Although the blaOXA-58 gene has been infrequently described in Brazil, contrasting with other bordering South American countries, we verified the maintenance of this resistance determinant over time among carbapenem-resistant Acinetobacter species isolates, not only in nosocomial settings but also in the environment. In addition, to the best of our knowledge, this is the first study to have used WPS analysis to evaluate the genetic surroundings of blaOXA-58 in Brazil. Moreover, the A. seifertii and A. baumannii clinical strains evaluated in this study were recovered 17 years apart in hospitals located in distinct Brazilian geographic regions.
KEYWORDS: Acinetobacter calcoaceticus-Acinetobacter baumannii complex, Gram-negative bacilli, carbapenem resistance, carbapenem-hydrolyzing class D β-lactamase, nosocomial infection
ABSTRACT
We characterize by whole-plasmid-sequence (WPS) two-plasmid-borne blaOXA-58 obtained from Acinetobacter seifertii (Asp-1069) and A. baumannii (Acb-45063) clinical strains recovered 17 years apart from distinct Brazilian regions. Multilocus sequence type (MLST) analysis showed that the Asp-1069 and Acb-45063 strains belong to ST551 and ST15/CC15, respectively. WPS analysis demonstrated that blaOXA-58 was located in two distinct plasmids named pAs1069_a (24,672 bp/44 open reading frames [ORFs]) and pAb45063_b (19,808 bp/24 ORFs), which belong to the GR8/GR23 (repAci23) and GR4 (repAci4) incompatibility groups, respectively. The genetic environments surrounding blaOXA-58 revealed that it was flanked by two intact ISAba3 copies on pAb45063_b, which differed from pAs1069_a. In the latter, the upstream ISAba3 copy was truncated by insertion of ISAba825 element. Although Re27-specific recombination sites were found adjacent to ISAba3-blaOXA-58-ISAba3 arrangement on pAb45063_b, such structures were absent on pAs1069_a. The conserved ISAba125-araC1-lysE arrangement was disrupted by TnaphA6 harboring the aminoglycosides resistance gene aphA6 on pAs1069_a, while an IS26-blaTEM-1-aac(3)-IIa-IS26 genetic structure was found upstream from ISAba3-blaOXA-58-ISAba3 on pAb45063_b. Other two plasmids, pAb45063_a (183,767 bp/209 ORFs) and pAs1069_b (13,129 bp/14 ORFs), were also found in the OXA-58-producing Acinetobacter species strains, harboring the strA and strB genes and the sul2 gene, which confer resistance to streptomycin and sulfonamides, respectively. The plasmid-mediated virulence factors corresponding to genes tonB, spl, glmM, ppa, sulP, and map were found in both strains, as well distinct toxin-antitoxin system-encoding genes stbD and relE (pAs1069_a), brnT and brnA (pAb45063_b), and xreE (pAb45063_a). Although infrequently reported in Brazil, plasmid-borne blaOXA-58 showed a complex and diverse genetic backbone that confers stability in different Acinetobacter species that have been isolated from nosocomial settings over time.
IMPORTANCE Although the blaOXA-58 gene has been infrequently described in Brazil, contrasting with other bordering South American countries, we verified the maintenance of this resistance determinant over time among carbapenem-resistant Acinetobacter species isolates, not only in nosocomial settings but also in the environment. In addition, to the best of our knowledge, this is the first study to have used WPS analysis to evaluate the genetic surroundings of blaOXA-58 in Brazil. Moreover, the A. seifertii and A. baumannii clinical strains evaluated in this study were recovered 17 years apart in hospitals located in distinct Brazilian geographic regions.
INTRODUCTION
Acinetobacter species are important pathogens frequently responsible for causing nosocomial infections, mainly in patients hospitalized at intensive care units (ICU) (1, 2). The spread of major carbapenem-resistant A. baumannii clones has been associated with the increasing frequency of the carbapenem resistance phenotype worldwide (1). The production of carbapenem-hydrolyzing class D β-lactamases (CHDLs) has been reported to be the main mechanism of carbapenem resistance (3). The spread of multidrug-resistant (MDR) A. baumannii sequence type 79 (ST79) isolates carrying blaOXA-23 and, more recently, blaOXA-72 has contributed to the high carbapenem resistance rates (85%) observed in Brazil (3). In contrast, blaOXA-58 has been rarely reported in this country, contrasting with the high frequency of this CHDL-encoding gene seen in neighboring countries, mainly in Argentina (4). To date, only six OXA-58-producing Acinetobacter species strains recovered from human (5–8) and environmental (9) sources have been reported in distinct Brazilian cities since the 90s (5–9). The diverse genetic structures surrounding blaOXA-58 documented worldwide (3, 10, 11) play a major role not only in the mobilization of this resistance determinant but also in driving its expression, decisively leading to the carbapenem resistance phenotype (3, 12).
