Supplemental Digital Content is available in the text
Keywords: Esophageal cancer, microRNA, Clinical features, Overall survival
Abstract
Background:
MicroRNAs (miRNAs) have played important roles in the regulation of gene expression in many cancers, but their roles in esophageal squamous cell carcinoma (ESCC) are still unclear. The aim of this study was to determine the potential ESCC-specific key miRNAs from a large sample dataset in The Cancer Genome Atlas (TCGA).
Methods:
Integrative bioinformatics analysis was used to identify key ESCC-specific miRNAs related to the ESCC patients’ tumor histological grade and lymphatic metastasis from TCGA. Next, these key miRNA potential gene regulatory functions and relationships with ESCC patients’ clinical characteristics and overall survival were analyzed. Finally, three key miRNAs were selected randomly and quantificational real-time polymerase chain reaction (qRT-PCR) was used to validate in 51 newly diagnosed ESCC patients’ tissues samples (collected from Nov. 2017 to Feb. 2019, in Wuwei, China) whether the bioinformatics analyses results were reliable and valid. Two-tailed Student's t test, Pearson Chi-squared test and Kaplan-Meier survival analysis were used in this study.
Results:
Thirty-five ESCC-specific miRNAs from TCGA database were investigated (fold-change > 2.0, P < 0.05), and 28 participated in the miRNAs-mRNAs co-expression network construction, while 17 were related with ESCC patients’ tumor histological grade, TNM stage, and lymphatic metastasis (P < 0.05). Meanwhile, six miRNAs (including miR-200b-3p, miR-31-5p, miR-15b-5p, miR-141-3p, miR-135b-5p, and miR-195-5p) were correlated with overall survival of ESCC patients (log-rank, P < 0.05). MiR-135b-5p, miR-15b-5p, and miR-195-5p were selected for verification of the expression levels in 51 ESCC patients’ tissue samples by using qRT-PCR. We found that the fold-changes between qRT-PCR and TCGA were completely consistent. The results also suggested that miR-135b-5p, miR-15b-5p, and miR-195-5p were significantly correlated with tumor differentiation degrees (P < 0.05), miR-195-5p was significantly correlated with tumor TNM stage (P < 0.05), and miR-135b-5p was significantly correlated with lymph-node metastasis (P < 0.05). MiR-135b-5p, miR-15b-5p, and miR-195-5p expression levels, ESCC patient clinical features association analysis results and the aforementioned TCGA bioinformatics analyses were similar.
Conclusion:
This study identified key ESCC-related miRNAs. The key miRNAs are worthy of further investigation as potential novel biomarkers for diagnosis, classification, and prognosis of ESCC.
Introduction
Esophageal squamous cell carcinoma (ESCC) is a common fatal malignant disease in humans, which causes more than 400,000 deaths per year.[1] In spite of the advances in multidisciplinary diagnosis and treatment, there are still important topics that need to be explored for the early diagnosis and prognosis biomarkers of ESCC.[2] The ESCC detection methods are based on the endoscopy examination, while this method was often affected by doctors’ individual experience and technology, with a high rate of missed diagnosis. Meanwhile, endoscopic diagnoses of ESCC cannot find the early stage of lymph node metastasis status in esophageal carcinoma.[3] In addition, conventional non-invasive serological biomarkers, such as carbohydrate antigen 19-9 (CA19-9) and carcinoembryonic antigen (CEA), are lack of specificity in the early diagnosis of ESCC.[4] Therefore, the significance of exploration of innovative and favorable biomarkers for ESCC diagnosis and prognosis should be emphasized.
MicroRNAs (miRNAs, 18–22 nucleotides) are small, non-coding RNAs that can bind to the messenger RNAs and negatively regulate the gene expression.[5] Studies have revealed that miRNAs function in many key biological roles in various disease progressions, including cancers. Therefore, further insights into the important roles of miRNAs in diseases, especially in cancers, have shown miRNAs as attractive tools and targets for novel diagnoses, prognoses, and therapeutic approaches. ESCC is a multistep-process disease, which involves multiple effects of gene-environmental factor interactions. The miRNAs play important regulatory roles in epigenetics and are closely related with many diseases progressions.[6] Further understanding of the ESCC tissue alterations and miRNA expression profiles may help us to identify better biomarkers for ESCC diagnosis, classification, and prognosis.
Further evidence suggested that aberrantly expressed miRNAs in ESCC tissues have a clinical influence on disease diagnosis and prognosis.[7] Recently, miRNA-associated biomarkers have been reported for the diagnosis and prognoses of ESCC.[8,9] However, the small sample sizes used for microarray detection often present a bias toward the identification of ESCC-associated miRNAs due to lack of RNA sequencing data, and therefore often generate errors. Thus far, The Cancer Genome Atlas (TCGA) (https://tcga-data.nci.nih.gov/tcga/) database has provided a resource for large sample size genome sequencing data from 33 human cancer types including more than 30,000 patients.[10] In the present study we downloaded and analyzed the ESCC patients’ RNA sequencing expression profiles and clinical pathology features from TCGA database using bioinformatics techniques. This way will improve the method to discover potential miRNA biomarkers for the diagnosis of ESCC, as well as its classification and prediction of prognosis.
Methods
Ethical approval
Informed consent forms and clinical information were obtained for all patients by the investigator and from medical records, respectively. The procedures used in this study were approved by the Wuwei Tumor Hospital of Gansu (Wuwei, China) and conformed to the Declaration of Helsinki and current legislation.
Data collection
The ESCC patient tumor tissues and normal esophageal epithelial tissue miRNA sequencing data set and patient personal information and clinical pathological features are included from the TCGA database (supplementary Table S1). RNA sequencing data were obtained from Illumina HiSeq platforms (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga/using-tcga/types). ESCC patient exclusion criteria included: (1) the admitting diagnosis was not ESCC; (2) suffering from two or more types of cancers including ESCC; (3) clinical information or RNA sequencing data were deficient; (4) patients treated with radiation and chemotherapy before specimen collection, and (5) overall survival time more than 5 years. In the present study, we obtained ESCC patients’ tissues samples including normal esophageal mucosa tissues 11 cases, pathologic Stage I 27 cases, Stage II 74 cases and Stage III-IV 60 cases. Before further analyses and processing, RNA-sequencing data normalization was performed in full compliance with the publication guidelines of TCGA; the approval of ethics committee was not required.
