Abstract
Plants in temperate areas evolved vernalisation requirement to avoid pre-winter flowering. In Brassicaceae, a period of extended cold reduces the expression of the flowering inhibitor FLOWERING LOCUS C (FLC) and paves the way for the expression of downstream flowering regulators. As with all polyploid species of the Brassicaceae, the model allotetraploid Brassica napus (rapeseed, canola) is highly duplicated and carries 9 annotated copies of Bna.FLC. To investigate whether these multiple homeologs and paralogs have retained their original function in vernalisation or undergone subfunctionalisation, we compared the expression patterns of all 9 copies between vernalisation-dependent (biennial, winter type) and vernalisation-independent (annual, spring type) accessions, using RT-qPCR with copy-specific primers and RNAseq data from a diversity set. Our results show that only 3 copies – Bna.FLC.A03b, Bna.FLC.A10 and to some extent Bna.FLC.C02 – are differentially expressed between the two growth types, showing that expression of the other 6 copies does not correlate with growth type. One of those 6 copies, Bna.FLC.C03b, was not expressed at all, indicating a pseudogene, while three further copies, Bna.FLC.C03a and Bna.FLC.C09ab, did not respond to cold treatment. Sequence variation at the COOLAIR binding site of Bna.FLC.A10 was found to explain most of the variation in gene expression. However, we also found that Bna.FLC.A10 expression is not fully predictive of growth type.
Subject terms: Plant evolution, Plant genetics
Introduction
Synchronization of reproduction with favorable environmental conditions is crucial for a species’ survival. For plants, timing of reproduction is mainly governed by flowering time regulation. In temperate climates, the two most important cues to induce flowering are a period of extended cold (the vernalisation pathway) and day length (the photoperiod pathway)1. In contrast to biennial or winter forms, annual or spring forms are lacking a functional vernalisation pathway and therefore flower without a period of extended cold.
The vernalisation pathway was found to be a complex regulatory system including transcriptional and epigenetic regulation mechanisms2. In Arabidopsis thaliana, the model Brassicaceae, a single copy of the transcriptional repressor FLOWERING LOCUS C (FLC) inhibits expression of the central flowering regulator FLOWERING LOCUS T (FT)3. In this state, FT repression via FLC overrides activating effects from the photoperiod pathway and other activating input signals4. FLC also represses the expression of further important flowering genes by binding to the promoter, like FLOWERING LOCUS D (FD), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1), and TEMPRANILLO 1 (TEM1)5,6. While the regulation of many FLC targets depends on complex formation with SHORT VEGETATIVE PHASE (SVP), FT and SOC1 expression can be independently regulated by either FLC or SVP7. The activating function of FT depends on dimer formation with FD8: FLC itself is regulated via the vernalisation pathway2 and via the autonomous pathway9. There is also cross-talk with the photoperiod pathway via the protein SENSITIVITY TO RED LIGHT REDUCED 1 (SRR1)10. FLC expression is regulated via chromatin conformation, which itself is governed via histone modifications11. Genetic and epigenetic regulation processes induced by extended cold stabilize a chromatin state which does not allow transcription of FLC mRNA12. This inactive state is mitotically, but not meiotically stable; therefore, the next generation starts in a non-vernalized state3,13. In the non-vernalized state, FLC expression is promoted by the FRI-C, a large protein complex built up by the scaffold protein FRIGIDA (FRI) in interaction with SUPPRESSOR OF FRIGIDA 1 (SUF4)14. FRI-C acts to recruit general transcription factors and specific chromatin modification factors to the FLC chromatin14. Cold induces the transcription of a noncoding antisense transcript from FLC called COOLAIR, which starts to decrease FLC mRNA levels12,15. Independently from that, but later, alternative splicing produces the noncoding sense RNA COLDAIR from the first intron of FLC itself12,15. COLDAIR recruits Polycomb Group proteins (PcG) proteins forming the PRC2-like complex to the FLC chromatin15, where they remove activating marks and add repressive marks to the histones in the FLC chromatin in interaction with the protein VERNALIZATION INSENSITIVE 3 (VIN3)16. This is further supported by the action of the Paf1 complex17. Those repressive marks are recognized and bound by the PRC2 complex containing the proteins VERNALISATION 2 (VRN2) and LIKE HETEROCHROMATIN 1 (LHP1) (also known as TFL2), keeping FLC repression stable until the next generation18–20.
