Abstract
Tea is the most popular non-alcoholic caffeine-containing and the oldest beverage in the world. In this study, we de novo assembled the chloroplast (cp) and mitochondrial (mt) genomes of C. sinensis var. assamica cv. Yunkang10 into a circular contig of 157,100 bp and two complete circular scaffolds (701719 bp and 177329 bp), respectively. We correspondingly annotated a total of 141 cp genes and 71 mt genes. Comparative analysis suggests repeat-rich nature of the mt genome compared to the cp genome, for example, with the characterization of 37,878 bp and 149 bp of long repeat sequences and 665 and 214 SSRs, respectively. We also detected 478 RNA-editing sites in 42 protein-coding mt genes, which are ~4.4-fold more than 54 RNA-editing sites detected in 21 protein-coding cp genes. The high-quality cp and mt genomes of C. sinensis var. assamica presented in this study will become an important resource for a range of genetic, functional, evolutionary and comparative genomic studies in tea tree and other Camellia species of the Theaceae family.
Subject terms: Plant breeding, DNA sequencing, Genome, Sequence annotation
| Measurement(s) | genome assembly |
| Technology Type(s) | DNA sequencing |
| Sample Characteristic - Organism | Camellia sinensis |
Machine-accessible metadata file describing the reported data: 10.6084/m9.figshare.9884729
Background & Summary
Tea is the most popular non-alcoholic caffeine-containing and the oldest beverage in the world since 3000 B. C.1,2. The production of tea made from the young leaves of Camellia sinensis var. sinensis and C. sinensis var. assamica, together with ornamentally well-known camellias (e.g., C. japonica, C. reticulata and C. sasanqua) and worldwide renowned wooden oil crop C. oleifera3 has made the genus Camellia possess huge economic values in Theaceae. Besides its industrial, cultural and medicinal values, botanists and evolutionary biologists have increasingly paid attention to this genus. As a result of frequent hybridization and polyploidization, Camellia is almost commonly regarded as one of the most taxonomically and phylogenetically difficult taxa in flowering plants4. Thus, it has long been problematic for the taxonomic classification of the Camellia species based on the morphological characteristics5. The chloroplast (cp) genomes are able to provide valuable information for taxonomic classification, tracing source populations6,7 and the reconstruction of phylogeny to resolve complex evolutionary relationships8–10 due to the conservation of genomic structure, maternal inheritance and a fairly low recombination rate. Genetically speaking, cp genomes are comparatively conserved than plant mitochondria (mt) genomes which are more heterogeneous in nature. However, the presence of NUPT (nuclear plastid DNA) into cp genomes argues that cp genomes assembled from WGS data may include the heterogeneity due to the nuclear cp DNA transferred to the nucleus, resulting in erroneous phylogenetic inferences11. It has long been acknowledged that mtDNA has the propensity to integrate DNA from various sources through intracellular and horizontal transfer12–14. Partially due to these reasons, the mt genomes vary from ~200 Kbp to ~11.3 Mbp in some living organisms15–17. The dynamic nature of mt genome structure has been recognized, and plant mt genomes can have a variety of different genomic configurations due to the recombination and differences in repeat content18,19. These characteristics make the plant mt genome a fascinating genetic system to investigate questions related to evolutionary biology. The first effort has been made to sequence the 13 representative Camellia chloroplast genomes using next-generation Illumina genome sequencing platform, which obtained novel insights into global patterns of structural variation across the Camellia cp genomes4. The reconstruction of phylogenetic relationships among these representative species of Camellia suggests that cp genomic resources are able to provide useful data to help to understand their evolutionary relationships and classify the ‘difficult taxa’. Increasing interest in the Camellia plants have made up to thirty-eight of cp genomes be sequenced up to date20–37. Recently, we decoded the first nuclear genome of C. sinensis var. assamica cv. Yunkang10, providing novel insights into genomic basis of tea flavors38. Besides the lack of the C. sinensis var. assamica cp genome among thirty-eight cp genomes that were sequenced in this genus4,20–37, up to data, none of mt genome has been determined in the genus Camellia.
In this study, we filtered cpDNA and mtDNA reads from the WGS genome sequence project38 and de novo assembled the mt genome and cp genome of C. sinensis var. assamica. The information of both cp and mt genomes will help to obtain a comprehensive understanding of the taxonomy and evolution of the genus Camellia. These genome sequences will also facilitate the genetic modification of these economically important plants, for example, through chloroplast genetic engineering technologies.
Methods
Plant materials, DNA extraction and genome sequencing
Young and healthy leaves of an individual plant of cultivar Yunkang10 of C. sinensis var. assamica were collected for genome sequencing in April, 2009, from Menghai County, Yunnan Province, China. Fresh leaves were harvested and immediately frozen in liquid nitrogen after collection, followed by the preservation at −80 °C in the laboratory prior to DNA extraction. High-quality genomic DNA was extracted from leaves using a modified CTAB method39. RNase A and proteinase K were separately used to remove RNA and protein contamination. The quality and quantity of the isolated DNA were separately checked by electrophoresis on a 0.8% agarose gel and a NanoDrop D-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). A total of eleven paired-end libraries, including four types of small-insert libraries (180 bp, 260 bp, 300 bp, 500 bp) and seven large-insert libraries (2 Kb, 3 Kb, 4 Kb, 5 Kb, 6 Kb, 8 Kb, 20 Kb), were prepared following the Illumina’s instructions, and sequenced using Illumina HiSeq. 2000 platform by following the standard Illumina protocols (Illumina, San Diego, CA). We totally generated ~707.88 Gb (~229.31×) of raw sequencing data38. Further reads quality control filtering processes yielded a total of ~492.15 Gb (~159.43×) high-quality data retained and used for subsequent genome assembly.
De novo chloroplast and mitochondria genome assemblies
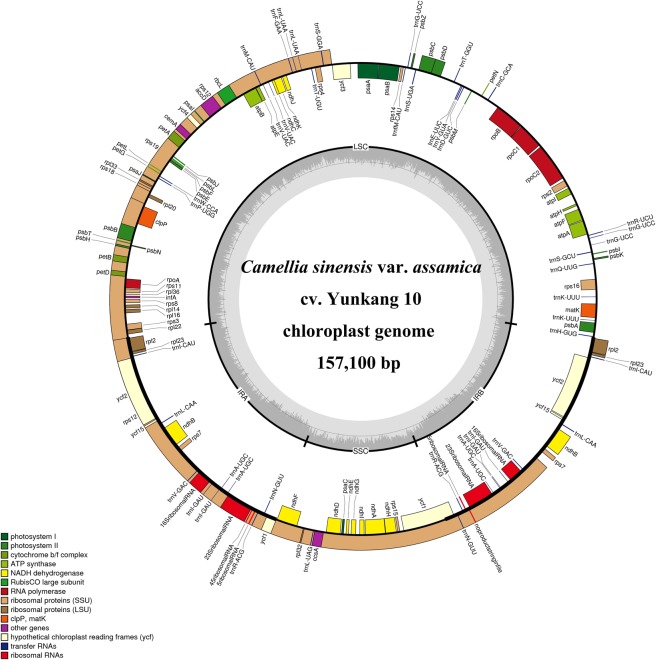
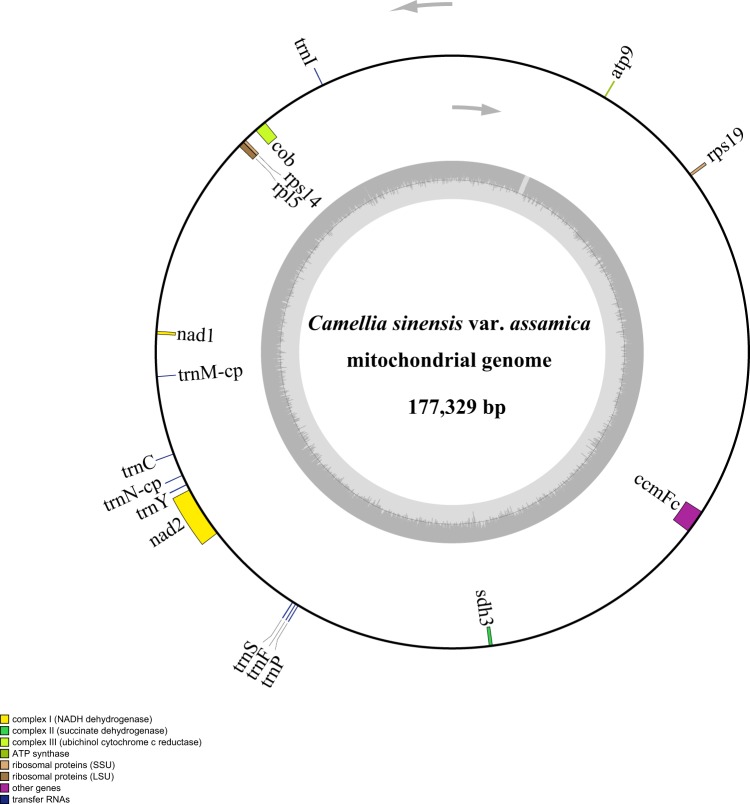
The chloroplast reads were filtered from whole genome Illumina sequencing data of C. sinensis var. assamica, we mapped all the sequencing reads to the reference genomes4 using bowtie2 (version 2.3.4.3)40. The mapped chloroplast reads were assembled into a circular contig of 157,100 bp in length with an overall GC content of 37.29% using CLC Genomics Workbench v. 3.6.1 (CLC Inc., Rarhus, Denmark) (Fig. 1). For mitochondria genome assembly, the PE and MP sequencing reads were used separately. Briefly, we first performed de novo assembly with VELVET v1.2.0841, which was previously described42,43. Scaffolds were constructed using SSPACE v.3.044. False connection was manually removed based on the coverage and distances of paired reads. Gaps between scaffolds were then filled with GapCloser (version 1.12)45,46 using all pair-end reads. We obtained the two complete circular scaffolds (701719 bp and 177329 bp) of the C. sinensis var. assamica mt genome from the de-novo assembly of the filtered mitochondrial reads (Figs 2–4). The two scaffolds of the mt genome had overall GC contents of 45.63% and 45.81%, respectively. The completed chloroplast and mitochondria genomes are publicly available in NCBI GenBank under accession numbers MH019307, MK574876 and MK574877 and BIG Genome Warehouse WGS000271, WGS000272.
Fig. 1.
Genome map of C. sinensis var. assamica cv. Yunkang10. Genes lying outside of the outer circle are transcribed in the clockwise direction whereas genes inside are transcribed in the counterclockwise direction. Genes belonging to different functional groups are color-coded. Area dashed darker gray in the inner circle indicates GC content while the lighter gray corresponds to AT content of the genome.
Fig. 2.
The assembly and annotation pipeline of the tea tree mitochondrial genome.
Fig. 4.
Circular map of scaffold 2 in the C. sinensis var. assamica cv. Yunkang10 mitochondrial genome. Gene map showing 17 annotated genes with different functional groups that are color-coded on outer circle as transcribed clock-wise (outside) and transcribed counter clock-wise (inside). The inner circle indicates the GC content as dark grey plot.
Genome annotation and visualization
The complete chloroplast genome of C. sinensis var. assamica was preliminarily annotated using the online program DOGMA47 (Dual Organellar Genome Annotator) followed by manual correction. A total of 141 genes were annotated, of which 87 were protein-coding genes, 46 were tRNA genes and eight were rRNA genes (Table 1). MITOFY15 was used to characterize the complement of protein-coding and rRNA genes in the mitochondrial genome. A tRNA gene search was carried out using the tRNA scan-SE software (version 1.3.1)48. We annotated a total of 71 genes, including 44 protein-coding genes, 24 tRNAs and 3 rRNAs (Table 2). Circular genome maps were drawn with OrganellarGenomeDRAW49 (Figs 3–4).
Table 1.
