Abstract
Triatomines are the vectors of Trypanosoma cruzi, the etiological agent of Chagas disease. Although Triatoma and Rhodnius are the most-studied vector genera, other triatomines, such as Panstrongylus, also transmit T. cruzi, creating new epidemiological scenarios. Panstrongylus has at least 13 reported species but there is limited information about its intraspecific genetic variation and patterns of diversification. Here, we begin to fill this gap by studying populations of P. geniculatus from Colombia and Venezuela and including other epidemiologically important species from the region. We examined the pattern of diversification of P. geniculatus in Colombia using mitochondrial and nuclear ribosomal data. Genetic diversity and differentiation were calculated within and among populations of P. geniculatus. Moreover, we constructed maximum likelihood and Bayesian inference phylogenies and haplotype networks using P. geniculatus and other species from the genus (P. megistus, P. lignarius, P. lutzi, P. tupynambai, P. chinai, P. rufotuberculatus and P. howardi). Using a coalescence framework, we also dated the P. geniculatus lineages. The total evidence tree showed that P. geniculatus is a monophyletic species, with four clades that are concordant with its geographic distribution and are partly explained by the Andes orogeny. However, other factors, including anthropogenic and eco-epidemiological effects must be investigated to explain the existence of recent geographic P. geniculatus lineages. The epidemiological dynamics in structured vector populations, such as those found here, warrant further investigation. Extending our knowledge of P. geniculatus is necessary for the accurate development of effective strategies for the control of Chagas disease vectors.
Introduction
Chagas disease affects about six million people in Latin America and is caused by the parasite Trypanosoma cruzi, which is mainly transmitted by insects of the subfamily Triatominae (Hemiptera: Reduviidae) [1]. The subfamily Triatominae is composed of five tribes, 19 genera, and 154 described species [2,3], but only a few genera are involved in the transmission of T. cruzi [4,5]. The genera Triatoma, Rhodnius, and Panstrongylus are the main vectors that transmit the parasite to humans [1,6]. After Triatoma and Rhodnius, Panstrongylus is the genus with the most species (currently 13), some of which appear to be involved in a domiciliation process (insofar as at least three developmental life stages can be found in homes) [5]. However, studies of T. cruzi transmission and control strategies have mainly focused on Rhodnius and Triatoma, and secondary vectors such as Panstrongylus remain unstudied. The species relationships, genetic diversity, and evolutionary trends of this genus are also rarely studied, in contrast to those of Rhodnius and Triatoma.
The incursion of P. geniculatus into the home is relevant because of the marked emergence of sylvatic T. cruzi strains, which are more virulent than domestic strains [7,8]. This change in vector behavior alters the transmission dynamics for T. cruzi and poses new challenges for the control of Chagas disease [2,5,9]. Panstrongylus geniculatus occurs in 18 countries in Latin America, from Mexico to Argentina, with a vast range of habitats, including dry, humid, rainy forest, and savannah environments [5]. This species is one of the principal vectors incriminated in the outbreaks of oral Chagas disease in Colombia and Venezuela [10–12], with reportedly higher parasitemia than other infection routes [2,7,10].
Panstrongylus geniculatus has a broad distribution in Colombia [13] where its domiciliation increases the risk of new Chagas disease cases. Previous studies have indicated that P. geniculatus has the highest frequency of T. cruzi infection in this country [1]. However, regardless of the epidemiological importance of this species, its current classification is difficult and based exclusively on morphological analyses [5,14–17]. Despite advances in molecular methods, few studies have used DNA markers to evaluate the phylogenetic status of this genus in the subfamily Triatominae, and the species relationships within Panstrongylus remain unclear. Previous studies have shown incongruence in the phylogeny of the species, suggesting polyphyly [18] and paraphyly [19]. As far as we know, there is no information about the processes and factors shaping the intraspecific and interspecific diversity in this genus. Here, we begin to address this lack of information using molecular data to document the genetic diversity of P. geniculatus. Our results extend our knowledge of this vector and our understanding of the biology of P. geniculatus.
Methods
Sampling
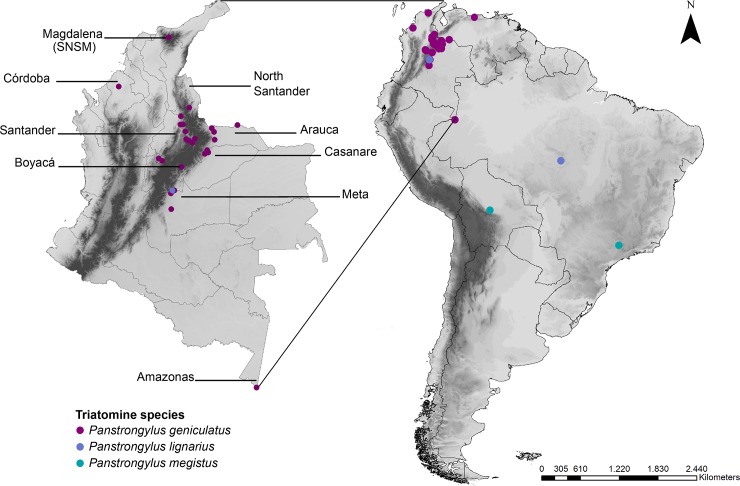
A total of 124 samples corresponding to three species (105 P. geniculatus, five P. lignarius, 14 P. megistus) were collected for this study. Panstrongylus geniculatus individuals were collected in Colombia and Venezuela (S1 Table; Fig 1). The remaining samples (P. lignarius and P. megistus) were collected in Brazil and Bolivia (S1 Table; Fig 1). Sylvatic ecotope individuals were collected using two techniques: manual search and modified Noireau baited chicken traps located in palm trees. The insects in the domestic ecotopes were collected by hand. All the specimens were identified with standard taxonomic keys [20], stored individually in plastic containers with 100% ethanol, and kept at −20°C until processing. UNIVERSIDAD DEL ROSARIO provided the field permit from ANLA (Autoridad Nacional de Licencias ambientales) 63257–2014. All collection was done on public land.
Fig 1. Map showing the locations of the samples collected in this study.
The size of the circle is not representative of the number of individuals collected at each location. The map was constructed using QGIS version 2.18.7.
DNA extraction, amplification, sequencing and alignment
DNA was extracted from the whole body (63 samples) and from the head, leg, and thorax (61 samples) using the DNeasy® Blood & Tissue kit (Qiagen, Germany) [21], but with double the amounts of buffer ATL, proteinase K, buffer AL, and ethanol. The final elution step was performed with only 150 μL of buffer AE. The concentration and quality of the DNA was measured as absorbance (A) ratios A260/A280 and A230/A260, respectively, with a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific Inc., MA, USA). The quality of the DNA was also verified on 2% agarose gels.
