Abstract
To date, a direct investigation of the early cellular changes induced by metastatic cells within the surrounding tissue is difficult to achieve. We present the strategy whereby metastatic cancer cells release a cell-penetrating fluorescent protein taken up by neighbouring cells, allowing spatial identification of the local metastatic cellular environment within the whole tissue. Hence, the presence of low represented niche cells can be detected and characterised among the bulk tissue. To highlight its potential, we have applied this system to study the lung metastatic environment of breast cancer. We report the unprecedented presence of cancer associated parenchymal cells (CAPs), showing stem cell-like features, expression of lung progenitor markers, multi-lineage differentiation potential and self-renewal activity. In ex vivo assays, lung epithelial cells acquire a CAP-like phenotype when co-cultured with cancer cells and support their growth. The data highlight the remarkable potential of this method as a platform for new discoveries.
Cancer cell behaviour is strongly influenced by the surrounding cells of the tumour microenvironment (TME). Various cell types are known in the TME to have a significant impact on cancer cell behaviour, namely mesenchymal cells such as activated fibroblasts, pericytes and endothelial cells, alongside with different types of inflammatory cells1.
During the early phase of metastatic growth, cancer cells generate a local tissue microenvironment (metastatic niche), which is very distinct from the normal tissue structure and key to support metastatic outgrowth2. However, a detailed analysis of the cellular composition of the metastatic niche, especially at early stages, is significantly constrained by the difficulty to spatially discriminate the niche cells within the bulk of the tissue. This hampers identification of those cells that might respond to early cancer cell colonization but remain less represented as metastases grow bigger.
In this study, we present a strategy to overcome these limitations whereby metastatic cancer cells mark their neighbouring cells identifying them in the tissue. We have applied this system to interrogate the early metastatic environment of breast cancer in the lung. After confirming that the system allows to quantitatively and qualitatively distinguish the subset of known metastatic niche cells among the entire tissue, we identified lung epithelial cells as a new component of metastatic TME, in which a regenerative-like program is activated. We show that those epithelial cells acquire multi-lineage differentiation potential when co-cultured with cancer cells and support their growth. The data support the notion that, in addition to the well characterized stromal activation, a parenchymal response might contribute to creating the metastatic microenvironment.
The Cherry-niche labelling system
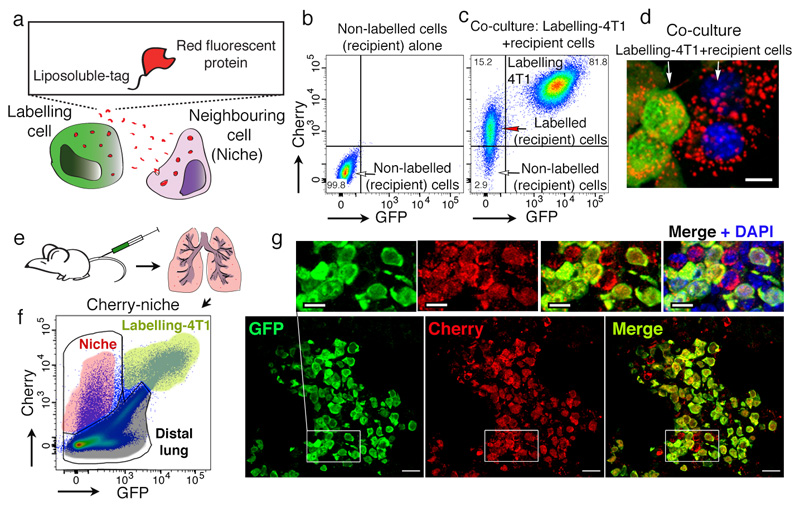
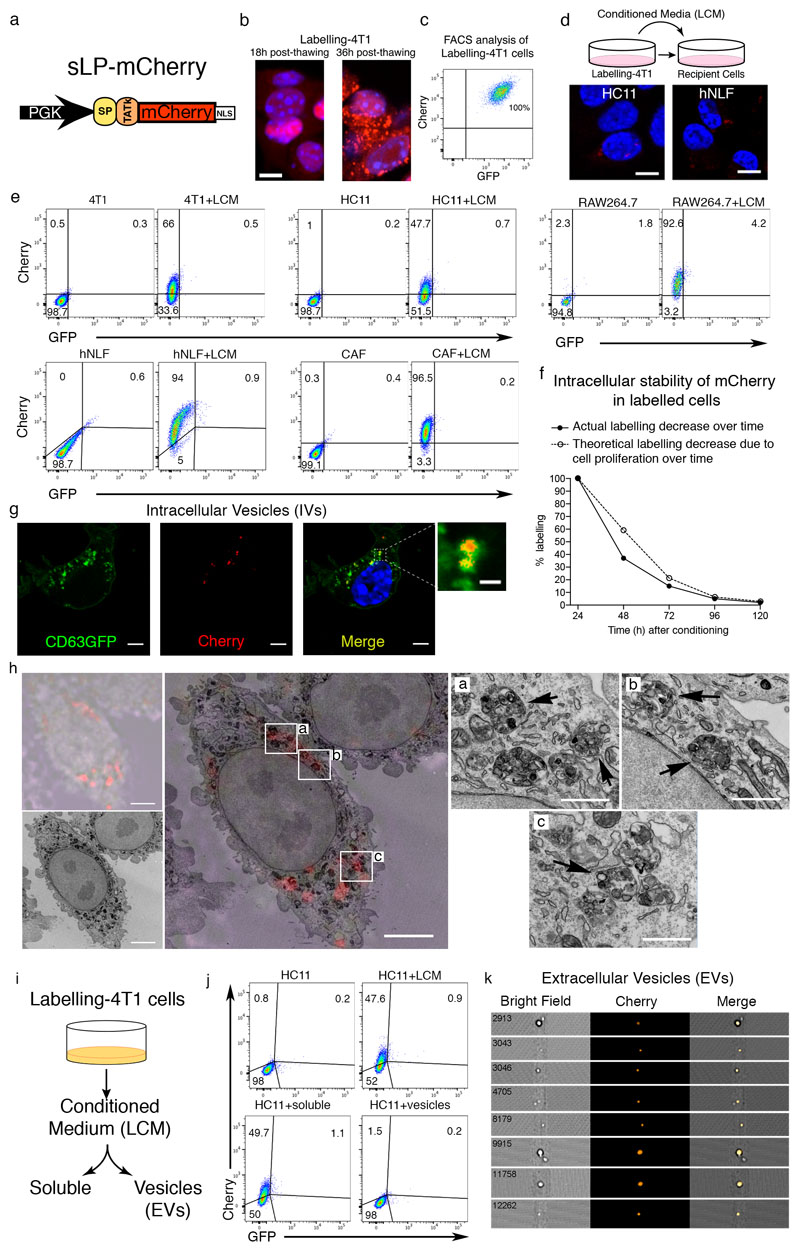
To develop a labelling system where metastatic cancer cells directly identify their neighbouring cells in vivo, we generated a version of a secreted monomeric Cherry red fluorescent protein (mCherry) containing a modified lipo-permeable Transactivator of Transcription (TATk) peptide3,4 (sLP-mCherry) (Figure 1a and Extended Data Figure 1a). We engineered 4T1 breast cancer cells to express the sLP-mCherry protein alongside a canonical cell-retained Green Fluorescent Protein (GFP), which we refer to as Labelling-4T1. In vitro, sLP-mCherry protein released by Labelling-4T1 is re-up-taken within producing cells as observed by changes in the intracellular localisation of the red fluorescence (Extended Data Figure 1b, c). Importantly, sLP-mCherry protein is also taken up by unlabelled cells both in co-culture (Figure 1b-d) and when cultured with Labelling-4T1 conditioned medium (LCM) (Extended Data Figure 1d-e). Upon uptake, sLP-mCherry fluorescence has an intracellular half-life of 43h (Extended Data Figure 1f) and is localized in CD63+ multi-lamellar bodies (lysosomal-like structures), where, due to its high photostability5, it retains high fluorescent intensity (Extended Data Figure 1g, h). LCM fractionation shows that only the soluble fraction displays labelling activity, while the extracellular vesicles (EVs), a portion of which contains sLP-mCherry, do not show in vitro labelling activity (Extended Data Figure 1i-k).
Figure 1. Cherry-niche labelling strategy.
a, Labelling design. b-c, Representative FACS plots of (b) naïve 4T1 cells alone or (c) co-cultured with Labelling-4T1. d, fluorescence image from co-culture (scale bar 10μm). Data representative from 2 independent experiments (b-d). e-g, In vivo labelling: (e) experimental scheme6; (f) representative FACS plot of a metastatic lung, n=50 mice. (g) representative images of Labelling-4T1 metastasis immuno-fluorescence staining (IF) (n=8 mice): cancer cells anti-GFP (green) and anti-Cherry (red), niche cells (Cherry only). DAPI (blue). Scale bars: main 20μm, inset 10μm. For gating strategy see Supplementary File 1.
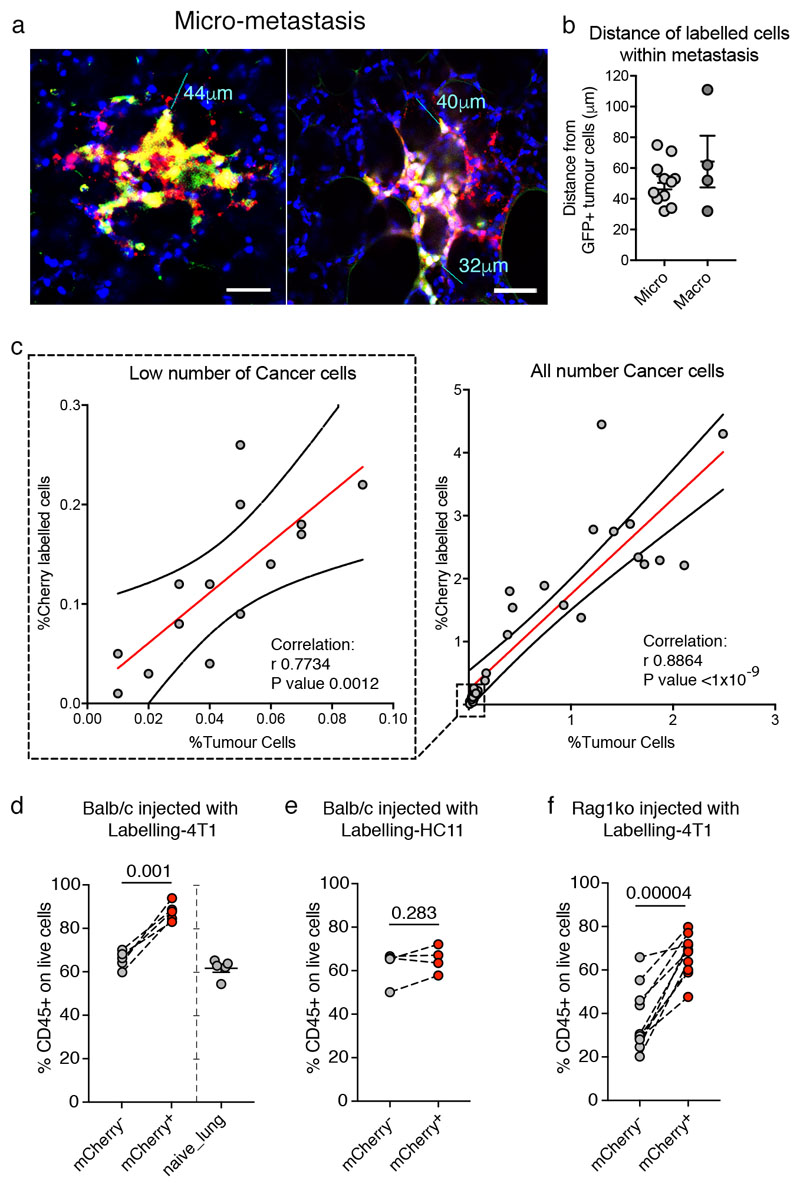
Critically, in vivo, Labelling-4T1 cells (GFP/Cherry double positive) intravenously injected into syngeneic BALB/c mice to induce lung metastases, efficiently label their surrounding host tissue cells (Cherry single positive), with a penetration of approximately five cell layers (Figure 1e6-g and Extended Data Figure 2a, b). This allows host cells in close proximity to cancer cells to be specifically discriminated from the distal lung (GFP/Cherry double negative) using fluorescent activated cell sorting (FACS) (Figure 1f). Notably, when metastases form, the number of mCherry+ niche cells in the tissue remains proportional to the growing metastatic cells (Extended Data Figure 2c). We detected no adaptive immunogenicity against sLP-mCherry and the local increase of CD45+ immune cells within the mCherry population was observed specifically as a response to cancer cells (Extended Data Figure 2d-f). Hence, the Cherry-niche marking system (Cherry-niche) enables the spatial reconstitution of the local metastatic niche within the whole tissue. This allows functional identification of labelled cells (Cherry-niche cells) and their direct comparison with the remaining unlabelled tissue cells within the same lung.
