Abstract
Necrotic enteritis (NE) causes huge economic losses to the poultry industry. Probiotics are used as potential alternatives to antibiotics to prevent NE. It is known that Clostridium butyricum can act as a probiotic that can prevent infection. However, whether or not it exerts a beneficial effect on NE in chickens remains elusive. Therefore, we investigated the impact of C. butyricum on immune response and intestinal microbiota during the development of NE in chickens, including experimental stages with basal diets, high-fishmeal-supplementation diets, and Clostridium perfringens challenge. Chickens were divided into two groups from day 1 to day 20: one group had its diet supplemented with C. butyricum supplementation and one did not. At day 20, the chickens were divided into four groups: C. perfringens challenged and unchallenged chickens with and without C. butyricum supplementation. All groups were fed a basal diet for 13 days and thereafter a basal diet with 50% fishmeal from day 14 to 24. Chickens were infected with C. perfringens from day 21 to 23. At days 13, 20 and 24, samples were collected for analysis of the relative expression of immune response and intestinal mucosa barrier-related genes and intestinal microbes. The results show that C. butyricum can inhibit the increase in IL-17A gene expression and the reduction in Claudin-1 gene induced-expression caused by C. perfringens challenge. Moreover, C. butyricum was found to increase the expression of anti-inflammatory IL-10 in infected chickens. Although C. butyricum was found to have a significant beneficial effect on the structure of intestinal bacteria in the basal diet groups and decrease the abundance of C. perfringens in the gut, it did not significantly affect the occurrence of intestinal lesions and did not significantly correct the shift in gut bacterial composition post C. perfringens infection. In conclusion, although C. butyricum promotes the expression of anti-inflammatory and tight junction protein genes and inhibits pro-inflammatory genes in C. perfringens-challenged chickens, it is not adequate to improve the structure of intestinal microbiota in NE chickens. Therefore, more effective schemes of C. butyricum supplementation to prevent and treat NE in chickens need to be identified.
Keywords: Clostridium perfringens, Clostridium butyricum, necrotic enteritis, gut microbiota, mucosal immunity, intestine barrier
Introduction
Necrotic enteritis (NE), which is caused by Clostridium perfringens in broiler chickens, is a common disease in the poultry industry, resulting in huge financial losses every year. It is estimated that NE causes losses of more than $US 6 billion per year and poses a substantial risk to public health (Wade Ben, 2015). However, C. perfringens infection alone does not cause clinical NE (Olkowski et al., 2006; Shojadoost et al., 2012). Predisposing factors, such as a high-fishmeal-supplementation diet or/and Eimeria coinfection, combined with C. perfringens infection, are required to induce NE in chickens (Wu et al., 2010, 2011; Fernandes da Costa et al., 2013). It has been reported that high-fishmeal diets that include high available nutrition cause an increase in C. perfringens-associated lesions in the chicken gut (Drew et al., 2004). We used a high-fishmeal-supplementation diet and C. perfringens challenge to induce NE in chickens. According to the findings of previous studies (Wu et al., 2010; Fernandes da Costa et al., 2013), the induction of NE in this study was divided into three main stages: (i) 1-13-day-old chickens on a basal diet, (ii) 14-20-day-old chickens on a high-fishmeal-supplementation diet, (iii) 21-24-day-old chickens on a high-fishmeal-supplementation diet and C. perfringens challenge.
Usually, NE is prevented through the use of antibiotics. However, many countries have restricted the use of antibiotics owing to the emergence of resistant strains, disruption of the host gut microbiota, and residual contamination of products and the environment. Probiotics, essential oils and organic acids are among the potential alternatives to antibiotics (Allaart et al., 2013). Clostridium butyricum is a butyric acid-producing gram-positive anaerobe that commonly exists in the intestines of humans and animals. In chickens, butyric acid inhibits pathogens and improves intestine barrier function (Meimandipour et al., 2010; Zhang et al., 2011), benefits intestinal microbiota, and stimulates immune factors (Kong et al., 2011; Zhang et al., 2011; Gao et al., 2012b; Yang et al., 2012). C. butyricum has been used as a probiotic that decreases the clinical signs of several diseases such as inflammatory bowel disease (IBD) and antibiotic-associated diarrhea (Seki et al., 2003; Yasueda et al., 2016). Moreover, previous studies have shown that C. butyricum could be a suitable alternative for reducing colonization by pathogens such as Salmonella and Escherichia coli (Zhang et al., 2016; Zhao et al., 2017).
However, the effect of C. butyricum on NE in chickens remains to be elucidated. Therefore, we aimed to explore the effects of C. butyricum on gut microbiota, immune response, and intestinal barrier function-related genes during the development of NE in chickens.
Materials and Methods
Ethics Statement
This study was carried out in accordance with the principles of the Basel Declaration and the recommendations of the Institutional Animal Care and Use Committee guidelines, Guangdong Academy of Agricultural Sciences. The study protocol was approved by the Guangdong Academy of Agricultural Sciences (No. SYXK (yue)-20180108).
Bacterial Strains
The pathogenic C. perfringens cpnetBF carrying the netB gene was isolated from the intestines of broiler chickens with NE in Guangdong Province, China (Huang et al., 2018). The strain was stored at −80°C in fluid thioglycollate broth (FT, Beckson, Dickinson, and Company) supplemented with 30% glycerol. C. butyricum YH 018 with a count of 1 × 109 cfu/g was obtained from Yihao Biotic Co., Ltd., China. The phylogenetic relationships based on 16S rRNA gene sequences are shown in Supplementary Figure S1. The concatenated sequences were aligned by MEGA 6 to infer a maximum-likelihood tree. C. butyricum YH 018 is clustered with type strains of species of the C. butyricum phylogroup including C. butyricum strain JCM 1391T and ATCC 19398T.
Animal Trial Design and Sample Collection
One-day-old Ross 308 commercial chickens (negative for C. perfringens) were purchased from a hatchery in Guangzhou City, Guangdong Province, China. The chickens were given food and water ad libitum and maintained in a 12 h light/12 h dark cycle. The animal trial was conducted at a poultry house located at the Institute of Animal Health, Guangdong Academy of Agricultural Sciences. A total of 120 chickens were randomly divided into two groups. All groups were fed an antibiotic-free basic chicken diet between the ages of 1 and 13 days (Supplementary Table S1) and then switched to a wheat-based diet containing 50% fishmeal from day 14 to 24. One group was supplemented with 1 × 109 cfu of C. butyricum/kg between the ages of 1 and 24 days. At an age of 20 days, each of the above-mentioned groups of chickens was further divided into two groups. From the ages of 21 days to 23 days, the C. butyricum supplemented and non-supplemented groups were challenged with C. perfringens cpnetBF-inoculated feed, as previously described (Stanley et al., 2012).
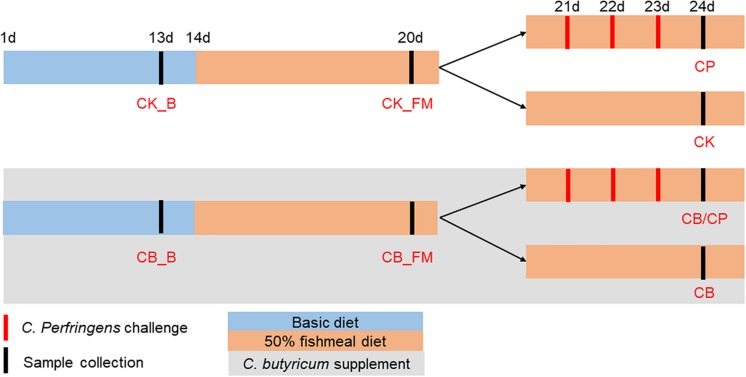
Chickens from each group were euthanized using cervical dislocation at an age of either 13, 20, or 24 days. Twelve replicates each of the jejunum-ileum contents of groups CB_B, CK_B, CB_FM, and CK_FM and six replicates each of the jejunum-ileum contents of groups CB/CP, CB, CP, and CK groups were collected for DNA extraction. The 12 ileal tissue replicates in each of the CB_B, CK_B, CB_FM and CK_FM groups and the six ileal tissue replicates in each of the CB/CP, CB, CP and CK groups were collected from a distance of 3 cm distal to the Meckel’s diverticulum and were immediately frozen in liquid nitrogen. The C. butyricum supplemented group was named CB_B (n = 60) and CB_FM (n = 48) at ages of 13 and 20 days, respectively, while the C. butyricum non-supplemented group was named CK_B (n = 60) and CK_FM (n = 48) at ages of 13 and 20, respectively. At an age of 24 days, the names of the four groups were as follows: CB/CP (n = 18): C. perfringens-challenged chickens with C. butyricum supplementation, CB (n = 18): C. perfringens-unchallenged chickens with C. butyricum supplementation, CP (n = 18): C. perfringens-challenged chickens without C. butyricum supplementation, and CK (n = 18): C. perfringens-unchallenged chickens without C. butyricum supplementation, as shown in Figure 1.