Gene annotation of the C. sinensis var. assamica cp genome.
| Category | Group | Genes |
|---|---|---|
| Photosynthesis related genes | Rubisco | rbcL |
| Photosystem I | psaA, psaB, psaC, psaI, psaJ | |
| Assembly/stability of Photosystem I | ycf3 | |
| Photosystem II | psbA, psbB, psbT, psbK, psbI, psbH, psbM, psbN, psbD, psbC, psbZ, psbJ, psbL, psbE, psbF | |
| ATP synthase | atpA, atpB, atpE, atpF, atpH, atpI | |
| Cytochrome b/f complex | petA, petB, petD, petN, petL, petG | |
| Cytochrome csynthesis | ccsA | |
| NADPH dehydrogenase | ndhA, ndhB (×2), ndhC, ndhD, ndhE, ndhF, ndhH, ndhG, ndhJ, ndhK, ndhI | |
| Transcription and translation related genes | Transcription | rpoA, rpoC2, rpoC1, rpoB |
| Ribosomal proteins | rps2, rps3, rps4, rps7 (×2), rps8, rps11, rps12, rps14, rps15, rps16, rps18, rps19, rpl2 (×2), rpl14, rpl16, rpl20, rpl22, rpl23 (×2), rpl32, rpl33, rpl36 | |
| Translation initiation factor | infA | |
| RNA genes | Ribosomal RNA | rrn16S (×2), rrn23S (×2), rrn4.5 (×2), rrn5 (×2) |
| Transfer RNA | trnH-GUG, trnK-UUU (×2), trnQ-UUG, trnS-GCU, trnG-UCC (×2), trnR-UCU, trnC-GCA, trnD-GUC, trnY-GUA, trnE-UUC, trnT-GGU, trnS-UGA, trnG-UCC, trnfM-CAU, trnS-GGA, trnT-UGU, trnL-UAA (×2), trnF-GAA, trnV-UAC (×2), trnM-CAU, trnW-CCA, trnP-UGG, trnI-CAU, trnL-CAA (×2), trnV-GAC, trnI-GAU (×3), trnA-UGC (×2), trnR-ACG (×2), trnN-GUU (×2), trnL-UAG, trnN-GUU, trnR-ACG, trnA-UGC (×2), trnV-GAC, trnI-CAU | |
| Other genes | RNA processing | matK |
| Carbon metabolism | cemA | |
| Fatty acid synthesis | accD | |
| Proteolysis | clpP | |
| Genes of unknown function | Conserved ORFs | ycf1 (×2), cf2, ycf4, ycf2, ycf15 (×2) |
Table 2.
Gene content of the C. sinensis var. assamica mt genome.
| Group of genes | Name of genes | |
|---|---|---|
| Scaffold 1 | Scaffold 2 | |
| Complex I | nad1, nad2, nad3, nad4, nad4L, nad5, nad6, nad7, nad9 (×2) | nad1, nad2 |
| Complex II | sdh3, sdh4 | sdh3 |
| Complex III | cob | |
| Complex IV | cox1, cox2, cox3 | |
| Complex V | atp1, atp4, atp6, atp8, atp9 | atp9 |
| Cytochrome c biogenesis | ccmFn, ccmB, ccmC | ccmFc |
| Ribosome large subunit | rpl2, rpl10, rpl16 | rpl5 |
| Ribosome small subunit | rps1, rps3, rps4, rps7, rps12, rps13, rps19 | rps14, rps19 |
| rRNA genes | rrn5, rrn18, rrn16 | |
| tRNA genes | trnS(Ser), trnD(Asp), trnK(Lys), trnfM(Met) (×2), trnI(Ile)-cp, trnE(Glu), trnH(His)-cp, trnP(Pro), trnW(Trp)-cp, trnG(Gly), trnQ(Gln), trnC(Cys), trnD(Asp), trnS(Ser), trnV(Val)-cp | trnI(Ile), trnM(Met)-cp, trnC(Cys), trnN(Asn)-cp, trnY(Tyr), trnS(Ser), trnF(Phe), trnP(Pro) |
| chloroplast-derived genes | trnI(Ile)-cp, trnH(His)-cp, trnW(Trp)-cp, trnV(Val)-cp | trnM(Met)-cp, trnN(Asn)-cp |
| Other proteins | matR, mttB | |
Fig. 3.
Circular map of scaffold 1 in the C. sinensis var. assamica cv. Yunkang10 mitochondrial genome. Gene map showing 54 annotated genes with different functional groups that are color-coded on outer circle as transcribed clock-wise (outside) and transcribed counter clock-wise (inside). The inner circle indicates the GC content as dark grey plot.
Simple sequence repeats (SSRs) were identified and located using MISA (http://pgrc.ipk-gatersleben.de/misa/). All the annotated SSRs were classified by the size and copy number of their tandemly repeated: monomer (one nucleotide, n ≥ 8), dimer (two nucleotides, n ≥ 4), trimer (three nucleotides, n ≥ 4), tetramer (four nucleotides, n ≥ 3), pentamer (five nucleotides, n ≥ 3), hexamer (six nucleotides, n ≥ 3). A total of 214 SSRs were identified in cp genome with 74.42% of which were monomers, 19.07% of dimers, 0.47% of trimers, 4.65% of tetramers and 0.93% of hexamers (Table 3). There were no pentamers found in the cp genome. In mt genome, we obtained 665 SSRs distributed into monomers, dimers, trimers, pentamers, tetramers and hexamers with 31.53%, 45.35%, 4.95%, 15.17%, 2.70% and 0.15%, respectively (Table 3). Repeat sequences including forward and palindromic repeats, were also searched by REPuter50 with the following parameters: minimal length 50 nt; mismatch 3 nt. Long repeat sequences (repeat unit > 50 bp) of forward and palindromic repeats were further annotated, resulting in 149 bp from 4 paired repeats in the cp genome (Table 4) and 37,878 bp from 58 paired repeats in the mt genome (Online-only Tables 1–2). Our repeat content analyses indicate that the mt genome is more abundant in repeat sequences and more variable than the cp genome of C. sinensis var. assamica (Table 4; Online-only Tables 1–2).
Table 3.
Statistics of SSR motifs in the C. sinensis var. assamica mt and cp genomes.
| SSR-Motif | mt Genome | cp Genome | ||
|---|---|---|---|---|
| SSR Number | SSR % | SSR Number | SSR % | |
| Monomer | 210 | 31.53 | 160 | 74.42 |
| Dimer | 302 | 45.35 | 41 | 19.07 |
| Trimer | 33 | 4.95 | 1 | 0.47 |
| Tetramer | 101 | 15.17 | 10 | 4.65 |
| Pentamer | 18 | 2.70 | 0 | 0.00 |
| Hexamer | 1 | 0.15 | 2 | 0.93 |
Table 4.
Long repeats (repeat unit > 50 bp) in the C. sinensis var. assamica cp genome.
| Repeat Length | Type* | Start of Copy 1 | Start of Copy 2 |
|---|---|---|---|
| 56 | F | 93938 | 93956 |
| 56 | P | 93938 | 149737 |
| 56 | P | 93956 | 149755 |
| 56 | F | 149737 | 149755 |
*P indicates palindromic repeats; F indicates forward repeats.
Overlapped repeats have been manually removed while calculating total length.
Online-only Table 1.
Long repeats (repeat unit > 50 bp) in Scaffold 1 of the C. sinensis var. assamica mt genome.
| Repeat Length | Type* | Start of Copy 2 | Start of Copy 1 |
|---|---|---|---|
| 5119 | F | 207173 | 443366 |
| 2191 | F | 389017 | 391244 |
| 1963 | F | 210330 | 212292 |
| 1962 | F | 212292 | 446523 |
| 1930 | F | 383226 | 385188 |
| 1650 | F | 205522 | 207173 |
| 1650 | F | 205522 | 443366 |
| 1469 | F | 538290 | 539780 |
| 814 | F | 496567 | 498047 |
| 705 | F | 619432 | 621461 |
| 665 | F | 497382 | 498862 |
| 255 | P | 151984 | 200526 |
| 228 | P | 448476 | 544136 |
| 204 | F | 277002 | 363807 |
| 131 | P | 73675 | 482324 |
| 125 | F | 301855 | 468834 |
| 104 | F | 297204 | 623713 |
| 88 | F | 228824 | 559689 |
| 87 | F | 594334 | 641398 |
| 84 | F | 530415 | 646532 |
| 82 | P | 224027 | 395044 |
| 82 | F | 509347 | 623862 |
| 81 | P | 152363 | 200041 |
| 80 | F | 304361 | 306020 |
| 78 | P | 299987 | 587603 |
| 74 | F | 165777 | 570981 |
| 70 | F | 165878 | 571083 |
| 69 | F | 123050 | 384677 |
| 69 | F | 123050 | 386639 |
| 67 | F | 18495 | 27472 |
| 66 | F | 299782 | 537227 |
| 66 | P | 364849 | 599005 |
| 66 | F | 684228 | 684285 |
| 65 | P | 508609 | 683320 |
| 64 | F | 542385 | 560020 |
| 63 | F | 605770 | 619261 |
| 62 | P | 70098 | 424512 |
| 62 | F | 151516 | 524252 |
| 62 | P | 156839 | 486845 |
| 61 | F | 123120 | 384747 |
| 61 | F | 123120 | 386709 |
| 61 | P | 142673 | 486240 |
| 60 | F | 302012 | 395122 |
| 59 | P | 265260 | 472040 |
| 58 | F | 285626 | 402303 |
| 57 | P | 152478 | 199950 |
| 57 | F | 276881 | 363698 |
| 56 | F | 402376 | 658389 |
| 55 | P | 41703 | 667438 |
| 55 | F | 258578 | 486959 |
*P indicates palindromic repeats; F indicates forward repeats. Overlapped repeats have been manually removed while calculating total length.
Online-only Table 2.
Long repeats (repeat unit > 50 bp) in Scaffold 2 of the C. sinensis var. assamica mt genome.
| Repeat Length | Type* | Start of Copy 1 | Start of Copy 2 |
|---|---|---|---|
| 704 | F | 30739 | 32294 |
| 156 | P | 29085 | 67620 |
| 86 | F | 67291 | 136332 |
| 67 | P | 4255 | 17574 |
| 67 | P | 23998 | 45730 |
| 62 | F | 67282 | 135282 |
| 55 | F | 120664 | 129253 |
| 53 | F | 135291 | 136332 |
*P indicates palindromic repeats; F indicates forward repeats. Overlapped repeats have been manually removed while calculating total length.
Prediction of RNA-editing sites
Putative RNA editing sites in protein-coding genes were predicted using the PREP-cp and PREP-mt Web-based program (http://prep.unl.edu/)51,52. To achieve a balanced trade-off between the number of false positive and false negative sites, the cutoff score (C-value) was set to 0.8 and 0.6, respectively53.
Almost all transcripts of protein encoding genes in the plant mitochondria are subject to RNA editing except the T-urf13 gene54. Our results showed that the extent of RNA editing varied by gene for both cp and mt genomes of C. sinensis var. assamica. In the C. sinensis var. assamica cp genome, we detected 54 RNA-editing sites in 21 protein-coding genes, ranging from one editing site in atpF, atpI, petB, psaI, psbE, psbF, rpoA, rps2 and rps8 to 8 editing sites in ndhB (Online-only Table 3). In the C. sinensis var. assamica mt genome, we predicted 478 RNA-editing sites in 42 protein-coding genes; they varied from two editing site in atp9 (of scaffold2), sdh3 (of scaffold1 and scaffold2, respectively) and rps14 (of scaffold2) to 35 editing sites in ccmFn (of scaffold1) (Online-only Table 4–5).
Online-only Table 3.