In A. thaliana, spring and winter forms have been shown to carry different allelic variants of FLC, FRI and VIN314,21–23. As the main vernalisation components were found to be conserved in the Brassicaceae24–31, their paralogs are also candidates for the difference between spring and winter forms in Brassica crops. Variation in paralogs of FLC was found to be associated to a change in flowering time for B. rapa32–35, B. oleracea25,36–38 and B. napus26,28,39–41. However, these crops have a paleopolyploid origin and were shown to carry several paralogs of FLC. The reference genome of B. napus carries nine annotated copies of Bna.FLC42. Early transformation studies revealed that at least five of them are able to delay flowering in A. thaliana, but to a varying extent28. The strongest effect was observed in plants transformed with BnFLC1 (Bna.FLC.A10)28. Later it was discovered that a transposon insertion in the upstream region of Bna.FLC.A10 is strongly associated to the vernalisation-dependent phenotype39. Depending on the genetic background investigated, different copies of Bna.FLC were repeatedly found within the confidence intervals of quantitative trait loci (QTL) for flowering time in B. napus43–48. Previously we found that a R10P mutation in the Bna.FLC.A10 (BnaA10g22080D) copy is a strong candidate for the winter-spring split in a diverse set of B. napus accessions41. More recently, it was found that the strongest selection signal separating spring and winter type populations was also found at Bna.FLC.A10, correlating with differential gene expression49. Although different data sets indicate that Bna.FLC.A10 might be the most decisive copy for vernalisation requirement28,39,41, the roles of the other copies remain unclear, mostly because specific primers for some copies could not be developed29,46. For example, the most extensive expression study on Bna.FLC copies to date was not able to resolve four of the nine copies specifically, and the study was only conducted in a winter and a semi-winter type, but not in a spring type29. In the present study, we conducted Reverse Transcription Quantitative Real-Time PCR (RT-qPCR) to measure the specific individual expression levels of all Bna.FLC copies. Comparing two winter-type and two spring-type B. napus accessions, along with a winter-hardy but not strictly vernalisation-dependent winter accession, we measured Bna.FLC expression levels with and without vernalisation, and in different tissues prior to vernalisation. Our results indicate that only Bna.FLC.A03b, Bna.FLC.A10, and Bna.FLC.C02 are differentially expressed between winter and spring type accessions, although tissue-specific differences exist. Comparisons with RNAseq data for a diversity set show that this differential expression is a general difference between winter and spring accessions before vernalisation. We then screened a diverse population of 53 winter and 48 spring accessions for Bna.FLC.A10 and Bna.FLC.C02 expression with RT-qPCR and found that only Bna.FLC.A10 was differentially expressed between winter and spring accessions. There were, however, exceptions in both winter and spring accession, and sequence analysis revealed that this was strongly associated to a sequence variant at the COOLAIR binding site and a broken reading frame in exon 1. We conclude that this sequence variant contributed to the difference between biennial and annual B. napus forms, whereas their remaining homologs appear to have undergone subfunctionalisation and pseudogenisation after polyploidisation.
Material and Methods
Sequence analysis
Genomic sequences were extracted from the respective reference genomes for B. napus (the Darmor-bzh reference genome, version 4.142), B. rapa (the ‘Chiifu-401-42’ reference genome, version 1.550) and B.oleracea (the ‘TO1000’ reference genome, version 2.151) using the respective annotation. A further copy of Bna.FLC was identified in41 and also included. Moreover, we identified four gene copies from an unpublished version of B. nigra genome which was made available by the courtesy of I. Parkin. The gene IDs for the single copies can be found in Table S1. The sequence from A. thaliana was retrieved from The Arabidopsis Information Resource (TAIR), www.arabidopsis.org, for gene model AT5G10140.1.
Sequence alignments were performed using CLUSTAL multiple sequence alignment by MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle/, version 3.8) with Default parameters. Based on this alignment, we constructed a Maximum likelihood tree and a neighbor joining tree using bootstrap analysis (100 replicates) using MEGA version 10.0.5. Exon-intron structure was determined by aligning the genomic sequences with the respective cDNA sequences from42. SNP variation for the five accessions also used in RT-qPCR was taken from41.
Promoter region analysis was performed using MEME (http://meme-suite.org/tools/meme) and JASPAR (http://jaspar.genereg.net/) using motifs for A. thaliana52.
Plant material, cultivation and sampling
Two B. napus winter accessions (Manitoba, Lisabeth) and two spring accessions (Girasol, Korall) were chosen along with a winter-hardy, vernalization-independent winter accession (Mansholt). Mansholt carries duplications for Bna.FLC.C09a and Bna.FLC.C09b, while the other accessions did not show copy number variations53. The plants were sown in 7 × 7 cm pots in 3 biological replicates per treatment and transplanted to 12 × 12 cm pots 4 weeks after sowing. Cultivation was performed in a greenhouse using a 16/8 h day/night rhythm with 20/17 °C. The first sampling was performed seven weeks after sowing when the plants reach BBCH stage 14 at midday (ZT 7) by cutting off the youngest fully developed leaf. Leaves were frozen in liquid nitrogen and stored at −80 °C until RNA extraction. After the first sampling, one set of plants was brought to the cool room for eight weeks using an 8/16 h day/night rhythm at constant 5 °C. Another set of plants remained in the greenhouse as control plants. After eight weeks of vernalisation, both sets were sampled again at BBCH20.
For the analysis of tissue-specific expression, Manitoba and Korall were grown in three replicates in the greenhouse under the same conditions. Ten weeks after sowing when the plants reach BBCH15, we sampled petioles, developed and emerging leaves separately and kept the samples at −80 °C until RNA extraction.
For the analysis of the time series, Manitoba was grown in 27 replicates in the greenhouse under the same conditions. From an age of 3 weeks up to 8 weeks, we took leaf samples from 3 biological replicates every week. The plants were then subjected to a cold treatment as described above, and we sampled again after 4, 6 and 8 weeks of cold treatment.
For the population screening, 3 biological replicates of 101 accessions (see Supplementary Table 2 for a list of accessions) were sown in quickpot plates and grown for 7 weeks until sampling.
Primer design, cDNA synthesis and quantitative PCR
Total RNA was extracted using the NucleoSpin miRNA kit (Macherey-Nagel) following manufacturer’s instructions, quantified using Qubit RNA Broad Range on a Qubit fluorimeter and stored at −80 °C until use.
cDNA synthesis was performed using the RevertAid cDNA synthesis kit (ThermoFisher) using 1 µg of total RNA and Oligo-DT primers. cDNA was quantified using the Qubit DNA High Sensitivity kit on a Qubit fluorimeter. Quantitative Real-time PCR was performed on a Real-Time PCR System ViiA7 cycler (Applied Biosystems) in 384-well plates. The reaction mix containing specific primers, template cDNA and FastStart Universal SYBR Green Master mix containing Rox (Roche) was pipetted by a robot (Biomek 4000, Beckman Coulter). As endogenous control, we used ubiquitin. The PCR program was as follows: initial denaturation (94 °C for 2 min), amplification and quantification (40 cycles, 95 °C for 20 sec, 60 °C for 30 sec, 72 °C for 30 sec), and final extension (72 °C for 5 min). A final melting curve was recorded between 55 and 95 °C. PCR efficiency was measured using a pool of all samples in a dilution series of 6 points. All samples were measured in 3 technical replicates. The normalized expression level was determined using the ΔCt method54. The primer sequences are shown in Supplementary Table 3. For comparisons between winter and spring, a Student’s t-test was performed, for comparisons of more than one factor, we calculated least significant distances in R using the package agricolae.