FIGURE 1.
Experimental design. CB_B (n = 60): C. butyricum supplemented chickens on a basal diet; CK_B (n = 60): C. butyricum non-supplemented chickens on a basal diet; CB_FM (n = 48): C. butyricum supplemented chickens on a high-fishmeal diet; CK_FM (n = 48): C. butyricum non-supplemented chickens on a high-fishmeal diet; CB/CP (n = 18): C. perfringens-challenged chickens with C. butyricum supplementation; CB (n = 18): C. perfringens-unchallenged chickens with C. butyricum supplementation; CP (n = 18): C. perfringens-challenged chickens without C. butyricum supplementation; CK (n = 18): C. perfringens-unchallenged chickens without C. butyricum supplementation.
Intestinal Lesion Score
At an age of 24 days, the lesions of the small intestine (duodenum to ileum) were scored on the basis of 10 replicates each from the four groups at the challenge stage, as previously described (Keyburn et al., 2006): 0, no gross lesions; 1, thin or friable walls; 2, focal necrosis or ulceration (one to five foci); 3, focal necrosis or ulceration (six to 15 foci); 4, focal necrosis or ulceration (16 or more foci); 5, patches of necrosis 2–3 cm long; 6, diffused necrosis typical of field cases.
DNA Extraction and the Abundance of C. perfringens
DNA from the jejunum-ileum contents was extracted using a QIAamp Fast DNA Stool Mini Kit (Qiagen, Valencia, CA, United States), according to the manufacturer’s instructions. Total DNA was quantified on a NanoDrop® ND-2000 UV spectrophotometer (NanoDrop Technologies, Wilmington, DE). Only DNA samples with an A260/A280 value >1.7 and A260/A230 value >1.8 were used for further analysis (Li et al., 2014). The extracts were stored at −20°C until used (Singh et al., 2015).
Quantitative PCR reaction was performed on the intestinal DNA template, in triplicate, using an SYBR Premix Ex Taq (Takara Bio, Otsushi, Japan). The expression of the C. perfringens genes was normalized using the 16S rRNA gene (Buffie et al., 2015). The relative abundance of C. perfringens in the intestine was calculated based on the value of the 16S rRNA gene using the 2–Δ Ct method. The primers used for the qPCR of C. perfringens and 16S rRNA gene are shown in Supplementary Table S2.
16s rRNA Gene Sequencing
The gene-specific sequences for the 16S V3-V4 region were amplified using primers 338F (5′- ACTCCTACGGGAGGCA GCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) using a thermocycler PCR system (GeneAmp 9700, ABI, United States). The PCR conditions were as follows: 3 min of denaturation at 95°C, 27 cycles of 30 s at 95°C, 30s for annealing at 55°C, 45s for elongation at 72°C, and a final extension step at 72°C for 10 min. PCR reactions were performed in triplicate using a 20-μL mixture containing 4 μL of 5 × FastPfu Buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of each primer (5 μM), 0.4 μL of FastPfu Polymerase, and 10 ng of template DNA. The resulting PCR products were extracted from the 2% agarose gel and further purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, United States) and quantified using a QuantiFluorTM-ST (Promega, United States) according to the manufacturer’s protocol. Bar-coded V3-V4 amplicons were sequenced using the 2 × 300 paired-end method by Illumina MiSeq with a 7-cycle index read. Sequence processing was performed using QIIME 2 to process the data. Operational taxonomic units (OTUs) were clustered using a 97% similarity cutoff in UPARSE (version 7.11), and chimeric sequences were identified and removed using UCHIME. The taxonomy of each 16S rRNA gene sequence was analyzed using an RDP Classifier algorithm2 against the Silva (SSU123) 16S rRNA database using a confidence threshold of 70%. Simpson and abundance-based coverage (ACE) indices were calculated for each time point to represent taxonomic alpha diversity. The Venn diagrams were calculated using R software. The microbial community structures of the different samples were compared using Bray-Curtis dissimilarity. Principal coordinate analysis (PCoA) was conducted to assess the relationships among the different groups. The linear discriminate analysis (LDA) effect size (LEfse) using the non-parametric factorial Kruskal-Wallis (KW) sum-rank test was used to identify significant differences in the relative abundance of bacteria between two groups. The functional potentials of the intestinal microbiotas were predicted using the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt).
RNA Isolation and RT-qPCR of the Immune Genes
Total RNA was isolated from ileal tissue using the EZNA® Total RNA Isolation Kit (Omega Bio-Tek) according to the manufacturer’s instructions. RNA was eluted in DEPC-treated water, quantified using a NanoDrop ND-2000 UV spectrophotometer (NanoDrop® Technologies, Wilmington, DE, United States), and stored at −80°C until further use.
Reverse transcription of the isolated RNA was performed using an M-MLV First Strand cDNA Synthesis Kit (Omega Bio-Tek) following the manufacturer’s instructions. RT-qPCR was performed with cDNA temple in triplicate using SYBR Premix Ex Taq (Takara Bio, Otsushi, Japan). The expressions of TLR2, IL-10, TNF, IL-1β, NF-κB, IL-17A, Claudin-1, Claudin-2, Occduin1, and IgA genes were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Zhang et al., 2017). The relative expression was calculated based on the expression of GAPDH, using the 2–Δ Ct method. The primers used for RT-qPCR are shown in Supplementary Table S2.
Statistical Analysis
Statistical analysis of the relative expression of mRNA, the relative abundance of C. perfringens and the alpha diversity indices of Simpson and ACE were performed using ANOVA. A p-value of <0.05 was considered to be statistically significant. For species abundance, comparisons were made using the Kruskal-Wallis test with Benjamini-Hochberg p-value correction.
Results
Clostridium perfringens Colonization
All chickens were negative for C. perfringens until the challenge, and the unchallenged groups remained negative throughout this study. In the challenged groups, the relative abundance of C. perfringens in chickens with C. butyricum supplementation was significantly lower than that of C. butyricum non-supplemented chickens (p < 0.05, Figure 2).
FIGURE 2.
Relative abundance of C. perfringens in the jejunum-ileum lumen post 1-day challenge. All chickens tested negative for C. perfringens until experimentally challenged, and the unchallenged groups tested negative for C. perfringens throughout the study. ∗p < 0.05, measured using ANOVA; n = 6.
Intestinal Lesion Score
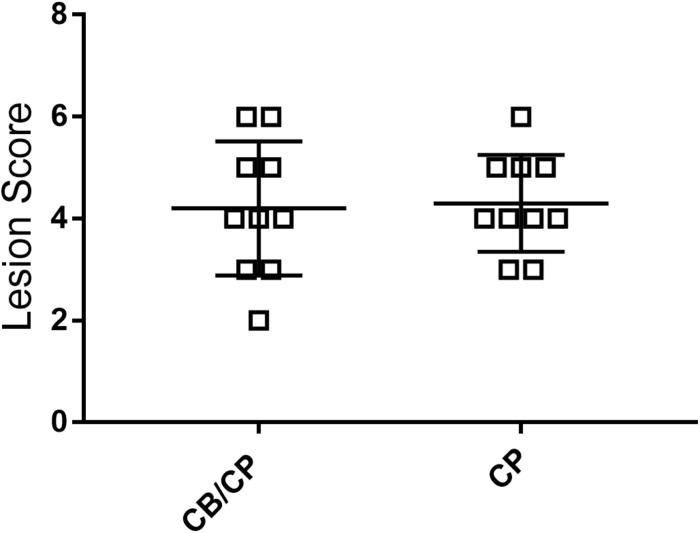
No intestinal lesions were observed in unchallenged chickens. The infection-induced intestinal lesion score was not influenced by the presence or absence of C. butyricum supplementation (p > 0.05, Figure 3).
FIGURE 3.
