Being uniparentally inherited and with a high mutation rate, mitochondrial DNA (mtDNA) should relentlessly acquire mutations, eventually destroying a species through “mutational meltdown”. Lieber et al. now show that mitochondrial fragmentation is an essential first step in Drosophila , preventing functional complementation by wild-type molecules, and packaging mutations into mitochondria before they are destroyed by mitophagy.
Although the majority of mitochondrial proteins are synthesized in the cytoplasm from nuclear gene transcripts, thirteen essential polypeptides are encoded by small circles of DNA within the mitochondria: the mitochondrial genome (mtDNA).1 Unlike the nuclear genome, which has two copies in each somatic cell, multi-cellular animal cells contain hundreds or thousands of mtDNA molecules that are uniparentally inherited, usually down the maternal line. It is intriguing that these independent genetic systems interact by contributing proteins that must work together in two fundamentally important cellular processes: the mitochondrial ribosome, which is required for intra-mitochondrial protein synthesis; and the multi-complex oxidative phosphorylation system, which is the principal source of cellular adenosine triphosphate (ATP). To add a further twist, unlike nuclear DNA, mtDNA is continuously replicated independently of the cell cycle (relaxed replication). Although the mtDNA polymerase (polΥ) has a proofreading capacity, the sheer intensity of relentless replication means that mtDNA has a high mutation rate, and the relative paucity of mtDNA repair does not help the situation. In theory, this creates a ticking time-bomb, first recognized by Herman Muller in 1960s for asexual genomes, where—unchecked—a constant mutation rate will eventually destroy a species through mutational meltdown (called “Mullers Ratchet”).2
Fortunately (for us), there is evidence of a process that mitigates against the mutational ratchet, with the key events occurring during the development of the female germ line.1 New mtDNA mutations only affect a proportion of the mtDNA molecules (called heteroplasmy). The percentage level of an heteroplasmic mutation can change when cells divide, if different proportions of mutant and wild-type alleles segregate into daughter cells. Alleles segregate fast when the genetic pool is small, and slowly when the pool is large. In several vertebrate and invertebrate species, recent work has shown a profound reduction in the amount of mtDNA contained within individual germ cells. This leads to a genetic bottleneck, accelerating the segregation of heteroplasmy. For synonymous genetic variants, which do not alter the protein sequence, heteroplasmy levels can increase or decrease by random genetic drift.3 However, variants with functional effects undergo selection, with evidence of both negative selection, which ‘purifies’ the germ line,4 and positive or ‘selfish’ selection seen in several species, including Drosophila5 and most recently in humans.6 Understanding this process is important—in part because it is fascinating biology, but also because deleterious mutations can cause severe incurable diseases affecting ~1 in 5000 people: harnessing the mechanisms of selection would open new doors to prevent or treat these diseases.
Although we now understand when most of the selection is occurring, precisely how it occurs has remained a mystery, and in particular, whether it was occurring at the level of the individual cell, the level of the organelle, or even directly on the mitochondrial genome itself.7 For example, mammalian germ cells containing very high levels of a deleterious mutation may have limited capacity to proliferate, migrate and populate the developing gonad, or they may undergo apoptotic cell death. At the other extreme, in fruit flies, mutations that directly affect mtDNA replication could be outcompeted by “selfish” replicator genomes.5 However, in a recent Nature letter, Lieber et al.8 cast the spotlight on mitochondrial dynamics, a physiological process remodeling the mitochondrial network to optimize different cellular functions,9 and mitochondrial selective autophagy.
