Abstract
Sensory systems such as the olfactory system detect chemical stimuli and thereby determine the relationships between the animal and its surroundings. Olfaction is one of the most conserved and ancient sensory systems in vertebrates. The vertebrate olfactory epithelium is colonized by complex microbial communities, but microbial contribution to host olfactory gene expression remains unknown. In this study, we show that colonization of germ-free zebrafish and mice with microbiota leads to widespread transcriptional responses in olfactory organs as measured in bulk tissue transcriptomics and RT-qPCR. Germ-free zebrafish olfactory epithelium showed defects in pseudostratification; however, the size of the olfactory pit and the length of the cilia were not different from that of colonized zebrafish. One of the mechanisms by which microbiota control host transcriptional programs is by differential expression and activity of specific transcription factors (TFs). REST (RE1 silencing transcription factor, also called NRSF) is a zinc finger TF that binds to the conserved motif repressor element 1 found in the promoter regions of many neuronal genes with functions in neuronal development and differentiation. Colonized zebrafish and mice showed increased nasal expression of REST, and genes with reduced expression in colonized animals were strongly enriched in REST-binding motifs. Nasal commensal bacteria promoted in vitro differentiation of Odora cells by regulating the kinetics of REST expression. REST knockdown resulted in decreased Odora cell differentiation in vitro. Our results identify a conserved mechanism by which microbiota regulate vertebrate olfactory transcriptional programs and reveal a new role for REST in sensory organs.
Keywords: germ-free, mice, olfactory receptors, olfactory sensory neurons, transcriptomics, zebrafish
Introduction
Sensory systems such as the olfactory system detect chemical stimuli present in the environment and thereby determine the relationships between the animal and its surroundings. Among the different sensory systems, olfaction is one of the most conserved and ancient (Hildebrand and Shepherd 1997; Ache and Young 2005). The teleost olfactory system consists of 1 sensory organ (the olfactory mucosa) that contains olfactory sensory neurons (OSNs) expressing both olfactory receptors (ORs) and vomeronasal receptors (VRs) (Hansen et al. 2004). In most higher vertebrates, from sarcopterygian fishes to mammals, 2 olfactory systems are found in anatomically distinct areas, the main olfactory system or olfactory mucosa and the accessory olfactory system comprised by the vomeronasal organ (Fleischer 2009). Despite this different anatomical organization, the general cellular and molecular mechanisms of vertebrate olfactory systems are considered to be highly conserved, as revealed in comparative studies in zebrafish (Danio rerio) and mammals (Saraiva et al. 2015).
Communities of commensal microorganisms (microbiota) colonize every animal mucosal surface. Microbiota influence diverse processes in animals including development, metabolism, immunity, disease status and, more recently, neural function and behavior via the gut-brain axis (Bercik et al. 2011; Cryan and Dinan 2012; Lyte 2013; Hand et al. 2016; Kaelberer et al. 2018; Koskinen et al. 2018). In accord with their function, vertebrate olfactory epithelia are constantly exposed to the external environment and are consequently colonized with diverse microbial communities (Bassis et al. 2014; Tacchi et al. 2014; Chaves-Moreno et al. 2015; Liu et al. 2015; Lowrey et al. 2015; François et al. 2016), although similarities of nasal microbial community composition across vertebrate taxa have not been explored. Similarly, the specific influences of microbiota on vertebrate olfactory systems are unclear. In humans, the nasal microbiota has been implicated in multiple disease states including allergies, rhinosinusitis, and susceptibility to Staphylococcus infection (Krismer et al. 2014; Liu et al. 2015). More recently, 2 studies have revealed that mice raised in the absence of microbiota have altered olfactory morphology and augmented electrophysiological responses to odorants (François et al. 2016; Jain et al. 2016). However, the broader impact of microbiota on olfactory transcriptional programs and underlying molecular mechanisms remain unknown.
One of the mechanisms by which microbiota control host transcriptional programs is by differential expression and activity of specific transcription factors (TFs) (Camp et al. 2014; Davison et al. 2017). Microbial regulation of host gene expression via TF regulation has been described in the gut of several model organisms including Drosophila, zebrafish, and mice (Rawls et al. 2004; Round and Mazmanian 2009; Dobson et al. 2016; Davison et al. 2017). TFs are also known to orchestrate the differentiation and maturation programs of neurons and glia in vertebrates (Ma et al. 2006); however, regulation of TF expression by microbiota in neuronal tissues has not yet been described. REST (RE1 silencing transcription factor, also called NRSF) is a zinc finger TF that binds to the conserved motif repressor element 1 (RE1) (Chong et al. 1995). REST may have arisen 420 million years ago, since it is found in teleost but not invertebrate genomes (Bruce et al. 2004). RE1 is found in the promoter regions of many neuronal genes with functions in neuronal development and differentiation such as ion channels, neurotransmitter receptors, cell adhesion molecules, or neurotrophins (Kraner et al. 1992; Mori et al. 1992; Kallunki et al. 1995; Kallunki et al. 1997; Mieda et al. 1997; Palm et al. 1998; Timmusk et al. 1999; Bruce et al. 2004). Further, identified REST target genes include cell surface identity molecules such as ORs and VRs (Qureshi et al. 2010). While REST is known to be expressed in the olfactory system of mammals, the role of REST in olfactory transcriptional regulation remained unresolved.
The goal of this study is to investigate the impact of microbiota on the transcriptional program of the olfactory organ of zebrafish and mouse. Our results indicate that microbiota colonization is a major determinant of olfactory transcriptional programs via modulation of key TFs including REST.
Materials and methods
Animal husbandry
All experiments using zebrafish and mice were performed using protocols approved by the Institutional Animal Care and Use Committees of Duke University and the University of North Carolina at Chapel Hill. Gnotobiotic zebrafish husbandry was performed at Duke University essentially as described (Pham et al. 2008; Kanther et al. 2011). Briefly, wild-type Tübingen zebrafish were mated naturally. Eggs were collected and incubated for approximately 6 h in gnotobiotic zebrafish media (GZM) containing ampicillin (100 μg/mL), kanamycin (10 μg/mL), gentamicin (50 μg/mL), and amphotericin B (0.25 μg/mL). Fertilized embryos were sorted, treated sequentially with polyvinyl pyrrolidone-iodine (0.1% [0.01% free iodine]) and bleach (0.003%), then transferred to tissue culture flasks containing sterile GZM. Zebrafish were reared at a density of ≤1 larvae/mL in an air incubator at 28.5 °C on a 14 h/10 h light/dark cycle. At 3 days post-fertilization (dpf), half of the flasks in each experiment were inoculated with water collected from a recirculating conventional aquaculture system to generate conventionalized (CV) cohorts as described (Pham et al. 2008). The remaining flasks were maintained germ-free (GF). Media was changed daily from 3 to 6 dpf (80% volume), and animals were fed autoclaved sterile ZM000 (ZM Ltd; 0.25% w/v final concentration). At 6 dpf, sterility of GF animals was assessed as described (Pham et al. 2008). GF and CV animals were euthanized with 4× tricaine (Sigma MS-222) and collected into RNAlater (Ambion) for gene expression analysis or fixed in 4% paraformaldehyde in PBS for immunofluorescence or fixed in 2.5% formaldehyde/2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (Electron Microscopy Sciences) for TEM.
All mouse husbandry was performed as described (Camp et al. 2014) at the University of North Carolina at Chapel Hill with the following exceptions. All mice used in this study were 10- to 12-week-old male C57BL/6J, housed on alpha-dri bedding (Shepherd) and fed autoclaved sterile 2020SX diet (Envigo) ad libitum. To generate CV mice, GF mice were colonized with a conventional microbiota from specific-pathogen free (SPF) mice by receiving a 200 μL oral gavage of 20% glycerol stock of 1:1 w/v fecal slurry which had been previously prepared from feces collected from SPF C57BL/6J mice and homogenized in reduced PBS. CV mice were euthanized 2 weeks after colonization. Mice were euthanized under CO2 and decapitated. The lower jaws were removed and the head was submerged in RNAlater (Thermo Fisher Scientific) for subsequent RNA isolation.
16S rRNA gene sequencing of zebrafish and mouse olfactory microbial communities
The bacterial community of the olfactory organ of adult zebrafish and 10- to 12-week-old male C57BL/6J mice was performed as explained elsewhere (Reid et al. 2017). Zebrafish were obtained from a New Mexico pet distributor (Clark’s). Mice were obtained from the University of New Mexico Health Sciences Animal Research Facility. Sterile 3-mm tungsten carbide beads (Qiagen) were used to homogenize the tissue samples in a TissueLyser II (Qiagen). For extraction, we followed the cetyltrimethylammonium bromide (CTAB) buffer method as previously described (Reid et al. 2017). DNA pellets were then resuspended in 30 μL of DNase- and RNase-free molecular biology grade water. Sample DNA concentration and purity were measured using the NanoDrop ND 1000 (Thermo Scientific). Total genomic DNA for each sample was amplified in triplicate using primers specific for the V1–V3 region of the prokaryotic 16S rDNA (28F 5′-GAGTTTGATCNTGGCTCAG-3′ and 519R, 5′-GTNTTACNGCGGCKGCTG-3′) and amplification was performed as previously described by Reid et al. (2017). PCR amplicons were purified using Axygen AxyPrep Mag PCR Clean-up Kit (Fisher Scientific) as per manufacturer’s instructions. Samples were then indexed by ligating index barcode to Illumina adapters onto the PCR amplicon using the Nextera XT Index Kit v2 Set A (Illumina). Samples were pooled and purified again using the Axygen AxyPrep Mag PCR Clean-up Kit and sequenced in Illumina MiSeq platform using the MiSeq Reagent Kit v3 (600 cycle). Sequence data were analyzed using Quantitative Insights Into Microbial Ecology (QIIME 1.9) pipeline (Caporaso et al. 2010) within the web-based platform Galaxy at the University of New Mexico (Giardine et al. 2005). Operational taxonomic units (OTUs) selected by the sumaclust method were aligned to the SILVA 16S/18S database with a percentage identity greater than 97%. Using the QIIME pipeline, core diversity analyses were performed considering several alpha diversity metrics (Reid et al. 2017).
For comparisons across vertebrate taxa, previously published 16S rRNA gene sequencing data sets from human (Bassis et al. 2014), rat (Chaves-Moreno et al. 2015), mouse (François et al. 2016), and trout (Lowrey et al. 2015) olfactory organs were retrieved and plotted together with the zebrafish and C57BL/6J data sets.