Predicted RNA-editing sites in the C. sinensis var. assamica cp genome. (*The cutoff score (C-value) was set to 0.8).
| No. | Gene | Nucleotide Pos | AA Pos | Effect | Score* |
|---|---|---|---|---|---|
| 1 | accD | 64 | 22 | CGG (R) => TGG (W) | 1 |
| 2 | accD | 1469 | 490 | CCT (P) => CTT (L) | 1 |
| 3 | atpA | 791 | 264 | CCA (P) => CTA (L) | 1 |
| 4 | atpA | 914 | 305 | TCA (S) => TTA (L) | 1 |
| 5 | atpF | 92 | 31 | CCA (P) => CTA (L) | 0.86 |
| 6 | atpI | 134 | 45 | GCT (A) => GTT (V) | 1 |
| 7 | matK | 445 | 149 | CAC (H) => TAC (Y) | 1 |
| 8 | matK | 467 | 156 | TCG (S) => TTG (L) | 1 |
| 9 | matK | 631 | 211 | CAT (H) => TAT (Y) | 1 |
| 10 | matK | 1234 | 412 | CAT (H) => TAT (Y) | 1 |
| 11 | ndhA | 341 | 114 | TCA (S) => TTA (L) | 1 |
| 12 | ndhA | 566 | 189 | TCA (S) => TTA (L) | 1 |
| 13 | ndhA | 1028 | 343 | TCT (S) => TTT (F) | 1 |
| 14 | ndhA | 1073 | 358 | TCT (S) => TTT (F) | 1 |
| 15 | ndhB | 149 | 50 | TCA (S) => TTA (L) | 1 |
| 16 | ndhB | 467 | 156 | CCA (P) => CTA (L) | 1 |
| 17 | ndhB | 586 | 196 | CAT (H) => TAT (Y) | 1 |
| 18 | ndhB | 611 | 204 | TCA (S) => TTA (L) | 0.8 |
| 19 | ndhB | 737 | 246 | CCA (P) => CTA (L) | 1 |
| 20 | ndhB | 746 | 249 | TCT (S) => TTT (F) | 1 |
| 21 | ndhB | 830 | 277 | TCA (S) => TTA (L) | 1 |
| 22 | ndhB | 1481 | 494 | CCA (P) => CTA (L) | 1 |
| 23 | ndhD | 20 | 7 | ACG (T) => ATG (M) | 1 |
| 24 | ndhD | 401 | 134 | TCA (S) => TTA (L) | 1 |
| 25 | ndhD | 692 | 231 | TCA (S) => TTA (L) | 1 |
| 26 | ndhD | 896 | 299 | TCA (S) => TTA (L) | 1 |
| 27 | ndhD | 905 | 302 | CCT (P) => CTT (L) | 1 |
| 28 | ndhD | 1328 | 443 | TCA (S) => TTA (L) | 0.8 |
| 29 | ndhF | 205 | 69 | CAT (H) => TAT (Y) | 0.8 |
| 30 | ndhF | 290 | 97 | TCA (S) => TTA (L) | 1 |
| 31 | ndhG | 166 | 56 | CAT (H) => TAT (Y) | 0.8 |
| 32 | ndhG | 314 | 105 | ACA (T) => ATA (I) | 0.8 |
| 33 | petB | 641 | 214 | CCA (P) => CTA (L) | 1 |
| 34 | psaI | 80 | 27 | TCT (S) => TTT (F) | 0.86 |
| 35 | psbE | 214 | 72 | CCT (P) => TCT (S) | 1 |
| 36 | psbF | 77 | 26 | TCT (S) => TTT (F) | 1 |
| 37 | rpoA | 368 | 123 | TCG (S) => TTG (L) | 1 |
| 38 | rpoB | 338 | 113 | TCT (S) => TTT (F) | 1 |
| 39 | rpoB | 473 | 158 | TCA (S) => TTA (L) | 0.86 |
| 40 | rpoB | 551 | 184 | TCA (S) => TTA (L) | 1 |
| 41 | rpoB | 566 | 189 | TCG (S) => TTG (L) | 1 |
| 42 | rpoB | 973 | 325 | CTT (L) => TTT (F) | 0.86 |
| 43 | rpoB | 2000 | 667 | TCT (S) => TTT (F) | 1 |
| 44 | rpoB | 2336 | 779 | ACA (T) => ATA (I) | 1 |
| 45 | rpoC1 | 41 | 14 | TCA (S) => TTA (L) | 1 |
| 46 | rpoC1 | 1556 | 519 | TCG (S) => TTG (L) | 1 |
| 47 | rpoC2 | 1505 | 502 | ACG (T) => ATG (M) | 0.86 |
| 48 | rpoC2 | 2290 | 764 | CGG (R) => TGG (W) | 1 |
| 49 | rpoC2 | 2726 | 909 | ACT (T) => ATT (I) | 1 |
| 50 | rpoC2 | 3728 | 1243 | TCA (S) => TTA (L) | 0.86 |
| 51 | rps2 | 248 | 83 | TCA (S) => TTA (L) | 1 |
| 52 | rps8 | 182 | 61 | TCA (S) => TTA (L) | 0.86 |
| 53 | rps14 | 80 | 27 | TCA (S) => TTA (L) | 1 |
| 54 | rps14 | 149 | 50 | CCA (P) => CTA (L) | 1 |
Online-only Table 4.
Predicted RNA-editing sites in Scaffold 1 of the C. sinensis var. assamica mt genome.
| No. | Gene | Nucleotide Position | AA Pos | Effect | Score* |
|---|---|---|---|---|---|
| 1 | matR | 32 | 11 | TCC (S) => TTC (F) | 0.62 |
| 2 | matR | 236 | 79 | TCC (S) => TTC (F) | 0.62 |
| 3 | matR | 326 | 109 | CCA (P) => CTA (L) | 1 |
| 4 | matR | 917 | 306 | TCA (S) => TTA (L) | 1 |
| 5 | matR | 1442 | 481 | GCC (A) => GTC (V) | 0.62 |
| 6 | matR | 1667 | 556 | TCC (S) => TTC (F) | 1 |
| 7 | matR | 1688 | 563 | CCT (P) => CTT (L) | 1 |
| 8 | matR | 1708 | 570 | CGC (R) => TGC (C) | 1 |
| 9 | matR | 1744 | 582 | CAC (H) => TAC (Y) | 1 |
| 10 | matR | 1775 | 592 | CCG (P) => CTG (L) | 1 |
| 11 | matR | 1814 | 605 | CCA (P) => CTA (L) | 0.88 |
| 12 | matR | 1832 | 611 | TCA (S) => TTA (L) | 0.88 |
| 13 | ccmFn | 38 | 13 | CCG (P) => CTG (L) | 1 |
| 14 | ccmFn | 98 | 33 | CCT (P) => CTT (L) | 1 |
| 15 | ccmFn | 137 | 46 | TCG (S) => TTG (L) | 1 |
| 16 | ccmFn | 142 | 48 | CGT (R) => TGT (C) | 1 |
| 17 | ccmFn | 151 | 51 | CCT (P) => TCT (S) | 0.83 |
| 18 | ccmFn | 248 | 83 | TCA (S) => TTA (L) | 1 |
| 19 | ccmFn | 256 | 86 | CGG (R) => TGG (W) | 1 |
| 20 | ccmFn | 283 | 95 | CTT (L) => TTT (F) | 0.83 |
| 21 | ccmFn | 334 | 112 | CAT (H) => TAT (Y) | 0.67 |
| 22 | ccmFn | 356 | 119 | TCC (S) => TTC (F) | 0.67 |
| 23 | ccmFn | 391 | 131 | CCT (P) => TCT (S) | 1 |
| 24 | ccmFn | 478 | 160 | CGT (R) => TGT (C) | 0.83 |
| 25 | ccmFn | 706 | 236 | CCT (P) => TTT (F) | 0.67 |
| 26 | ccmFn | 707 | 236 | CCT (P) => TTT (F) | 0.67 |
| 27 | ccmFn | 716 | 239 | TCA (S) => TTA (L) | 0.83 |
| 28 | ccmFn | 754 | 252 | CGT (R) => TGT (C) | 1 |
| 29 | ccmFn | 776 | 259 | TCA (S) => TTA (L) | 1 |
| 30 | ccmFn | 788 | 263 | CCA (P) => CTA (L) | 1 |
| 31 | ccmFn | 803 | 268 | TCA (S) => TTA (L) | 1 |
| 32 | ccmFn | 893 | 298 | GCG (A) => GTG (V) | 1 |
| 33 | ccmFn | 952 | 318 | CGC (R) => TGC (C) | 1 |
| 34 | ccmFn | 1270 | 424 | CGG (R) => TGG (W) | 1 |
| 35 | ccmFn | 1298 | 433 | CCA (P) => CTA (L) | 1 |
| 36 | ccmFn | 1315 | 439 | CAT (H) => TAT (Y) | 1 |
| 37 | ccmFn | 1330 | 444 | CGG (R) => TGG (W) | 1 |
| 38 | ccmFn | 1348 | 450 | CGG (R) => TGG (W) | 1 |
| 39 | ccmFn | 1381 | 461 | CGG (R) => TGG (W) | 1 |
| 40 | ccmFn | 1399 | 467 | CGT (R) => TGT (C) | 1 |
| 41 | ccmFn | 1442 | 481 | TCG (S) => TTG (L) | 1 |
| 42 | ccmFn | 1462 | 488 | CTT (L) => TTT (F) | 1 |
| 43 | ccmFn | 1466 | 489 | CCA (P) => CTA (L) | 1 |
| 44 | ccmFn | 1478 | 493 | TCA (S) => TTA (L) | 1 |
| 45 | ccmFn | 1487 | 496 | TCT (S) => TTT (F) | 1 |
| 46 | ccmFn | 1513 | 505 | CCC (P) => TCC (S) | 1 |
| 47 | ccmFn | 1561 | 521 | CGG (R) => TGG (W) | 0.67 |
| 48 | nad5 | 155 | 52 | CCG (P) => CTG (L) | 1 |
| 49 | nad5 | 238 | 80 | CCG (P) => TCG (S) | 0.8 |
| 50 | nad5 | 269 | 90 | TCC (S) => TTC (F) | 0.7 |
| 51 | nad5 | 355 | 119 | CCT (P) => TTT (F) | 1 |
| 52 | nad5 | 356 | 119 | CCT (P) => TTT (F) | 1 |
| 53 | nad5 | 371 | 124 | CCA (P) => CTA (L) | 0.9 |
| 54 | nad5 | 395 | 132 | TCT (S) => TTT (F) | 0.9 |
| 55 | nad5 | 503 | 168 | CCT (P) => CTT (L) | 1 |
| 56 | nad5 | 536 | 179 | CCT (P) => CTT (L) | 1 |
| 57 | nad5 | 626 | 209 | TCT (S) => TTT (F) | 0.9 |
| 58 | nad5 | 628 | 210 | CGC (R) => TGC (C) | 0.9 |
| 59 | nad5 | 673 | 225 | CTT (L) => TTT (F) | 0.9 |
| 60 | nad5 | 710 | 237 | TCG (S) => TTG (L) | 1 |
| 61 | nad5 | 722 | 241 | TCA (S) => TTA (L) | 1 |
| 62 | nad5 | 832 | 278 | CCA (P) => TCA (S) | 0.9 |
| 63 | nad5 | 872 | 291 | ACG (T) => ATG (M) | 1 |
| 64 | nad5 | 1307 | 436 | TCA (S) => TTA (L) | 1 |
| 65 | nad4 | 29 | 10 | TCC (S) => TTC (F) | 0.67 |
| 66 | nad4 | 74 | 25 | ACT (T) => ATT (I) | 0.89 |
| 67 | nad4 | 77 | 26 | CCT (P) => CTT (L) | 0.78 |
| 68 | nad4 | 107 | 36 | CCG (P) => CTG (L) | 1 |
| 69 | nad4 | 154 | 52 | CCC (P) => TCC (S) | 1 |
| 70 | nad4 | 158 | 53 | CCT (P) => CTT (L) | 1 |
| 71 | nad4 | 166 | 56 | CGG (R) => TGG (W) | 1 |
| 72 | nad4 | 197 | 66 | TCT (S) => TTT (F) | 1 |
| 73 | nad4 | 362 | 121 | ACA (T) => ATA (I) | 0.89 |
| 74 | nad4 | 368 | 123 | TCT (S) => TTT (F) | 1 |
| 75 | nad4 | 376 | 126 | CGT (R) => TGT (C) | 0.78 |
| 76 | nad4 | 403 | 135 | CGC (R) => TGC (C) | 1 |
| 77 | nad4 | 416 | 139 | CCT (P) => CTT (L) | 0.89 |
| 78 | nad4 | 433 | 145 | CTT (L) => TTT (F) | 1 |
| 79 | nad4 | 436 | 146 | CCC (P) => TTC (F) | 0.89 |
| 80 | nad4 | 437 | 146 | CCC (P) => TTC (F) | 0.89 |
| 81 | nad4 | 449 | 150 | CCA (P) => CTA (L) | 1 |
| 82 | nad4 | 547 | 183 | CTC (L) => TTC (F) | 0.67 |
| 83 | nad4 | 1336 | 446 | CAC (H) => TAC (Y) | 1 |
| 84 | nad4 | 1352 | 451 | CCG (P) => CTG (L) | 1 |
| 85 | nad4 | 1357 | 453 | CGC (R) => TGC (C) | 1 |
| 86 | atp6 | 37 | 13 | CCA (P) => TCA (S) | 0.75 |
| 87 | atp6 | 116 | 39 | TCA (S) => TTA (L) | 1 |
| 88 | atp6 | 167 | 56 | CCG (P) => CTG (L) | 1 |
| 89 | atp6 | 173 | 58 | CCG (P) => CTG (L) | 1 |
| 90 | atp6 | 224 | 75 | TCC (S) => TTC (F) | 1 |
| 91 | atp6 | 229 | 77 | CGC (R) => TGC (C) | 0.75 |
| 92 | atp6 | 236 | 79 | TCG (S) => TTG (L) | 0.67 |
| 93 | atp6 | 254 | 85 | TCG (S) => TTG (L) | 1 |
| 94 | atp6 | 262 | 88 | CGT (R) => TGT (C) | 1 |
| 95 | atp6 | 269 | 90 | CCC (P) => CTC (L) | 1 |
| 96 | atp6 | 401 | 134 | TCA (S) => TTA (L) | 1 |
| 97 | atp6 | 460 | 154 | CCT (P) => TCT (S) | 1 |