RNAseq analysis
For validation of the results from RT-qPCR, we downloaded a data set from NCBI Sequence Read Archive (SRA, SRP069066) published in55. These represent RNAseq data from Illumina HiSeq2500 in single-end mode for a publically available B. napus diversity set (ASSYST set56,57) for 3 weeks old leaf samples without vernalization. Unfortunately, no biological replicates are given, so the data can only be used for comparison or pooled data analysis. We selected (1) the same 5 accessions which were used in our RT-qPCR experiment for comparison (2) 30 randomly selected spring accessions and 30 randomly selected winter accessions for pooled data analysis (see Supplementary Table 4).
Another publically available data set was downloaded to analyze the time course in gene expression for several vernalisation genes before, during and after cold treatment in the spring cultivar Westar from the NCBI SRA (SRP132445). Each sample was a pool of 6–10 individual plants. We only analyzed time points with two replicates, which were 22 days old plants (before vernalisation), 43 and 64 old plants (during vernalization), and 67 and 72 days old plants (after vernalization). For each time point, both leaves and shoot apex were sampled.
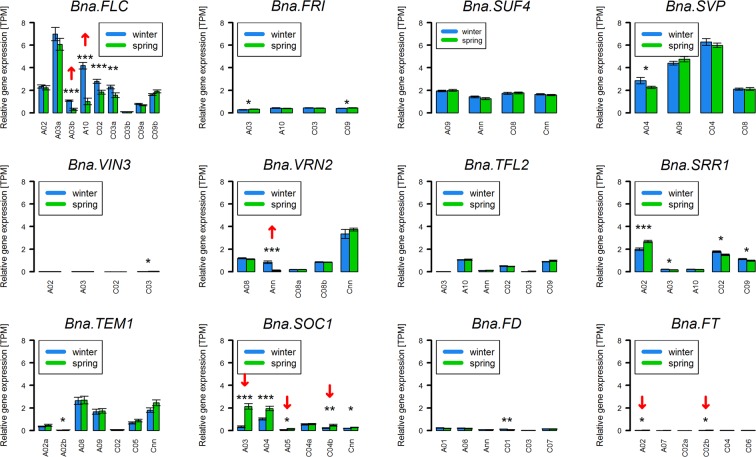
Quality control was performed using FastQC, version 0.10.1 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Accordingly, adapter removal and trimming was performed using Trimmomatic version 0.3858 by first removing TruSeq adapters followed by head cropping the first 9 bases. Clean reads were mapped onto version 4.1 of the B. napus ‘Darmor-Bzh’ reference genome using Bowtie259, alignment mode “very-sensitive”. Removal of duplicates, sorting and indexing was carried out with samtools version 0.1.1960. We then calculated transcripts per million (TPM) using bedtools software with multiBamCov61 for raw coverage calling, followed by normalization in R (version 3.1.2) according to62 for the sum of all exons of every gene. From this, we selected a list of important vernalisation gene copies, and calculated |log2(mean(TPM(winter))/mean(TPM(spring))|. Besides FLC, we included other important vernalisation regulators (FRI, SUF4, TFL2, VIN3, VRN2, SVP, SRR1) and genes directly regulated by vernalisation (SOC1, FD, FT, TEM1).
Results
Phylogenetic analysis
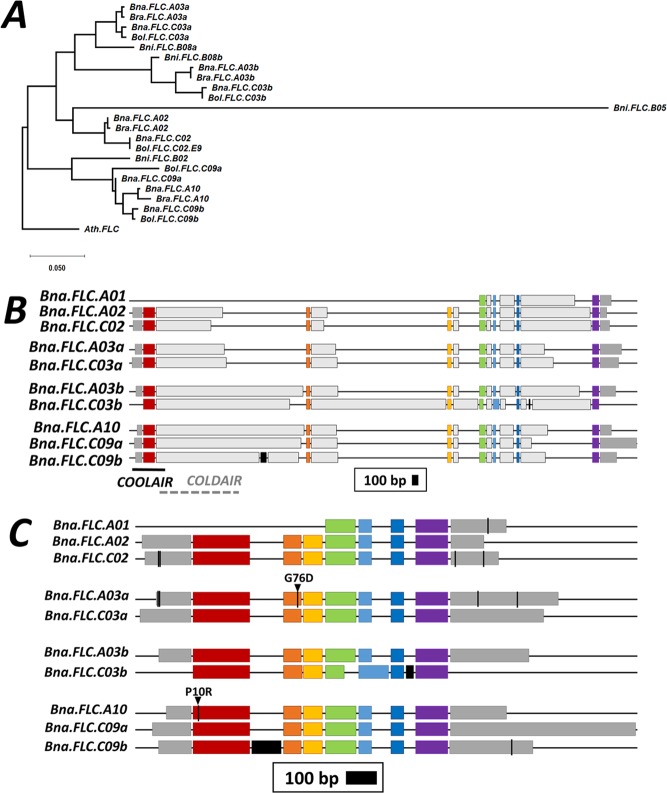
The coding sequences of the respective homeologous copies are more related to each other than to their inter-subgenome paralogs (Fig. 1A, Supplementary Fig. 1). This is also evident from the gene structure. Most Bna.FLC copies have 7 exons, while the truncated pseudogene on A01 only carries exons 4–7. Bna.FLC.C03b and Bna.FLC.C09b both carry an additional exon between exons 1/2 and 6/7, respectively (Fig. 1B,C). With the exception of Bna.FLC.A01, similar relationships were reported elsewhere28,29,63. When comparing all Bna.FLC copies with their progenitors from B. rapa and B. oleracea, there is mostly low divergence between the progenitor and the polyploid (Fig. 1A). The largest distance was found between Bra.FLC.A02 and Bna.FLC.A02, followed by Bol.FLC.C09b and Bna.FLC.C09b. The only copy without a respective homolog in the progenitors was Bna.FLC.A01, suggesting that it has been duplicated after the recent interspecific hybridization leading to B. napus (<7500 years ago42), most likely from Bna.FLC.A02 (Supplementary Fig. 2). In contrast, both Bna.FLC.C09 copies have a closely related homolog in the progenitor species B. oleracea, indicating that they have been duplicated earlier. The related Brassica species B. nigra was found to carry 4 FLC copies, however, two of those copies (Bni.FLC.B02 and Bni.FLC.B08b) were truncated in our version of the B. nigra reference genome, which possibly is due to incomplete annotation, but might also indicate pseudogenization. The respective copies cluster with the same main four branches than the AC species copies, but more distantly, reflecting the higher divergence of the B genome from the AC genomes. Bni.FLC.B05 clusters distantly with the A02/C02 clade, Bni.FLC.B08ab cluster with the respective A03ab/C03ab clades, and Bni.FLC.B02 cluster with the A10/C09 clade (Fig. 1A).