Study design and data preprocessing
Hierarchical cluster analyses were performed using R package heat maps. Then, according to the z-score values to deduce the expression levels of samples RNA sequencing data, we analyzed the gene pair Euclidean distances to detect gene clusters. Subsequently, the significantly different miRNA and mRNA expression profiles in ESCC were analyzed separately, and we determined the significantly different genes of the normal vs. cancerous tissues of ESCC patients. We compared the differentially expressed miRNAs and mRNA of four stage levels, including ESCC (G I stage) non-lymphatic metastases vs. non-tumor esophageal tissues, ESCC (G I stage) lymphatic metastases vs. non-tumor esophageal tissues, ESCC (G II stage) lymphatic metastases vs. non-tumor esophageal tissues, and ESCC (G III-IV stage) lymphatic metastases vs. non-tumor esophageal tissues, respectively. The intersections of the miRNAs and mRNAs were selected in the above four stage levels for further analyses using bioinformatics methods. Figure 1 is the flow diagram of this study design.
Figure 1.
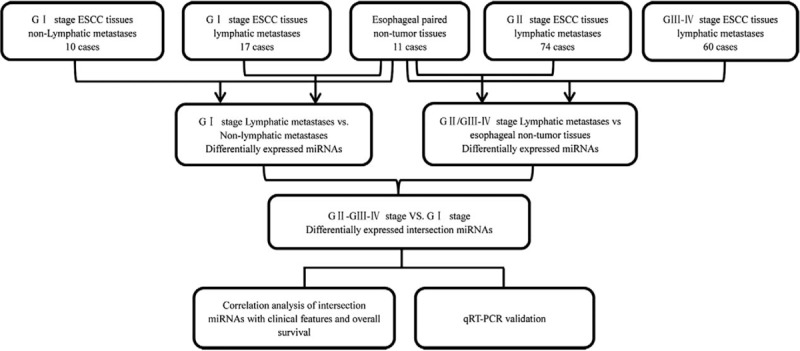
Flow diagram of the study. Data preparing, processing, and bioinformatics analysis flowchart.
Co-expression network of miRNAs and mRNAs
For the current study, according to the findings that miRNAs can bind to the messenger RNAs and negatively regulate gene expression, a miRNA-mRNA network was built. Significantly differentially expressed intersecting miRNA and mRNA were used to build a co-expression network, which showed fold change >2.0 and P < 0.05. Target genes of miRNAs were predicted by miRanda (http://www.microrna.org/microrna/home.do) and Targetscan (http://www.targetscan.org/). The combined miRNA predicted target gene information and significantly differentially expressed intersecting mRNAs of TCGA ESCC patients were used to build the miRNA-mRNA network.
The miRNAs-mRNAs co-expression network was constructed using significantly differential gene expression profiles using R package software (The University of Auckland, Auckland, New Zealand). Pearson correlation matrices models were used to find the potential relevance of all pair-wise genes. Subsequently, based on the weighted adjacency matrix, we measured the network connectivity of a gene with other gene network generations, and built the miRNAs-mRNAs co-expression network by using Cytoscape 3.0 software (National Institute of General Medical Sciences, MD, USA).
Analysis of the association between key miRNAs and clinical status
According to the co-expression of miRNAs and mRNAs, the key miRNAs involved in co-expression networks were selected as our targets to determine if they positively correlated with ESCC. Subsequently, we further analyzed the potential associations between co-expression network key miRNAs and TCGA ESCC patients’ clinical features, including sex, TNM stage, tumor grade, lymphatic metastases status, and pathological stage by using multiple linear regression analyses.
Kaplan-Meier survival analyses
To analyze the correlation between the changes of the selected key miRNA expression levels and ESCC patients’ prognostic survival, correlations were determined by Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/). Publicly available data and GEPIA tools were prepared to calculate the Kaplan-Meier survival analysis parameters using TCGA ESCC patients’ datasets. All the RNA sequencing data probe sets or records were averaged by the quantile normalized data. Finally, the survival distributions equality of different TCGA ESCC patients and the key miRNA expression level changes were analyzed using the Kaplan-Meier, log-rank, and the hazard ratio (HR) tests.
Preparation for human ESCC samples
Next, 51 ESCC patients’ tumor tissues samples and paired non-tumor esophagus tissues (aged 40–69 years) were collected from Gansu Wuwei Tumor Hospital, China, for RT-qPCR validation. All of these patients were diagnosed with ESCC according to their histopathology. Samples were collected and stored in RNAlater (Ambion, Foster City, CA, USA) at −80°C. Collection and use of ESCC patients’ tumor tissue samples were approved by the ethics committee of Gansu Wuwei Tumor Hospital.
Quantitative real time PCR (qRT-PCR) validation of bioinformatics data
Total RNA of ESCC patients’ tissues samples were isolated by TRIzol® reagent (Invitrogen, Carlsbad, CA, USA). Total RNA quantity was evaluated by a NanoDrop® ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). A Reverse Transcription Kit (Promega, Madison, WI, USA) and GoTaq® qPCR Master Mix of Power SYBR® Green (Promega) were used to synthesized cDNA and for qRT-PCR detection. Finally, qRT-PCR reactions were performed using the Step One PlusTM PCR System (Applied Biosystems, Foster City, CA, USA).
Statistical analyses
Data were analyzed using the SPSS Statistics 21.0 (International Business Machines Corporation, Armonk, NY, USA) and expressed as mean ± standard deviation (SD). All analyses were performed three times and represent data from three individual experiments. Two-tailed Student's t test was used to significance of differences between subgroups. Pearson Chi-squared test was used to find the association between the expression of miRNAs and ESCC patients’ clinicopathological characteristics. Kaplan-Meier survival analysis was used to investigate the correlation between the changes of miRNAs expression levels and patients’ prognostic overall survival times. Statistical significance was set at P < 0.05. The qRT-PCR results were calculated and evaluated by the 2−ΔΔCt method.