Tissue spatial resolution
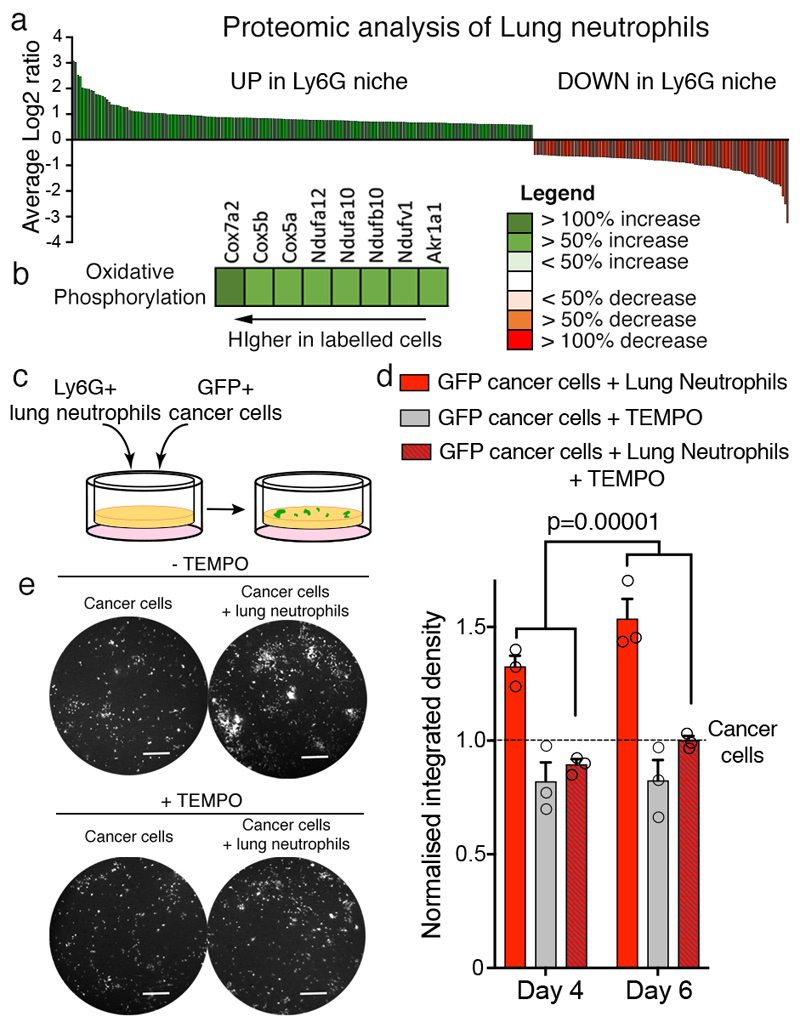
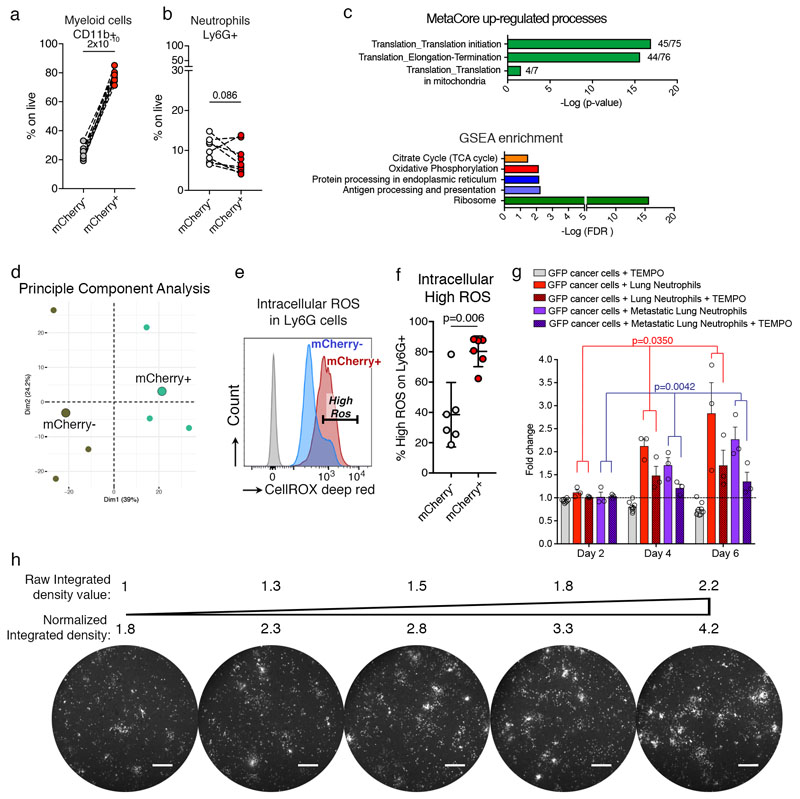
To demonstrate the utility of Cherry-niche to specifically interrogate the local early changes induced by cancer cells, we seeded 4T1-labelling cells in the lung via the tail vein. Here the lung tissue distant from micro-metastases remains unperturbed by primary tumour-derived systemic changes7. To validate the Cherry-niche strategy, we first examined known components involved in metastatic niche formation. CD45+ immune cells are very abundant within Cherry-niche and nearly exclusively derive from the myeloid lineage (CD11b+) (Extended Data Figure 2d and 3a). Lung neutrophils are widely reported to enhance metastatic growth of cancer cells8,9, and were indeed detected within the Cherry-niche (Extended Data Figure 3b). Since abnormalities in lung neutrophils associated with cancer are broadly found10, we isolated Cherry-niche neutrophils (Ly6G+) and compared their proteome to unlabelled neutrophils from the same lungs (Figure 2a). We found the sub-pool of Cherry-niche neutrophils to have distinct features, with an increase in translational and oxidative phosphorylation activities, as well as higher levels of intracellular Reactive Oxygen Species (ROS) determined by FACS analysis (Figure 2b, Extended Data Figure 3c-f and Supplementary File 2). To validate the functional relevance of specific niche cell features identified using Cherry-niche, we developed a three-dimensional (3D) scaffold (Alvetex) co-culture system, mimicking complex tissue-like cell-cell interactions. When monitoring actin-GFP+ Mouse Mammary Tumour Virus (MMTV) Polyoma virus Middle T antigen (PyMT) breast cancer cells, we found lung neutrophils to boost their growth in a ROS dependent manner (Figure 2c-e and Extended Data Figure 3g, h). Collectively, these data highlight the potential of Cherry-niche to detect in vivo changes spatially restricted to the metastatic environment.
Figure 2. Cherry-niche allows detection of niche neutrophils.
a,b, Proteomic analysis of Ly6G+ FACS-sorted cells: (a) all differentially detected proteins and (b) oxidative phosphorylation associated proteins. c-e, 3D co-culture of MMTV-PyMT-GFP+ cancer cells and Ly6G+ MACS-sorted cells with or without the ROS inhibitor TEMPO: (c) co-culture scheme; (d) GFP signal quantification (n=3 independent experiments, each with 3 to 10 technical replicates). Data normalised to cancer cell growth and represented as mean ±SEM. Statistical analysis on biological replicates by Two-way ANOVA; (e) representative images from 3 independent experiments (day 6, scale bar 400μm). See Source Data.
The non-immune Cherry-niche signature
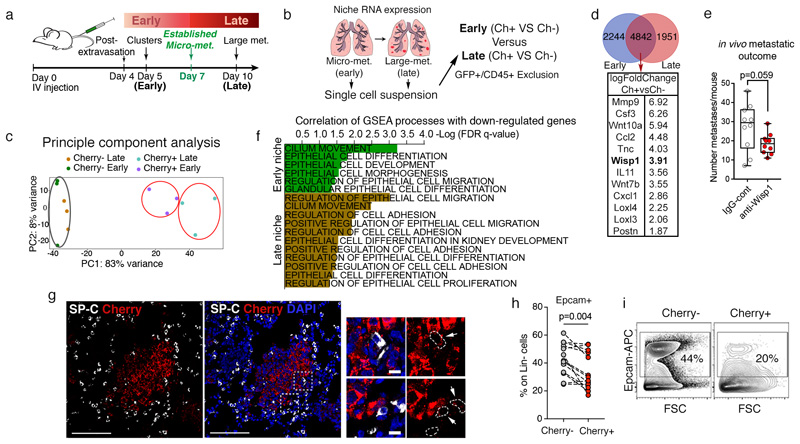
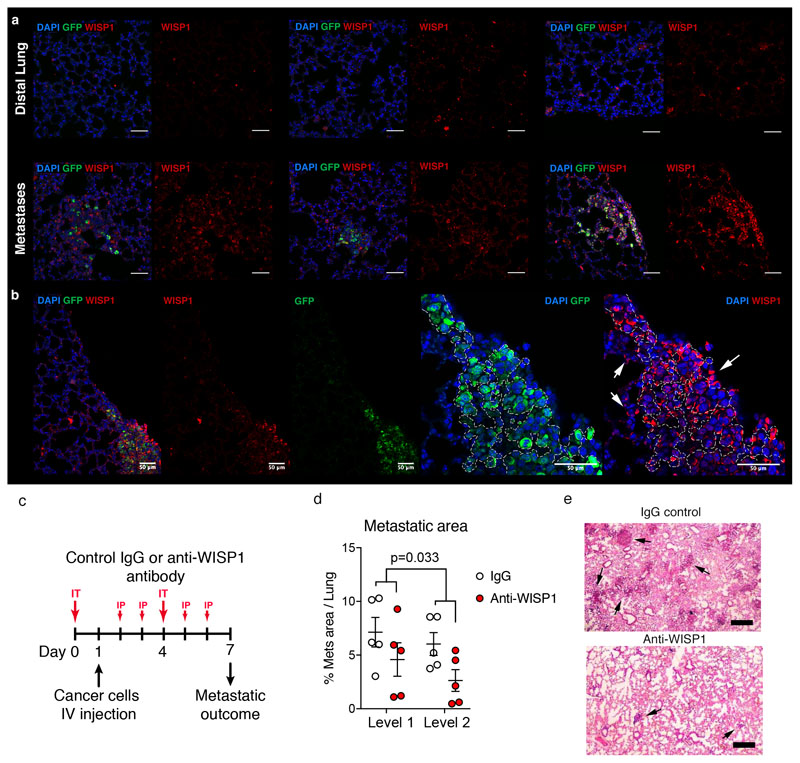
Whereas the contribution of immune cells to metastatic outgrowth has often been investigated11, less is known about the role of other TME cell types during metastatic nesting. Notably, Cherry-niche labelling can be used to provide spatio-temporal information, by applying it to different stages of metastatic progression. We generated the gene expression profile of non-immune (CD45-ve) Cherry-niche cells at the time point directly preceding micro-metastases as well as at an advanced metastatic stage (Figure 3a, b6). The majority of alterations were detected at the early stage, but additional changes subsequently discriminated the niche of macro-metastases (Figure 3c and Extended Data Figure 4a, b), confirming the evolution of the metastatic TME over time. MetaCore dataset enrichment and Gene Set Enrichment Analysis (GSEA) highlighted changes in pathways related to proliferation, inflammation and tissue remodelling (Extended Data Figure 4b, c). We next focused on the upregulated (>2) genes encoding for soluble factors in the niche at both time points (Figure 3d, Supplementary File 3). Again, as validation of the ability of our labelling system to faithfully capture the in vivo niche, we could find many previously reported tumour promoting factors12–19. We also found Wnt1 induced protein (Wisp1), previously suggested to act as an oncogene in breast cancer20, to be an abundant niche factor (Figure 3d). Indeed, upregulation of Wisp1 in both cancer and niche cells was detected and its pro-metastatic activity was confirmed by exogenous inhibition in vivo (Figure 3e and Extended Data Figure 5a-e).
Figure 3. Cherry-niche identifies epithelial component of metastatic TME.
a, Schematic of metastatic progression using labelling-4T1 cells6. b, Experimental design for RNA-seq6. c, Principle Component Analysis (PCA) diagram of CD45-Ter119- cell signatures from metastatic lungs at early (n=3, 10 mice each) and late (n=3, 5 mice each) time points. d, Venn diagram of differentially expressed genes in Cherry-niche from RNA-seq and selected factors common at early and late stages. e, Anti-Wisp1 blocking antibody treatment in vivo (n=10, from two independent experiments; data shown on a Tukey plot: box from the 25th to 75th percentiles, the bar is the median and the whiskers from smallest to largest value). f, GSEA correlation from RNA-seq data comparing early (n=3) or late (n=3) Cherry+ samples vs their respective Cherry-controls. g, Representative IF image of lung tissue (n=3 mice): mCherry-labelled micro-metastasis (red), Surfactant protein C (SP-C) (white) and DAPI (blue). Scale bars: main 100μm, inset 10μm (white arrows: mCherry labelled SP-C+ cells). h, Epcam+ cell frequency on Lin-(CD45-CD31-Ter119-) cells in distal lung (Ch-) and Cherry-niche (Ch+) estimated by FACS (n=13). i, Representative FACS plots from (h). Statistical analysis by unpaired two-tailed t-test with Welch’s correction (e), weighted Kolmogorov–Smirnov-like statistic with Benjamini-Hochberg correction (f) and paired two-tailed t-test (h). See Source Data.