The carriage of blaOXA-58 by distinct Acinetobacter species recovered over a long period of time—3 decades—suggests that this CHDL-encoding gene has been mobilized by horizontal gene transfer (HGT), although the hypothesis of clonal spread could not be completely discarded. In order to reveal the genetic environment of blaOXA-58 and the corresponding implications for the mobilization and maintenance of this resistance determinant over time, we molecularly characterize two distinct plasmids harboring blaOXA-58 obtained from A. seifertii and A. baumannii clinical strains recovered in the years 1993 and 2010, respectively, from distinct Brazilian geographic regions.
RESULTS
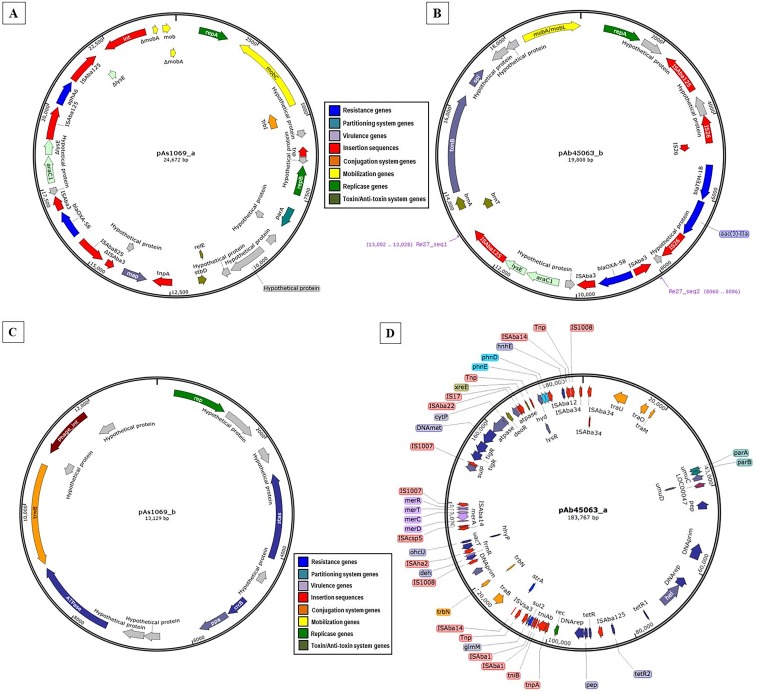
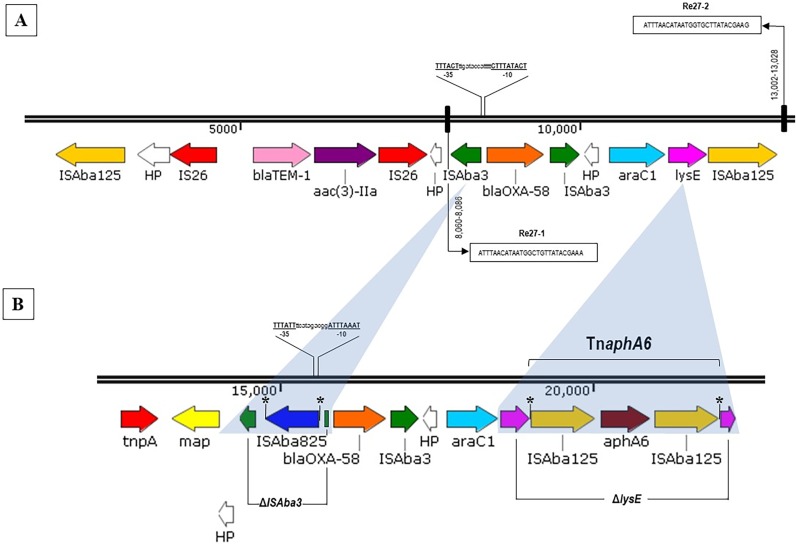
Two distinct plasmids were obtained for each strain according to WPS analysis, and the plasmids were named pAs1069_a (24,672 bp/44 open reading frames [ORFs]) and pAs1069_b (13,129 bp/14 ORFs) for the A. seifertii Asp-1069 strain that belongs to ST551IP (Institut Pasteur scheme). For A. baumannii Acb-45063 strain ST15/CC15IP, two plasmids were also detected and were named pAb45063_a (183,767 bp/209 ORFs) and pAb45063_b (19,808 bp/24 ORFs) (Table 1). Plasmids of similar sizes were observed by alkaline lysis gel analysis (data not shown), considering an accepted variation range of ±10%, as follows: ∼155-kb and ∼32-kb plasmids for strain Acb-45063, corresponding to pAb45063_a and pAb45063_b, respectively, and ∼32-kb and ∼21-kb plasmids for strain Asp-1069, corresponding to pAs1069_a and pAs1069_b, respectively. WPS analysis demonstrated that blaOXA-58 genes were carried by pAs1069_a/24,672-bp (Fig. 1A) and pAb45063_b/19,808-bp (Fig. 1B) plasmids, which belong to the A. baumannii replicon type group (AbGR) GR8/GR23 (repAci23) and GR4 (repAci4), respectively (Table 1). On pAb45063_b, blaOXA-58 was flanked by two intact copies of ISAba3 (Fig. 2A). In contrast, the genetic environment surrounding blaOXA-58 on pAs1069_a revealed an imperfect 