ABSTRACT
Attempts for bioremediation of toxic organohalogens resulted in the identification of organohalide-respiring bacteria harbouring reductive dehalogenases (RDases) enzymes. RDases consist of the catalytic subunit (RdhA, encoded by rdhA) that does not have membrane-integral domains, and a small putative membrane anchor (RdhB, encoded by rdhB) that (presumably) locates the A subunit to the outside of the cytoplasmic membrane. Recent genomic studies identified a putative rdh gene in an uncultured deltaproteobacterial genome that was not accompanied by an rdhB gene, but contained transmembrane helixes in N-terminus. Therefore, rather than having a separate membrane anchor protein, this putative RDase is likely a hybrid of RdhA and RdhB, and directly connected to the membrane with transmembrane helixes. However, functionality of the hybrid putative RDase remains unknown. Further analysis showed that the hybrid putative rdh genes are present in the genomes of pure cultures and uncultured members of Bacteriodetes and Deltaproteobacteria, but also in the genomes of the candidate divisions. The encoded hybrid putative RDases have cytoplasmic or exoplasmic C-terminus localization, and cluster phylogenetically separately from the existing RDase groups. With increasing availability of (meta)genomes, more diverse and likely novel rdh genes are expected, but questions regarding their functionality and ecological roles remain open.
Keywords: reductive dehalogenase, organohalide respiration, transmembrane helix
Recent genomic analysis revealed putative reductive dehalogenase genes from extreme subsurface environments that unlike known reductive dehalogenases have membrane integral domains.
INTRODUCTION
With the advent of the Industrial Revolution, human impacts on the environment increased dramatically. Hazardous halogenated organic compounds, organohalogens, were widely distributed in the natural environment through careless use and indiscriminate disposal, and caused major public concerns due to possible effects on human and environmental health (Häggblom 1992). In attempts for organohalogen bioremediation, a hallmark discovery was the identification of microbes that could use organohalogens as electron acceptors and reductively dehalogenate them (Suflita et al. 1982). This new metabolism, later termed organohalide respiration (OHR), has found great practical application in bioremediation. Accordingly, bioaugmentation with microbial consortia containing organohalide-respiring bacteria (OHRB) has become a showcase of successful engineered remediation of contaminated environments (Ellis et al. 2000; Stroo, Leeson and Ward 2012).
Over the past three decades, a wealth of knowledge has been obtained about the ecophysiology, biochemistry and environmental distribution of OHRB (Häggblom and Bossert 2003; Adrian and Löffler 2016). Using biochemical, PCR-based and (meta)genomic analysis, reductive dehalogenases (RDases) have been identified as the key enzymes of OHR (Lu et al. 2015; Hug 2016). The RDase-encoding genes (rdh) have a conserved operon structure that consists of rdhA, coding for the catalytic subunit (RdhA); rdhB, coding for a small putative membrane anchor (RdhB) that (presumably) locates the A subunit to the outside of the cytoplasmic membrane; and a variable set of accessory genes (e.g. rdhCTKZED) (Kruse, Smidt and Lechner 2016). The catalytic subunits (RdhAs) are characterized by two iron-sulfur clusters (FeS1: CXXCXXCXXXCP; FeS2: CXXCXXXCP) and an N-terminus twin-arginine translocation motif (TAT: RRXFXK) (Holliger, Wohlfarth and Diekert 1998). This signal peptide is necessary for secretion of the mature RdhA protein through the cell membrane to the outer side of the cytoplasmic membrane (Smidt and de Vos 2004).
A second type of rdhA genes were discovered that lacked TAT motif, were located in the cytoplasm, and lacked respiratory function. This group was termed as ‘catabolic’ reductive dehalogenase that are used to convert organohalogens to non-halogenated compounds to be used as carbon sources (Chen et al. 2013; Payne et al. 2015). These types of rdhA genes were mostly found in marine than terrestrial environments (Reviewed in Atashgahi, Häggblom and Smidt 2018a).