Figure 1.
(A) Phylogenetic tree constructed with Maximum Likelihood of Brassica FLC from three diploid (B. rapa, B. nigra, B. oleracea) and one polyploid (B. napus) species with Ath.FLC as outgroup. cDNA sequences were extracted from the respective reference genomes, the sequence from A. thaliana was retrieved from TAIR. Sequence alignment was performed using CLUSTAL multiple sequence alignment by MUSCLE with Default parameters. Based on this alignment, a Maximum Likelihood tree was constructed using MEGA version 10.0.5. (B) Full gene structure including UTR (dark grey boxes), introns (light grey boxes) and exons (colored boxes). Black boxes represent extra exons. Box length is proportional to length in bp, the legend shows the length of 100 bp. The solid line in black shows the localization of COOLAIR, the dotted line in grey represents the approximate position of COLDAIR. (C) Exon structure including the position of sequence variants within the 5 accessions used for the initial RT-qPCR. Dark grey boxes represent UTRs, colored boxes represent exons in the same color code than for B, black boxes represent extra exons. Black lines represent a variant, black lines with a black triangle represent a non-synonymous SNP, annotated with the respective amino acid mutation.
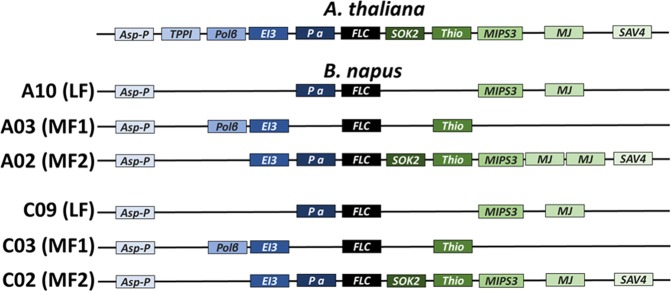
The Brassica A and C (sub)genomes can be subdivided into a least fractionated (LF) and two more fractionated (MF1, MF2) partitions64. Using synteny analysis64, we found that Bna.FLC.A10 (BnaA10g22080D) and Bna.FLC.C09a (BnaC09g46500D) are located in LF, Bna.FLC.A03a (BnaA03g02820D) and Bna.FLC.C03a (BnaC03g04170D) are located in MF1, and Bna.FLC.A02 (BnaA02g00370D) and Bna.FLC.C02 (BnaC02g00490D) are located in MF2 (Fig. 2). Bna.FLC.A03b (BnaA03g13630D), Bna.FLC.C03b (BnaC03g16530D) and Bna.FLC.C09b (BnaC09g46540D) are non-syntenic copies, indicating that they arose after the hexaploidization of the common Brassica ancestor.
Figure 2.
Graphical representation of local synteny for the syntenic Bna.FLC copies. The figure shows the gene order in the vicinity of Ath.FLC, Bna.FLC.A10, Bna.FLC.A03a, Bna.FLC.A02, Bna.FLC.C09a, Bna.FLC.C03a and Bna.FLC.C02, respectively. LF, MF1 and MF2 stand for least fractionated, more fractionated 1 and more fractionated 2, respectively. Synteny was derived from BRAD, gene names were derived from TAIR: Asp-P: aspartyl protease family protein, TPPI: TREHALOSE-6-PHOSPHATE PHOSPHATASE I, Polβ: DNA-directed RNA polymerase subunit beta, EI3: Ethylene insensitive 3 family protein, P a: Pollen Ole e 1 allergen and extensin family protein, SOK2 (no synonym given), Thio: Thioesterase superfamily protein, MIPS3: MYO-INOSITOL-1-PHOSPHATE SYNTHASE 3, MJ: Major facilitator superfamily protein, SAV4: SHADE AVOIDANCE 4.
Analysis of regulatory regions
We compared the sequences of three different regulatory regions using42: the promoter regions, intron 1 known to contain the long non-coding RNA COLDAIR, and the regions containing the antisense RNA COOLAIR.