Intestinal lesion scores of chickens after C. perfringens infection. Measured using ANOVA; n = 10. No intestinal lesion was found in the unchallenged groups.
Expression of Immune Response Genes
The expressions of the immune and intestinal barrier-related genes detected in the different groups are shown in Figure 4. In the basal diet chickens, only IL-17A expression was significantly different between the C. butyricum supplemented and non- supplemented groups. The relative expressions of TLR2, IL-10, Claudin-1, and Claudin-2 were significantly higher in the CB_FM group than in the CK_FM group (p < 0.05). Compared with the other three groups, C. perfringens infection induced the highest expression of TNF in the CB/CP group. The expression of IL-10 showed no significant difference between the CP and CK group; however, it was significantly higher in the CB/CP group than in the CP group. The relative expression of IL-17A increased significantly in the CP group compared in with the CK group and significantly decreased in the CB/CP and CB groups compared in with the CP and CK groups. Conversely, the expression of Claudin-1 significantly decreased in the CP group compared with in the CK group and increased significantly in the CB/CP group compared in with the CP group (p < 0.05). The C. perfringens challenge significantly increased the relative expression of IgA, but C. butyricum had no effect on the level of IgA. There were no significant differences between the relative expressions of the IL-1β, NF-κB, and occludin-1 genes of the groups at different stages.
FIGURE 4.
Relative expression of immune and intestinal mucosa barrier-related genes in the ileum. Relative gene expression is represented as log10 2–Δ Ct. Significant differences are indicated by an asterisk (ANOVA, ∗p < 0.05). CB_B, CK_B, CB_FM and CK_FM: n = 12; CB/CP, CB, CP and CK: n = 6.
Intestinal Bacterial Diversity Analysis
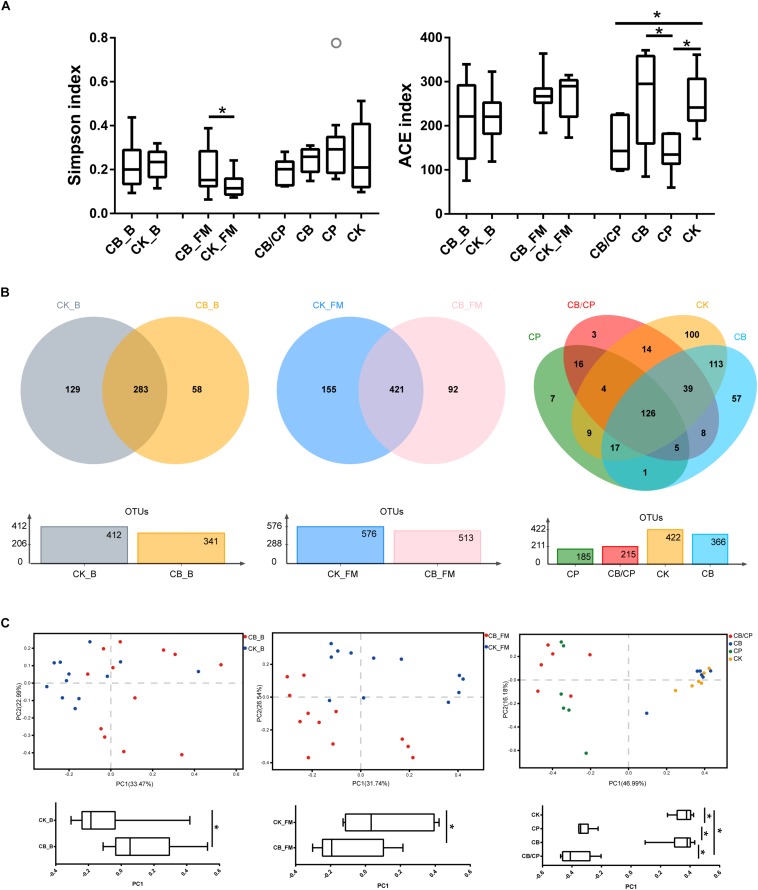
A total of 856,602, 1,034,832, and 916,190 high-quality 16S rRNA sequences were obtained from 13-, 20-, and 24-day-old intestinal content samples, respectively. The Simpson and ACE indices were used as measures of the alpha diversity (evenness and richness) of the gut microbiota (Figure 5A). In the basal diet groups, no significant differences were found between the ACE and Simpson index values of the CB_B and CK_B groups. During the stage of high-fishmeal supplementation, C. butyricum significantly increased the Simpson indices in the CB_FM group compared with in the non-supplemented CK_FM group (p < 0.05). Infection by C. perfringens significantly decreased the ACE indices compared with the unchallenged groups, CB and CK (p < 0.05). However, there was no significant difference between the CB/CP and CP groups.
FIGURE 5.
Diversity of the intestinal bacteria community at different stages. (A) Alpha diversity was estimated using Simpson and ACE indices values. ∗p < 0.05, measured using ANOVA. (B) Unique and shared intestinal OTUs shown in Venn diagrams. (C) PCoA plot of Bray-Curtis indices for the gut microbiota. PC1 comparisons were made using ANOVA. ∗p < 0.05. CB_B, CK_B, CB_FM and CK_FM: n = 12; CB/CP, CB, CP and CK: n = 6.
A Venn diagram reveals the shared and specific OTUs of the different groups (Figure 5B). Dietary C. butyricum decreased the number of unique OTUs in the CB_B (58) and CB_FM (92) groups compared with the CK_B (129) and CK_FM (155) groups, respectively. At the stage of C. perfringens challenge, the four groups shared 126 OTUs. In addition, compared with the CB/CP group, the CP group had a less unique OTU and shared less OTUs with the CK group.
Principal coordinate analysis (PCoA) of Bray-Curtis indices demonstrates the diversity of bacteria among the different groups (Figure 5C). Before C. perfringens challenge, the gut bacterial structure of the chickens with C. butyricum supplementation (CB_B and CB_FM) was found to be distinct from that of the groups without C. butyricum (CK_B and CK_FM). After C. perfringens challenge, the samples from the CB/CP and CP groups were found to be distinct from those of the CB and CK groups. However, the microbial structure of the CB/CP group was not significantly different from that of the CP group (p < 0.05).
Structure and Composition of Intestinal Bacteria
At the order level, the predominant microorganisms in the chicken intestine were Lactobacillales and Clostridiales (Figure 6 and Supplementary Figure S2). Following high-fishmeal supplementation, Corynebacteriales decreased dramatically in the CB_FM group compared with in the CK_FM group (Supplementary Figure S2). After C. perfringens challenge, Clostridiales significantly increased in the CP group compared with in the CK group, whereas Bacillales decreased. Moreover, these microbiotic changes were significantly alleviated in the CB/CP group. In addition, LEfSe showed that high-fishmeal supplementation specifically increased the overrepresented bacteria, Clostridiales, in the CB_FM group (Figure 6), while similar results were also noted in the CB/CP group. These observations reveal that dietary C. butyricum increases the relative proportion of Clostridiales at different stages of NE.
FIGURE 6.
Differential enrichment of intestinal microbiota between groups measured using LEfSe (LDA score = 3). Bars represent significant abundance from phylum to genus between (A) the CB_B and CK_B groups, (B) the CB_FM and CK_FM groups, (C) and the CB/CP and CP groups. CB_B, CK_B, CB_FM and CK_FM: n = 12; CB/CP, CB, CP and CK: n = 6.
For further investigation of the composition of bacteria at different stages, the significance of differences in microbial species abundance was analyzed (Supplementary Table S3). C. butyricum increased significantly in the C. butyricum-supplemented groups, especially at the stage of high-fishmeal supplementation. Moreover, the abundance of Lactobacillus_sp._KC45b significantly increased and Candidatus Arthromitus and unclassified_g__Brachybacterium significantly decreased in the CB_B group compared with in the CK_B group. In the C. perfringens-challenged groups, CB/CP and CP, the abundance of Lactobacillus salivarius decreased significantly compared with in the CK group. Notably, the abundances of Weissella thailandensis, unclassified_g__Weissella, and Pediococcus acidilactici in the CB/CP group were higher than in the CK group. However, no significant differences between the CB/CP and CP groups or between the CB and CK groups were found using cutoff values of p < 0.05 and FDR < 0.1.