Studying Drosophila melanogaster and Drosophila yakuba, Lieber et al.8 observed a dramatic change in mitochondrial morphology during a critical period when selection is thought to occur, raising the possibility that mitochondrial fission was a key player. A reduction in mitochondrial size and interconnectivity, specifically observed in the germline cyst, was explained by a drastic reduction in both the transcript and protein levels of the outer mitochondrial membrane pro-fusion protein, mitofusin (mfn). Harnessing the power of genetics, they investigated the segregation of a heat-sensitive mtDNA mutation in the gene coding for cytochrome c oxidase (COX, complex IV of the respiratory chain) subunit I, which is known to undergo selection in the female germ line.10 Using fluorescent in situ hybridization (FISH), they then studied segregation of mtDNA heteroplasmy at the single cell level throughout oocyte development. By manipulating key genes that regulate mitochondrial dynamics (silencing mfn or overexpressing the main proponent of mitochondrial division, Dynamin-related protein 1, Drp1), they showed that the fragmentation of mitochondria is a key first step in the selection against the COX I variant. This mitochondrial size reduction leads to the compartmentalization of mutant mtDNA into smaller organelles, which prevents their mixing with wild-type mtDNA and exposes them specifically to destruction by selective autophagy, called mitophagy. The authors proposed that this mitochondrial quality control was triggered by a decrease in ATP synthesis from mitochondria containing mutant mtDNA leading to the activation of mitophagy, dependent of Atg1 (the fly ortholog of mammalian ULK1), regulator of the autophagosome biogenesis, and the mitophagic receptor, BNIP3 (a close ortholog of mammalian BNIP3L).
The findings of Lieber et al.8 take us an important step forward, but several important questions need to be addressed. How does BNIP3 target mitochondria based on organelle-specific defect in ATP synthesis? Are other mitophagic regulators involved, e.g., PINK1, as proposed by some?11 How many mtDNA molecules are present in each targeted mitochondrion (could it be one?), and is the packaging of mutations into organelles completely random, or does the mitochondrial network ‘select’ mutations for compartmentalization and destruction? These are difficult questions to answer, and high-resolution subcellular analyses will be required. Importantly, Lieber et al.8 move us forward in the right direction, by demonstrating heterogeneity within the cellular compartment imaging mtDNA heteroplasmy within mitochondria (Fig. 1).
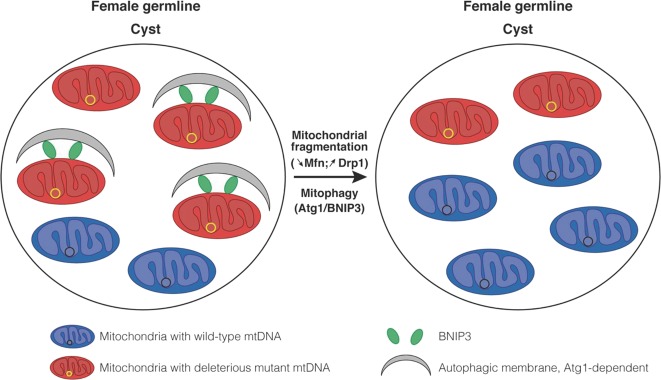
Fig. 1.
Proposed roles of mitochondrial fragmentation and quality control in mutant mtDNA selection in Drosophila germline. Selection of deleterious mutant mtDNA occurs during oocyte development, specifically in the cyst germline. Lieber et al.8 have found that compared to other germ and somatic cells, cyst harbored small, independent and fragmented mitochondria resulting from the lack of the pro-fusion factor Mitofusin (mfn). This mitochondrial size reduction leads to the compartmentalization of mutant mtDNA into smaller organelles, which prevents their mixing with wild-type mtDNA. Mitochondria containing deleterious mtDNA are marked for selection due to a decreased capacity of ATP synthesis. This leads to the mitochondrial recruitment of the mitophagy receptor, BNIP3, and the selective degradation of these mitochondria by autophagy in an Atg1-dependent mechanism, allowing mitochondria containing wild-type mtDNA to re-populate the germline. This new mechanism of mtDNA segregation can be positively or negatively regulated by manipulating key proteins which control either mitochondrial morphology (mfn and Drp1) or quality (BNIP3 and Atg1)
