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dontu G, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.