Abstract
Sensorineural hearing loss (SNHL) is the most common sensory disorder. Its underlying etiologies include a broad spectrum of genetic and environmental factors that can lead to hearing loss that is congenital or late onset, stable or progressive, drug related, noise induced, age related, traumatic or post-infectious. Habilitation options typically focus on amplification using wearable or implantable devices; however exciting new gene-therapy-based strategies to restore and prevent SNHL are actively under investigation. Recent proof-of-principle studies demonstrate the potential therapeutic potential of molecular agents delivered to the inner ear to ameliorate different types of SNHL. Correcting or preventing underlying genetic forms of hearing loss is poised to become a reality. Herein, we review molecular therapies for hearing loss such as gene replacement, antisense oligonucleotides, RNA interference and CRISPR-based gene editing. We discuss delivery methods, techniques and viral vectors employed for inner ear gene therapy and the advancements in this field that are paving the way for basic science research discoveries to transition to clinical trials.
Introduction
Hearing loss is the most common sensorineural deficit. It affects approximately 466 million people worldwide, 34 million of whom are children (http://www.searo.who.int/bangladesh/infographicworldhearingday2018.pdf?ua=1). By 2030, this number will have increased to nearly 630 million people, and by 2050, over 900 million people will have some degree of hearing loss (http://www.searo.who.int/bangladesh/infographicworldhearingday2018.pdf?ua=1). Its underlying etiologies are not surprisingly varied and, with advancing age, become increasingly more complex. By way of example, both genetic and environmental factors can lead to hearing loss that is congenital or late onset, stable or progressive, drug related, noise induced, age related, traumatic or post-infectious. On the otherwise healthy newborn with hearing loss, in which noise-induced, age-related and traumatic deafness do not occur, the etiology is most often genetic or infectious secondary to a prenatal cytomegalovirus infection.
Genetic hearing loss impacts about 1 in 500 newborns and over the course of a lifetime predisposes to or is directly responsible for ~ 50–60% of all deafness, with percentages higher in developed countries (1). In the pre-lingual and young pediatric population, hearing loss has a profound impact on communication and language acquisition and carries a social stigma that leads to economic and educational disadvantages and isolation throughout life. Early diagnosis and intervention remediate much of this effect, and as a result, universal physiologic newborn hearing screening has been widely implemented across the United States in order to identify deaf or hard-of-hearing newborns and reduce time-to-diagnosis and intervention (2).
In the past decade, genetic testing has emerged as the most important diagnostic test for evaluation of children with deafness. Establishing a genetic diagnosis provides essential information for understanding the underlying pathophysiology, and with the advent of comprehensive genetic testing, it is trivial to screen all genes known to be implicated in hearing loss simultaneously (3,4). The accumulation of genetic, genomic and clinical data is readily accessible through extensive databases such as ClinVar and Human Gene Mutation Database, both of which continuously curate the rapidly increasing volume of reported genetic variants, and the Deafness Variation Database, a deafness-specific open-access resource that integrates all available genetic, genomic and clinical data, together with expert curation to generate a single resource for clinical and research use (5).
Spurred by advances in our genetic understanding of sensorineural hearing loss (SNHL) and the notable successes in other specialties, interest in gene therapy for hearing loss has grown. Current habilitation options focus on hearing aids and cochlear implants, both of which bypass the biologic deficit by amplifying sounds in the case of hearing aids or by encoding them as electrical impulses that are transmitted to the auditory nerve through an electrode array in the case of cochlear implants. While these devices are very effective, they do not actually restore ‘normal’ hearing, making the development of novel therapeutics to restore or prevent hearing loss an important goal to enhance quality of life (6).
Genetic hearing loss is viewed as a relatively circumscribed and comparatively straightforward therapeutic ‘target’, as an established genetic diagnosis defines both the underlying pathophysiology and the essential problem to the ‘corrected’. In addition, the inner ear is an isolated site that can be accessed safely surgically and into which therapeutics can be delivered with minimal off-target systemic effects. In this review, we focus on recent advances in inner ear gene therapies for SNHL.
Methods for gene therapy—gene specific
Gene replacement
Gene replacement is arguably the most ‘straight forward’ form of gene therapy (Table 1) (7–26). Based on identifying and replacing the defective gene with a normal or wild-type copy, notable successes have been achieved treating patients with Leber’s congenital amaurosis and with hemophilia (27,28). Perhaps portending eventual success in the treatment of persons with hearing loss are a number of studies on murine models of Usher syndrome (12–15,17,18,29), Jervell and Lange–Nielsen syndrome (23) and a type of hearing loss caused by absence of vesicular glutamate transporter-3 (VGLUT3) (26).
Table 1.
Gene-specific reports of gene therapy in mouse models of deafness
| Gene | Gene therapy method | Genotype | Age at intervention | Length of follow up | Age at best ABR results | Click-evoked best ABR results | Tone-burst best ABR results | Reference | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 8kHz | 16kHz | 24kHz | 32kHz | ||||||||
| Tmc1 | GR | Homo | P0–2 | P84–90 | P28–30 | NR | 65 | 65 | 85 | 100 | Nist-Lund et al., 2019 (7) |
| Homo | P0–2 | P60 | P25–30 | NR | 85 | 100 | 120 | 120 | Askew et al., 2015 (8) | ||
| RNAi | Hetero | P15–16 | P140–146 | P56–62 | 40 | 50 | 85 | NR | 90 | Yoshimura et al., 2019 (9) | |
| Hetero | P0–2 | P245–251 | P28 | 40 | 20 | 30 | NR | 90 | Shibata et al., 2016 (10) | ||
| CRISPR/Cas9 | Hetero | P1 | P28–34 | P28–34 | NR | 55 | 60 | 65 | 80 | Gao et al., 2018 (11) | |
| Clrn1 | GR | Homo | P1 | P28–34 | P28–34 | NR | 85 | 90 | 95 | 95 | Gyorgy et al., 2019 (12) |
| Homo | P1–3 | P60 | P22–24 | NR | 30 | 30 | 35 | 50 | Dulon et al., 2018 (13) | ||
| Homo | P1–3 | P70 | P22 | 45 | 35 | 35 | NR | 40 | Geng et al., 2017 (14) | ||
| Ush1c c.216G>A | GR | Homo | P1 | P168–174 | P42–48 | NR | 50 | 50 | 80 | 95 | Pan et al., 2017 (15) |
| ASOs | Homo | P5 | P84–90 | P28–34 | 45 | 45 | 45 | NR | 70 | Lentz et al., 2013 (16) | |
| Whrn | GR | Homo | P1–5 | P90 | P30–90 | NR | 95 | 95 | NR | 95 | Chien et al., 2016 (17) |
| Homo | P1–5 | P120 | P30 | NR | 60 | 80 | NR | 80 | Isgrig et al., 2017 (18) | ||
| Otof | GR | Homo | P10 | P210–216 | P28–34 | 35 | 65 | 65 | NR | 60 | Akil et al., 2019 (19) |
| Homo | P6–7 | P30 | P23–30 | 50 | 70 | 90 | 90 | 90 | Al-Moyed et al., 2019 (20) | ||
| Lhfpl5 | GR | Homo | P0–1 | P28–34 | P28–29 | NR | 70 | 90 | 85 | 100 | Gyorgy et al., 2017 (21) |
| MsrB3 | GR | Homo | E12.5 | P56–62 | P28 | 25 | 25 | 30 | 40 | 40 | Kim et al., 2016 (22) |
| Kcnq1 | GR | Homo | P0–2 | P210–216 | NR | 45 | 55 | 65 | 75 | 80 | Chang et al., 2015 (23) |
| Gjb2 | GR | Homo | P0 | P84–90 | P70–84 | NR | NR | NR | 65 | NR | Iizuka et al., 2015 (24) |
| Homo | P0–1 | P28–34 | P28–34 | NR | 85 | 85 | 85 | 85 | Yu et al., 2014 (25) | ||
| Vglut3 | GR | Homo | P1 | P483–489 | P50–52 | 40 | 65 | 55 | NR | 65 | Akil et al., 2012 (26) |
ABR, auditory brainstem response; GR; gene replacement; NR, not recorded/no report.
The first successful inner-ear gene therapy study treated mice homozygous for the targeted deletion of VGLUT3. These mice are born deaf but exogenous replacement of VGLUT3 and its overexpression in inner hair cells (IHCs) mediated by adeno-associated virus 1 (AAV1) leads to sustained hearing recovery, partial restoration of ribbon synapse morphology and a startle response. Interestingly, although transgene expression of VGLUT3 within the inner ear was not specific solely to IHCs, the observed phenotypic rescue reflected improved IHC function, suggesting that cell-specific transduction may not be an absolute necessity in all instances. Two important limitations of this study are the fact that autosomal recessive non-syndromic hearing loss caused by VGLUT3 has not yet been described in humans. The observed utility of this approach, which was successful at P1 for the duration of the study but showed a variable level of rescue at later time points such as P10–12, remains to be established. This temporal difference is relevant because, in the P1 mouse, inner ear maturation is occurring and auditory function does not fully mature until about P15. In contrast, humans are born with mature inner ears. This difference confounds any inferences murine results may have for human deafness unless gene therapy is delivered to the mature murine ear.
Gene suppression—antisense oligonucleotides
Antisense oligonucleotides (ASOs) are modified nucleic acid sequences that bind to complementary RNA sequences by Watson–Crick base pairing. They regulate gene expression by two primary mechanisms that are dependent on their chemical properties and target (30). The first, ASO knock down, occurs with ribonuclease-H (RNase-H) cleavage of the RNA strand from the RNA–DNA duplex, with resultant degradation of the mRNA. The second, splice site switching, occurs when ASOs interfere with alternative splicing by targeting splice sites, exons or introns, resulting in exclusion or inclusion of specific exons (31). To date, five ASOs are approved by the US Food and Drug Administration and many clinical trials are underway. The first-approved ASO, fomivirsen, is used for the treatment of cytomegalovirus-induced retinitis in patients with acquired immune deficiency syndrome (32).