If this mechanism is a fundamental process that has evolved to counteract Muller’s ratchet, it stands to reason that it will also be important in mammals, as previously proposed for humans.7 However, several important questions remain. First, oocyte development in vertebrates is very different from flies, where the progenitors retain the capacity for self-renewal throughout life. By contrast, the full complement of vertebrate female germ cells are formed during in utero development, and they remain suspended in meiosis until selected for ovulation. Thus, there will be considerably more mtDNA replication in the fly ovary throughout life than in vertebrates, leading to a higher mutation burden in the female germ line. It is therefore possible that species-specific mechanisms have evolved to counteract this. On the other hand, similar discrete mitochondria have been observed around the time of the mtDNA genetic bottleneck in human embryos, each containing ~5 mtDNA,7 raising the possibility of a similar mechanism of selection. Second, how does inhibition of autophagy (Atg1 and BNIP3) lead to the selective decrease in wild-type mtDNA that they also observed, and without this affecting the mutational load? This implies a mechanism linking autophagic degradation with mtDNA replication, but only for the wild-type genome. This is difficult to explain based on current evidence. Finally, other mechanisms could still come in to play, including impaired protein translation in mitochondria containing mutant mtDNA tagged by PINK1.11
Although the endogenous selective mechanism appeared to be specific to the female germ line, Lieber et al.8 were able to reduce the mutational burden in somatic cells by solely inducing mitochondrial fragmentation. This led them to suggest that a similar strategy could ‘cleanse’ somatic cells of deleterious mutations, and thus provide a new treatment for mtDNA diseases. However, manipulating mitochondrial morphology and promoting mitochondrial fission will also trigger detrimental effects at both the mitochondrial and cellular levels. In addition, their data in flies—flies in the face of the human clinical evidence. There are several examples where patients harboring fragmented mitochondria (e.g., due to mutations in the inner mitochondrial membrane fusion protein OPA1), accumulate mtDNA mutations throughout life, leading to multi-system neurological phenotypes.12 This could be because, as discussed earlier, heteroplasmic mutations clonally expand more rapidly when the mtDNA pool is small. However, complete segregation into small units would prevent any one mutation from repopulating all of the mtDNA in the cell. Clearly, the relationship between mtDNA mutations and the dynamic fusing and dividing mitochondrial network is complex. That said, at least in Drosophila, de-fusing mitochondria appears to defuse Herman Muller’s mtDNA time-bomb.
Competing interests
The authors declare no competing interests.
References
- 1.Stewart JB, Chinnery PF. Nat. Rev. Genet. 2015;16:530–542. doi: 10.1038/nrg3966. [DOI] [PubMed] [Google Scholar]
- 2.Muller HJ. Mutat. Res. 1964;1:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- 3.Jenuth J, Peterson AC, Fu K, Shoubridge EA. Nat. Genet. 1996;14:146–151. doi: 10.1038/ng1096-146. [DOI] [PubMed] [Google Scholar]
- 4.Stewart JB, Freyer C, Elson JL, Larsson NG. Nat. Rev. Genet. 2008;9:657–662. doi: 10.1038/nrg2396. [DOI] [PubMed] [Google Scholar]
- 5.Ma H, O’Farrell PH. Nat. Genet. 2016;48:798–802. doi: 10.1038/ng.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei W, et al. Science. 2019;364:eaau6520. doi: 10.1126/science.aau6520. [DOI] [PubMed] [Google Scholar]
- 7.Floros VI, et al. Nat. Cell Biol. 2018;20:144–151. doi: 10.1038/s41556-017-0017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lieber Toby, Jeedigunta Swathi P., Palozzi Jonathan M., Lehmann Ruth, Hurd Thomas R. Mitochondrial fragmentation drives selective removal of deleterious mtDNA in the germline. Nature. 2019;570(7761):380–384. doi: 10.1038/s41586-019-1213-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tilokani L, Nagashima S, Paupe V, Prudent J. Essays Biochem. 2018;62:341–360. doi: 10.1042/EBC20170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hill JH, Chen Z, Xu H. Nat. Genet. 2014;46:389–392. doi: 10.1038/ng.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, et al. Mol. Cell. 2019;73:1127–1137. doi: 10.1016/j.molcel.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hudson G, et al. Brain. 2008;131:329–337. doi: 10.1093/brain/awm272. [DOI] [PubMed] [Google Scholar]