Putative rdh genes with N-terminus transmembrane helixes
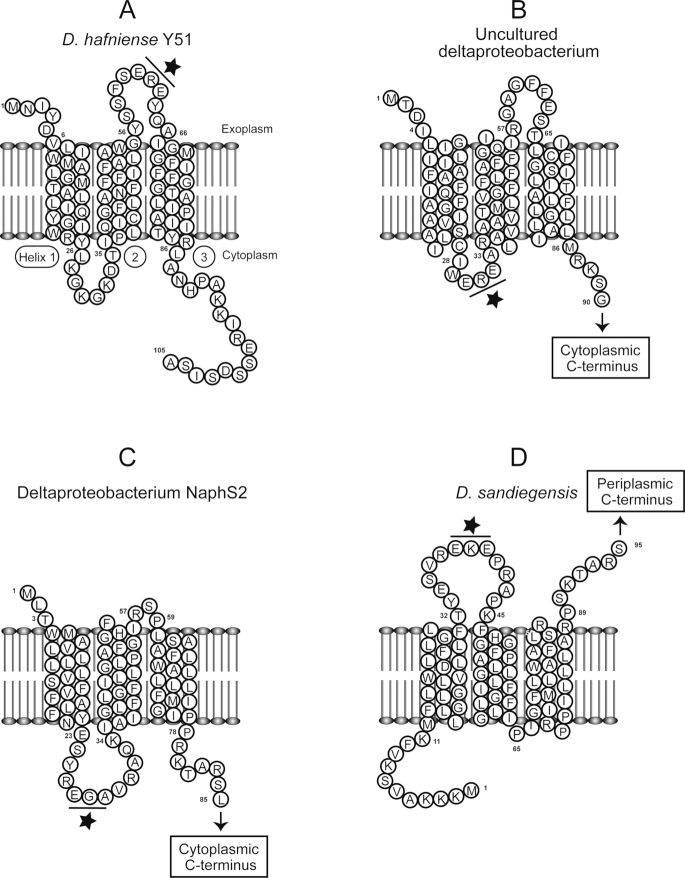
A recent single-cell genomic study from marine sediments in the Aarhus Bay discovered a third type of potential RDases in uncultured Desulfatiglans-related deltaproteobacterium (Jochum et al. 2018). A single-cell genome (SAG2) contained a putative rdh gene that is not accompanied by an rdhB, does not encode a TAT signal peptide, and as a unique feature, encodes three transmembrane helices (TMHs) in the N-terminus. Whereas the known respiratory RDases do not have membrane-integral domains, most RdhBs have three TMHs (Fig. 1). For instance, similar to the RdhB of Desulfitobacterium hafniense Y51 (Fig. 1A), the putative RDase from the uncultured Desulfatiglans-related deltaproteobacterium (Fig. 1B) has an exoplasmic N-terminus, followed by three TMHs. The remaining C-terminus contains the two binding motifs for FeS clusters, features of the known RDases. However, as the possible catalytic site, the C-terminus is facing the inner side of the cytoplasmic membrane (Fig. 1B) which is a likely localization in absence of the TAT signal peptide. The short cytoplasmic loop between helix 1 and 2 contains the two conserved glutamic acid residues (EXE motif) (Fig. 1B), proposed to play a role in the RdhA–RdhB interaction (Schubert et al. 2018). Similar cytoplasmic localization of the C-terminus of the putative RDase may enable such an interaction with this loop. Therefore, rather than having a separate membrane anchor protein, this putative RDase is predicted to act like a hybrid of RdhB and RdhA, and likely directly connected to the membrane with the TMHs.
Figure 1.
Predicted topology of the PceB protein of D. hafniense Y51 (A), and N-terminus TMHs of the hybrid putative RDases from uncultured deltaproteobacterium (SAG2) obtained from the Aarhus Bay (B), deltaproteobacterium strain NaphS2 (C), and D. sandiegensis(D). The position of the EXE motif is indicated by a star. Note that in panel B, C and D, only partial sequences of the hybrid putative RDases containing N-terminus TMHs were shown. TMHs were detected using TMHMM Server v. 2.0 (Sonnhammer, Von Heijne and Krogh 1998). Permission to reprint panel A was obtained from (Schubert et al. 2018).
The study of Jochum et al. further revealed that the hybrid putative rdh is similar to the putative rdh of two deltaproteobacterial pure cultures, i.e. deltaproteobacterium strain NaphS2 and Dethiosulfatarculus sandiegensis (Jochum et al. 2018). Indeed the putative rdh genes of these bacteria are not accompanied by an rdhB gene, lack TAT motif and contain three N-terminus TMHs. Similar to the putative RDase of the uncultured Desulfatiglans-related proteobacterium obtained from the Aarhus Bay (Fig. 1B), the putative RDase of the strain NaphS2 (Fig. 1C) has cytoplasmic C-terminus. In contrast, the putative RDase of D. sandiegensis has exoplasmic C-terminus (Fig. 1D), similar to the known RDases. The EXE motif in the loop between helix 1 and 2 is facing exoplasm, enabling potential interactions with the exoplasmic C-terminus (Fig. 1D). The three putative RDase share 46%–58% amino acid identity to each other, but share lower identity to the known RDases, e.g. 26%–29% identity to the TceA of Dehalococcoides mccartyi strain195 (DET0079). Although the existence of rdh genes lacking the TAT motif and rdhB were reported in the genomes of strain NaphS2 and D. sandiegensis (Sanford, Chowdhary and Löffler 2016; Liu and Häggblom 2018), the existence of TMHs in their putative RDase proteins were not reported. However, functionality of the hybrid putative RDases remains unknown.
The hybrid putative rdh genes are widespread
The sequence of the putative RDase of the uncultured proteobacterium obtained from the Aarhus Bay (Jochum et al. 2018) was used as a query in blastp searches against the NCBI non-redundant protein database in December 2018. The results showed that beyond the three identified proteobacterial hybrid putative rdh (Jochum et al. 2018), many other similar genes exist in the genomes of pure cultures as well as metagenome-assembled genomes (MAGs) that have gone unrecognized so far (Table 1). The majority of the sequences have three TMHs (detected using TMHMM Server v. 2.0 (Sonnhammer, Von Heijne and Krogh 1998)), the EXE motifs in their N-terminus, and either cytoplasmic or exoplasmic C-terminus containing the two FeS motifs (Table 1, Fig. 2). The C1–C5 regions from known the RDases are also conserved among the hybrid putative RDases (Fig. S1, Supporting Information), however, they are clustered phylogenetically separately from the existing RDase groups (Hug et al. 2013; Hug 2016) (Fig. S2, Supporting Information). Notably, the majority of the putative RDases are annotated as hypothetical proteins during automated annotation of the genomes.
Table 1.