Results
Differentially expressed miRNAs screening in ESCC patients
We used ESCC patients’ RNA-seq datasets collected from 161 cancerous and 11 normal specimens for further analyses. There were 232 miRNAs that were significantly differentially expressed between ESCC patients’ tumor vs. normal esophageal tissues samples in the Level 3 ESCC RNA-Sequencing (RNA-Seq) data from TCGA database. Next, we integrated and analyzed the relevance between the 232 miRNA expression levels and ESCC patients’ differential clinical stage and pathologic grade. There were 103 miRNAs that showed significant differential changes between G I stage (non-lymphatic metastasis) ESCC tumors and normal esophageal tissues samples. In addition, 119 miRNAs were significantly differentially changed between G I stage (lymphatic metastasis) ESCC tumors and normal esophageal tissues samples; 125 miRNAs were found to be significantly differentially changed between G II stage (lymphatic metastasis) ESCC tumors and normal esophageal tissue samples; and 150 miRNAs were found to be significantly differentially changed between G III-IV stage (lymphatic metastasis) ESCC tumors and normal esophageal tissues samples. Subsequently, we used Venn diagrams (http://bioinfogp.cnb.csic.es/tools/venny/index.html) to analyze the overlapping subclasses of miRNAs, and found there were 35 key intersecting miRNAs [Figure 2]. These 35 intersecting miRNAs included 25 miRNAs that were overexpressed and 10 miRNAs that were downregulated (supplementary Table S2).
Figure 2.
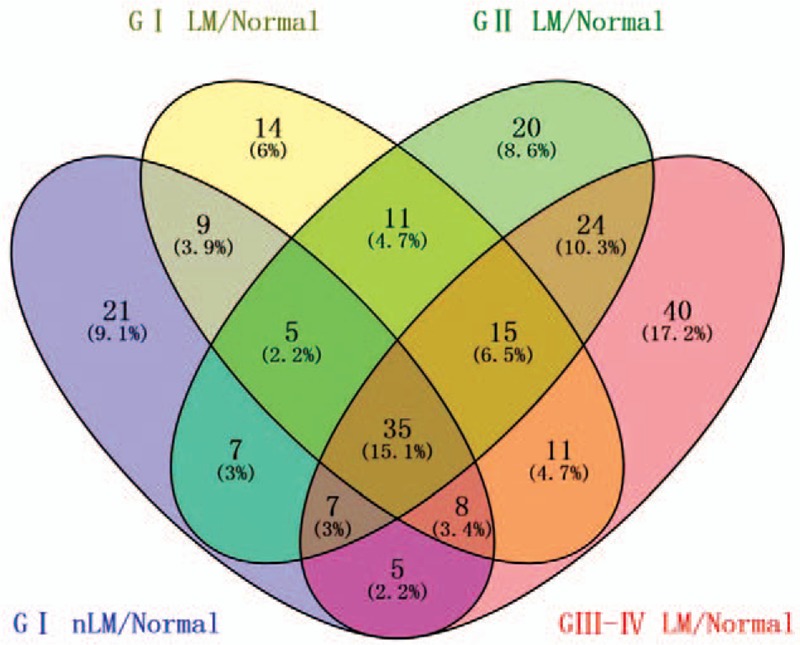
Venn diagram analysis of differentially expressed miRNAs between G I nLM/Normal, G I LM/Normal, G II LM/Normal, G III-IV LM/Normal. (LM represents lymphatic metastasis; nLM represent non-lymphatic metastasis; Normal represents normal esophageal tissues).
MiRNAs-mRNAs co-expression network construction and potential regulatory function analyses
We chose the abovementioned 35 intersecting miRNAs from 232 ESCC associated miRNAs by using a Venn diagram overlapping subclass analysis to construct the miRNAs-mRNAs network. Next, to build the miRNA-mRNA co-expression network, we also predicted the mRNAs targeted by the miRNAs. According to these 35 intersecting miRNAs (supplementary Table S2), we predicted that these miRNAs targeted mRNAs by using the miRanda and Targetscan databases. Based on the bioinformatics analysis, we found that there were 376 intersecting mRNAs and 2235 significantly differentially expressed mRNAs in ESCC cancer tissues from TCGA database. Finally, based on the 35 intersecting miRNAs and 376 intersecting mRNAs, we constructed a miRNAs-mRNAs co-expression network. The relationships between miRNAs and mRNAs were analyzed by their negative regulation mechanisms and the co-expression network was drawn using Cytoscape 3.0. There were 28 targeted miRNAs that potentially regulated 72 mRNAs and were involved in the miRNA-mRNA co-expression network [Figure 3]. To further analyze these 72 negatively regulated mRNAs in the co-expression network, we found that some played important roles in cancer development, such as E2F2, FGF2, COL4A3, ERBB4, FGFR1, RELN, and TCF3.
Figure 3.
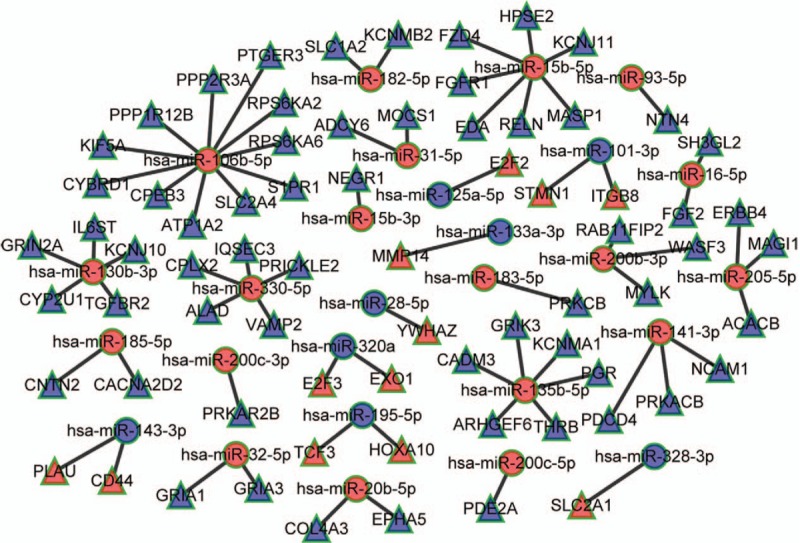
The miRNAs-mRNAs competing endogenous RNAs network. Red balls represent upregulated miRNAs, blue balls, downregulated miRNAs; Red triangles represent upregulated mRNAs, blue triangles, downregulated mRNAs.