Lentz et al. (16) have reported on the utility of ASO treatment in the murine inner ear to rescue the USH1C 216G>A (216A) mutation in a mouse model of USH1C. This founder mutation in the Acadian population leads to a cryptic 5′ splice site, which is used in preference of the authentic 5′ splicing site of exon 3. The result is a frameshift and truncated harmonin protein. The Usher syndrome mouse model used in the study was a knock-in based on the human 216A mutation—Ush1c c.216G>A. The group designed an ASO-29 to redirect cryptic splicing of 216A pre-mRNA to the authentic site.
ASO-29 injected into homozygous (216AA) and heterozygous (216GA) knock-in mice intraperitoneally at P3 and P10 led to near-normal hearing thresholds in broad band noise and 8 and 16 kHz pure tones in 216AA mice treated between P3 and P5. Hearing threshold at 32 kHz could not be rescued, and over the course of 3 months there was a gradual decline in thresholds at 8 and 16 kHz. The vestibular dysfunction was also rescued. 216AA Mice treated at P10 showed significantly higher thresholds than animals treated at P3–5 but significantly better thresholds than untreated or control-ASO-treated animals in both broad band noise and at 8 kHz. Histological assessment of ASO-29-treated inner ears demonstrated rescue of outer hair calls (OHCs) and IHCs at P1 but not at P5 (33).
These observations may reflect two important limitations of ASO at least in this animal model and for this indication—a therapeutic time window in the USH1C mouse before which therapy must be delivered to be effective and lack of a sustained response.
Gene suppression—RNA interference
RNA interference (RNAi) is a biological process in which RNA molecules inhibit gene expression or translation by neutralizing specifically targeted messenger RNA (mRNA) molecules. Its therapeutic use typically focuses on one of two types of small RNA molecules—small interfering RNA (siRNA) or microRNA (miRNA)—which represent a novel class of therapeutic agents with the potential to treat a wide range of disorders, including cancers and infections. Several clinical trials using siRNA- and miRNA-based drugs have been initiated, and one RNAi-based therapeutic is now available for the treatment of hereditary transthyretin-mediated amyloidosis in adults (34).
While siRNAs and miRNAs share many similarities—for example, both are short duplex RNA molecules that exert gene silencing effects at the post-transcriptional level—their mechanisms of action and clinical applications are different. siRNAs tend to be more specific than miRNAs. Once an siRNA enters a cell, it associates with a ribonucleoprotein complex known as RISC (RNA-induced silencing complex), which uses one strand of the siRNA molecule as a template to recognize a specific mRNA transcript. The identified mRNA molecule is then cleaved by Argonaute 2, a protein in the RISC complex, thereby preventing it from serving as a translational template (35,36).
miRNAs are in general less specific and typically regulate the expression of multiple genes (37). They are derived from a non-coding RNA primary transcript (pri-miRNA) that is processed in the cell nucleus into a stem-loop pre-miRNA structure by Drosha, an RNase III enzyme, and DGCR8, a double-stranded RNA-binding protein. The pre-miRNA is then exported to the cytoplasm by Exportin 5 using GTP bound to the Ran protein. In the cytoplasm, the dsRNA portion of pre-miRNA is cleaved by Dicer to produce a mature miRNA molecule that can be integrated into the RISC complex. At this point, miRNA and siRNA share the same downstream intracellular machinery.
The breadth of miRNA activity is derived from its partially complementarity to multiple mRNAs (37–39). However, artificial miRNAs can be designed to base pair perfectly with selected RNA targets, thereby inducing cleavage of specific mRNA molecules. These artificial miRNAs, like designer siRNAs, provide RNAi-based gene-specific and even allele-specific mRNA silencing (40). In proof-of-concept studies relevant to hearing loss, Maeda et al. (41) used siRNAs to suppress expression of an exogenous deafness-inducing pathogenic variant of GJB2 thereby preventing hearing loss. Building on these results, Shibata et al. (10) designed an artificial miRNA to specifically target the mutation-carrying TMC1 allele in the Beethoven mouse, a murine model of human autosomal dominant non-syndromic hearing loss at the DFNA36 locus, and slowed the progressive hearing-loss phenotype in mice treated at P1–2. A follow-up study by Yoshimura et al. (9) treated older animals, and although progression of hearing loss could also be slowed in animals treated at P15 and P30, the results were not as dramatic, and in animals treated at P60, no effect was observed. These findings suggest that, for TMC1-related deafness, the opportunity to intervene using RNAi is temporally defined and, beyond a specific time point, targeted allele suppression has no effect.
Gene editing: CRISPR/Cas9
Targeted genome editing has emerged as a powerful tool for biological research. Although there are three major programmable nucleases—ZFNs (zinc finger nucleases), TALENs (transcriptional activator-like effector nucleases) and CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 (CRISPR-associated protein 9)–ZFNs, and TALENs require large DNA fragments (500–1500 bp), while Cas9 can identify a target sequence with only 20 bp of guide RNA (gRNA). CRISPR/Cas9 can also be used for multiplexing by delivering multiple gRNAs to target multiple genes in the same cell at the same time (42). This efficiency and flexibility has propelled CRISPR/Cas9 to the forefront as a tool for prevention and treatment of human disease.
Recognized challenges include off-target effects, mutations, editing efficiency and uncertainty of single-site target selectivity in complex genomes; however, several methods have been developed to address these challenges (43–49). For example, Hsu et al. (44) have demonstrated that the concentration of single-gRNA and Cas9 can be limited to improve the on-target effect and Kim et al. (47) have developed RNA-guided engineered nucleases to reduce the frequency of producing off-target indels.
As applied to auditory research, Gao et al. (11) injected Cas9–gRNA–lipid complexes targeting the mutant TMC1 allele into P1 Beethoven mice and substantially reduced progression of hearing loss. Significant hearing preservation was detected from 8 to 23 kHz with average ABR thresholds 15 dB lower for treated ears as compared to untreated contralateral ears. This study was the first of its kind to demonstrate the potential of CRISPR/Cas9 for the treatment of autosomal dominant hearing loss related to hair cell dysfunction.
Similar to the aforementioned gene therapy methods, the therapeutic time window in this mouse model is in the early postnatal period for this therapy to be effective. Further longitudinal studies will be necessary to assess the longevity of the treatment effect.
Methods for gene therapy—non-specific
Cell replacement: stem cell-based therapy
While mammalian cochlear hair cells lack regenerative capacity, avian cochlea hair cells do not and following acoustic trauma can be replaced by mitosis of supporting cells (50–53) (Table 2) (50,51,54–75). Attempts to regenerate hair cells using progenitor cells have explored trans-differentiation of supporting cells into HCs and mitosis of supporting cells (54,55). Both strategies depend on the condition of the extant supporting cells, with concomitant intracochlear drug delivery required to direct outcome (76,77). Studies in newborn mice suggest that ‘naïve’ cells can be used to ‘jump-start’ the formation of supporting cells and their transformation into hair cells, as naïve cells respond better to molecular cues; however, the potential for trans-differentiation is temporally limited (78,79). Hair cell loss months or years earlier precludes this possibility perhaps in part due to loss of molecular interactions between hair cells and supporting cells (80).
Table 2.
Stem-cell based therapies
| Methods | Sub methods | Cells used | End result | In vivo experiment | References |
|---|---|---|---|---|---|
| Inner ear stem cell induction | Direct transdifferentiation | SCs | 1 HC | Adler et al.,1997 (54); Roberson et al., 2004 (55) | |
| Mitosis of SCs | SCs, daughter cells | 1 SC and 1 HC | Corwin and Contanche, 1988 (50); Ryals and Rubel, 1988 (51) | ||
| Cell transplantation | Embryonic stem cells | mESCs | Inner HCLC | Li et al., 2003 (56) | |
| mESCs | Mechanosensitive inner ear HCLC | Oshima et al., 2010 (57) | |||
| mESCs | SCs, mechanosensitive HCLC, neurons | Koehler et al., 2013 (58) | |||
| mESCs | iHCs with HC specific markers and protrusions reminiscent of stereociliary bundles | Costa et al., 2015 (59) | |||
| hESCs | ONPs, OEPs | Transplanted into adult gerbils;ABR improved | Chen et al., 2012 (60) | ||
| hESCs | iHCs with HC specific markers and protrusions reminiscent of stereociliary bundles | Ronaghi et al., 2014 (61) | |||
| hESCs, hiPSCs | Neurons, SCs, functional HCLCs in 3D culture | Koehler et al., 2017 (62) | |||
| ASCs | BM-MSCs | SGNs | Transplanted into adult gerbils | Matsuoka et al., 2007 (63) | |
| BM-MSCs | Fibrocyte-like cells | Transplanted into young and adult mice; migrated MSCs seen only in young mice | Kasagi et al., 2013 (64) | ||
| Adult human MSCs | SGNs | Bas et al., 2014(65) | |||
| HUMSCs | Unknown | Transplanted into albino deaf pig; new ABR waves appeared after injection | Ma et al., 2016 (66) | ||
| ratMSCs | HCs | Pre-injected by deferoxamine to home MSCs in noise induced injured cochlea | Peyvandi et al., 2018 (67) | ||
| BM-MSCs | Assess adverse effect | No effect on ABR or DPOAE in rats | Mittal et al., 2019 (68) | ||
| HL-MSCs | Cochlear nerves | Transplanted into intracranial cochlear nerve trunk; ABR improved | Chen et al., 2019 (69) | ||
| Induced pluripotent stem cells | miPSCs | SGNs | Transplanted into mouse cochlea | Nishimura et al., 2009 (70) | |
| miPSCs | Mechanosensitive inner ear HCLC | Oshima et al., 2010 (57) | |||
| hiPSCs | HCLCs | Ohnishi et al., 2015(71) | |||
| hiPSCs from human MYO15A patients | HCLCs | Corrected mutation by CRISPR/Cas9; functional and morphological rescue | Chen et al., 2016 (72) | ||
| hiPSCs from human MYO7A patients | HCLCs | Same as above | Tang et al., 2016 (73) | ||
| miPSCs | Outer sulcus cell-like cells | Transplanted into mouse otocysts at E11.5 | Takeda et al., 2017 (74) | ||
| hiPSCs | OEPs, HCLCs | Transplanted into mouse cochlea; synaptic connection with SGNs | Chen et al., 2018 (75) |
SC, Supporting cell; HC, Hair cell; HCLC, Hair cell-like cell; mESC, mouse embryonic stem cell; hESC, human embryonic stem cell; iHC, induced hair cell; ONP, otic neural progenitor; MSC, mesenchymal stem cell; BM-MSC, bone marrow-derived MSC; HL-MSC, human limbus-MSC; HUMSC, human umbilical cord MSC; ABR, auditory brainstem response; DPOAE, distortion product otoacoustic emissions; PSC, pluripotent stem cells; hiPSC, human-induced PSC; miPSC, mouse-induced PSC.