We used two sets of primers to amplify the nuclear ribosomal RNA (rRNA) genes, 18S rRNA (823 bp) [19] and 28S rRNA (696 bp) [22], and three sets of primers to amplify mitochondrial gene fragments: 630 bp of the NADH dehydrogenase subunit 4 gene, ND4 [23], 552 bp of the cytochrome b gene, Cytb [24,25], and 508 bp of 16S rRNA [19] (See S2 Table). The PCRs for all gene fragments were conducted in a final volume of 12.5 μl containing 1.5 μl of DNA template, 6.25 μl of GoTaq Green Master Mix (2×), 1.25 μl of each primer (10 μM), and 2.25 μl of nuclease-free water. The thermal cycling conditions have been reported elsewhere (see references in S2 Table). For all loci, we visualized 2 μL of the PCR product on a 1.5% agarose gel stained with SYBR® Safe DNA Gel Stain to verify the success of the PCR. Samples that showed a solid band of the expected size were purified with ExoSAP-IT and sequenced with Sanger sequencing (both strands) by Macrogen (Seoul, South Korea) using an ABI Prism 3100 Genetic Analyzer. The output sequences were read and assembled into contigs with Geneious 11.0.5 [26]. After base calling and editing, we used the same program to obtain a consensus sequence for each sample. A Geneious plug-in was used to align each locus with Muscle [27]. The resulting alignments were manually inspected for misalignments and ambiguities (S3 Table). In cases where ambiguities of bases were present, the different haplotypes were inferred for each locus with the PHASE algorithm implemented in DnaSP v5.10 [28], with 1000 iterations per simulation.
Phylogenetic analyses and species tree reconstruction
We only used the mitochondrial gene fragments for these analyses because the nuclear ribosomal loci did not resolve P. geniculatus intraspecifically (see results; S1 Fig). To determine the phylogenetic position of P. geniculatus within the genus Panstrongylus, we included additional mitochondrial gene fragment sequences (i.e., 16S rRNA, Cytb) available in GenBank for the following eight species: P. geniculatus, P. megistus, P. lignarius, P. lutzi, P. tupynambai, P. chinai, P. rufotuberculatus and P. howardi (see S4 Table for accession numbers).
We estimated the phylogenetic relationships using two tree inference criteria: maximum likelihood (ML) and Bayesian inference (BI). For the ML tree, we used the concatenated alignment (individual analyses per gene fragment were also performed [see S2 Fig]) with an unlinked substitution model per locus in IQ-Tree [29]. The best-fit nucleotide substitution model for each gene fragment was obtained with IQ-tree model selection based on the Bayesian information criterion [30], and the node supports were calculated with 10,000 ultrafast bootstrap pseudoreplicates. Ultrafast bootstrap values > 95% were deemed trustworthy [31]. For Cytb, we included sequences from Venezuela previously used in [32].
The BI reconstruction was implemented in MrBayes v. 3.2.6 [33]. We performed one run with four Markov chains (MCMC) for 100 million generations, sampling every 1000 generations and discarding the first 10% as burn-in. We calculated the posterior probability (PP) of the nodes based on the 50% majority consensus of the best tree distribution. All tree distributions were checked for convergence using Tracer v. 1.7.1 [34]. Both the best ML and BI topologies were rooted to the mid-point because we were looking at the relationships within Panstrongylus. The trees were visualized and edited with FigTree v.1.4.4 [35].
The species tree and time of divergence estimates were based on the monophyletic clades from the phylogenetic reconstructions. We identified four possible independent lineages of P. geniculatus and the other six lineages represented the rest of the Panstrongylus species included in this study. We estimated a species tree with three partitions, one for each gene fragment, using a coalescence-based approach implemented in StarBEAST2 v 2.4 [36,37]. We performed three independent runs of 1 × 108 generations, sampled every 1000 generations, and discarded the first 15% as the burn-in period. The models of substitutions (estimated in IQ-tree) and priors were specified as follow (otherwise as default): Cytb, TIM2 + F + I + G4; 16S rRNA, TPM2 + F + G4; ND4, HKY + F + I + G4 (models); relaxed uncorrelated lognormal clock (estimate); Yule process of speciation; and gene tree ploidy were mitochondrial (priors). We calibrated the root node of the species tree using the crown fossil P. hispaniolae (PaleoDatabase 49461, early Miocene), and used a uniform prior distribution with the following upper and lower bound range (20.44–13.82 Ma) [38]. We assessed the convergence of the model by examining the trace files in Tracer v.1.7.1 [34], after obtaining an effective sample size of > 200 for all parameters. We combined the tree files (two runs) using LogCombiner v.1.10.4 [37], and the uncertainty of the trees was visualized with DensiTree v.2.1 [37]. However, the maximum credibility trees with the coalescence-based divergence times and 95% highest probability densities were produced in Tree Annotator v.1.10.4 [36]. The species tree and divergence times were visualized with FigTree v.1.4.4 [35].
Population genetics
We calculated haplotype networks for each locus using the available Panstrongylus species to visualize the haplotype diversity and clustering. We estimated these networks with a minimum distance algorithm in the program PopArt [39]. The haplotypic structure within P. geniculatus was assessed with the same approach.
Genetic diversity statistics were calculated for the clusters determined with the haplotype networks: nucleotide diversity (π), substitution rate (θ), number of segregating sites (S), and Tajima’s D. These statistics were calculated with DNASP v5.10 [28]. The genetic structure between the species and geographic groups was estimated with FST [40], DXY, and Da [41]. The effect of isolation by distance on the population structure was evaluated with the Mantel test [42] using the R package vegan [43]. To do so, a matrix of genetic distances was estimated by linearizing the FST values [44], and the pairwise geographic distances among localities were processed with the function distm in the R package geosphere [45]. We also examined the linear correlation between the geographic distances and genetic distances as recommended by Legendre et al. [46], with the entire dataset, but without extreme points. Because we were mainly interested in the diversification of P. geniculatus, all the summary statistics and structure analyses were performed with the samples collected for this study, including P. megistus and P. lignarius.
Results
Phylogenetic analyses and species tree reconstruction
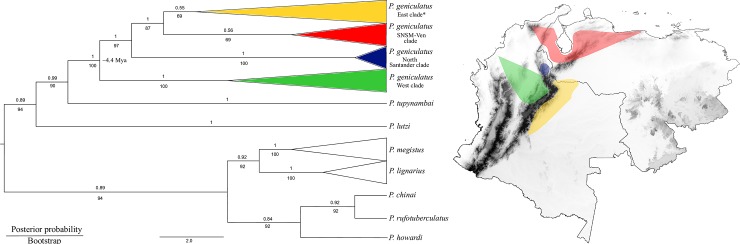
The ML and BI topologies for the mitochondrial gene fragments were completely concordant and revealed eight well-supported clades, corresponding to eight species (Fig 2). Panstrongylus geniculatus was monophyletic and had four clades (Fig 2), which were associated, to some extent, with its geographical range: 1) East of the Eastern Cordillera of the Colombian Andes clade (bootstrap support [BS] = 69, PP = 0.55), which included the departments Arauca, Casanare, Meta, Amazonas, and Córdoba, and some individuals from Santander, Boyacá, and Magdalena; 2) Magdalena (Sierra Nevada of Santa Marta, SNSM) with Venezuela clade (SNSM-Ven; BS = 69, PP = 0.56), which included all individuals from Venezuela and Magdalena; 3) North Santander clade (NSant; BS = 100, PP = 1), which only included the samples collected there; and 4) West of the Eastern Cordillera of the Colombian Andes clade (BS = 100, PP = 1), which included the rest of the Santander and Boyacá samples. However, the first two clades were not well-supported. Panstrongylus geniculatus diverged from P. tupynambai somewhere around 9.28 Mya (Confident Intervals, here after CI: 14.13–4.51 Mya, Fig 3), and the diversification within P. geniculatus occurred ~4.35 Mya (CI: 6.58–2.20 Mya, Fig 3). Finally, ML reconstruction for Cytb fragment including Venezuelan sequences obtained from [32] were recovered in the SNSM-Ven clade (S2 Fig) supporting the pattern previously described in the concatenated data.