We next probed for the presence of previously uncharacterized niche cells, which might be difficult to resolve by standard techniques due to a lower frequency. Interestingly, we found pathways associated with lung epithelial cells in the metastatic niche signature (Figure 3f). Micro-metastases grow embedded within the alveolar compartment of the lung, and alveolar type II cells (AT2) expressing Surfactant protein C (SP-C) were found in the metastatic niche (Figure 3g). Indeed, further confirming the presence of cells of parenchymal origin, using the epithelial cell adhesion molecule (Epcam) marker, we found Cherry-niche epithelial cells predominantly as low/mid-Epcam cells (Figure 3h, i).
Cancer associated parenchymal cells (CAPs)
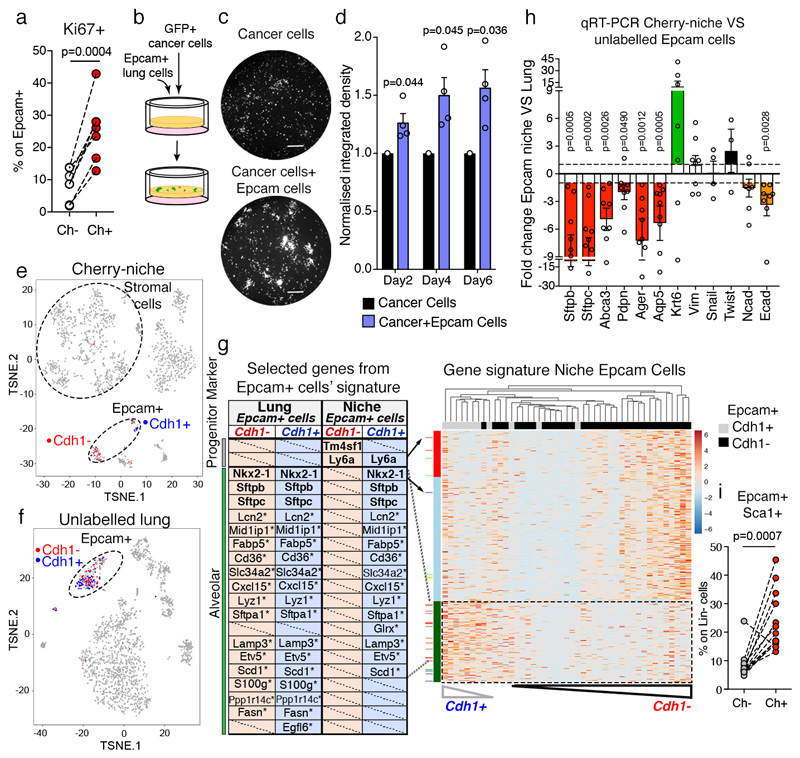
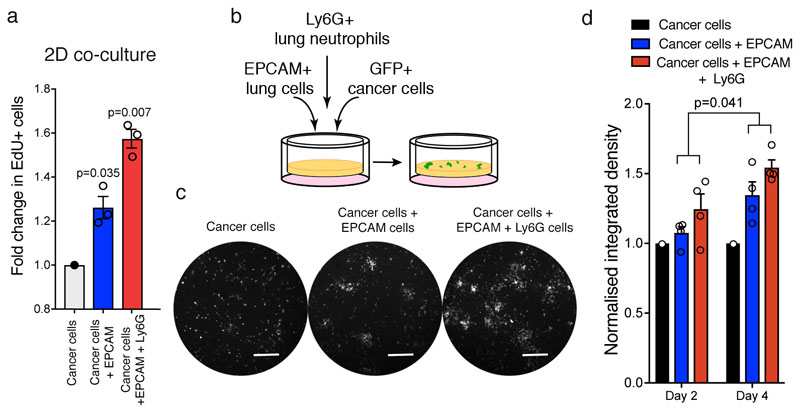
We found Cherry-niche epithelial cells to have a higher proliferative activity compared to their normal lung counterpart (Figure 4a). Concordantly, we found the presence of alveolar cell clusters with increased proliferative activity at the metastatic borders of human breast cancer lung metastases, suggesting that a lung parenchyma response to metastatic growth may occur in both mouse and human (Extended Data Figure 6a-f). Cancer cells profit from the presence of a lung parenchymal response, as freshly isolated Epcam+ cells from naïve lungs supported the growth of MMTV-PyMT/actin-GFP tumour cells in our 3D scaffold co-culture system (Figure 4b-d). Moreover, in line with the previous data (Figure 2c-e), the presence of both lung neutrophils and epithelial cells further enhanced tumour growth (Extended Data Figure 7a-d), highlighting the cellular complexity of the metastatic niche.
Figure 4. Metastatic niche lung epithelial cells display progenitor phenotype.
a, Scatter plot of Epcam+ cell proliferation by Ki67 staining on FACS-sorted cells (n=7 from independent sorts). b-d, MMTV-PyMT-GFP+ cancer cell growth in 3D co-culture with MACS-sorted Epcam+ cells: (b) co-culture scheme, (c) representative images from 4 independent experiments (day 6, scale bar 400μm), (d) GFP signal quantification (n=4, each with 3 technical replicates, statistical analysis on biological replicates). Data normalised to cancer cell growth. e-g, scRNA-seq analysis: tSNE plots of CD45- cells from (e) Cherry-niche (n=1473) or (f) distal lung (n=1996); (g) (right) heatmap of niche Epcam+ cells; ordered genes in rows and hierarchically clustered cells in columns; (left) table shows established lineage markers (bold) and putative alveolar markers25 (*). h qRT-PCR analysis of Epcam+ FACS-sorted cells (n=9 Sftpc, Aqp5; n=8 Sftpb, Abca3, Pdpn, Ager, Vim, Ecad; n=7 Krt6, Ncad; n=4 Snail, n=3 Twist). Data represented as fold change to Cherry- Lung Epcam cells (Statistical analysis on the DCt values). i, Epcam+Sca1+ cell frequency on Lin-(CD45-CD31-Ter119-) cells by FACS (n=13). Statistical analysis by paired two-tailed t-test (a, h, i), one sample two-tailed t-test (d). Data represented as mean ±SEM. See Source Data.
We then aimed to better define the perturbation occurring in lung epithelial cells when they are in the proximity of cancer cells. To contextualize their presence among the other niche cellular components, we performed single cell RNA sequencing of CD45-ve cells. tSNE analysis of Cherry-niche cells identified a large stromal cluster, where different stromal cells can be distinguished (Figure 4e and Extended Data Figure 8a-c). This is in agreement with the various known mesenchymal cell components of TME, as well as the characterization of different fibroblast subsets21–24. Notably, specifically in the niche, Epcam-expressing epithelial cells are distributed in two clusters distinguished by the expression of E-Cadherin (Cdh1) (Fig 4e). We found that only niche Epcam+Cdh1+ cells share the expression of alveolar genes25 with unlabelled distant lung Epcam+ cells (Fig 4f, g). Conversely, niche Epcam+Cdh1- cells express both the Sca1 (Ly6a) and Tm4sm1 progenitor markers26–28 (Fig 4g). As validation of this de-differentiated signature observed in epithelial cells in the niche, qPCR of Epcam sorted Cherry-niche cells also show an overall reduction in expression of alveolar lineage markers (Figure 4h). Moreover, the enrichment of Epcam+Sca1+ cells was confirmed by FACS in the lung Cherry-niche of different metastatic cell types (Figure 4i and Extended Data Figure 9a-c). Similarly, the presence of epithelial cells expressing another lung progenitor marker, integrin 4 (CD104)29, was increased in the niche as well as in ex vivo co-cultures (Extended Data Figure 9d-i).
In summary, we describe a parenchymal response to lung metastasis involving de-differentiated pools of epithelial cells in the niche, which we define as cancer associated parenchymal cells (CAPs).
CAPs are activated AT2 cells
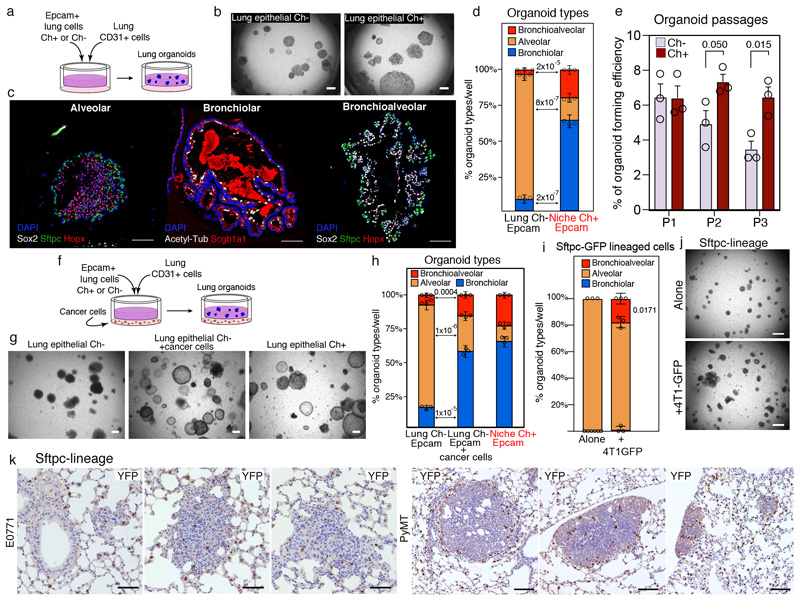
To functionally characterize CAP cells, we tested their lineage differentiation potential ex vivo using a 3D Matrigel-based organoid co-culture system27 (Figure 5a). Unlabelled resident lung Epcam+ cells are predominantly alveolar, as previously shown27, and formed mainly alveolar organoids when co-cultured with CD31+ cells (Figure 5b-d). Strikingly, Cherry-niche Epcam+ cells favoured the bronchiolar lineage and showed a remarkable capacity to generate multi-lineage bronchioalveolar organoids (Figure 5d). Despite the bias in organoid formation towards the bronchial lineage, there were no visible signs of Cherry labelled bronchial cells in vivo (Extended Data Figure 10a). CAPs also retained high self-renewal capacity over multiple passages (Figure 5e).
Figure 5. CAPs show multi-lineage differentiation potential.
a-e, Lung organoids: (a) co-culture scheme; (b) representative bright-field images (scale bar 100μm); (c) representative IF of organoid sections stained with the indicated markers (scale bar 50μm); (d) quantification; (e) organoid formation efficiency over passages. f-h, Lung organoids with or without Labelling-4T1: (f) co-culture scheme, (g) representative bright-field images (scale bar 100μm) and (h) quantification. i, j, Lung organoids with Sftpc-CreERT2 lineage cells with or without 4T1-GFP: (i) quantification and (j) representative bright-field images, scale bar 150μm. Images are representative of six (b, c, g) and three (j) organoid cultures. Data generated from independent sorts (d, h, i) and represented as cumulative percentage using the mean ±SD of three co-cultures per sorting. k, Representative staining of lineage cells in metastatic lungs from Sftpc-CreERT2 mice injected with cancer cells, either E0771 (n=3; scale bar 50μm) or MMTV-PyMT (n=3; scale bar 100μm). Statistical analysis by unpaired two-tailed t-test (d, e, h) and one sample two-tailed t-test (i), on original non-cumulative values (see Source Data).
Next, we tested whether tumour cells could directly induce the CAP phenotype. When Epcam+ cells from either unlabelled distal micro-metastatic lungs or naïve lungs were co-cultured with metastatic cells, they generated a higher proportion of bronchiolar and bronchioalveolar organoids (Figure 5f-h and Extended Data Figure 10b, c). Similar alterations were induced by cancer cells when the assay was performed using mouse lung fibroblasts (MLg cells) instead of CD31+ cells (Extended Data Figure 10b, c).
Certainly, lung Epcam+ cells are predominantly alveolar, but they also contain epithelial progenitors that could be enriched by cancer cells to generate an increased plasticity27,30. Therefore, we performed organoid cultures using lineage-labelled AT2 cells (Sftpc-lineage). Remarkably, Sftpc-lineaged cells, which show no plasticity in co-culture with CD31+ cells, when exposed to cancer cells generated a notable amount of multi-lineage bronchioalveolar organoids, supporting the idea of a reprogramming activity driven by cancer cell-derived factors ex vivo (Figure 5i, j). Despite the potential of cancer cells to modulate the organoid formation ability of lineage-labelled club cells (Scgb1a1-lineage), only rare single Scgb1a1-lineage cells were found in proximity to lung metastases (Extended Data Figure 10d-f). Conversely, metastases growing in Sftpc-lineage lungs demonstrated the alveolar (AT2) origin of the CAPs (Figure 5k).