| 98 | atp6 | 463 | 155 | CAT (H) => TAT (Y) | 1 |
| 99 | atp6 | 485 | 162 | CCA (P) => CTA (L) | 1 |
| 100 | atp6 | 527 | 176 | TCA (S) => TTA (L) | 1 |
| 101 | atp6 | 548 | 183 | TCC (S) => TTC (F) | 1 |
| 102 | atp6 | 635 | 212 | CCG (P) => CTG (L) | 1 |
| 103 | atp6 | 656 | 219 | TCA (S) => TTA (L) | 1 |
| 104 | atp6 | 664 | 222 | CAT (H) => TAT (Y) | 1 |
| 105 | atp6 | 671 | 224 | TCT (S) => TTT (F) | 1 |
| 106 | atp6 | 680 | 227 | TCA (S) => TTA (L) | 1 |
| 107 | atp6 | 707 | 236 | ACA (T) => ATA (I) | 0.92 |
| 108 | atp6 | 718 | 240 | CAA (Q) => TAA (X) | 1 |
| 109 | mttB | 58 | 20 | CAT (H) => TAT (Y) | 0.88 |
| 110 | mttB | 83 | 28 | TCG (S) => TTG (L) | 0.88 |
| 111 | mttB | 91 | 31 | CCA (P) => TCA (S) | 1 |
| 112 | mttB | 127 | 43 | CGT (R) => TGT (C) | 0.88 |
| 113 | mttB | 134 | 45 | CCA (P) => CTA (L) | 0.62 |
| 114 | mttB | 164 | 55 | TCC (S) => TTC (F) | 0.75 |
| 115 | mttB | 196 | 66 | CCG (P) => TCG (S) | 1 |
| 116 | mttB | 253 | 85 | CGT (R) => TGT (C) | 0.62 |
| 117 | mttB | 290 | 97 | TCT (S) => TTT (F) | 1 |
| 118 | mttB | 299 | 100 | TCG (S) => TTG (L) | 0.75 |
| 119 | ccmB | 28 | 10 | CAT (H) => TAT (Y) | 0.89 |
| 120 | ccmB | 43 | 15 | CCC (P) => TCC (S) | 0.67 |
| 121 | ccmB | 71 | 24 | CCA (P) => CTA (L) | 1 |
| 122 | ccmB | 80 | 27 | TCG (S) => TTG (L) | 1 |
| 123 | ccmB | 128 | 43 | TCA (S) => TTA (L) | 1 |
| 124 | ccmB | 137 | 46 | TCC (S) => TTC (F) | 1 |
| 125 | ccmB | 149 | 50 | CCG (P) => CTG (L) | 1 |
| 126 | ccmB | 154 | 52 | CGG (R) => TGG (W) | 1 |
| 127 | ccmB | 160 | 54 | CCT (P) => TCT (S) | 0.67 |
| 128 | ccmB | 164 | 55 | CCG (P) =>=> CTG (L) | 0.89 |
| 129 | ccmB | 172 | 58 | CCT (P) => TCT (S) | 0.89 |
| 130 | ccmB | 179 | 60 | CCT (P) => CTT (L) | 1 |
| 131 | ccmB | 193 | 65 | CCT (P) => TTT (F) | 0.89 |
| 132 | ccmB | 194 | 65 | CCT (P) => TTT (F) | 0.89 |
| 133 | ccmB | 286 | 96 | CGG (R) => TGG (W) | 1 |
| 134 | ccmB | 304 | 102 | CGT (R) => TGT (C) | 0.78 |
| 135 | ccmB | 313 | 105 | CGT (R) => TGT (C) | 0.89 |
| 136 | ccmB | 338 | 113 | CCG (P) => CTG (L) | 1 |
| 137 | ccmB | 367 | 123 | CGG (R) => TGG (W) | 0.78 |
| 138 | ccmB | 424 | 142 | CGT (R) => TGT (C) | 0.89 |
| 139 | ccmB | 428 | 143 | TCG (S) => TTG (L) | 1 |
| 140 | ccmB | 467 | 156 | TCG (S) => TTG (L) | 0.89 |
| 141 | ccmB | 476 | 159 | CCA (P) => CTA (L) | 0.89 |
| 142 | ccmB | 485 | 162 | TCA (S) => TTA (L) | 1 |
| 143 | ccmB | 494 | 165 | TCA (S) => TTA (L) | 1 |
| 144 | ccmB | 503 | 168 | CCA (P) => CTA (L) | 1 |
| 145 | ccmB | 512 | 171 | TCT (S) => TTT (F) | 1 |
| 146 | ccmB | 514 | 172 | CGT (R) => TGT (C) | 1 |
| 147 | ccmB | 551 | 184 | TCA (S) => TTA (L) | 1 |
| 148 | ccmB | 554 | 185 | TCG (S) => TTG (L) | 0.89 |
| 149 | ccmB | 566 | 189 | TCC (S) => TTC (F) | 0.78 |
| 150 | ccmB | 569 | 190 | TCT (S) => TTT (F) | 0.78 |
| 151 | ccmB | 572 | 191 | CCG (P) => CTG (L) | 1 |
| 152 | ccmB | 596 | 199 | TCG (S) => TTG (L) | 0.89 |
| 153 | rpl10 | 101 | 34 | TCG (S) => TTG (L) | 0.83 |
| 154 | rpl10 | 239 | 80 | TCG (S) => TTG (L) | 0.83 |
| 155 | rpl10 | 314 | 105 | TCA (S) => TTA (L) | 0.83 |
| 156 | rps7 | 152 | 51 | CCA (P) => CTA (L) | 0.75 |
| 157 | rps7 | 343 | 115 | CAC (H) => TAC (Y) | 0.62 |
| 158 | rps7 | 368 | 123 | TCA (S) => TTA (L) | 0.88 |
| 159 | atp1 | 1039 | 347 | CCC (P) => TCC (S) | 1 |
| 160 | atp1 | 1064 | 355 | TCG (S) => TTG (L) | 1 |
| 161 | atp1 | 1178 | 393 | TCA (S) => TTA (L) | 0.9 |
| 162 | atp1 | 1216 | 406 | CTT (L) => TTT (F) | 1 |
| 163 | atp1 | 1292 | 431 | CCG (P) => CTG (L) | 0.8 |
| 164 | atp1 | 1415 | 472 | CCA (P) => CTA (L) | 1 |
| 165 | atp1 | 1490 | 497 | CCA (P) => CTA (L) | 0.9 |
| 166 | atp9 | 20 | 7 | TCA (S) => TTA (L) | 1 |
| 167 | atp9 | 50 | 17 | TCA (S) => TTA (L) | 1 |
| 168 | atp9 | 82 | 28 | CTT (L) => TTT (F) | 1 |
| 169 | atp9 | 92 | 31 | TCG (S) => TTG (L) | 1 |
| 170 | atp9 | 134 | 45 | TCA (S) => TTA (L) | 1 |
| 171 | atp9 | 182 | 61 | TCG (S) => TTG (L) | 1 |
| 172 | atp9 | 191 | 64 | CCA (P) => CTA (L) | 1 |
| 173 | atp9 | 212 | 71 | TCA (S) => TTA (L) | 1 |
| 174 | atp9 | 215 | 72 | TCC (S) => TTC (F) | 1 |
| 175 | atp9 | 223 | 75 | CGA (R) => TGA (X) | 1 |
| 176 | sdh3 | 67 | 23 | CCC (P) => TCC (S) | 1 |
| 177 | sdh3 | 376 | 126 | CTC (L) => TTC (F) | 0.83 |
| 178 | rpl16 | 79 | 27 | CAG (Q) => TAG (X) | 1 |
| 179 | rpl16 | 227 | 76 | ACT (T) => ATT (I) | 1 |
| 180 | rpl16 | 355 | 119 | CTC (L) => TTC (F) | 0.89 |
| 181 | rpl16 | 524 | 175 | CCA (P) => CTA (L) | 1 |
| 182 | rpl16 | 530 | 177 | TCG (S) => TTG (L) | 0.75 |
| 183 | rps3 | 314 | 105 | CCA (P) => CTA (L) | 0.86 |
| 184 | rps3 | 647 | 216 | CCG (P) => CTG (L) | 1 |
| 185 | rps3 | 674 | 225 | CCG (P) => CTG (L) | 0.86 |
| 186 | rps3 | 785 | 262 | TCA (S) => TTA (L) | 1 |
| 187 | rps3 | 838 | 280 | CGT (R) => TGT (C) | 1 |
| 188 | rps3 | 902 | 301 | TCA (S) => TTA (L) | 0.86 |
| 189 | rps19 | 62 | 21 | TCG (S) => TTG (L) | 1 |
| 190 | rps19 | 109 | 37 | CCT (P) => TTT (F) | 1 |
| 191 | rps19 | 110 | 37 | CCT (P) => TTT (F) | 1 |
| 192 | rpl2 | 215 | 72 | CCA (P) => CTA (L) | 0.75 |
| 193 | rpl2 | 329 | 110 | CCA (P) => CTA (L) | 1 |
| 194 | rpl2 | 494 | 165 | GCG (A) => GTG (V) | 0.67 |
| 195 | rpl2 | 517 | 173 | CTC (L) => TTC (F) | 1 |
| 196 | rpl2 | 550 | 184 | CCC (P) => TCC (S) | 1 |
| 197 | atp8 | 47 | 16 | TCA (S) => TTA (L) | 1 |
| 198 | atp8 | 58 | 20 | CTC (L) => TTC (F) | 1 |
| 199 | atp8 | 452 | 151 | CCA (P) => CTA (L) | 0.75 |
| 200 | cox3 | 289 | 97 | CTT (L) => TTT (F) | 0.92 |
| 201 | cox3 | 304 | 102 | CGG (R) => TGG (W) | 1 |
| 202 | cox3 | 311 | 104 | TCT (S) => TTT (F) | 0.92 |
| 203 | cox3 | 314 | 105 | TCT (S) => TTT (F) | 0.92 |
| 204 | cox3 | 419 | 140 | CCC (P) => CTC (L) | 1 |
| 205 | cox3 | 422 | 141 | CCT (P) => CTT (L) | 0.92 |
| 206 | cox3 | 512 | 171 | TCA (S) => TTA (L) | 0.75 |
| 207 | cox3 | 653 | 218 | TCG (S) => TTG (L) | 1 |
| 208 | cox3 | 754 | 252 | CGG (R) => TGG (W) | 0.92 |
| 209 | cox3 | 764 | 255 | CCA (P) => CTA (L) | 0.92 |
| 210 | sdh4 | 155 | 52 | CCA (P) => CTA (L) | 0.88 |
| 211 | sdh4 | 203 | 68 | CCA (P) => CTA (L) | 0.75 |
| 212 | sdh4 | 259 | 87 | CAT (H) => TAT (Y) | 0.88 |
| 213 | cox1 | 155 | 52 | TCT (S) => TTT (F) | 1 |
| 214 | cox1 | 167 | 56 | TCT (S) => TTT (F) | 1 |
| 215 | cox1 | 265 | 89 | CCA (P) => TCA (S) | 1 |
| 216 | cox1 | 356 | 119 | TCA (S) => TTA (L) | 1 |
| 217 | cox1 | 365 | 122 | TCT (S) => TTT (F) | 1 |
| 218 | cox1 | 428 | 143 | TCC (S) => TTC (F) | 1 |
| 219 | cox1 | 464 | 155 | TCA (S) => TTA (L) | 1 |
| 220 | cox1 | 503 | 168 | CCA (P) => CTA (L) | 1 |
| 221 | cox1 | 581 | 194 | TCT (S) => TTT (F) | 1 |
| 222 | cox1 | 628 | 210 | CGG (R) => TGG (W) | 1 |
| 223 | cox1 | 659 | 220 | CCC (P) => CTC (L) | 1 |
| 224 | cox1 | 674 | 225 | TCC (S) => TTC (F) | 1 |
| 225 | cox1 | 758 | 253 | ACA (T) => ATA (I) | 1 |
| 226 | cox1 | 773 | 258 | TCT (S) => TTT (F) | 1 |
| 227 | cox1 | 950 | 317 | TCC (S) => TTC (F) | 1 |
| 228 | cox1 | 1099 | 367 | CAC (H) => TAC (Y) | 1 |
| 229 | cox1 | 1187 | 396 | CCG (P) => CTG (L) | 0.89 |
| 230 | cox1 | 1318 | 440 | CGT (R) => TGT (C) | 0.78 |
| 231 | cox1 | 1346 | 449 | TCA (S) => TTA (L) | 1 |
| 232 | cox1 | 1402 | 468 | CCA (P) => TCA (S) | 1 |
| 233 | cox1 | 1412 | 471 | TCG (S) => TTG (L) | 1 |
| 234 | nad7 | 38 | 13 | TCG (S) => TTG (L) | 0.75 |
| 235 | nad7 | 77 | 26 | TCA (S) => TTA (L) | 1 |
| 236 | nad7 | 83 | 28 | TCA (S) => TTA (L) | 1 |
| 237 | nad7 | 137 | 46 | TCA (S) => TTA (L) | 1 |
| 238 | nad7 | 205 | 69 | CAT (H) => TAT (Y) | 1 |
| 239 | nad7 | 212 | 71 | TCA (S) => TTA (L) | 1 |
| 240 | nad7 | 277 | 93 | CGT (R) => TGT (C) | 1 |
| 241 | nad7 | 296 | 99 | TCA (S) => TTA (L) | 0.88 |
| 242 | nad7 | 305 | 102 | TCA (S) => TTA (L) | 1 |
| 243 | nad7 | 344 | 115 | TCA (S) => TTA (L) | 1 |
| 244 | nad7 | 494 | 165 | TCC (S) => TTC (F) | 1 |
| 245 | nad7 | 539 | 180 | TCA (S) => TTA (L) | 0.88 |
| 246 | nad7 | 812 | 271 | TCA (S) => TTA (L) | 0.88 |
| 247 | nad7 | 859 | 287 | CCT (P) => TCT (S) | 0.88 |
| 248 | nad7 | 943 | 315 | CGT (R) => TGT (C) | 1 |
| 249 | nad7 | 965 | 322 | TCT (S) => TTT (F) | 1 |
| 250 | nad7 | 989 | 330 | TCT (S) => TTT (F) | 1 |
| 251 | nad7 | 1010 | 337 | CCA (P) => CTA (L) | 1 |
| 252 | nad7 | 1052 | 351 | TCT (S) => TTT (F) | 1 |
| 253 | nad9 | 428 | 143 | TCC (S) => TTC (F) | 0.73 |