As an alternative, stem cell transplantation has been explored using embryonic stem cells (ESCs), adult-derived stem cells (ASCs) or induced pluripotent stem cells (iPSCs) as seeds from which to generate HCs. ESCs have the advantage of pluripotency and can be differentiated into hair cell-like cells (57), otic sensory neurons (81) and spiral ganglion neuron (SGN)-like cells (82); however, the available pool is limited and their use raises ethical issues. Although ASCs obviate these ethical considerations, they are of limited pluripotency, and as a result, most studies have focused on iPSCs.
Several laboratories have reported stunning successes using iPSCs to generate hair cells that are even responsive to mechanical stimulation. Chen et al. (75), for example, used human iPSCs to generate otic epithelial progenitors (OEPs) and investigated their migration, differentiation and synaptic connections in mouse cochlea. In vitro, OEP-derived hair cell-like cells formed synaptic connections with SGNs in co-culture. In vivo, a few OEPs migrated into the organ of Corti and differentiated into hair cells that established connections with native SGNs (75).
While these results offer the potential promise for cell-based therapies for hearing loss in the future, there are major challenges to consider. First, regenerated hair cells more closely resemble vestibular rather than cochlear hair cells (58,62,83–86); second, after culture, the cells must be introduced into the inner ear and then self-insert into the proper location in the membranous labyrinth (87,88); and third, the carcinogenic potential of stem cells must be carefully followed (89,90). To put these challenges into perspective, it is worth noting that mice are deaf even if all hair cells form but are disorganized.
Goals and timing of therapy
The goal of gene therapy is hearing preservation or restoration. To achieve this goal, an exquisite understanding of normal and abnormal inner ear development and function is required. Included in this understanding is cross-talk between HCs and SGNs, the complexities of which are only beginning to be appreciated. Sun et al. (91), for example, have recently shown that active mechanotransduction channels in HCs are critical to shape the spontaneous firing patterns in SGNs prior to the hearing onset. This activity is initiated at least in part by the spontaneous release of ATP from supporting cells, which causes HCs to depolarize and release glutamate, triggering discrete bursts of action potentials in primary auditory neurons. Subtype specification of SGNs is thus initiated in the pre-hearing period coincident with the timeframe in which spontaneous activity in SGNs is observed. Refinement of SGN subtypes continues into the fourth week after birth, suggesting that sensory input drives some aspects of SGN specification.
Maturation of ribbon synapses also continues until the fourth postnatal week (92–94), overlapping the period when murine SGNs refine their firing properties from an immature to a mature state in which there is a range of fibers with different spontaneous firing rates (95), indicating an intricate interplay between molecular and functional diversification (91). This study raises questions about cochlear-directed gene therapy and whether studies focused solely on the organ of Corti may fail because of an unrecognized need to address SGN function. This interrelationship is especially apt to be important with age-related and noise-induced hearing loss.
Viral vectors
Viral vectors are the workhorse for cochlear gene therapy, with studies exploring the use of adenovirus (96–99), AAV (7–9,12,15,17–23,25,77,100–117). helper-dependent adenovirus (118), herpes simplex virus (119–121), vaccinia virus (121), sendai virus (122) and lentivirus (111,114,123,124). Of all these choices, AAV has emerged as the most attractive vector for cochlear gene delivery. Belonging to the Parvoviridae family genus Dependovirus, AAV is a small virus (25 nm) that lacks pathogenicity and has minimal immunogenicity (125). It transduces both non-dividing and dividing cells to provide stable long-term gene expression by persisting as an episome without chromosomal integration (125).
The major disadvantage of AAV is its low viral capacity (4.7 kb), which is halved when using a self-complementary AAV (125). This limitation can be overcome by dual injection methods at the expense of transduction efficiency (116). In a recent report, this approach was used to deliver Otof (6 kb) into the inner ear of mice lacking this gene, with restoration of hearing (19).
In the inner ear, AAVs demonstrate broad tropism, stable gene expression and little to no ototoxicity (126). Transgene expression is affected by several factors including serotype, age at treatment, method of delivery, titer, promoter type and the presence or absence of enhancers (such as WPRE). Significant effort has defined the tropism of AAV subtypes relative to different cell types in the murine cochlea (Table 3).
Table 3.
AAV transduction in vivo
| Reference | Mouse model | AAV model | Delivery method | Age at delivery | Transduction rate of IHCs (%) | Transduction rate of OHCs (%) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Apex | Middle | Base | Apex | Middle | Base | |||||
| Gyorgy et al., 2019 (12) | BL6 | 9PHP | RWM | P1 | 70 | 70 | 50 | 40 | 35 | 35 |
| CAA | 4 weeks | NR | 0 | 0 | 0 | |||||
| CD1 | RWM | P1 | 70 | 60 | 70 | 50 | 40 | 40 | ||
| Clrn | 63 | 54 | 39 | 36 | 31 | 9 | ||||
| Gu et al., 2019 (100) | ICR | 2/2 | CO | P2–3 | Few | few | ||||
| 2/9 | 39.6 ± 16.3 | 52.7 ± 5.7 | 78.3 ± 7.2 | 14.4 ± 0.9 | 15.0 ± 3.0 | 100 | ||||
| Anc80 | 100 | 100 | 100 | 93.5 ± 7.2 | 95.6 ± 4.4 | 81.6 ± 11.4 | ||||
| Kim et al., 2019 (101) | ICR | 2/DJ | RWM | P2 | 52 | 41 | 37 | 90 | 88 | 37 |
| 2/DJ8 | 59 | 24 | 10 | <10 | <10 | <10 | ||||
| 2PHP | 86 | 81 | 62 | 63 | 31 | 16 | ||||
| Akil et al., 2019 (19)a | Otof −/− | AAV2 quadY-F capsid | RWM | P2 | 78 ± 6 | 0 | ||||
| P10 | 64 ± 6 | 0 | ||||||||
| P17 | 82 ± 9 | 0 | ||||||||
| P30 | 85 ± 7 | 0 | ||||||||
| Isgrig et al., 2019 (102) | CBA | 2.7m8 | CAA | P0–5 | 84.1 ± 5.66 | 83.1 ± 6.17 | ||||
| 1–6months | 84.5 ± 4.91 | 74.9 ± 6.53 | ||||||||
| 8BP2 | P0–5 | 55.7 ± 9.53 | 44.1 ± 7.94 | |||||||
| Akil et al., 2019 (103) | FVB | 5 | RWM | P1–3 | 80 | <1 | ||||
| Yoshimura et al., 2018 (104) | C3H | 2/9 | RWM +CF |
P15–16 | 94.6 | 96.8 | 94.2 | NR | ||
| P56–60 | 89.7 | 92.2 | 98.1 | NR | ||||||
| P15–16 | 16.7 | 17.4 | 18 | NR | ||||||
| Anc80 | 84.5 | 90.8 | 91.9 | NR | ||||||
| Tao et al., 2018 (105) | BL6 | 1 | CO | 10 weeks | 7.9 ± 2.0 | 11.8 ± 4.7 | 7.0 ± 0.6 | 0 | 0 | 0 |
| 2 | 95.4 ± 8.0 | 86.5 ± 13.9 | 85.5 ± 17.0 | 12.1 ± 12.3 | 7.1 ± 3.0 | 1.6 ± 1.6 | ||||
| 6.2 | 11.9 ± 1.7 | 3.1 ± 0.8 | 2.6 ± 0.6 | 0 | 0 | 0 | ||||
| 8 | 61.0 ± 18.9 | NR | 72.6 ± 27.5 | 0 | 0 | 0 | ||||