Recently, a rare population of AT2 cells expressing Axin2 with stem cell and repair activity (AT2stem), was described in the lung alveoli31. While a small proportion of Axin2+ cells was found in the unlabelled epithelial cluster, Axin2 was undetectable in the niche Epcam clusters (data not shown). Therefore, even if cancer cell seeding could trigger lung injury, this phenomenon does not appear to specifically maintain an Axin2+ AT2 population in the niche.
Collectively, these data show the alveolar origin of CAPs and the potential of cancer cells to induce multi-lineage potential of epithelial cells ex vivo.
Discussion
This study proposes a novel labelling system and demonstrates its ability to resolve the host tissue cellular environment spatially restricted to regions surrounding cancer cells. Remarkably, we report the presence of a lung epithelial compartment within the metastatic niche, which originates from Alveolar Type II cells. We define this novel TME component as CAPs and describe their activated regenerative state by showing a de-differentiated signature, tissue stem cell-like features, multi-lineage differentiation potential and increased self-renewal activity.
Parenchymal cells have been previously described to trigger a tissue-wide pro-tumourigenic inflammatory response to systemic primary tumour signals32,33. In addition to these systemic effects, here we collectively show that a regenerative-like activation in the lung parenchyma occurs as a direct local response during breast cancer metastasis. This parenchymal response, alongside with the stromal activation, might be important to orchestrate tumour niche formation.
Taken together, these results consolidate Cherry-niche as a platform for new discoveries with the potential to identify, isolate and functionally test tissue cells from the metastatic niche with unprecedented spatial resolution.
Online Content
Data availability information, Methods, along with any additional Extended Data display items in the online version of the paper; references unique to these sections appear only in the online paper. Source Data are available in the indicated legends
Methods (online)
Statistical Analysis
Statistical analyses were performed using Prism software (version 7.0c, GraphPad Software, USA). P values were obtained from two-tailed Student t-tests with paired or unpaired adjustment. When needed, unpaired t-test were adjusted using Welch’s correction for unequal variance. In one instance (Fig. 4i) data in one of the groups did not pass D'Agostino & Pearson normality test, therefore a Wilcoxon matched-pairs signed rank test was performed. Single-sample tests were also used for comparisons of co-cultured cancer cell growth on scaffolds to the normalized value of cancer cells alone. For comparisons between two scaffold conditions of growth over time or to perform multiple analysis between experimental groups, Two-way ANOVA was used.
Mouse strains
All mice used are available from Jackson Laboratory. MMTV-PyMT mice34 are on FVB and C57BL/6 background, actin-GFP35 mice and Rag1KO are on FVB background (gift from J. Huelsken laboratory (EPFL, Lausanne, Switzerland)). Sftpc-CreERT236, Rosa26R-YFP37 (Sftpc-CreERT2;R26R-YFP) are on a C57BL/6 background. Balb/cj mice and the above-mentioned lines were bred and maintained under specific-pathogen-free conditions by The Francis Crick Biological Research Facility and female mice were used between 6 to 10 weeks of age. Breeding and all animal procedures were performed at the Francis Crick in accordance with UK Home Office regulations under project license P83B37B3C.
For ex-vivo organoid lineage tracing experiments, Scgb1a1-CreERT2 and Rosa26R-fGFP38, Sftpc-CreERT2 (Sftpc-CreERT2;R26R-fGFP and Scgb1a1-CreERT2;R26R-fGFP) mice on a C57BL/6 background were bred and maintained under specific-pathogen-free conditions at the Gurdon Institute of University of Cambridge in accordance with UK Home Office project licence PC7F8AE82.
Tamoxifen administration
Tamoxifen (Merck Sigma-Aldrich, Germany) was dissolved in Mazola corn oil (Merck Sigma-Aldrich, Germany) in a 20mg/ml stock solution. Two doses of tamoxifen (0.2mg/g body weight) were given via oral gavage every other day and lung tissues were collected two days after tamoxifen administration to isolate cells for lung organoids. For in vivo lineage tracing three doses of tamoxifen (0.2mg/g body weight) were given via oral gavage over consecutive days and mice were injected two weeks later.
Cells
MLg cells (murine normal lung fibroblasts) were purchased from ATCC (USA). CAF (cancer associated fibroblasts) isolated from MMTV-PyMT tumours and human normal fibroblast (hNLF) were a gift from E.Sahai. MMTV-PyMT cells were isolated from MMTV-PyMT tumours as previously described19. All other cell lines were provided the Cell Services Unit of The Francis Crick Institute. All cell lines were authenticated and mycoplasma tested by the Cell Services Unit of The Francis Crick Institute. MMTV-PyMT cells were cultured on collagen solution coated dishes in MEM medium (DMEM/F12 (ThermoFisher Scientific, USA) with 2% fetal bovine serum (FBS; Labtech, UK), 100U/ml penicillin-streptomycin (ThermoFisher Scientific, USA), 20ng/ml EGF (ThermoFisher Scientific, USA) and 10μg/ml insulin (Merck Sigma-Aldrich, Germany)). Collagen solution is made by 30μg/ml PureCol collagen (Advanced Biomatrix, USA), 0.1% bovine serum albumin (BSA), 20mM HEPES in HBSS (ThermoFisher Scientific, USA). HC11 cells were cultured in RPMI (ThermoFisher Scientific, USA) supplemented with 10% FBS, 100U/ml penicillin-streptomycin, 10ng/ml EGF (ThermoFisher Scientific, USA) and 5μg/ml insulin. All other cell lines were cultured in DMEM (ThermoFisher Scientific, USA) supplemented with 10% FBS and 100U/ml penicillin-streptomycin. All cells were cultured at 37ºC and 5% CO2.
Human Samples
Human pulmonary breast cancer metastases from independent patients were obtained from the Grampian Biorepository, Aberdeen Royal Infirmary (REC approval: 16/NS/0055). Four samples were stained by Immunohistochemistry and Immunofluorescence and epithelial cells proliferation was quantified.
Information about the human samples used can be found in Supplementary File 5.
Labelling system
A soluble peptide (SP)3 and a modified TAT peptide4 were cloned upstream of the mCherry cDNA, under the control of a mouse PGK promoter (sLP-Cherry, see Supplementary File 6 for sequence). The sLP-Cherry sequence was cloned into a pRRL lentiviral backbone. 4T1, Renca, CT26 and HC11 cells were stably infected with sLP-Cherry and pLentiGFP lentiviral particles and subsequently sorted to isolate Cherry+GFP+ cells.
Induction of experimental metastases
Procedures were performed at the Francis Crick in accordance with UK Home Office regulations under project license P83B37B3C. Cancer cells were injected intravenously to generate metastasis in the lung: 4T1 (1,000,000 cells), Renca (500,000 cells), CT26 (200,000 cells) were re-suspended in 100μl PBS and tail-vein injected in Balb/cJ mice. Mice were sacrificed on the basis of a time period rather than based on their clinical signs. Therefore, the experimental end point (time controlled, seven days unless otherwise specified), most likely occurred before a humane end point (as determined by deterioration of heath conditions). All animals were monitored daily for unexpected clinical signs following the P83B37B3C licence guidelines and the principles set out in the NCRI Guidelines for the Welfare and Use of Animals in Cancer Research (UK). Deterioration of health conditions such as reduction in food and water consumption and changes on the animal’s general appearance or weight loss of 10% over a 24hr period would result in animals being sacrificed prior to the experimental end point.
In vivo lineage tracing experiments
Sftpc-CreERT2 and Scgb1a1-CreERT2 mice on C57BL/6 background were tail-vein injected either with 175,000 MMTV-PyMT C57BL/6 cells and lungs collected 4 weeks later or with 700,000 E0771 cells and lungs collected 12 days later.
Tissue digestion for cell isolation or analysis
Lung tissues were dissociated as previously described19. Briefly, lungs were removed at day 7 after tumour cell injection (unless otherwise specified), minced manually and then digested for 30 min in a shaker at 37ºC with a mixture of DNase I (Merck Sigma-Aldrich, Germany) and Liberase TM and TH (Roche Diagnostics, Switzerland) in HBSS solution. Samples were then washed, passed through a 100μm filter and incubated in Red Blood Cell Lysis buffer (Miltenyi Biotec, Germany) for 3-5 min at room temperature. After a wash with MACS buffer (0.5% BSA and 250mM EDTA in PBS), samples were passed through a 40μm filter and a 20μm strainer-capped flow cytometry tube to generate a single cell suspension to use for flow cytometric analysis or further purification.
FACS analysis and cell sorting
Prepared single-cell suspensions of mouse lung tissues and in vitro cell lines were incubated with mouse FcR Blocking Reagent (Miltenyi Biotec, Germany) for 10 min at 4ºC followed by an incubation with a mix of pre-labelled antibodies (antibody information is provided in Supplementary file 4) for 30 min at 4ºC. After two washes with MACS buffer, dead cells were stained with 4′,6-diamidino-2-phenylindole (DAPI). Flow cytometry analyses were carried out on a BD LSR-Fortessa (BD Biosciences, USA) and FlowJo 10.4.2 (FlowJO, LCC 2006-2018, USA) was used for further analysis. All cell-sorting experiments were carried out on a BD Influx cell sorter (BD Biosciences, USA).
Tissue digestion and FACS analysis in ex-vivo lineage tracing experiments
Lung tissues were dissociated with a collagenase/dispase solution as previously described27. Briefly, after lungs were cleared by perfusion with cold PBS through the right ventricle, 2ml of dispase (50U/ml, BD Biosciences, USA) was instilled into the lungs through the trachea until the lungs inflated, followed by instillation of 1% low melting agarose (Bio-Rad Laboratories, USA) through the trachea to prevent leakage of dispase. Each lobe was dissected and minced into small pieces in a conical tube containing 3ml of PBS, 60μL of collagenase/dispase (Roche, Switzerland), and 7.5μL of 1% DNase I (Merck Sigma-Aldrich, Germany) followed by rotating incubation for 45 min at 37°C. The cells were then filtered sequentially through 100- and 40-μm strainers and centrifuged at 1000rpm for 5 min at 4°C. The cell pellet was resuspended in 1ml of ACK lysis buffer (0.15M NH4Cl, 10mM KHCO3, 0.1mM EDTA) and lysed for 90 s at room temperature. 6ml basic F12 media (ThermoFisher Scientific, USA) was added and 500μl of FBS (Fisher Scientific, USA) was slowly added in the bottom of tube. Cells were centrifuged at 1000 rpm for 5 min at 4°C. The cell pellet was resuspended in PF10 buffer (PBS with 10% FBS) for further staining. The antibodies used were as follows: CD45 (30-F11)-APC (BD Biosciences, USA), CD31 (MEC13.3)-APC (BD Biosciences, USA), and EpCAM (G8.8)-PE-Cy7 (BioLegend, USA). For antibody list see Supplementary file 4. MOFLO system (Beckman Coulter, USA) was used for the sorting at Wellcome-MRC Stem Cell Institute Flow Cytometry Facility.
Lung organoid assay
Lung organoid co-culture assays were previously reported27,39. Briefly, freshly sorted epithelial cells (Epcam+CD45-CD31-Ter119-GFP-) from either the metastatic niche or the distal lung were resuspended in 3D basic media (DMEM/F12, supplemented with 10% FBS, penicillin/streptomycin, 1mM HEPES, and insulin/transferrin/selenium (ITS) (Merck Sigma-Aldrich, Germany), and mixed with MACS-sorted CD31+ lung stromal cells or MLg cells followed by resuspension in growth factor-reduced (GFR) Matrigel (BD Biosciences, USA) at a ratio of 1:1. 100μl of mixture was then placed in a 24-well transwell insert with a 0.4μm pore (Corning, USA). 1-2.5 x103 distal lung or niche epithelial cells and 25,000 CD31+ or MLg cells were seeded in each insert. 500μl of 3D basic media was placed in the lower chamber and media was changed every other day. In addition, freshly sorted lineage-labelled Scgb1a1+ club cells or Sftpc+ AT2 cells were resuspended in 3D basic media followed by mixing with GFR matrigel retaining CD31+ stromal cells as described above. For co-culture of lung epithelial cells with tumour cells, a mixture of 1-2.5 x103 distal lung epithelial cells and 25,000 CD31+ cells in Matrigel was placed in the transwell insert, and 2,000 tumour cells FACS-sorted from metastatic lungs were seeded in the lower chamber. Plates were scored for colony number after 14 days. Organoid-forming efficiency was calculated as number of organoids formed/number of cells plated per well as a percentage. Quantification of distinct types of differentiated organoids was performed by scoring the organoids expressing Sox2 or SP-C/Hopx by IF staining from at least five step sections (20μm apart) per individual well. Bright-field images were acquired after 14 days using an EVOS microscope (ThermoFisher Scientific, USA).