Oligomicroarray
GF and CV 6 dpf zebrafish larvae (N = 4) were dissected under a stereomicroscope. First the heads were separated from the rest of the body, then the lower jaw was removed and finally the portion of the head anterior to the eyes was dissected using a sterile needle. Samples were stored in TRIzol for subsequent total RNA extraction. A total of 12 fish olfactory rosettes were collected for each of the 4 biological replicates. The RNA was extracted from the 4 pools from the CV and GF group and used for transcriptomic analysis. A zebrafish oligomicroarray (Agilent Zebrafish Gene Expression Microarray v3 4x44K) was used to study the expression of a total of 44K features. The olfactory and vomeronasal organs from GF and CV mice (N = 4) were dissected as explained elsewhere (Dunston et al. 2013) and total RNA was extracted. The Agilent 4x44K mouse oligomicroarray (Agilent mouse Gene Expression Microarray v2 4x44K) was used to detect changes in gene expression. For microarray hybridization, a common reference design was employed. In particular, the RNA reference sample was produced by adding an equimolar mix of total RNAs extracted from pools of olfactory organs of both GF and CV zebrafish or mice. Each experimental sample (Cy3 labeled) was hybridized against this reference sample (Cy5 labeled) in a 2-color experiment. To generate fluorescently labeled RNA for hybridizations, a MessageAMP aRNA Amplification Kit (Ambion) was used for initial amplification of mRNA. Briefly, 1 μg total RNA was reverse transcribed and the cDNA was used as a template for in vitro transcription in the presence of amino allyl-modified dUTP, which generated amplified antisense RNA (aRNA). For labeling, aRNA (3 μg) was denatured at 70 °C for 2 min in a volume of 10 μL to which 3 μL of 0.5 M NaHCO3 and 2 μL Cy dye (dye Cy3 or Cy5 mono-reactive dye pack, GE Healthcare) were added. Incorporation of dyes was performed for 1 h in the dark and the excess label was removed by DyeEx 2.0 spin column purification kit (Qiagen). The level of dye incorporation was checked by spectrophotometry (Nanodrop ND1000, LabTech). Prior to hybridization, 825 ng of each labeled template was fragmented in the presence of blocking agent and fragmentation buffer (Agilent). Fragmentation progressed in the dark at 60 °C for 30 min and samples were stored on ice until ready to load onto each microarray. The hybridizations were performed overnight at 65 °C. Following hybridization, the slides were rinsed in gene expression wash buffers 1 and 2 (Agilent) following the manufacturer’s instructions. The slides were then scanned using Agilent Scanner (Model # G2505C) at the resolution of 5 μm and Agilent Scan Control software (version A.8.1.3). Data were extracted using Agilent Feature Extraction software (version 10.5.1.1).
Bioinformatic and statistical analysis of microarrays results
A Lowess normalization of background-corrected data was next conducted and all intensity values <1.0 were set to 1.0. Statistical analysis of the arrays was performed using the GeneSpring GX analysis platform (version 12.6.1, Agilent Technologies). All quality control features were excluded from subsequent analyses. Further data filtering involved removal of saturated probe features, nonuniform features, population outliers, and those features showing intensities not significantly different from background in Cy3 or Cy5 channels. Fold-change differences between GF and CV samples were calculated using cutoff of 2-fold. Significant differential expression in the zebrafish and mouse microarray data sets was established by 2-sample (unpaired) t-test (P < 0.05) with unequal variance for genes with a fold change of at least 2. The Benjamini–Hochberg false discovery rate correction was applied within Genespring software (Agilent, version 12.6.1). In order to determine specific genes that comprise a conserved microbiota response, we identified one-to-one and one-to-many orthologs between zebrafish and mouse using Ensembl Gene IDs from Ensembl Biomart (http://www.ensembl.org/biomart/) based on homology type from Ensembl Genes 85. To compare across microarray platforms, associated microarray probe identifiers (Ensembl Transcript IDs and official gene symbols) from both zebrafish and mouse were converted to Ensembl Gene IDs uniquely using the genome assembly from which the corresponding microarray was designed; Zv9 (Ensembl Genes 79) and mm9 (Ensembl Genes 67). One-to-one and one-to-many orthologs that were differentially expressed in both zebrafish and mouse in GF versus CV olfactory tissue were then used for subsequent analyses.
Enrichment for GO biological processes was performed on all features that had GO identifiers associated using the GOEAST program (Zheng and Wang 2008). Fisher’s exact test was performed within the GOEAST program to determine whether GO identifiers occurred more often in a group than would appear by chance. For GO analysis, only biological process GO identifiers were considered if they were associated with at least 4 differentially expressed genes.
Searches in DAVID bioinformatics database 6.7 (https://david.ncifcrf.gov) (Huang et al. 2009) were performed to identify relevant biological pathways in the microarray data sets as well the data set resulting from the ortholog and homolog analyses. Eighty-six percentage of all differentially expressed mouse genes were considered by the software, whereas only 60% of the genes differentially expressed in zebrafish were considered.
Identification of common TF-binding site motifs in promoters of genes differentially regulated by microbiota
Enriched TF-binding site motifs were identified in promoters that were up- or downregulated in response to microbes in zebrafish and mouse using Homer v4.9 (http://homer.ucsd.edu/homer/index.html). For each organism, 1000 bp upstream of the transcription start site (TSS) for genes that were up- or downregulated by microbiota were utilized as input with the 1000 bp upstream of the TSS genes that were not differentially regulated by microbiota used as a background. Genes that were not differentially regulated by microbiota were defined as having a log2 fold-change value of between −0.25 and 0.25 (8743 Genes). TSS coordinates were downloaded from Ensembl (http://www.ensembl.org/index.html) using Ensembl Genes 67 for mouse and Ensembl Genes 79 for zebrafish. The default Homer enrichment threshold was used and the −log10P-value was graphed to compare motif enrichment for the different microbiota responses in the 2 organisms.
RT-qPCR confirmation of oligomicroarrays
cDNA synthesis was performed using 1 µg of total RNA as previously described (Tacchi et al. 2014). The resultant cDNA was stored at −20 °C. To validate the microarrays, the expression of 9 different genes including 3 odorant/OR genes was measured by RT-qPCR using specific primers for zebrafish and mouse (Supplementary Table S1). The qPCR was performed using 3 μL of a diluted cDNA template, 12.5 µL of Absolute blue SYBR Green ROX PCR master mix (Thermo Scientific), and 150 nM forward and reverse primers in a 25 µL reaction volume. The reaction was performed using the ABI Prism 7000 (Applied Biosystems) sequence detection system and the PCR cycling program was as follows: denaturation step at 95 °C for 10 min, then 35 cycles of 95 °C for 15 s and 62 °C for 1 min followed by melting (dissociation stage) from 72 °C to 95 °C (Tacchi et al. 2013).
Transmission electron microscopy
For transmission electron microscopy, the olfactory organs from GF and CV 6 dpf zebrafish larvae (N = 3 per condition) were fixed for 4 h at 4 °C in 2.5% (v/v) glutaraldehyde in PBS, then transferred to 1% osmium tetroxide (w/v) in PBS for 2 h at 4 °C. After washing in PBS (3 times, 10 min), samples were dehydrated in a graded series of ethanol (10–100%) through changes of propylene oxide. Samples were then embedded in Epon resin, sectioned and stained with uranyl acetate and lead citrate before being examined in a PHILIPS TECNAI 12 transmission electron microscope. Sections were examined in a JEM-1011 and images were acquired using an ORIUS SC200 camera. Vesicles present in the nasal epithelia as well as the proximal ciliar maximum length were quantified using Leica QWin image analysis software (Leica Microsystems). Manual color segmentation of cytoplasm and cell nuclei of nasal epithelia was conducted using Amira software version 6.0.
Gel electrophoresis and western blot
In order to confirm expression of REST protein in the olfactory organ of mouse and zebrafish we dissected the olfactory epithelium of a CV 4-week-old mouse and a 6-month-old mouse under sterile conditions. Additionally, the olfactory epithelium of 3 CV adult zebrafish and of CV and GF 6 dpf zebrafish larvae (N = 50) were also dissected. Brain tissue from juvenile and adult mice and zebrafish were also collected and used as positive controls. All samples were sonicated in 100 μL RIPA buffer (Sigma). The protein lysates were centrifuged at 10 000 rpm for 20 min, supernatants were collected, and the protein concentration of each sample was quantified using the Pierce 660nm reagent. Subsequently and equal amount of Laemmli sample buffer (Bio-Rad) was added to each sample and boiled at 97 °C for 5 min. Samples were loaded onto a mini Protean TGX precast gel (Bio-Rad), transferred to a PVDF membrane, and incubated for 2 h at room temperature with either the primary antibody rabbit anti-human NRSF/REST (1:500, GeneTex, GTX37363), which recognizes 2 bands (1 of 120 KDa and 1 of 55 KDa) or with the primary rabbit polyclonal GAPDH antibody (1:1000, GeneTex, GTX100118). After washing 3 times with PBT, the membrane was incubated with a secondary antibody HRP goat anti-rabbit IgG (Jackson Immunoresearch, 1:7500) for an hour. Blots were developed using the Pierce ECL western blotting reagents (Thermo Scientific) in a Bio-Rad ChemiDoc Touch Imaging system v1.2.0.12. To further evaluate the specificity of REST antibody in zebrafish, 6 dpf wild-type and REST mutant zebrafish larvae containing a truncated version of REST (N = 30) (Moravec et al. 2015) were also dissected and a western blot was performed as above.
Bacterial culture
The olfactory epithelia from a healthy laboratory-reared adult SPF rat was dissected with sterile instruments, scraped, and placed in a Petri dish with 2 mL of PBS. The solution was then centrifuged at 400 × g for 5 min to separate bacteria from any host cells. To pellet bacteria, a second centrifugation (13 000 × g for 15 min) was performed and the supernatant was substituted with 1 mL of fresh sterile PBS. The bacterial suspension was then plated on a Luria Bertani (LB) (BD, Amresco) agar plate and incubated at 37 °C for 2 days under aerobic conditions. Total genomic DNA was extracted from a selected colony as previously explained (Lowrey et al. 2015). Subsequently, DNA was amplified using the 16S rDNA primers 787F and 1492R (Reid et al. 2017) and a standard PCR protocol with an initial activation step at 94 °C for 2 min, followed by 30 cycles of 94 °C for 45 s, annealing at 55 °C for 45 s, and 72 °C for 1 min. Final extension for 10 min at 72 °C. PCR products were inserted into pGem-T-Easy vector, cloned, purified, and then sequenced. Results were nBlasted against the NCBI database, identifying this clone as Staphylococcus aureus. GenBank accession number KX197204 has been assigned to the sequence obtained in this manuscript. Staphylococcus aureus was grown in LB under aerobic conditions at 37 °C. Comamonas denitrificans was purchased from American Type Culture Collection (ATCC 700940). Comamonas denitrificans was grown overnight according to manufacturer’s instructions in LB under aerobic conditions at 37 °C. Finally, the cecal contents of a 12-week-old C57BL/6J male mouse were plated onto LB agar plates and incubated at 37 °C for 2 days under aerobic conditions. One of the colonies was picked and used for DNA extraction, PCR amplification, and cloning as explained above. The commensal strain was identified as Acinetobacter radioresistens (accession number MF772494) and was grown overnight in LB under continuous shaking in aerobic conditions at 37 °C. Pure S. aureus, C. denitrificans, and A. radioresistens overnight cultures were pelleted and washed 3 times in Dulbecco’s Modified Eagle’s Medium (DMEM) prior to being used for the in vitro experiments.