| 9 | 58.8 ± 7.6 | 61.8 ± 19.0 | 49.3 ± 11.9 | 0 | 0 | 0 | ||||
| rh.39 | 42.3 ± 12.0 | 36.2 ± 4.7 | 61.1 ± 7.9 | 0 | 0 | 0 | ||||
| rh.43 | 93.2 ± 3.1 | 92.7 ± 6.1 | 94.3 ± 3.9 | 0 | 0 | 0 | ||||
| Anc80 | 98.4 ± 2.7 | 98.4 ± 2.7 | 89.3 ± 18.5 | 67.2 ± 28.5 | 39.7 ± 31.7 | 10.4 ± 18.1 | ||||
| Shibata et al., 2017 (106) | C3H | 2/9 | Systemic injection | P0–1 | 70–90 | 40–70 | 20–40 | sparse | ||
| 20–30 | 5–20 | 0–5 | ||||||||
| 70 | NR | 10 | ||||||||
| 1 | 3 | NR | 3 | |||||||
| Landegger et al., 2017 (107) | BL6 | Anc80 | RWM | P1 | 100 | NR | 100 | 95 | NR | 95 |
| Suzuki et al., 2017 (108) | CBA | Anc80 | CAA | 7 weeks | 100 | 100 | 100 | 80–90 | 35–75 | 20–35 |
| Gyorgy et al., 2017 (21) | CD1 | 2/1 | RWM | P0–1 | 50 | 70 | 70 | 10 | 10 | 20 |
| CO | 25 | 30 | 45 | 10 | 10 | 25 | ||||
| e2/1 | RWM | 85 | 90 | 90 | 30 | 25 | 25 | |||
| CO | 50 | 65 | 70 | 15 | 20–30 | 40 | ||||
| 2/9 | CO | NR | NR | |||||||
| e2/9 | 60 | 25 | ||||||||
| Lhfpl5 | e2/1 | CO | >95 | >85 | ||||||
| Pan et al., 2017 (15)a | BL6 | Anc80 | RWM | P1 | 69 (Total of all IHCs and OHCs) | 65 (co-transfection of total of all IHCs+OHCs) | ||||
| Anc80 | 74 (total of all IHCs and OHCs) | |||||||||
| Isgrig et al., 2017 (18) | Whrn | 2/8 | CA | P1–5 | 71.7 ± 26.0 | 81.2 ± 15.3 | 75.2 ± 17.6 | 10.4 ± 6.38 | 8.64 ± 13.2 | 3.21 ± 5.95 |
| Chien et al., 2016 (17) | Whrn | 2/8 | RWM | P1–5 | 15.3 | 16.2 | 11.8 | 0 | 0 | 0 |
| Kim et al., 2016 (22) | ICR | 2/1 | Trans uterine injection | E12.5 | >90 | 83 | ||||
| MsrB3 | 89–91 | 84–92 | ||||||||
| Shu et al., 2016 (109) | CD1 | 1 | CO | P1–2 | 2.6 ± 0.6 | 9.6 ± 4.1 | 14.9 ± 2.6 | 6.3 ± 1.1 | 15 ± 6.3 | 18.3 ± 6.5 |
| 2 | 11.8 ± 2.1 | 15 ± 2.9 | 22.6 ± 4.8 | 4.1 ± 1.1 | 28 ± 4.5 | 39.5 ± 8.5 | ||||
| 5 | 0 | 11.2 ± 2.9 | 28.1 ± 3.4 | 0 | 0 | 0 | ||||
| 6.2 | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| 7 | 3.1 ± 0.8 | 16.2 ± 2.6 | 20.5 ± 2.5 | 0 | 0 | 0 | ||||
| 8 | 4.2 ± 0.9 | 14.2 ± 2.1 | 15 ± 3.1 | 6.1 ± 1.3 | 18.7 ± 1.7 | 21 ± 2.5 | ||||
| 9 | 0 | 0 | 0 | 4.2 ± 0.9 | 16.2 ± 2.3 | 21 ± 3.1 | ||||
| rh.10 | 8.1 ± 2 | 24 ± 4.7 | 34 ± 5.7 | 0 | 0 | 0 | ||||
| rh.39 | NR | NR | ||||||||
| rh.43 | 0 | 3 ± 1.1 | 5.3 ± 2.1 | 0 | 0 | 0 | ||||
| CBA | 1 | CO | 6 weeks | 12.2 ± 2.3 | 24.1 ± 6.2 | 45.8 ± 7.3 | NR | |||
| 2 | 13.2 ± 2.1 | 27.2 ± 4.5 | 35.2 ± 6.3 | |||||||
| 5 | NR | |||||||||
| 6.2 | 10.5 ± 1.5 | 18 ± 2.1 | 28 ± 4.8 | |||||||
| 7 | NR | |||||||||
| 8 | NR | |||||||||
| 9 | 9.1 ± 1.4 | 35.1 ± 3.2 | 61.6 ± 8 | |||||||
| rh.8 | NR | |||||||||
| rh.10 | NR | |||||||||
| rh.39 | 5.6 ± 1.6 | 15 ± 2.3 | 20 ± 4.7 | |||||||
| rh.43 | NR | |||||||||
| Chien et al., 2015 (110) | CBA | 2/8 | RWM | 1–2 months | 12 | 12 | 31 | NR | ||
| CO | 4.3 | 5.3 | 28 | NR | ||||||
| Askew et al., 2015 (8) | BL/6 | 2/1 | RWM | P0–2 | 59 ± 2 | NR | ||||
| 70 ± 9 | NR | |||||||||
| Yu et al., 2014 (25) | Gjb2 | 2/1 | RWM | P1 | Apex:13 ± 2, Middle:32 ± 4, Base:44 ± 3 (total of all IHCs and OHCs) | |||||
| Wang et al., 2013 (111) | BL6 | 2/1 | CO | P1 | NR | NR | 0 | NR | NR | 0 |
| 10.2 ± 1.9 | 32 ± 4.3 | |||||||||
| 2/7 | 82.1 ± 9.3 | 0 | ||||||||
| Akil et al., 2012 (26) | VGLUT3 | 1 | RWM | P1-P3 | 100 | 100 | 100 | NR | ||
| P10–12 | 40 | 40 | 40 | NR | ||||||
| P1–3 | NR | NR | ||||||||
| P10–12 | 100 | 100 | 100 | NR | ||||||
| CO | P10–12 | 100 | 100 | 100 | NR | |||||
| P10–12 | 40 | 40 | 40 | NR | ||||||
| Xia et al., 2012 (112) | BL6 | 8 | RWM | P7 | 58 ± 4 | 19 ± 4 | ||||
| RWMPD | 47 ± 7 | 17 ± 4 | ||||||||
| Kilpatrick et al., 2011 (113) | CBA | 1 | CO | P7 | AAV2 and AAV8 had robust transduction | AAV8 had robust transduction | ||||
| 2 | ||||||||||
| 5 | ||||||||||
| 6 | ||||||||||
| 8 | ||||||||||
| Bedrosian et al., 2006 (114) | BALB/c | 2/1 | Ex-utero injection | E12 | 82.54 | NR | 79.4 | 63.7 | NR | 64.67 |
| 2/2 | NR | NR | ||||||||
| 2/5 | 0 | 0 | ||||||||
| 2/6 | 0 | 0 | ||||||||
| 2/7 | 0 | 0 | ||||||||
| 2/8 | Second best | Second best | ||||||||
| 2/9 | 0 | 0 | ||||||||
| CBA | 2/1 | Trans uterine injection | E12 | Best | Best | |||||
| 2/2 | Second best | Second best | ||||||||
| 2/5 | 0 | 0 | ||||||||
| 2/6 | 0 | 0 | ||||||||
| 2/7 | 0 | 0 | ||||||||
| 2/8 | Second best | Second best | ||||||||
| 2/9 | 0 | 0 | ||||||||
| Liu et al., 2005 (115) | BL6 and ICR | 3 | RWM | 4 | 100 | 100 | 0 | 0 | ||
Anc80, AAV2/Anc80l65; 2/DJ, rAAV2/DJ; 2/DJ8, rAAV2/DJ8; 2.7m8, AAV2.7m8; 8BP2, AAV8BP2; 2PHP, rAAV2/PHP.B; 9PHP, AAV9/PHP.B; rh.39, AAVrh.39; rh.43, AAVrh.43; e2/1, exo-AAV2/1; e2/9, exo-AAV2/9; rh.8, AAVrh.8; rh.10, AAVrh.10; BL6, C57BL/6; C3H, C3HeB/FeJ; CBA, CBA/CaJ; RWM, round window membrane; RWM + CF, round window membrane combined with canal fenestration; RWMPD, round window membrane partial digestion; CA, canalostomy; CAA, canalostomy to ampulla; CO, cochleostomy
aThey injected dual vectors.
In general, AAV1, 2, 5, 6, 8, 9 and rh10 consistently transduce IHCs in both neonatal and adult mice; however, transduction of OHCs and supporting cells is variable and may be impacted by route of delivery. For example, when AAV1 is injected into the endolymph in neonatal mice, it transfects supporting cells (i.e. Deiters and Hensen’s cells) but not when it is injected into the perilymphatic space (111). Likewise, when AAV8 is injected into the endolymph in adult mice, OHCs, IHCs and supporting cells are transduced, but when injected into the perilymphatic space, transduction is limited to IHCs (113,115). Most studies have utilized ubiquitous promoters such as CMV and CBA; however, cell-specific promoters permit cell-specific expression, which may reduce the risk of off-target effects (127).
New synthetic vectors have emerged as an alternative to conventional AAV vectors and have demonstrated superior transduction in the inner ear (15,21,100–102,104,105,107,108). Most widely used is AAV2/Anc80L65, a novel designer AAV in which the main capsid proteins approximate the imputed ancestral sequence of AAV1, 2, 8 and 9 (128). AAV2/Anc80L65 has shown promising potential to transduce IHCs and OHCs when injected through either perilymph or endolymph in both neonatal and adult mice (100,108).
Delivery of gene therapy
The membranous labyrinth, which includes the cochlear duct, semi-circular ducts, utricle and saccule, lies within the bony labyrinth in the temporal bone. It is relatively isolated, has minimal lymphatic circulation and is separated from blood by the blood–labyrinthine barrier, three factors that limit efficacious systemic delivery of therapeutics to only neonatal mice (106,129). Direct local injection of viral vectors into the inner ear is necessary to order to achieve viral titers appropriate for gene therapy. Established injection routes include (1) round window membrane (RWM), (2) canalostomy, (3) cochleostomy into either the endolymph or perilymph and (4) RWM combined with canal fenestration (CF) (Figure 1).
Figure 1.