3D Cell culture
Primary MMTV-PyMT actin-GFP cells were seeded at a density of 5,000 cells/well in a collagen solution coated Alvetex Scaffold 96-well plate (ReproCELL, Europe). The following day, Ly6G+ lung cells and/or Epcam+ lung epithelial cells were MACS sorted and seeded on top of the cancer cells at a density of 50,000 cells per well. In selected experiments, wells were supplemented with 4-Hydroxy-TEMPO (200μM, Merck Sigma-Aldrich, Germany) or mouse anti-Wisp1 (250ng/ml, MAB1680, R&D, USA). The growth of GFP+ cells was monitored daily for 6 days using the SteREO Lumar.V12 stereomicroscope (Zeiss, Germany), and images were quantified using ImageJ (NIH, USA). For quantification, the Li’s Minimum Cross Entropy thresholding algorithm was performed on the stacked images.
For the CD104 staining experiment, Epcam+ lung cells were harvested from mouse lung tissue via MACS sorting and seeded at a density of 1,500,000 cells per well on collagen solution coated Alvetex Scaffold 12-well inserts. After 48 h, MMTV-PyMT actin-GFP cells were seeded on top of the Epcam+ cells at a density of 2,000 cells per scaffold insert.
Immunofluorescence and immunohistochemistry
Mouse lungs were fixed in 4% PFA in PBS for 24 h and embedded in paraffin blocks. 4μm thick tissue sections were cut, deparaffinised and rehydrated using standard methods. After heat-mediated antigen retrieval in citrate buffer (unless stated otherwise), sections were blocked with a solution of 1% BSA, 10% Donkey serum in PBS. For antibody list see Supplementary file 4.
mCherry and GFP staining
An overnight incubation at 4ºC with goat anti-GFP and rabbit anti-mCherry antibodies was followed by 1 h incubation at room temperature with anti-goat AlexaFluor 488 and anti-rabbit AlexaFluor 555 (both secondary antibodies were purchased from ThermoFisher Scientific (USA) and used at 1:400). Next, the slides were incubated with Sudan Black B for 20 min and mounted with Vectashield Mounting Medium with DAPI (Vector Laboratories, USA).
Lineage staining
An overnight incubation at 4ºC with goat anti-GFP antibody was followed by 45 min incubation at room temperature with secondary biotinylated-conjugated antibodies. Next, the VECTASTAIN Elite ABC kit (Vector Laboratories, USA) was used according to the manufacturer’s instructions. The visualization of cell nuclei was performed with hematoxylin and analysis employed the Nikon Eclipse 90i light microscope and NIS-elements software (Nikon, Japan).
WISP1 staining
An overnight incubation at 4ºC with goat anti-GFP and rabbit anti-WISP1 antibodies was followed by 30 min incubation at room temperature with anti-goat AlexaFluor 488 and anti-rabbit AlexaFluor 555 (both secondary antibodies were purchased from ThermoFisher Scientific (USA) and used at 1:500). Next, the slides were incubated with Sudan Black B for 20 minutes and mounted with Vectashield Mounting Medium with DAPI (Vector Laboratories, USA).
Ki67 staining
Epcam+CD45-CD31-Ter119-GFP- cells were sorted from lung suspensions, plated on poly-lysine glass coverslips for 15 min at room temperature and fixed in 4% PFA in PBS for 10 min. After fixation, cells were permeabilized with 0.1% Triton-X-100 in PBS for 5 min and incubated with a blocking solution (1% BSA, 10% goat serum, 0.3M glycine, 0.1% Tween in PBS) for 1 h at room temperature. Next, cells were incubated overnight with an anti-mouse Ki67 antibody diluted in blocking solution followed by a 1 h incubation with a goat anti-rabbit AlexaFluor 488 (1:500, ThermoFisher Scientific (USA)). Finally, cells were mounted with Vectashield Mounting Medium with DAPI for imaging.
E-cadherin staining
CD49f+CD104+CD45-CD31-Ter119-GFP- cells were sorted from lung suspensions, cytospun on glass slides and fixed in 4% PFA in PBS for 10 min. Next, cells were permeabilized with 0.5% TritonX-100 for 30 min and incubated in blocking solution (4% BSA, 0.05% Tween20 in PBS) for 45 min at room temperature. Then, cells were incubated with a rat anti-E-cadherin antibody in blocking solution overnight at 4ºC followed by an incubation with a goat anti-rat AlexaFluor 647 (1:500, ThermoFisher Scientific (USA)). Finally, cells were mounted with Vectashield Mounting Medium with DAPI for imaging.
CD104 staining
Epcam+ cells were MACS sorted and plated on Alvetex scaffold inserts as described above. 7 days after plating the whole scaffold was collected, washed with PBS and incubated in blocking solution (10% goat serum in PBS) for 1 h at room temperature. Next, the samples were incubated with a conjugated anti-CD104-eFluor660 antibody (1:100 in PBS with 1:10 FcR blocking (Miltenyi Biotec, Germany)) for 1 h at room temperature. Then, the samples were fixed with 4% PFA in PBS for 10 min and mounted with Vectashield Mounting Medium with DAPI. Pictures were captured with the Axio Scan.Z1 slide scanner (Zeiss, Germany).
Lung organoid staining
Cultured organoids were fixed with 4% PFA in PBS for 2-4 h at room temperature followed by immobilization with Histogel (ThermoFisher Scientific, USA) for paraffin embedding. At least five step sections (20μm apart) per individual well were stained. Fluorescence images were acquired using a confocal microscope Leica TCS SP5 (Leica Microsystems, Germany). All the images were further processed with Fiji software.
TTF1 and Ki67 co-staining
Target retrieval solution pH9 (Agilent DAKO, USA) was used as antigen retrieval. For histology, 1h incubation at room temperature with mouse anti-TTF1 was followed by 45 min incubation at room temperature with secondary biotinylated-conjugated antibodies. Next, the VECTASTAIN Elite ABC kit (Vector Laboratories, USA) was used according to the manufacturer’s instructions. The visualization of cell nuclei was performed with hematoxylin and analysis employed the Nikon Eclipse 90i light microscope and NIS-elements software (Nikon, Japan). For immune-fluorescence, 1h incubation at room temperature with mouse anti-TTF1 and rabbit anti-Ki67 was followed by 45 min incubation at room temperature with anti-mouse AlexaFluor 555 and anti-rabbit AlexaFluor 488 (both secondary antibodies were purchased from ThermoFisher Scientific (USA) and used at 1:250). Next, the slides were incubated with Sudan Black B for 20 min and mounted with Vectashield Mounting Medium with DAPI (Vector Laboratories, USA).
All pictures were captured with either a Zeiss Upright710 confocal microscope or a Zeiss Upright780 confocal microscope unless differently stated.
Quantitative real time PCR
RNA preparation was performed using the MagMax-96 Total RNA Isolation Kit (ThermoFisher Scientific, USA). cDNA synthesis was performed using a SuperScript III First-Strand Synthesis System (ThermoFisher Scientific, USA), according to the manufacturer’s protocol. Quantitative real-time PCR samples were prepared with 50-100ng total cDNA for each PCR reaction. The PCR, data collection and data analysis were performed on a 7500 FAST Real-Time PCR System (ThermoFisher Scientific, USA). GAPDH was used as internal expression reference. List of primers used can be found in Supplementary File 4.
Anti-Wisp1 treatment in vivo
BALB/cJ female mice (6-8 weeks old) were administered with anti-Wisp1 (5μg AF1680 and 5μg MAB1680, R&D, USA) or a control-IgG antibody via an intra-tracheal injection (50μl/mouse). The following day, mice were intravenously injected with 250,000 4T1 cells. Anti-Wisp1/control-IgG treatment was repeated daily, via a second intra-tracheal injection on day 4, and intra-peritoneal injections on days 2,3,5 and 6. Mice were harvested 7 days after the first treatment and lungs were embedded, cut and H&E stained. The lung metastatic burden was assessed by counting number of metastases on four levels (100μm interval) from two lung lobes (n=10 per group).
EdU in vitro proliferation assay
MMTV-PyMT actin-GFP cells were seeded at a density of 10,000 cells per well into collagen solution coated 6-well plates. The following day, Ly6G+ lung cells and/or Epcam+ lung cells were isolated via MACS sorting and added to the wells at a density of 100,000 cells/well. After 60h, wells were supplemented with 20μM EdU (5-ethynyl-2´-deoxyuridine). Cells were harvested 6h later, and EdU incorporation was assessed using the Click-iT Plus EdU Flow Cytometry Assay Kit (ThermoFisher Scientific, USA), according to the manufacturer’s instructions. Sample data were acquired on a BD LSR-Fortessa flow cytometer and analysed using FlowJo 10 software.
Conditioned media preparation and vesicle isolation
4T1-sLP-mCherry-GFP cells (Labelling-4T1) were plated on 10cm petri dishes. When cells were 80% confluent, 10ml of DMEM with 10% FCS was added to be conditioned for 48 h. The conditioned media preparation and vesicles isolation were performed as previously described40. Briefly, the media was collected and spun at 300g for 10 min. Next, the supernatant was collected and spun at 2,000g for 10 min. The supernatant after this second centrifugation was collected and used as conditioned media. For vesicle isolation, the conditioned media was subsequently ultra-centrifuged at 10,000g for 30 min and at 100,000g for 70 min. The vesicle pellet at this stage was washed with PBS, spun at 100,000g for 70 min and resuspended again in PBS for in vitro uptake experiments.
ImageStream analysis
Image stream analyses were carried out on an ImageStream Mark X II Imaging Flow Cytometer (Amnis Merck, USA). The acquired data were analysed using IDEA software (Amnis Merck, USA).
Electron Microscopy (EM)
Experiments were performed on glass bottom dishes with a numbered grid (MatTek, USA) to enable subsequent location of the same cell imaged by confocal microscopy. After confocal imaging, cells were fixed in 8% formaldehyde in 0.1M phosphate buffer (pH 7.4) added in equal quantities to cell media for 15 min and then further fixed in 2.5% glutaraldehyde and 4% formaldehyde in 0.1M phosphate buffer (pH 7.4) for 1 h and then processed using the National Center for Microscopy and Imaging Research (NCMIR) protocol (Deerinck, T.J., et al., NCMIR methods for 3D EM: a new protocol for preparation of biological specimens for serial block face scanning electron microscopy. National Center for Microscopy and Imaging Research (2010) (available from https://ncmir.ucsd.edu/sbem-protocol)).
For transmission electron microscopy (TEM), 70nm serial sections were cut using a UC6 ultramicrotome (Leica Microsystems, Germany) and collected on formvar-coated slot grids. No post-staining was required due to the density of metal deposited using the NCMIR protocol. Images were acquired using a 120 kV Tecnai G2 Spirit TEM (FEI Company ThermoFisher Scientific, USA) and an Orius CCD camera (Gatan, USA).
RNA sequencing sample preparation
Bulk RNA sequencing
CD45-Ter119- (CD45-ve) cells were sorted from single cell suspensions of metastatic lungs stained with anti-mouse CD45 and Ter119 antibodies and DAPI. RNA isolation was performed using the MagMax-96 Total RNA Isolation Kit (ThermoFisher Scientific, USA) that allows high quality RNA extraction from samples with low cell numbers (<10,000 cells). RNA quality for each sample was assessed using the Agilent RNA 6000 Pico Kit (Agilent Technologies, USA). RNA was amplified and analysed at the Barts and London Genome Centre.
Single cell RNA sequencing
CD45-Ter119- cells were sorted from single cell suspensions of metastatic lungs stained with anti-mouse CD45 and Ter119 antibodies and DAPI. Library generation for 10× Genomics were performed following the Chromium Single Cell 3′ Reagents Kits (10X Genomics, USA) and sequenced on an Hiseq4000 (Illumina, USA), to achieve an average of 50,000 reads per cell.
Determination of intracellular ROS levels
Single cell suspensions from mouse lungs were incubated with mouse FcR Blocking Reagent for 5 min on ice and subsequently incubated with CellROX® Deep Red Reagent (ThermoFisher Scientific, USA) for 30 min at 37ºC following manufacturer’s recommendations. Next, cells were washed twice with MACS buffer, stained with DAPI and analysed by flow cytometry.
Quantitative proteomic analysis of Ly6G cells
Neutrophils were FACS-sorted from single cell suspensions of metastatic lungs stained with a conjugated anti-mouse Ly6G-APC antibody (3 samples from independent sorts). Ly6G cells from the metastatic niche (Ch+) and the distal lung (Ch-) were digested into peptides using a previously described protocol (http://dx.doi.org/10.1101/220343) and analysed by Data Independent Acquisition (DIA) mass spectrometry41 on a Orbitrap Fusion Lumos instrument (ThermoFisher Scientific, USA). A hybrid spectral library was generated using the search engine Pulsar in Spectronaut Professional+ (version 11.0.15038, Biognosys AG, Switzerland) by combing Data Dependent Acquisition (DDA) runs obtained from a pooled sample of Ly6G cells, and the DIA data. Data analysis and differential protein expression was performed using Spectronaut Professional+. A detailed description of sample processing, data acquisition and processing are provided upon request.