| 254 | nad9 | 506 | 169 | TCT (S) => TTT (F) | 0.75 |
| 255 | nad9 | 527 | 176 | CCA (P) => CTA (L) | 0.92 |
| 256 | nad9 | 581 | 194 | TCG (S) => TTG (L) | 0.92 |
| 257 | nad9 | 604 | 202 | CAT (H) => TAT (Y) | 1 |
| 258 | nad9 | 712 | 238 | CCG (P) => TCG (S) | 0.83 |
| 259 | nad9 | 742 | 248 | CGG (R) => TGG (W) | 1 |
| 260 | nad9 | 782 | 261 | TCC (S) => TTC (F) | 1 |
| 261 | nad9 | 812 | 271 | TCA (S) => TTA (L) | 1 |
| 262 | nad9 | 853 | 285 | CTT (L) => TTT (F) | 1 |
| 263 | nad9 | 953 | 318 | TCT (S) => TTT (F) | 1 |
| 264 | nad4L | 11 | 4 | TCT (S) => TTT (F) | 1 |
| 265 | nad4L | 17 | 6 | TCA (S) => TTA (L) | 1 |
| 266 | nad4L | 25 | 9 | CGG (R) => TGG (W) | 1 |
| 267 | nad4L | 56 | 19 | CCT (P) => CTT (L) | 1 |
| 268 | nad4L | 65 | 22 | TCA (S) => TTA (L) | 1 |
| 269 | nad4L | 70 | 24 | CCA (P) => TCA (S) | 1 |
| 270 | nad4L | 80 | 27 | TCA (S) => TTA (L) | 1 |
| 271 | nad4L | 101 | 34 | TCG (S) => TTG (L) | 0.88 |
| 272 | nad4L | 128 | 43 | TCG (S) => TTG (L) | 1 |
| 273 | nad4L | 149 | 50 | TCA (S) => TTA (L) | 0.75 |
| 274 | nad4L | 158 | 53 | TCA (S) => TTA (L) | 0.88 |
| 275 | nad4L | 167 | 56 | CCA (P) => CTA (L) | 0.88 |
| 276 | nad4L | 200 | 67 | TCA (S) => TTA (L) | 1 |
| 277 | nad4L | 251 | 84 | TCT (S) => TTT (F) | 0.88 |
| 278 | atp4 | 71 | 24 | TCA (S) => TTA (L) | 1 |
| 279 | atp4 | 89 | 30 | TCA (S) => TTA (L) | 1 |
| 280 | atp4 | 118 | 40 | CGT (R) => TGT (C) | 0.71 |
| 281 | atp4 | 215 | 72 | TCG (S) => TTG (L) | 1 |
| 282 | atp4 | 248 | 83 | CCT (P) => CTT (L) | 1 |
| 283 | atp4 | 395 | 132 | TCA (S) => TTA (L) | 1 |
| 284 | atp4 | 407 | 136 | CCA (P) => CTA (L) | 0.71 |
| 285 | atp4 | 416 | 139 | ACT (T) => ATT (I) | 0.86 |
| 286 | ccmC | 76 | 26 | CGG (R) => TGG (W) | 0.78 |
| 287 | ccmC | 103 | 35 | CAT (H) => TAT (Y) | 1 |
| 288 | ccmC | 115 | 39 | CGG (R) => TGG (W) | 0.78 |
| 289 | ccmC | 133 | 45 | CTT (L) => TTT (F) | 0.67 |
| 290 | ccmC | 161 | 54 | CCG (P) => CTG (L) | 0.78 |
| 291 | ccmC | 179 | 60 | GCG (A) => GTG (V) | 0.78 |
| 292 | ccmC | 184 | 62 | CGG (R) => TGG (W) | 1 |
| 293 | ccmC | 299 | 100 | TCT (S) => TTT (F) | 1 |
| 294 | ccmC | 331 | 111 | CGG (R) => TGG (W) | 1 |
| 295 | ccmC | 395 | 132 | TCG (S) => TTG (L) | 1 |
| 296 | ccmC | 400 | 134 | CTT (L) => TTT (F) | 0.89 |
| 297 | ccmC | 421 | 141 | CGT (R) => TGT (C) | 0.78 |
| 298 | ccmC | 436 | 146 | CCT (P) => TCT (S) | 0.89 |
| 299 | ccmC | 446 | 149 | CCG (P) => CTG (L) | 0.78 |
| 300 | ccmC | 451 | 151 | CCT (P) => TCT (S) | 1 |
| 301 | ccmC | 458 | 153 | TCA (S) => TTA (L) | 0.78 |
| 302 | ccmC | 463 | 155 | CGT (R) => TGT (C) | 1 |
| 303 | ccmC | 467 | 156 | GCT (A) => GTT (V) | 0.78 |
| 304 | ccmC | 473 | 158 | CCG (P) => CTG (L) | 1 |
| 305 | ccmC | 497 | 166 | TCT (S) => TTT (F) | 1 |
| 306 | ccmC | 521 | 174 | TCG (S) => TTG (L) | 1 |
| 307 | ccmC | 548 | 183 | TCT (S) => TTT (F) | 1 |
| 308 | ccmC | 568 | 190 | CCT (P) => TCT (S) | 1 |
| 309 | ccmC | 575 | 192 | CCC (P) => CTC (L) | 1 |
| 310 | ccmC | 605 | 202 | TCC (S) => TTC (F) | 1 |
| 311 | ccmC | 608 | 203 | CCC (P) => CTC (L) | 0.89 |
| 312 | ccmC | 614 | 205 | TCA (S) => TTA (L) | 0.78 |
| 313 | ccmC | 619 | 207 | CGT (R) => TGT (C) | 0.78 |
| 314 | ccmC | 650 | 217 | CCT (P) => CTT (L) | 0.78 |
| 315 | ccmC | 656 | 219 | CCA (P) => CTA (L) | 0.89 |
| 316 | ccmC | 673 | 225 | CCT (P) => TCT (S) | 0.78 |
| 317 | cox2 | 71 | 24 | TCT (S) => TTT (F) | 1 |
| 318 | cox2 | 161 | 54 | TCA (S) => TTA (L) | 0.95 |
| 319 | cox2 | 163 | 55 | CGG (R) => TGG (W) | 1 |
| 320 | cox2 | 253 | 85 | CGG (R) => TGG (W) | 1 |
| 321 | cox2 | 278 | 93 | CCG (P) => CTG (L) | 1 |
| 322 | cox2 | 379 | 127 | CGG (R) => TGG (W) | 1 |
| 323 | cox2 | 443 | 148 | ACG (T) => ATG (M) | 1 |
| 324 | cox2 | 461 | 154 | CCA (P) => CTA (L) | 1 |
| 325 | cox2 | 476 | 159 | TCA (S) => TTA (L) | 1 |
| 326 | cox2 | 544 | 182 | CCT (P) => TCT (S) | 1 |
| 327 | cox2 | 557 | 186 | CCT (P) => CTT (L) | 1 |
| 328 | cox2 | 581 | 194 | TCA (S) => TTA (L) | 1 |
| 329 | cox2 | 632 | 211 | TCG (S) => TTG (L) | 0.84 |
| 330 | cox2 | 698 | 233 | ACG (T) => ATG (M) | 1 |
| 331 | cox2 | 742 | 248 | CGG (R) => TGG (W) | 1 |
| 332 | rps13 | 5 | 2 | TCA (S) => TTA (L) | 0.6 |
| 333 | rps13 | 26 | 9 | TCA (S) => TTA (L) | 0.9 |
| 334 | rps13 | 56 | 19 | TCA (S) => TTA (L) | 0.9 |
| 335 | rps13 | 100 | 34 | CGT (R) => TGT (C) | 0.9 |
| 336 | rps13 | 287 | 96 | TCG (S) => TTG (L) | 1 |
| 337 | rps4 | 133 | 45 | CCG (P) => TCG (S) | 0.67 |
| 338 | rps4 | 164 | 55 | TCA (S) => TTA (L) | 1 |
| 339 | rps4 | 184 | 62 | CCC (P) => TCC (S) | 0.83 |
| 340 | rps4 | 193 | 65 | CAT (H) => TAT (Y) | 1 |
| 341 | rps4 | 257 | 86 | CCA (P) => CTA (L) | 1 |
| 342 | rps4 | 266 | 89 | CCA (P) => CTA (L) | 0.83 |
| 343 | rps4 | 278 | 93 | TCG (S) => TTG (L) | 0.67 |
| 344 | rps4 | 290 | 97 | CCG (P) => CTG (L) | 0.83 |
| 345 | rps4 | 335 | 112 | CCG (P) => CTG (L) | 1 |
| 346 | rps4 | 482 | 161 | TCA (S) => TTA (L) | 1 |
| 347 | rps4 | 914 | 305 | TCG (S) => TTG (L) | 0.83 |
| 348 | rps4 | 925 | 309 | CAT (H) => TAT (Y) | 0.83 |
| 349 | rps4 | 935 | 312 | CCA (P) => CTA (L) | 0.67 |
| 350 | rps4 | 950 | 317 | TCT (S) => TTT (F) | 1 |
| 351 | rps4 | 1001 | 334 | CCA (P) => CTA (L) | 0.83 |
| 352 | rps4 | 1010 | 337 | CCT (P) => CTT (L) | 1 |
| 353 | rps4 | 1015 | 339 | CGG (R) => TGG (W) | 1 |
| 354 | nad1 | 8 | 3 | CCT (P) => CTT (L) | 0.9 |
| 355 | nad1 | 65 | 22 | TCC (S) => TTC (F) | 1 |
| 356 | nad1 | 100 | 34 | CCT (P) => TCT (S) | 0.9 |
| 357 | nad1 | 149 | 50 | GCG (A) => GTG (V) | 0.9 |
| 358 | nad1 | 209 | 70 | TCC (S) => TTC (F) | 1 |
| 359 | nad1 | 308 | 103 | TCA (S) => TTA (L) | 1 |
| 360 | nad1 | 434 | 145 | ACT (T) => ATT (I) | 1 |
| 361 | nad6 | 7 | 3 | CTT (L) => TTT (F) | 1 |
| 362 | nad6 | 83 | 28 | TCG (S) => TTG (L) | 1 |
| 363 | nad6 | 88 | 30 | CCC (P) => TTC (F) | 0.7 |
| 364 | nad6 | 89 | 30 | CCC (P) => TTC (F) | 0.7 |
| 365 | nad6 | 95 | 32 | CCA (P) => CTA (L) | 1 |
| 366 | nad6 | 103 | 35 | CGC (R) => TGC (C) | 1 |
| 367 | nad6 | 161 | 54 | CCA (P) => CTA (L) | 1 |
| 368 | nad6 | 169 | 57 | CAT (H) => TAT (Y) | 1 |
| 369 | nad6 | 191 | 64 | TCA (S) => TTA (L) | 1 |
| 370 | nad6 | 446 | 149 | TCC (S) => TTC (F) | 1 |
| 371 | nad6 | 463 | 155 | CCT (P) => TCT (S) | 0.8 |
| 372 | nad6 | 569 | 190 | TCT (S) => TTT (F) | 1 |
| 373 | nad2 | 26 | 9 | TCC (S) => TTC (F) | 0.89 |
| 374 | nad2 | 203 | 68 | TCT (S) => TTT (F) | 0.67 |
| 375 | nad2 | 206 | 69 | TCC (S) => TTC (F) | 1 |
| 376 | nad2 | 230 | 77 | TCT (S) => TTT (F) | 1 |
| 377 | nad2 | 236 | 79 | TCC (S) => TTC (F) | 0.67 |
| 378 | nad2 | 251 | 84 | CCA (P) => CTA (L) | 1 |
| 379 | nad2 | 262 | 88 | CGC (R) => TGC (C) | 1 |
| 380 | nad2 | 289 | 97 | CAT (H) => TAT (Y) | 1 |
| 381 | nad2 | 296 | 99 | TCA (S) => TTA (L) | 1 |
| 382 | nad2 | 323 | 108 | CCT (P) => CTT (L) | 1 |
| 383 | nad2 | 392 | 131 | TCG (S) => TTG (L) | 1 |
| 384 | rps12 | 71 | 24 | TCG (S) => TTG (L) | 0.94 |
| 385 | rps12 | 100 | 34 | CGC (R) => TGC (C) | 1 |
| 386 | rps12 | 104 | 35 | CCG (P) => CTG (L) | 1 |
| 387 | rps12 | 196 | 66 | CAC (H) => TAC (Y) | 0.94 |
| 388 | rps12 | 221 | 74 | TCG (S) => TTG (L) | 0.88 |
| 389 | rps12 | 269 | 90 | TCG (S) => TTG (L) | 0.94 |
| 390 | rps12 | 284 | 95 | TCC (S) => TTC (F) | 0.76 |
| 391 | nad3 | 5 | 2 | TCA (S) => TTA (L) | 0.79 |
| 392 | nad3 | 44 | 15 | CCG (P) => CTG (L) | 1 |
| 393 | nad3 | 62 | 21 | CCA (P) => CTA (L) | 0.95 |
| 394 | nad3 | 80 | 27 | CCA (P) => CTA (L) | 1 |
| 395 | nad3 | 146 | 49 | TCC (S) => TTC (F) | 1 |
| 396 | nad3 | 208 | 70 | CCT (P) => TTT (F) | 0.95 |
| 397 | nad3 | 209 | 70 | CCT (P) => TTT (F) | 0.95 |
| 398 | nad3 | 215 | 72 | CCG (P) => CTG (L) | 1 |
| 399 | nad3 | 230 | 77 | TCC (S) => TTC (F) | 0.86 |
| 400 | nad3 | 247 | 83 | CCT (P) => TCT (S) | 1 |
| 401 | nad3 | 251 | 84 | CCC (P) => CTC (L) | 0.91 |
| 402 | nad3 | 266 | 89 | CCG (P) => CTG (L) | 1 |
| 403 | nad3 | 275 | 92 | TCT (S) => TTT (F) | 1 |
| 404 | nad3 | 317 | 106 | TCT (S) => TTT (F) | 0.95 |
| 405 | nad3 | 344 | 115 | TCG (S) => TTG (L) | 1 |
| 406 | nad3 | 349 | 117 | CGG (R) => TGG (W) | 1 |
| 407 | rps1 | 23 | 8 | CCT (P) => CTT (L) | 0.67 |
| 408 | rps1 | 56 | 19 | CCT (P) => CTT (L) | 0.67 |
| 409 | rps1 | 380 | 127 | TCA (S) => TTA (L) | 0.67 |
*The cutoff score (C-value) was set to 0.6.