5′ ISAba3 that was disrupted by an ISAba825 and an intact copy of ISAba3 downstream (Fig. 2B). Putative promoter regions of blaOXA-58 were predicted for pAb45063_b and for pAs1069_a, conferred by ISAba3 and by ISAba825, respectively (Fig. 2A and B). ISAba825 generates a 4-bp duplication (AACT) upon transposition (Fig. 2B).
TABLE 1.
Microbiological data and plasmid characterization of two OXA-58-producing Acinetobacter species clinical isolates recovered in Brazila
| Strain | Species | Yr of Isolation |
Clinical specimen |
MLST | MIC (μg/ml) |
Plasmid | Size (bp) |
ORF | G+C (%) |
AbGR | Genetic marker(s) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAZ | CRO | FEP | IPM | MEM | AMK | GEN | CIP | TIG | MIN | PMB | SUT | Resistance | Virulence | ||||||||||
| Asp-1069 | A. seifertii | 1993 | Tracheal aspirate | ST551 | 256 | 128 | 128 | 32 | 32 | 256 | 512 | 1 | 0.5 | 0.25 | 4 | >32 | pAs1069_a | 24,672 | 44 | 36.62 | GR8 | aphA6, blaOXA-58 | map |
| pAs1069_b | 13,129 | 14 | 35.65 | NT | ppa | ||||||||||||||||||
| Acb-45063 | A. baumannii | 2010 | Blood | ST15 | 256 | 512 | 64 | 32 | 16 | 8 | 512 | 64 | 1 | 0.25 | 0.06 | >32 | pAb45063_a | 183,767 | 209 | 37.61 | NT | strA, strB, sul2 | sulP, glmM |
| pAb45063_b | 19,808 | 24 | 38.51 | GR4 | aac(3)-IIa, blaTEM-1B, blaOXA-58 | tonB, sep | |||||||||||||||||
MLST, multilocus sequence typing; CAZ, ceftazidime; CRO, ceftriaxone; FEP, cefepime; IPM, imipenem; MEM, meropenem; AMK, amikacin; GEN, gentamicin; CIP, ciprofloxacin; TIG, tigecycline; MIN, minocycline; PMB, polymyxin B; SUT, trimethoprim-sulfamethoxazole; ORF, open reading frame; G+C, guanine-cytosine content; AbGR, Acinetobacter replicon type group; NT, nontypeable.
FIG 1.
Schematic representation of circular plasmid maps found in A. seifertii Asp-1069 (A and C) and A. baumannii Acb-45063 (B and D) strains. Arrows designate transcription directions of genes and ORFs. Genes were grouped according to their predictive functions as indicated by the color coding key.
FIG 2.