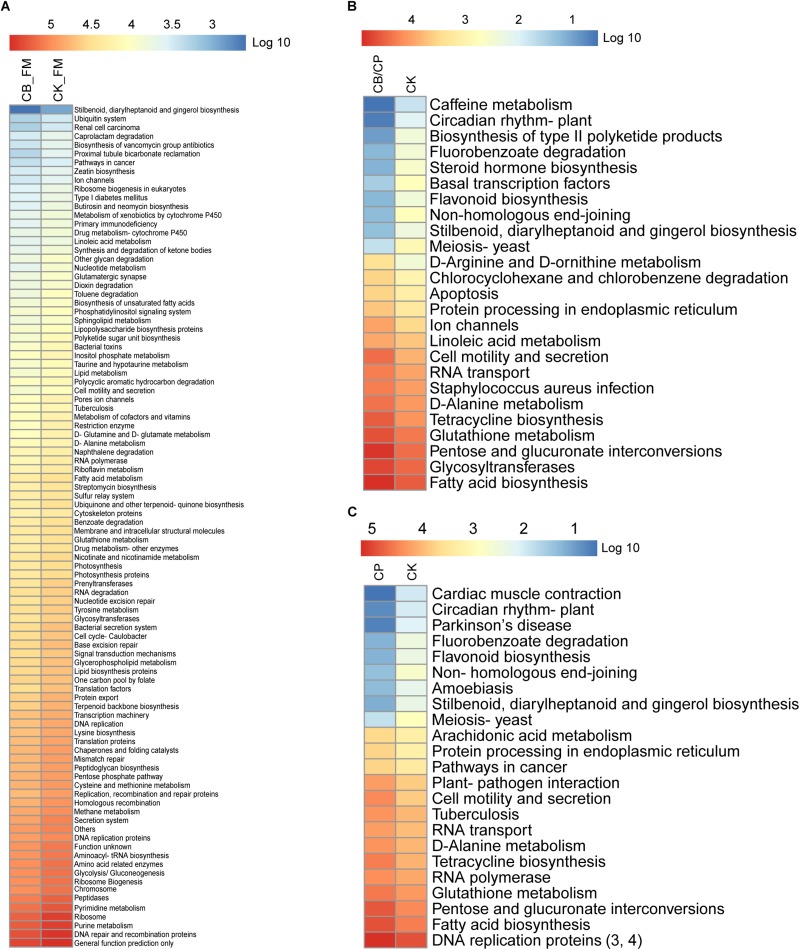
Predicting the Function of Intestinal Bacteria
The functions of the intestinal microbiota metagenome were predicted using PICRUSt from the Kyoto Encyclopedia of Genes and Genomes pathways (Figure 7). A total of 281 functions were detected in all of the groups. No significant differences were found between the CB_B and CK_B groups and the CB/CP and CP groups. However, there were significant differences (Mann-Whitney U test, p < 0.05, FDR < 0.1) between the CB_FM and CK_FM groups in 96 functions, comprising 34.16% of the total functions, including purine metabolism and pyrimidine metabolism. Moreover, there were 16 and 14 significant differences between the CB/CP and CK groups and the CP and CK groups, respectively, for example, fatty acid biosynthesis, pentose and glucuronate interconversions, and glutathione metabolism.
FIGURE 7.
Heatmap of predicted KEGG pathways of the gut bacteria in each group. The gene copy numbers of each group are shown as mean values. The significant differences between two groups at different stages were measured using the Mann-Whitney U test with a cutoff of p < 0.05 and FDR < 0.1. No significant difference was found between the CB_B and CK_B groups, the CB/CP and CP groups, and the CB and CK groups. Comparisons between the CB/CP and CB groups and the CB and CP groups were not performed. CB_B, CK_B, CB_FM, and CK_FM: n = 12; CB/CP, CB, CP, and CK: n = 6. Comparisons between CB_FM and CK_FM groups, CB/CP and CK groups and CP and CK groups are presented in panel (A), (B), and (C) respectively.
All raw sequences were submitted to the Sequence Read Archive database at NCBI (accession no. SRP159781).
Discussion
Previous reports have shown that NE is caused by factors including a fishmeal diet, Eimeria, and C. perfringens and is associated with a shift in gut microbiota structures and inflammation (Stanley et al., 2012, 2014). C. butyricum is a probiotic that has been proven to alleviate certain infections. Hence, we investigated the effect of C. butyricum on gut bacterial community composition and immune factors during the development of the NE caused by a high-fishmeal diet and C. perfringens infection.
The immune system plays a critical role in fighting infections. It has been reported that dietary C. butyricum can promote the expression of Claudin and Occludin to maintain intestinal barrier function, activate the expression of NF-κB, TLR2, and IL-10 and decrease pro-inflammatory cytokines, IL-1β, and TNF expression to modulate inflammatory response (Gao et al., 2012b; Hayashi et al., 2013; Zhang et al., 2016; Chen et al., 2018; Li H. et al., 2018; Liu et al., 2018). However, other studies have reported that C. butyricum has no influence on the immune system of neonatal mice and pigs (Miao et al., 2018; Peeters et al., 2019). In line with this finding, in this study, during the basal diet stage, no significant differences were found between C. butyricum supplemented and non-supplemented groups in the expression of every intestinal barrier function and inflammatory gene apart from IL-17A. A previous study reported that the composition of the diet is associated with the efficacy of C. butyricum. A C. butyricum supplemented diet improves the immune response and promotes intestinal barrier function with a low digestibility diet, whereas C. butyricum has no effect with a highly digestible diet (Chen et al., 2018). Based on these findings, we speculate that intestinal barrier function and inflammatory genes are probably not affected by C. butyricum in the findings of this study due to the type of diet.
Intestinal dysbiosis can elevate the production of pro-inflammatory cytokines, IL-1β, TNF, and NF-κB, and C. butyricum can decrease the expression of these factors in piglets, suggesting that C. butyricum can alleviate the inflammatory response in piglets (Chen et al., 2018; Cao et al., 2019; Wang et al., 2019). Zhang et al. reported that C. butyricum promotes the expression of TNF in chickens with E. coli K88 challenge (Zhang et al., 2016). Similar results were obtained in the current study, where C. butyricum was found to increase the expression of TNF in response to C. perfringens infection. A TNF-induced inflammatory response is common in chickens infected with pathogens such as Salmonella (Feng et al., 2016). Similarly, the NetB toxin of C. perfringens increases the TNF mRNA level in the intestinal lymphocytes of chickens (Jang et al., 2012). However, in this study, the expressions of TNF and TRL2 were not affected by C. perfringens challenge, which is consistent with the results of previous studies (Lu et al., 2009; Du et al., 2016). The high expression of IL-17A is associated with the initiation of several inflammatory diseases (Kolls and Linden, 2004). As previously mentioned, the results of this study demonstrate that C. perfringens challenge can upregulate the expression of IL-17 (Fasina and Lillehoj, 2019). Previous research has shown that a decrease in IL-17A expression can ameliorate inflammation (Kanai et al., 2015). Various mechanisms have been proposed to explain the anti-inflammatory effects of probiotics on intestinal inflammation, one of which is suppression of the expression of IL-17 (Tanabe, 2013). At the same time, it has been proven that the intake of Bacillus subtilis by mice with inflammatory bowel disease can downregulate the expression of Il-17 in order to decrease intestinal damage (Gong et al., 2016). We observed that C. butyricum could inhibit the upregulation of IL-17A expression induced by C. perfringens. Moreover, C. butyricum increases the expression of anti-inflammatory IL-10 mRNA, which plays a vital role in preventing many inflammatory diseases (Gao et al., 2012a). For instance, Bifidobacterium-induced IL-10 production benefits patients with colitis (Imaoka et al., 2008). In the same manner, we found that C. butyricum increases the expression of IL-10 mRNA levels in C. perfringens-challenged chickens, a situation that may indicate the prevention of NE.
IgA is the predominant intestinal immunoglobulin that contributes to intestinal barrier function (Pabst, 2012). Previous studies have reported that dietary C. butyricum supplementation can promote the expression of IgA in rabbits, broiler chickens, and mice (Murayama et al., 1995; Yang et al., 2012; Liu et al., 2018). However, we observed that dietary C. butyricum has no significant effect on the expression of IgA in the intestine. This is similar to the results of other studies, which demonstrate that C. butyricum supplementation does not affect the levels of IgA in neonatal mice, broilers, and laying hens (Liao et al., 2015; Miao et al., 2018; Zhan et al., 2019). Perdigon et al. (1995) show that Lactobacillus casei and Lactobacillus acidophilus improve the level of IgA in a dose-dependent manner. Therefore, we assume that the different effects of C. butyricum on the level of IgA may be related to the dose of C. butyricum.