List of the hybrid putative RDases with TMHs in their N-terminus. Sequence information and the predicted functions by the automated annotation for each sequence are included in Supporting Information.
| Organism | Length (aa) | TMH | C-terminus orientation | GenBank accession number | Sample source used for (meta)genome sequencing | Reference |
|---|---|---|---|---|---|---|
| Deltaproteobacteria bacterium | 482 | 3 | Cytoplasmic | - a | Marine sediment from Aarhus Bay | (Jochum et al. 2018) |
| Dethiosulfatarculus sandiegensis | 487 | 3 | Exoplasmic | WP_08 246 4279 | Pure deltaproteobacterial culture isolated from a methanogenic long-chain paraffins degrading consortium obtained from marine sediments | (Davidova et al. 2016) |
| Deltaproteobacterium NaphS2 | 478 | 3 | Cytoplasmic | EFK11122 | Pure deltaproteobacterial culture isolated from naphthalene-degrading enrichment obtained from marine sediments | (Galushko et al. 1999; Didonato Jr et al. 2010) |
| Marinifilaceae bacterium strain SPP2 | 459 | 3 | Exoplasmic | WP_09 642 9615 | Pure Marinilabiliales culture isolated from the Antarctic marine sediment | (Watanabe, Kojima and Fukui 2018) |
| Marinifilum fragile | 456 | 3 | Exoplasmic | WP_05 471 5848 | Pure Marinilabiliales culture isolated from tidal flat sediment in Korea | (Na et al. 2009) |
| Marinifilum breve | 457 | 3 | Cytoplasmic | WP_110 360 576 | Pure Marinilabiliales culture isolated from the Yongle Blue Hole in the South China Sea | (Fu et al. 2018) |
| Marinifilum flexuosum | 454 | 3 | Cytoplasmic | WP_120 240 634 | Pure Marinilabiliales culture isolated from coastal Mediterranean Sea water | (Ruvira et al. 2013) |
| Ancylomarina sp. M1P | 450 | 3 | Cytoplasmic | WP_125 029 802 | Pure Marinilabiliales culture isolated from Black Sea water | Unpublished |
| Labilibaculum filiforme | 454 | 3 | Exoplasmic | WP_101 260 201 | Pure Marinilabiliales culture isolated from the subsurface sediments of the Baltic Sea | (Vandieken et al. 2018) |
| Labilibacter marinus | 444 | 3 | Cytoplasmic | WP_06 663 2432 | Pure Marinilabiliales culture isolated from marine sediment at Weihai in China | (Liu et al. 2015; Lu et al. 2017) |
| Salinivirga cyanobacteriivorans | 453 | 3 | Cytoplasmic | WP_05 795 4221 | Pure Marinilabiliales culture isolated from the suboxic zone of a hypersaline cyanobacterial mat | (Ben Hania et al. 2017) |
| Caldithrix abyssi | 444 | 3 | Cytoplasmic | WP_0 069 30498 | Pure Calditrichales culture isolated from Mid-Atlantic Ridge hydrothermal vent | (Miroshnichenko et al. 2003; Kublanov et al. 2017) |
| Deltaproteobacteria bacterium | 491 | 3 | Exoplasmic | RLB29679 | Hydrothermal sediments | (Dombrowski, Teske and Baker 2018) |
| Deltaproteobacteria bacterium | 451 | 3 | Exoplasmic | RLB34449 | Hydrothermal sediments | (Dombrowski, Teske and Baker 2018) |
| Deltaproteobacteria bacterium | 455 | 3 | Exoplasmic | RLC06278 | Hydrothermal sediments | (Dombrowski, Teske and Baker 2018) |
| Deltaproteobacteria bacterium | 456 | 3 | Exoplasmic | RLB93792 | Hydrothermal sediments | (Dombrowski, Teske and Baker 2018) |
| Deltaproteobacteria bacterium | 455 | 3 | Exoplasmic | RLC22838 | Hydrothermal sediments | (Dombrowski, Teske and Baker 2018) |
| Deltaproteobacteria bacterium | 414 | 3 | Cytoplasmic | RLC21098 | Hydrothermal sediments | (Dombrowski, Teske and Baker 2018) |
| Deltaproteobacteria bacterium | 497 | 3 | Cytoplasmic | RLB22016 | Hydrothermal sediments | (Dombrowski, Teske and Baker 2018) |