Genetic contexts surrounding blaOXA-58 found in plasmids pAb45063_b (A) and pAs1069_a (B). Genes and their transcriptional orientations are represented by horizontal arrows. Identical genes found in both genetic structures are represented with the same colors. Genes of no predicted functions (HP [hypothetical proteins]) are represented in white. The putative original promoters driving the expression of blaOXA-58 genes are highlighted. Direct repeat sequences are represented by an asterisk (*). Gene names preceded by an uppercase Greek delta (Δ) represent truncated genes, and the corresponding regions are shaded in gray. The Re27 regions are boxed. Promoter prediction for blaOXA-58 was performed using BPROM (SoftBerry).
Two Re27 sequences were found adjacent to an ISAba3-blaOXA-58-ISAba3 arrangement on pAb45063_b, with Re27-1 located upstream of 5′-ISAba3 and Re27-2 adjacent to ISAba125 located downstream of araC1 and lysE genes (Fig. 2A), which coded for a threonine efflux protein and a transcriptional regulator, respectively. In contrast, we failed to identify Re27-like regions on pAs1069_a. In this plasmid, lysE was disrupted by TnaphA6, which harbored aminoglycoside modifying enzyme (AME)-encoding gene aphA6. This AME confers resistance to gentamicin and amikacin. aphA6 was flanked by two copies of ISAba125 in the same orientation (Fig. 2B), while TnaphA6 generates a 7-bp duplication (ATTCGCC) upon transposition (Fig. 2B). The production of aminoglycoside O-phosphotransferase AphA6 by A. seifertii Asp-1069 justified the high MICs for amikacin (256 μg/ml) and gentamicin (512 μg/ml) verified in such strain (Table 1). In addition, a genetic arrangement composed of two copies of IS26 and the narrow-spectrum-β-lactamase-encoding gene blaTEM-1 and the AME-encoding gene aac(3)-IIa was also found upstream of ISAba3-blaOXA-58-ISAba3 on pAb45063_b (Fig. 2A). AAC(3)-IIa confers a high level of resistance to gentamicin but not amikacin, justifying the phenotype observed for A. baumannii Acb-45063 (MICs of 512 and 8 μg/ml for gentamicin and amikacin, respectively; Table 1). Other two plasmids (pAb45063_a and pAs1069_b) were also detected in the OXA-58-producing Acinetobacter species strains evaluated in the present study (Fig. 1C and D). The 183,767-bp plasmid Ab45063_a carried the streptomycin resistance genes strA and strB and the sulfonamide resistance gene sul2 (Fig. 1D), contrasting with the small plasmid pAs1069_b of 13,129 bp (Fig. 1C). Although the two OXA-58-producing Acinetobacter species strains showed similar profiles of susceptibility to β-lactams, A. baumannii Acb-45063 MICs were 0.06 μg/ml and 64 μg/ml for polymyxin B and ciprofloxacin, respectively, contrasting with those presented by A. seifertii Asp-1069 strains (MICs of 4 and 1 μg/ml for polymyxin B and ciprofloxacin, respectively) (Table 1).
Distinct virulence factors were observed in all four plasmids (Table 1) as follows: outer membrane protein-encoding gene tonB, septicolysin-encoding gene spl, phosphoglucosamine mutase-encoding gene glmM, inorganic pyrophosphatase-encoding gene ppa, sulfate permease-encoding gene sulP, and methionine aminopeptidase type I-encoding gene map. In addition, distinct toxin-antitoxin system-encoding genes were also detected in the two blaOXA-58-harboring plasmids (Fig. 1A and B), such as stbD and relE (pAs1069_a) and brnT and brnA (pAb45063_b). Although xreE was found in the large (183-kb) pAb45063_a plasmid (Fig. 1D), no toxin-antitoxin systems were found in the 13-kb pAs1069_b plasmid (Fig. 1C).
DISCUSSION
The plasmid carrying blaOXA-58, pAs1069_a, recovered from A. seifertii, shares 99% identity with the plasmid harboring blaOXA-58 pAb242_25 described in a MDR A. baumannii clinical strain (Ab242) isolated in the city of Rosario, Argentina (4). Interestingly, Ab242 strain was recovered in 1997 (11), 4 years later than the two clonally related OXA-58-producing A. seifertii clinical strains isolated in Brazil (8). Also, Narciso and colleagues described an OXA-58-producing A. seifertii strain (Ac-12.1) recovered in 2012 from a cloaca of a black-necked swan residing in the lakes of the São Paulo Zoo (9). This strain was clonally related to both A. seifertii clinical strains—including the Asp-1069 evaluated in the present study—isolated 19 years earlier in a tertiary hospital located in the city of São Paulo (8, 9). Since the genetic environment surrounding blaOXA-58 of Ac-12.1 was identical to that of the corresponding gene in the Asp-1069 strain, except for a truncated copy of 3′-ISAba3 (8, 9), it reinforces the idea of the capability of rearrangement and the complexity of transposable elements among plasmid-borne blaOXA-58 genes (3).
