Abstract
Purpose
Oestrogen receptor-positive (ER+) and human epidermal receptor 2-negative (HER2–) breast cancers are classified as Luminal A or B based on gene expression, but immunohistochemical markers are used for surrogate subtyping. The aims of this study were to examine the agreement between molecular subtyping (MS) and surrogate subtyping and to identify subgroups consisting mainly of Luminal A or B tumours.
Methods
The cohort consisted of 2063 patients diagnosed between 2013–2017, with primary ER+/HER2– breast cancer, analysed by RNA sequencing. Surrogate subtyping was performed according to three algorithms (St. Gallen 2013, Maisonneuve and our proposed Grade-based classification). Agreement (%) and kappa statistics (κ) were used as concordance measures and ROC analysis for luminal distinction. Ki67, progesterone receptor (PR) and histological grade (HG) were further investigated as surrogate markers.
Results
The agreement rates between the MS and St. Gallen 2013, Maisonneuve and Grade-based classifications were 62% (κ = 0.30), 66% (κ = 0.35) and 70% (κ = 0.41), respectively. PR did not contribute to distinguishing Luminal A from B tumours (auROC = 0.56). By classifying HG1-2 tumours as Luminal A-like and HG3 as Luminal B-like, agreement with MS was 80% (κ = 0.46). Moreover, by combining HG and Ki67 status, a large subgroup of patients (51% of the cohort) having > 90% Luminal A tumours could be identified.
Conclusions
Agreement between MS and surrogate classifications was generally poor. However, a post hoc analysis showed that a combination of HG and Ki67 could identify patients very likely to have Luminal A tumours according to MS.
Keywords: Breast cancer, Intrinsic subtype, Molecular subtyping, Surrogate marker, Gene expression
Background
Almost 20 years have passed since the breast cancer molecular intrinsic subtypes, based on patterns of gene expression, were first presented [1, 2]. The majority of oestrogen receptor (ER)-positive and human epidermal receptor 2 (HER2)-negative (ER+/HER2–) breast cancers are classified as Luminal A or Luminal B. An important difference between these groups is that patients with Luminal B tumours have a higher risk of relapse [2] and are therefore often recommended adjuvant chemotherapy in addition to endocrine therapy [3]. During the last decade, several commercial multigene assays have been introduced as tools for estimating the risk of recurrence (ROR) and for selecting patients for whom adjuvant chemotherapy can be omitted [4–8]. One of these tests, PAM50/Prosigna®, provides information on both intrinsic subtype and a ROR score, based on gene expression and tumour size [4, 9].
Surrogate subgroups based on hormone receptor expression, proliferation and HER2 status were introduced a decade ago as a tool for risk stratification and guidance of adjuvant therapy [10]. In the St. Gallen 2013 surrogate subtype classification, a combination of the routine pathological markers ER, PR and HER2 and the proliferation marker Ki67 is used to classify tumours into the intrinsic subtypes. Ki67 (high/low) and PR (high/low) are used to separate Luminal A-like from Luminal B-like tumours [11]. The following year, Maisonneuve and colleagues proposed a new surrogate definition for the luminal intrinsic subtypes by introducing an intermediate Ki67 group [12]. They also showed that PR was a discriminator for prognosis only in tumours with intermediate Ki67. In both St. Gallen 2013 and Maisonneuve classifications, the cut-off for high PR was set at ≥ 20%. The classification proposed by Maisonneuve showed improved stratification compared with St. Gallen 2013 in terms of long-term outcome (distant disease-free survival). Furthermore, in their large cohort of almost 10 000 breast cancer patients, they found that patients with poorly differentiated [histological grade (HG) 3] Luminal A-like tumours had a prognosis similar to that of patients with Luminal B-like tumours, whereas patients with well-differentiated (HG1) Luminal B-like tumours had a prognosis similar to that of Luminal A-like tumours. Similar results were obtained in a study (n = 671) by our research group, where HG was shown to add prognostic information in terms of distant disease-free survival to the St. Gallen 2013 classification [13]. In the St. Gallen consensus statement 2017, the surrogate classification was not described in detail; Luminal A-like tumours are defined as having high ER/PR, clearly low Ki67, HG1, whereas Luminal B-like tumours have lower ER/PR, clearly high Ki67, HG3. There is no clear definition for the intermediate group and the use of molecular assays for these tumours has been suggested as a tool for improved risk stratification [3].