| Deltaproteobacteria bacterium | 359 | 3 | Cytoplasmic | RLC02598 | Hydrothermal sediments | (Dombrowski, Teske and Baker 2018) |
| Desulfobacteraceae bacterium 4572_187 | 478 | 3 | Exoplasmic | OQY12990 | Hydrothermal sediment | (Dombrowski et al. 2017) |
| Desulfobacteraceae bacterium 4572_89 | 454 | 3 | Exoplasmic | OQY53460 | Hydrothermal sediments | (Dombrowski et al. 2017) |
| Bacteroidetes bacterium | 457 | 3 | Exoplasmic | RLD45891 | Hydrothermal sediments | (Dombrowski, Teske and Baker 2018) |
| Bacteroidetes bacterium | 447 | 3 | Exoplasmic | RLD65038 | Hydrothermal sediments | (Dombrowski, Teske and Baker 2018) |
| Bacteroidetes bacterium | 457 | 3 | Exoplasmic | RLD32997 | Hydrothermal sediments | (Dombrowski, Teske and Baker 2018) |
| Bacteroidetes bacterium | 402 | 1 | Exoplasmic | RLD55593 | Hydrothermal sediments | (Dombrowski, Teske and Baker 2018) |
| Bacteroidetes bacterium | 469 | 3 | Cytoplasmic | RLD42118 | Hydrothermal sediments | (Dombrowski, Teske and Baker 2018) |
| Bacteroidetes bacterium 4484_249 | 446 | 2 | Cytoplasmic | OQX80664 | Hydrothermal sediments | (Dombrowski et al. 2017) |
| Bacteroidetes bacterium | 476 | 4 | Cytoplasmic | RLD38167 | Hydrothermal sediments | (Dombrowski, Teske and Baker 2018) |
| Bacteroidetes bacterium | 454 | 3 | Cytoplasmic | RLD75418 | Hydrothermal sediments | (Dombrowski, Teske and Baker 2018) |
| Acidobacteria bacterium | 450 | 3 | Cytoplasmic | RLE20106 | Hydrothermal sediments | (Dombrowski, Teske and Baker 2018) |
| Chloroflexi bacterium | 453 | 3 | Exoplasmic | RLD03862 | Hydrothermal sediments | (Dombrowski, Teske and Baker 2018) |
| Chloroflexi bacterium | 457 | 3 | Exoplasmic | RLD00869 | Hydrothermal sediments | (Dombrowski, Teske and Baker 2018) |
| Chloroflexi bacterium | 453 | 3 | Cytoplasmic | RLD11393 | Hydrothermal sediments | (Dombrowski, Teske and Baker 2018) |
| Bacterium | 453 | 3 | Cytoplasmic | RKZ14043 | Hydrothermal sediments | (Dombrowski, Teske and Baker 2018) |
| Bacterium | 448 | 3 | Cytoplasmic | RKZ19839 | Hydrothermal sediments | (Dombrowski, Teske and Baker 2018) |
| Candidate division KSB1 bacterium | 457 | 3 | Exoplasmic | RKY76530 | Hydrothermal sediments | (Dombrowski, Teske and Baker 2018) |
| Candidate division KSB1 bacterium 4572_119 | 417 | 1 | Exoplasmic | OQX94610 | Hydrothermal sediments | (Dombrowski et al. 2017) |
| Candidate division KSB1 bacterium 4484_87 | 458 | 3 | Exoplasmic | OQX85480 | Hydrothermal sediments | (Dombrowski et al. 2017) |
| Candidate division Zixibacteria bacterium | 501 | 3 | Exoplasmic | RKX26209 | Hydrothermal sediments | (Dombrowski, Teske and Baker 2018) |
| Candidate division Zixibacteria bacterium | 461 | 3 | Cytoplasmic | RKX27199 | Hydrothermal sediments | (Dombrowski, Teske and Baker 2018) |
| Candidatus Aminicenantes bacterium 4484_214 | 511 | 3 | Cytoplasmic | OQX52307 | Hydrothermal sediments | (Dombrowski et al. 2017) |
| Candidatus Aminicenantes bacterium | 469 | 3 | Cytoplasmic | RLE02852 | Hydrothermal sediments | (Dombrowski, Teske and Baker 2018) |
| Candidatus Omnitrophica bacterium | 389 | 1 | Exoplasmic | RKY41132 | Hydrothermal sediments | (Dombrowski, Teske and Baker 2018) |
| Deltaproteobacteria bacterium | 463 | 3 | Exoplasmic | PLX41189 | Perchlorate-reducing communities | (Barnum et al. 2018) |
| Salinivirgaceae bacterium | 497 | 4 | Exoplasmic | PLX17815 | Perchlorate-reducing communities | (Barnum et al. 2018) |