Occludin and Claudin are tight junction proteins that play critical roles in regulating intestinal epithelial barrier function and preventing macromolecular transmission (Ballard et al., 1995; Fanning et al., 1998). We found that C. perfringens and a high-fishmeal diet decrease levels of Claudin-1 without influencing the expression of Occludin and Claudin-2, which is contrary to the report by Liu et al. (2012). However, C. butyricum supplementation was found to elevate the expression of Claudin-1 in C. perfringens-infected chickens. This finding suggests that C. butyricum may ameliorate damage to the intestinal barrier induced by C. perfringens.
In addition, we found that dietary C. butyricum could decrease the abundance of C. perfringens in the intestine of challenged chickens, consistent with previous reports that probiotics can eradicate pathogens (Gaggia et al., 2010; Yang et al., 2012). However, C. butyricum did not have any effect on intestinal lesion.
With regard to the alpha diversity of the gut microbiota, we observed that during the basal diet stage, C. butyricum had no significant effect on the ACE and Simpson indices, which is consistent with previous reports that the alpha diversity of intestinal bacteria is not affected by C. butyricum (Zhang et al., 2018). However, in the high-fishmeal-supplementation groups, C. butyricum supplementation was found to increase the Simpson indices, indicating that C. butyricum could reduce the diversity of intestinal bacteria. We speculate that high-fishmeal diets provide an opportunity for the proliferation of C. butyricum, which then competes with other bacteria for nutrients and niches and hence reduces the microbial diversity of the chicken intestine under a high-fishmeal diet. In addition, C. perfringens causes a decrease in the ACE index, which cannot be improved with C. butyricum supplementation.
The results of the PCoA indicate that in the uninfected groups, C. butyricum supplementation significantly alters the structure of intestinal microbiota, independent of a high-fishmeal diet, which is consistent with the results of a previous study (Kong et al., 2011). Nevertheless, no significant difference was found between the PCoA results of the C. butyricum supplemented and non-supplemented groups after infection. This indicates that C. butyricum is unable to alter the disruption of the gut bacterial structure caused by C. perfringens infection.
In the basal diet groups, dietary C. butyricum could decrease the abundance of harmful bacteria, such as Candidatus Arthromitus and unclassified_g__Brachybacterium, and increase the abundance of beneficial bacteria, such as Lactobacillus_sp._KC45b. Candidatus Arthromitus, one group of segmented filamentous bacteria (SFB), has been found in the intestinal bacteria of vertebrates such as chickens, rodents, and fish. It has been reported that the presence of SFB is correlated with diarrhea in poultry (Goodwin and Krabill, 1989; Angel et al., 1990; Morishita et al., 1992). Brachybacterium spp. were first isolated from poultry deep litter (Collins et al., 1988) and have previously been considered non-harmful bacteria. However, a recent study shows that Brachybacterium spp. are associated with blood infections in humans (Tamai et al., 2018). Lactobacillus spp. exerts a beneficial effect of resistance to pathogens such as Campylobacter (Stern et al., 2001) and Clostridium populations (Decroos et al., 2004). This result is consistent with previous studies that indicate that C. butyricum benefits the composition of intestinal bacteria (Yang et al., 2012). However, similar results were not found in the high-fishmeal-supplemented groups, indicating that although a high-fishmeal diet could facilitate the abundance of C. butyricum compared with that of basal diet groups, the structure and composition of intestinal bacteria in high-fishmeal-supplemented chickens is not better than that of basal diet chickens.
C. perfringens can disrupt the balance of chicken intestinal bacteria. For example, we observed that C. perfringens challenge decreases the abundance of the beneficial bacterium Lactobacillus salivarius, and this finding is consistent with those of previous studies (Lin et al., 2017; Huang et al., 2018). The results of the PCoA and species composition show that there are no significant differences between the intestinal microbiota structures in the CB/CP and CP groups, indicating that the beneficial effect of C. butyricum is less than the harmful effect of C. perfringens on the structure of the intestinal flora. However, compared with the CK group, the abundance of beneficial bacteria Weissella thailandensis, unclassified_g__Weissella, and Pediococcus acidilactici increased in the CB/CP group. Weissella thailandensis, a class of lactic acid bacteria (LAB), are most commonly used as probiotics and play key roles in disease resistance (Rolfe, 2000). Previous studies have shown that Pediococcus acidilactici can act as a potential probiotic as it produces lactic acid and bacteriocins against other enteric pathogens (Daeschel and Klaenhammer, 1985; Klaenhammer, 1993). These results suggest that C. butyricum has a certain beneficial effect on the intestinal microbiota of NE chickens.
It has been reported that the probiotics Lactobacillus acidophilus and Bacillus coagulans (Li Z. et al., 2018; Wu et al., 2018) significantly decrease the lesion score and have a significant beneficial influence on the intestinal bacteria of NE chickens. It has also been demonstrated that C. butyricum exerts preventive and therapeutic effects on bacterial infections such as enterohemorrhagic Escherichia coli, Clostridium difficile, and Salmonella enteritidis (Takahashi et al., 2004; Zhao et al., 2017; Oka et al., 2018). However, another study reported that C. butyricum could not prevent Salmonella typhimurium infection (Peeters et al., 2019). In line with this finding, we observed that C. butyricum has no significant effect on the lesion score and the structure of intestinal microbiota after C. perfringens challenge. The reason we speculate is that C. butyricum could not improve the gut microbiota disturbance induced by NE. In other words, the beneficial effect of C. butyricum on the structure of intestinal bacteria is not as strong as the harmful effect of NE. In addition, the dose of C. butyricum also has an effect. Different doses of C. butyricum have different influences on the expression of intestinal barrier function and inflammatory genes. High-dose C. butyricum can promote the expression of Claudin and Occludin, whereas low-dose C. butyricum decreases the mRNA levels of these genes (Liu et al., 2018). Symbiotic products and the time-point at which administration takes place also affect its efficacy against NE in chicken. In general, mixed products, such as probiotics and prebiotics, have a greater influence than individual components alone. To the best of our knowledge, probiotics, such as Bifidobacterium, Bacillus subtilis, Bacillus licheniformis, Lactobacillus salivarius, and Lactobacillus plantarum (Murry et al., 2004; Teo and Tan, 2005; Schoster et al., 2013; Lin et al., 2017), and prebiotics, such as chicory fructans, can inhibit C. perfringens growth (Kleessen et al., 2003). In addition, the time-point at which it is administered is also important for probiotic effectiveness. Nakphaichit et al. (2011) reported that dietary Lactobacillus reuteri supplementation during the first week of life has a beneficial effect on the composition of intestinal bacteria and that the effects last for up to 6 weeks. Therefore, it is difficult to predict the specific reason for the failure of the CB treatment based only on this single experimental trial. Therefore, further studies will be conducted to find a more reasonable scheme of administration of probiotics to prevent and treat NE in chickens.
In conclusion, C. butyricum can inhibit the increase of IL-17A gene expression and the decrease of Claudin-1 gene expression induced by C. perfringens challenge. Moreover, dietary C. butyricum promotes the expression of IL-10 in infected chickens. This finding suggests that C. butyricum has a beneficial effect on the immune response and intestinal barrier function of NE chickens. C. butyricum can significantly shift the structure and composition of intestinal bacteria during the stages of basal diet and high-fishmeal-supplementation diet. C. butyricum has a particularly large beneficial effect on the structure of intestinal bacteria during the basal diet stage. Meanwhile, C. butyricum decreases the abundance of C. perfringens in the gut. However, C. butyricum cannot significantly alter the structure of intestinal microbiota in C. perfringens-challenged chickens and does not have a significant effect on the occurrence of intestinal lesions induced by NE. Therefore, schemes that are more effective than C. butyricum supplementation need to be identified to prevent and treat NE in chickens.
Data Availability Statement
The datasets generated for this study can be found in Sequence Read Archive database, SRP159781.
Ethics Statement
This study was carried out in accordance with the principles of the Basel Declaration and recommendations of the Institutional Animal Care and Use Committee guidelines, Guangdong Academy of Agricultural Sciences. The protocol was approved by the Guangdong Academy of Agricultural Sciences (No. SYXK (yue)-20180108).