Fig 2. BI consensus phylogenetic tree inferred from mitochondrial DNA and distribution of the geographical clades.
Numbers at the nodes are the bootstrap/posterior probability support values. Distributions were calculated with minimum convex polygon and are color coded based on the tree clades. The map was constructed using QGIS version 2.18.7.
Fig 3. Dated coalescent Bayesian species tree estimated in *BEAST2.
Numbers at the nodes are the estimated times of divergence. Each blue bar corresponds to the 95% highest posterior density (HDP) interval.
Population genetics
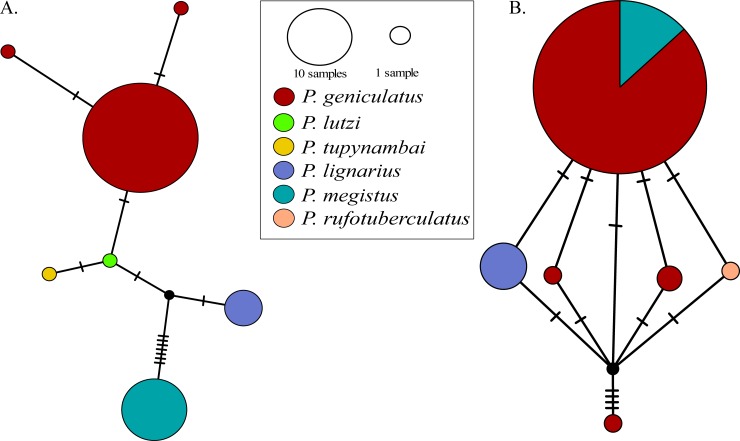
The two nuclear ribosomal loci showed low haplotype diversity (Fig 4). Specifically, the 28S rRNA gene fragment showed that P. geniculatus, P. megistus, P. lignarius, P. lutzi, and P. tupynambai were separated clusters, although there were few mutational steps between each group (Fig 4), whereas the 18S rRNA gene showed that P. geniculatus and P. megistus were intermixed.
Fig 4.
Haplotype networks inferred with nuclear rRNA: A. 28S and B. 18S. Each tick on the haplotype network represents a mutational step.
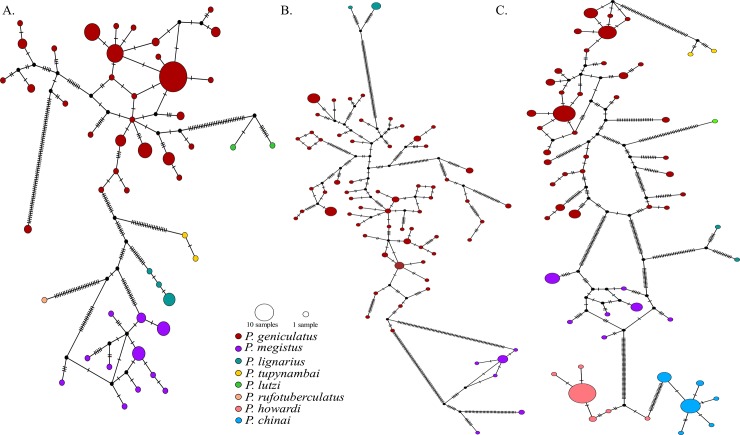
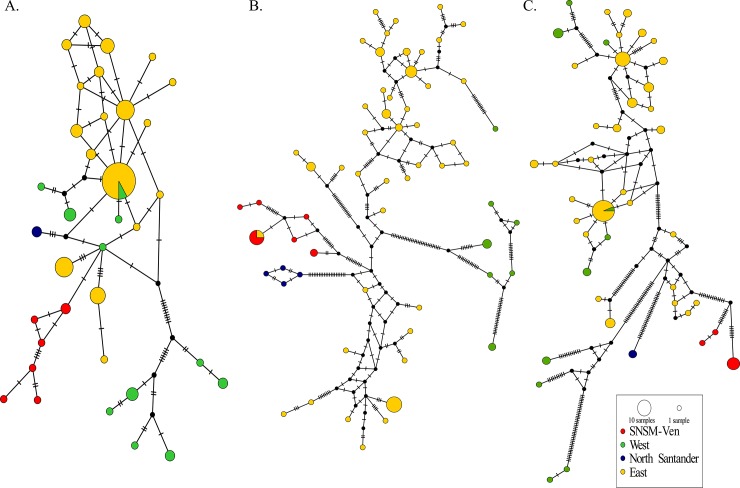
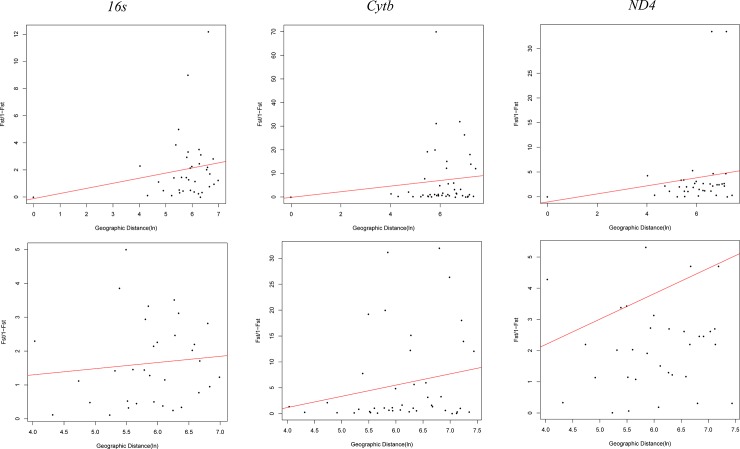
The mitochondrial networks clustered the haplotypes by species, with many mutational steps among them (Fig 5). Consistent with this, we detected high genetic differentiation between the species (Table 1). However, nucleotide diversity was higher in P. geniculatus than in the other species (Table 2). A close examination of the P. geniculatus haplotype networks (Fig 6) confirmed that the previously described geographic groups (Fig 2) were genetically differentiated (Table 1). Isolation by distance was ruled out as a causal factor contributing to the geographic genetic structure (Table 3, Fig 7), and even though we found significant correlation between the geographic and genetic distances in some cases, the coefficient of determination of the linear model was poor and the Mantel test result was not significant (Table 3, Fig 7). We also found shared haplotypes in samples collected from both sides of the Eastern Cordillera (Fig 6 and S3 Fig). Specifically, these shared haplotypes involved individuals from the western localities of Santander, Boyacá, and Cordoba, which grouped with the samples from the east of the Eastern Cordillera. Our data also showed that the population in northwestern Colombia (SNSM) contained mitochondrial DNA (mtDNA) from east of the Eastern Cordillera.
Fig 5.
Haplotype networks for mitochondrial loci grouped by species: A. 16S, B. ND4 and C. Cytb. Each tick on the branches represents a mutational step.