| Marinilabiliales bacterium | 456 | 3 | Exoplasmic | PLW95329 | Perchlorate-reducing communities | (Barnum et al. 2018) |
| Marinilabiliales bacterium | 446 | 3 | Exoplasmic | PLW99613 | Perchlorate-reducing communities | (Barnum et al. 2018) |
| Marinilabiliales bacterium | 455 | 3 | Cytoplasmic | PLW92978 | Perchlorate-reducing communities | (Barnum et al. 2018) |
| Marinilabiliales bacterium | 454 | 3 | Cytoplasmic | PLX09622 | Perchlorate-reducing communities | (Barnum et al. 2018) |
| Marinilabiliales bacterium | 458 | 3 | Cytoplasmic | PLX19442 | Perchlorate-reducing communities | (Barnum et al. 2018) |
| Marinilabiliales bacterium | 452 | 3 | Cytoplasmic | PLX02242 | Perchlorate-reducing communities | (Barnum et al. 2018) |
| Bacteroidetes bacterium GWE2_32_14 | 432 | 2 | Exoplasmic | OFX85901 | Aquifers | (Anantharaman et al. 2016) |
| Bacteroidetes bacterium GWE2_40_15 | 462 | 3 | Exoplasmic | OFX81662 | Aquifers | (Anantharaman et al. 2016) |
| Candidatus Fischerbacteria bacterium RBG_13_37_8 | 447 | 3 | Cytoplasmic | OGF65237 | Aquifers | (Anantharaman et al. 2016) |
| Desulfobacterales bacterium RIFOXYA12_FULL_46_15 | 456 | 3 | Cytoplasmic | OGR28476 | Aquifers | (Anantharaman et al. 2016) |
| Desulfobacteraceae bacterium | 476 | 3 | Exoplasmic | RPI80002 | Wetlands | (Martins et al. 2018) |
| Deltaproteobacteria bacterium | 468 | 3 | Exoplasmic | RPJ06807 | Wetlands | (Martins et al. 2018) |
| Bacteroidales bacterium | 454 | 3 | Exoplasmic | RPH31952 | Wetlands | (Martins et al. 2018) |
| Bacterium SM23_31 | 446 | 3 | Cytoplasmic | KPK88368 | Estuary sediments | (Baker et al. 2015) |
| Candidate division Zixibacteria bacterium SM23_73_2 | 441 | 3 | Cytoplasmic | KPL04245 | Estuary sediments | (Baker et al. 2015) |
| Latescibacteria bacterium DG_63 | 453 | 3 | Cytoplasmic | KPJ61247 | Estuary sediments | (Baker et al. 2015) |
| Deltaproteobacteria bacterium HGW-Deltaproteobacteria-15 | 542 | 3 | Exoplasmic | PKN64391 | Deep terrestrial subsurface sediments | (Hernsdorf et al. 2017) |
| Candidate division Zixibacteria bacterium HGW-Zixibacteria-1 | 459 | 3 | Exoplasmic | PKK82132 | Deep terrestrial subsurface sediments | (Hernsdorf et al. 2017) |
| Desulfobacteraceae bacterium | 489 | 3 | Cytoplasmic | RJR39500 | Deep terrestrial subsurface fluids | (Momper et al. 2017) |
| Marinimicrobia bacterium 46_43 | 453 | 3 | Exoplasmic | KUK91590 | Oil Reservoirs | (Hu et al. 2016) |
| Candidatus Korarchaeota archaeon | 452 | 3 | Exoplasmic | PMB78244 | Hot springs | (Wilkins et al. 2018) |
| Desulfobacterales bacterium S5133MH16 | 488 | 3 | Exoplasmic | OEU64681 | Marine sediments | Unpublished |
| Candidate division KSB1 bacterium | 432 | 3 | Cytoplasmic | RQW00415 | - b | Unpublished |
Not available; sequence information provided in Supporting Information
Not available
Figure 2.
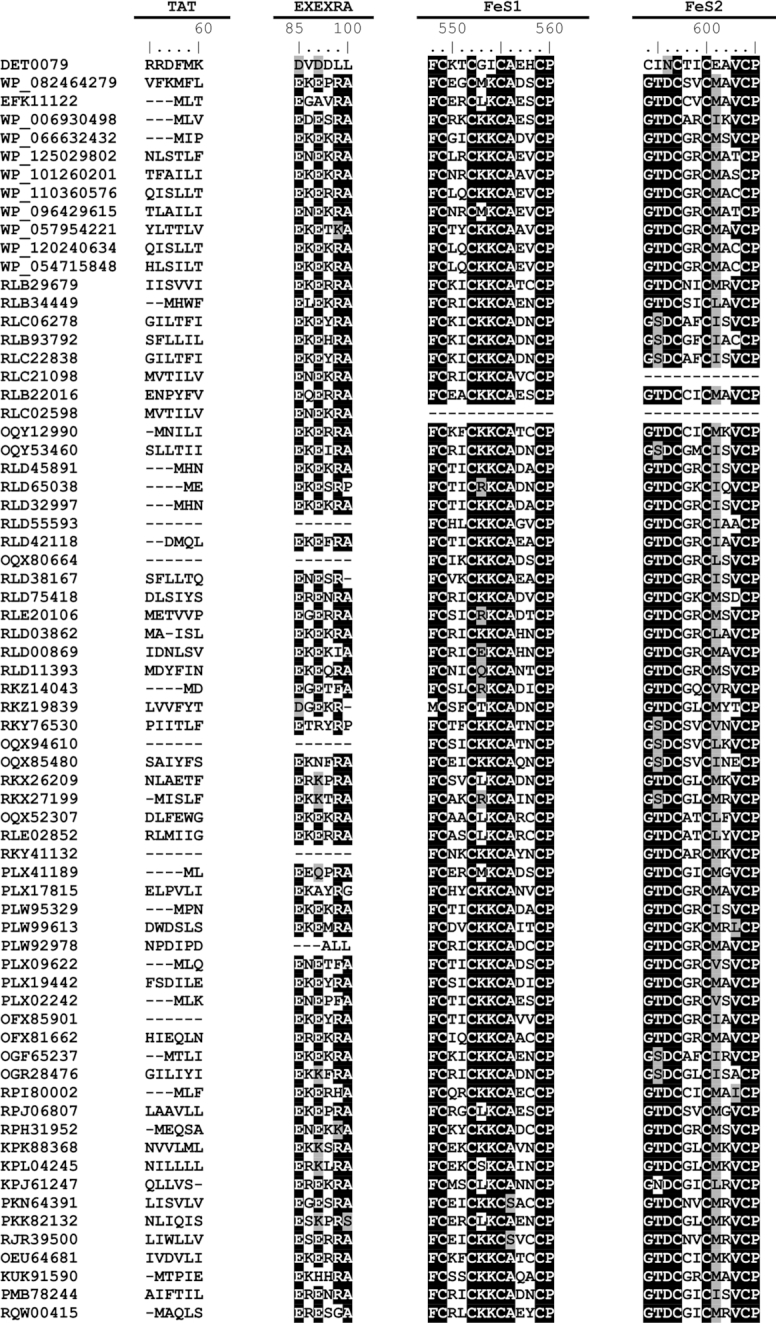
Sequence alignment of the hybrid putative RDases. Only conserved sequence motifs among experimentally characterized RDases (TAT, FeS1, FeS2), and the conserved glutamic acid residues (EXE) are included. The accession numbers are ordered according to Table 1, except the first accession number that belongs to TceA of Dehalococcoides mccartyi strain 195. ClustalW (Thompson, Higgins and Gibson 1994) multiple sequence alignment was conducted using BioEdit version 7.2.5 (http:/bioedit.software.informer.com/).