Odora cell line maintenance, cell differentiation, and olfactory marker protein staining
Undifferentiated rat Odora cells were cultured as previously described (Murrell and Hunter 1999). To induce the differentiation, 1 mg/mL insulin solution from bovine pancreas, 20 mM dopamine and 100 mM ascorbic acid (both from Sigma) were added into the DMEM high glucose media normally supplemented with 10% fetal bovine serum (FBS, Hyclone), 100 mg streptomycin, 100 U/mL penicillin, 200 mM of L-glutamine (Life Tech), and 2.5 mg/mL of fungizone (Gibco). Differentiated cells were incubated at 39 °C with 7% of CO2 until 90% of the cells reached the typical neuronal phenotype.
In order to evaluate the contribution of commensal bacteria to the Odora cells differentiation, undifferentiated cells were cultured in 24-well plates and incubated with normal DMEM (undifferentiating media), or differentiation media, or undifferentiating media containing 105 cfu/mL of live S. aureus, C. denitrificans, and A. radioresistens. The experiment was repeated 3 independent times. Cells were incubated at 39 °C with 7% of CO2 for 72 h. Brightfield images were taken daily using a ×20 objective under a Nikon Eclipse Ti microscope. Subsequently, Odora cells were rinsed twice in PBS and fixed with 4% PFA. Three additional washes in PBT (1% BSA + 0.5% Tween-20 in PBS) were performed, before and after blocking for 15 min at RT with starting block T20 (Thermo Fisher). Then, cells were incubated at 4 °C overnight with mouse anti-human olfactory marker protein (OMP) monoclonal antibody (1:200, sc-365818, Santa Cruz Biotechnology) or the specific isotype control mouse IgG2a followed by the secondary antibody (1:250, Jackson ImmunoResearch, Cy3 AffiniPure Goat Anti-Mouse IgG (H+L)). Nuclei were stained with DAPI. Ten different images (×20) were captured under a Nikon Eclipse Ti microscope using the Advanced Elements Research software v4. The total number of OMP positive cells per field was counted. Expression of other OSN differentiation markers was also measured by RT-qPCR by measuring the expression levels of OMP, olfactory cyclic nucleotide-gated channel beta (oCNGb), and REST. RT-qPCR was performed as described above using rat specific primers shown in Supplementary Table S1.
Silencing of REST
Silencing of REST gene in undifferentiated Odora cells was performed via MATra-Si transfection reagent (PromoKine) following the manufacturer’s instruction. The REST silencing oligos set (Supplementary Table S1) was purchased from ABM company (https://www.abmgood.com/Rest-siRNA-Oligo-i500115.html). A silencing solution containing 1.5 µg of the 3 siREST oligos or positive (siGAPDH) or negative (siNeg) controls, 97 µL of serum-free DMEM medium, and 1.5 µL of MATra-Si reagent was incubated at room temperature for 20 min. Subsequently, these solutions were added into the 12-well plate prepared with 1 × 106 undifferentiated Odora cells (4 replicate wells per each treatment). The plate was then positioned onto a magnetic surface and stirred constantly for 15 min, prior incubation for 72 h at 39°C with 7% of CO2. RT-qPCR was used to monitor the knockdown of REST at the mRNA level.
Statistical analysis
Data were analyzed in Prism version 5.0 (GraphPad). Data are expressed as mean ± SE. In the RT-qPCR experiments the relative expression level of the genes was determined using the Pfaffl method (Pfaffl 2001). Using this method, the fold-change difference in expression in the GF compared to the CV animals was quantified considering the CV samples as controls. The RT-qPCR measurements were analyzed by the unpaired t-test. For fluorescence microscopy experiments on Odora cells, results are shown as mean % of OMP+ cells ± SE and multiple comparisons analysis between experimental treatments was performed with 1-way ANOVA. P-values lower than 0.05 were considered statistically significant.
Availability of data and materials
The data sets generated during the current study are at the European Bioinformatics Institute archived under accession number E-MTAB-5045 and E-MTAB-5046 for zebrafish and mouse, respectively. Additionally, array data were submitted to ArrayExpress in compliance with MIAME. The zebrafish and mouse nasal 16S rRNA gene sequence results are deposited in NCBI under BioProject # PRJNA389614. Further information and requests for data and materials should be directed to and will be fulfilled by Irene Salinas (isalinas@unm.edu).
Results
Defining composition of microbiota associated with olfactory organs in multiple vertebrate species
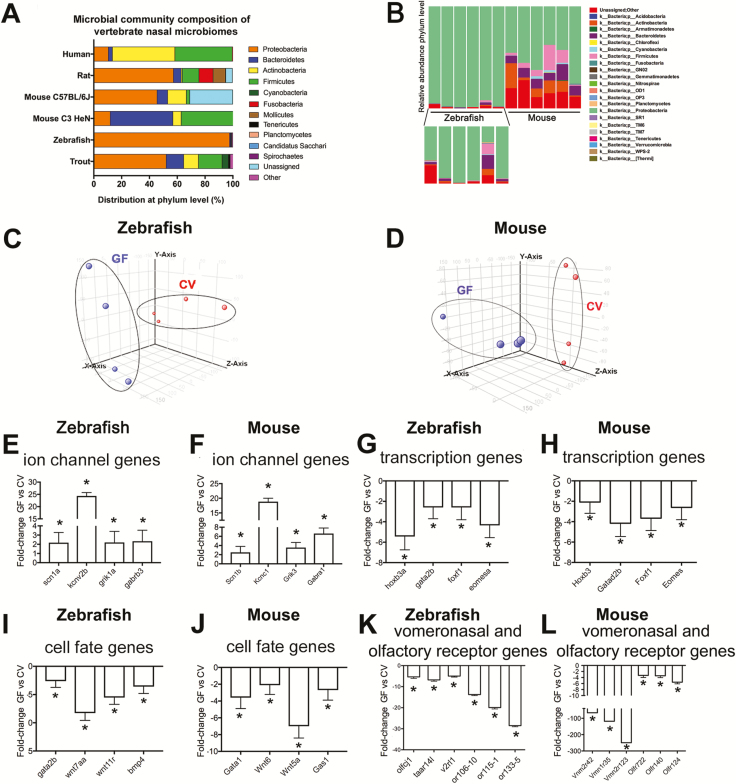
We used 16S rDNA gene sequencing to define the membership of the bacterial communities colonizing the olfactory organs in adult zebrafish and C57BL/6J mice. We found that zebrafish olfactory organs were colonized by members of 7 bacterial phyla, all of which were also represented among the 8 phyla detected in C57BL/6J mice. In all zebrafish and mice surveyed, nasal microbiomes were dominated by members of the Proteobacteria phylum (Figure 1). However, fewer bacterial taxa were shared between zebrafish and mice at the order or genus levels (Supplementary Tables S2). At the order level, zebrafish nasal microbiomes show high degree of interindividual variation and unresolved taxonomies (Supplementary Table S2). Comamonas sp. was the most abundant genus observed in mice (Supplementary Table S2). We compared our results with previously available data sets in rainbow trout (Oncorhynchus mykiss) (Lowrey et al. 2015), C3H/HeN mice (François et al. 2016), cotton rat (Chaves-Moreno et al. 2015), and human (Bassis et al. 2014) (Figure 1). Similar to our zebrafish and C57BL/6J mouse results, Proteobacteria was also the dominant phylum in rainbow trout and cotton rat, whereas Actinobacteria and Firmicutes dominate the nasal microbial community of humans, and Bacteriodetes and Firmicutes dominated in C3H/HeN mice.
Figure 1.
The nasal microbial community of zebrafish and other vertebrates is dominated by Proteobacteria and influences the transcriptional profile of the olfactory organ. (A) Relative abundance of bacteria at the phylum level present in the olfactory organ of zebrafish (this study), C57BL/6J mice (this study), rainbow trout (Lowrey et al. 2015), C3H/HeN mice (François et al. 2016), cotton rat (Chaves-Moreno et al. 2015), and human (Bassis et al. 2014). Only phyla with a relative abundance >0.1 are represented. Note that different V regions of the 16S rDNA were targeted in different studies and different sequencing platforms (454 or Illumina) and sampling depths were used in different studies. (B) Bacterial community composition of adult zebrafish (also magnified in the lower inset) and mice nasal microbiomes at the phylum level. Each bar represents an individual animal (N = 6). (C) PCA of individual biological microarray replicates for zebrafish samples. (D) PCA of individual biological microarray replicates for mouse samples. Bar plot showing the fold change in expression (mean ± SE) of representative genes within the GO category “ion transport” showing significant increased expression in the olfactory organ of GF compared to CV zebrafish (E) and GF compared to CV mice (F). Bar plot showing the fold change in expression (mean ± SE) of representative genes within the GO category “transcription, DNA-templated” showing consistent decreased expression in the olfactory organ of GF compared to CV zebrafish (G) and in the olfactory organ of GF compared to CV mice (H). Bar plot showing the fold change in expression (mean ± SE) of representative genes within the GO category “cell fate commitment” showing significant decreased expression in the olfactory organ of GF compared to CV zebrafish (I) and in the olfactory organ of GF compared to CV mice (J). Bar plot showing fold change in expression of selected OR genes in the olfactory organ of GF compared to CV zebrafish (K) and mice (L). Asterisk indicates statistically significant differences when P < 0.05. In zebrafish, selected genes were sodium channel, voltage-gated, type I, alpha (scn1a), potassium channel, subfamily V, member 2b (kcnv2b), glutamate receptor, ionotropic, kainate 1a (grik1a), gamma-aminobutyric acid (GABA) A receptor, beta 3 (gabrb3), homeobox B3a (hoxb3a), forkhead box F1 (foxf1), beta 3 Eomesodermin homolog a (eomesa), GATA binding protein 2b (gata2b), wingless-type MMTV integration site family, member 7Aa (wnt7aa), wingless-type MMTV integration site family, member 11, related (wnt11r), bone morphogenetic protein 4 (bmp4), olfactory receptor C family, j1 (olfcj1), trace amine associated receptor 14l (taar14l), vomeronasal 2 receptor, l1 (v2rl1), odorant receptor, family G, subfamily 106, member 10 (or106-10), odorant receptor, family F, subfamily 115, member 1 (or115-1) and odorant receptor, family H, subfamily 133, member 5 (or133-5). In mouse, selected genes were sodium channel, voltage-gated, type I, beta (Scn1b), potassium voltage gated channel, Shaw-related subfamily, member 1 (Kcnc1), transcript variant 1, glutamate receptor, ionotropic, kainate 3 (Grik3), gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 (Gabra1), homeobox B3 (Hoxb3), transcript variant 1, GATA zinc finger domain containing 2B (Gatad2b), forkhead box F1 (Foxf1), eomesodermin homolog (Xenopus laevis) (Eomes), transcript variant 1, GATA binding protein 1 (Gata1), wingless-related MMTV integration site 6 (Wnt6), wingless-related MMTV integration site 5A (Wnt5a), growth arrest specific 1 (Gas1), vomeronasal 2, receptor 42 (Vmn2r42), vomeronasal 1, receptor 35 (Vmn1r35), vomeronasal 2, receptor 123 (Vmn2r123), olfactory receptor 722 (Olfr722), olfactory receptor 140 (Olf140), olfactory receptor 124 (Olf124).