Table 1. Measures of population structure between species and geographic clusters of P. geniculatus at mtDNA loci.
| Population1 | P. geniculatus | P. geniculatus | P. megistus | SNSM-Ven | SNSM-Ven | SNSM-Ven | West | West | NSant | |
|---|---|---|---|---|---|---|---|---|---|---|
| Population2 | P. megistus | P. lignarius | P. lignarius | West | NSant | East | NSant | East | East | |
| Cytb | FST | 0.932 | ---- | ---- | 0.459 | 0.863 | 0.676 | 0.599 | 0.157 | 0.853 |
| DXY | 0.161 | ---- | ---- | 0.084 | 0.082 | 0.068 | 0.082 | 0.052 | 0.075 | |
| Da | 0.123 | ---- | ---- | 0.039 | 0.071 | 0.047 | 0.049 | 0.008 | 0.064 | |
| P value | 0.00001*** | ---- | ---- | 0.00001*** | 0.039* | 0.00001*** | 0.028* | 0.00001*** | 0.001** | |
| ND4 | FST | 0.756 | 0.823 | 0.863 | 0.641 | 0.874 | 0.550 | 0.701 | 0.538 | 0.744 |
| DXY | 0.158 | 0.159 | 0.155 | 0.085 | 0.061 | 0.045 | 0.093 | 0.088 | 0.062 | |
| Da | 0.119 | 0.131 | 0.134 | 0.057 | 0.053 | 0.025 | 0.065 | 0.047 | 0.046 | |
| P value | 0.00001*** | 0.00001*** | 0.00001*** | 0.00001*** | 0.002** | 0.00001*** | 0.006** | 0.00001*** | 0.00001*** | |
| 16S rRNA | FST | 0.911 | 0.910 | 0.969 | 0.417 | 0.756 | 0.456 | 0.539 | 0.328 | 0.713 |
| DXY | 0.084 | 0.095 | 0.075 | 0.029 | 0.015 | 0.013 | 0.025 | 0.025 | 0.011 | |
| Da | 0.077 | 0.086 | 0.073 | 0.012 | 0.011 | 0.006 | 0.008 | 0.008 | 0.008 | |
| P value | 0.00001*** | 0.00001*** | 0.00001*** | 0.00001*** | 0.037* | 0.00001*** | 0.005** | 0.00001*** | 0.00001*** | |
| Concatenated Data | FST | 0.768 | 0.845 | 0.861 | 0.671 | 0.882 | 0.642 | 0.598 | 0.523 | 0.744 |
| DXY | 0.143 | 0.125 | 0.142 | 0.074 | 0.039 | 0.041 | 0.063 | 0.049 | 0.053 | |
| Da | 0.112 | 0.104 | 0.102 | 0.033 | 0.039 | 0.022 | 0.047 | 0.023 | 0.041 | |
| P value | 0.00001*** | 0.00001*** | 0.00001*** | 0.00001*** | 0.0027** | 0.00001*** | 0.002** | 0.00001*** | 0.00001*** |
Probability calculated with the Hudson permutation test, with 1000 replicates.
*0.01 < P < 0.05
**0.001 < P < 0.01
***P < 0.001. “----”: some statistics could not be calculated due to low number of haplotypes per population.
Table 2. Genetic summary statistics for the three species analyzed in this study and for the Eastern, Western, North Santander (NSant) and Magdalena-Venezuela (SNSM-Ven) groups at each mitochondrial locus.
| Genomic marker | Species/lineage | n | h | S | π ± SD | D |
|---|---|---|---|---|---|---|
| Cytb | P. geniculatus | 134 | 61 | 136 | 0.051±0.002 | −0.233 |
| P. megistus | 13 | 2 | 21 | 0.020±0.003 | 2.539 | |
| P. lignarius | 1 | 1 | 0 | 0 | ---------- | |
| East | 61 | 30 | 54 | 0.022±0.002 | 0.185 | |
| NSant | 2 | 1 | 0 | 0 | --------- | |
| SNSM-Ven | 57 | 25 | 70 | 0.022±0.003 | -1.099 | |
| West | 14 | 10 | 99 | 0.066±0.010 | 0.734 | |
| ND4 | P. geniculatus | 92 | 60 | 145 | 0.045±0.003 | −0.26792 |
| P. megistus | 12 | 7 | 62 | 0.031±0.012 | −0.28969 | |
| P. lignarius | 6 | 2 | 20 | 0.012±0.007 | −1.49247 | |
| East | 66 | 44 | 76 | 0.028±0.002 | 0.081 | |
| NSant | 4 | 4 | 3 | 0.003±0.001 | 2.012 | |
| SNSM-Ven | 12 | 6 | 23 | 0.013±0.003 | −0.050 | |
| West | 10 | 7 | 92 | 0.054±0.011 | 0.039 | |
| 16S rRNA | P. geniculatus | 92 | 35 | 48 | 0.014±0.002 | −0.84369 |
| P. megistus | 14 | 4 | 4 | 0.003±0.001 | 1.05159 | |
| P. lignarius | 6 | 2 | 2 | 0.001±0.001 | −1.13197 | |
| East | 65 | 18 | 16 | 0.006±0.001 | -0.185 | |
| NSant | 2 | 1 | 0 | 0 | --------- | |
| SNSM-Ven | 7 | 6 | 1 | 0.007±0.001 | 1.381 | |
| West | 18 | 11 | 34 | 0.027±0.002 | 1.400 | |
| Concatenated Data | East | 70 | 61 | 497 | 0.005±0.003 | -0.677 |
| NSant | 4 | 4 | 3 | 0.002±0.001 | 2.011 | |
| SNSM-Ven | 12 | 7 | 11 | 0.016±0.01 | 1.032 | |
| West | 18 | 11 | 34 | 0.027±0.002 | 1.400 |
n: number of sequences; h: number of haplotypes; S: number of segregating sites; π: nucleotide diversity; D: Tajima’s D. None of the loci showed Tajima’s D values that departed from neutral expectations.
Fig 6.
Haplotype networks for mitochondrial loci grouped by geographic location: A. 16S, B. ND4 and C. Cytb. Each tick on the branches represents a mutational step.
Table 3. Isolation by distance analysis.
Mantel and correlation tests were performed with all the samples and without the extreme points.
| Entire Data | Excluding extreme points | |||||||
|---|---|---|---|---|---|---|---|---|
| Locus | R | R2 | P value | Mantel r | P value | R | R2 | P value |
| 16S rRNA | 0.3121 | 0.0974 | 0.0046** | 0.1442 | 0.2548 | 0.8107 | 0.6572 | 0.5143 |
| Cytb | 0.1935 | 0.0374 | 0.0538 | 0.0672 | 0.3373 | 0.2025 | 0.041 | 0.0616 |
| ND4 | 0.2422 | 0.0587 | 0.0294* | 0.0998 | 0.2328 | 0.0813 | 0.0066 | 0.5164 |
P value:
*0.01 < P < 0.05
**0.001 < P < 0.01
Fig 7. Isolation by distance plots for mitochondrial loci.
Two tests of correlation were performed: with all the data (top) and without the extreme points (bottom).
Discussion
Increasing evidence suggests that some species of Panstrongylus should be considered primary vectors of Chagas disease [1,5]. Despite this, few studies have reported the genetic diversity of these species. Panstrongylus geniculatus has become relevant over the years for three main reasons: its geographic distribution, its record of domiciliation, and its association with oral outbreaks of Chagas disease, mainly in Colombia and Venezuela. The phenotypic variability in this species has been studied at the morphological level [14–17], and only one karyotype study of P. geniculatus is known [47]. The observed intraspecific karyotypic and morphological diversity coupled to the wide distribution of P. geniculatus prompted these authors to propose that it is a complex of species [47]. However, this proposition remained to be tested at the molecular level.