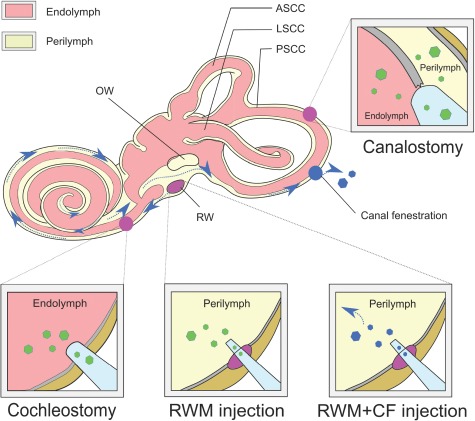
Inner ear delivery of gene therapy. Shown are the major routes by which gene therapy can be delivered to the inner ear. With the RWM approach, a small needle is used to pierce the RWM and deliver vector directly into the perilymph. A ‘canalostomy’ is done by drilling a hole into a semicircular canal (usually the posterior canal) and inserting a canula into the hole. Vector is delivered into both endolymph and perilymph in one of two directions depending on the orientation of the cannula. A cochleostomy requires a hole in the bony labyrinth between round window and basal turn of cochlea. Vector is delivered into endolymph. The RWM + CF technique combines RWM injection with a CF, which functions as a vent to allow egress of fluid (red, endolymph; yellow, perilymph; blue arrow, expected flow of injected vectors through cochlea when RWM + CF is done; PSCC, posterior semicircular canal; ASCC, anterior semicircular canal; LSCC, lateral semicircular canal; OW, oval window; RW, round window).
Round window membrane
The RWM is a three-layered membranous opening into the perilymphatic space of the scala tympani. This approach is well established and used clinically for cochlear implantation and, to date, is the most commonly utilized method of introducing transgenes into the inner ear in animal models (Table 1). Concerns for hearing loss secondary to perilymphatic leakage have been raised but can be obviated by plugging the RWM perforation with fascia (10,104). One disadvantage of the RWM approach is that distribution of the viral vector throughout the cochlear duct is challenging and, as a result, transduction tends to occur in a base-to-apex gradient in adult mice (104).
Canalostomy
Kawamoto et al. (130) developed the canalostomy approach for mouse cochlear gene therapy as an alternative to RWM injection. They injected an adenovirus expressing bacterial lacZ through a fenestration in the posterior semicircular canal, directing the cannula toward the crus commune. Hearing was preserved, with transduction mostly restricted to vestibular organs; minimal cochlear transduction was achieved. Suzuki et al. (108) modified the technique by using AAV2/Anc80L65 and targeting injection toward the ampulla. Hearing was not compromised, and cochlear transduction was 100% in IHCs and 80–90%, 35–70% and 20–35% in apical, mid and basal OHCs, respectively.
Cochleostomy
Cochleostomy directly delivers transgenes to the scala media, which can be accessed via a hole drilled through the basal portion of cochlea into the cochlear endolymphatic space near the round window. Chien et al. (110) compared cochleostomy to RWM injection in adult mice and demonstrated similar transduction efficiency although significant hearing loss was noted following the cochleostomy. In contrast, Kilpatrick et al. (113) showed that IHC and OHC transduction with AAV8 were superior with a cochleostomy and that hearing loss was minimal at high frequencies (≥32 kHz) and absent at low to middle frequencies (<32 kHz) 1 month after surgery. However, this approach is technically challenging. Its clinical application may be as an approach for stem cell transplantation or hair cell regeneration.
RWM with CF
Both the RWM and canalostomy injections in adult mice demonstrate transduction biases in either a base-to-apex or apex-to-base gradient, which cannot be overcome without increasing injection volume (18,108,130). Unfortunately, increased injection volume leads to hearing loss. To improve injection efficiency while maintaining hearing, Yoshimura et al. modified the RWM approach by adding a CF (104). The fenestration serves as a vent and permits longitudinal flow throughout the cochlea resulting in even distribution of the injected vector. Delivery is into perilymph, hearing is preserved and near total IHC transduction is possible. One disadvantage of this approach is the short-lived vestibular dysfunction associated with creation of a venting hole in the posterior semicircular canal. Treated mice have nystagmus in the acute recovery phase, which abates by the next day, although they do not have abnormal circling behavior.
Conclusion
Rapid development in diagnostics and therapy for SNHL has been made in recent years, and there have been multiple reports describing variably successful gene therapy in neonatal and adult mice models of human deafness. In addition, a three-part, multicenter, open label, single dose study is listed under ClinicalTrials.gov (https://clinicaltrials.gov/ct2/show/NCT02132130) to assess the safety, tolerability and efficacy of intra-labyrinthine CGF166, a recombinant adenovirus 5 vector containing the human atonal transcription factor cDNA, in patients with severe-to-profound hearing loss. However, for inner-ear gene therapy to enter the clinical realm with the goal of preventing or restoring hearing, important questions remain to the addressed in both mouse models of deafness and in nonhuman primates.
Funding
The National Institute on Deafness and Other Communication Disorders (R01DC003544, R01DC002842 and R01DC012049 to R.J.H.S. and R01DC015052 to C.C.M.) and National Institute for Health Research Manchester Biomedical Research Centre (to C.C.M.)
References
- 1. Smith R.J., Bale J.F. Jr. and White K.R. (2005) Sensorineural hearing loss in children. Lancet, 365, 879–890. [DOI] [PubMed] [Google Scholar]
- 2. Sugaya A., Fukushima K., Kasai N., Kataoka Y., Maeda Y., Nagayasu R., Toida N., Ohmori S., Fujiyoshi A., Taguchi T. et al. (2015) Impact of early intervention on comprehensive language and academic achievement in Japanese hearing-impaired children with cochlear implants. Int. J. Pediatr. Otorhinolaryngol., 79, 2142–2146. [DOI] [PubMed] [Google Scholar]
- 3. Shearer A.E., Hildebrand M.S., Sloan C.M. and Smith R.J. (2011) Deafness in the genomics era. Hear. Res., 282, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sloan-Heggen C.M., Bierer A.O., Shearer A.E., Kolbe D.L., Nishimura C.J., Frees K.L., Ephraim S.S., Shibata S.B., Booth K.T., Campbell C.A. et al. (2016) Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum. Genet., 135, 441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Azaiez H., Booth K.T., Ephraim S.S., Crone B., Black-Ziegelbein E.A., Marini R.J., Shearer A.E., Sloan-Heggen C.M., Kolbe D., Casavant T. et al. (2018) Genomic landscape and mutational signatures of deafness-associated genes. Am. J. Hum. Genet., 103, 484–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Geleoc G.S. and Holt J.R. (2014) Sound strategies for hearing restoration. Science, 344, 1241062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nist-Lund C.A., Pan B., Patterson A., Asai Y., Chen T., Zhou W., Zhu H., Romero S., Resnik J., Polley D.B. et al. (2019) Improved TMC1 gene therapy restores hearing and balance in mice with genetic inner ear disorders. Nat. Commun., 10, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Askew C., Rochat C., Pan B., Asai Y., Ahmed H., Child E., Schneider B.L., Aebischer P. and Holt J.R. (2015) Tmc gene therapy restores auditory function in deaf mice. Sci. Transl. Med., 7, 295ra108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yoshimura H., Shibata S.B., Ranum P.T., Moteki H. and Smith R.J.H. (2019) Targeted allele suppression prevents progressive hearing loss in the mature murine model of human TMC1 deafness. Mol. Ther., 27, 681–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shibata S.B., Ranum P.T., Moteki H., Pan B., Goodwin A.T., Goodman S.S., Abbas P.J., Holt J.R. and Smith R.J.H. (2016) RNA interference prevents autosomal-dominant hearing loss. Am. J. Hum. Genet., 98, 1101–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gao X., Tao Y., Lamas V., Huang M., Yeh W.H., Pan B., Hu Y.J., Hu J.H., Thompson D.B., Shu Y. et al. (2018) Treatment of autosomal dominant hearing loss by in vivo delivery of genome editing agents. Nature, 553, 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gyorgy B., Meijer E.J., Ivanchenko M.V., Tenneson K., Emond F., Hanlon K.S., Indzhykulian A.A., Volak A., Karavitaki K.D., Tamvakologos P.I. et al. (2019) Gene transfer with AAV9-PHP.B rescues hearing in a mouse model of Usher syndrome 3A and transduces hair cells in a non-human primate. Mol. Ther. Methods Clin. Dev., 13, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dulon D., Papal S., Patni P., Cortese M., Vincent P.F., Tertrais M., Emptoz A., Tlili A., Bouleau Y., Michel V. et al. (2018) Clarin-1 gene transfer rescues auditory synaptopathy in model of Usher syndrome. J. Clin. Invest., 128, 3382–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Geng R., Omar A., Gopal S.R., Chen D.H., Stepanyan R., Basch M.L., Dinculescu A., Furness D.N., Saperstein D., Hauswirth W. et al. (2017) Modeling and preventing progressive hearing loss in Usher syndrome III. Sci. Rep., 7, 13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pan B., Askew C., Galvin A., Heman-Ackah S., Asai Y., Indzhykulian A.A., Jodelka F.M., Hastings M.L., Lentz J.J., Vandenberghe L.H. et al. (2017) Gene therapy restores auditory and vestibular function in a mouse model of Usher syndrome type 1c. Nat. Biotechnol., 35, 264–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lentz J.J., Jodelka F.M., Hinrich A.J., McCaffrey