Of the 11 pure cultures containing hybrid putative rdh in their genomes, eight belong to the Marinilabiliales order within Bacteroidetes, that have been isolated from water or sediment samples in marine environment (Table 1). Among these, three strains belong to the genus Marinifilum, Gram-negative facultative anaerobes that can tolerate moderate salt concentrations (Na et al. 2009; Ruvira et al. 2013; Fu et al. 2018). Interestingly, hybrid putative rdh genes were also found in the MAGs of uncultured Marinilabiliales obtained from perchlorate-reducing enrichment cultures originating from marine sediments (Barnum et al. 2018). These genomes mostly lacked respiratory perchlorate, chlorate, oxygen and sulfur reductases and were proposed to be specialized for the fermentation of dead cells (Barnum et al. 2018). These finding indicate an important role of the hybrid putative rdh genes in Marinilabiliales members. Another pure culture harbouring the hybrid putative rdh in its genome is Caldithrix abyssi, a thermophilic anaerobic bacterium isolated from a Mid-Atlantic Ridge hydrothermal vent (Miroshnichenko et al. 2003). Calditrichaeota are abundant seabed microbes with genomic potential to degrade detrital proteins through the use of extracellular peptidases (Marshall et al. 2017).
Except the MAGs obtained from the marine perchlorate-reducing enrichment cultures (Barnum et al. 2018), all other MAGs-containing hybrid putative rdh were obtained from harsh environments such as hydrothermal vents (Dombrowski et al. 2017; Dombrowski, Teske and Baker 2018), hot springs (Wilkins et al. 2018), wetlands with extremely high concentrations of dissolved organic carbon and diverse sulfur species (Martins et al. 2018), deep terrestrial environments (Hernsdorf et al. 2017; Momper et al. 2017), etc (Table 1). Most of the sequences from the MAGs were obtained from hydrothermal vent sediments in Guaymas Basin (Gulf of California) with fluctuating temperature and chemical gradients (Dombrowski et al. 2017; Dombrowski, Teske and Baker 2018). The MAGs are mostly from uncultured Bacteriodetes and Deltaproteobacteria, but also from the candidate divisions (Table 1). Members of all these phyla have known/proposed diverse metabolic potential, and may not be restricted to reductive dehalogenation. However, physiological proofs for OHR have only been obtained for deltaproteobacterial members with classic rdh gene operon i.e. rdhA, rdhB and one or more transcriptional regulatory genes (Sanford, Chowdhary and Löffler 2016; Liu and Häggblom 2018).
Outstanding questions
Genomics and allied technologies have greatly increased the diversity of putative rdh genes in recent years, and extended their distribution from contaminated environments to deep subsurface (Table 1), Antarctic soils (Zlamal et al. 2017), and even human and animal intestinal tract (Atashgahi et al. 2018b). With the expanding availability of the bacterial genomes and increasing application of deep sequencing in diverse environments, much more diverse and likely novel rdh genes are expected in future. This brings forward major open questions:
Do the newly discovered genes encode RDases? If they indeed encode RDases, what are their functions? Three roles have been shown for the known RDases: energy conservation by OHR, and facilitated fermentation of organic substrates (e.g. pyruvate, lactate or yeast extract) by reoxidation of respiratory cofactors for membrane-bound RDases, and catabolic reductive dehalogenation for cytoplasmic RDases (Fincker and Spormann 2017). Can the hybrid putative RDases with cytoplasmic C-terminus be involved in catabolic reductive dehalogenation, facilitated fermentation or both? In turn, how are the hybrid putative RDases with exoplasmic C-terminus secreted through the cell membrane in absence of TAT signal peptide?
If indeed involved in reductive dehalogenation, what are the physiological organohalogen substrates of the hybrid putative RDases? The lack of correlation between the rdh sequences and their organohalogen substrates has precluded the ability to predict substrates for novel genes, and to test their functionality using the predicted organohalogens.
Why the majority of the environmental hybrid putative rdh sequences and rdh-containing pure cultures have been obtained from harsh environments? Can it be that their physiological organohalogen substrates are found in these environments?
What are the ecological functions of the microbes containing (the hybrid putative) RDases? Detoxification of organohalogens and thereby securing a hospitable environments for themselves and the nearby organisms? Providing carbon sources for themselves (catabolic RDase) or others (respiratory RDase)?
Can (the hybrid putative) RDases be involved in the production of halogenated bioactive compounds as was shown for biosynthesis of marine bacterial pyrroles mediated by a reductive debrominase that utilizes a redox thiol mechanism (El Gamal et al. 2016)? Likewise, can the RDases participate in in the production of halogenated bioactive compounds in Eukaryotes such as sponges that are known to harbour Deltaproteobacteria with rdh genes (Wilson et al. 2014; Liu et al. 2017)?
ACKNOWLEDGEMENTS
I thank Kasper U. Kjeldsen for providing sequence information, Torsten Schubert and Peng Peng for assistance in creating figures, and two anonymous reviewers for their constructive comments.
FUNDING
This work was supported by the SIAM Gravitation grant ‘Microbes for Health and the Environment’ (Project 024.002.002) of the Netherlands Ministry of Education, Culture and Science, and the Netherlands Science Foundation (NWO).
Conflict of interest. None declared.
