Strain PM004 is a cultured representative of human microbial taxon 955, a bacterium from the phylum Saccharibacteria. It is an obligate parasite with a genome of <0.9 Mb and can be grown in coculture with its host, Pseudopropionibacterium propionicum. The complete genome sequence is presented here.
ABSTRACT
Strain PM004 is a cultured representative of human microbial taxon 955, a bacterium from the phylum Saccharibacteria. It is an obligate parasite with a genome of <0.9 Mb and can be grown in coculture with its host, Pseudopropionibacterium propionicum. The complete genome sequence is presented here.
ANNOUNCEMENT
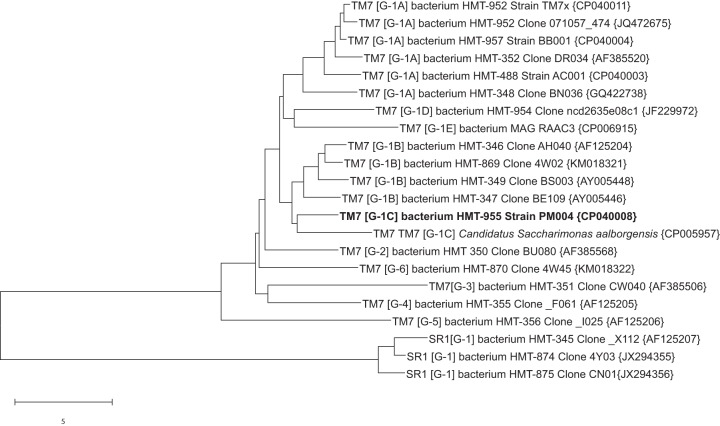
The human oral cavity contains many previously uncultivated bacteria, including members of the candidate phyla radiation (CPR) (1, 2). Oral isolate TM7x (3, 4) was the first CPR bacterium cultured. This member of the candidate phylum Saccharibacteria was isolated in coculture with Actinomyces odontolyticus strain XH001 and is thought to be an obligate parasitic epibiont (5, 6). Strain TM7x is a member of human microbiome taxon (HMT) 952 in the Human Oral Microbial Database (7, 8). The human oral Saccharibacteria strain PM004 was isolated in coculture with Pseudopropionibacterium propionicum strain F0700. HMT 955 strain PM004 differs sufficiently from HMT 952 strain TM7x by both 16S rRNA (93.7% identity) and genome comparisons to represent a novel genus and species. A phylogenetic tree for human oral and important environmental Saccharibacteria is presented in Fig. 1. Strain PM004 was isolated in Cambridge, MA, from the subgingival plaque of a 47-year-old Asian male smoker with periodontitis. The sample was dispersed and filtered through a 0.2-μm filter to remove all but ultrasmall bacteria and was added to broth cultures of P. propionicum. The novel Saccharibacteria bacterium was recovered growing in coculture with P. propionicum.
FIG 1.
Neighbor-joining tree (10) for Saccharibacteria (TM7) bacterium HMT 955 strain PM004. The tree was produced from aligned full-length 16S rRNA sequences (∼1,450 bp) using MEGA X (11). The designations in square brackets are the TM7 class level groups as described in reference 12. GenBank accession numbers for 16S rRNA or genome sequences are given in curly brackets. The scale bar shows a 5% difference in sequence similarity. For trees showing broader TM7 diversity, see references 12, to ,14.
DNA for genome sequencing was isolated from a 200-ml coculture of strain PM004 and P. propionicum in Trypticase soy broth with 1% yeast extract. The cocultured PM004 cells were filtered through a 0.45-μm filter and pelleted by centrifugation. DNA was isolated from cells by bead beating and through the use of a MasterPure DNA isolation kit (Lucigen). Library preparation and DNA sequencing were performed at the Johns Hopkins Deep Sequencing and Microarray Core. Genomic DNA was sheared to 10 to 20 kb using a Covaris g-TUBE and purified using AMPure XP beads (Agencourt Bioscience). Size selection and further cleanup were performed using BluePippin (Sage Science). The library was sequenced on a PacBio RS II instrument on one single-molecule real-time (SMRT) cell per library. Default parameters were used for all software. The reads were assembled using the SMRT Analysis software version 2.3.0 HGAP3 pipeline. Methylation motifs were detected using the SMRT Analysis software version 2.3.0 Base_Modification_and_Motif_ Analysis pipeline. Genes were annotated using the NCBI Prokaryotic Genome Annotation Pipeline using the best-placed reference protein set (GeneMarkS-2+ v4.8) (9).