Although the OXA-58-producing A. baumannii Acb-45063 strain was included in ST15IP, it belongs to same clonal complex (CC15IP/CC103OX [Oxford scheme]) as the Ab242 ST104OX A. baumannii strain recovered in Argentina (4). Note that the city of Porto Alegre, where the Acb-45063 strain was recovered, is located in a Brazilian state next to the Argentinian border, where blaOXA-58 is prevalent and of public health concern (4, 10). However, the plasmids carrying blaOXA-58 detected in Argentinean and Brazilian A. baumannii strains showed distinct genetic backbones. Although plasmids carrying blaOXA-58 that belonged to GR8/GR23 were found among distinct Acinetobacter species in South American countries in the 1990s, a distinct genetic backbone surrounding blaOXA-58 was found in a GR4 plasmid from a A. baumannii Acb-45063 strain recovered in Brazil at 2010. According to Ravasi and colleagues, the presence of ISAba825 upstream of blaOXA-58, as observed in pAb45063_b (ΔISAba3/ISAba825-blaOXA-58-ISAba3), results in a hybrid promoter that overexpresses this CHDL, leading to 16-fold and 8-fold increases in the MICs for imipenem and meropenem, respectively (13). Re27-like sites found in pAb45063_b, but not in pAs1069_a, are short genomic sequences implicated in site specific recombination processes involved in the evolution of plasmids, many of them carrying CHDL-encoding genes (4, 10, 14, 15). These sequences have been identified bordering ISAba3-like elements, allowing the occurrence of multiple recombination processes that promote different arrangements and acquisition of blaOXA-58 by Acinetobacter species (4, 10–12, 14, 15).
Although it has been previously suggested that the presence of virulence-encoding genes does not guarantee the expression of virulence factors and/or bacterial pathogenicity (16), curiously, our study revealed the presence of distinct virulence factors in all plasmids evaluated, some of which had never been described before in Acinetobacter spp. (13, 15–17). The TonB outer membrane protein is associated with iron uptake, and its expression may be related to the survival of the bacterial cell in the lungs and blood (13, 16, 17). The spl gene encodes a septicolysin with cytolytic activity related to the invasion of tissues or cells, while glmM codes for a phosphoglucosamine mutase that has been related as a highly sensitive predictor of several clinical outcomes (13, 16, 17). Other three genes found, ppa, sulP, and map, have been associated with bacterial pathogenicity (4). In addition, distinct toxin-antitoxin system-encoding genes found in three of four plasmids evaluated ensure the stability of transferable genetic elements in the bacterial host cell (4, 15, 16).
In conclusion, a complex and dynamic backbones were found surrounding the blaOXA-58 carried by distinct plasmids from A. seifertii and A. baumannii strains recovered 17 years apart in Brazil. Such data demonstrated that although this CHDL-encoding gene has rarely been reported in Brazil, genetic plasticity has occurred over time, composed of a variety of resistance and virulence markers associated with the stability conferred by toxin-antitoxin systems. These findings accounted in part for the success of efforts that have kept plasmids carrying blaOXA-58 from escaping nosocomial settings for a long period of time.
MATERIALS AND METHODS
Ethical approval.
Ethical approval for this study was obtained from Research Ethics Committee from Federal University of São Paulo—UNIFESP/São Paulo Hospital (process number 5158010817).
Bacterial isolates.
Two OXA-58-producing Acinetobacter species clinical strains, Asp-1069 and Acb-45063, were selected for this study. The A. seifertii Asp-1069 strain was previously characterized (8) and is considered to be the most ancient Acinetobacter species carrying blaOXA-58 reported worldwide to date. Asp-1069 was recovered in 1993 from a tracheal aspirate of a patient hospitalized in the city of São Paulo, southeastern Brazilian region (8). The Acb-45063 strain was isolated in 2010 from a blood culture drawn from a patient hospitalized in the city of Porto Alegre, southern Brazilian region. For this study, the Acb-45063 strain was identified at the species level as A. baumannii by sequencing of partial regions of the RNA polymerase β subunit (rpoB) gene (18). The CHDL-encoding genes were confirmed by PCR followed by DNA sequencing using specific primers (2, 8, 9). MICs of 12 antimicrobial agents (Sigma-Aldrich, St. Louis, USA) were determined by cation-adjusted broth microdilution and interpreted according to European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines (http://www.eucast.org/clinical_breakpoints).