AUTHOR CONTRIBUTIONS
X-YP and Z-HX conceived the study and participated in its design and coordination. TH designed the experiments and drafted the manuscript. TH, BG, Q-LW, M-GY, and RX carried out the animal experiments. TH and BG carried out the amplicon sequencing and qPCR. TH participated in the statistical analyses. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This study was supported by the Guangzhou Science and Technology Program, China (Grant No. 201604020138), Special Fund for Agro-scientific Research in the Public Interest Ministry of Agriculture of the People’s Republic of China (Grant No. 201303040-16), Science and Technology Planning Project of Guangdong Province, China (Grant No. 2014A020208133), and Agricultural Science and Technology Innovation and Major Projects of Guangdong Province (2018), President Fund of Guangdong Academy of Agricultural Sciences (Grant No. 201801B).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02309/full#supplementary-material
Maximum-likelihood tree based on 16S rRNA gene sequences showing the phylogenetic relationships between strain C. butyricum YH 018 and closely related species of the genus Clostridium. The tree was constructed using the program MEGA 6 with bootstrap values based on 1000 replications. Filled circles mark nodes that were also present in respective maximum-parsimony and neighbor-joining trees. Scale bar, 0.01 nucleotide substitutions per position.
Shifts in gut bacterial composition in different groups at order level. ∗p < 0.05 measured using the Kruskal-Wallis test. CB_B, CK_B, CB_FM, and CK_FM: n = 12; CB/CP, CB, CP, and CK: n = 6.
Composition and nutritive levels of the diets (air-dry basis, %).
Primers used for qPCR.
Relative intestinal microbiota abundances (%) of significantly different species on three different stage of chicken with NE.
References
- Allaart J. G., Van Asten A. J., Grone A. (2013). Predisposing factors and prevention of Clostridium perfringens-associated enteritis. Comp. Immunol. Microbiol. Infect. Dis 36 449–464. 10.1016/j.cimid.2013.05.001 [DOI] [PubMed] [Google Scholar]
- Angel C. R., Sell J. L., Fagerland J. A., Reynolds D. L., Trampel D. W. (1990). Long-segmented filamentous organisms observed in poults experimentally infected with stunting syndrome agent. Avian. Dis. 34 994–1001. 10.2307/1591395 [DOI] [PubMed] [Google Scholar]
- Ballard S. T., Hunter J. H., Taylor A. E. (1995). Regulation of tight-junction permeability during nutrient absorption across the intestinal epithelium. Annu. Rev. Nutr. 15 35–55. 10.1146/annurev.nu.15.070195.000343 [DOI] [PubMed] [Google Scholar]
- Buffie C. G., Bucci V., Stein R. R., Mckenney P. T., Ling L., Gobourne A., et al. (2015). Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517 205–208. 10.1038/nature13828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G., Tao F., Hu Y., Li Z., Zhang Y., Deng B., et al. (2019). Positive effects of a Clostridium butyricum-based compound probiotic on growth performance, immune responses, intestinal morphology, hypothalamic neurotransmitters, and colonic microbiota in weaned piglets. Food Funct. 10 2926–2934. 10.1039/c8fo02370k [DOI] [PubMed] [Google Scholar]
- Chen L., Li S., Zheng J., Li W., Jiang X., Zhao X., et al. (2018). Effects of dietary Clostridium butyricum supplementation on growth performance, intestinal development, and immune response of weaned piglets challenged with lipopolysaccharide. J. Anim. Sci. Biotechnol. 9:62. 10.1186/s40104-018-0275-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins M. D., Brown J., Jones D. (1988). Brachybacterium faecium gen. nov., sp. nov., a Coryneform bacterium from poultry deep litter. Int. J. Syst. Bacteriol. 38 45–48. 10.1099/00207713-38-1-45 [DOI] [Google Scholar]
- Daeschel M. A., Klaenhammer T. R. (1985). Association of a 13.6-megadalton plasmid in Pediococcus pentosaceus with bacteriocin activity. Appl. Environ. Microbiol. 50 1538–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroos K., Vercauteren T., Werquin G., Verstraete W. (2004). Repression of Clostridium population in young broiler chickens after administration of a probiotic mixture. Commun. Agric. Appl. Biol. Sci. 69 5–13. [PubMed] [Google Scholar]
- Drew M. D., Syed N. A., Goldade B. G., Laarveld B., Van Kessel A. G. (2004). Effects of dietary protein source and level on intestinal populations of Clostridium perfringens in broiler chickens. Poult. Sci. 83 414–420. 10.1093/ps/83.3.414 [DOI] [PubMed] [Google Scholar]
- Du E., Wang W., Gan L., Li Z., Guo S., Guo Y. (2016). Effects of thymol and carvacrol supplementation on intestinal integrity and immune responses of broiler chickens challenged with Clostridium perfringens. J. Anim. Sci. Biotechnol. 7:19. 10.1186/s40104-016-0079-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning A. S., Jameson B. J., Jesaitis L. A., Anderson J. M. (1998). The tight junction protein ZO-1 establishes a link between the transmembrane protein occludin and the actin cytoskeleton. J. Biol. Chem. 273 29745–29753. 10.1074/jbc.273.45.29745 [DOI] [PubMed] [Google Scholar]
- Fasina Y. O., Lillehoj H. S. (2019). Characterization of intestinal immune response to Clostridium perfringens infection in broiler chickens. Poult. Sci. 98 188–198. 10.3382/ps/pey390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Wang L., Zhou L., Yang X., Zhao X. (2016). Using in vitro immunomodulatory properties of lactic acid bacteria for selection of probiotics against Salmonella infection in broiler chicks. PLoS One 11:e0147630. 10.1371/journal.pone.0147630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes da Costa S. P., Mot D., Bokori-Brown M., Savva C. G., Basak A. K., Van Immerseel F., et al. (2013). Protection against avian necrotic enteritis after immunisation with NetB genetic or formaldehyde toxoids. Vaccine 31 4003–4008. 10.1016/j.vaccine.2013.05.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaggia F., Mattarelli P., Biavati B. (2010). Probiotics and prebiotics in animal feeding for safe food production. Int. J. Food Microbiol. 141(Suppl. 1), S15–S28. 10.1016/j.ijfoodmicro.2010.02.031 [DOI] [PubMed] [Google Scholar]
- Gao Q., Qi L., Wu T., Wang J. (2012a). An important role of interleukin-10 in counteracting excessive immune response in HT-29 cells exposed to Clostridium butyricum. BMC Microbiol. 12:100. 10.1186/1471-2180-12-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Qi L., Wu T., Wang J. (2012b). Clostridium butyricum activates TLR2-mediated MyD88-independent signaling pathway in HT-29 cells. Mol. Cell. Biochem. 361 31–37. 10.1007/s11010-011-1084-y [DOI] [PubMed] [Google Scholar]
- Gong Y., Li H., Li Y. (2016). Effects of Bacillus subtilis on epithelial tight junctions of mice with inflammatory bowel disease. J. Interferon Cytokine Res. 36 75–85. 10.1089/jir.2015.0030 [DOI] [PubMed] [Google Scholar]
- Goodwin M. A., Krabill V. A. (1989). Diarrhea associated with small-intestinal cryptosporidiosis in a budgerigar and in a cockatiel. Avian. Dis. 33 829–833. 10.2307/1591170 [DOI] [PubMed] [Google Scholar]
- Hayashi A., Sato T., Kamada N., Mikami Y., Matsuoka K., Hisamatsu T., et al. (2013). A single strain of Clostridium butyricum induces intestinal IL-10-producing macrophages to suppress acute experimental colitis in mice. Cell Host Microbe 13 711–722. 10.1016/j.chom.2013.05.013 [DOI] [PubMed] [Google Scholar]