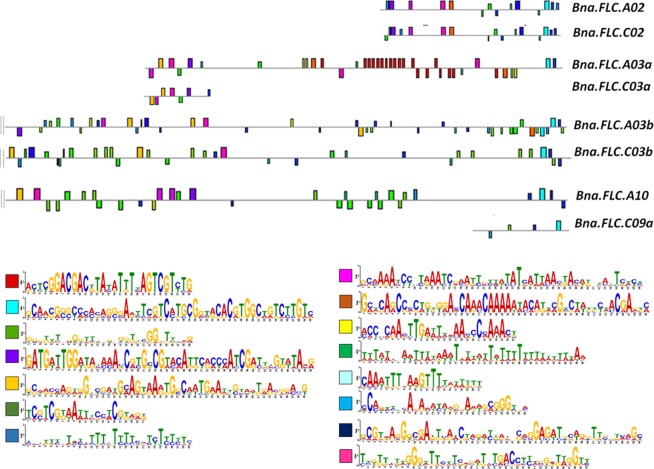
The promoter regions (the upstream region of the gene extending until the next annotated gene) ranged from 736 bp (Bna.FLC.C03a) to 10,928 bp (Bna.FLC.A03a). The Bna.FLC.C09b promoter region was excluded from the analysis due to a significant amount of missing sequence data. Only a 26 bp motif (consensus CMTGCGRYRCACRTGGCWRTCYTSTM), located shortly before the 5′UTR of the gene, was highly conserved between the promoter regions of all 8 analysed copies, annotated as MA1359.1, the binding site for the transcription factor BIGPETAL (BPEp).The motif also overlapped with the COOLAIR binding site. Apart from this, motif search using MEME did not reveal a conclusive pattern. While the promoter regions of Bna.FLC.A02 and Bna.FLC.C02 were highly conserved both in length and motif content, the promoter regions of Bna.FLC.A03a and Bna.FLC.A10 were remarkably longer than their C subgenome counterparts and motif composition was different. The promoter regions of Bna.FLC.A03b and Bna.FLC.C03b were comparable in length, but different in motif composition (Fig. 3). The promoter region of Bna.FLC.A03b carries a highly repetitive 29 bp motif (consensus AYTCGGACGACKTATATTTHAGTCGTYTG) which is specific among all copies. No other motif was specific to a single promoter region. We have also retrieved the respective promoter regions from the diploid Brassica species B. rapa, B. nigra and B.oleracea as well as the promoter region of A. thaliana. The alignment shows that conservation is generally low among all 5 species, also given the strong variation in length (736 bp in Bna.FLC.C03a – 16946 bp in Bni.FLC.B08b). However, some few AT rich regions seem to be fairly conserved (Supplementary Fig. 3). When we aligned subgroups of promoter regions to the A. thaliana promoter region, we found that different parts of those regions were conserved within those subgroups (Supplementary Fig. 4). While the A02/C02 subgroup is more conserved towards the beginning and the end of the promoter region, the subgroup A03a/C03a is most conserved in the central part, and the subgroups A03b/C03b and A10/C09a towards the end of the region. The promoter regions from B. nigra showed the lowest conservation with A. thaliana, while the regions from the A10/C09a group showed the highest conservation with A. thaliana. Within Bna.FLC.A10, a region with a BLAST hit to the MITE transposon in front of Tapidor-Bna.FLC.A10 reported by39 was found to be unique among the promoter regions. When comparing motif composition in the orthologous copies using MEME excluding B. nigra, but including Bna.FLC.C09b, we found that motif composition is generally conserved between the orthologous promoter regions. Motif composition was fully conserved for the A02, the A03b, C03b and A10 copies, although distances and order were slightly varying. The A03a, C03a, C09a and C09b promoter regions varied slightly in motif composition, for example, the repetitive sequence found in Bna.FLC.A03a was not contained in Bra.FLC.A03a, indicating a later origin. Moreover, divergence between homeologs was also absent (A03b/C03b) or low (A02/C02, A03a/C03a), but high for A10/C09ab, indicating stronger divergence in regulatory motifs between the A10 and the C09 copies. The 26 bp motif overlapping with COOLAIR was also conserved among almost all species and copies, with the exception of the C03b promoter regions and Bol.FLC.C02. (Supplementary Fig. 5). The lengths of intron 1 ranged from 912 bp (Bna.FLC.C02) to 2,470 bp (Bna.FLC.A10) being considerably longer on A10/C09a and A03b/C03b, but intermediate on C09b. On A02/C02 and A03a/C03a, respectively, the first intron was missing large fragments of 1,000–1,500 bp at the 3′ end. When we aligned those to A. thaliana COLDAIR (HG975388.1), alignment was generally poor, making a direct comparison difficult.
Figure 3.
Motif composition of the upstream (promoter) regions of 8 analysed Bna.FLC gene copies done by MEME. The promoter region for Bna.FLC.C09b was excluded due to missing sequence information. Every motif is represented by a distinct color (see legend).
Finally, we aligned the sequence of the long non-coding antisense transcript COOLAIR (GQ352646.1) to Bna.FLC and compared the overlapping regions. COOLAIR is supposed to cover parts of the 5′upstream region, exon 1, parts of intron 1, parts of intron 6, exon 7 and parts of the 3′downstream region. However, we were only able to match the 5′upstream region, exon 1 and parts of intron 1 to our sequences. Considerable variation was found at the 5′ end (3′ end of the antisense RNA) with Bna.FLC.C02 showing an insert of 43 bp, while Bna.FLC.A10 showed more sequence variation at this site. Both copies on C09, in contrast, were approximately 75 bp shorter at this end (Fig. 1B). While RNAseq data (see below) before vernalisation did not show clear evidence for COOLAIR expression, there is a pattern of directional reads in the time series of the spring type at Bna.FLC.A10, but coverage was too low for quantification.