By using molecular assays, more patients can be spared adjuvant chemotherapy, but since these tests are associated with significant costs, the routinely used pathological morphology and immunohistochemical (IHC) markers still form the basis for the adjuvant treatment decision for most patients. In an ongoing Swedish population-based observational study, SCAN-B (Sweden Cancerome Analysis Network—Breast) [14, 15], primary breast cancer tissue samples are classified into molecular intrinsic subtypes by RNA sequencing, using the PAM50 genes and an implementation of the nearest-centroid method.
We hypothesised that the concordance of the molecular subtypes (Luminal A and Luminal B) to the clinicopathological surrogate subtypes (Luminal A-like and Luminal B-like) could be improved by prioritising HG when subtyping tumours and only including the other markers in HG2 tumours. In this Grade-based classification, HG1 tumours were therefore classified as Luminal A-like and HG3 tumours as Luminal B-like. Ki67 and PR were then evaluated as discriminators in HG2 tumours. For definition of the different surrogate classifications, see Appendix Table 4.
Table 4.
Definition of different surrogate subtyping classifications for ER+/HER2– breast cancer tumours
| Clinicopathological surrogate definition | Characteristics |
|---|---|
| St. Gallen 2013 [11] | |
| Luminal A-like | Low Ki67 (< 20%) and high PR (≥ 20%) |
| Luminal B-like | High Ki67 (≥ 20%) and/or low PR (< 20%) |
| Maisonneuve [12] | |
| Luminal A-like |
Low Ki67 (< 14%) or Intermediate Ki67 (14–19%) and high PR (≥ 20%) |
| Luminal B-like |
High Ki67 (≥ 20%) or Intermediate Ki67 (14–19%) and low PR (< 20%) |
| St. Gallen 2017 [3] | |
| Luminal A-like | High ER/PR, clearly low Ki67, HG1 |
| Intermediate | Uncertainties persist about risk and degree of responsiveness to endocrine and cytotoxic therapies |
| Luminal B-like | Lower ER/PR, clearly high Ki67, HG3 |
| Grade-based classification | |
| Luminal A-like |
HG1 or HG2 and low Ki67 (< 14%) or HG2 and intermediate Ki67 (14–19%) and high PR (≥ 20%) |
| Luminal B-like |
HG3 or HG2 and high Ki67 (≥ 20%) or HG2 and intermediate Ki67 (14–19%) and low PR (< 20%) |
ER oestrogen receptor, HER2 human epidermal growth factor receptor 2, HG histological grade, PR progesterone receptor
The primary aim of this study was to compare the concordance between MS classification (as assessed by RNA sequencing within the SCAN-B project) and different surrogate classifications, namely St. Gallen 2013, Maisonneuve and our proposed Grade-based classification, for ER+/HER2– tumours. Secondary aims were to investigate the discriminatory value of each of the markers Ki67, PR and HG, and moreover to conduct an exploratory analysis to define subgroups consisting mainly of Luminal A or Luminal B tumours.
Materials and methods
Patients
The study cohort consists of patients consecutively included in SCAN-B (Sweden Cancerome Analysis Network—Breast, ClinicalTrials.gov ID: NCT02306096), a multicentre study that was initiated in 2010 with the long-term aim of prospectively analysing breast tumour tissue by genetic methods for translational research and for the development and implementation of molecular assays. There were eight participating hospitals during the years 2013–2015 and nine from 2015 and onwards. These hospitals cover approximately 25% of all patients diagnosed with breast cancer in Sweden [16]. The genomic analyses were all centrally assessed at one institution (Lund, Sweden). All patients who are considered to be able to provide an informed consent are asked to participate and > 90% of invited patients accept to be included in the trial, hence the SCAN-B cohort is considered to be population-based. For about 75% of the participating patients, tumour tissue is available for further gene expression and biomarker analyses [14, 15]. Fresh breast cancer tumour tissue is collected during the routine pathology preparation and analysed by RNA sequencing. Clinicopathological variables and patient-related characteristics for all patients diagnosed in Sweden are recorded in the Swedish National Quality Register for Breast Cancer (NKBC), which has a coverage rate of almost 100% [16]. Data from this register are made available for enrolled patients and transferred to SCAN-B.