To obtain further insight into the potential regulatory functions of the 28 miRNAs on the 72 mRNAs involved in miRNAs-mRNAs networks, the up- and down-regulated mRNAs were analyzed using the DAVID database. Gene Ontology (GO) analyses revealed upregulated mRNAs enriched in ‘regulation of cellular process, regulation of biological process and protein binding’ and downregulated mRNAs transcripts enriched in ‘single-organism cellular process, single-organism process, and response to stimulus’ [Figure 4A]. These significantly enriched GO functions provided a better understanding of the roles of mRNAs in regulatory functions. In addition, pathway analysis was performed for all 72 mRNAs, and the results suggested that the enriched pathways targeted by upregulated mRNAs transcripts were ‘microRNAs in cancer, human T-cell leukaemia virus type I (HTLV-I) infection, cell cycle and hepatitis B.’ The enriched pathways of downregulated transcripts were ‘insulin secretion, glutamatergic synapse, proteoglycans in cancer, and vascular smooth muscle contraction’ [Figure 4B].
Figure 4.
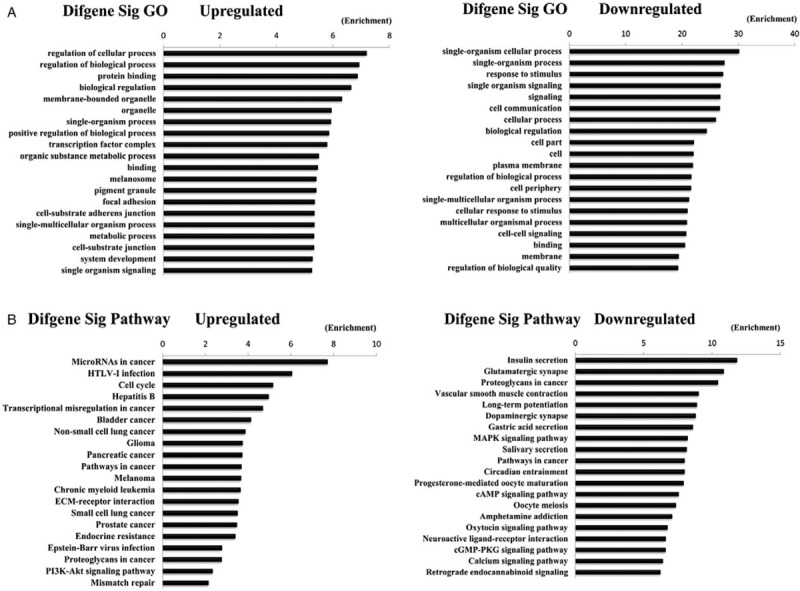
Top 20 enrichments of Gene Ontology (GO) terms for differentially expressed mRNAs and pathways for differentially expressed intersection mRNAs. (A) Top 20 enrichments of GO terms for differentially expressed mRNAs (the bar plot shows the enrichment scores of the significant enrichment GO terms). (B) Top 20 enrichments of pathways for differentially expressed intersection mRNAs (the bar plot shows the enrichment scores of the significant enrichment pathways).
Correlations between miRNA signatures and clinical characteristics
We selected the 28 miRNAs, which were involved in miRNA-mRNA co-expression networks, and further analyzed the correlation between these miRNAs and TCGA database of 161 ESCC patients’ clinical pathology features. The ESCC clinical/demographic features included patients’ sex, race, tumor grade, TNM stage, and lymphatic metastasis taken from TCGA database. Multiple linear regression analysis results suggested that there were 17 miRNAs differentially expressed with different clinical pathology features (P < 0.05). We showed that miR-32-5p, miR-330-5p, miR-135b-5p, miR-195-5p, and miR-145-5p were associated with ESCC patients’ tumor grade, miR-16-5p, miR-32-5p, miR-93-5p, miR-185-5p, miR-15b-5p, miR-135b-5p, miR133a-3p, miR-28-5p, and miR-195-5p were associated with TNM stages, and miR-32-5p, miR-93-5p, miR-15b-5p, miR-135b-5p, miR-330-5p, miR-106b-5p, miR-182-5p, miR-20b-5p, miR-141-3p, miR-28-5p, miR-195-5p, and miR-320a were associated with lymphatic metastasis. Meanwhile, miR-16-5p and miR-183-5p were found associated with racial/ethnic background [Table 1].
Table 1.
Correlations between ESCC specific intersection miRNAs and clinical features.

Prognostic analysis of miRNA expression levels and ESCC patient survival
A Kaplan-Meier survival analysis was used to investigate the relationships between the miRNAs-mRNAs co-expression network 28 key miRNAs and ESCC patients’ survival time. The 28 key miRNA expression levels and the ESCC patient overall survival information in TCGA database were calculated using univariate Cox proportional hazard regression model analyses. We found six miRNAs closely related with ESCC patient overall survival time (log-rank, P < 0.05). Among these six miRNAs, there were five miRNAs (miR-200b-3p, miR-31-5p, miR-15b-5p, miR-141-3p, and miR-135b-5p) that were negatively related with ESCC patient overall survival time (P < 0.05), and miR-195-5p was positively correlated with ESCC patient overall survival time (P < 0.05) [Figure 5].
Figure 5.