Multilocus sequence typing (MLST).
MLST analyses of Acb-45063 and Asp-1069 strains were performed by double-stranded DNA sequencing of internal regions of seven housekeeping genes (cpn60, fusA, gltA, pyrG, recA, rplB, and rpoB) following the Institute Pasteur scheme. Determination of the sequence type (ST) was performed through the A. baumannii MLST website (http://pubmlst.org/abaumannii/). The relationship between novel STs and existing STs was surveyed using the eBURST program (http://eburst.mlst.net/).
Plasmid DNA extraction using the alkaline lysis method.
For the calculation of the mean size of plasmids, the plasmid DNA extraction was performed by the alkaline lysis method according to the Birnboim and Doly protocol with a few modifications. One colony per bacterial isolate was inoculated into 3 ml of Trypticase soy broth (TSB) (Oxoid, Basingstoke, United Kingdom) in a tube and incubated at 37°C for 20 to 24 h. Aliquots of 1 ml each were subjected to centrifugation at 12,000 rpm for 3 min, and the pellet was resuspended in 100 μl of solution I (2 mg/ml lysozyme, 2% glucose, 10 mM EDTA, 25 mM Tris-HCl [pH 8.0], 1 mg/ml RNase). After 30 min of incubation in an ice bath, 200 μl of solution II (0.2 N NaOH and 1% SDS) was added. The supernatants were then homogenized by inversion and kept in an ice bath for 7 min. Then, 150 μl of solution III (3 M sodium acetate, pH 4.8) was added to the supernatants and homogenized by inversion and kept in an ice bath for 90 min for the sedimentation of chromosomal DNA. After that, the supernatants were centrifuged at 12,000 rpm for 10 min and transferred to new tubes, and 1 ml of ice-cold ethanol was added. The solution was homogenized by inversion to precipitate the plasmid DNA and incubated at –20°C overnight. The supernatants were centrifuged at 12,000 rpm for 10 min and resuspended in 100 μl of solution IV (100 mM sodium acetate, pH 8.0). Plasmid DNA was precipitated again by the addition of 200 μl of ice-cold ethanol and incubated at –20°C overnight. Finally, a new centrifugation was performed at 12,000 rpm for 10 min. The supernatants were discharged, and pellets were air dried and resuspended in 20 μl of sterile Mili-Q water. The plasmid DNA extractions were stored at –20°C. Electrophoresis was performed on 0.8% agarose (110 V/50 mA) for 2 h and stained with ethidium bromide. The calculation of the estimated plasmid sizes was based on a standard strain with known plasmid sizes running on the same agarose gel using a logarithmic curve.
Plasmid extraction and whole-plasmid sequencing (WPS).
For WPS, the pool of plasmids was extracted using a QIAPrep Spin MiniPrep extraction kit (Qiagen, Hilden, Germany), concentrated in a Concentrator Plus evaporator (Eppendorf, Hamburg, Germany), and then quantified on a digital Nanovue Plus spectrophotometer (GE Healthcare, Canada). For library preparation, the extractions were quantified again in a Qubit 3.0 Fluorometer (Thermo Fisher Scientific, DE, USA). Libraries were constructed using an Illumina TruSeq Nano DNA LT library preparation kit—set A (Illumina, CA, USA) generating ∼550-bp fragments. WPS analysis was performed in a MiSeq platform (Illumina, CA, USA) (2 × 300 bp) in paired-end mode. The quality and quantification of the libraries were evaluated by quantitative real-time PCR (qRT-PCR).
Plasmid assembly, automatic annotation, and manual validation.