All patients with ER+/HER2– breast cancers enrolled in SCAN-B during 2013–2017 (n = 3196) were identified. Patients diagnosed before 2013 were not included as register data on Ki67 for these are scarce. Patients who did not undergo primary surgery (n = 134) were excluded. The rationale for this was that Ki67 assessed on core biopsies is not comparable to Ki67 assessed on surgical specimens [17, 18]. Moreover, histological grade should not be assessed on core biopsies according to Swedish guidelines. The register data for multifocal tumours are known to be associated with uncertainties, so these were excluded (n = 645), as were tumours with missing data for any of Ki67, PR and HG (n = 66). Because we aimed to investigate agreement only for luminal tumours, tumours with a non-luminal molecular subtype were excluded (n = 288), resulting in a study cohort of 2063 patients (Fig. 1). Tumour and patient characteristics are summarised in Table 1.
Fig. 1.
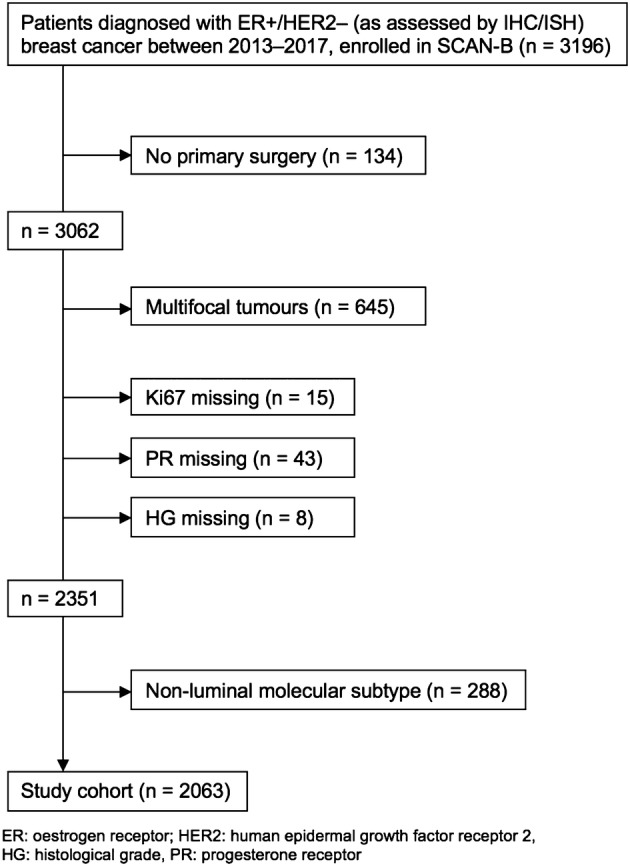
Flow chart of the study cohort
Table 1.
Tumour and patient characteristics of the included patients diagnosed with ER+/HER2– tumours by IHC/ISH with a molecular luminal profile (n = 2063)
| Characteristics | Number of patients n (%) |
|---|---|
| Tumour size (mm) | |
| ≤ 20 | 1451 (71) |
| > 20 but ≤ 50 | 557 (27) |
| > 50 | 46 (2) |
| Missing | 9 |
| Number of positive nodes | |
| 0 | 1440 (71) |
| 1–3 | 509 (25) |
| 4–9 | 66 (3) |
| ≥ 10 | 29 (1) |
| Missing | 19 |
| PRa | |
| Positive | 1780 (86) |
| Negative | 283 (14) |
| Histological grade | |
| 1 | 458 (22) |
| 2 | 1202 (58) |
| 3 | 403 (20) |
| Ki67 | |
| Low (< 14%) | 523 (25) |
| Intermediate (14–19%) | 443 (22) |
| High (≥ 20%) | 1097 (53) |
| Age | |
| < 40 | 14 (1) |
| ≥ 40 but < 50 | 106 (5) |
| ≥ 50 but < 60 | 356 (17) |
| ≥ 60 | 1587 (77) |
| Histopathological tumour type | |
| Ductal/no special type | 1619 (79) |
| Lobular | 295 (14) |
| Other | 148 (7) |
| Missing | 1 |
aPR progesterone receptor. Regarded as positive if defined as positive in the Swedish National Quality Register for Breast Cancer, or a value of PR > 10%
Tumour processing and subtype classification
Fresh tumour tissue was collected for SCAN-B at local pathology departments in conjunction with the regular clinical routines of preparing formalin-fixed specimens for routine histopathology analyses. Fresh tissue was preserved in RNAlater and continuously sent for further processing at the SCAN-B laboratory (Lund, Sweden) where RNA/DNA was extracted. In general, RNA was sequenced within a week after surgery. The intrinsic subtype was determined by a nearest-centroid implementation using the PAM50 genes and centroids as described by Parker et al. [4]. To avoid cohort dependence when assigning PAM50 subtype, fixed reference cohorts for gene centring were selected to match the original training population used by Parker et al. Tumour specimens were then assigned to subtype, namely Luminal A, Luminal B, HER2-enriched, Basal-like or Normal-like, according to the most frequent nearest centroid. Tumours were denoted as Unclassified when the correlation coefficients were below 0.2 to all of the subtype centroids. Methods of tissue preparation and analyses have been described elsewhere [14, 19].