This data combined indicate that the nasal microbial communities of vertebrates can be colonized by diverse bacterial taxa and that these communities have predicted common functional attributes.
Microbiota regulate gene expression in zebrafish and mouse olfactory organ
Microbiota are known to induce conserved responses in the transcriptional program of zebrafish and mouse digestive tract (Rawls et al. 2004; Rawls et al. 2006; Davison et al. 2017). Two recent studies have revealed that mice raised in the absence of microbiota have altered olfactory morphology and augmented electrophysiological responses to odorants (François et al. 2016; Jain et al. 2016). However, the broader impact of microbiota on olfactory transcriptional programs and underlying molecular mechanisms remain unknown. Here we sought to define the impact of microbiota colonization on gene expression in zebrafish and mice olfactory systems.
The general cellular and molecular mechanisms of vertebrate olfactory systems are considered to be highly conserved, as revealed in comparative studies in zebrafish (D. rerio) and mammals (Saraiva et al. 2015). In this study, we extracted RNA from dissected olfactory organs from zebrafish larvae and the comparable olfactory epithelium and vomeronasal organ from adult mice reared GF or colonized with their respective normal microbiotas (a process called conventionalization; CV) and evaluated gene expression using DNA microarrays. Principal component analysis (PCA) showed that the variability of the individual zebrafish and mouse replicates grouped based on GF and CV conditions (Figure 1C and D). In accord, we found 5198 and 2946 genes that were differentially expressed in the olfactory organs of GF compared to CV zebrafish and mice, respectively (Supplementary Table S3). In zebrafish, 2703 genes or 3569 probes (69% of all the significantly modified probes) displayed lower expression in GF compared to CV olfactory organs, whereas 1336 genes or 1629 probes (31% of the significantly modified probes) were elevated in GF compared to CV zebrafish olfactory organs. In mice, 1359 genes or 1508 probes (51% of the significantly modified probes) showed decreased expression in GF olfactory and vomeronasal organs, whereas 1327 genes or 1438 probes (49% of the significantly modified probes) were elevated in GF olfactory and vomeronasal organs. These microarray results were validated by RT-qPCR in both zebrafish and mouse (Supplementary Figure S1).
To broadly define biological processes affected by microbiota colonization, we performed Gene Ontology (GO) enrichment analysis on genes significantly increased or decreased by microbial colonization in zebrafish (Supplementary Table S4) and mouse (Supplementary Table S5) olfactory organs. This analysis revealed that genes involved in ion transport (Figure 1E and F) and synapse organization were more highly expressed in the olfactory organ of GF compared to CV animals in both hosts, whereas genes involved in transcriptional regulation (Figure 1G and H), development and cell fate commitment (Figure 1I and J) had lower expression in GF compared to CV zebrafish and mice. Since odorant receptors are key determinants in olfactory neuron axonal guidance and formation of olfactory glomeruli (Feinstein et al. 2004), we examined whether these genes were differentially expressed in both experimental groups. A large number of odorant receptor genes were differentially regulated in response to microbes in zebrafish (61 out of 197) and mouse (91 out of 1395) based on the microarray experiment (Table 1). A breakdown of the significantly modified odorant receptor genes in zebrafish and mouse by class of receptor is shown in Table 1. While the complexity of OR superfamily evolution makes comparison of overlapping functional gene products from zebrafish to mouse difficult, surprisingly, of OR genes that were differentially regulated by microbiota colonization >90% and 95% were higher in the presence of microbiota in zebrafish and mouse, respectively (Table 1). The decreased expression of OR genes in GF zebrafish and mouse was confirmed by RT-qPCR (Supplementary Figure S1). Thus, our results suggest a common conserved transcriptional increase of OR genes in zebrafish and mouse in response to microbes.
Table 1.
Olfactory mRNAs significantly modified in the olfactory organ of GF compared to CV in zebrafish and mice
| Total number of olfactory/odorant genes significantly modified | Lower | Higher |
|---|---|---|
| Zebrafish | 55 | 6 |
| Mouse | 87 | 4 |
| Number of olfactory/odorant genes per receptor class significantly modified | Zebrafish | Mouse |
| ORA/V1R genes | 2/3 | 21/166 |
| TAAR genes | 10/43 | 1/14 |
| OlfC/V2R genes | 8/15 | 31/97 |
| OR genes | 40/131 | 37/1115 |
| FPR genes | 1/1 | 1/5 |
To search for further evidence of conserved transcriptional responses to microbiota, we compared our lists of zebrafish and mouse genes that are differentially regulated upon microbiota colonization. We identified 152 nonredundant one-to-one orthologs that were differential in both zebrafish and mouse data sets, 83 of which are regulated in the same direction in both animals. Additionally, 167 one-to-many homolog pairs were found from which 76 were regulated in the same direction in both animals (Supplementary Table S6). The 83 one-to-one orthologs and the 76 one-to-many homologs were then combined into a single list of 159 genes and subjected to GO and KEGG pathway analyses (Supplementary Table S7). The enriched pathways shared in zebrafish and mice included the MAPK signaling pathway which is critical in odorant recognition and olfactory neuron survival in animals (Hirotsu et al. 2000; Watt and Storm 2000), and the adherens junctions pathway which is critical for the mosaic cellular pattern of the olfactory epithelium (Katsunuma et al. 2016). Among the GO terms, embryonic organ development was the only term common to zebrafish and mice. These results indicate that microbiota regulate common biological pathways and gene orthologs in the olfactory organ of zebrafish and mouse, and suggest potential impacts on organ development as well as potential conservation in upstream transcriptional regulators.
Microbiota control the differentiation and maturation of the zebrafish olfactory organ
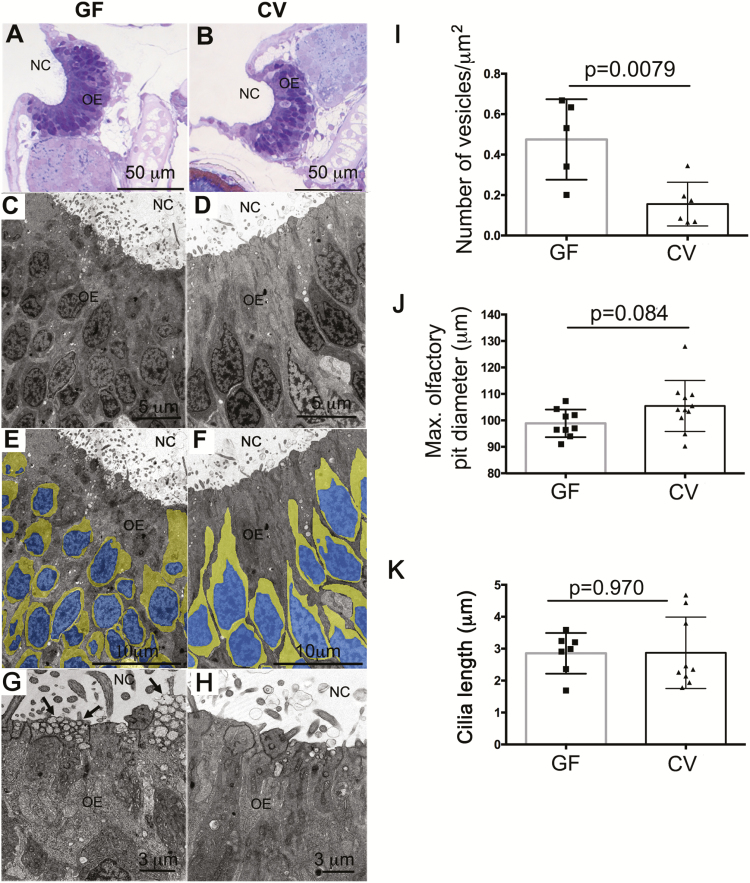
In the zebrafish (Saraiva et al. 2015) and mouse (Ibarra-Soria et al. 2017), OR mRNA levels obtained by transcriptomic profiling can be used to quantify the diversity of OSN subtypes that express each receptor (Ibarra-Soria et al. 2017). Thus, differences in OR expression levels detected in our data sets may reflect differences in OSN subpopulations. We observed by light microscopy that the olfactory neurons of GF zebrafish larvae had altered cell morphologies and lower cytoplasm to nuclei aspect ratios than those of CV animals (Figure 2A and B). TEM studies confirmed that GF zebrafish possess a less differentiated olfactory epithelium compared to CV animals (Figure 2C–F). Specifically, in GF zebrafish, the pseudostratification of the olfactory epithelium was less defined compared to CV animals. Additionally, significantly more transport vesicles were observed in the apical cytoplasmic region of supporting (or sustentacular) cells in GF zebrafish olfactory organs compared to CV (Figure 2G–I). However, when we measured the olfactory pit maximum diameter of GF versus CV zebrafish, no significant differences were found (Figure 2J). Quantification of the maximum ciliar length did not revealed any differences between groups (Figure 2K).
Figure 2.