Online-only Table 5.
Predicted RNA-editing sites in Scaffold 2 of the C. sinensis var. assamica mt genome.
| No. | Gene | Nucleotide Position | AA Pos | Effect | Score* |
|---|---|---|---|---|---|
| 1 | rps19 | 116 | 39 | TCG (S) => TTG (L) | 1 |
| 2 | rps19 | 163 | 55 | CCT (P) => TTT (F) | 1 |
| 3 | rps19 | 164 | 55 | CCT (P) => TTT (F) | 1 |
| 4 | atp9 | 53 | 18 | TCA (S) => TTA (L) | 1 |
| 5 | atp9 | 83 | 28 | TCA (S) => TTA (L) | 1 |
| 6 | cob | 118 | 40 | CCG (P) => TCG (S) | 0.92 |
| 7 | cob | 178 | 60 | CAC (H) => TAC (Y) | 1 |
| 8 | cob | 286 | 96 | CTC (L) => TTC (F) | 1 |
| 9 | cob | 298 | 100 | CAC (H) => TAC (Y) | 1 |
| 10 | cob | 325 | 109 | CAT (H) => TAT (Y) | 1 |
| 11 | cob | 358 | 120 | CGG (R) => TGG (W) | 1 |
| 12 | cob | 419 | 140 | CCA (P) => CTA (L) | 1 |
| 13 | cob | 568 | 190 | CAT (H) => TAT (Y) | 0.92 |
| 14 | cob | 680 | 227 | TCT (S) => TTT (F) | 1 |
| 15 | cob | 808 | 270 | CCC (P) => TCC (S) | 1 |
| 16 | cob | 853 | 285 | CAT (H) => TAT (Y) | 1 |
| 17 | cob | 908 | 303 | CCA (P) => CTA (L) | 1 |
| 18 | cob | 914 | 305 | TCT (S) => TTT (F) | 1 |
| 19 | cob | 982 | 328 | CAC (H) => TAC (Y) | 0.85 |
| 20 | cob | 1015 | 339 | CGC (R) => TGC (C) | 1 |
| 21 | cob | 1084 | 362 | CCT (P) => TCT (S) | 1 |
| 22 | cob | 1124 | 375 | CCG (P) => CTG (L) | 1 |
| 23 | rps14 | 47 | 16 | GCG (A) => GTG (V) | 0.6 |
| 24 | rps14 | 271 | 91 | CCT (P) => TCT (S) | 0.6 |
| 25 | rpl5 | 35 | 12 | TCA (S) => TTA (L) | 0.78 |
| 26 | rpl5 | 47 | 16 | CCG (P) => CTG (L) | 1 |
| 27 | rpl5 | 59 | 20 | CCG (P) => CTG (L) | 0.89 |
| 28 | rpl5 | 64 | 22 | CAC (H) => TAC (Y) | 1 |
| 29 | rpl5 | 92 | 31 | TCG (S) => TTG (L) | 1 |
| 30 | rpl5 | 172 | 58 | CGC (R) => TGC (C) | 0.89 |
| 31 | rpl5 | 518 | 173 | CCA (P) => CTA (L) | 0.89 |
| 32 | rpl5 | 521 | 174 | CCG (P) => CTG (L) | 1 |
| 33 | nad2 | 110 | 37 | TCT (S) => TTT (F) | 1 |
| 34 | nad2 | 125 | 42 | TCC (S) => TTC (F) | 1 |
| 35 | nad2 | 272 | 91 | TCT (S) => TTT (F) | 0.67 |
| 36 | nad2 | 284 | 95 | TCA (S) => TTA (L) | 1 |
| 37 | nad2 | 293 | 98 | TCT (S) => TTT (F) | 1 |
| 38 | nad2 | 412 | 138 | CAT (H) => TAT (Y) | 1 |
| 39 | nad2 | 442 | 148 | CGT (R) => TGT (C) | 0.78 |
| 40 | nad2 | 446 | 149 | ACT (T) => ATT (I) | 1 |
| 41 | nad2 | 512 | 171 | TCA (S) => TTA (L) | 0.78 |
| 42 | nad2 | 542 | 181 | TCA (S) => TTA (L) | 1 |
| 43 | nad2 | 611 | 204 | TCG (S) => TTG (L) | 1 |
| 44 | nad2 | 731 | 244 | CCA (P) => CTA (L) | 0.67 |
| 45 | nad2 | 760 | 254 | CGT (R) => TGT (C) | 1 |
| 46 | nad2 | 932 | 311 | TCA (S) => TTA (L) | 0.67 |
| 47 | nad2 | 941 | 314 | CCA (P) => CTA (L) | 1 |
| 48 | nad2 | 989 | 330 | TCA (S) => TTA (L) | 1 |
| 49 | sdh3 | 67 | 23 | CCA (P) => TCA (S) | 1 |
| 50 | sdh3 | 74 | 25 | TCC (S) => TTC (F) | 1 |
| 51 | ccmFc | 38 | 13 | TCC (S) => TTC (F) | 0.83 |
| 52 | ccmFc | 50 | 17 | CCT (P) => CTT (L) | 1 |
| 53 | ccmFc | 52 | 18 | CGT (R) => TGT (C) | 1 |
| 54 | ccmFc | 103 | 35 | CCC (P) => TCC (S) | 1 |
| 55 | ccmFc | 119 | 40 | TCT (S) => TTT (F) | 1 |
| 56 | ccmFc | 122 | 41 | TCC (S) => TTC (F) | 1 |
| 57 | ccmFc | 146 | 49 | CCT (P) => CTT (L) | 1 |
| 58 | ccmFc | 151 | 51 | CCT (P) => TCT (S) | 0.83 |
| 59 | ccmFc | 155 | 52 | TCA (S) => TTA (L) | 1 |
| 60 | ccmFc | 160 | 54 | CCT (P) => TCT (S) | 0.67 |
| 61 | ccmFc | 203 | 68 | ACG (T) => ATG (M) | 1 |
| 62 | ccmFc | 305 | 102 | TCA (S) => TTA (L) | 0.83 |
| 63 | ccmFc | 391 | 131 | CGT (R) => TGT (C) | 1 |
| 64 | ccmFc | 406 | 136 | CGT (R) => TGT (C) | 0.83 |
| 65 | ccmFc | 620 | 207 | GCG (A) => GTG (V) | 1 |
| 66 | ccmFc | 704 | 235 | GCT (A) => GTT (V) | 0.83 |
| 67 | ccmFc | 1100 | 367 | CCA (P) => CTA (L) | 1 |
| 68 | ccmFc | 1121 | 374 | TCG (S) => TTG (L) | 1 |
| 69 | ccmFc | 1276 | 426 | CGA (R) => TGA (X) | 1 |
*The cutoff score (C-value) was set to 0.6.
Phylogenetic analyses
To further determine the phylogenetic position of C. sinensis var. assamica we performed phylogenomic analysis of 20 complete cp genomes using the GTR + R + I model under the maximum likelihood (ML) inference in MEGA v.7.055. Besides C. sinensis var. assamica cv. Yunkang 10, we selected cp genomes from the eighteen Camelia species (C. oleifera, C. crapnelliana, C. szechuanensis, C. mairei, C. elongata, C. grandibracteata, C. leptophylla, C. petelotii, C. pubicosta, C. reticulata, C. azalea, C. japonica, C. cuspidata, C. danzaiensis, C. impressinervis, C. pitardii, C. yunnanensis and C. taliensis) using Apterosperm oblata as outgroup. Our results showed that C. sinensis var. assamica was grouped with C. grandibracteata with 100% bootstrap support (Fig. 5).
Fig. 5.
Phylogenetic relationships of 20 complete chloroplast genomes. Maximum likelihood phylogenetic tree of C. sinensis var. assamica cv. Yunkang 10 with 18 species in the genus Camellia based on complete chloroplast genome sequences. The chloroplast sequence of Apterosperma oblata was set as outgroup. The position of C. sinensis var. assamica cv. Yunkang 10 is shown in bold and bootstrap values are shown for each node.
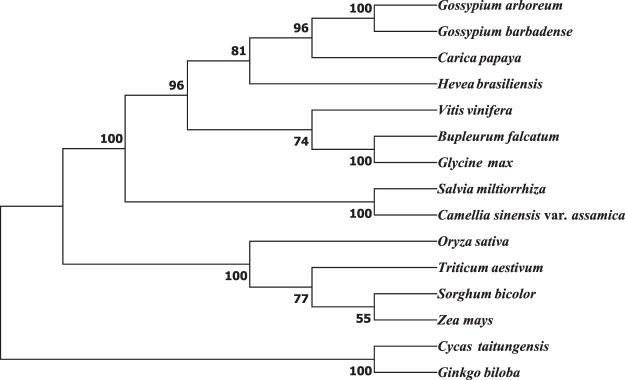
The same method was used for phylogenetic analysis with mt genome. A total of thirteen conserved mt protein-coding genes among C. sinensis var. assamica and 14 other plant species were individually aligned with ClustalW56, and then concatenated to construct a contiguous sequence in the order of cob, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, nad7 and nad9. The selected 14 species includes Cycas taitungensis, Ginkgo biloba, Triticum aestivum, Oryza sativa, Sorghum bicolor, Zea mays, Gossypium arboretum, G. barbadense, Carica papaya, Vitis vinifera, Hevea brasiliensis, Bupleurum falcatum, Glycine max and Salvia miltiorrhiza. The alignment file was used for the construction of Neighbor-Joining Tree at 1000 bootstrap replicates with MEGA 7.0.2655. Our results showed that C. sinensis var. assamica is clearly grouped with other dicots that were separated from monocots of the angiosperms while the two gymnosperms (Cycas taitungensis and Ginkgo biloba) were formed the basal clade (Fig. 6).