K.E., Farris H.E., Spalitta M.J., Bazan N.G., Duelli D.M., Rigo F. and Hastings M.L. (2013) Rescue of hearing and vestibular function by antisense oligonucleotides in a mouse model of human deafness. Nat. Med., 19, 345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chien W.W., Isgrig K., Roy S., Belyantseva I.A., Drummond M.C., May L.A., Fitzgerald T.S., Friedman T.B. and Cunningham L.L. (2016) Gene therapy restores hair cell stereocilia morphology in inner ears of deaf whirler mice. Mol. Ther., 24, 17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Isgrig K., Shteamer J.W., Belyantseva I.A., Drummond M.C., Fitzgerald T.S., Vijayakumar S., Jones S.M., Griffith A.J., Friedman T.B., Cunningham L.L. et al. (2017) Gene therapy restores balance and auditory functions in a mouse model of Usher syndrome. Mol. Ther., 25, 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Akil O., Dyka F., Calvet C., Emptoz A., Lahlou G., Nouaille S., Boutet de Monvel J., Hardelin J.P., Hauswirth W.W., Avan P. et al. (2019) Dual AAV-mediated gene therapy restores hearing in a DFNB9 mouse model. Proc. Natl. Acad. Sci. U. S. A., Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Al-Moyed H., Cepeda A.P., Jung S., Moser T., Kugler S. and Reisinger E. (2019) A dual-AAV approach restores fast exocytosis and partially rescues auditory function in deaf otoferlin knock-out mice. EMBO Mol. Med., 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gyorgy B., Sage C., Indzhykulian A.A., Scheffer D.I., Brisson A.R., Tan S., Wu X., Volak A., Mu D., Tamvakologos P.I. et al. (2017) Rescue of hearing by gene delivery to inner-ear hair cells using exosome-associated AAV. Mol. Ther., 25, 379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim M.A., Cho H.J., Bae S.H., Lee B., Oh S.K., Kwon T.J., Ryoo Z.Y., Kim H.Y., Cho J.H., Kim U.K. et al. (2016) Methionine sulfoxide reductase B3-targeted in utero gene therapy rescues hearing function in a mouse model of congenital sensorineural hearing loss. Antioxid. Redox Signal., 24, 590–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chang Q., Wang J., Li Q., Kim Y., Zhou B., Wang Y., Li H. and Lin X. (2015) Virally mediated Kcnq1 gene replacement therapy in the immature scala media restores hearing in a mouse model of human Jervell and Lange-Nielsen deafness syndrome. EMBO Mol. Med., 7, 1077–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Iizuka T., Kamiya K., Gotoh S., Sugitani Y., Suzuki M., Noda T., Minowa O. and Ikeda K. (2015) Perinatal Gjb2 gene transfer rescues hearing in a mouse model of hereditary deafness. Hum. Mol. Genet., 24, 3651–3661. [DOI] [PubMed] [Google Scholar]
- 25. Yu Q., Wang Y., Chang Q., Wang J., Gong S., Li H. and Lin X. (2014) Virally expressed connexin26 restores gap junction function in the cochlea of conditional Gjb2 knockout mice. Gene Ther., 21, 71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Akil O., Seal R.P., Burke K., Wang C., Alemi A., During M., Edwards R.H. and Lustig L.R. (2012) Restoration of hearing in the VGLUT3 knockout mouse using virally mediated gene therapy. Neuron, 75, 283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bennett J., Wellman J., Marshall K.A., McCague S., Ashtari M., DiStefano-Pappas J., Elci O.U., Chung D.C., Sun J., Wright J.F. et al. (2016) Safety and durability of effect of contralateral-eye administration of AAV2 gene therapy in patients with childhood-onset blindness caused by RPE65 mutations: a follow-on phase 1 trial. Lancet, 388, 661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pipe S.W. (2018) Gene therapy for hemophilia. Pediatr. Blood Cancer, 65. [DOI] [PubMed] [Google Scholar]
- 29. Hashimoto T., Gibbs D., Lillo C., Azarian S.M., Legacki E., Zhang X.M., Yang X.J. and Williams D.S. (2007) Lentiviral gene replacement therapy of retinas in a mouse model for Usher syndrome type 1B. Gene Ther., 14, 584–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Studzinska S. (2018) Review on investigations of antisense oligonucleotides with the use of mass spectrometry. Talanta, 176, 329–343. [DOI] [PubMed] [Google Scholar]
- 31. Goyal N. and Narayanaswami P. (2018) Making sense of antisense oligonucleotides: a narrative review. Muscle Nerve, 57, 356–370. [DOI] [PubMed] [Google Scholar]
- 32. Urban E. and Noe C.R. (2003) Structural modifications of antisense oligonucleotides. Farmaco, 58, 243–258. [DOI] [PubMed] [Google Scholar]
- 33. Ponnath A., Depreux F.F., Jodelka F.M., Rigo F., Farris H.E., Hastings M.L. and Lentz J.J. (2018) Rescue of outer hair cells with antisense oligonucleotides in Usher mice is dependent on age of treatment. J. Assoc. Res. Otolaryngol., 19, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Adams D., Gonzalez-Duarte A., O'Riordan W.D., Yang C.C., Ueda M., Kristen A.V., Tournev I., Schmidt H.H., Coelho T., Berk J.L. et al. (2018) Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N. Engl. J. Med., 379, 11–21. [DOI] [PubMed] [Google Scholar]
- 35. Zhang H., Kolb F.A., Jaskiewicz L., Westhof E. and Filipowicz W. (2004) Single processing center models for human dicer and bacterial RNase III. Cell, 118, 57–68. [DOI] [PubMed] [Google Scholar]
- 36. Martinez J., Patkaniowska A., Urlaub H., Luhrmann R. and Tuschl T. (2002) Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell, 110, 563–574. [DOI] [PubMed] [Google Scholar]
- 37. Bartel D.P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell, 116, 281–297. [DOI] [PubMed] [Google Scholar]
- 38. Ha M. and Kim V.N. (2014) Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol., 15, 509–524. [DOI] [PubMed] [Google Scholar]
- 39. Lam J.K., Chow M.Y., Zhang Y. and Leung S.W. (2015) siRNA versus miRNA as therapeutics for gene silencing. Mol. Ther. Methods Clin. Dev., 4, e252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim V.N., Han J. and Siomi M.C. (2009) Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol., 10, 126–139. [DOI] [PubMed] [Google Scholar]
- 41. Maeda Y., Fukushima K., Nishizaki K. and Smith R.J.H. (2005) In vitro and in vivo suppression of GJB2 expression by RNA interference. Hum. Mol. Genet., 14, 1641–1650. [DOI] [PubMed] [Google Scholar]
- 42. Zou B., Mittal R., Grati M., Lu Z., Shu Y., Tao Y., Feng Y., Xie D., Kong W., Yang S. et al. (2015) The application of genome editing in studying hearing loss. Hear. Res., 327, 102–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K. and Sander J.D. (2013) High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol., 31, 822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. et al. (2013) DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol., 31, 827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pattanayak V., Lin S., Guilinger J.P., Ma E., Doudna J.A. and Liu D.R. (2013) High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol., 31, 839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Antico C.J., Fine E.J., Cradick T.J. and Bao G. (2013) CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res., 41, 9584–9592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kim S., Kim D., Cho S.W., Kim J. and Kim J.S. (2014) Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res., 24, 1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cho S.W., Kim S., Kim Y., Kweon J., Kim H.S., Bae S. and Kim J.S. (2014) Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res., 24, 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liao H.K., Hatanaka F., Araoka T., Reddy P., Wu M.Z., Sui Y., Yamauchi T., Sakurai M., O'Keefe D.D., Nunez-Delicado E. et al. (2017) In vivo target gene activation via CRISPR/Cas9-mediated trans-epigenetic modulation. Cell, 171, 1495–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Corwin J.T. and Cotanche D.A. (1988) Regeneration of sensory hair cells after acoustic trauma. Science, 240, 1772–1774. [DOI] [PubMed] [Google Scholar]
- 51. Ryals B.M. and Rubel E.W. (1988) Hair cell regeneration after acoustic trauma in adult Coturnix quail. Science, 240, 1774–1776. [DOI] [PubMed] [Google Scholar]
- 52. Cruz R.M., Lambert P.R. and Rubel E.W. (1987) Light microscopic evidence of hair cell regeneration after gentamicin toxicity in chick cochlea. Arch. Otolaryngol. Head Neck Surg., 113, 1058–1062. [DOI] [PubMed] [Google Scholar]
- 53. Cotanche D.A. (1987) Regeneration of hair cell stereociliary bundles in the chick cochlea following severe acoustic trauma. Hear. Res., 30, 181–195. [DOI] [PubMed] [Google Scholar]
- 54. Adler H.J., Komeda M. and Raphael Y. (1997) Further evidence for supporting cell conversion in the damaged avian basilar papilla. Int. J. Dev. Neurosci., 15, 375–385. [DOI] [PubMed] [Google Scholar]
- 55. Roberson D.W., Alosi J.A. and Cotanche D.A. (2004) Direct transdifferentiation gives rise to the earliest new hair cells in regenerating avian auditory epithelium. J. Neurosci. Res., 78, 461–471. [DOI] [PubMed] [Google Scholar]