First, plasmid reads were assembled using Newbler 3.0 (Kartchner, AZ, USA) and Ray 2.3.1 software (Université Laval, QC, Canada). The System for Automated Bacterial Integration of Annotation pipeline (SABIA; available in http://www.sabia.lncc.br) was used for gene prediction and automatic annotation with 90% coverage, 90% similarity, and an E value of <10−5. For manual validation, the following platforms were used: NCBI BLAST, UniProt, ISFinder, ResFinder 2.1, Plasmid Finder 1.3, MLSTFinder, and VirulenceFinder 1.5. Creation of the illustration of the circularized plasmids and in silico analysis of A. baumannii replicon type group (AbGR) (19) were performed using Snap Gene software 3.3.3 (GSL Biotech LLC, Chicago, USA). BPROM (Softberry Inc, New York, USA) was applied for predicting promoter sequences. The genetic structures surrounding blaOXA-58 genes were analyzed according to a study previously published by Poirel and Nordmann (20).
Data accessibility.
The complete nucleotide sequences of pAs1069_a, pAs1069_b, pAb45063_a, and pAb45063_b have been submitted to GenBank under accession numbers MK323040, MK323041, MK323042, and MK323043, respectively.
ACKNOWLEDGMENTS
We thank the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for providing grants to A.P.M., A.P.S., C.S.N., W.M.B.S.M., and A.C.N. We also thank the National Council for Science and Technological Development (CNPq) for providing a grant to A.C.G. (Process number 305535/2014-5). A.C.G. recently received research funding and/or consultation fees from Bayer, Eurofarma, Pfizer, MSD, and Zambon.
The rest of us have no conflicts of interest to declare. This study was not financially supported by any diagnostic/pharmaceutical company.
REFERENCES
- 1.Mugnier PD, Poirel L, Naas T, Nordmann P. 2010. Worldwide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii. Emerg Infect Dis 16:35–40. doi: 10.3201/eid1601.090852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vasconcelos AT, Barth AL, Zavascki AP, Gales AC, Levin AS, Lucarevschi BR, Cabral BG, Brasiliense DM, Rossi F, Furtado GH, Carneiro IC, da Silva JO, Ribeiro J, Lima KV, Correa L, Britto MH, Silva MT, da Conceição ML, Moreira M, Martino MD, de Freitas MR, Oliveira MS, Dalben MF, Guzman RD, Cayô R, Morais R, Santos SA, Martins WM. 2015. The changing epidemiology of Acinetobacter spp. producing OXA carbapenemases causing bloodstream infections in Brazil: a BrasNet report. Diagn Microbiol Infect Dis 83:382–385. doi: 10.1016/j.diagmicrobio.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Evans B, Amyes S. 2014. Oxa β-lactamase. Clin Microbiol Rev 27:241–263. doi: 10.1128/CMR.00117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cameranesi MM, Morán-Barrio J, Limansky AS, Repizo GD, Viale AM. 2018. Site-specific recombination at XerC/D sites mediates the formation and resolution of plasmid co-integrates carrying a bla(OXA-58)- and TnaphA6-resistance module in Acinetobacter baumannii. Front Microbiol 9:66. doi: 10.3389/fmicb.2018.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antonio CS, Neves PR, Medeiros M, Mamizuka EM, Elmor de Araújo MR, Lincopan N. 2011. High prevalence of carbapenem-resistant Acinetobacter baumannii carrying the blaOXA-143 gene in Brazilian hospitals. Antimicrob Agents Chemother 55:1322–1323. doi: 10.1128/AAC.01102-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Figueiredo DQ, Santos KR, Pereira EM, Schuenck RP, Mendonça-Souza CR, Teixeira LM, Mondino SS. 2011. First report of the bla(OXA-58) gene in a clinical isolate of Acinetobacter baumannii in Rio de Janeiro, Brazil. Mem Inst Oswaldo Cruz 106:368–370. doi: 10.1590/s0074-02762011000300019. [DOI] [PubMed] [Google Scholar]