In accordance with Swedish guidelines, ER/PR-positivity was defined as > 10% of positively stained tumour cells of the tissue section assessed by IHC. Ki67 was assessed by counting the percentage of positively stained nuclei in at least 200 cells in hotspots. HER2 was assessed by IHC and scored as 0, 1+ , 2+ or 3+ and 2+ cases were further analysed by in situ hybridisation (ISH). HER2+ was defined as score 3+ alone or score 2+ in combination with her2 gene amplification by ISH [20]. Histological grade was determined in accordance with the work of Elston-Ellis [21]. The tumours were classified according to the surrogate definitions of St. Gallen 2013, Maisonneuve and our proposed Grade-based classification (Appendix Table 4).
Statistical analyses and ethics
Percentage agreement and kappa statistics were used for concordance analyses. Kappa (κ) is a measure of concordance between categorical variables, which adjusts for the amount of agreement that would be expected by chance. The most commonly used interpretation of κ is as follows: ≤ 0.40 poor/fair agreement; 0.41–0.60 moderate agreement; 0.61–0.80 substantial agreement and > 0.80 almost perfect agreement [22]. To evaluate the capacity to distinguish between Luminal A and Luminal B, ROC analysis was performed, and the area under the ROC curve (auROC) was calculated. For kappa and auROC, 95% confidence intervals (CIs) were reported. McNemar’s test was performed to evaluate the difference among the luminal distributions between MS and surrogate classification (p value < 0.05 considered statistically significant). Statistical analyses were performed using IBM SPSS Statistics for Macintosh, Version 25.0.
Results
In the present cohort, 39% (n = 808) and 61% (n = 1255) of the tumours were classified as Luminal A-like and Luminal B-like, respectively, according to St. Gallen 2013. The corresponding figures were 43% (n = 894) and 57% (n = 1169) for the Maisonneuve classification, and 49% (n = 1004) and 51% (n = 1059) for the Grade-based classification. By MS, 71% (n = 1458) of the tumours were assessed as Luminal A and 29% (n = 605) as Luminal B. There were significant differences of the luminal distributions of MS compared with the St. Gallen 2013, Maisonneuve and Grade-based classifications, respectively (all p values < 0.001).
Agreement between MS and the three surrogate classifications
The agreements between MS and the different surrogate classifications were as follows: St. Gallen 2013: 62% [κ = 0.30, (95% CI 0.27–0.34)], Maisonneuve: 66% [κ = 0.35, (95% CI 0.32–0.38)] and Grade-based: 70% [κ = 0.41, (95% CI 0.37–0.44)].
Ki67, PR and histological grade (HG) as surrogate markers
The rates of Luminal A (MS) in tumours with low (< 14%), intermediate (14–19%) and high (≥ 20%) Ki67 were 96%, 86% and 53%, respectively (Table 2). The proportions of low, intermediate and high Ki67 in this cohort were 25%, 22% and 53%, which differ from the distribution in the work of Maisonneuve et al. (34%, 24% and 42%) [12]. We therefore conducted an exploratory analysis using the same Ki67 percentiles, resulting in a slight change of the cut-offs: low: < 16%, intermediate: 16–23% and high: ≥ 24%. After this adjustment, the agreement between the Maisonneuve surrogate classification based on the new Ki67 cut-offs and MS increased to 73% [κ = 0.44, (95% CI 0.40–0.48)]. The corresponding figure for the Grade-based classification was 75% [κ = 0.46, (95% CI 0.43–0.50)].
Table 2.