- 56. Li H., Roblin G., Liu H. and Heller S. (2003) Generation of hair cells by stepwise differentiation of embryonic stem cells. Proc. Natl. Acad. Sci. U. S. A., 100, 13495–13500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Oshima K., Shin K., Diensthuber M., Peng A.W., Ricci A.J. and Heller S. (2010) Mechanosensitive hair cell-like cells from embryonic and induced pluripotent stem cells. Cell, 141, 704–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Koehler K.R., Mikosz A.M., Molosh A.I., Patel D. and Hashino E. (2013) Generation of inner ear sensory epithelia from pluripotent stem cells in 3D culture. Nature, 500, 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Costa A., Sanchez-Guardado L., Juniat S., Gale J.E., Daudet N. and Henrique D. (2015) Generation of sensory hair cells by genetic programming with a combination of transcription factors. Development, 142, 1948–1959. [DOI] [PubMed] [Google Scholar]
- 60. Chen W., Jongkamonwiwat N., Abbas L., Eshtan S.J., Johnson S.L., Kuhn S., Milo M., Thurlow J.K., Andrews P.W., Marcotti W. et al. (2012) Restoration of auditory evoked responses by human ES-cell-derived otic progenitors. Nature, 490, 278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ronaghi M., Nasr M., Ealy M., Durruthy-Durruthy R., Waldhaus J., Diaz G.H., Joubert L.M., Oshima K. and Heller S. (2014) Inner ear hair cell-like cells from human embryonic stem cells. Stem Cells Dev., 23, 1275–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Koehler K.R., Nie J., Longworth-Mills E., Liu X.P., Lee J., Holt J.R. and Hashino E. (2017) Generation of inner ear organoids containing functional hair cells from human pluripotent stem cells. Nat. Biotechnol., 35, 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Matsuoka A.J., Kondo T., Miyamoto R.T. and Hashino E. (2007) Enhanced survival of bone-marrow-derived pluripotent stem cells in an animal model of auditory neuropathy. Laryngoscope, 117, 1629–1635. [DOI] [PubMed] [Google Scholar]
- 64. Kasagi H., Kuhara T., Okada H., Sueyoshi N. and Kurihara H. (2013) Mesenchymal stem cell transplantation to the mouse cochlea as a treatment for childhood sensorineural hearing loss. Int. J. Pediatr. Otorhinolaryngol., 77, 936–942. [DOI] [PubMed] [Google Scholar]
- 65. Bas E., Van De Water T.R., Lumbreras V., Rajguru S., Goss G., Hare J.M. and Goldstein B.J. (2014) Adult human nasal mesenchymal-like stem cells restore cochlear spiral ganglion neurons after experimental lesion. Stem Cells Dev., 23, 502–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ma Y., Guo W., Yi H., Ren L., Zhao L., Zhang Y., Yuan S., Liu R., Xu L., Cong T. et al. (2016) Transplantation of human umbilical cord mesenchymal stem cells in cochlea to repair sensorineural hearing. Am. J. Transl. Res., 8, 5235–5245. [PMC free article] [PubMed] [Google Scholar]
- 67. Peyvandi A.A., Abbaszadeh H.A., Roozbahany N.A., Pourbakht A., Khoshsirat S., Niri H.H., Peyvandi H. and Niknazar S. (2018) Deferoxamine promotes mesenchymal stem cell homing in noise-induced injured cochlea through PI3K/AKT pathway. Cell Prolif., 51, e12434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mittal R., Ocak E., Zhu A., Perdomo M.M., Pena S.A., Mittal J., Bohorquez J. and Eshraghi A.A. (2019) Effect of bone marrow-derived mesenchymal stem cells on cochlear function in an experimental rat model. Anat. Rec. (Hoboken), doi: 10.1002/ar.24065 . [DOI] [PubMed] [Google Scholar]
- 69. Chen H.C., Liang C.M., Wang C.H., Huang M.Y., Lin Y.Y., Shih C.P., Kuo C.Y., Lin Y.C. and Chen H.K. (2019) Transplantation of human limbus-derived mesenchymal stromal cells via occipital approach improves hearing in animal auditory neuropathy. Int. J. Pediatr. Otorhinolaryngol., 117, 67–72. [DOI] [PubMed] [Google Scholar]
- 70. Nishimura K., Nakagawa T., Ono K., Ogita H., Sakamoto T., Yamamoto N., Okita K., Yamanaka S. and Ito J. (2009) Transplantation of mouse induced pluripotent stem cells into the cochlea. Neuroreport, 20, 1250–1254. [DOI] [PubMed] [Google Scholar]
- 71. Ohnishi H., Skerleva D., Kitajiri S., Sakamoto T., Yamamoto N., Ito J. and Nakagawa T. (2015) Limited hair cell induction from human induced pluripotent stem cells using a simple stepwise method. Neurosci. Lett., 599, 49–54. [DOI] [PubMed] [Google Scholar]
- 72. Chen J.R., Tang Z.H., Zheng J., Shi H.S., Ding J., Qian X.D., Zhang C., Chen J.L., Wang C.C., Li L. et al. (2016) Effects of genetic correction on the differentiation of hair cell-like cells from iPSCs with MYO15A mutation. Cell Death Differ., 23, 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tang Z.H., Chen J.R., Zheng J., Shi H.S., Ding J., Qian X.D., Zhang C., Chen J.L., Wang C.C., Li L. et al. (2016) Genetic correction of induced pluripotent stem cells from a deaf patient with MYO7A mutation results in morphologic and functional recovery of the derived hair cell-like cells. Stem Cells Transl. Med., 5, 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Takeda H., Minoda R., Miwa T., Yamada T. and Ise M. (2017) Transplanting mouse induced pluripotent stem cells into mouse otocysts in vivo. Neurosci. Lett., 647, 153–158. [DOI] [PubMed] [Google Scholar]
- 75. Chen J., Hong F., Zhang C., Li L., Wang C., Shi H., Fu Y. and Wang J. (2018) Differentiation and transplantation of human induced pluripotent stem cell-derived otic epithelial progenitors in mouse cochlea. Stem Cell Res. Ther., 9, 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Nakagawa T. and Ito J. (2011) Local drug delivery to the inner ear using biodegradable materials. Ther. Deliv., 2, 807–814. [DOI] [PubMed] [Google Scholar]
- 77. Staecker H. and Rodgers B. (2013) Developments in delivery of medications for inner ear disease. Expert Opin. Drug Deliv., 10, 639–650. [DOI] [PubMed] [Google Scholar]
- 78. Joshi O., Wang S.Y., Kuznetsova T., Atlasi Y., Peng T., Fabre P.J., Habibi E., Shaik J., Saeed S., Handoko L. et al. (2015) Dynamic reorganization of extremely long-range promoter-promoter interactions between two states of pluripotency. Cell Stem Cell, 17, 748–757. [DOI] [PubMed] [Google Scholar]
- 79. Martin Gonzalez J., Morgani S.M., Bone R.A., Bonderup K., Abelchian S., Brakebusch C. and Brickman J.M. (2016) Embryonic stem cell culture conditions support distinct states associated with different developmental stages and potency. Stem Cell Reports, 7, 177–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Choi J., Huebner A.J., Clement K., Walsh R.M., Savol A., Lin K., Gu H., Di Stefano B., Brumbaugh J., Kim S.Y. et al. (2017) Prolonged Mek1/2 suppression impairs the developmental potential of embryonic stem cells. Nature, 548, 219–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Perny M., Ting C.C., Kleinlogel S., Senn P. and Roccio M. (2017) Generation of otic sensory neurons from mouse embryonic stem cells in 3D culture. Front. Cell. Neurosci., 11, 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Matsuoka A.J., Morrissey Z.D., Zhang C., Homma K., Belmadani A., Miller C.A., Chadly D.M., Kobayashi S., Edelbrock A.N., Tanaka-Matakatsu M. et al. (2017) Directed differentiation of human embryonic stem cells toward placode-derived spiral ganglion-like sensory neurons. Stem Cells Transl. Med., 6, 923–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Nie J., Koehler K.R. and Hashino E. (2017) Directed differentiation of mouse embryonic stem cells into inner ear sensory epithelia in 3D culture. Methods Mol. Biol., 1597, 67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Longworth-Mills E., Koehler K.R. and Hashino E. (2016) Generating inner ear organoids from mouse embryonic stem cells. Methods Mol. Biol., 1341, 391–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Liu X.P., Koehler K.R., Mikosz A.M., Hashino E. and Holt J.R. (2016) Functional development of mechanosensitive hair cells in stem cell-derived organoids parallels native vestibular hair cells. Nat. Commun., 7, 11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Koehler K.R. and Hashino E. (2014) 3D mouse embryonic stem cell culture for generating inner ear organoids. Nat. Protoc., 9, 1229–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hu Z. and Ulfendahl M. (2013) The potential of stem cells for the restoration of auditory function in humans. Regen. Med., 8, 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Okano T. and Kelley M.W. (2012) Stem cell therapy for the inner ear: recent advances and future directions. Trends Amplif., 16, 4–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sugai K., Fukuzawa R., Shofuda T., Fukusumi H., Kawabata S., Nishiyama Y., Higuchi Y., Kawai K., Isoda M., Kanematsu D. et al. (2016) Pathological classification of human iPSC-derived neural stem/progenitor cells towards safety assessment of transplantation therapy for CNS diseases. Mol. Brain, 9, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lee A.S., Tang C., Rao M.S., Weissman I.L. and Wu J.C. (2013) Tumorigenicity as a clinical hurdle for pluripotent stem cell therapies. Nat. Med., 19, 998–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sun S., Babola T., Pregernig G., So K.S., Nguyen M., Su S.M., Palermo A.T., Bergles D.E., Burns J.C. and Muller U. (2018) Hair cell mechanotransduction regulates spontaneous activity and spiral ganglion subtype specification in the auditory system. Cell, 174, 1247–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Liberman L.D. and Liberman M.C. (2016) Postnatal maturation of auditory-nerve heterogeneity, as seen in spatial gradients of synapse morphology in the inner hair cell area. Hear. Res., 339, 12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Huang L.C., Barclay M., Lee K., Peter S., Housley G.D., Thorne P.R. and Montgomery J.M. (2012) Synaptic profiles during neurite extension, refinement and retraction in the developing cochlea. Neural Dev., 7, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Sobkowicz H.M., Rose J.E., Scott G.L. and Levenick C.V. (1986) Distribution of synaptic ribbons in the developing organ of Corti. J. Neurocytol., 15, 693–714. [DOI] [PubMed] [Google Scholar]