- 7.de Souza Gusatti C, Bertholdo LM, Otton LM, Marchetti DP, Ferreira AE, Corção G. 2012. First occurrence of blaOXA-58 in Acinetobacter baumannii isolated from a clinical sample in southern Brazil. Braz J Microbiol 43:243–246. doi: 10.1590/S1517-838220120001000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cayô R, Rodrigues-Costa F, Pereira Matos A, Godoy Carvalhaes C, Dijkshoorn L, Gales AC. 2016. Old clinical isolates of Acinetobacter seifertii in Brazil producing OXA-58. Antimicrob Agents Chemother 60:2589–2591. doi: 10.1128/AAC.01957-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Narciso AC, Martins W, Cayô R, Pereira de Matos A, Santos SV, Ramos PL, Batista da Cruz J, Gales AC. 2017. Detection of OXA-58-producing Acinetobacter seifertii recovered from a black-necked swan at a zoo lake. Antimicrob Agents Chemother 61:e01360-17. doi: 10.1128/AAC.01360-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou S, Chen X, Meng X, Zhang G, Wang J, Zhou D, Guo X. 2015. “Roar” of blaNDM-1 and “silence” of blaOXA-58 co-exist in Acinetobacter pittii. Sci Rep 5:8976. doi: 10.1038/srep08976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopes BS, Al-Hassan L, Amyes SG. 2012. ISAba825 controls the expression of the chromosomal bla(OXA-51-like) and the plasmid borne bla(OXA-58) gene in clinical isolates of Acinetobacter baumannii isolated from the USA. Clin Microbiol Infect 18:E446–E451. doi: 10.1111/j.1469-0691.2012.03979.x. [DOI] [PubMed] [Google Scholar]
- 12.Villalón P, Valdezate S, Medina-Pascual MJ, Carrasco G, Vindel A, Saez-Nieto JA. 2013. Epidemiology of the Acinetobacter-derived cephalosporinase, carbapenem-hydrolysing oxacillinase and metallo-β-lactamase genes, and of common insertion sequences, in epidemic clones of Acinetobacter baumannii from Spain. J Antimicrob Chemother 68:550–553. doi: 10.1093/jac/dks448. [DOI] [PubMed] [Google Scholar]
- 13.Ravasi P, Limansky AS, Rodriguez RE, Viale AM, Mussi MA. 2011. ISAba825, a functional insertion sequence modulating genomic plasticity and blaOXA-58 expression in Acinetobacter baumannii. Antimicrob Agents Chemother 55:917–920. doi: 10.1128/AAC.00491-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silva L, Mourão J, Grosso F, Peixe L. 2018. Uncommon carbapenemase-encoding plasmids in the clinically emergent Acinetobacter pittii. J Antimicrob Chemother 73:52–56. doi: 10.1093/jac/dkx364. [DOI] [PubMed] [Google Scholar]
- 15.Fu Y, Jiang J, Zhou H, Jiang Y, Fu Y, Yu Y, Zhou J. 2014. Characterization of a novel plasmid type and various genetic contexts of blaOXA-58 in Acinetobacter spp. from multiple cities in China. PLoS One 9:e84680. doi: 10.1371/journal.pone.0084680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hujer AM, Higgins PG, Rudin SD, Buser GL, Marshall SH, Xanthopoulou K, Seifert H, Rojas LJ, Domitrovic TN, Cassidy PM, Cunningham MC, Vega R, Furuno JP, Pfeiffer CD, Beldavs ZG, Wright MS, Jacobs MR, Adams MD, Bonomo RA. 2017. Nosocomial outbreak of extensively drug-resistant Acinetobacter baumannii isolates containing blaOXA-237 carried on a plasmid. Antimicrob Agents Chemother 61:e00797-17. doi: 10.1128/AAC.00797-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Acosta J, Merino M, Viedma E, Poza M, Sanz F, Otero JR, Chaves F, Bou G. 2011. Multidrug-resistant Acinetobacter baumannii harboring OXA-24 carbapenemase, Spain. Emerg Infect Dis 17:1064–1067. doi: 10.3201/eid/1706.091866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.La Scola B, Gundi VA, Khamis A, Raoult D. 2006. Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species. J Clin Microbiol 44:827–832. doi: 10.1128/JCM.44.3.827-832.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bertini A, Poirel L, Mugnier PD, Villa L, Nordmann P, Carattoli A. 2010. Characterization and PCR-based replicon typing of resistance plasmids in Acinetobacter baumannii. Antimicrob Agents Chemother 54:4168–4177. doi: 10.1128/AAC.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poirel L, Nordmann P. 2006. Genetic structures at the origin of acquisition and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-58 in Acinetobacter baumannii. Antimicrob Agents Chemother 50:1442–1448. doi: 10.1128/AAC.50.4.1442-1448.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete nucleotide sequences of pAs1069_a, pAs1069_b, pAb45063_a, and pAb45063_b have been submitted to GenBank under accession numbers MK323040, MK323041, MK323042, and MK323043, respectively.