Phylogenetic relationships among Panstrongylus species
The phylogenetic relationships among Panstrongylus species are controversial. Few representatives of this genus were included in studies, which primary focus was to resolve the phylogenetic relationships within the subfamily Triatominae [19,48–51]. In these analyses, Panstrongylus was recovered as polyphyletic with Triatoma species [18–19,48,50–51]. Although, the aim of our study was to establish the intraspecific variation of P. geniculatus, we were able reconstruct the relationships of the Panstrongylus species included here from mtDNA analyses (Fig 2 and Fig 5). Two main clades were detected: the first one included P. lutzi and P. tupynambai, and this clade was sister to P. geniculatus (geniculatus group [48]). The second one contained P. lignarius and P. megistus, and this clade was sister to P. chinai, P. rufotuberculatus and P. howardi, as previously reported (megistus group [48]). This result contrasts with the previous lack of resolution obtained in other studies [18,49–51].
Genetic variation and phylogeography of P. geniculatus
Our estimates of intraspecific genetic diversity were higher than the previously reported in two Caracas municipalities within Venezuela [32], due to a broader geographical sampling included in this study (Table 2). Genetic differences estimated between our geographical clades were higher than the values observed between populations occurring in a continuous space in Venezuela (without barriers) [32]. This could suggest that geographical barriers (such as the Andes) might promote stronger divergence than other eco-epidemiological factors. However, the measure of absolute divergence among the clades obtained in this study (DXY) was too low, suggesting that P. geniculatus is not a complex of species (Tables 1 and 2). In contrast, our nuclear rRNA dataset did not show the genetic structure detected with P. geniculatus mtDNA (Fig 4, S1 Fig). This can be explained by the differences in the coalescence times and effective population sizes in the nuclear rRNA and mtDNA analyses [52–54]. Our results based on mtDNA data suggest that the Andean orogeny might contributed to the structured geographic populations of P. geniculatus, as has been observed for other members of the Triatominae [48,55–57]. We found that the West and East clades of P. geniculatus diverged (Fig 3; 4.35 Mya [CI: 6.58–2.20 Mya]) within the period of the Andean uplift of the Colombian Eastern Cordillera, which achieved its final elevation ~2.5 Mya, during the Pliocene [58].
The rapid uplift of the Andes created a wide range of new ecological niches, with opportunities for colonization [58]. Interestingly, the shared haplotypes, with no evidence of isolation by distance, can be attributed to widely dispersed gene flow (Table 3, Fig 6, Fig 7, and S3 Fig). Thus, long-distance dispersal coupled to niche colonization could facilitate the admixtures of P. geniculatus populations that occur on opposite sides of the Andes. Interestingly, at the border between the Colombia East Andes and Mérida Andes in Venezuela, there are altitudinal depressions (e.g. Yaracuy and Táchira passes) that could facilitate genetic exchange among the P. geniculatus populations that occur on both sides of the Andes [59,60]. Geographic barriers, in this case the Andes uplift, have driven the diversification of many animals, including insects such as butterflies [61–63], bees [64], other triatomines [22,65–67], and arachnid species [68], among others. Moreover, gene flow by organismal dispersal through corridors has been documented in other arthropods [68–70].
Vector importance of the P. geniculatus variability
Many studies have sought to explain the high morphological variability of certain species within the subfamily Triatominae. One of the most studied species is Triatoma dimidiata because of the high variance in its phenotypic and genetic data [23,71–72]. An investigation conducted in Colombia revealed a similar genetic structure [73] to the one found in this study. That structure correlated with eco-epidemiological and morphological traits, but not with geographic events that would favor the divergence of populations. Contrary to the lack of geographic association in that study, another research into different populations has suggested a major role for geological variation in the divergence within T. dimidiata [65]. This reflects the different ways in which the divergence of a population can occur. Geographic variation may explain its diversification, but so can orogenic and eco-epidemiological changes, among other factors [65–66,73]. Panstrongylus geniculatus could be experiencing a similar situation, insofar as geographic variation is not the only factor modeling the population structure. Mountains can also promote diversification through processes such as niche partitioning, altitudinal gradients, climatic variations, and long-distance dispersal, among other factors [22,65–67]. Other eco-epidemiological factors involving vector adaptability may also be shaping the divergence between P. geniculatus populations [65,74]. Further studies are required, with more markers or even genomes, and with more broadly sampled triatomine specimens, especially from other countries, to test the contributions of these factors to the diversification of this species.
The pattern of shared genetic variation between P. geniculatus geographical clades described before, could be explained using passive dispersion (e.g. vertebrate hosts) [75], considering that the adults of these species fly poorly [9, 72]. Panstrongylus geniculatus is commonly associated with opossums, armadillos, and bats [76], but more recent studies have demonstrated that this species has a wide range of feeding sources, including rodents, canines, ruminants and primates [1]. Its passive dispersal by human activity and with its carriage on animals is important for its long-distance migration, as previously described [74].
The broad spectrum of its ecological niches, coupled to its many feeding sources and its high level of genetic diversity demonstrated here (Table 1 and 2), increases the possibility of niche specialization in P. geniculatus and therefore the divergence of its populations. This is relevant because changes in the interaction between the vector and the host affect the dynamics of a disease [77]. If P. geniculatus is undergoing diversification, there will be new epidemiological challenges in the effective control of Chagas disease. The dispersal of the species is also highly relevant because of the incursion of sylvatic T. cruzi strains, which have been reported to be more virulent than domestic strains [7,8]. Mixed infections with the TcI, TcIII, and TcIV strains of T. cruzi, which are associated with sylvatic foci, have been reported [9], and there have also been reports of infection with the rest of the discrete typing units (DTUs) [1].
The success of vector control programs requires the accurate identification and incrimination of suspected vector species [78] and an understanding of the factors affecting transmission. These factors include the biological diversity, population dynamics, and spatial distribution of vector populations [77]. Specifically, the divergence of a species with a population structure can modify disease transmission and therefore alter the disease dynamics [77]. Panstrongylus geniculatus forms several vector populations, which could exploit several host populations. This research was a preliminary study to clarify the biology of P. geniculatus and may allow the development of effective strategies for its control, which should vary with the vector implicated [77].
Conclusion
Our study represents the first multilocus approach to estimate the intraspecific relationships of P. geniculatus from northern South America. We identified four genetically differentiated clades, which could be associated with geographic events that shaped the demography of the species, including the uplift of the Eastern Cordillera of the Colombian Andes. Further studies with samples drawn from more widely spread locations, which consider vector adaptability factors, are required to extend our knowledge of this pathogen–vector interaction. This information should then be considered in the design of vector control strategies aimed to reduce the prevalence of Chagas disease.
Supporting information
Last two letters of the ID correspond to species: Pg: Panstrongylus geniculatus, Pl: P. lignarius and Pm: P. megistus. Accession numbers: MK829849—MK 829943 (Cytb), MK829944—MK830032 (ND4), MK612546—MK6126555 (16S), MK612656—MK612759 (18S) and MK632224—MK632304 (28S).
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Numbers at the nodes are the bootstrap support values after 10,000 bootstrap replicates.
(TIF)
Numbers at the nodes are the bootstrap support values after 10,000 bootstrap replicates.
(TIFF)
Numbers on the nodes are support values (bootstrap for maximum likelihood and posterior probability for Bayesian inference). Colors of individuals indicate their geographic origin (i.e., they belong to the clade of the same color). Tip labels correspond to the identifiers (IDs) described in S1 Table.
(TIFF)
Acknowledgments
We thank Janine Miller, PhD, from Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript. We also thank the anonymously reviewers for their significant contribution to improve this paper.