REFERENCES
- Adrian L, Löffler FE. Organohalide-Respiring Bacteria. Springer-Verlag, Berlin, Germany; 2016. [Google Scholar]
- Anantharaman K, Brown CT, Hug LA et al.. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat Com. 2016;7:13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atashgahi S, Häggblom MM, Smidt H. Organohalide respiration in pristine environments: implications for the natural halogen cycle. Environ Microbiol. 2018. a;20:934–48. [DOI] [PubMed] [Google Scholar]
- Atashgahi S, Shetty SA, Smidt H et al.. Flux, impact and fate of halogenated xenobiotic compounds in the gut. Front Physiol. 2018b;9:888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BJ, Lazar CS, Teske AP et al.. Genomic resolution of linkages in carbon, nitrogen, and sulfur cycling among widespread estuary sediment bacteria. Microbiome. 2015;3:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnum TP, Figueroa IA, Carlström CI et al.. Genome-resolved metagenomics identifies genetic mobility, metabolic interactions, and unexpected diversity in perchlorate-reducing communities. ISME J. 2018;12:1568–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Hania W, Joseph M, Bunk B et al.. Characterization of the first cultured representative of a Bacteroidetes clade specialized on the scavenging of cyanobacteria. Environ Microbiol. 2017;19:1134–48. [DOI] [PubMed] [Google Scholar]
- Chen K, Huang L, Xu C et al.. Molecular characterization of the enzymes involved in the degradation of a brominated aromatic herbicide. Mol Microbiol. 2013;89:1121–39. [DOI] [PubMed] [Google Scholar]
- Davidova IA, Wawrik B, Callaghan AV et al.. Dethiosulfatarculus sandiegensis gen. nov., sp. nov., isolated from a methanogenic paraffin-degrading enrichment culture and emended description of the family Desulfarculaceae. Int J Syst Evol Microbiol. 2016;66:1242–8. [DOI] [PubMed] [Google Scholar]
- Didonato Jr RJ, Young ND, Butler JE et al.. Genome sequence of the deltaproteobacterial strain NaphS2 and analysis of differential gene expression during anaerobic growth on naphthalene. PLoS One. 2010;5:e14072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrowski N, Seitz KW, Teske AP et al.. Genomic insights into potential interdependencies in microbial hydrocarbon and nutrient cycling in hydrothermal sediments. Microbiome. 2017;5:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrowski N, Teske AP, Baker BJ. Expansive microbial metabolic versatility and biodiversity in dynamic Guaymas Basin hydrothermal sediments. Nat Com. 2018;9:4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Gamal A, Agarwal V, Rahman I et al.. Enzymatic reductive dehalogenation controls the biosynthesis of marine bacterial pyrroles. J Am Chem Soc. 2016;138:13167–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis DE, Lutz EJ, Odom JM et al.. Bioaugmentation for accelerated in situ anaerobic bioremediation. Environ Sci Technol. 2000;34:2254–60. [Google Scholar]
- Fincker M, Spormann AM. Biochemistry of catabolic reductive dehalogenation. Annu Rev Biochem. 2017;86:357–86. [DOI] [PubMed] [Google Scholar]
- Fu T, Jia C, Fu L et al.. Marinifilum breve sp. nov., a marine bacterium isolated from the Yongle Blue Hole in the South China Sea and emended description of the genus Marinifilum. Int J Syst Evol Microbiol. 2018;68:3540–5. [DOI] [PubMed] [Google Scholar]
- Galushko A, Minz D, Schink B et al.. Anaerobic degradation of naphthalene by a pure culture of a novel type of marine sulphate-reducing bacterium. Environ Microbiol. 1999;1:415–20. [DOI] [PubMed] [Google Scholar]
- Hernsdorf AW, Amano Y, Miyakawa K et al.. Potential for microbial H2 and metal transformations associated with novel bacteria and archaea in deep terrestrial subsurface sediments. ISME J. 2017;11:1915–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliger C, Wohlfarth G, Diekert G. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol Rev. 1998;22:383–98. [Google Scholar]
- Hug LA, Maphosa F, Leys D et al.. Overview of organohalide-respiring bacteria and a proposal for a classification system for reductive dehalogenases. Philos Trans R Soc B Biol Sci. 2013;368:20120322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug LA. Diversity, evolution, and environmental distribution of reductive dehalogenase genes. In: Adrian Land Löffler F(eds). Organohalide-Respiring Bacteria. Springer-Verlag, Berlin, Germany. 2016, 377–93. [Google Scholar]
- Hu P, Tom L, Singh A et al.. Genome-resolved metagenomic analysis reveals roles for candidate phyla and other microbial community members in biogeochemical transformations in oil reservoirs. MBio. 2016;7:e01669–01615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häggblom MM, Bossert ID. Dehalogenation: Microbial Processes and Environmental Applications. Kluwer Academic Publisher Group, Boston, USA; 2003. [Google Scholar]
- Häggblom MM. Microbial breakdown of halogenated aromatic pesticides and related compounds. FEMS Microbiol Rev. 1992;103:29–71. [DOI] [PubMed] [Google Scholar]