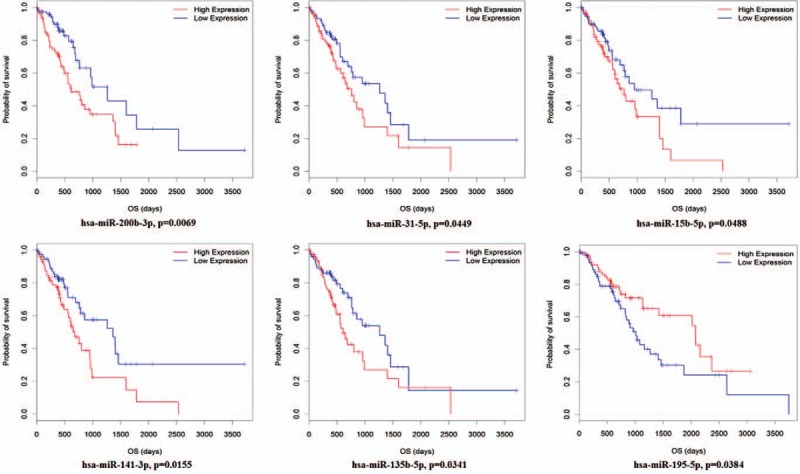
Kaplan-Meier survival curves for six miRNAs associated with ESCC patients overall survival time (Horizontal axis overall survival time: days, Vertical axis survival function). hsa: homo sapiens; OS: overall survival.
Verification of selected miRNAs expression levels in ESCC tissues by qRT-PCR
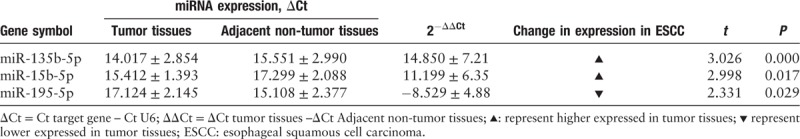
Based on the above bioinformatics analyses, three miRNAs (miR-135b-5p, miR-15b-5p, and miR-195-5p) were randomly selected. The three key miRNA expression levels in newly diagnosed 51 ESCC patients’ tumor samples and their self-paired normal esophagus epithelial tissues samples were compared to previous TCGA bioinformatics analysis results for reliability and validity. The expression levels of miR-135b-5p and miR-15b-5p were consistently upregulated in 51 ESCC patient tumor tissue samples compared with normal esophageal epithelial tissues (P < 0.05); however, miR-195-5p expression levels were consistently downregulated (P < 0.05) [Table 2]. Subsequently, we also compared the increased or decreased fold-changes between these three miRNAs verified by qRT-PCR and TCGA ESCC RNA fold-changes. The fold-changes measured by qRT-PCR of the 51 collected ESCC patients’ tissue samples and TCGA ESCC RNA sequencing results were consistent (supplementary Table S2 and Table 2).
Table 2.
Relative expression of miRNAs in 51 pairs of ESCC tumor and non-tumor esophagus tissues (mean ± SD).
Relationships between the three selected miRNAs and ESCC clinical pathological features
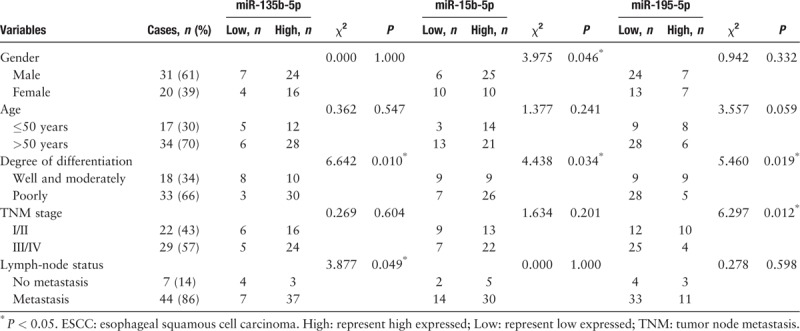
We found that miR-135b-5p, miR-15b-5p, and miR-195-5p expression levels were significantly related with the 51 ESCC patients tumor differentiation degrees (P < 0.05) [Table 3]; miR-195-5p was significantly related with tumor TNM stage (P < 0.05), and miR-135b-5p was significantly related with lymph-node metastasis (P < 0.05). The differential expression of miR-15b-5p was significantly associated with the 51 ESCC patients’ sex (P < 0.05). These three miRNAs from ESCC patients’ correlated with the above TCGA bioinformatics comprehensive analysis [Table 1] and were in good agreement, suggesting that TCGA bioinformatics analysis was reliable.
Table 3.
Association between the expression of miR-135b-5p, miR-15b-5p, miR-195-5p and clinicopathological characteristics of ESCC patients.
Discussion
ESCC is the main type of esophageal cancer and is a common cause of cancer-related death worldwide.[11,12] ESCC development and progression is a complex process, which includes a variety of obvious changes in gene expression levels and subsequent physiological changes. MiRNAs are small, non-coding regulatory RNAs that inhibit target mRNAs.[13] Recent advances in miRNA expression profiling have suggested new directions to study the pathological changes of ESCC, and also indicate that miRNAs have great potential as novel biomarkers in ESCC diagnosis and prognosis.[14,15] Recent studies mainly concentrated on small sample size RNA sequencing or microarray data detection and, therefore, may not accurately reflect the dysregulated miRNAs that link to tumorigenesis and progression of ESCC.[16,17] Therefore, the ESCC related significant differences of miRNAs should be identified in a large sample size to enhance accuracy and reliability of predictive value.
In this study, we analyzed the dysregulated miRNAs in ESCC from TCGA database. According to the ESCC patient tissue sample miRNA sequencing data sets in TCGA, we further explored the significant differences of miRNAs related to patients’ clinical features and overall survival time by using integrated bioinformatics analysis. Three miRNAs were selected randomly and we detected their expression levels in 51 newly collected ESCC patients’ tumor samples and their self-paired normal esophageal epithelial tissue samples by qRT-PCR. We further analyzed the correlations between the three miRNAs and ESCC patients’ clinical pathological characteristics to confirm the accuracy and reliability of the bioinformatics analysis results.