Bna.FLC expression levels differentiate before vernalization
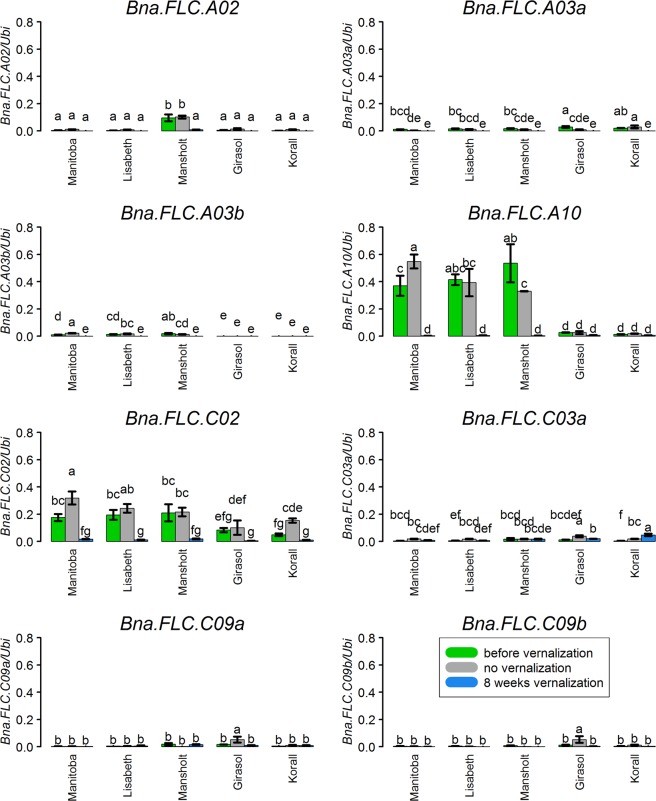
We then studied the gene expression patterns of all Bna.FLC copies in five different accessions (3 winter and 2 spring cultivars) and found that only eight copies were expressed in leaves seven weeks after sowing (BBCH 14). Bna.FLC.C03b was never expressed. The highest expression levels were detected for Bna.FLC.A10 and Bna.FLC.C02 in winter accessions. Both copies showed a differential expression pattern between winter and spring accessions, whereby the average ratio of winter/spring expression level was 19.0 for Bna.FLC.A10 and 2.8 for Bna.FLC.C02. The copy Bna.FLC.A03b showed no expression in the spring accessions, whereas Bna.FLC.A02 showed considerably higher expression in Mansholt, a winter type without strict vernalisation requirement (Fig. 4).
Figure 4.
Relative gene expression of all eight expressed Bna.FLC copies in leaves before vernalisation 7 weeks after sowing (BBCH 14), and without and with vernalisation 15 weeks after sowing in the accessions Manitoba, Lisabeth (winter-type), Mansholt (winter-hardy, but not vernalisation-dependent), Girasol and Korall (both spring-type). The values were calculated from RT-qPCR using the ΔCt method and represent 3 biological replicates. Whiskers show SEM. “NE” stands for “not expressed”. Small letters represent group by LSD test. The scale is the same for all 3 plots.
We assessed Bna.FLC expression in petioles, emerging and developed leaves of 10 week old plants (BBCH 15) before vernalisation in a winter (Manitoba) and a spring accession (Korall) to detect tissue-specific patterns (Supplementary Fig. 6). Bna.FLC.A03a and Bna.FLC.A10 were differentially expressed between winter and spring accessions in all tissues. Bna.FLC.C02 expression was only significantly different in emerging leaves (p = 0.021) and petioles (p = 0.008), indicating differential expression dynamics in winter and spring material. Additionally, in petioles, Bna.FLC.A03a showed a significantly higher expression in Korall (p = 0.037), while Bna.FLC.C09b showed a significantly lower expression in Korall (p = 0.019).
For validation, we downloaded available RNAseq data published in55. Three week old leaves were sampled before vernalisation for the same accessions we have used here. Only Bna.FLC.A03b and Bna.FLC.A10 were differentially expressed between both growth types. Bna.FLC.C03b expression was extremely low. In contrast to RT-qPCR, the expression of Bna.FLC.A02, Bna.FLC.A03a, Bna.FLC.C02, Bna.FLC.C03a and Bna.FLC.C09b was comparable to Bna.FLC.A10, but did not show differential expression between both growth types (Supplementary Fig. 7). Only Bna.FLC.C09b had a lower expression level. Those differences may partly be due to the different developmental stage and due to the high sequence similarity of the copies which may confound mapping.
We then analyzed a larger data set from the same study containing 30 spring and 30 winter accessions, using all winter accessions as biological replicates for “winter” and respectively so for “spring”. We included other important vernalisation regulators (FRI, SUF4, TFL2, VIN3, VRN2, SVP, SRR1) and genes directly regulated by vernalisation (SOC1, FD, FT, TEM1). We found 3 gene copies which were strongly (|log2(FC)| >1) and significantly (p < 0.05) upregulated in winter accessions. These were Bna.FLC.A03b, Bna.FLC.A10 and Bna.VRN2.Ann. Three copies of the downstream gene Bna.SOC1 (on A03, A05 and C04) and two copies of the central flowering time gene Bna.FT (on A02 and C02) were strongly and significantly downregulated in winter accessions compared to spring accessions. Bna.VIN3, Bna.FT and Bna.FD were expressed at an extremely low level, as expected (Fig. 5).
Figure 5.
Normalized gene expressed levels (TPM) from RNAseq data for 12 different gene copy groups for genes involved in vernalisation (FLC, FRI, SUF4, SVP, VIN3, VRN2, TFL2), affecting FLC expression (SRR1) or affected by vernalisation (TEM1, SOC1, FD, FT) in a diverse set of 30 winter and 30 spring accessions. Whiskers represent standard errors of the mean (SEM). Asterisks show the level of significance based on the Student’s t-test (*p-value < 0.05, **p-value < 0.01, ***p-value < 0.001). Red arrows indicate copies which were strongly (|log2(FC)| >1) up or downregulated comparing winter and spring accessions. Arrows pointing upwards indicate augmented expression in winter accessions, arrows pointing downwards indicate lower expression in winter accessions.