Bioinformatic analysis
Bulk RNA sequencing
the sequencing was performed on biological triplicates for each condition generating approximately 35 million 76bp paired end reads. The RSEM package (version 1.2.29)42 and Bowtie2 were used to align reads to the mouse mm10 transcriptome, taken from known Gene reference table available at UCSC (https://genome.ucsc.edu/). For RSEM, all parameters were run as default except “--forward-prob” which was set to “0.5”. Differential expression analysis was carried out with DESeq2 package43 (version 1.12.4) within R version 3.3.1 (https://www.r-project.org/). Genes were considered to be differentially expressed if the adjusted p value was less than 0.05. Differentially expressed genes were taken forward and their pathway and process enrichments were analysed using Metacore (https://portal.genego.com). Hypergeometric test was used to determine statistical enriched pathways and processes and the associated p-value corrected using Benjamini-Hochberg method.
Gene Set Enrichment Analysis, GSEA, (version 2.2.3)44,45 was carried out using ranked gene lists using the Wald statistic and the gene sets of C2 canonical pathways and C5 biological processes. All parameters were kept as default except for enrichment statistic (classic) and max size which was changed to 5000 respectively. Gene signatures with FDR q-value equal or less than 0.05 were considered statistically significant. A weighted Kolmogorov–Smirnov-like statistic was performed and the associated p-value corrected with Benjamini-Hochberg method.
Single cell RNA sequencing
raw reads were initially processed by the Cell Ranger v2.1.1 pipeline, which deconvolved reads to their cell of origin using the UMI tags, aligned these to the mm10 transcriptome using STAR (v2.5.1b) and reported cell-specific gene expression count estimates. All subsequent analyses were performed in R-3.4.1 using the cellrangerRkit, monocle and pheatmap packages. Genes were considered to be “expressed” if the estimated (log10) count was at least 0.1. Primary filtering was then performed by removing from consideration: genes expressed in fewer than 20 cells; cells expressing fewer than 50 genes; cells for which the total yield (i.e. sum of expression across all genes) was more than 2 standard deviations from the mean across all cells in that sample; cells for which mitochondrial genes made up greater than 10% of all expressed genes. PCA decomposition was performed and, after consideration of the eigenvalue “elbow-plots”, the first 25 components were used to construct t-SNE plots for both samples. Niche cells expressing Epcam were subdivided into those also expressing Cdh1 and those not expressing Cdh1. Other genes expressed in at least 50% of cells in a given group were said to be co-expressed and the set of genes co-expressed in one or more groups was presented as a heatmap, with the columns (cells) clustered using the standard Euclidean hierarchical method.
Extended Data
Extended Data Figure 1. Cherry-niche in vitro.
a, sLP-mCherry design. b, Fluorescence images of Labelling-4T1 cells post-thawing. Scale bar 10μm. c, Representative FACS plot of Labelling-4T1 cells. d, In vitro cultures of the indicated cell types with Labelling-4T1 cell conditioned media (LCM): culture scheme and representative fluorescence images of HC11 (murine mammary epithelial cells) and hNLF (human normal lung fibroblasts) with LCM (scale bar 10μm). e, FACS plots of 4T1, HC11, RAW264.7 (murine macrophages), hNLF, murine breast Carcinoma Associated Fibroblasts (CAF) cultured with LCM. f, FACS analysis of 293T cells cultured with LCM, at different time-points after LCM removal (black dots); white dots show the theoretical decrease considering the cell proliferation rate only (the amount of 293T cells mCherry labelled after 24h incubation with LCM was set to 100%). g, Representative fluorescence image of 4T1-CD63GFP cells cultured with LCM. Scale bars: main 5μm, inset 1μm. h, Representative Correlative Light and Electron Microscopy (CLEM) of Labelling-4T1 cells re-up-taking sLP-mCherry (n=5 different cells analysed): upper-left panel shows bright-field image overlaid mCherry IF (~700nm optical section); lower-left panel shows EM of same cell (~70nm section thickness); large central panel shows best approximation of IF/bright-field/EM overlay (scale bar 5μm); right panels show EM insets from indicated areas in the large central panel (black arrows point at vesicular structures containing the mCherry, scale bar 1μm). i, j, Analysis of in vitro labelling potential of soluble fraction and extracellular vesicles (EVs) isolated from LCM by FACS: (i) schematic representation of LCM fractionation; (j) HC11 cells cultured with either LCM, soluble fraction after EVs depletion (soluble) or purified EVs. k, ImageStream analysis of Cherry+ EVs in LCM (16% of total EVs are Cherry+). Data are representative one of three (b), ten (c), two (d, e-g, j, k) independent experiments. See Source Data.
Extended Data Figure 2. Cherry-niche in vivo.
a, b, Distance of labelled cells within metastases: (a) representative fluorescence images (lines measure the maximum distance of labelled cells (mCherry) from Labelling-4T1 cells (mCherry/GFP); scale bar 50μm); (b) quantification of labelling distance in micro-metastases (n=11) and macro-metastases (n=4). c, Correlation between the percentage of mCherry labelled niche cells and the percentage of cancer cells in metastatic lungs analysed by FACS: only low number of cancer cells (left, n=14 mice) and all cancer cell frequencies (right, n=31 mice). Statistical analysis by Pearson correlation. d-f, CD45+ cell frequency on live cells in distal lung, Cherry-niche and naïve lungs (collected from mice which were not injected) by FACS: (d) Balb/c mice injected with Labelling-4T1 cells (n=5 per group); (e) Balb/c mice injected with Labelling-HC11 cells (n=4); (f) Rag1ko mice injected with Labelling-4T1 cells (n=10). Statistical analysis by paired two-tailed t-test. Data are represented as mean ±SEM. See Source Data.
Extended Data Figure 3. Cherry-niche neutrophils increase ROS production.
a, b, (a) CD11b+ and (b) Ly6G+cell frequencies on live cells in distal lung and Cherry-niche by FACS (n=9 per group). c, Enriched processes by MetaCore analysis and GSEA on proteomic data (by comparing Cherry-niche (n=3) and distal lung (n=3) neutrophils; Cherry-niche dominant proteins obtained by using WebGestalt (http://www.webgestalt.org/option.php). d, PCA of proteins found in unlabelled or Cherry-niche neutrophils (n=3, each with 10 mice, small circles; large circles represent the average of the triplicates). e, Representative FACS plot and f, scatter plot of intrinsic ROS in Ly6G+ cells (n=6). g, GFP signal quantification of 3D co-culture with MMTV-PyMT-GFP+ cancer cells, Ly6G+ MACS-sorted cells from either naïve or metastatic lungs with or without the ROS inhibitor TEMPO (n=3, each with 3 wells). Data is normalised to cancer cell growth (Statistical analysis on biological replicates). h, Representative cancer cell growth on the scaffold (from 14 independent experiments): integrated density of the GFP signal measured on the scaffold using ImageJ and the corresponding fluorescent image of GFP+ cancer cell growth (scale bar 400μm). Statistical analysis by paired two-tailed t-test (a, b, f), hypergeometric test with Benjamini-Hochberg correction (c, Metacore), weighted Kolmogorov–Smirnov-like statistic with Benjamini-Hochberg correction (c, GSEA) and Two-way ANOVA (g). Data represented as mean ±SD (f) and ±SEM (g). See Source Data.
Extended Data Figure 4. RNA sequencing of non-immune Cherry-niche cells.
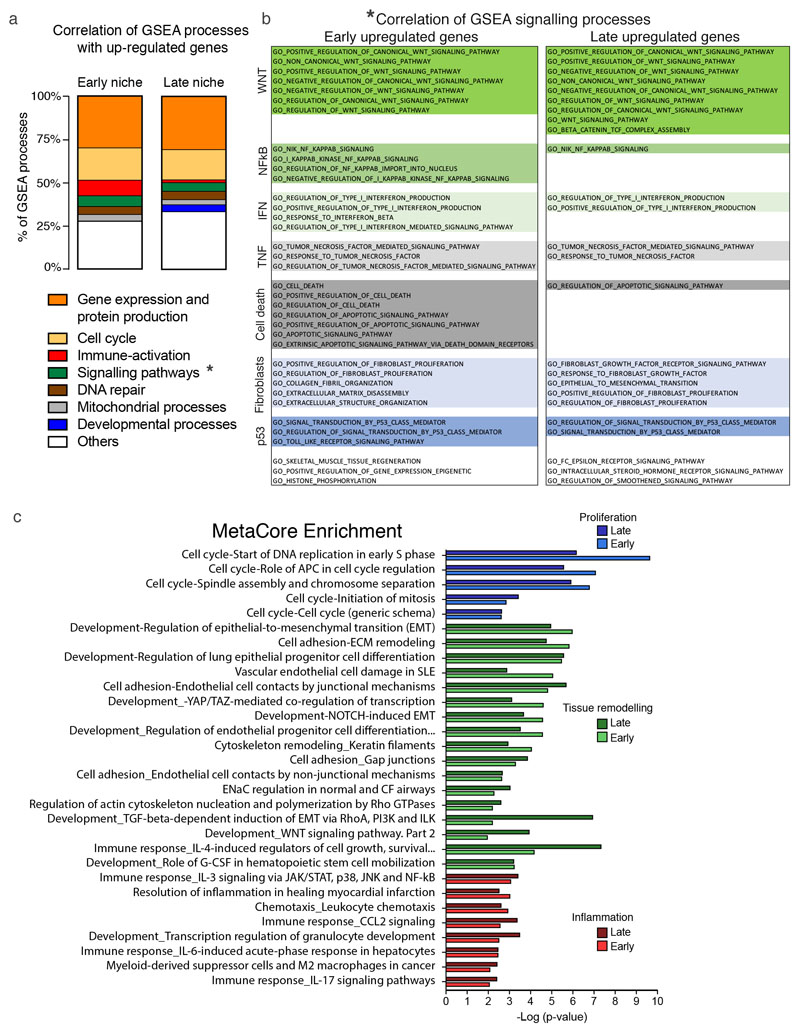
a, b, GSEA on Cherry-niche upregulated genes: (a) percentage of correlating processes related to the indicated activity and (b) specific signalling pathways (indicated by the * in (a) either at early or late time point). c, MetaCore analysis on genes differentially expressed from RNA-seq data, comparing early (n=3) or late (n=3) Cherry+ samples vs the respective Cherry- (see Fig.3a, b). Statistical analysis by hypergeometric test with Benjamini-Hochberg correction.
Extended Data Figure 5. Wisp1 supports metastatic growth.
a, b, Representative IF images of lung metastatic tissues (n=2 mice) stained with GFP (green) to detect Labelling-4T1 cells, WISP1 (red) and DAPI (blue) showing distal lung and metastatic areas, scale bar 50 μm; (b) a representative image showing the enrichment of Wisp1+ cells within lung metastasis including niche cells (white arrows); scale bar 50μm. c-e, Anti-WISP1 blocking antibody treatment in vivo: (c) experimental design (IT, intratracheal injection; IP, intraperitoneal injection); (d) metastatic outcome measured as the percentage of lung area covered by metastases (quantification was performed on two lung levels 100μm apart); (e) representative H&E images (n=5 mice each group; black arrows show metastatic foci); scale bar 500μm. Two experiments with lower overall metastatic frequency are quantified in Figure 3e. Statistical analysis by Two-way Anova (d). Data represented as mean ±SEM. See Source Data.
Extended Data Figure 6. Lung pneumocytes react to cancer cells in human breast pulmonary metastases.