According to the integrated bioinformatics overlapping subclasses analysis results we found 35 dysregulated miRNAs in ESCC tumor tissues and significant correlation between ESCC histological grades and lymphatic metastasis status. Among these 35 dysregulated miRNAs, some have been detected that showed significant differences expressed in esophagus cancer, including upregulated miR-205, which may act as an oncogene in ESCC.[18] Alder et al[19] reported that miR-31 and miR-21 were dysregulated in ESCC and their expression levels were associated with zinc deficiency promoted esophageal cancer. Gu et al[20] investigated the miRNA diagnostic values in ESCC pathogenesis and prognosis, and found that in ESCC upregulation of miR-93, miR-106, and miR-205 were closely related with ESCC pathogenesis and prognosis. In addition, upregulated miR-20b-5p, miR-93-5p, miR-16-5p, miR-330-5p, miR-106b-5p, and miR-484 were reported in esophageal cancer and were involved in regulating the progression of this disease.[21–23] The downregulation of miR-133a expression showed changes from Barrett esophagus to esophageal cancer.[24] Derouet et al reported that miR-145 expression effected the progression of esophageal cancer by enhancing cell invasion and apoptosis.[25] MiR-143-3p, miR-125a-5p, and miR-101-3p levels also were reported to be significantly different in esophageal cancer tissues and cell lines.[20,26–28] For the remaining 16 dysregulated miRNAs, including 10 upregulated and six downregulated miRNAs, their functions in the development and progression of ESCC have not been reported.
Based on the theory that mRNA transcripts bind by single-stranded miRNAs,[29] we constructed a miRNAs-mRNAs network according to their negative regulatory relationship. Among these ESCC related 35 intersecting miRNAs, there were 28 key miRNAs involved in the miRNAs-mRNAs co-expression network. The miRNAs-mRNAs co-expression network revealed potential regulatory relationships between miRNAs and mRNAs in ESCC. In the miRNAs-mRNAs co-expression network, we also found some of these 28 key miRNAs were reported as potential diagnostic and prognostic biomarkers in ESCC, including miR-101-3p, miR-125a-5p, miR-16-5p, and miR-20b-5p.[22,30–32] To further analyze these negatively regulated 72 mRNAs in the co-expression network, we showed that some played crucial roles in the development and progression of ESCC, such as E2F2, FGF2, and TCF3.[33–35] Subsequently, we analyzed these 28 key miRNAs in the miRNAs-mRNAs co-expression network indirectly by GO enrichment analyses and they were involved in cell signaling pathway regulation. GO analysis results suggested that the enriched GOs mainly targeted cell molecules and biological processes. The pathway analysis also revealed that some pathways were related with cancer, such as the PI3K-AKT signaling pathway and MAPK signaling pathway.[36,37] Ishibashi et al also reported that the overexpressed miR-141-3p targeted PHLPP-2 gene expression and affected the activity of the PI3K/AKT pathway in ESCC.[38] Therefore, our bioinformatics analysis results suggested that these 28 key miRNAs may play crucial roles in the progression of ESCC and cancer pathways.
To investigate the correlation between the 28 key miRNA expression levels and the 161 ESCC patients’ clinical pathology features, we identified 17 miRNAs that were related with 161 ESCC patients’ clinical pathology features, including miR-32-5p, miR-16-5p, miR-32-5p, and miR-183-5p. These miRNAs mainly related with ESCC patients’ tumor grade, TNM stages, and lymphatic metastasis status. Among these 17 miRNAs, miR-16-5p, miR-330-5p, miR-145-5p, miR-93-5p, miR-93-5p, miR133a-3p, miR-28-5p, miR-15b-5p, miR-20b-5p, miR-141-3p, and miR-320a were previously reported to be important indicators in lymphatic metastasis, invasion, and as diagnostic biomarkers of ESCC.[22,32,39–43] However, the functions of other miRNAs have not been correlated with ESCC clinical pathology features. We further analyzed the relationships between the 28 key miRNAs and ESCC patients’ overall survival time. There were six miRNAs that correlated with ESCC patients’ overall survival in TCGA. Among these six miRNAs, only miR-15b-5p has been reported with a correlation to ESCC survival,[22] and other miRNAs have not been studied. Our bioinformatics analyses revealed the potential of using miRNAs as biomarkers for diagnosis, classification, and prognosis of ESCC.
Finally, miR-135b-5p, miR-15b-5p, and miR-195-5p were selected and validated using TCGA bioinformatics analysis results from 51 newly collected ESCC patients’ tissue samples. The correlation analyses between these three miRNA expression levels and the ESCC patients’ clinical pathology features also were investigated. The results suggested that the ESCC patients’ clinical pathology correlated with TCGA bioinformatics analyses results.
In summary, the present study successfully identified the correlation of ESCC-specific miRNAs from a large-scale sample in TCGA database by using integrated bioinformatics analyses. We investigated the ESCC related specific miRNAs in different clinical pathology features and patients’ overall survival time. Finally, we validate the reliability and validity of TCGA database ESCC patients’ RNA sequencing integrated bioinformatics results by qRT-PCR. We found that miRNAs (miR-16-5p, miR-183-5p, miR-32-5p, miR-330-5p, miR-135b-5p, miR-195-5p, miR-145-5p, miR-93-5p, miR-185-5p, miR133a-3p, miR-28-5p, miR-15b-5p, miR-106b-5p, miR-182-5p, miR-20b-5p, miR-141-3p, and miR-320a) and (miR-200b-3p, miR-31-5p, miR-15b-5p, miR-141-3p, miR-135b-5p and miR-195-5p) were related with ESCC patients’ clinical pathology features and patients’ overall survival time, respectively. Taken together, above TCGA database ESCC patients’ clinical pathology features and overall survival time related miRNAs are worthy of further study about their application functions as biomarkers indicators in diagnosis, clinical pathology classification, and prognosis estimation of ESCC patients.
Acknowledgements
We thank Dong-Lin Cheng who supported the bioinformatics analysis technical assistance in the project.
Funding
This study was supported by grants from the Fundamental Research Funds for the Central Universities (No. lzujbky-2018-13), the National Natural Science Foundation of China (No. 81803188), and the Gansu Province Science and Technology Project (No. 1606RJYA270).
Conflicts of interest
None.