- Huang T., Gao B., Chen W. L., Xiang R., Yuan M. G., Xu Z. H., et al. (2018). Temporal effects of high fishmeal diet on gut microbiota and immune response in Clostridium perfringens-challenged chickens. Front. Microbiol. 9:2754. 10.3389/fmicb.2018.02754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaoka A., Shima T., Kato K., Mizuno S., Uehara T., Matsumoto S., et al. (2008). Anti-inflammatory activity of probiotic Bifidobacterium: enhancement of IL-10 production in peripheral blood mononuclear cells from ulcerative colitis patients and inhibition of IL-8 secretion in HT-29 cells. World J. Gastroenterol. 14 2511–2516. 10.3748/wjg.14.2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S. I., Lillehoj H. S., Lee S. H., Lee K. W., Lillehoj E. P., Hong Y. H., et al. (2012). Vaccination with Clostridium perfringens recombinant proteins in combination with Montanide ISA 71 VG adjuvant increases protection against experimental necrotic enteritis in commercial broiler chickens. Vaccine 30 5401–5406. 10.1016/j.vaccine.2012.06.007 [DOI] [PubMed] [Google Scholar]
- Kanai T., Mikami Y., Hayashi A. (2015). A breakthrough in probiotics: Clostridium butyricum regulates gut homeostasis and anti-inflammatory response in inflammatory bowel disease. J. Gastroenterol. 50 928–939. 10.1007/s00535-015-1084-x [DOI] [PubMed] [Google Scholar]
- Keyburn A. L., Sheedy S. A., Ford M. E., Williamson M. M., Awad M. M., Rood J. I., et al. (2006). Alpha-toxin of Clostridium perfringens is not an essential virulence factor in necrotic enteritis in chickens. Infect. Immun. 74 6496–6500. 10.1128/IAI.00806-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaenhammer T. R. (1993). Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol. Rev. 12 39–85. 10.1111/j.1574-6976.1993.tb00012.x [DOI] [PubMed] [Google Scholar]
- Kleessen B., Elsayed N. A., Loehren U., Schroedl W., Krueger M. (2003). Jerusalem artichokes stimulate growth of broiler chickens and protect them against endotoxins and potential cecal pathogens. J. Food Prot. 66 2171–2175. 10.4315/0362-028X-66.11.2171 [DOI] [PubMed] [Google Scholar]
- Kolls J. K., Linden A. (2004). Interleukin-17 family members and inflammation. Immunity 21 467–476. 10.1016/j.immuni.2004.08.018 [DOI] [PubMed] [Google Scholar]
- Kong Q., He G. Q., Jia J. L., Zhu Q. L., Ruan H. (2011). Oral administration of Clostridium butyricum for modulating gastrointestinal microflora in mice. Curr. Microbiol. 62 512–517. 10.1007/s00284-010-9737-8 [DOI] [PubMed] [Google Scholar]
- Li H., Gong Y., Xie Y., Sun Q., Li Y. (2018). Clostridium butyricum protects the epithelial barrier by maintaining tight junction protein expression and regulating microflora in a murine model of dextran sodium sulfate-induced colitis. Scand. J. Gastroenterol. 53 1031–1042. 10.1080/00365521.2016.1192678 [DOI] [PubMed] [Google Scholar]
- Li Z., Wang W., Liu D., Guo Y. (2018). Effects of Lactobacillus acidophilus on the growth performance and intestinal health of broilers challenged with Clostridium perfringens. J. Anim. Sci. Biotechnol. 9:25. 10.1186/s40104-018-0243-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., He J., He Z., Zhou Y., Yuan M., Xu X., et al. (2014). Phylogenetic and functional gene structure shifts of the oral microbiomes in periodontitis patients. ISME J. 8 1879–1891. 10.1038/ismej.2014.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X. D., Ma G., Cai J., Fu Y., Yan X. Y., Wei X. B., et al. (2015). Effects of Clostridium butyricum on growth performance, antioxidation, and immune function of broilers. Poult. Sci. 94 662–667. 10.3382/ps/pev038 [DOI] [PubMed] [Google Scholar]
- Lin Y., Xu S., Zeng D., Ni X., Zhou M., Zeng Y., et al. (2017). Disruption in the cecal microbiota of chickens challenged with Clostridium perfringens and other factors was alleviated by Bacillus licheniformis supplementation. PLoS One 12:e0182426. 10.1371/journal.pone.0182426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Guo S., Guo Y. (2012). Xylanase supplementation to a wheat-based diet alleviated the intestinal mucosal barrier impairment of broiler chickens challenged by Clostridium perfringens. Avian. Pathol. 41 291–298. 10.1080/03079457.2012.684089 [DOI] [PubMed] [Google Scholar]
- Liu L., Zeng D., Yang M., Wen B., Lai J., Zhou Y., et al. (2018). Probiotic Clostridium butyricum improves the growth performance, immune function, and gut microbiota of weaning rex rabbits. Probiotics Antimicrob Proteins 10.1007/s12602-018-9476-x [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Lu Y., Sarson A. J., Gong J., Zhou H., Zhu W., Kang Z., et al. (2009). Expression profiles of genes in Toll-like receptor-mediated signaling of broilers infected with Clostridium perfringens. Clin. Vaccine Immunol. 16 1639–1647. 10.1128/CVI.00254-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meimandipour A., Shuhaimi M., Soleimani A. F., Azhar K., Hair-Bejo M., Kabeir B. M., et al. (2010). Selected microbial groups and short-chain fatty acids profile in a simulated chicken cecum supplemented with two strains of Lactobacillus. Poult. Sci. 89 470–476. 10.3382/ps.2009-00495 [DOI] [PubMed] [Google Scholar]
- Miao R. X., Zhu X. X., Wan C. M., Wang Z. L., Wen Y., Li Y. Y. (2018). Effect of Clostridium butyricum supplementation on the development of intestinal flora and the immune system of neonatal mice. Exp. Ther. Med. 15 1081–1086. 10.3892/etm.2017.5461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita T. Y., Lam K. M., Mccapes R. H. (1992). Research note: isolation of two filamentous bacteria associated with enteritis in turkey poults. Poult. Sci. 71 203–207. 10.3382/ps.0710203 [DOI] [PubMed] [Google Scholar]
- Murayama T., Mita N., Tanaka M., Kitajo T., Asano T., Mizuochi K., et al. (1995). Effects of orally administered Clostridium butyricum MIYAIRI 588 on mucosal immunity in mice. Vet. Immunol. Immunopathol. 48 333–342. 10.1016/0165-2427(95)05437-B [DOI] [PubMed] [Google Scholar]
- Murry A. C., Jr., Hinton A., Jr., Morrison H. (2004). Inhibition of growth of Escherichia coli, Salmonella typhimurium, and Clostridia perfringens on chicken feed media by Lactobacillus salivarius and Lactobacillus plantarum. Int. J. Poult. Sci. 3 603–607. 10.3923/ijps.2004.603.607 [DOI] [Google Scholar]
- Nakphaichit M., Thanomwongwattana S., Phraephaisarn C., Sakamoto N., Keawsompong S., Nakayama J., et al. (2011). The effect of including Lactobacillus reuteri KUB-AC5 during post-hatch feeding on the growth and ileum microbiota of broiler chickens. Poult. Sci. 90 2753–2765. 10.3382/ps.2011-01637 [DOI] [PubMed] [Google Scholar]
- Oka K., Osaki T., Hanawa T., Kurata S., Sugiyama E., Takahashi M., et al. (2018). Establishment of an endogenous Clostridium difficile rat infection model and evaluation of the effects of Clostridium butyricum MIYAIRI 588 probiotic strain. Front. Microbiol. 9:1264. 10.3389/fmicb.2018.01264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olkowski A. A., Wojnarowicz C., Chirino-Trejo M., Drew M. D. (2006). Responses of broiler chickens orally challenged with Clostridium perfringens isolated from field cases of necrotic enteritis. Res. Vet. Sci. 81 99–108. 10.1016/j.rvsc.2005.10.006 [DOI] [PubMed] [Google Scholar]
- Pabst O. (2012). New concepts in the generation and functions of IgA. Nat. Rev. Immunol. 12 821–832. 10.1038/nri3322 [DOI] [PubMed] [Google Scholar]
- Peeters L., Mostin L., Wattiau P., Boyen F., Dewulf J., Maes D. (2019). Efficacy of Clostridium butyricum as probiotic feed additive against experimental Salmonella Typhimurium infection in pigs. Livest. Sci. 221 82–85. 10.1016/j.livsci.2018.12.019 [DOI] [Google Scholar]
- Perdigon G., Alvarez S., Rachid M., Aguero G., Gobbato N. (1995). Immune system stimulation by probiotics. J. Dairy Sci. 78 1597–1606. 10.3168/jds.S0022-0302(95)76784-4 [DOI] [PubMed] [Google Scholar]
- Rolfe R. D. (2000). The role of probiotic cultures in the control of gastrointestinal health. J. Nutr. 130 396S–402S. 10.1093/jn/130.2.396S [DOI] [PubMed] [Google Scholar]