- Jochum LM, Schreiber L, Marshall IP et al.. Single-cell genomics reveals a diverse metabolic potential of uncultivated Desulfatiglans-related Deltaproteobacteria widely distributed in marine sediment. Front Microbiol. 2018;9:2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse T, Smidt H, Lechner U. Comparative genomics and transcriptomics of organohalide-respiring bacteria and regulation of rdh gene transcription. In: Adrian Land Löffler F(eds). Organohalide-Respiring Bacteria. Springer-Verlag, Berlin, Germany; 2016,345–76. [Google Scholar]
- Kublanov IV, Sigalova OM, Gavrilov SN et al.. Genomic analysis of Caldithrix abyssi, the thermophilic anaerobic bacterium of the novel bacterial phylum Calditrichaeota. Front Microbiol. 2017;8:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Häggblom MM. Genome-guided identification of organohalide-respiring Deltaproteobacteria from the marine environment. mBio. 2018;9:e02471–02418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lopez N, Ahn YB et al.. Novel reductive dehalogenases from the Mmrine sponge associated bacterium Desulfoluna spongiiphila. Env Microbiol Rep. 2017;9:537–49. [DOI] [PubMed] [Google Scholar]
- Liu Q-Q, Li J, Xiao D et al.. Saccharicrinis marinus sp. nov., isolated from marine sediment. Int J Syst Evol Microbiol. 2015;65:3427–32. [DOI] [PubMed] [Google Scholar]
- Lu D-C, Zhao J-X, Wang F-Q et al.. Labilibacter aurantiacus gen. nov., sp. nov., isolated from sea squirt (Styela clava) and reclassification of Saccharicrinis marinus as Labilibacter marinus comb. nov. Int J Syst Evol Microbiol. 2017;67:441–6. [DOI] [PubMed] [Google Scholar]
- Lu Y, Atashgahi S, Hug LA et al.. Primers that target functional genes of organohalide-respiring bacteria. In: McGenity TJandTimmis KN B N(eds). Hydrocarbon and Lipid Microbiology Protocols, Springer Protocols Handbooks. Springer, Berlin, Germany, 2015, 177–205. [Google Scholar]
- Marshall IP, Starnawski P, Cupit C et al.. The novel bacterial phylum Calditrichaeota is diverse, widespread and abundant in marine sediments and has the capacity to degrade detrital proteins. Env Microbiol Rep. 2017;9:397–403. [DOI] [PubMed] [Google Scholar]
- Martins PD, Danczak RE, Roux S et al.. Viral and metabolic controls on high rates of microbial sulfur and carbon cycling in wetland ecosystems. Microbiome. 2018;6:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miroshnichenko ML, Kostrikina NA, Chernyh NA et al.. Caldithrix abyssi gen. nov., sp. nov., a nitrate-reducing, thermophilic, anaerobic bacterium isolated from a Mid-Atlantic Ridge hydrothermal vent, represents a novel bacterial lineage. Int J Syst Evol Microbiol. 2003;53:323–9. [DOI] [PubMed] [Google Scholar]
- Momper L, Jungbluth SP, Lee MD et al.. Energy and carbon metabolisms in a deep terrestrial subsurface fluid microbial community. ISME J. 2017;11:2319–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na H, Kim S, Moon EY et al.. Marinifilum fragile gen. nov., sp. nov., isolated from tidal flat sediment. Int J Syst Evol Microbiol. 2009;59:2241–6. [DOI] [PubMed] [Google Scholar]
- Payne KA, Quezada CP, Fisher K et al.. Reductive dehalogenase structure suggests a mechanism for B12-dependent dehalogenation. Nature. 2015;517:513–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvira MA, Lucena T, Pujalte MJ et al.. Marinifilum flexuosum sp. nov., a new Bacteroidetes isolated from coastal Mediterranean Sea water and emended description of the genus Marinifilum Na et al., 2009. Syst Appl Microbiol. 2013;36:155–9. [DOI] [PubMed] [Google Scholar]
- Sanford RA, Chowdhary J, Löffler FE. Organohalide-respiring deltaproteobacteria. In:Adrian Land Löffler FE(eds). Organohalide-Respiring Bacteria. Springer-Verlag, Berlin, Germany; 2016, 235–258. [Google Scholar]
- Schubert T, Adrian L, Sawers RG et al.. Organohalide respiratory chains: composition, topology and key enzymes. FEMS Microbiol Ecol. 2018;94:fiy035. [DOI] [PubMed] [Google Scholar]
- Smidt H, de Vos WM. Anaerobic microbial dehalogenation. Annu Rev Microbiol. 2004;58:43–73. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, Von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–82. [PubMed] [Google Scholar]
- Stroo HF, Leeson A, Ward CH. Bioaugmentation for Groundwater Remediation. Springer, New York, USA. 2012. [Google Scholar]
- Suflita JM, Horowitz A, Shelton DR et al.. Dehalogenation: a novel pathway for the anaerobic biodegradation of haloaromatic compounds. Science. 1982;218:1115–7. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandieken V, Marshall IP, Niemann H et al.. Labilibaculum manganireducens gen. nov., sp. nov. and Labilibaculum filiforme sp. nov., novel Bacteroidetes isolated from subsurface sediments of the Baltic Sea. Front Microbiol. 2018;8:2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Kojima H, Fukui M. Complete genome sequence of Marinifilaceae bacterium strain SPP2, isolated from the Antarctic marine sediment. Mar Genom. 2018;39:1–2. [Google Scholar]
- Wilkins L, Ettinger C, Jospin G et al.. There and back again: metagenome-assembled genomes provide new insights into two thermal pools in Kamchatka, Russia. bioRxiv. 2018:392308. [Google Scholar]
- Wilson MC, Mori T, Rückert C et al.. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature. 2014;506:58–62. [DOI] [PubMed] [Google Scholar]
- Zlamal JE, Raab TK, Little M et al.. Biological chlorine cycling in the Arctic Coastal Plain. Biogeochemistry. 2017;134:243–60. [Google Scholar]