Supplementary Material
Footnotes
How to cite this article: Li CY, Zhang WW, Xiang JL, Wang XH, Li J, Wang JL. Identification of microRNAs as novel biomarkers for esophageal squamous cell carcinoma: A study based on The Cancer Genome Atlas (TCGA) and bioinformatics. Chin Med J 2019;00–00. doi: 10.1097/CM9.0000000000000427
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Wang B, Liang T, Li J. Long noncoding RNA LINC01296 is associated with poor prognosis in ESCC and promotes ESCC cell proliferation, migration and invasion. Eur Rev Med Pharmacol Sci 2018; 22:4524–4531. doi: 10.26355/eurrev_201807_15507. [DOI] [PubMed] [Google Scholar]
- 3.Wang JW, Guan CT, Wang LL, Chang LY, Hao CQ, Li BY, et al. Natural history analysis of 101 severe dysplasia and esophageal carcinoma cases by endoscopy. Gastroenterol Res Pract 2017; 2017:9612854.doi: 10.1155/2017/9612854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scarpa M, Noaro G, Saadeh L, Cavallin F, Cagol M, Alfieri R, et al. Esophageal cancer management: preoperative CA19.9 and CEA serum levels may identify occult advanced adenocarcinoma. World J Surg 2015; 39:424–432. doi: 10.1007/s00268-014-2835-1. [DOI] [PubMed] [Google Scholar]
- 5.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017; 16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 6.Bracken CP, Scott HS, Goodall GJ. A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genet 2016; 17:719–732. doi: 10.1038/nrg.2016.134. [DOI] [PubMed] [Google Scholar]
- 7.He B, Yin B, Wang B, Xia Z, Chen C, Tang J. MicroRNAs in esophageal cancer (review). Mol Med Rep 2012; 6:459–465. doi: 10.3892/mmr.2012.975. [DOI] [PubMed] [Google Scholar]
- 8.Cai X, Yang X, Jin C, Li L, Cui Q, Guo Y, et al. Identification and verification of differentially expressed microRNAs and their target genes for the diagnosis of esophageal cancer. Oncol Lett 2018; 16:3642–3650. doi: 10.3892/ol.2018.9066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wan J, Wu W, Che Y, Kang N, Zhang R. Insights into the potential use of microRNAs as a novel class of biomarkers in esophageal cancer. Dis Esophagus 2016; 29:412–420. doi: 10.1111/dote.12338. [DOI] [PubMed] [Google Scholar]
- 10.Mao S, Li Y, Lu Z, Che Y, Sun S, Huang J, et al. Survival-associated alternative splicing signatures in esophageal carcinoma. Carcinogenesis 2018; doi: 10.1093/carcin/bgy123. [DOI] [PubMed] [Google Scholar]
- 11.Zheng X, Song X, Shao Y, Xu B, Hu W, Zhou Q, et al. Prognostic role of tumor-infiltrating lymphocytes in esophagus cancer: a meta-analysis. Cell Physiol Biochem 2018; 45:720–732. doi: 10.1159/000487164. [DOI] [PubMed] [Google Scholar]
- 12.Xu D, Li G, Li H, Jia F. Comparison of IMRT versus 3D-CRT in the treatment of esophagus cancer: a systematic review and meta-analysis. Medicine (Baltimore) 2017; 96:e7685.doi: 10.1097/MD.0000000000007685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet 2016; 17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 14.Zhang K, Chen J, Song H, Chen LB. SNHG16/miR-140-5p axis promotes esophagus cancer cell proliferation, migration and EMT formation through regulating ZEB1. Oncotarget 2018; 9:1028–1040. doi: 10.18632/oncotarget.23178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang HB, Jiang ZB, Li M. Research on the typical miRNA and target genes in squamous cell carcinoma and adenocarcinoma of esophagus cancer with DNA microarray. Pathol Oncol Res 2014; 20:245–252. doi: 10.1007/s12253-013-9688-z. [DOI] [PubMed] [Google Scholar]
- 16.Guraya S. Prognostic significance of circulating microRNA-21 expression in esophageal, pancreatic and colorectal cancers; a systematic review and meta-analysis. Int J Surg 2018; 60:41–47. doi: 10.1016/j.ijsu.2018.10.030. [DOI] [PubMed] [Google Scholar]
- 17.Streppel MM, Pai S, Campbell NR, Hu C, Yabuuchi S, Canto MI, et al. MicroRNA 223 is upregulated in the multistep progression of Barrett's esophagus and modulates sensitivity to chemotherapy by targeting PARP1. Clin Cancer Res 2013; 19:4067–4078. doi: 10.1158/1078-0432.CCR-13-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hezova R, Kovarikova A, Srovnal J, Zemanova M, Harustiak T, Ehrmann J, et al. MiR-205 functions as a tumor suppressor in adenocarcinoma and an oncogene in squamous cell carcinoma of esophagus. Tumour Biol 2016; 37:8007–8018. doi: 10.1007/s13277-015-4656-8. [DOI] [PubMed] [Google Scholar]
- 19.Alder H, Taccioli C, Chen H, Jiang Y, Smalley KJ, Fadda P, et al. Dysregulation of miR-31 and miR-21 induced by zinc deficiency promotes esophageal cancer. Carcinogenesis 2012; 33:1736–1744. doi: 10.1093/carcin/bgs204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu J, Wang Y, Wu X. MicroRNA in the pathogenesis and prognosis of esophageal cancer. Curr Pharm Des 2013; 19:1292–1300. doi: 10.2174/138161213804805775. [DOI] [PubMed] [Google Scholar]
- 21.Slotta-Huspenina J, Drecoll E, Feith M, Habermehl D, Combs S, Weichert W, et al. MicroRNA expression profiling for the prediction of resistance to neoadjuvant radiochemotherapy in squamous cell carcinoma of the esophagus. J Transl Med 2018; 16:109.doi: 10.1186/s12967-018-1492-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiam K, Wang T, Watson DI, Mayne GC, Irvine TS, Bright T, et al. Circulating serum exosomal miRNAs as potential biomarkers for esophageal adenocarcinoma. J Gastrointest Surg 2015; 19:1208–1215. doi: 10.1007/s11605-015-2829-9. [DOI] [PubMed] [Google Scholar]
- 23.Liu R, Liao J, Yang M, Shi Y, Peng Y, Wang Y, et al. Circulating miR-155 expression in plasma: a potential biomarker for early diagnosis of esophageal cancer in humans. J Toxicol Environ Health A 2012; 75:1154–1162. doi: 10.1080/15287394.2012.699856. [DOI] [PubMed] [Google Scholar]