Ki67 subgroup distribution and proportion of molecular luminal subtypes for all tumours and for tumours with HG1, HG2 and HG3
| Molecular subtype | Ki67 < 14%, n (%) | Ki67 14–19%, n (%) | Ki67 ≥ 20%, n (%) | Total, n (%) |
|---|---|---|---|---|
| All tumours (n = 2063) | ||||
| Luminal A | 500 (96) | 379 (86) | 579 (53) | 1458 (71) |
| Luminal B | 23 (4) | 64 (14) | 518 (47) | 605 (29) |
| Total | 523 (25) | 443 (22) | 1097 (53) | 2063 (100) |
| HG1 tumours (n = 458) | ||||
| Luminal A | 211 (97) | 116 (89) | 96 (87) | 423 (92) |
| Luminal B | 7 (3) | 14 (11) | 14 (13) | 35 (8) |
| Total | 218 (48) | 130 (28) | 110 (24) | 458 (100) |
| HG2 tumours (n = 1202) | ||||
| Luminal A | 282 (95) | 255 (86) | 389 (64) | 926 (77) |
| Luminal B | 15 (5) | 43 (14) | 218 (36) | 276 (23) |
| Total | 297 (25) | 298 (25) | 607 (51) | 1202 (100) |
| HG3 tumours (n = 403) | ||||
| Luminal A | 7 (88) | 8 (53) | 94 (25) | 109 (27) |
| Luminal B | 1 (13) | 7 (47) | 286 (75) | 294 (73) |
| Total | 8 (2) | 15 (4) | 380 (94) | 403 (100) |
In total, 16% (n = 330) of the tumours were PR-low (cut-off < 20%) and 59% (n = 194) of these were Luminal A by MS, compared with 73% (n = 1264) in PR-high (cut-off ≥ 20%) tumours (n = 1733). To evaluate the capacity of PR to distinguish between Luminal A and B tumours, ROC analyses were performed. In the surrogate algorithms presented in this paper, PR is used as a luminal discriminator in the following groups: tumours with low Ki67 (St. Gallen 2013, n = 523), intermediate Ki67 (Maisonneuve, n = 443) and intermediate Ki67 and HG2 (Grade-based, n = 298). The auROC values for PR (percentages from 0 to 100%) for these three groups were 0.51 (95% CI 0.39–0.63), 0.56 (95% CI 0.48–0.63) and 0.56 (95% CI 0.46–0.65), respectively.
The proportions of HG1, HG2 and HG3 in the overall cohort were 22%, 58% and 20%, respectively, and the distribution of Luminal A (MS) in the different HG categories was as follows: HG1: 92%, HG2: 77% and HG3: 27% (Table 2). In an exploratory analysis, we classified HG1-2 tumours as Luminal A-like and HG3 tumours as Luminal B-like, and the agreement with the corresponding MS increased to 80% [κ = 0.46 (95% CI 0.41–0.50)]. When including only HG1 and HG3 tumours, the agreement was 83% [κ = 0.66 (95% CI 0.61–0.71)] (n = 861).
Exploratory analysis of combining HG and Ki67 as a surrogate classification
To identify subgroups in which MS may be omitted due to a high proportion of molecular subtypes of either Luminal A or B, tumours were divided into nine subgroups based on HG (1–3) and Ki67 [low (< 14%), intermediate (14–19%), high (≥ 20%)]. PR was not included in this refined algorithm due to its low capacity to distinguish between the luminal tumours presented above. Six of the subgroups (HG1 with any Ki67 category, HG2 with low and intermediate Ki67 and HG3 with low Ki67), comprising 51% (n = 1061) of the tumours, were found to consist of 91% Luminal A tumours, as assessed by MS. The distribution of Luminal A in the nine subgroups defined by Ki67 and HG is presented in Table 3.
Table 3.
Proportion of Luminal A tumours, according to molecular subtyping, in subgroups generated by combining histological grade (HG1–3) and Ki67 in three categories (according to Maisonneuve et al. [12])
| Ki67 | HG1 | HG2 | HG3 |
|---|---|---|---|
| Low (< 14%) |
97% Luminal A (n = 211 of 218) |
95% Luminal A (n = 282 of 297) |
88% Luminal A (n = 7 of 8) |
| Intermediate (14–19%) |
89% Luminal A (n = 116 of 130) |
86% Luminal A (n = 255 of 298) |
53% Luminal A (n = 8 of 15) |
| High (≥ 20%) |
87% Luminal A (n = 96 of 110) |
64% Luminal A (n = 389 of 607) |
25% Luminal A (n = 94 of 380) |
| Total |
92% Luminal A (n = 423 of 458) |
77% Luminal A (n = 926 of 1202) |
27% Luminal A (n = 109 of 403) |
Discussion
In this population-based contemporary study of breast cancer, including more than 2000 prospectively assessed breast tumours, we show that there was poor agreement between different surrogate definitions of luminal tumour subtypes and molecular PAM50 (Parker algorithm) [4] subtyping. Considerably more tumours were Luminal A as determined by MS than by the surrogate classifications. However, one should not expect to find perfect surrogates for the molecular subtypes since the surrogate algorithms are based on IHC assessment of protein levels, whereas the MS is based on the measurement of mRNA transcript levels of corresponding genes (ESR1, PGR and MKI67) as well as of additional genes within the PAM50 panel, depicting the underlying biological signalling pathways. By simply designating HG1-2 as Luminal A-like and HG3 as Luminal B-like, the agreement with MS was superior to all of the surrogate classifications presented in this paper. The discriminatory importance of HG is not unexpected, being reported both in a previous study from our research group and by Maisonneuve et al. [12, 13].