- 95. Wu J.S., Young E.D. and Glowatzki E. (2016) Maturation of spontaneous firing properties after hearing onset in rat auditory nerve fibers: spontaneous rates, refractoriness, and interfiber correlations. J. Neurosci., 36, 10584–10597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Raphael Y., Frisancho J.C. and Roessler B.J. (1996) Adenoviral-mediated gene transfer into Guinea pig cochlear cells in vivo. Neurosci. Lett., 207, 137–141. [DOI] [PubMed] [Google Scholar]
- 97. Weiss M.A., Frisancho J.C., Roessler B.J. and Raphael Y. (1997) Viral-mediated gene transfer in the cochlea. Int. J. Dev. Neurosci., 15, 577–583. [DOI] [PubMed] [Google Scholar]
- 98. Ishimoto S., Kawamoto K., Kanzaki S. and Raphael Y. (2002) Gene transfer into supporting cells of the organ of Corti. Hear. Res., 173, 187–197. [DOI] [PubMed] [Google Scholar]
- 99. Husseman J. and Raphael Y. (2009) Gene therapy in the inner ear using adenovirus vectors. Adv Otorhinolaryngol., 66, 37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Gu X., Chai R., Guo L., Dong B., Li W., Shu Y., Huang X. and Li H. (2019) Transduction of Adeno-associated virus vectors targeting hair cells and supporting cells in the neonatal mouse cochlea. Front. Cell. Neurosci., 13, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kim M.A., Ryu N., Kim H.M., Kim Y.R., Lee B., Kwon T.J., Bok J. and Kim U.K. (2019) Targeted gene delivery into the mammalian inner ear using synthetic serotypes of adeno-associated virus vectors. Mol. Ther. Methods Clin. Dev., 13, 197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Isgrig K., McDougald D.S., Zhu J., Wang H.J., Bennett J. and Chien W.W. (2019) AAV2.7m8 is a powerful viral vector for inner ear gene therapy. Nat. Commun., 10, 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Akil O., Blits B., Lustig L.R. and Leake P.A. (2019) Virally mediated overexpression of glial-derived neurotrophic factor elicits age- and dose-dependent neuronal toxicity and hearing loss. Hum. Gene Ther., 30, 88–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Yoshimura H., Shibata S.B., Ranum P.T. and Smith R.J.H. (2018) Enhanced viral-mediated cochlear gene delivery in adult mice by combining canal fenestration with round window membrane inoculation. Sci. Rep., 8, 2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Tao Y., Huang M., Shu Y., Ruprecht A., Wang H., Tang Y., Vandenberghe L.H., Wang Q., Gao G., Kong W.J. et al. (2018) Delivery of adeno-associated virus vectors in adult mammalian inner-ear cell subtypes without auditory dysfunction. Hum. Gene Ther., 29, 492–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Shibata S.B., Yoshimura H., Ranum P.T., Goodwin A.T. and Smith R.J.H. (2017) Intravenous rAAV2/9 injection for murine cochlear gene delivery. Sci. Rep., 7, 9609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Landegger L.D., Pan B., Askew C., Wassmer S.J., Gluck S.D., Galvin A., Taylor R., Forge A., Stankovic K.M., Holt J.R. et al. (2017) A synthetic AAV vector enables safe and efficient gene transfer to the mammalian inner ear. Nat. Biotechnol., 35, 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Suzuki J., Hashimoto K., Xiao R., Vandenberghe L.H. and Liberman M.C. (2017) Cochlear gene therapy with ancestral AAV in adult mice: complete transduction of inner hair cells without cochlear dysfunction. Sci. Rep., 7, 45524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Shu Y., Tao Y., Li W., Shen J., Wang Z. and Chen Z.Y. (2016) Adenovirus vectors target several cell subtypes of mammalian inner ear in vivo. Neural Plast., 2016, 9409846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Chien W.W., McDougald D.S., Roy S., Fitzgerald T.S. and Cunningham L.L. (2015) Cochlear gene transfer mediated by adeno-associated virus: comparison of two surgical approaches. Laryngoscope, 125, 2557–2564. [DOI] [PubMed] [Google Scholar]
- 111. Wang Y., Sun Y., Chang Q., Ahmad S., Zhou B., Kim Y., Li H. and Lin X. (2013) Early postnatal virus inoculation into the scala media achieved extensive expression of exogenous green fluorescent protein in the inner ear and preserved auditory brainstem response thresholds. J. Gene Med., 15, 123–133. [DOI] [PubMed] [Google Scholar]
- 112. Xia L., Yin S. and Wang J. (2012) Inner ear gene transfection in neonatal mice using adeno-associated viral vector: a comparison of two approaches. PLoS One, 7, e43218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Kilpatrick L.A., Li Q., Yang J., Goddard J.C., Fekete D.M. and Lang H. (2011) Adeno-associated virus-mediated gene delivery into the scala media of the normal and deafened adult mouse ear. Gene Ther., 18, 569–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Bedrosian J.C., Gratton M.A., Brigande J.V., Tang W., Landau J. and Bennett J. (2006) In vivo delivery of recombinant viruses to the fetal murine cochlea: transduction characteristics and long-term effects on auditory function. Mol. Ther., 14, 328–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Liu Y., Okada T., Sheykholeslami K., Shimazaki K., Nomoto T., Muramatsu S., Kanazawa T., Takeuchi K., Ajalli R., Mizukami H. et al. (2005) Specific and efficient transduction of cochlear inner hair cells with recombinant adeno-associated virus type 3 vector. Mol. Ther., 12, 725–733. [DOI] [PubMed] [Google Scholar]
- 116. Trapani I., Colella P., Sommella A., Iodice C., Cesi G., de Simone S., Marrocco E., Rossi S., Giunti M., Palfi A. et al. (2014) Effective delivery of large genes to the retina by dual AAV vectors. EMBO Mol. Med., 6, 194–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Umansky A.M., Jeffe D.B. and Lieu J.E. (2011) The HEAR-QL: quality of life questionnaire for children with hearing loss. J. Am. Acad. Audiol., 22, 644–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Wenzel G.I., Xia A., Funk E., Evans M.B., Palmer D.J., Ng P., Pereira F.A. and Oghalai J.S. (2007) Helper-dependent adenovirus-mediated gene transfer into the adult mouse cochlea. Otol. Neurotol., 28, 1100–1108. [DOI] [PubMed] [Google Scholar]
- 119. Bowers W.J., Chen X., Guo H., Frisina D.R., Federoff H.J. and Frisina R.D. (2002) Neurotrophin-3 transduction attenuates cisplatin spiral ganglion neuron ototoxicity in the cochlea. Mol. Ther., 6, 12–18. [DOI] [PubMed] [Google Scholar]
- 120. Chen X., Frisina R.D., Bowers W.J., Frisina D.R. and Federoff H.J. (2001) HSV amplicon-mediated neurotrophin-3 expression protects murine spiral ganglion neurons from cisplatin-induced damage. Mol. Ther., 3, 958–963. [DOI] [PubMed] [Google Scholar]
- 121. Derby M.L., Sena-Esteves M., Breakefield X.O. and Corey D.P. (1999) Gene transfer into the mammalian inner ear using HSV-1 and vaccinia virus vectors. Hear. Res., 134, 1–8. [DOI] [PubMed] [Google Scholar]
- 122. Kanzaki S., Shiotani A., Inoue M., Hasegawa M. and Ogawa K. (2007) Sendai virus vector-mediated transgene expression in the cochlea in vivo. Audiol. Neurootol., 12, 119–126. [DOI] [PubMed] [Google Scholar]
- 123. Duan M. and Mi Q. (2010) Local delivery of reporter gene to the cochlea does not spread to brain tissue in an animal model. Acta Otolaryngol., 130, 25–30. [DOI] [PubMed] [Google Scholar]
- 124. Han J.J., Mhatre A.N., Wareing M., Pettis R., Gao W.Q., Zufferey R.N., Trono D. and Lalwani A.K. (1999) Transgene expression in the Guinea pig cochlea mediated by a lentivirus-derived gene transfer vector. Hum. Gene Ther., 10, 1867–1873. [DOI] [PubMed] [Google Scholar]
- 125. Colella P., Ronzitti G. and Mingozzi F. (2018) Emerging issues in AAV-mediated in vivo gene therapy. Molecular therapy. Methods & clinical development, 8, 87–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Chien W.W., Monzack E.L., McDougald D.S. and Cunningham L.L. (2015) Gene therapy for sensorineural hearing loss. Ear Hear., 36, 1–7. [DOI] [PubMed] [Google Scholar]
- 127. Liu Y., Okada T., Nomoto T., Ke X., Kume A., Ozawa K. and Xiao S. (2007) Promoter effects of adeno-associated viral vector for transgene expression in the cochlea in vivo. Exp. Mol. Med., 39, 170–175. [DOI] [PubMed] [Google Scholar]
- 128. Zinn E., Pacouret S., Khaychuk V., Turunen H.T., Carvalho L.S., Andres-Mateos E., Shah S., Shelke R., Maurer A.C., Plovie E. et al. (2015) In silico reconstruction of the viral evolutionary lineage yields a potent gene therapy vector. Cell Rep., 12, 1056–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Saito T., Zhang Z.J., Tokuriki M., Ohtsubo T., Noda I., Shibamori Y., Yamamoto T. and Saito H. (2001) Expression of p-glycoprotein is associated with that of multidrug resistance protein 1 (MRP1) in the vestibular labyrinth and endolymphatic sac of the Guinea pig. Neurosci. Lett., 303, 189–192. [DOI] [PubMed] [Google Scholar]
- 130. Kawamoto K., Oh S.H., Kanzaki S., Brown N. and Raphael Y. (2001) The functional and structural outcome of inner ear gene transfer via the vestibular and cochlear fluids in mice. Mol. Ther., 4, 575–585. [DOI] [PubMed] [Google Scholar]