Ultrastructural and cellular changes in the zebrafish olfactory organ in the absence of microbiota. (A) Light micrograph of a semithin section of a GF zebrafish olfactory organ stained in toluidine blue. (B) Light micrograph of a semithin section of a CV zebrafish olfactory organ stained in toluidine blue. (C, E, and G) Transmission electron micrograph (TEM) of a GF zebrafish olfactory epithelium. (D, F, and H) TEM of a CV zebrafish olfactory epithelium. Images in (E) and (F) were pseudocolored as explained in the materials in methods to highlight the nucleus area (blue) and the cytoplasm (yellow) of OSNs. Arrows in (G) indicate transport vesicles in the apical pole of sustentacular cells. NC: nasal cavity; OE: olfactory epithelium. (I) Number of apical vesicles quantified in TEM images from CV and GF zebrafish larvae (N = 3 larvae/group). (J) Mean maximum olfactory pit diameter measured in CV and GF zebrafish larvae (N = 10). (K) Quantification of the maximum ciliar length in CV and GF zebrafish (N = 8).
In teleost fishes, Gαo is a marker for microvillous OSNs similar to vomeronasal neurons in mammals, whereas Gαolf is a marker for ciliated OSNs in both fish and mammal (Tacchi et al. 2014; Chaves-Moreno et al. 2015; Liu et al. 2015; Lowrey et al. 2015; François et al. 2016). Regarding the expression of these 2 markers, microarray results showed a nonsignificant trend toward lower Gαo expression in the olfactory organs of GF compared to CV zebrafish. Additionally, in the zebrafish and mouse microarrays, Gαo (Gnao1a) was not significantly modified. Interesting, in zebrafish, a second isoform (Gnao1b) showed an opposite trend (a significant increase in GF compared to the CV larvae) (Supplementary Table S3). Gαolf (Gnal) was significantly downregulated (2.5-fold) in GF mouse but not zebrafish (Supplementary Table S3). Combined, these results are inconclusive but suggest that defects in odorant receptor gene expression in GF animals are not accompanied by defects in these 2 gene markers for microvillous and ciliated OSNs, respectively.
NRSF/REST-binding sites are commonly enriched at genes that are downregulated in olfactory tissue in response to microbiota
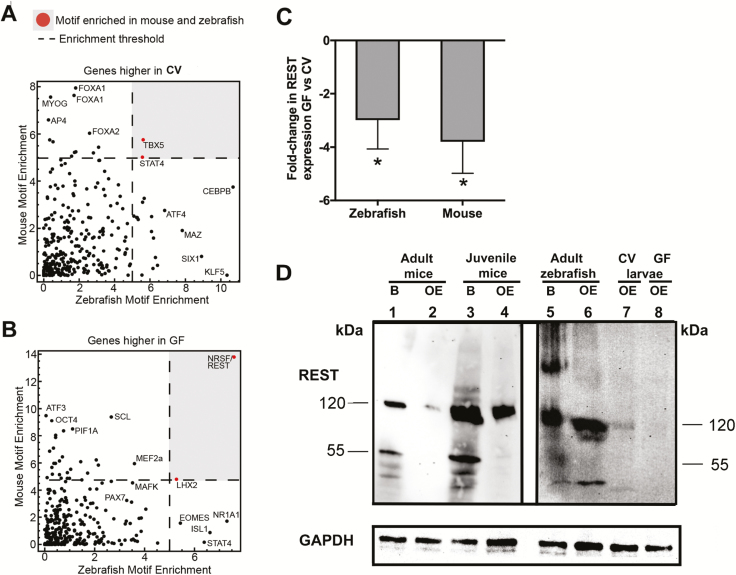
TFs are known to orchestrate the differentiation and maturation programs of neurons and glia in vertebrates (Ma et al. 2006); however, regulation of TF expression by microbiota in neuronal tissues has not yet been described. To determine if common regulators underlie aspects of the common response to microbes, we performed TF motif enrichment analyses using the 1000 bp upstream of the TSS of genes that were differentially expressed in zebrafish and mice (Supplementary Table S3). We found that TBX5 and STAT4 motifs are enriched in genes expressed lower in GF compared to CV in both zebrafish and mice (Figure 3A). This suggests microbially induced genes in the olfactory system may be regulated by TFs in the STAT family which are known to regulate aspects of inflammation and repair in response to microbes (Knights et al. 2014; Villarino et al. 2017), control gliogenesis (Bonni 1997; He et al. 2005), and even directly bind the OMP (a marker for mature OSNs) promoter in OSNs (Ibarra-Soria et al. 2002).
Figure 3.
NRSF/REST-binding sites are commonly enriched at genes that are downregulated in olfactory tissue in response to microbiota. (A) TF motif enrichment (−log10P-value) using the 1000 bp upstream of the TSS of genes that are significantly more highly expressed in CV conditions in zebrafish and mouse olfactory tissue. A dashed line represents the threshold for motif enrichment. TBX5 and STAT4 motifs that are enriched in both zebrafish and mouse are marked in red and within the gray area. (B) Same as (A) for genes that were significantly higher in GF conditions. NRSF/REST and LHX2 motifs that are enriched in both zebrafish and mouse are marked in red and within the gray area (P < 0.05). (C) Gene expression levels of REST in GF zebrafish and mouse compared to CV animals (N = 4). Asterisk indicates statistically significant differences when P < 0.05. (D) Protein expression of REST in brain and olfactory organ of adult mice (lanes 1–2), juvenile mice (lanes 3–4), adult zebrafish brain and olfactory organ (lanes 5–6), and olfactory organs of CV and GF zebrafish larvae (lanes 7–8). Blots probed with an isotype control instead of anti-REST antibody produced no bands. GAPDH was used as a loading control.
Interestingly, genes with higher expression in GF mice and zebrafish were enriched in LHX2 and NRSF/REST motifs (Figure 3B). LHX2 is known to regulate differentiation and identity of OSNs (Hirota and Mombaerts 2004; Kolterud et al. 2004; Zhang et al. 2016; Monahan et al. 2017). Furthermore, NRSF/REST is a critical transcriptional repressor that prevents inappropriate expression of neuronal genes and controls maturation of neurons (Parlato et al. 2014; Zhao et al. 2016). REST target genes include cell identity surface molecules such as ORs and VRs. REST is also known to be expressed in the olfactory epithelium of mammals. The present study reveals a conserved role for REST in vertebrate olfactory systems and indicates that microbiota are critical regulators of REST expression. In support, we observed reduction of REST expression in GF versus CV in both mouse and zebrafish olfactory organs at the gene level by RT-qPCR (Figure 3C). This was in agreement with the microarray results for mouse but not for zebrafish, where the change in expression did not reach statistical significance by microarray (Supplementary Table S3; Figure 3C).
At the protein level, REST was well expressed in the CNS of the juvenile and adult mouse and zebrafish as expected (Figure 3D). Moreover, we detected REST protein expression in the adult mouse olfactory organ although at lower levels than those detected in the adult mouse CNS, especially for the 55 kDa band. In the juvenile mouse and adult zebrafish olfactory organ, REST protein levels were lower than in the CNS. The high molecular bands detected by the anti-REST antibody in the zebrafish brain were not present in any other sample and do not correspond with the expected 120 and 55 kDa bands that this antibody recognizes according to the manufacturer (GeneTex). Therefore, these bands were considered not specific (Figure 3D). In order to confirm the specificity of the REST antibody in zebrafish, a western blot comparing wild-type and REST mutants was performed. In support, the intensity of the 120 KDa band was lower in the mutant compared to the wild-type olfactory organ and a new band of 60 KDa was observed. Additionally, the 55 KDa band was absent in the REST mutant (Supplementary Figure S2). In agreement with the gene expression data, we detected greater REST expression (120 kDa band) in the CV zebrafish larvae olfactory organ compared to the GF samples. These results confirm that REST is expressed not only in the CNS but also in the peripheral nervous system (olfactory organ) of both juveniles and adult stages of mice and zebrafish. Combined, this data indicate that microbiota control the transcriptional program of vertebrate olfactory systems by regulating the expression of key TFs like REST and this may directly influence olfactory development across vertebrates.
In vitro differentiation of rat Odora cells displays bacterial specificity
We next used Odora cells, a rat OSN cell line, to test whether distinct bacteria can drive differentiation of these cells in vitro. This cell line is the only established primary cell culture of vertebrate OSNs. Odora cells can be grown as undifferentiated epithelial-like cells using DMEM culture media in control conditions (5% CO2 and 33 °C) and differentiate into differentiated OSNs when grown with 7% CO2 at 39 °C in the presence of insulin, dopamine, and ascorbic acid (Murrell and Hunter 1999). Upon differentiation, OMP expression increases in Odora cells; however, these cells are not fully functional OSNs (Murrell and Hunter 1999). We hypothesized that different bacterial species may have distinct capacities to stimulate Odora cell differentiation in vitro. We tested 2 nasal bacteria strains: S. aureus (strain NBRC 100910; phylum Firmicutes) which we isolated from the olfactory organ of a laboratory-reared SPF rat; and C. denitrificans (ATCC 700940; phylum Proteobacteria) originally isolated from sludge in Sweden (Dalhammer G, personal communication). Staphylococcus aureus is a common member of the mammalian nasal microbiota (Bassis et al. 2014; Liu et al. 2015; Mulcahy and McLoughlin 2016), and the Comamonas genus is one of the most abundant in the mouse olfactory organ by 16S rDNA sequencing (Supplementary Table S2). We also tested A. radioresistens (strain NBRC 102413; phylum Proteobacteria) which we isolated from mouse cecal contents.