Most Bna.FLC copies are downregulated by cold
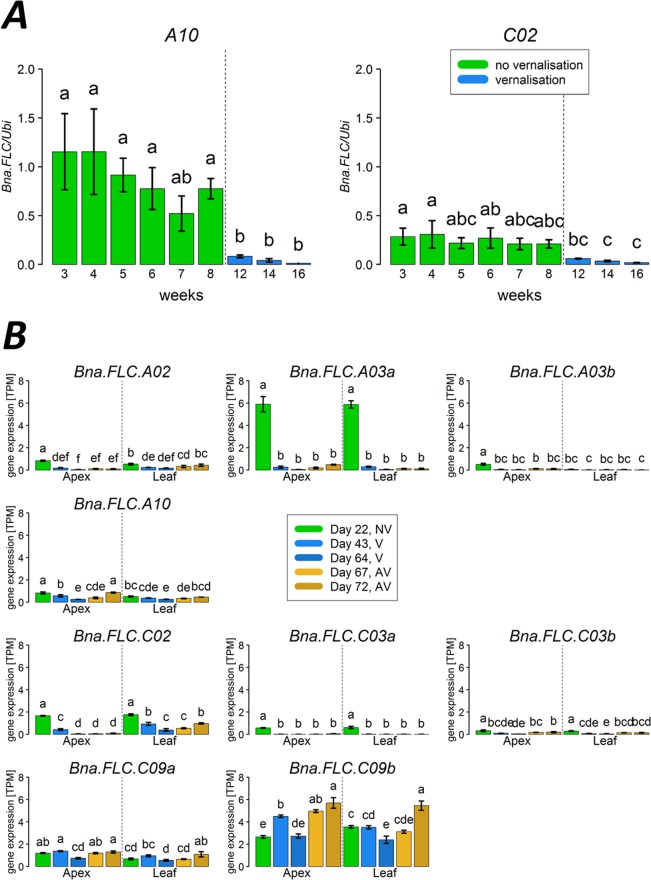
Without cold treatment, 15 weeks after sowing (BBCH20), Bna.FLC expression patterns did not show a clear pattern. Expression remained mostly constant, although single accessions show slightly decreased expression (Bna.FLC.A03a, Bna.FLC.A03b, Bna.FLC.A10) or slightly increased expression (Bna.FLC.A03b, Bna.FLC.A10, Bna.FLC.C02, Bna.FLC.C03a). However, the changes did not affect the differential expression pattern between winter and spring types for Bna.FLC.A10 (Fig. 4). We also used our primer set to study a time course in the winter type Manitoba for the two copies Bna.FLC.A10 and Bna.FLC.C02. We have sampled independent plants of the same genotype every week from an age of 3 weeks to 8 weeks, when they were subjected to vernalisation. During vernalisation, we sampled at 4, 6 and 8 weeks of vernalisation. The results show clearly that the cold treatment is the only factor influencing gene expression for those copies, while the effect of age can be neglected (Fig. 6A). Longer vernalization seems to further decrease Bna.FLC levels, although those effects are only significant for Bna.FLC.C02.
Figure 6.
Time series of Bna.FLC expression. (A) Relative gene expression for Bna.FLC.A10 and Bna.FLC.C02 for a time series of 3–16 weeks in the winter cultivar Manitoba. (B) Normalized gene expressed levels (TPM) values from RNAseq from a publically available data set for the spring cultivar Westar. Whiskers show SEM. Small letters represent group by LSD test.
After eight weeks of cold treatment (BBCH 20), most Bna.FLC copies were downregulated. Exceptions were Bna.FLC.C03a, which either did not change or even increased, and both C09 copies, where downregulation was not significant. Downregulation was generally stronger in winter accessions than in spring accessions, partly due to the higher expression level before vernalisation (Fig. 4).
We also determined Bna.FLC and other vernalisation regulators’ expression patterns in a time series in both leaf and apex in a publically available data set from the spring cultivar Westar. For each tissue, we analyzed five time points: 22 days (before vernalization), 43 and 64 days (during vernalization), and 67 and 72 days (after vernalization). The first sampling point was comparable to the sampling point of the other RNAseq data set, and the expression of all copies lay in the same range than the spring accessions from this diversity set. With the exception of Bna.FLC.C02, the expression patterns over time were similar in both tissue types. All copies with the exception of Bna.FLC.C09a were downregulated during cold. After cold, Bna.FLC.A10 expression increased again to reach its original state, while both C09 copies even exceeded their original expression (Fig. 6B).
Analyzing the expression patterns of other vernalization regulators in this spring type cultivar, we found that all four copies of Bna.FRI got slightly upregulated and all four copies of Bna.VIN3 got strongly upregulated during cold. Bna.VIN3 levels returned to their original state after cold, while Bna.FRI levels remained almost constant. This indicates that Bna.VIN3 expression reacts normally in spring types. Interestingly, four out of six Bna.FT copies were already fairly well expressed before vernalization, reflecting the spring type behavior, but got completely downregulated during cold treatment and strongly upregulated afterwards. This could possibly be due to a short day length regime in the cold room (Supplementary Figs 8–10).