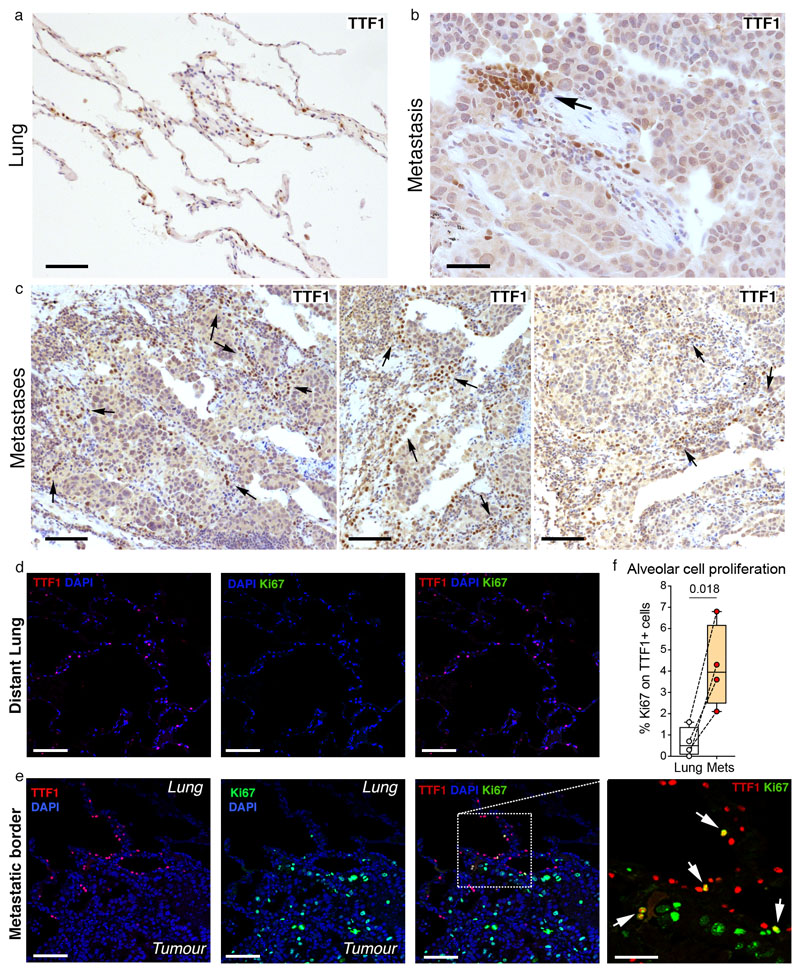
a-c, Histology on human breast tumour lung metastases sections: (a) representative image of distal lung (scale bar 100μm) and (b) image from the tumour-lung interface showing the pneumocyte marker Thyroid Transcription Factor 1 (TTF1) expression (scale bar 50μm); (c) representative histology images from metastatic border (scale bar 100μm). d-f, Alveolar cell proliferation in human breast tumour lung metastases analysed by IF: representative images from (d) distal lung and (e) metastatic border showing TTF1 (red), Ki67 (green) and DAPI (blue), scale bars: main 100μm, inset 50μm; (f) quantification shown on a Tukey plot: box from the 25th to 75th percentiles, the bar is the median and the whiskers from smallest to largest value. Statistical analysis by paired two-tailed t-test. Tissue sections from n=4 independent patients were analysed. See Source Data.
Extended Data Figure 7. Epithelial cells support cancer cell growth ex vivo.
a, MMTV-PyMT-GFP+ cancer cell proliferation in 2D co-culture with MACS-sorted Epcam+ and Ly6G+ cells stained with EdU and analysed by FACS (n=3 independent experiments). Data normalised to cancer cell proliferation. b-d, 3D co-culture of MMTV-PyMT-GFP+ cancer cell with MACS-sorted Epcam+ and Ly6G+ cells: (b) co-culture scheme; (c) representative images from 4 independent experiments (day 4, scale bar 400μm); (d) GFP signal quantification. Data normalised to cancer cell growth (n=4 independent experiments (dots), each with 3-4 technical replicates). Statistical analysis on biological replicates by one sample two-tailed t-test (a) and Two-way ANOVA (d). Data are represented as mean ±SEM. See Source Data.
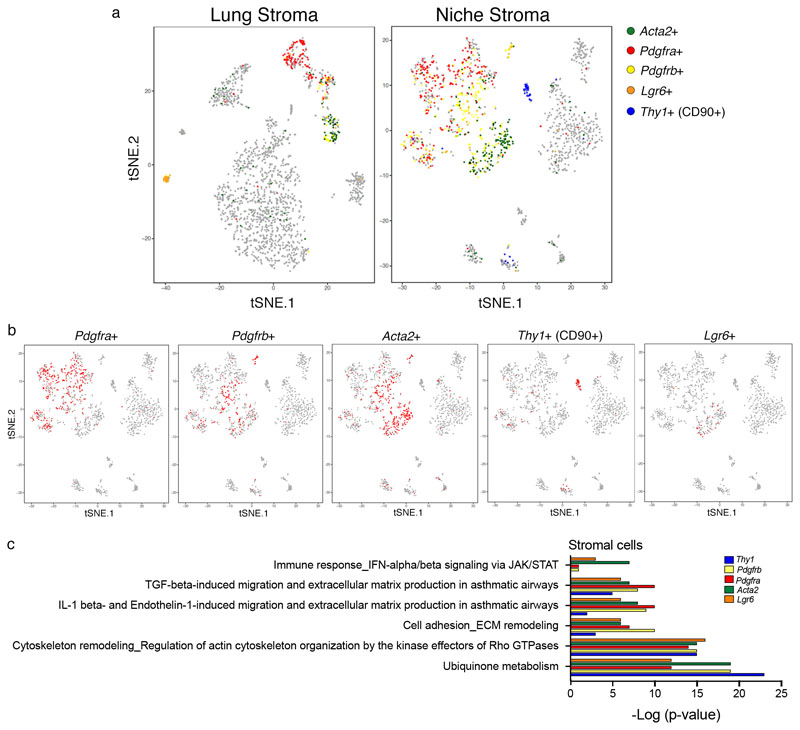
Extended Data Figure 8. scRNA-seq analysis reveals different sub-pools of stromal cells in the niche.
a, tSNE plots of CD45- cells isolated from distal lung (n=1996) or Cherry-niche (n=1473) after scRNA-seq analysis: the CAFs are coloured based on the expression levels of the indicated genes. b, tSNE niche plots (a), where each plot shows in red the cells expressing the indicated stromal marker. c, MetaCore pathway enrichment analysis using the list of genes detected in at least 50% of the indicated marker defined cells (n=66 Thy1+ cells, n=175 Pdgfrb1+ cells, n=322 Pdgfra+ cells, n=330 Acta2+ cells, n=25 Lgr6+ cells). Statistical analysis by hypergeometric test with Benjamini-Hochberg correction.
Extended Data Figure 9. Cherry-niche epithelial cells are enriched for stem cell markers.
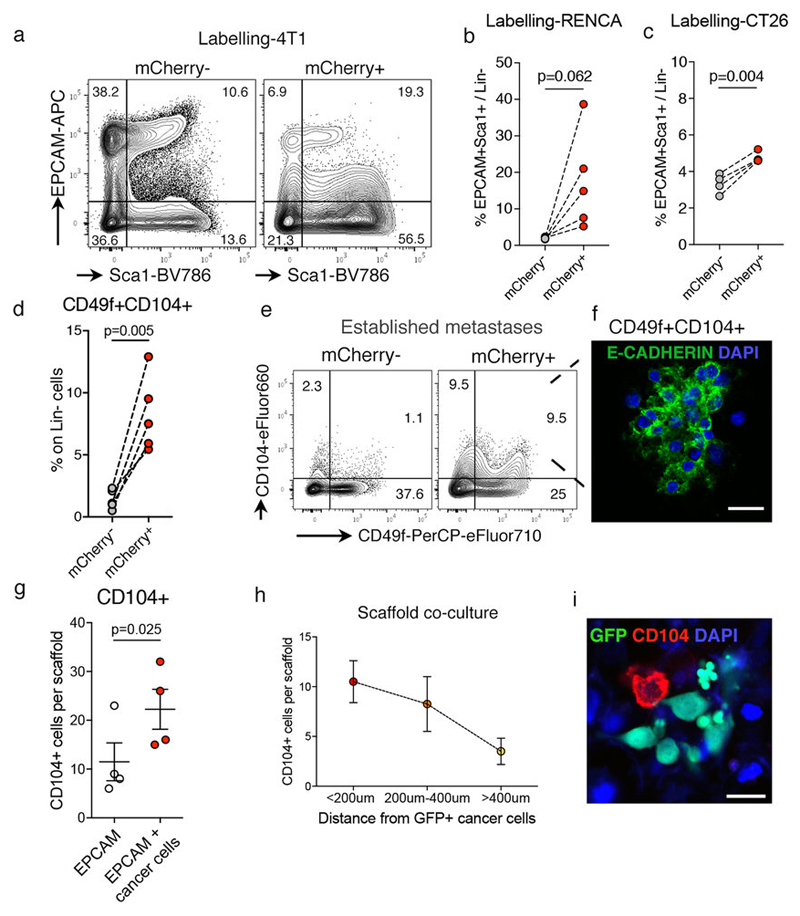
a, Representative FACS plots showing Lin- (CD45-CD31-Ter119-) cells in distal lung and Cherry-niche from Labelling-4T1 injected mice (quantification in Fig.4i). b, c, Scatter plots showing FACS quantification of Epcam+Sca1+ cell frequency on Lin- (CD45-CD31-Ter119-) cells in distal lung and Cherry-niche with (b) Labelling-RENCA (n=5) and (c) Labelling-CT26 (n=4). d-f, (d) Scatter plot of CD49f+CD104+ cell frequency on Lin- (CD45-CD31-Ter119-) cells in distal lung and Cherry-niche by FACS (n=5); (e) representative FACS plots; (f) representative IF image of FACS-sorted Cherry-niche CD49f+CD104+ cells using E-cadherin (green) and DAPI (blue); scale bar 20μm. g-i, 3D co-culture of MMTV-PyMT-GFP+ cancer cells with MACS-sorted Epcam+ cells: (g) quantification of integrin β4 (CD104) expression on Epcam+ cells; (h) number of CD104+ cells proximal to cancer cells (n=4 from three independent sorts); (i) representative IF image from the co-culture stained with CD104 (red); GFP+ cancer cells (green) and DAPI (blue); scale bar 20μm. Statistical analysis on biological replicates (g-h) by paired two-tailed t-test (b-d, g). Data represented as mean ±SEM. See Source Data.
Extended Data Figure 10. Cancer cells change lung epithelial cell lineage commitment ex-vivo.
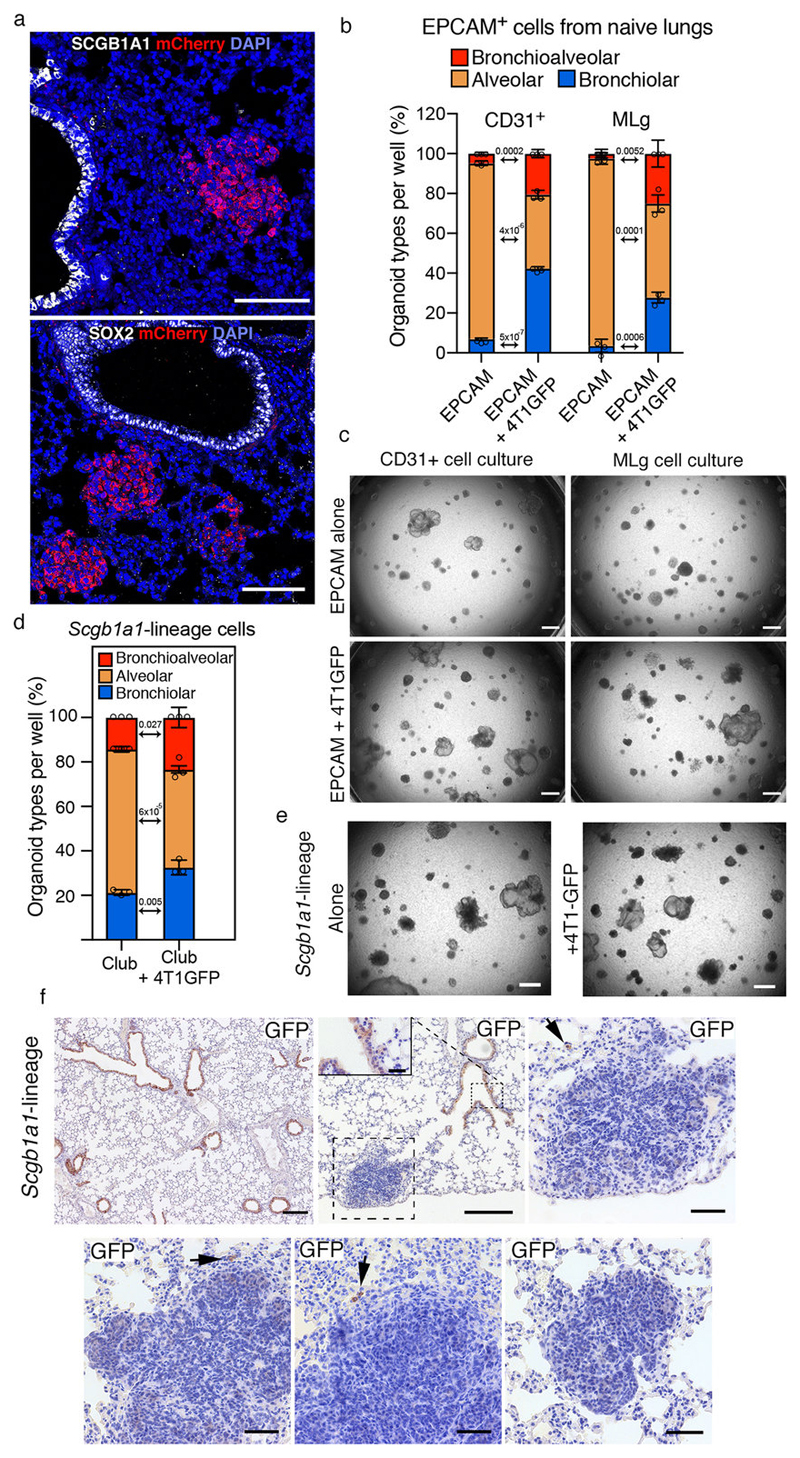
a, Representative IF images of lung metastatic sections (n=3 mice) co-stained with airway markers, either (a) Scgb1a1 (white) or (b) Sox2 (white), Cherry (red) and DAPI (blue); scale bar 100μm. b, c, Lung organoids with Epcam+ FACS-sorted cells in co-culture with either lung stromal CD31+ cells or MLg fibroblasts alone or in presence 4T1-GFP cells from metastatic lungs in the lower chamber: (b) quantification and (c) representative bright-field images of organoids, scale bar 150μm. d-e, Lung organoids with Scgb1a1-CreERT2 lineage cells with or without 4T1-GFP: quantification (d) and representative bright-field pictures (e), scale bar 150μm. f, Representative staining of lineage cells in metastatic lungs from Scgb1a1-CreERT2 mice injected with MMTV-PyMT cancer cells. Scale bars: main 50μm, apart from the first 2 panels where it’s 200μm (inset 25μm). Data generated with sorted Epcam (b) or club-lineage cells (d) and represented as cumulative percentage indicating the mean ±SD of three co-cultures per sorting. Statistical analysis by two-tailed t-test (b, d), for original non-cumulative values see accompanying Source Data. Images are representative of three organoid cultures (c, e). See Source Data.