Data Availability
All relevant data are within the manuscript and its Supporting Information files. GenBank accession numbers are in S4 Table.
Funding Statement
This work was funded by DIRECCION DE INVESTIGACION E INNOVACION from UNIVERSIDAD DEL ROSARIO "Big grant: Genómica, evolución y biogeografía de especies del género Rhodnius: vectores de la enfermedad de Chagas—IV-FGD002". CS and FS were funded by DIRECCION DE INVESTIGACION E INNOVACION FONDOS CONCURSABLES 2018, Universidad del Rosario—MODALIDAD SEMILLEROS-IV-ACA008. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hernández C, Salazar C, Brochero H, Teherán A, Buitrago LS, Vera M, et al. Untangling the transmission dynamics of primary and secondary vectors of Trypanosoma cruzi in Colombia: parasite infection, feeding sources and discrete typing units. Parasit Vectors. 2016;9: 620 10.1186/s13071-016-1907-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coura JR, Borges-Pereira J. Chagas disease: 100 years after its discovery. A systemic review. Acta Trop. 2010;115(1–2): 5–13. 10.1016/j.actatropica.2010.03.008 [DOI] [PubMed] [Google Scholar]
- 3.da Rosa JA, Justino HHG, Nascimento JD, Mendonça VJ, Rocha CS, de Carvalho DB, et al. A new species of Rhodnius from Brazil (Hemiptera, Reduviidae, Triatominae). ZooKeys. 2017;675(675): 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galvão C, Carcavallo R, Rocha DDS, Jurberg J. A checklist of the current valid species of the subfamily Triatominae Jeannel, 1919 (Hemiptera, Reduviidae) and their geographical distribution, with nomenclatural and taxonomic notes. Zootaxa. 2003;202(1): 1. [Google Scholar]
- 5.Patterson JS, Barbosa SE, Feliciangeli MD. On the genus Panstrongylus Berg 1879: Evolution, ecology and epidemiological significance. Acta Trop. 2009;110(2–3): 187–199. 10.1016/j.actatropica.2008.09.008 [DOI] [PubMed] [Google Scholar]
- 6.Molina JA, Gualdrón LE, Brochero HL, Olano V, Barrios D, Guhl F. Distribución actual e importancia epidemiológica de las especies de triatominos (Reduviidae: Triatominae) en Colombia. Biomédica. 2000;20: 344–360. [Google Scholar]
- 7.Valente VC, Valente SA, Noireau F, Carrasco HJ, Miles MA. Chagas disease in the Amazon Basin: association of Panstrongylus geniculatus (Hemiptera: Reduviidae) with domestic pigs. J Med Entomol. 1998;35(2): 99–103. 10.1093/jmedent/35.2.99 [DOI] [PubMed] [Google Scholar]
- 8.Sampson-Ward L, Urdaneta-Morales S. Urban Trypanosoma cruzi: biological characterization of isolates from Panstrongylus geniculatus. Ann Soc Belg Med Trop. 1988;68(2):95–106. [PubMed] [Google Scholar]
- 9.Jácome-Pinilla D, Hincapie-Peñaloza E, Ortiz MI, Ramírez JD, Guhl F, Molina J. Risks associated with dispersive nocturnal flights of sylvatic Triatominae to artificial lights in a model house in the northeastern plains of Colombia. Parasit Vectors. 2015;8: 600 10.1186/s13071-015-1209-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carrasco HJ, Segovia M, Londoño JC, Ortegoza J, Rodríguez M, Martínez CE. Panstrongylus geniculatus and four other species of triatomine bug involved in the Trypanosoma cruzi enzootic cycle: high risk factors for Chagas’ disease transmission in the Metropolitan District of Caracas, Venezuela. Parasit Vectors. 2014;7: 602 10.1186/s13071-014-0602-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guhl F, Restrepo M, Angulo VM, Antunes CMF, Campbell-Lendrum D, Davies CR. Lessons from a national survey of Chagas disease transmission risk in Colombia. Trends Parasitol. 2005;21(6): 259–262. 10.1016/j.pt.2005.04.011 [DOI] [PubMed] [Google Scholar]
- 12.Carrasco HJ, Torrellas A, García C, Segovia M, Feliciangeli MD. Risk of Trypanosoma cruzi I (Kinetoplastida: Trypanosomatidae) transmission by Panstrongylus geniculatus (Hemiptera: Reduviidae) in Caracas (metropolitan district) and neighboring states, Venezuela. Int J Parasitol. 2005;35(13): 1379–1384. 10.1016/j.ijpara.2005.05.003 [DOI] [PubMed] [Google Scholar]
- 13.Guhl F, Aguilera G, Pinto NA, Vergara D. Actualización de la distribución geográfica y ecoepidemiología de la fauna de triatominos (Reduviidae: Triatominae) en Colombia. Biomédica. 2007;27: 143–162. [PubMed] [Google Scholar]
- 14.Aldana E, Heredia-Coronado E, Avendaño-Rangel F, Lizano E, Concepcióm JL, Bonfante-Cabarcas R, et al. Análisis morfométrico de Panstrongylus geniculatus de Caracas, Venezuela. Biomédica. 2011;31: 108–117. 10.1590/S0120-41572011000100013 [DOI] [PubMed] [Google Scholar]
- 15.Avendaño-Rangel F, Sandoval CM, Aldana E, Aldana EJ. Descripción de setas cuticulares externas de cabeza, tórax, patas, abdomen y genitalias en cuatro especies de Triatominae. Biomédica. 2016;36(3): 354–358. 10.7705/biomedica.v36i3.3122 [DOI] [PubMed] [Google Scholar]
- 16.Santos C, Jurberg J, Galvão C, Rocha D da S, Fernandez JIR. Morphometric study of the genus Panstrongylus Berg, 1879 (Hemiptera, Reduviidae, Triatominae). Mem Inst Oswaldo Cruz. 2003;98: 939–944. 10.1590/s0074-02762003000700014 [DOI] [PubMed] [Google Scholar]
- 17.Lorosa ES, Dos Santos CM, Jurberg J. Foco de doença de Chagas em São Fidélis, no Estado do Rio de Janeiro. Rev Soc Bras Med Trop. 2008;41(4):419–20. 10.1590/s0037-86822008000400020 [DOI] [PubMed] [Google Scholar]
- 18.Marcilla A, Bargues MD, Abad-Franch F, Panzera F, Carcavallo RU, Noireau F, et al. Nuclear rDNA ITS-2 sequences reveal polyphyly of Panstrongylus species (Hemiptera: Reduviidae: Triatominae), vectors of Trypanosoma cruzi. Infect Genet Evol. 2002;1(3): 225–235. [DOI] [PubMed] [Google Scholar]
- 19.Justi SA, Russo CAM, Mallet JRDS, Obara MT, Galvão C. Molecular phylogeny of Triatomini (Hemiptera: Reduviidae: Triatominae). Parasit Vectors. 2014;7(1): 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lent H, Wygodzinsky PW. Revision of the Triatominae (Hemiptera, Reduviidae), and their significance as vectors of Chagas disease. Bull Am Mus Nat Hist. 1979; 163:125–520. [Google Scholar]
- 21.Qiagen. DNeasy® Blood & Tissue Handbook. For purification of total DNA from animal blood animal tissue. DNeasy® Blood Tissue Handb Purif Total DNA from Anim blood Anim tissue. 2006; 1–59.