- Schoster A., Kokotovic B., Permin A., Pedersen P. D., Dal Bello F., Guardabassi L. (2013). In vitro inhibition of Clostridium difficile and Clostridium perfringens by commercial probiotic strains. Anaerobe 20 36–41. 10.1016/j.anaerobe.2013.02.006 [DOI] [PubMed] [Google Scholar]
- Seki H., Shiohara M., Matsumura T., Miyagawa N., Tanaka M., Komiyama A., et al. (2003). Prevention of antibiotic-associated diarrhea in children by Clostridium butyricum MIYAIRI. Pediatr. Int. 45 86–90. 10.1046/j.1442-200X.2003.01671.x [DOI] [PubMed] [Google Scholar]
- Shojadoost B., Vince A. R., Prescott J. F. (2012). The successful experimental induction of necrotic enteritis in chickens by Clostridium perfringens: a critical review. Vet. Res. 43:74. 10.1186/1297-9716-43-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., Teal T. K., Marsh T. L., Tiedje J. M., Mosci R., Jernigan K., et al. (2015). Intestinal microbial communities associated with acute enteric infections and disease recovery. Microbiome 3:45. 10.1186/s40168-015-0109-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley D., Keyburn A. L., Denman S. E., Moore R. J. (2012). Changes in the caecal microflora of chickens following Clostridium perfringens challenge to induce necrotic enteritis. Vet. Microbiol. 159 155–162. 10.1016/j.vetmic.2012.03.032 [DOI] [PubMed] [Google Scholar]
- Stanley D., Wu S. B., Rodgers N., Swick R. A., Moore R. J. (2014). Differential responses of cecal microbiota to fishmeal, Eimeria and Clostridium perfringens in a necrotic enteritis challenge model in chickens. PLoS One 9:e104739. 10.1371/journal.pone.0104739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern N. J., Cox N. A., Bailey J. S., Berrang M. E., Musgrove M. T. (2001). Comparison of mucosal competitive exclusion and competitive exclusion treatment to reduce Salmonella and Campylobacter spp. colonization in broiler chickens. Poult. Sci. 80 156–160. 10.1093/ps/80.2.156 [DOI] [PubMed] [Google Scholar]
- Takahashi M., Taguchi H., Yamaguchi H., Osaki T., Komatsu A., Kamiya S. (2004). The effect of probiotic treatment with Clostridium butyricum on enterohemorrhagic Escherichia coli O157:H7 infection in mice. FEMS Immunol. Med. Microbiol. 41 219–226. 10.1016/j.femsim.2004.03.010 [DOI] [PubMed] [Google Scholar]
- Tamai K., Akashi Y., Yoshimoto Y., Yaguchi Y., Takeuchi Y., Shiigai M., et al. (2018). First case of a bloodstream infection caused by the genus Brachybacterium. J. Infect. Chemother. 24 998–1003. 10.1016/j.jiac.2018.06.005 [DOI] [PubMed] [Google Scholar]
- Tanabe S. (2013). The effect of probiotics and gut microbiota on Th17 cells. Int. Rev. Immunol. 32 511–525. 10.3109/08830185.2013.839665 [DOI] [PubMed] [Google Scholar]
- Teo A. Y., Tan H. M. (2005). Inhibition of Clostridium perfringens by a novel strain of Bacillus subtilis isolated from the gastrointestinal tracts of healthy chickens. Appl. Environ. Microbiol. 71 4185–4190. 10.1128/AEM.71.8.4185-4190.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade Ben K. A. (2015). The true cost of necrotic enteritis. World Poult. 31 16–17. [Google Scholar]
- Wang K. L., Chen G. Y., Cao G. T., Xu Y. L., Wang Y. X., Yang C. M. (2019). Effects of Clostridium butyricum and Enterococcus faecalis on growth performance, intestinal structure and inflammation in lipopolysaccharide-challenged weaned piglets. J. Anim. Sci. 10.1093/jas/skz235 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S. B., Rodgers N., Choct M. (2010). Optimized necrotic enteritis model producing clinical and subclinical infection of Clostridium perfringens in broiler chickens. Avian. Dis. 54 1058–1065. 10.1637/9338-032910-Reg.1 [DOI] [PubMed] [Google Scholar]
- Wu S. B., Rodgers N., Choct M. (2011). Real-time PCR assay for Clostridium perfringens in broiler chickens in a challenge model of necrotic enteritis. Appl. Environ. Microbiol. 77 1135–1139. 10.1128/AEM.01803-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Shao Y., Song B., Zhen W., Wang Z., Guo Y., et al. (2018). Effects of Bacillus coagulans supplementation on the growth performance and gut health of broiler chickens with Clostridium perfringens-induced necrotic enteritis. J. Anim. Sci. Biotechnol. 9:9. 10.1186/s40104-017-0220-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. M., Cao G. T., Ferket P. R., Liu T. T., Zhou L., Zhang L., et al. (2012). Effects of probiotic, Clostridium butyricum, on growth performance, immune function, and cecal microflora in broiler chickens. Poult. Sci. 91 2121–2129. 10.3382/ps.2011-02131 [DOI] [PubMed] [Google Scholar]
- Yasueda A., Mizushima T., Nezu R., Sumi R., Tanaka M., Nishimura J., et al. (2016). The effect of Clostridium butyricum MIYAIRI on the prevention of pouchitis and alteration of the microbiota profile in patients with ulcerative colitis. Surg. Today 46 939–949. 10.1007/s00595-015-1261-9 [DOI] [PubMed] [Google Scholar]
- Zhan H. Q., Dong X. Y., Li L. L., Zheng Y. X., Gong Y. J., Zou X. T. (2019). Effects of dietary supplementation with Clostridium butyricum on laying performance, egg quality, serum parameters, and cecal microflora of laying hens in the late phase of production. Poult. Sci. 98 896–903. 10.3382/ps/pey436 [DOI] [PubMed] [Google Scholar]
- Zhang B., Lv Z., Li H., Guo S., Liu D., Guo Y. (2017). Dietary l-arginine inhibits intestinal Clostridium perfringens colonisation and attenuates intestinal mucosal injury in broiler chickens. Br. J. Nutr. 118 321–332. 10.1017/S0007114517002094 [DOI] [PubMed] [Google Scholar]
- Zhang B., Yang X., Guo Y., Long F. (2011). Effects of dietary lipids and Clostridium butyricum on the performance and the digestive tract of broiler chickens. Arch. Anim. Nutr. 65 329–339. 10.1080/1745039X.2011.568274 [DOI] [PubMed] [Google Scholar]
- Zhang J., Chen X., Liu P., Zhao J., Sun J., Guan W., et al. (2018). Dietary Clostridium butyricum induces a phased shift in fecal microbiota structure and increases the acetic acid-producing bacteria in a weaned piglet model. J. Agric. Food Chem. 66 5157–5166. 10.1021/acs.jafc.8b01253 [DOI] [PubMed] [Google Scholar]
- Zhang L., Zhang L., Zhan X., Zeng X., Zhou L., Cao G., et al. (2016). Effects of dietary supplementation of probiotic, Clostridium butyricum, on growth performance, immune response, intestinal barrier function, and digestive enzyme activity in broiler chickens challenged with Escherichia coli K88. J Anim. Sci. Biotechnol. 7:3. 10.1186/s40104-016-0061-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Yang J., Wang L., Lin H., Sun S. (2017). Protection mechanism of Clostridium butyricum against Salmonella enteritidis infection in broilers. Front. Microbiol. 8:1523. 10.3389/fmicb.2017.01523 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Maximum-likelihood tree based on 16S rRNA gene sequences showing the phylogenetic relationships between strain C. butyricum YH 018 and closely related species of the genus Clostridium. The tree was constructed using the program MEGA 6 with bootstrap values based on 1000 replications. Filled circles mark nodes that were also present in respective maximum-parsimony and neighbor-joining trees. Scale bar, 0.01 nucleotide substitutions per position.
Shifts in gut bacterial composition in different groups at order level. ∗p < 0.05 measured using the Kruskal-Wallis test. CB_B, CK_B, CB_FM, and CK_FM: n = 12; CB/CP, CB, CP, and CK: n = 6.
Composition and nutritive levels of the diets (air-dry basis, %).
Primers used for qPCR.
Relative intestinal microbiota abundances (%) of significantly different species on three different stage of chicken with NE.
Data Availability Statement
The datasets generated for this study can be found in Sequence Read Archive database, SRP159781.