The reproducibility of HG has, however, been shown to be only moderate [23]. Regarding Ki67, the issues of inter-laboratory variability and cut-off levels are also well known [24, 25]. In an exploratory analysis, we used the same percentiles for low, intermediate and high Ki67 as in the study by Maisonneuve et al. [12], by which the agreement with MS improved (from 66 to 73%). This emphasises the importance of critical reflection over Ki67 cut-offs in the local laboratory. Moreover, only 47% of the tumours in the high-Ki67 category were shown to be Luminal B according to MS, which raises the question of whether a cut-off of 20% is too low to be able to identify Luminal B-like tumours and thereby identify patients with hormone receptor-positive tumours who might benefit from additional adjuvant chemotherapy. We found no added value of PR in the surrogate classifications. However, since there were no follow-up data available for this cohort, we were not able to evaluate PR as a prognostic marker.
When considering the lack of agreement in the results presented here, one should also be aware of the concerns regarding agreement between different multigene tests. Bartlett et al. compared different multiparameter tests regarding risk classification and intrinsic subtyping and found a rate of discordant results among Prosigna®, Blueprint® and MammaTyper® as high as 41% for tumour subtyping [26]. Similar discordant results from the application and comparison of different commercial gene signatures on RNA sequencing data were found in our own study of the SCAN-B cohort (Vallon-Christersson et al. submitted).
As current commercially available variants of molecular assays are associated with considerable cost, it would be desirable to identify patients who could be spared chemotherapy based on less costly molecular tests or from routine pathology only. We found that 91% of the tumours with HG1 (irrespective of Ki67) and HG2 tumours with Ki67 < 20% were identified to be Luminal A by MS. These comprised slightly more than half of the overall cohort; the cost-effective use of commercial molecular assays for further stratification, in the absence of negative prognostic features such as age, nodal involvement or large tumours, may therefore be limited in this group. Tumours with HG3 and low Ki67 were also found to be mostly Luminal A; however, owing to the small numbers in this subgroup (n = 8), this result is considered unreliable.
The strengths of this study are the large cohort of more than 2000 patients, the fact that patients were prospectively included and that the study cohort can be regarded as population-based. A limitation is that the MS definition in the SCAN-B project has not yet been clinically validated. However, as pointed out by others, a nearest-centroid classification that appropriately addresses cohort composition to match the original study population is expected to be almost equivalent to the commercial assay [27]. Moreover, because of the short follow-up, no evaluation of the prognostic value of the presented results was performed.
Conclusion
The present study indicates poor agreement between surrogate classifications and MS of luminal breast cancer tumours. By combining HG and Ki67, a large subgroup of the patients could be identified as Luminal A assessed by MS. This group of patients may not benefit from the use of molecular assays, especially if other clinicopathological factors indicate a low risk of recurrence. We are aware of the issue regarding the poor reproducibility of Ki67 and HG assessments, favouring MS. Nonetheless, the results of this study offer new insights into how to use MS in combination with histopathological markers in a clinical context, but further studies including adequate follow-up data are needed to correlate these findings with patient outcome.
Acknowledgements
Open access funding provided by Lund University. The authors thank all patients who participated in the SCAN-B study, the staff of the SCAN-B laboratory, collaborators at Hallands Hospital Halmstad, Helsingborg Hospital, Blekinge County Hospital, Central Hospital Kristianstad, Skåne University Hospital Lund/Malmö, Central Hospital Växjö, County Hospital Ryhov Jönköping and Uppsala University Hospital, the South Swedish Breast Cancer Group, the Regional Cancer Centre South and the Swedish National Quality Register for Breast Cancer. Special thanks go to the Department of Oncology, Region Jönköping County, for making this study possible. Thanks also go to Bo Rolander at Futurum—the Academy for Health and Care, Region Jönköping County, for support with the database construction.