There were 80,150 postfilter reads covering 1,166,362,784 sequenced bases. The mean read length was 14,552 bases, and the N50 read length was 21,336 bases. The reads were assembled into 21 contigs. The genome of HMT 955 strain PM004 was present in a single contig of 864,547 bp with average reference coverage of 918×. It was circularized to 842,202 by removing a duplicated 22-kbp segment at the ends of the initial contig. Twenty other contigs of less than 54 kbp and 40× coverage were identified as low-level contamination of host bacteria. The GC content of the DNA was 46.8%. Genome annotation identified a total 875 genes, of which 825 were predicted to be coding sequences (CDSs), 50 were RNAs, and 10 were pseudogenes.
Data availability.
The genome sequence was deposited at GenBank under accession number CP040008 and SRA accession number SRR9734868. Base modification files were submitted with the GenBank accession.
ACKNOWLEDGMENTS
The research reported in this publication was supported by the National Institute of Dental and Craniofacial Research of the National Institutes of Health under award numbers R37 DE016937 and R01 DE024468.
The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Brown CT, Hug LA, Thomas BC, Sharon I, Castelle CJ, Singh A, Wilkins MJ, Wrighton KC, Williams KH, Banfield JF. 2015. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523:208–211. doi: 10.1038/nature14486. [DOI] [PubMed] [Google Scholar]
- 2.Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, Butterfield CN, Hernsdorf AW, Amano Y, Ise K, Suzuki Y, Dudek N, Relman DA, Finstad KM, Amundson R, Thomas BC, Banfield JF. 2016. A new view of the tree of life. Nat Microbiol 1:16048. doi: 10.1038/nmicrobiol.2016.48. [DOI] [PubMed] [Google Scholar]
- 3.He X, McLean JS, Edlund A, Yooseph S, Hall AP, Liu SY, Dorrestein PC, Esquenazi E, Hunter RC, Cheng G, Nelson KE, Lux R, Shi W. 2015. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc Natl Acad Sci U S A 112:244–249. doi: 10.1073/pnas.1419038112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McLean JS, Bor B, To TT, Liu Q, Kerns KA, Solden L, Wrighton K, He X, Shi W. 2018. Evidence of independent acquisition and adaption of ultra-small bacteria to human hosts across the highly diverse yet reduced genomes of the phylum Saccharibacteria. bioRxiv. doi: 10.1101/258137. [DOI]
- 5.Bor B, Poweleit N, Bois JS, Cen L, Bedree JK, Zhou ZH, Gunsalus RP, Lux R, McLean JS, He XS, Shi WY. 2016. Phenotypic and physiological characterization of the epibiotic interaction between TM7x and its basibiont Actinomyces. Microb Ecol 71:243–255. doi: 10.1007/s00248-015-0711-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bor B, McLean JS, Foster KR, Cen L, To TT, Serrato-Guillen A, Dewhirst FE, Shi W, He X. 2018. Rapid evolution of decreased host susceptibility drives a stable relationship between ultrasmall parasite TM7x and its bacterial host. Proc Natl Acad Sci U S A 115:12277–12282. doi: 10.1073/pnas.1810625115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. 2010. The human oral microbiome. J Bacteriol 192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Escapa IF, Chen T, Huang Y, Gajare P, Dewhirst FE, Lemon KP. 2018. New insights into human nostril microbiome from the Expanded Human Oral Microbiome Database (eHOMD): a resource for the microbiome of the human aerodigestive tract. mSystems 3:e00187-18. doi: 10.1128/mSystems.00187-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 11.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol 35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Camanocha A, Dewhirst FE. 2014. Host-associated bacterial taxa from Chlorobi, Chloroflexi, GN02, Synergistetes, SR1, TM7, and WPS-2 phyla/candidate divisions. J Oral Microbiol 6:25468. doi: 10.3402/jom.v6.25468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hugenholtz P, Tyson GW, Webb RI, Wagner AM, Blackall LL. 2001. Investigation of candidate division TM7, a recently recognized major lineage of the domain Bacteria with no known pure-culture representatives. Appl Environ Microbiol 67:411–419. doi: 10.1128/AEM.67.1.411-419.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dinis JM, Barton DE, Ghadiri J, Surendar D, Reddy K, Velasquez F, Chaffee CL, Lee MC, Gavrilova H, Ozuna H, Smits SA, Ouverney CC. 2011. In search of an uncultured human-associated TM7 bacterium in the environment. PLoS One 6:e21280. doi: 10.1371/journal.pone.0021280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequence was deposited at GenBank under accession number CP040008 and SRA accession number SRR9734868. Base modification files were submitted with the GenBank accession.