- 22.Díaz S, Panzera F, Jaramillo-O N, Pérez R, Fernández R, Vallejo G, et al. Genetic, cytogenetic and morphological trends in the evolution of the Rhodnius (Triatominae: Rhodniini) trans-Andean group. PLoS One. 2014;9(2): e87493 10.1371/journal.pone.0087493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gómez-Palacio A, Triana O. Molecular evidence of demographic expansion of the Chagas disease vector Triatoma dimidiata (Hemiptera, Reduviidae, Triatominae) in Colombia. PLoS Negl Trop Dis. 2014;8(3): e2734 10.1371/journal.pntd.0002734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monteiro FA, Barrett TV, Fitzpatrick S, Cordon-Rosales C, Feliciangeli D, Berad CD. Molecular Phylogeography of the Amazonian Chagas disease vectors Rhodnius prolixus and R. robustus. Mol Ecol 2003, 12:997–1006. 10.1046/j.1365-294x.2003.01802.x [DOI] [PubMed] [Google Scholar]
- 25.Patterson PS, Gaunt MW. Phylogenetic multilocus codon models and molecular clocks reveal the monophyly of hematophagous reduviid bugs and their evolution at the formation of South America. Mol Phyl Evol 2010, 56:608–621. [DOI] [PubMed] [Google Scholar]
- 26.Geneious. Geneious 11.0.5 [software]. 2018 Jan 24. [cited 2018 Sep 17]. Available from: https://www.geneious.com
- 27.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21): 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 28.Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11): 1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- 29.Trifinopoulos J, Nguyen L-T, von Haeseler A, Minh BQ. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44: W232–235. 10.1093/nar/gkw256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berchtold A. Sequence analysis and transition models. Encycl Anim Behav. 2010: 139–145. [Google Scholar]
- 31.Minh BQ, Nguyen MAT, von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 2013;30(5): 1188–1195. 10.1093/molbev/mst024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bechara CCN, Londoño JC, Segovia M, Sanchez MAL, Martínez CE, Rodríguez MM, et al. Genetic variability of Panstrongylus geniculatus (Reduviidae: Triatominae) in the Metropolitan District of Caracas, Venezuela. Infect Genet Evol 2018; 66:236–244. 10.1016/j.meegid.2018.09.011 [DOI] [PubMed] [Google Scholar]
- 33.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3): 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tracer. Tracer v1.7.1. [software]. 2018 May 1 [cited 2018 Sep 17]. Available from: http://tree.bio.ed.ac.uk/software/tracer/
- 35.Rambaut A. FigTree v1.4.4. [software]. 2018 Nov 25. [Cited 2019 Feb 12]. Available from: http://tree.bio.ed.ac.uk/software/figtree/.
- 36.Heled J, Drummond AJ. Bayesian inference of species trees from multilocus data. Mol Biol Evol. 2010;27(3): 570–580. 10.1093/molbev/msp274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, et al. BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 2014;10(4): e1003537 10.1371/journal.pcbi.1003537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poinar G. Panstrongylus hispaniolae sp. n. (Hemiptera: Reduviidae: Triatominae), a new fossil triatomine in Dominican amber, with evidence of gut flagellates. Palaeodiversity. 2013;6:1–8. [Google Scholar]
- 39.Leigh JW, Bryant D. POPART: Full-feature software for haplotype network construction. Methods Ecol Evol. 2015;6(9): 1110–1116. [Google Scholar]
- 40.Wright S. The genetical structure of populations. Ann Eugen. 1949;15(1): 323–354. [DOI] [PubMed] [Google Scholar]
- 41.Nei M, Miller JC. A simple method for estimating average number of nucleotide substitutions within and between populations from restriction data. Genet Soc Am. 1990. 125:873–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27(1): 209–220. [PubMed] [Google Scholar]
- 43.Dixon P. VEGAN, a package of R functions for community ecology. J Veg Sci. 2003;14:927–30. [Google Scholar]
- 44.Rousset F. GENEPOP’007: A complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour. 2008;8(1): 103–106. 10.1111/j.1471-8286.2007.01931.x [DOI] [PubMed] [Google Scholar]
- 45.Hijmans RJ. Spherical trigonometry: R package geosphere version 1.5–10 [software]. 2017 Nov 5 [cited 2018 Sep 17] Available from: https://cran.r-project.org/web/packages/geosphere/index.html
- 46.Legendre P, Fortin MJ. Comparison of the Mantel test and alternative approaches for detecting complex multivariate relationships in the spatial analysis of genetic data. Mol Ecol Resour. 2010;10(5): 831–844. 10.1111/j.1755-0998.2010.02866.x [DOI] [PubMed] [Google Scholar]
- 47.Crossa RP, Hernández M, Caraccio MN, Rose V, Valente SAS, Valente VC, et al. Chromosomal evolution trends of the genus Panstrongylus (Hemiptera, Reduviidae), vectors of Chagas disease. Infect Genet Evol. 2002;2(1): 47–56 [DOI] [PubMed] [Google Scholar]
- 48.Justi SA, Galvão C, Schrago CG. Geological changes of the Americas and their influence on the diversification of the neotropical kissing bugs (Hemiptera: Reduviidae: Triatominae). PLoS Negl Trop Dis. 2016;10(4): e0004527 10.1371/journal.pntd.0004527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Monteiro FA, Weirauch C, Felix M, Lazoski C, Abad-Franch F. Evolution, Systematics, and Biogeography of the Triatominae, Vectors of Chagas Disease. Adv Parasitol. 2018;99:265–344 10.1016/bs.apar.2017.12.002 [DOI] [PubMed] [Google Scholar]
- 50.Hypsa V, Tietz DF, Zrzavý J, Rego R, Galvao C, Jurberg J. Phylogeny and biogeography of Triatominae (Hemiptera: Reduviidae): molecular evidence of a New World origin of the Asiatic clade a clav Hyp. Mol Phylogenet Evol. 2002;23:447–57. [DOI] [PubMed] [Google Scholar]
- 51.de Paula A, Diotaiuti L, John C. Testing the sister-group relationship of the Rhodniini and Triatomini (Insecta: Hemiptera: Reduviidae: Triatominae). Mol Phylogenet Evol. 2005;35:712–8. 10.1016/j.ympev.2005.03.003 [DOI] [PubMed] [Google Scholar]
- 52.Zhang D-X, Hewitt GM. Nuclear DNA analyses in genetic studies of populations: practice, problems and prospects. Mol Ecol. 2003;12(3): 563–584. 10.1046/j.1365-294x.2003.01773.x [DOI] [PubMed] [Google Scholar]
- 53.Ballard JWO, Whitlock MC. The incomplete natural history of mitochondria. Mol Ecol. 2004;13(4): 729–744. 10.1046/j.1365-294x.2003.02063.x [DOI] [PubMed] [Google Scholar]
- 54.Eo SH, DeWoody JA. Evolutionary rates of mitochondrial genomes correspond to diversification rates and to contemporary species richness in birds and reptiles. Proc Biol Sci. 2010;277(1700): 3587–3592. 10.1098/rspb.2010.0965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Justi SA, Galvão C. The evolutionary origin of diversity in Chagas disease vectors. Trends Parasitol. 2017;33(1): 42–52. 10.1016/j.pt.2016.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Justi SA, Cahan S, Stevens L, Monroy C, Lima-Cordón R, Dorn PL. Vectors of diversity: Genome wide diversity across the geographic range of the Chagas disease vector Triatoma dimidiata sensu lato (Hemiptera: Reduviidae). Mol Phylogenet Evol. 2018;120:144–50. 10.1016/j.ympev.2017.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abad-Franch F, Monteiro FA. Biogeography and evolution of Amazonian triatomines (Heteroptera: Reduviidae): Implications for Chagas disease surveillance in humid forest ecoregions. Mem. Inst. Oswaldo Cruz.; 2007: 57–69. 10.1590/s0074-02762007005000108 [DOI] [PubMed] [Google Scholar]