Appendix
See Table 4.
Funding
This study was funded by grants from the Swedish Breast Cancer Association, the Swedish Breast Cancer Group and Futurum—the Academy for Health and Care, Region Jönköping County.
Compliance with ethical standards
Conflict of interest
Author Anna Ehinger has received remuneration from Roche, Amgen and Novartis. Author Lao H. Saal has received remuneration and has stock ownership in SAGA Diagnostics AB. Author Maria Ekholm has had a consultant/advisory role in Pfizer and Novartis. The other authors declare that they have no conflict of interest.
Ethical approval
This study is covered by previous ethical approvals from the Ethics Board in Lund (Dnr: 2009/658, 2010/383, 2012/58, 2013/459, 2014/521, 2015/277, 2016/944). It is a prospective study, but no procedures or interventions were performed.
Informed consent
Informed consent has previously been obtained from all individual participants included in the SCAN-B study.
Research involving human and animal participants
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lonning PE, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Curigliano G, Burstein HJ, Winer PE. De-escalating and escalating treatments for early-stage breast cancer: the St. Gallen International Expert Consensus Conference on the Primary Therapy of Early Breast Cancer 2017. Ann Oncol. 2017;28(8):1700–1712. doi: 10.1093/annonc/mdx308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, Quackenbush JF, Stijleman IJ, Palazzo J, Marron JS, Nobel AB, Mardis E, Nielsen TO, Ellis MJ, Perou CM, Bernard PS. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–1167. doi: 10.1200/jco.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 6.van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 7.Sparano JA, Gray RJ, Makower DF, et al. Adjuvant chemotherapy guided by a 21-gene expression assay in breast cancer. N Engl J Med. 2018;379(2):111–121. doi: 10.1056/NEJMoa1804710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cardoso F, van’t Veer LJ, Bogaerts J, et al. 70-Gene signature as an aid to treatment decisions in early-stage breast cancer. N Engl J Med. 2016;375(8):717–729. doi: 10.1056/NEJMoa1602253. [DOI] [PubMed] [Google Scholar]
- 9.NanoString Technologies (2018) Prosigna-Breast cancer prognostic gene signature assay. https://www.nanostring.com/diagnostics/prosigna-uk. Accessed 15 Dec 2018
- 10.Gnant M, Harbeck N, Thomssen C. St. Gallen 2011: summary of the consensus discussion. Breast Care (Basel) 2011;6(2):136–141. doi: 10.1159/000328054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldhirsch A, Winer EP, Coates AS, et al. Personalizing the treatment of women with early breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol. 2013;24(9):2206–2223. doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maisonneuve P, Disalvatore D, Rotmensz N, Curigliano G, Colleoni M, Dellapasqua S, Pruneri G, Mastropasqua MG, Luini A, Bassi F, Pagani G, Viale G, Goldhirsch A. Proposed new clinicopathological surrogate definitions of luminal A and luminal B (HER2-negative) intrinsic breast cancer subtypes. Breast Cancer Res. 2014;16(3):R65. doi: 10.1186/bcr3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehinger A, Malmström P, Bendahl P-O, Elston CW, Falck A-K, Forsare C, Grabau D, Rydén L, Stål O, Fernö M. Histological grade provides significant prognostic information in addition to breast cancer subtypes defined according to St. Gallen 2013. Acta Oncol. 2017;56(1):68–74. doi: 10.1080/0284186X.2016.1237778. [DOI] [PubMed] [Google Scholar]