- 24.Wu X, Ajani JA, Gu J, Chang DW, Tan W, Hildebrandt MA, et al. MicroRNA expression signatures during malignant progression from Barrett's esophagus to esophageal adenocarcinoma. Cancer Prev Res (Phila) 2013; 6:196–205. doi: 10.1158/1940-6207.CAPR-12-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Derouet MF, Liu G, Darling GE. MiR-145 expression accelerates esophageal adenocarcinoma progression by enhancing cell invasion and anoikis resistance. PLoS One 2014; 9:e115589.doi: 10.1371/journal.pone.0115589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wijnhoven BP, Hussey DJ, Watson DI, Tsykin A, Smith CM, Michael MZ. MicroRNA profiling of Barrett's oesophagus and oesophageal adenocarcinoma. Br J Surg 2010; 97:853–861. doi: 10.1002/bjs.7000. [DOI] [PubMed] [Google Scholar]
- 27.Drahos J, Schwameis K, Orzolek LD, Hao H, Birner P, Taylor PR, et al. MicroRNA profiles of Barrett's esophagus and esophageal adenocarcinoma: differences in glandular non-native epithelium. Cancer Epidemiol Biomarkers Prev 2016; 25:429–437. doi: 10.1158/1055-9965.EPI-15-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Li M, Wang Z, Han S, Tang X, Ge Y, et al. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J Biol Chem 2015; 290:3925–3935. doi: 10.1074/jbc.M114.596866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maltby S, Plank M, Tay HL, Collison A, Foster PS. Targeting MicroRNA function in respiratory diseases: mini-review. Front Physiol 2016; 7:21.doi: 10.3389/fphys.2016.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gong J, Chu Y, Xu M, Huo J, Lv L. Esophageal squamous cell carcinoma cell proliferation induced by exposure to low concentration of cigarette smoke extract is mediated via targeting miR-101-3p/COX-2 pathway. Oncol Rep 2016; 35:463–471. doi: 10.3892/or.2015.4379. [DOI] [PubMed] [Google Scholar]
- 31.Fassan M, Pizzi M, Realdon S, Balistreri M, Guzzardo V, Zagonel V, et al. The HER2-miR125a5p/miR125b loop in gastric and esophageal carcinogenesis. Hum Pathol 2013; 44:1804–1810. doi: 10.1016/j.humpath.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 32.Huang Z, Zhang L, Zhu D, Shan X, Zhou X, Qi LW, et al. A novel serum microRNA signature to screen esophageal squamous cell carcinoma. Cancer Med 2017; 6:109–119. doi: 10.1002/cam4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Evangelou K, Havaki S, Kotsinas A. E2F transcription factors and digestive system malignancies: how much do we know? World J Gastroenterol 2014; 20:10212–10216. doi: 10.3748/wjg.v20.i29.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maehara O, Suda G, Natsuizaka M, Ohnishi S, Komatsu Y, Sato F, et al. Fibroblast growth factor-2-mediated FGFR/Erk signaling supports maintenance of cancer stem-like cells in esophageal squamous cell carcinoma. Carcinogenesis 2017; 38:1073–1083. doi: 10.1093/carcin/bgx095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma J, Wang XB, Li R, Xuan SH, Wang F, Li XH, et al. RNAi-mediated TCF-3 gene silencing inhibits proliferation of Eca-109 esophageal cancer cells by inducing apoptosis. Biosci Rep 2017; 37.doi: 10.1042/BSR20170799. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Shi H, Xu J, Zhao R, Wu H, Gu L, Chen Y. FGF2 regulates proliferation, migration, and invasion of ECA109 cells through PI3K/Akt signalling pathway in vitro. Cell Biol Int 2016; 40:524–533. doi: 10.1002/cbin.10588. [DOI] [PubMed] [Google Scholar]
- 37.Han S, Lee CW, Trevino JG, Hughes SJ, Sarosi GJ. Autocrine extra-pancreatic trypsin 3 secretion promotes cell proliferation and survival in esophageal adenocarcinoma. PLoS One 2013; 8:e76667.doi: 10.1371/journal.pone.0076667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishibashi O, Akagi I, Ogawa Y, Inui T. MiR-141-3p is upregulated in esophageal squamous cell carcinoma and targets pleckstrin homology domain leucine-rich repeat protein phosphatase-2, a negative regulator of the PI3K/AKT pathway. Biochem Biophys Res Commun 2018; 501:507–513. doi: 10.1016/j.bbrc.2018.05.025. [DOI] [PubMed] [Google Scholar]
- 39.Bibby BA, Reynolds JV, Maher SG. MicroRNA-330-5p as a putative modulator of neoadjuvant chemoradiotherapy sensitivity in oesophageal adenocarcinoma. PLoS One 2015; 10:e134180.doi: 10.1371/journal.pone.0134180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mei LL, Wang WJ, Qiu YT, Xie XF, Bai J. Shi ZZ. miR-145-5p suppresses tumor cell migration, invasion and epithelial to mesenchymal transition by regulating the Sp1/NF-kappaB signaling pathway in esophageal squamous cell carcinoma. Int J Mol Sci 2017; 18.doi: 10.3390/ijms18091833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu MX, Liao J, Xie M, Gao ZK, Wang XH, Zhang Y, et al. miR-93-5p transferred by exosomes promotes the proliferation of esophageal cancer cells via intercellular communication by targeting PTEN. Biomed Environ Sci 2018; 31:171–185. doi: 10.3967/bes2018.023. [DOI] [PubMed] [Google Scholar]
- 42.Zaidi AH, Saldin LT, Kelly LA, Bergal L, Londono R, Kosovec JE, et al. MicroRNA signature characterizes primary tumors that metastasize in an esophageal adenocarcinoma rat model. PLoS One 2015; 10:e122375.doi: 10.1371/journal.pone.0122375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen L, Jin Y, Wang L, Sun F, Yang X, Shi M, et al. Identification of reference genes and miRNAs for qRT-PCR in human esophageal squamous cell carcinoma. Med Oncol 2017; 34:2.doi: 10.1007/s12032-016-0860-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