Fig. 6.
Phylogeny inferred from 13 genes common in the 15 plant mitochondrial genomes. Neighbor-joining tree of C. sinensis var. assamica cv. Yunkang 10 with other 14 species based on 13 conserved protein-coding gene sequences with bootstrap support values on each node. The mt sequence of Cycas taitungensis and Ginkgo biloba were set as outgroup.
Data Records
Raw reads from Illumina are deposited in the NCBI Sequence Read Archive (SRA)57–62 and BIG Genome Warehouse63. Assembled cp genome sequences and accompanying gene annotations of C. sinensis var. assamica are deposited in the NCBI GenBank64 and BIG Genome Warehouse65. The mt genome final assembly and accompanying gene annotations are deposited at NCBI GenBank66,67 and BIG Genome Warehouse68. The alignment and tree files of the chloroplast genome and mitochondrial genome form the Camellia genus were deposited in Figshare database69.
Technical Validation
Quality filtering of raw reads
The initially generated raw sequencing reads were evaluated in terms of the average quality score at each position, GC content distribution, quality distribution, base composition, and other metrics. Furthermore, the sequencing reads with low quality were also filtered out before the genome assembly and annotation of gene structure.
Assembly and validation
The chloroplast reads were filtered from whole genome Illumina sequencing data of C. sinensis var. assamica. We mapped all the cleaned reads to the reference chloroplast sequence4 using bowtie2 (version 2.3.4.3)40 with default parameters. The mapped chloroplast reads were de novo assembled into the complete chloroplast genome.
For mitochondria genome assembly, the PE and MP sequencing reads were used separately. Briefly, we first performed de novo assembly with VELVET v1.2.0841, which was previously described42,43. Scaffolds were constructed using SSPACE v.3.044. False connection was manually removed based on the coverage and distances of paired reads. Gaps between scaffolds were then filled with GapCloser (version 1.12)45,46 using all pair-end reads.
Acknowledgements
We would thank Yunnan Tea Research Institute for providing tea plant materials in this study. We are grateful An-dan Zhu for technical support and anonymous reviewers for valuable comments on the manuscript. This work was supported by the Project of Innovation Team of Yunnan Province and Ten Thousands Talents Program of China (to L. Z. Gao).
Online-only Tables
Author Contributions
Li-zhi Gao designed the study; Fen Zhang, Wei Li and Dan Zhang assembled, annotated and analyzed the mt genome; Cheng-wen Gao assembled, annotated and analyzed the cp genome; Fen Zhang, Wei Li and Cheng-wen Gao drafted the manuscript; Li-zhi Gao revised the manuscript.
Code Availability
The following bioinformatic tools and versions were used for generating all results as described in the main text:
1. Bowtie2, version 2.3.4.3, was used for aligning sequencing reads to long reference sequences with default parameters: http://bowtie-bio.sourceforge.net/bowtie2/index.shtml
2. CLC Genomics Workbench, version 3.6.1, was used for genome assembly with default parameters: https://www.qiagenbioinformatics.com/products/clc-genomics-workbench/
3. Velvet, version 1.2.08, was used for genome de novo assembly, which was previously described: https://www.ebi.ac.uk/~zerbino/velvet/
4. SSPACE, version 3.0, was used for genome scaffolds assembly with default parameters: https://www.baseclear.com/services/bioinformatics/basetools/sspace-standard/
5. GapCloser, version 1.12, was used to fill the gaps between scaffolds with default parameters: https://sourceforge.net/projects/soapdenovo2/files/GapCloser/
6. DOGMA (an online tool), accessed at 12/2018, was used for annotating cp genomes with default parameters: http://dogma.ccbb.utexas.edu/
7. Mitofy (an online tool), accessed at 12/2018, was used for annotating plant mt genomes with default parameters: http://dogma.ccbb.utexas.edu/mitofy/
8. tRNAscanSE, VERSION 1.3.1, was used to search tRNA with default parameters: http://lowelab.ucsc.edu/tRNAscan-SE/
9. Organellar Genome DRAW (an online tool), accessed at 12/2018, was used for creating high quality visual representation of cp gemome with default parameters: https://chlorobox.mpimp-golm.mpg.de/OGDraw.html
10. MISA,version 1.0, was used for annotating SSR with monomer (one nucleotide, n ≥ 8), dimer (two nucleotides, n ≥ 4), trimer (three nucleotides, n ≥ 4), tetramer (four nucleotides, n ≥ 3), pentamer (five nucleotides, n ≥ 3), hexamer (six nucleotides, n ≥ 3): http://pgrc.ipk-gatersleben.de/misa/misa.html
11. REPuter (an online tool), accessed at 1/2019, was used for annotating long repeated sequences with the following parameters: minimal length 50 nt; mis match 3 nt: https://bibiserv.cebitec.uni-bielefeld.de/reputer/
12. PREP-cp (an online tool), accessed at 1/2019, was used for predicting RNA editor for plant cp genes with the cutoff score (C-value) setting to 0.8: http://prep.unl.edu/
13. PREP-mt (an online tool), accessed at 1/2019, was used for predicting RNA editor for plant mt genes with the cutoff score (C-value) setting to 0.6: http://prep.unl.edu/
14. MEGA, version 7.0.26, was used for phylogenomics and phylomedicine at 1000 bootstrap: https://www.megasoftware.net/
15. ClustalW, version 2, was used for multiple sequence alignment with default parameters: https://www.ebi.ac.uk/Tools/msa/clustalw2/
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Fen Zhang, Wei Li and Cheng-wen Gao.
References
- 1.Mondal TK, Bhattacharya A, Laxmikumaran M, Singh Ahuja P. Recent Advances of Tea (Camellia Sinensis) Biotechnology. Plant Cell, Tissue and Organ Culture. 2004;76:195–254. doi: 10.1023/B:TICU.0000009254.87882.71. [DOI] [Google Scholar]
- 2.Banerjee B. Tea. Dordrecht: Springer Netherlands; 1992. Botanical classification of tea; pp. 25–51. [Google Scholar]
- 3.Ming, T. & Bartholomew, B. Theaceae. In Flora of China. (Beijing and St. Louis: Science Press and Missouri Botanical Garden, 2007).
- 4.Huang H, Shi C, Liu Y, Mao SY, Gao LZ. Thirteen Camellia Chloroplast Genome Sequences Determined by High-Throughput Sequencing: Genome Structure and Phylogenetic Relationships. BMC Evol Biol. 2014;14:151. doi: 10.1186/1471-2148-14-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu H, Jiang W, Ghiassi M, Lee S, Nitin M. Classification of Camellia (Theaceae) Species Using Leaf Architecture Variations and Pattern Recognition Techniques. PloS one. 2012;7:e29704. doi: 10.1371/journal.pone.0029704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mccauley DE, Stevens JE, Peroni PA, Raveill JA. The Spatial Distribution of Chloroplast DNA and Allozyme Polymorphisms within a Population of Silene alba (Caryophyllaceae) American Journal of Botany. 1996;83:727–731. doi: 10.1002/j.1537-2197.1996.tb12761.x. [DOI] [Google Scholar]
- 7.Small RL, Wendel RCCJ. Use of Nuclear Genes for Phylogeny Reconstruction in Plants. Australian Systematic Botany. 2004;17:145–170. doi: 10.1071/SB03015. [DOI] [Google Scholar]
- 8.Jansen RK, et al. Analysis of 81 Genes From 64 Plastid Genomes Resolves Relationships in Angiosperms and Identifies Genome-Scale Evolutionary Patterns. Proceedings of the National Academy of Sciences. 2007;104:19369. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parks M, Cronn R, Liston A. Increasing Phylogenetic Resolution at Low Taxonomic Levels Using Massively Parallel Sequencing of Chloroplast Genomes. Bmc Biology. 2009;7:84. doi: 10.1186/1741-7007-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE. Phylogenetic Analysis of 83 Plastid Genes Further Resolves the Early Diversification of Eudicots. Proceedings of the National Academy of Sciences. 2010;107:4623. doi: 10.1073/pnas.0907801107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richly E, Leister D. NUPTs in Sequenced Eukaryotes and their Genomic Organization in Relation to NUMTs. Molecular Biology and Evolution. 2004;21:1972–1980. doi: 10.1093/molbev/msh210. [DOI] [PubMed] [Google Scholar]
- 12.Schuster W, Brennicke A. Plastid, Nuclear and Reverse Transcriptase Sequences in the Mitochondrial Genome of Oenothera: Is Genetic Information Transferred Between Organelles Via RNA? EMBO J. 1987;6:2857–2863. doi: 10.1002/j.1460-2075.1987.tb02587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stern DB, Lonsdale DM. Mitochondrial and Chloroplast Genomes of Maize Have a 12-Kilobase DNA Sequence in Common. Nature. 1982;299:698–702. doi: 10.1038/299698a0. [DOI] [PubMed] [Google Scholar]
- 14.Vaughn JC, Mason MT, Sper-Whitis GL, Kuhlman P, Palmer JD. Fungal Origin by Horizontal Transfer of a Plant Mitochondrial Group I Intron in the Chimeric CoxI Gene of Peperomia. Journal of molecular evolution. 1995;41:563. doi: 10.1007/BF00175814. [DOI] [PubMed] [Google Scholar]
- 15.Alverson AJ, et al. Insights Into the Evolution of Mitochondrial Genome Size From Complete Sequences of Citrullus Lanatus and Cucurbita Pepo (Cucurbitaceae) Mol Biol Evol. 2010;27:1436–1448. doi: 10.1093/molbev/msq029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ward BL, Anderson RS, Bendich AJ. The Mitochondrial Genome is Large and Variable in a Family of Plants (Cucurbitaceae) Cell. 1981;25:793–803. doi: 10.1016/0092-8674(81)90187-2. [DOI] [PubMed] [Google Scholar]
- 17.Sloan DB, et al. Rapid Evolution of Enormous, Multichromosomal Genomes in Flowering Plant Mitochondria with Exceptionally High Mutation Rates. PLoS Biol. 2012;10:e1001241. doi: 10.1371/journal.pbio.1001241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmer JD, Herbon LA. Plant Mitochondrial DNA Evolves Rapidly in Structure, but Slowly in Sequence. J Mol Evol. 1988;28:87–97. doi: 10.1007/BF02143500. [DOI] [PubMed] [Google Scholar]
- 19.Marechal A, Brisson N. Recombination and the Maintenance of Plant Organelle Genome Stability. New Phytol. 2010;186:299–317. doi: 10.1111/j.1469-8137.2010.03195.x. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Q, et al. The Complete Chloroplast Genome Sequence of Camellia Mingii (Theaceae), a Critically Endangered Yellow Camellia Species Endemic to China. Mitochondrial DNA Part B. 2019;4:1338–1340. doi: 10.1080/23802359.2019.1596765. [DOI] [Google Scholar]
- 21.Lin Y, et al. Characterization of the Complete Chloroplast Genome of Camellia Renshanxiangiae (Theaceae) Mitochondrial DNA Part B. 2019;4:1490–1491. doi: 10.1080/23802359.2019.1601041. [DOI] [Google Scholar]
- 22.Li W, Zhang C, Guo X, Liu Q, Wang K. Complete Chloroplast Genome of Camellia Japonica Genome Structures, Comparative and Phylogenetic Analysis. PLOS ONE. 2019;14:e216645. doi: 10.1371/journal.pone.0216645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park J, et al. The Complete Chloroplast Genome of Common Camellia Tree, Camellia Japonica L. (Theaceae), Adapted to Cold Environment in Korea. Mitochondrial DNA Part B. 2019;4:1038–1040. doi: 10.1080/23802359.2019.1580164. [DOI] [Google Scholar]
- 24.Park J, et al. The Complete Chloroplast Genome of Common Camellia Tree in Jeju Island, Korea, Camellia Japonica L. (Theaceae): Intraspecies Variations On Common Camellia Chloroplast Genomes. Mitochondrial DNA Part B. 2019;4:1292–1293. doi: 10.1080/23802359.2019.1591214. [DOI] [Google Scholar]