- 14.Saal LH, Vallon-Christersson J, Häkkinen J, Hegardt C, Grabau D, Winter C, Brueffer C, Tang MHE, Reuterswärd C, Schulz R, Karlsson A, Ehinger A, Malina J, Manjer J, Malmberg M, Larsson C, Rydén L, Loman N, Borg Å. The Sweden Cancerome Analysis Network—Breast (SCAN-B) Initiative: a large-scale multicenter infrastructure towards implementation of breast cancer genomic analyses in the clinical routine. Genome Med. 2015;7(1):20. doi: 10.1186/s13073-015-0131-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rydén LA-O, Loman N, Larsson C, Hegardt C, Vallon-Christersson J, Malmberg M, Lindman H, Ehinger A, Saal LH, Borg Å. Minimizing inequality in access to precision medicine in breast cancer by real-time population-based molecular analysis in the SCAN-B initiative. Br J Surg. 2018;105(2):e158–e168. doi: 10.1002/bjs.10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Regionala Cancercentrum i Samverkan (2019) Nationellt kvalitetsregister för bröstcancer och bröstrekonstruktion. https://www.cancercentrum.se/samverkan/cancerdiagnoser/brost/kvalitetsregister/. Accessed 14 Apr 2019
- 17.Romero Q, Bendahl PO, Klintman M, Loman N, Ingvar C, Ryden L, Rose C, Grabau D, Borgquist S. Ki67 proliferation in core biopsies versus surgical samples—a model for neo-adjuvant breast cancer studies. BMC Cancer. 2011;11:341. doi: 10.1186/1471-2407-11-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knutsvik G, Stefansson IM, Aziz S, Arnes J, Eide J, Collett K, Akslen LA. Evaluation of Ki67 expression across distinct categories of breast cancer specimens: a population-based study of matched surgical specimens, core needle biopsies and tissue microarrays. PLoS ONE. 2014;9(11):e112121. doi: 10.1371/journal.pone.0112121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brueffer C, Vallon-Christersson J, Grabau D, Ehinger A, Häkkinen J, Hegardt C, Malina J, Chen Y, Bendahl P-O, Manjer J, Malmberg M, Larsson C, Loman N, Rydén L, Borg Å, Saal LH. Clinical value of RNA sequencing-based classifiers for prediction of the five conventional breast cancer biomarkers: a report from the population-based multicenter Sweden Cancerome Analysis Network—Breast Initiative. JCO Precis Oncol. 2018;2:1–18. doi: 10.1200/PO.17.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Regionala Cancercentrum i Samverkan (2018) Kvalitetsdokument för patologi. http://www.svfp.se/foreningar/uploads/L15178/kvast/brostpatologi/KVASTbrostcancer2018.pdf. Accessed 30 Nov 2018
- 21.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19(5):403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 22.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33(1):159–174. doi: 10.2307/2529310. [DOI] [PubMed] [Google Scholar]
- 23.Boiesen P, Bendahl PO, Anagnostaki L, Domanski H, Holm E, Idvall I, Johansson S, Ljungberg O, Ringberg A, Ostberg G, Fernö M. Histologic grading in breast cancer: reproducibility between seven pathologic departments. Acta Oncol. 2000;39(1):41–45. doi: 10.1080/028418600430950. [DOI] [PubMed] [Google Scholar]
- 24.Ekholm M, Grabau D, Bendahl PO, Bergh J, Elmberger G, Olsson H, Russo L, Viale G, Ferno M. Highly reproducible results of breast cancer biomarkers when analysed in accordance with national guidelines—a Swedish survey with central re-assessment. Acta Oncol. 2015;54(7):1040–1048. doi: 10.3109/0284186X.2015.1037012. [DOI] [PubMed] [Google Scholar]
- 25.Focke CM, Burger H, van Diest PJ, Finsterbusch K, Glaser D, Korsching E, Decker T. Interlaboratory variability of Ki67 staining in breast cancer. Eur J Cancer. 2017;84:219–227. doi: 10.1016/j.ejca.2017.07.041. [DOI] [PubMed] [Google Scholar]
- 26.Bartlett JM, Bayani J, Marshall A, Dunn JA, Campbell A, Cunningham C, Sobol, Hall PS, Poole CJ, Cameron DA, Earl HM, Rea DW, Macpherson IR, Canney P, Francis A, McCabe C, Pinder SE, Hughes-Davies L, Makris A, Stein RC. Comparing breast cancer multiparameter tests in the OPTIMA Prelim Trial: No test is more equal than the others. J Natl Cancer Inst. 2016;108(9):djw050. doi: 10.1093/jnci/djw050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wallden B, Storhoff J, Nielsen T, Dowidar N, Schaper C, Ferree S, Liu S, Leung S, Geiss G, Snider J, Vickery T, Davies SR, Mardis ER, Gnant M, Sestak I, Ellis MJ, Perou CM, Bernard PS, Parker JS. Development and verification of the PAM50-based Prosigna breast cancer gene signature assay. BMC Med Genomics. 2015;8:54. doi: 10.1186/s12920-015-0129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]