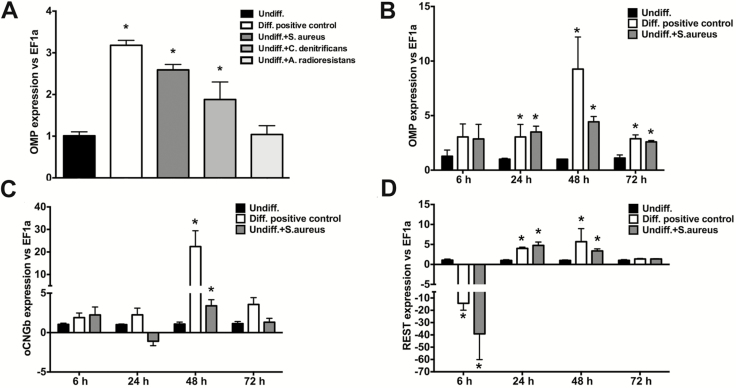
Undifferentiated Odora cells were cultured in control DMEM, DMEM containing all the differentiation agents (Diff. positive control), or in control DMEM to which live S. aureus, C. denitrificans, or A. radioresistens were added. OMP expression was upregulated in differentiated control Odora cells as well as undifferentiated cells treated with nasal strain S. aureus or C. denitrificans but not with A. radioresistens or the undifferentiated controls (Figure 4A). This result was confirmed by immunofluorescence microscopy (Supplementary Figure S3A–D). Multiple bacterial concentrations were tested and the 105 cfu/mL dose was selected based on optimal cell viability and differentiation markers (Supplementary Figure S3E). Analysis of kinetic expression revealed that transcript levels of OMP were significantly higher in differentiated Odora cells (Diff. positive control) as well as undifferentiated cells treated with S. aureus compared to undifferentiated cells at 24, 48, and 72 h (Figure 4B). Expression levels of oCNGb, another marker for mature OSNs (Murrell and Hunter 1999), were only significantly higher in positive control differentiated cells as well as S. aureus differentiated cells at 48 h (Figure 4C). Importantly, REST expression showed dynamic changes at different time points. At 6 h, both Diff. positive control cells and Undiff. + S. aureus cells showed significant downregulation in REST expression (~15- and 38-fold, respectively) (Figure 4D). Following this downregulation, we recorded significant and sustained increases in REST expression at 24 and 48 h in both Diff. positive control and Undiff. + S. aureus compared to Undiff. control cells (Figure 4D). These results show that bacterial stimulation of Odora cell differentiation shows specificity in vitro and that this is accompanied by simultaneous dynamic control of REST expression in OSNs.
Figure 4.
Nasal commensal bacteria promote rat Odora cell differentiation in vitro in a REST dependent manner. (A) Undifferentiated Odora cells were incubated with the nasal bacterial strain S. aureus, the nasal bacterial strain C. denitrificans, or the gut commensal strain (A. radioresistens). OMP expression levels were measured 72 h later by RT-qPCR. Data are shown as the mean fold change ± SE in expression compared to undifferentiated controls. (B) Kinetics of OMP gene expression; (C) oCNGb gene expression; and (D) REST gene expression in undifferentiated Odora cells, control differentiated Odora cells, or Odora cells differentiated in the presence of live S. aureus at 6, 24, 48, and 72 hours. Data are shown as the mean fold change ± SE in expression compared to undifferentiated controls measured by RT-qPCR. Asterisks denote statistically significant differences (P < 0.05). Results are representative of 3 independent experiments with N = 5 per experiment.
Silencing REST results in decreased differentiation of Odora cells in vitro
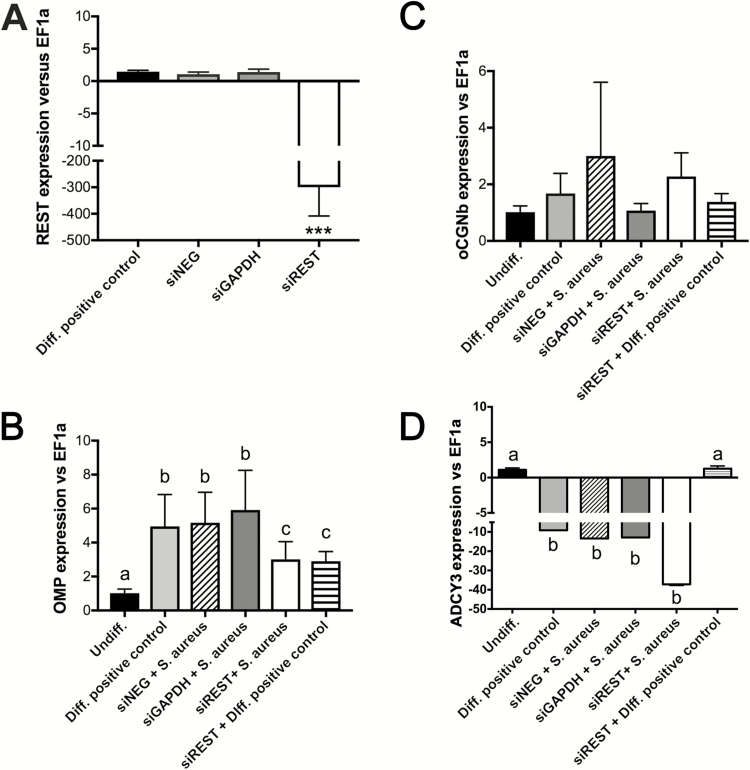
Our in vivo and in vitro results suggested that microbial regulation of REST expression in the olfactory system may be one of the mechanisms by which OSNs differentiate. To test this possibility, we knocked down REST function in Odora cells using siRNA technology. Successful silencing of REST (~200-fold decrease in expression after 48 h) in the rat Odora cell line was confirmed by RT-qPCR analysis (Figure 5A). Importantly, knockdown of REST (siREST) resulted in decreased levels of OMP expression in S. aureus differentiated Odora cells as well as Diff. positive control cells compared to negative controls after 72 h in culture (Figure 5B). oCNGb expression levels, on the other hand, were highly variable and not significantly different among treatments (Figure 5C). Adenylate cyclase 3 (ADCY3) expression, another marker of neuronal differentiation, was downregulated in the Diff. positive control cells, as well as in the S. aureus differentiated neurons. REST silencing, however, showed opposite effects in ADCY3 expression in the Diff. positive control and the S. aureus treatment (Figure 5D). These data establish that REST plays a role in Odora cell differentiation, and that commensal bacteria promote Odora cell differentiation using a REST-dependent mechanism.
Figure 5.
Silencing NRSF/REST results in decreased differentiation of Odora cells in vitro. (A) REST mRNA levels in differentiated Odora cells, Odora cells treated with siNEG oligo (negative control), siGAPDH oligo (positive control), and a cocktail of 3 different siREST oligos. Gene expression levels were measured by RT-qPCR. Relative expression levels are shown as the ratio between REST expression and the housekeeping gene EF1a. ***P < 0.001. (B) OMP mRNA levels; (C) oCNGb mRNA levels; and (D) ADCY3 mRNA levels in undifferentiated Odora cells, differentiated Odora cells (Diff. positive control), Odora cells differentiated in the presence of S. aureus and the negative control silencing probe (siNEG), Odora cells differentiated in the presence of S. aureus and the GAPDH silencing probe, differentiated Odora cells and siREST probe (Diff. positive control + siREST), Odora cells differentiated in the presence of S. aureus and the siREST probe. Different letters denote statistically significant differences among treatments following 1-way ANOVA and Tukey’s post hoc test (P < 0.05).
Discussion
Detection of chemical cues in the environment is key to the survival and fitness of all vertebrate species. As a consequence, olfaction is one of the most ancient and conserved sensory systems in the vertebrate lineage (Treloar et al. 2010). Olfactory organs consist of millions of OSNs, the only neurons in the vertebrate body that are in direct contact with the external environment. Apart from receiving chemical stimuli, the olfactory epithelium of all vertebrates is exposed to complex microbial signals, such as those derived from the microbial communities residing on the mucosal surface of the olfactory epithelium.
Microbiota are known to influence diverse physiological systems in their hosts. Notably, microbiota have been shown to regulate host transcriptional programs in the intestinal mucosa (Rawls et al. 2006; Larsson et al. 2012; Camp et al. 2014; Dobson et al. 2016; Davison et al. 2017). A recent study in mice had shown that the expression of a few selected genes in the mouse olfactory epithelium was affected in the absence of microbiota (François et al. 2016). However, the global effects of microbiota on the olfactory system of vertebrates remained poorly understood. Here, we hypothesized that microbiota play a conserved role in the regulation of gene expression in olfactory systems of vertebrates from teleosts to mammals.
We first characterized the composition of the bacterial communities present in the zebrafish and mouse olfactory organs and compared them to a number of previously published studies from other vertebrates. We found that Proteobacteria dominated the nasal bacterial communities although similarities were not observed at shallower taxonomic resolution. We observed marked differences between nasal microbiota in C57Bl/6J mice (dominated by Proteobacteria, in particular Comamonadaceae) compared to C3H/HeN mice (dominated by Bacteroidetes and Firmicutes) (François et al. 2016). These differences in microbiota composition could be explained by differences in age and sex of the mice used in each of the 2 studies. Additionally, genetic evolutionary divergence between the 2 strains may play a role in the microbial colonization of mucosal surfaces (Hugenholtz and de Vos 2018). Further studies using identical housing conditions should be conducted in order to dissect the influence of the host genetic background on the nasal microbial communities.
Despite the differences in the nasal microbiota of zebrafish and mice, we show that both communities induce widespread transcriptional responses in the olfactory organs of their respective hosts. Interestingly, this common response was recorded even if larval zebrafish and adult mice were used in our study. Our comparative transcriptomic approach identified pathways and orthologs regulated similarly in zebrafish and mice, setting the stage for further mechanistic studies. Our analysis of 1-1 orthologs regulated by microbiota in both zebrafish and mouse identified 83 ortholog pairs that were regulated in the same direction, identifying them as a potential core host response. However, this was only about half of all orthologs differentially expressed in both hosts and therefore further studies will be needed to determine the basis for this partial overlap.
Among the transcriptome changes observed, GF animals had marked deficiencies in OR and VR expression. All vertebrates express OR but the OR multigene family is highly variable among different vertebrate species (Niimura 2012). Gene expression studies in zebrafish were coupled to ultrastructural observations and anatomical measurements. While we observed defects in the pseudostratification of the olfactory epithelium in GF zebrafish compared to CV controls, we did not detect changes in the overall size of the olfactory pit or the thickness of the ciliar layer. This is in disagreement with the architectural changes in the olfactory epithelium of GF mice previously reported, including a thinner ciliar layer and decreased proliferation rates (François et al. 2016). Since we did not quantify numbers of different OSN types in this study, further in vivo studies are necessary to ascertain whether the lowered OR gene expression levels detected in the absence of microbiota are due to defects in neuronal differentiation/maturation (and therefore different proportions of neuronal types) or overall decreased expression in OSNs. Additionally, further studies are needed to determine how these host–microbe interactions are affected by developmental stage, diet, gender, and maternal factors.
Increasing evidence indicates that regulation of TFs is a key mechanism by which microbiota influences host transcriptional programs. A number of studies have identified TFs regulated by the gut microbiota that control gene expression in intestinal epithelial cells and immune cells (Camp et al. 2014; Davison et al. 2017). The present study suggests that nasal microbiota also regulates olfactory transcriptomic responses via specific host TFs. Specifically, the TFs implicated here, including REST, are well known to play role in neuronal cell development.