- 25.Li W, et al. Characterization of the Complete Chloroplast Genome of Camellia Granthamiana (Theaceae), a Vulnerable Species Endemic to China. Mitochondrial DNA Part B. 2018;3:1139–1140. doi: 10.1080/23802359.2018.1521310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Meng-Meng, Cao Ze-Peng, Zhang Jun, Zhang Da-Wei, Huo Xiao-Wei, Zhang Gang. Characterization of the complete chloroplast genome of the Camellia nitidissima, an endangered and medicinally important tree species endemic to Southwest China. Mitochondrial DNA Part B. 2018;3(2):884–885. doi: 10.1080/23802359.2018.1501304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, Han Y. The Complete Chloroplast Genome Sequence of Endangered Camellias (Camellia Pubifurfuracea) Conservation Genetics Resources. 2018;10:843–845. doi: 10.1007/s12686-017-0944-5. [DOI] [Google Scholar]
- 28.Dong M, et al. The Complete Chloroplast Genome of an Economic Plant, Camellia Sinensis Cultivar Anhua, China. Mitochondrial DNA Part B. 2018;3:558–559. doi: 10.1080/23802359.2018.1462124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W, Xing F, Ng WL, Zhou Y, Shi X. The Complete Chloroplast Genome Sequence of Camellia Ptilophylla (Theaceae): A Natural Caffeine-Free Tea Plant Endemic to China. Mitochondrial DNA Part B. 2018;3:426–427. doi: 10.1080/23802359.2018.1457996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, Han Y. The Complete Chloroplast Genome Sequence of Camellias (Camellia Fangchengensis) Mitochondrial DNA Part B. 2018;3:34–35. doi: 10.1080/23802359.2017.1419086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu X, Zheng W, Wen J. The Complete Chloroplast Genome of the Long Blooming and Critically Endangered Camellia Azalea. Conservation Genetics Resources. 2018;10:5–7. doi: 10.1007/s12686-017-0749-6. [DOI] [Google Scholar]
- 32.Zhang W, Zhao Y, Yang G, Tang Y, Xu Z. Characterization of the Complete Chloroplast Genome Sequence of Camellia Oleifera in Hainan, China. Mitochondrial DNA Part B. 2017;2:843–844. doi: 10.1080/23802359.2017.1407687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim S, Cho CH, Yang M, Kim S. The Complete Chloroplast Genome Sequence of the Japanese Camellia (Camellia Japonica L.) Mitochondrial DNA Part B. 2017;2:583–584. doi: 10.1080/23802359.2017.1372719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang G, Luo Y, Hou N, Deng L. The Complete Chloroplast Genomes of Three Rare and Endangered Camellias (Camellia Huana, C. Liberofilamenta and C. Luteoflora) Endemic to Southwest China. Conservation Genetics Resources. 2017;9:583–585. doi: 10.1007/s12686-017-0727-z. [DOI] [Google Scholar]
- 35.Tong Y, Wu C, Gao L. Characterization of Chloroplast Microsatellite Loci From Whole Chloroplast Genome of Camellia Taliensis and their Utilization for Evaluating Genetic Diversity of Camellia Reticulata (Theaceae) Biochemical Systematics and Ecology. 2013;50:207–211. doi: 10.1016/j.bse.2013.04.003. [DOI] [Google Scholar]
- 36.Yang JB, Yang SX, Li HT, Yang J, Li DZ. Comparative Chloroplast Genomes of Camellia Species. PLoS One. 2013;8:e73053. doi: 10.1371/journal.pone.0073053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaundun SS, Matsumoto S. Molecular Evidence for Maternal Inheritance of the Chloroplast Genome in Tea, Camellia Sinensis (L.) O. Kuntze. Journal of the Science of Food and Agriculture. 2011;91:2660–2663. doi: 10.1002/jsfa.4508. [DOI] [PubMed] [Google Scholar]
- 38.Xia E, et al. The Tea Tree Genome Provides Insights into Tea Flavor and Independent Evolution of Caffeine Biosynthesis. Molecular Plant. 2017;10:866–877. doi: 10.1016/j.molp.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Porebski S, Bailey LG, Baum BR. Modification of a CTAB DNA Extraction Protocol for Plants Containing High Polysaccharide and Polyphenol Components. Plant Molecular Biology Reporter. 1997;15:8–15. doi: 10.1007/BF02772108. [DOI] [Google Scholar]
- 40.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and Memory-Efficient Alignment of Short DNA Sequences to the Human Genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zerbino DR, Birney E. Velvet: Algorithms for De Novo Short Read Assembly Using De Bruijn Graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu A, Guo W, Jain K, Mower JP. Unprecedented Heterogeneity in the Synonymous Substitution Rate within a Plant Genome. Mol Biol Evol. 2014;31:1228–1236. doi: 10.1093/molbev/msu079. [DOI] [PubMed] [Google Scholar]
- 43.Grewe F, et al. Comparative Analysis of 11 Brassicales Mitochondrial Genomes and the Mitochondrial Transcriptome of Brassica Oleracea. Mitochondrion. 2014;19 Pt B:135–143. doi: 10.1016/j.mito.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 44.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding Pre-Assembled Contigs Using SSPACE. Bioinformatics. 2011;27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 45.Nadalin F, Vezzi F, Policriti A. GapFiller: A De Novo Assembly Approach to Fill the Gap within Paired Reads. BMC Bioinformatics. 2012;13(Suppl 14):S8. doi: 10.1186/1471-2105-13-S14-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo R, et al. SOAPdenovo2: An Empirically Improved Memory-Efficient Short-Read De Novo Assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wyman SK, Jansen RK, Boore JL. Automatic Annotation of Organellar Genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- 48.Lowe TM, Eddy SR. TRNAscan-SE: A Program for Improved Detection of Transfer RNA Genes in Genomic Sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW): A Tool for the Easy Generation of High-Quality Custom Graphical Maps of Plastid and Mitochondrial Genomes. Curr Genet. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- 50.Kurtz S, et al. REPuter: The Manifold Applications of Repeat Analysis On a Genomic Scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mower JP. PREP-Mt: Predictive RNAEditor for Plant Mitochondrial Genes. BMC Bioinformatics. 2005;6:96. doi: 10.1186/1471-2105-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mower JP. The PREP Suite: Predictive RNA Editors for Plant Mitochondrial Genes, Chloroplast Genes and User-Defined Alignments. Nucleic Acids Res. 2009;37:W253–W259. doi: 10.1093/nar/gkp337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chaw SM, et al. The Mitochondrial Genome of the Gymnosperm Cycas Taitungensis Contains a Novel Family of Short Interspersed Elements, Bpu Sequences, and Abundant RNA Editing Sites. Mol Biol Evol. 2008;25:603–615. doi: 10.1093/molbev/msn009. [DOI] [PubMed] [Google Scholar]
- 54.Ward GC, Levings CR. The Protein-Encoding Gene T-urf13 is Not Edited in Maize Mitochondria. Plant Mol Biol. 1991;17:1083–1088. doi: 10.1007/BF00037148. [DOI] [PubMed] [Google Scholar]
- 55.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Larkin MA, et al. Clustal W and Clustal X Version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 57.2017. NCBI Sequence Read Archive. SRX2708522
- 58.2017. NCBI Sequence Read Archive. SRX2708523
- 59.2017. NCBI Sequence Read Archive. SRX2708528
- 60.2017. NCBI Sequence Read Archive. SRX2708529
- 61.2017. NCBI Sequence Read Archive. SRX2708545
- 62.2017. NCBI Sequence Read Archive. SRX2708546
- 63.2019. BIGD Genome Sequence Archive. http://bigd.big.ac.cn/gsa/browse/CRA001582
- 64.Gao C-W, Gao L-Z. 2018. Camellia sinensis var. assamica cultivar Yunkang 10 plastid, complete genome. GenBank. MH019307
- 65.2019. BIGD Genome Warehouse. http://bigd.big.ac.cn/search?dbId=gwh&q=GWHAAIB00000000
- 66.Zhang F. 2019. Camellia sinensis var. assamica mitochondrion, complete genome. GenBank. MK574876
- 67.Zhang F. 2019. Camellia sinensis var. assamica mitochondrion, complete genome. GenBank. MK574877
- 68.2019. BIGD Genome Warehouse. http://bigd.big.ac.cn/search?dbId=gwh&q=GWHAAIC00000000
- 69.Zhang F. 2019. Deciphering tea tree chloroplast and mitochondrial genomes of Camellia sinensis var. assamica. figshare. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- 2017. NCBI Sequence Read Archive. SRX2708522
- 2017. NCBI Sequence Read Archive. SRX2708523
- 2017. NCBI Sequence Read Archive. SRX2708528
- 2017. NCBI Sequence Read Archive. SRX2708529
- 2017. NCBI Sequence Read Archive. SRX2708545
- 2017. NCBI Sequence Read Archive. SRX2708546
- 2019. BIGD Genome Sequence Archive. http://bigd.big.ac.cn/gsa/browse/CRA001582
- Gao C-W, Gao L-Z. 2018. Camellia sinensis var. assamica cultivar Yunkang 10 plastid, complete genome. GenBank. MH019307
- 2019. BIGD Genome Warehouse. http://bigd.big.ac.cn/search?dbId=gwh&q=GWHAAIB00000000
- Zhang F. 2019. Camellia sinensis var. assamica mitochondrion, complete genome. GenBank. MK574876
- Zhang F. 2019. Camellia sinensis var. assamica mitochondrion, complete genome. GenBank. MK574877
- 2019. BIGD Genome Warehouse. http://bigd.big.ac.cn/search?dbId=gwh&q=GWHAAIC00000000
- Zhang F. 2019. Deciphering tea tree chloroplast and mitochondrial genomes of Camellia sinensis var. assamica. figshare. [DOI] [PMC free article] [PubMed]
Data Availability Statement
The following bioinformatic tools and versions were used for generating all results as described in the main text:
1. Bowtie2, version 2.3.4.3, was used for aligning sequencing reads to long reference sequences with default parameters: http://bowtie-bio.sourceforge.net/bowtie2/index.shtml
2. CLC Genomics Workbench, version 3.6.1, was used for genome assembly with default parameters: https://www.qiagenbioinformatics.com/products/clc-genomics-workbench/
3. Velvet, version 1.2.08, was used for genome de novo assembly, which was previously described: https://www.ebi.ac.uk/~zerbino/velvet/
4. SSPACE, version 3.0, was used for genome scaffolds assembly with default parameters: https://www.baseclear.com/services/bioinformatics/basetools/sspace-standard/
5. GapCloser, version 1.12, was used to fill the gaps between scaffolds with default parameters: https://sourceforge.net/projects/soapdenovo2/files/GapCloser/
6. DOGMA (an online tool), accessed at 12/2018, was used for annotating cp genomes with default parameters: http://dogma.ccbb.utexas.edu/
7. Mitofy (an online tool), accessed at 12/2018, was used for annotating plant mt genomes with default parameters: http://dogma.ccbb.utexas.edu/mitofy/
8. tRNAscanSE, VERSION 1.3.1, was used to search tRNA with default parameters: http://lowelab.ucsc.edu/tRNAscan-SE/
9. Organellar Genome DRAW (an online tool), accessed at 12/2018, was used for creating high quality visual representation of cp gemome with default parameters: https://chlorobox.mpimp-golm.mpg.de/OGDraw.html
10. MISA,version 1.0, was used for annotating SSR with monomer (one nucleotide, n ≥ 8), dimer (two nucleotides, n ≥ 4), trimer (three nucleotides, n ≥ 4), tetramer (four nucleotides, n ≥ 3), pentamer (five nucleotides, n ≥ 3), hexamer (six nucleotides, n ≥ 3): http://pgrc.ipk-gatersleben.de/misa/misa.html
11. REPuter (an online tool), accessed at 1/2019, was used for annotating long repeated sequences with the following parameters: minimal length 50 nt; mis match 3 nt: https://bibiserv.cebitec.uni-bielefeld.de/reputer/
12. PREP-cp (an online tool), accessed at 1/2019, was used for predicting RNA editor for plant cp genes with the cutoff score (C-value) setting to 0.8: http://prep.unl.edu/
13. PREP-mt (an online tool), accessed at 1/2019, was used for predicting RNA editor for plant mt genes with the cutoff score (C-value) setting to 0.6: http://prep.unl.edu/
14. MEGA, version 7.0.26, was used for phylogenomics and phylomedicine at 1000 bootstrap: https://www.megasoftware.net/
15. ClustalW, version 2, was used for multiple sequence alignment with default parameters: https://www.ebi.ac.uk/Tools/msa/clustalw2/