- 58.Cuervo AM. Evolutionary Assembly of the Neotropical Montane Avifauna [Internet]. LSU Doctoral Dissertations; 2013. [cited 2018 Sep 22]. Available from: https://digitalcommons.lsu.edu/gradschool_dissertations/275/ [Google Scholar]
- 59.Cadena CD, Pedraza CA, Brumfield RT. Climate, habitat associations and the potential distributions of Neotropical birds: Implications for diversification across the Andes. Rev Acad Colomb Cienc Ex Fis Nat. 2016;40(155): 275–287. [Google Scholar]
- 60.Haffer J. In: Wolters H. E., editor. Avifauna of Northwestern Colombia, South America. Germany: Bonner Universitäts-Buchdruckerei;1975. p. 22–30 [Google Scholar]
- 61.Chazot N, De-Silva DL, Willmott KR, Freitas AVL, Lamas G, Mallet J, et al. Contrasting patterns of Andean diversification among three diverse clades of Neotropical clearwing butterflies. Ecol Evol. 2018;8(8): 3965–3982. 10.1002/ece3.3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.De-Silva DL, Elias M, Willmott K, Mallet J, Day JJ. Diversification of clearwing butterflies with the rise of the Andes. J Biogeogr. 2016;43(1): 44–58. 10.1111/jbi.12611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De-Silva DL, Mota LL, Chazot N, Mallarino R, Silva-Brandão KL, Piñerez LMG, et al. North Andean origin and diversification of the largest ithomiine butterfly genus. Sci Rep. 2017;7: 45966 10.1038/srep45966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dick CW, Roubik DW, Gruber KF, Bermingham E. Long-distance gene flow and cross-Andean dispersal of lowland rainforest bees (Apidae: Euglossini) revealed by comparative mitochondrial DNA phylogeography. Mol Ecol. 2004;13(12): 3775–3785. 10.1111/j.1365-294X.2004.02374.x [DOI] [PubMed] [Google Scholar]
- 65.Bargues MD, Klisiowicz DR, Gonzalez-Candelas F, Ramsey JM, Monroy C, Ponce C, et al. Phylogeography and genetic variation of Triatoma dimidiata, the main Chagas disease vector in Central America, and its position within the genus Triatoma. PLoS Negl Trop Dis. 2008;2(5): e233 10.1371/journal.pntd.0000233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Campos R, Torres-Pérez F, Botto-Mahan C, Coronado X, Solari A. High phylogeographic structure in sylvatic vectors of Chagas disease of the genus Mepraia (Hemiptera: Reduviidae). Infect Genet Evol. 2013;19: 280–286. 10.1016/j.meegid.2013.04.036 [DOI] [PubMed] [Google Scholar]
- 67.Monteiro FA, Peretolchina T, Lazoski C, Harris K, Dotson EM, Abad-Franch F, et al. Phylogeographic pattern and extensive mitochondrial DNA divergence disclose a species complex within the Chagas disease vector Triatoma dimidiata. PLoS One. 2013;8(8): e70974 10.1371/journal.pone.0070974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Salgado-Roa FC, Pardo-Diaz C, Lasso E, Arias CF, Solferini VN, Salazar C. Gene flow and Andean uplift shape the diversification of Gasteracantha cancriformis (Araneae: Araneidae) in northern South America. Ecol Evol. 2018;8(14): 7131–7142. 10.1002/ece3.4237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dasmahapatra KK, Lamas G, Simpson F, Mallet J. The anatomy of a ‘suture zone’ in Amazonian butterflies: a coalescent-based test for vicariant geographic divergence and speciation. Mol Ecol. 2010;19(19): 4283–4301. 10.1111/j.1365-294X.2010.04802.x [DOI] [PubMed] [Google Scholar]
- 70.Nadeau NJ, Ruiz M, Salazar P, Counterman B, Medina JA, Ortiz-Zuazaga H, et al. Population genomics of parallel hybrid zones in the mimetic butterflies, H. melpomene and H. erato. Genome Res. 2014; 24(8): 1316–1333. 10.1101/gr.169292.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dorn PL, de la Rúa NM, Axen H, Smith N, Richards BR, Charabati J, et al. Hypothesis testing clarifies the systematics of the main Central American Chagas disease vector, Triatoma dimidiata (Latreille, 1811), across its geographic range. Infect Genet Evol. 2016;44: 431–443. 10.1016/j.meegid.2016.07.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ramírez CJ, Jaramillo CA, Delgado M del P, Pinto NA, Aguilera G, Guhl F. Genetic structure of sylvatic, peridomestic and domestic populations of Triatoma dimidiata (Hemiptera: Reduviidae) from an endemic zone of Boyacá, Colombia. Acta Trop. 2005;93(1): 23–29. 10.1016/j.actatropica.2004.09.001 [DOI] [PubMed] [Google Scholar]
- 73.Gómez-Palacio A, Triana O, Jaramillo-O N, Dotson EM, Marcet PL. Eco-geographical differentiation among Colombian populations of the Chagas disease vector Triatoma dimidiata (Hemiptera: Reduviidae). Infect Genet Evol. 2013;20: 352–361. 10.1016/j.meegid.2013.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gourbière S, Dorn P, Tripet F, Dumonteil E. Genetics and evolution of triatomines: From phylogeny to vector control. Heredity. 2012;108: 190–202. 10.1038/hdy.2011.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.World Health Organization. Chagas disease in Latin America: an epidemiological update based on 2010 estimates. Weekly epidemiological record. 2015. 90(06),33–44. [PubMed] [Google Scholar]
- 76.Omahmaharaj I. Studies on vectors of Trypanosoma cruzi in Trinidad, West-Indies. Med Vet Entomol. 1992;6(2): 115–120. [DOI] [PubMed] [Google Scholar]
- 77.McCoy KD. The population genetic structure of vectors and our understanding of disease epidemiology. Parasite. 2008;15(3): 444–448. 10.1051/parasite/2008153444 [DOI] [PubMed] [Google Scholar]
- 78.Monteiro FA, Escalante AA, Beard CB. Molecular tools and triatomine systematics: a public health perspective. Trends Parasitol. 2001;17(7): 344–347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Last two letters of the ID correspond to species: Pg: Panstrongylus geniculatus, Pl: P. lignarius and Pm: P. megistus. Accession numbers: MK829849—MK 829943 (Cytb), MK829944—MK830032 (ND4), MK612546—MK6126555 (16S), MK612656—MK612759 (18S) and MK632224—MK632304 (28S).
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Numbers at the nodes are the bootstrap support values after 10,000 bootstrap replicates.
(TIF)
Numbers at the nodes are the bootstrap support values after 10,000 bootstrap replicates.
(TIFF)
Numbers on the nodes are support values (bootstrap for maximum likelihood and posterior probability for Bayesian inference). Colors of individuals indicate their geographic origin (i.e., they belong to the clade of the same color). Tip labels correspond to the identifiers (IDs) described in S1 Table.
(TIFF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files. GenBank accession numbers are in S4 Table.