The specific roles of REST in neuronal differentiation are complex—though initially described as a repressor of genes involved in neuronal differentiation, subsequent studies identified additional roles for REST suggesting it may also act as a transcriptional activator in differentiation (Tang 2009). Yet, no previous studies have investigated the control of REST by microbiota. We focused on REST due to the strong enrichment for REST-binding motifs found in putative cis-regulatory regions near genes downregulated by microbial colonization. In accord, REST expression was significantly reduced in both zebrafish and mouse olfactory organs. Importantly, we showed that REST is expressed in the olfactory system of both juvenile and adult mice and zebrafish. Since the present study uses 2 different developmental stages of the GF models (adult mice and larval zebrafish), detection of REST in olfactory tissues of both species at different developmental stages is important in order to interpret our results. Interestingly, we detected lower REST expression in the adult mouse olfactory organ compared to its CNS counterpart but comparable levels of REST expression in adult zebrafish olfactory organ compared to its CNS counterpart. These differences may account for the differences here reported regarding the effects that microbiota have on the olfactory organ transcriptional programs of zebrafish and mice. Further studies will address how microbiota regulate olfactory systems at different life stages of vertebrates.
The impact of bacterial exposure on REST expression in cultured Odora cells was more complicated. Odora cell differentiation induced by the nasal commensal S. aureus or standard differentiation media was accompanied by short-term suppression of REST expression. Future studies are needed to identify the specific bacterial stimuli that promote OSN differentiation in vivo. For instance, microbial-associated molecular patterns or other microbiota products may be detected by olfactory cells inducing secondary signals or processes that alter development; or microbes themselves may translocate into the tissue to evoke diverse immune and other host responses. The host signal transduction mechanisms that perceive and integrate those stimuli to regulate REST expression and function, and how those pathways are dynamically modified during differentiation of OSNs and the larger olfactory organ also deserve further investigation.
In the present study, we did not evaluate the functional downstream effects of GF conditions on the function of the olfactory organ. Electro-olfactogram studies of GF mice recently showed a global increase in the amplitude of responses to selected odorants as well as altered response kinetics (François et al. 2016). It is important to highlight that the latter study used different mice strains and methods to reconstitute the microbiota than the ones used in the present study. Thus, further studies are needed to determine the relationship between microbe-induced alterations in odorant receptor expression and physiological responses to odorants, and how those responses might be affected by variation in microbial community composition.
In summary, this work establishes that the transcriptional programs utilized by olfactory systems are under the influence of commensal microbiota, and that this is a common theme in vertebrate lineages that last shared a common ancestor approximately 420 million years ago. We implicate several TFs in this conserved response and provide functional evidence that microbial stimulation of Odora cell differentiation is dependent on REST, a TF known to have key biological roles in neuronal and glial development. Our results suggest that the ability of vertebrates to detect and discriminate odors and other chemicals in their environment through the olfactory organ is shaped by their microbiota. Further, the present study predicts that the alterations in microbiota that often occur as a function of host age, genotype, health, and other environmental exposures might influence olfactory perception and downstream physiologies and behavior.
Supplemental material
Supplementary data are available at Chemical Senses online.
Figure S1. Confirmation of microarray studies by qRT-PCR of mRNAs of 9 genes in the olfactory organ of GF and CV zebrafish (A) and mice (B). In zebrafish, selected genes were: dopamine receptor D4b (drd4b), transforming growth factor alpha (tgfa), DnaJ (Hsp40) homolog, subfamily B, member 12b (dnajb12b), Transient receptor potential cation channel, subfamily M, member 5 (trpm5), Odorant receptor, family F, subfamily 115, member 1 (or115-1), and G protein-coupled receptor 78 (gpr78b). Gene selection in mouse is the following: Trace amine-associated receptor 5 (Taar5), Vomeronasal type 2 receptor 123 (Vmn2r123), Nuclear factor of activated T-cells 3 (NF-ATc3), C-C motif chemokine receptor 2 (CCR2), Calmodulin 4 (Calm4), and Calreticulin 4 (Calr4). Asterisks denote statistically significant differences (P < 0.05).
Figure S2. Western blot showing defects in REST protein expression in the olfactory organ of 6 dpf REST−/− zebrafish larvae (REST mutant, lane 1) and the olfactory organ of 6 dpf wild-type zebrafish (lane 2) (1 pool N = 50).
Figure S3. Increased expression of OMP protein in differentiated Odora cells in response to S. aureus as revealed by anti-OMP antibody staining (red). (A) Undifferentiated Odora cells. (B) Odora cells differentiated with positive control medium (Diff. positive control). (C) Odora cells differentiated by incubating with live S. aureus cells. Cell nuclei were stained with DAPI (blue). (D) Mean percentage of OMP+ cells in each experimental group. Different letters denote statistically significant groups (P < 0.05). Results are representative of 2 independent experiments with N = 4 wells per experiment. (E) Dilution series experiment showing the increased percentage of OMP+ cells in Odora cells in response to several concentrations of S. aureus as assessed by immunofluorescence microscopy. Different letters denote statistically significant groups (P < 0.05). Results are representative of 2 independent experiments with N = 4 wells per experiment.
Table S1. Primers used in the present study.
Table S2. Bacterial community composition of adult zebrafish and mouse nasal microbiomes at the order level and at the genus level. Each column represents 1 individual. Only taxa with abundances >0.1% in at least 1 individual are shown.
Table S3. Gene list showing all mRNAs modified in the olfactory organ of GF compared to CV zebrafish and GF compared to CV mice. In this table the genes are ordered by fold change in expression level and genes shown were significant at P < 0.05 following unpaired t-test (P < 0.05) with Benjamini–Hochberg false discovery rate (FDR) correction and greater than 2-fold change. Asterisk next to the gene symbol indicates that the NRSF/REST transcription factor motif was detected in the promoter region of that gene.
Table S4. List of GO terms for biological process representing the features with significantly higher expression in the olfactory organ of GF compared to CV zebrafish (sheet 1), or features with significantly lower expression in the olfactory organ of GF compared to CV zebrafish (sheet 2). In this table the GOs are ordered by P-value in an ascending manner.
Table S5. List of GO terms for biological process representing the features with significantly higher expression in the olfactory organ of GF compared to CV mice (sheet 1), or features with significantly higher expression in the olfactory organ of GF compared to CV mice (sheet 2). In this table the GOs are ordered by P-value in an ascending manner.
Table S6. List of all ortholog genes identified in the one-to-one and one-to-many analyses in GF versus CV zebrafish and GF versus CV mouse olfactory organs.
Table S7. List of GO terms for biological process and KEGG pathways representing the orthologs 1 to 1 (152 genes) analysis from the olfactory organ of GF compared to CV zebrafish and mouse. In this table the GO and pathways are ordered by P-value in an ascending manner.
Acknowledgements
The authors are grateful to Balfour Sartor and the National Gnotobiotic Rodent Resource Center at the University of North Carolina at Chapel Hill for assistance with gnotobiotic mouse experiments. We thank Dr Mary Beth Genter for providing the Odora cell line, Dr Sirotkin for providing the 6 dpf REST mutant zebrafish larvae, and to Dr Hiroaki Matsunami and Dr Kevin Zhu for critical reading of the manuscript.
Funding
This work was supported by grants from the National Institutes of Health [P01-DK094779, R01-DK081426, P30-DK034987, P20-GM103452].
Conflict of interests
The authors declare no conflict of interests.
References
- Ache BW, Young JM. 2005. Olfaction: diverse species, conserved principles. Neuron. 48:417–430. [DOI] [PubMed] [Google Scholar]
- Bassis CM, Tang AL, Young VB, Pynnonen MA. 2014. The nasal cavity microbiota of healthy adults. Microbiome. 2:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercik P, Denou E, Collins J, Jackson W, Lu J, Jury J, Deng Y, Blennerhassett P, Macri J, McCoy KD, et al. 2011. The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology. 141:599–609. [DOI] [PubMed] [Google Scholar]
- Bonni A. 1997. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science. 278:477–483. [DOI] [PubMed] [Google Scholar]
- Bruce AW, Donaldson IJ, Wood IC, Yerbury SA, Sadowski MI, Chapman M, Göttgens B, Buckley NJ. 2004. Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc Natl Acad Sci USA. 101:10458–10463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp JG, Frank CL, Lickwar CR, Guturu H, Rube T, Wenger AM, Chen J, Bejerano G, Crawford GE, Rawls JF. 2014. Microbiota modulate transcription in the intestinal epithelium without remodeling the accessible chromatin landscape. Genome Res. 24:1504–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 7:335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves-Moreno D, Plumeier I, Kahl S, Krismer B, Peschel A, Oxley AP, Jauregui R, Pieper DH. 2015. The microbial community structure of the cotton rat nose. Environ Microbiol Rep. 7:929–935. [DOI] [PubMed] [Google Scholar]
- Chong JA, Tapia-Ramírez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G. 1995. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 80:949–957. [DOI] [PubMed] [Google Scholar]
- Cryan JF, Dinan TG. 2012. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 13:701–712. [DOI] [PubMed] [Google Scholar]
- Davison JM, Lickwar CR, Song L, Breton G, Crawford GE, Rawls JF. 2017. Microbiota regulate intestinal epithelial gene expression by suppressing the transcription factor hepatocyte nuclear factor 4 alpha. Genome Res. 27:1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson AJ, Chaston JM, Douglas AE. 2016. The Drosophila transcriptional network is structured by microbiota. BMC Genomics. 17:975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunston D, Ashby S, Krosnowski K, Ogura T, Lin W. 2013. An effective manual deboning method to prepare intact mouse nasal tissue with preserved anatomical organization. J Vis Exp. 78:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein P, Bozza T, Rodriguez I, Vassalli A, Mombaerts P. 2004. Axon guidance of mouse olfactory sensory neurons by odorant receptors and the beta2 adrenergic receptor. Cell. 117:833–846. [DOI] [PubMed] [Google Scholar]