Supplementary Material
sLP-Cherry sequence.
Patient information on pulmonary breast cancer metastases samples.
List of the antibodies used and list of the primers for the qRT-PCR targets.
Upregulated genes (>2 fold) in non-immune-mCherry-niche compared to unlabelled lung tissue at both 5 and 10 days post cancer cells seeding.
The entire list of differentially detected proteins (AVG log2 ratio) in mCherry-niche versus unlabelled Ly6G+ neutrophils. Differences were considered when changes were >0.58 and <−0.58.
FACS gating strategy examples, a list of antibodies used for FACS and immune-staining, a list of primers used for the qRT–PCRs (Fig. 4h) and information about the human pulmonary breast cancer metastases from four patients.
Acknowledgments
We thank E. Sahai, P. Scaffidi (The Francis Crick Institute, London) and V. Sanz-Moreno (Barts Cancer Institute, London) for scientific discussions, critical reading of the manuscript and sharing cell lines and mouse strains. We thank M. Izquierdo (CSIC, Madrid) for sharing the CD63-GFP plasmid. We are grateful to E. Nye and the pathologists G. Stamp and E. Herbert from from the Experimental Histopathology Unit for histological processing and analysis support. We thank J. Bee from the Biological Resources Unit at the Francis Crick Institute for technical support with mice and mouse tissues. Thanks to R. Goldstone and A. Edwards from the Advanced Sequencing Facility at the Francis Crick Institute for technical support. We thank M. Llorian-Sopena from the Bioinformatics & Biostatistics Unit at the Francis Crick Institute for helping with the RNA sequencing analysis. We also thank the Flow Cytometry Unit at the Francis Crick Institute, particularly S. Purewal and J. Cerveira, for invaluable technical help, and Cell Services Unit at the Francis Crick Institute. We are grateful to C. Moore (The Francis Crick Institute, London) for intra-tracheal injections. We are also grateful to I. Pshenichnaya, P. Humphreys, S. McCallum and Cambridge Stem Cell Institute core facilities for technical assistance. We gratefully acknowledge support from the FLI Core Facility Proteomics, which is a member of the Leibniz Association and is financially supported by the Federal Government of Germany and the State of Thuringia. This work was supported by the Francis Crick Institute which receives its core funding from Cancer Research UK (FC001112), the UK Medical Research Council (FC001112), and the Wellcome Trust (FC001112) and the European Research Council grant (ERC CoG-H2020-725492); and by the Wellcome Trust – MRC Stem Cell Institute which receives funding from the Sir Henry Dale Fellowship from Wellcome and the Royal Society (107633/Z/15/Z) and the European Research Council Starting Grant (679411).
Footnotes
Data availability
The RNA sequencing datasets (GSE117930) and the single cell RNA sequencing datasets (GSE131508) are deposited in the Gene Expression Omnibus (GEO, NCBI) repository. The proteomic datasets are deposited in PRoteomics IDEntifications (PRIDE) repository (PXD010597).
Author Contribution L.O. designed and performed most of the experiments, analysed and interpreted the data and contributed to the manuscript preparation. E.N. assisted with data collection, performed all the 3D-scaffold co-culture experiments, the in vivo Wisp1 experiments, the scRNA sequencing, interpreted and analysed the data and contributed to the manuscript preparation. I.K. performed the qPCR analysis, some of the tissue IF staining and analysed the data. A.M. and J.H.L. performed some of the tissue IF staining, all the lung organoid experiments, interpreted and analysed the data. V.B. performed some of the tissue IF staining. P.C. and S. H. performed bioinformatics analysis. I.H., J.K. and A.O. performed the proteomic and analysed the data. E.G.G. helped with the collection of Ly6G+ cells for proteomics. G.M. performed the 3D-scaffold co-culture to analyse CD104+ cells. A.W. and L.C. performed the electron microscopy experiments. E.H. and V.S. provided human samples. L.O., E.N., I.K., V.B. and J.H.L., critically reviewed the manuscript. J.H.L., supervised the lung organoid experiments. I.M. designed and supervised the study, interpreted the data and wrote the manuscript.
Author Information Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. All animal work was conducted under UK Home Office regulations, project licenses P83B37B3C and PC7F8AE82.
References
- 1.Hanahan D, Coussens LM. Accessories to the Crime: Functions of Cells Recruited to the Tumor Microenvironment. CCELL. 2012;21:309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 2.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nature Medicine. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barash S, Wang W, Shi Y. Human secretory signal peptide description by hidden Markov model and generation of a strong artificial signal peptide for secreted protein expression. Biochemical and Biophysical Research Communications. 2002;294:835–842. doi: 10.1016/S0006-291X(02)00566-1. [DOI] [PubMed] [Google Scholar]
- 4.Flinterman M, et al. Delivery of Therapeutic Proteins as Secretable TAT Fusion Products. Mol Ther. 2009;17:334–342. doi: 10.1038/mt.2008.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Meth. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 6.del Pozo Martin Y, et al. Mesenchymal Cancer Cell-Stroma Crosstalk Promotes Niche Activation, Epithelial Reversion, and Metastatic Colonization. CellReports. 2015;22:2456–2469. doi: 10.1016/j.celrep.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peinado H, et al. Pre-metastatic niches: organ-specific homes for metastases. Nat Rev Cancer. 2017;17:302–317. doi: 10.1038/nrc.2017.6. [DOI] [PubMed] [Google Scholar]
- 8.Wculek SK, Malanchi I. Neutrophils support lung colonization of metastasis-initiating breast cancer cells. Nature. 2015;17:413–417. doi: 10.1038/nature16140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coffelt SB, Wellenstein MD, de Visser KE. Neutrophils in cancer: neutral no more. Nature review cancer. 2016;16:431–446. doi: 10.1038/nrc.2016.52. [DOI] [PubMed] [Google Scholar]
- 10.Singhal S, et al. Origin and Role of a Subset of Tumor-Associated Neutrophils with Antigen-Presenting Cell Features in Early-Stage Human Lung Cancer. CCELL. 2016;30:120–135. doi: 10.1016/j.ccell.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blomberg OS, Spagnuolo L, de Visser KE. Immune regulation of metastasis: mechanistic insights and therapeutic opportunities. Dis Model Mech. 2018;11 doi: 10.1242/dmm.036236. dmm036236–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kessenbrock K, Plaks V, Werb Z. Matrix Metalloproteinases: Regulators of the Tumor Microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kowanetz M, et al. Granulocyte-colony stimulating factor promotes lung metastasis through mobilization of Ly6G+Ly6C+ granulocytes. Proc Natl Acad Sci USA. 2010;107:21248–21255. doi: 10.1073/pnas.1015855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qian B-Z, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature review cancer. 2011;475:222–225. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Acharyya S, et al. A CXCL1 Paracrine Network Links Cancer Chemoresistance and Metastasis. Cell. 2012;150:165–178. doi: 10.1016/j.cell.2012.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oskarsson T, et al. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nature Medicine. 2011;17:867–874. doi: 10.1038/nm.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erez N. Opening LOX to metastasis. Nature. 2015;522:41–42. doi: 10.1038/nature14529. [DOI] [PubMed] [Google Scholar]
- 18.Onnis B, Fer N, Rapisarda A, Perez VS, Melillo G. Autocrine production of IL-11 mediates tumorigenicity in hypoxic cancer cells. J Clin Invest. 2013;123:1615–1629. doi: 10.1172/JCI59623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malanchi I, et al. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature. 2011;481:85–89. doi: 10.1038/nature10694. [DOI] [PubMed] [Google Scholar]
- 20.Su F, Overholtzer M, Besser D, Levine AJ. WISP-1 attenuates p53-mediated apoptosis in response to DNA damage through activation of the Akt kinase. Genes Dev. 2002;16:46–57. doi: 10.1101/gad.942902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Costa A, et al. Fibroblast Heterogeneity and Immunosuppressive Environment in Human Breast Cancer. CCELL. 2018;33:463–479.e10. doi: 10.1016/j.ccell.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 22.Karnoub AE, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 23.Hosaka K, et al. Pericyte–fibroblast transition promotes tumor growth and metastasis. Proc Natl Acad Sci USA. 2016;113:E5618–E5627. doi: 10.1073/pnas.1608384113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murgai M, et al. KLF4-dependent perivascular cell plasticity mediates pre-metastatic niche formation and metastasis. Nature Medicine. 2017;23:1176–1190. doi: 10.1038/nm.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Treutlein B, et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim CFB, et al. Identification of Bronchioalveolar Stem Cells in Normal Lung and Lung Cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 27.Lee J-H, et al. Lung Stem Cell Differentiation in Mice Directed by Endothelial Cells via a BMP4-NFATc1-Thrombospondin-1 Axis. Cell. 2014;156:440–455. doi: 10.1016/j.cell.2013.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zacharias WJ, et al. Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature. 2018;555:251–255. doi: 10.1038/nature25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chapman HA, et al. Integrin α6β4 identifies an adult distal lung epithelial population with regenerative potential in mice. J Clin Invest. 2011;121:2855–2862. doi: 10.1172/JCI57673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McQualter JL, Yuen K, Williams B, Bertoncello I. Evidence of an epithelial stem/progenitor cell hierarchy in the adult mouse lung. Proc Natl Acad Sci USA. 2010;107:1414–1419. doi: 10.1073/pnas.0909207107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nabhan AN, Brownfield DG, Harbury PB, Krasnow MA, Desai TJ. Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science. 2018;359:1118–1123. doi: 10.1126/science.aam6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, et al. Tumor Exosomal RNAs Promote Lung Pre-metastatic Niche Formation by Activating Alveolar Epithelial TLR3 to Recruit Neutrophils. CCELL. 2016;30:243–256. doi: 10.1016/j.ccell.2016.06.021. [DOI] [PubMed] [Google Scholar]
- 33.Lee JW, et al. Hepatocytes direct the formation of a pro-metastatic niche in the liver. Nature. 2019;567:249–252. doi: 10.1038/s41586-019-1004-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. `Green mice' as a source of ubiquitous green cells. FEBS Letters. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 36.Rock JR, et al. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc Natl Acad Sci USA. 2011;108:E1475–83. doi: 10.1073/pnas.1117988108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rawlins EL, et al. The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell. 2009;4:525–534. doi: 10.1016/j.stem.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee J-H, et al. Anatomically and Functionally Distinct Lung Mesenchymal Populations Marked by Lgr5 and Lgr6. Cell. 2017;170:1149–1156.e12. doi: 10.1016/j.cell.2017.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;Chapter 3:Unit 3.22–3.22.29. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 41.Bruderer R, et al. Optimization of Experimental Parameters in Data-Independent Mass Spectrometry Significantly Increases Depth and Reproducibility of Results. Mol Cell Proteomics. 2017;16:2296–2309. doi: 10.1074/mcp.RA117.000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
sLP-Cherry sequence.
Patient information on pulmonary breast cancer metastases samples.
List of the antibodies used and list of the primers for the qRT-PCR targets.
Upregulated genes (>2 fold) in non-immune-mCherry-niche compared to unlabelled lung tissue at both 5 and 10 days post cancer cells seeding.
The entire list of differentially detected proteins (AVG log2 ratio) in mCherry-niche versus unlabelled Ly6G+ neutrophils. Differences were considered when changes were >0.58 and <−0.58.
FACS gating strategy examples, a list of antibodies used for FACS and immune-staining, a list of primers used for the qRT–PCRs (Fig. 4h) and information about the human pulmonary breast cancer metastases from four patients.