- Fleischer J. 2009. Mammalian olfactory receptors. Front Cell Neurosci. 3:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- François A, Grebert D, Rhimi M, Mariadassou M, Naudon L, Rabot S, Meunier N. 2016. Olfactory epithelium changes in germfree mice. Sci Rep. 6:24687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, et al. 2005. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 15:1451–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand TW, Vujkovic-Cvijin I, Ridaura VK, Belkaid Y. 2016. Linking the microbiota, chronic disease, and the immune system. Trends Endocrinol Metab. 27:831–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen A, Anderson KT, Finger TE. 2004. Differential distribution of olfactory receptor neurons in goldfish: structural and molecular correlates. J Comp Neurol. 477:347–359. [DOI] [PubMed] [Google Scholar]
- He F, Ge W, Martinowich K, Becker-Catania S, Coskun V, Zhu W, Wu H, Castro D, Guillemot F, Fan G, et al. 2005. A positive autoregulatory loop of Jak-STAT signaling controls the onset of astrogliogenesis. Nat Neurosci. 8:616–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand JG, Shepherd GM. 1997. Mechanisms of olfactory discrimination: converging evidence for common principles across phyla. Annu Rev Neurosci. 20:595–631. [DOI] [PubMed] [Google Scholar]
- Hirota J, Mombaerts P. 2004. The LIM-homeodomain protein Lhx2 is required for complete development of mouse olfactory sensory neurons. Proc Natl Acad Sci USA. 101:8751–8755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu T, Saeki S, Yamamoto M, Iino Y. 2000. The Ras-MAPK pathway is important for olfaction in Caenorhabditis elegans. Nature. 404:289–293. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4:44–57. [DOI] [PubMed] [Google Scholar]
- Hugenholtz F, de Vos WM. 2018. Mouse models for human intestinal microbiota research: a critical evaluation. Cell Mol Life Sci. 75:149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra-Soria X, Nakahara TS, Lilue J, Jiang Y, Trimmer C, Souza MAA, Netto PH, Ikegami K, Murphy NR, Kusma M, et al. 2017. Variation in olfactory neuron repertoires is genetically controlled and environmentally modulated. eLife. 6:1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Waldvogel-Thurlow S, Darveau R, Douglas R. 2016. Differences in the paranasal sinuses between germ-free and pathogen-free mice. Int Forum Allergy Rhinol. 6:631–637. [DOI] [PubMed] [Google Scholar]
- Kaelberer MM, Buchanan KL, Klein ME, Barth BB, Montoya MM, Shen X, Bohórquez DV. 2018. A gut-brain neural circuit for nutrient sensory transduction. Science. 361: eaat5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallunki P, Edelman GM, Jones FS. 1997. Tissue-specific expression of the L1 cell adhesion molecule is modulated by the neural restrictive silencer element. J Cell Biol. 138:1343–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallunki P, Jenkinson S, Edelman GM, Jones FS. 1995. Silencer elements modulate the expression of the gene for the neuron-glia cell adhesion molecule, Ng-CAM. J Biol Chem. 270:21291–21298. [DOI] [PubMed] [Google Scholar]
- Kanther M, Sun X, Mühlbauer M, Mackey LC, Flynn EJ, Bagnat M, Jobin C, Rawls JF. 2011. Microbial colonization induces dynamic temporal and spatial patterns of NF-κB activation in the zebrafish digestive tract. Gastroenterology. 141:197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsunuma S, Honda H, Shinoda T, Ishimoto Y, Miyata T, Kiyonari H, Abe T, Nibu K, Takai Y, Togashi H. 2016. Synergistic action of nectins and cadherins generates the mosaic cellular pattern of the olfactory epithelium. J Cell Biol. 212:561–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knights D, Silverberg MS, Weersma RK, Gevers D, Dijkstra G, Huang H, Tyler AD, van Sommeren S, Imhann F, Stempak JM, et al. 2014. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med. 6:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolterud A, Alenius M, Carlsson L, Bohm S. 2004. The Lim homeobox gene Lhx2 is required for olfactory sensory neuron identity. Development. 131:5319–5326. [DOI] [PubMed] [Google Scholar]
- Koskinen K, Reichert JL, Hoier S, Schachenreiter J, Duller S, Moissl-Eichinger C, Schöpf V. 2018. The nasal microbiome mirrors and potentially shapes olfactory function. Sci Rep. 8:1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraner SD, Chong JA, Tsay HJ, Mandel G. 1992. Silencing the type II sodium channel gene: a model for neural-specific gene regulation. Neuron. 9:37–44. [DOI] [PubMed] [Google Scholar]
- Krismer B, Liebeke M, Janek D, Nega M, Rautenberg M, Hornig G, Unger C, Weidenmaier C, Lalk M, Peschel A. 2014. Nutrient limitation governs Staphylococcus aureus metabolism and niche adaptation in the human nose. PLoS Pathog. 10:e1003862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson E, Tremaroli V, Lee YS, Koren O, Nookaew I, Fricker A, Nielsen J, Ley RE, Bäckhed F. 2012. Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut. 61:1124–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CM, Price LB, Hungate BA, Abraham AG, Larsen LA, Christensen K, Stegger M, Skov R, Andersen PS. 2015. Staphylococcus aureus and the ecology of the nasal microbiome. Sci Adv. 1:e1400216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey L, Woodhams DC, Tacchi L, Salinas I. 2015. Topographical mapping of the rainbow trout (Oncorhynchus mykiss) microbiome reveals a diverse bacterial community with antifungal properties in the skin. Appl Environ Microbiol. 81:6915–6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyte M. 2013. Microbial endocrinology in the microbiome-gut-brain axis: how bacterial production and utilization of neurochemicals influence behavior. PLoS Pathog. 9:e1003726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K, Zheng S, Zuo Z. 2006. The transcription factor regulatory factor X1 increases the expression of neuronal glutamate transporter type 3. J Biol Chem. 281:21250–21255. [DOI] [PubMed] [Google Scholar]
- Mieda M, Haga T, Saffen DW. 1997. Expression of the rat m4 muscarinic acetylcholine receptor gene is regulated by the neuron-restrictive silencer element/repressor element 1. J Biol Chem. 272:5854–5860. [DOI] [PubMed] [Google Scholar]
- Monahan K, Schieren I, Cheung J, Mumbey-Wafula A, Monuki E, Lomvardas S. 2017. Cooperative interactions enable singular olfactory receptor expression. bioRxiv. 1:1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moravec CE, Li E, Maaswinkel H, Kritzer MF, Weng W, Sirotkin HI. 2015. Rest mutant zebrafish swim erratically and display atypical spatial preferences. Behav Brain Res. 284:238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori N, Schoenherr C, Vandenbergh DJ, Anderson DJ. 1992. A common silencer element in the SCG10 and type II Na+ channel genes binds a factor present in nonneuronal cells but not in neuronal cells. Neuron. 9:45–54. [DOI] [PubMed] [Google Scholar]
- Mulcahy ME, McLoughlin RM. 2016. Host-bacterial crosstalk determines Staphylococcus aureus nasal colonization. Trends Microbiol. 24:872–886. [DOI] [PubMed] [Google Scholar]
- Murrell JR, Hunter DD. 1999. An olfactory sensory neuron line, odora, properly targets olfactory proteins and responds to odorants. J Neurosci. 19:8260–8270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y. 2012. Olfactory receptor multigene family in vertebrates: from the viewpoint of evolutionary genomics. Curr Genomics. 13:103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm K, Belluardo N, Metsis M, Timmusk T. 1998. Neuronal expression of zinc finger transcription factor REST/NRSF/XBR gene. J Neurosci. 18:1280–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlato R, Mandl C, Hölzl-Wenig G, Liss B, Tucker KL, Ciccolini F. 2014. Regulation of proliferation and histone acetylation in embryonic neural precursors by CREB/CREM signaling. Neurogenesis. 1:e970883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham LN, Kanther M, Semova I, Rawls JF. 2008. Methods for generating and colonizing gnotobiotic zebrafish. Nat Protoc. 3:1862–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi IA, Gokhan S, Mehler MF. 2010. REST and CoREST are transcriptional and epigenetic regulators of seminal neural fate decisions. Cell Cycle. 9:4477–4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls JF, Mahowald MA, Ley RE, Gordon JI. 2006. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 127:423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls JF, Samuel BS, Gordon JI. 2004. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci USA. 101:4596–4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid KM, Patel S, Robinson AJ, Bu L, Jarungsriapisit J, Moore LJ, Salinas I. 2017. Salmonid alphavirus infection causes skin dysbiosis in Atlantic salmon (Salmo salar L.) post-smolts. PLoS One. 12:e0172856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. 2009. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 9:313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiva LR, Ahuja G, Ivandic I, Syed AS, Marioni JC, Korsching SI, Logan DW. 2015. Molecular and neuronal homology between the olfactory systems of zebrafish and mouse. Sci Rep. 5:11487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacchi L, Larragoite E, Salinas I. 2013. Discovery of J chain in African lungfish (Protopterus dolloi, Sarcopterygii) using high throughput transcriptome sequencing: implications in mucosal immunity. PLoS One. 8:e70650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacchi L, Musharrafieh R, Larragoite ET, Crossey K, Erhardt EB, Martin SAM, LaPatra SE, Salinas I. 2014. Nasal immunity is an ancient arm of the mucosal immune system of vertebrates. Nat Commun. 5:5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang BL. 2009. REST regulation of neural development: from outside-in? Cell Adh Migr. 3:1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmusk T, Palm K, Lendahl U, Metsis M. 1999. Brain-derived neurotrophic factor expression in vivo is under the control of neuron-restrictive silencer element. J Biol Chem. 274:1078–1084. [PubMed] [Google Scholar]
- Treloar HB, Miller AM, Arundhati R, Greer CA. 2010. Chapter 5 Development of the olfactory system. In: Menini A, editor. The neurobiology of olfaction. Boca Raton (FL): CRC Press/Taylor & Francis, 1–41. [Google Scholar]
- Villarino AV, Kanno Y, O’Shea JJ. 2017. Mechanisms and consequences of Jak-STAT signaling in the immune system. Nat Immunol. 18:374–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt WC, Storm DR. 2000. Odorants stimulate the ERK/mitogen-activated protein kinase pathway and activate cAMP-response element-mediated transcription in olfactory sensory neurons. J Biol Chem. 276:2047–2052. [DOI] [PubMed] [Google Scholar]
- Zhang G, Titlow WB, Biecker SM, Stromberg AJ, McClintock TS. 2016. Lhx2 determines odorant receptor expression frequency in mature olfactory sensory neurons. eNeuro. 3:ENEURO.0230-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Zhu M, Yu Y, Qiu L, Zhang Y, He L, Zhang J. 2016. Brain REST/NRSF is not only a silent repressor but also an active protector. Mol Neurobiol. 54:541–550. [DOI] [PubMed] [Google Scholar]
- Zheng Q, Wang XJ. 2008. GOEAST: a web-based software toolkit for gene ontology enrichment analysis. Nucleic Acids Res. 36:W358–W363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets generated during the current study are at the European Bioinformatics Institute archived under accession number E-MTAB-5045 and E-MTAB-5046 for zebrafish and mouse, respectively. Additionally, array data were submitted to ArrayExpress in compliance with MIAME. The zebrafish and mouse nasal 16S rRNA gene sequence results are deposited in NCBI under BioProject # PRJNA389614. Further information and requests for data and materials should be directed to and will be fulfilled by Irene Salinas (isalinas@unm.edu).