Population-wide patterns for Bna.FLC.A10 and Bna.FLC.C02
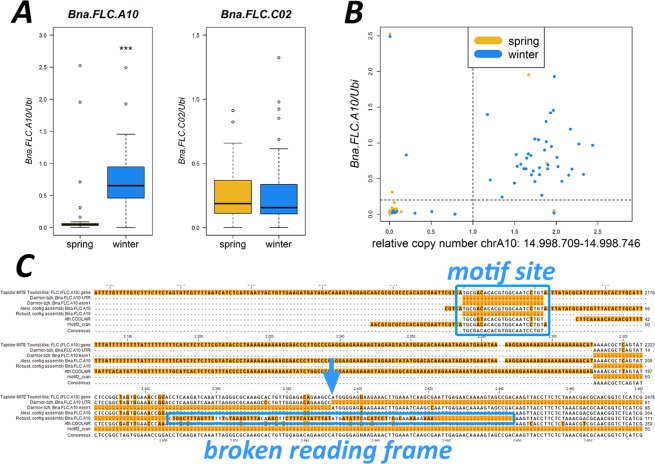
We then wanted to assess if the two most expressed copies (Bna.FLC.A10 and Bna.FLC.C02) could be associated to growth type on a population-wide scale. For this, we have screened a large set of 101 accessions (53 winter and 48 spring accessions) for gene expression with RT-qPCR. We found that Bna.FLC.A10 gene expression was highly significantly different between winter and spring accessions (p < 1.8 × 10−7), but Bna.FLC.C02 gene expression was not. For Bna.FLC.A10, although the overall pattern was clear, some exceptions were observed in both winter and spring accessions (Fig. 7A). We found four spring accessions with a high Bna.FLC.A10 expression, while we found ten winter accessions with low Bna.FLC.A10 expression. Although the population has been selected to reflect both early and late flowering accessions, there is no bias towards Bna.FLC.A10 expression in early or late flowering accessions. To understand how vernalisation may be upheld or circumvent, we checked the RNAseq diversity set and found four accessions with unusual FLC pattern in both the spring and the winter pool. We found that in a winter background, accessions with lower Bna.FLC.A10 have increased Bna.SOC1 expression for a copy on A03 (still lower than spring types) and a copy on A05 (higher than spring types) and increased expression of Bna.FT.A02 (but still considerably lower than spring types). At the same time, they have less Bna.VIN3.A02, even lower than spring accessions. In a spring background, we found increased expression of Bna.TEM1.C02 (higher than winter), but decreased expression of Bna.FT.A02 (even lower than in winter), Bna.VIN3.C02 (to 0) and Bna.TFL2.C03 (approximately winter level).
Figure 7.
Relationships between the broken reading frame found to be correlating with Bna.FLC.A10 expression. (A) Boxplots for relative gene expression of Bna.FLC.A10 and Bna.FLC.C02 in 48 spring and 53 winter accessions in leaves of 7 week old plants. (B) Relative copy number (RCN) of the region containing the sequence variant as a proxy for the presence of the sequence variant plotted against Bna.FLC.A10 relative gene expression. The dotted lines separate “low” and “high” RCN/ gene expression. (C) Sequence alignment of the unaffected (accession Alesi) and affected (accession Robust) contig assembly with the Darmor-bzh-5′UTR, the Darmor-bzh exon 1, the Tapidor full length sequence from39, the A. thaliana COOLAIR sequence and the sequence of the conserved 26 bp motif (indicated by blue frame). The site of the broken reading frame is also indicated. The blue arrow shows the ATG start site.
To understand which causes could be responsible for this pattern, we went back to the resequencing data published in41. Here, we realized that a short fragment encompassing the R10P SNP was only covered by non-uniquely mapping reads in certain accessions, while perfectly covered with uniquely mapping reads in others. This points to a previously undiscovered short InDel. InDels of a size between 20–50 bp are hard to detect with short read sequencing, because the respective reads will be too divergent to be mapped. Therefore, we have extracted the reads from all Bna.FLC copies and reassembled them for both an affected and a non-affected accession. Aligning the resulting contigs with both the Tapidor and the Darmor-bzh version reveals that the start of the coding sequence is severely distorted in the affected accession, effectively breaking the open reading frame. The start codon is still present, but both the 5′UTR and the first exon are completely divergent from the non-affected sequence. This also affects the presence of the conserved 26 bp motif in front of the 5′UTR, which is missing in the affected accession, and the COOLAIR binding site. The first exon misses a fragment of 23 bp, representing a frameshift deletion (Fig. 7C). To assess this pattern for the total population, we took normalized coverage in the respective region (38 bp) as a proxy for this pattern and found that this is almost fully (two exceptions) explaining the observed expression pattern, indicating that either promoter variation or a broken reading frame of Bna.FLC.A10 contributes to the winter-spring split (Fig. 7B).
Discussion
Regulatory diversification among Bna.FLC copies predates interspecific hybridization
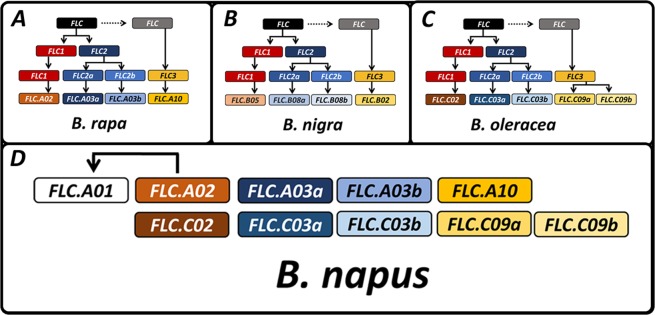
FLC is a MADS-box transcription factor. The MADS-box genes represent a family of transcription factors regulating different developmental transitions, including flowering and fruit development65. Duplication and diversification played an important role in the evolution of the different clades in the MADS-box family65,66. One of the most important sources of duplication is polyploidy. Here, we analysed the expression patterns of all FLC homologs in B. napus, a recent allotetraploid which arose from two mesohexaploid ancestors. Due to the complex genetic history of B. napus including polyploidy and gene loss, a copy number between 4 and 5 is expected for single-copy A. thaliana27,67. Bna.FLC, however, was not affected by gene loss, but has rather undergone copy number amplification, as 10 loci could be identified as homologs of Ath.FLC. Based on our analysis, we now propose an evolutionary path for Bna.FLC (Fig. 8). Therein, we conclude that the duplication of the A03 and C03 copies predates the differentiation of B. rapa, B.nigra and B. oleracea, while the duplication of the C09 copies occurred after the AC split during B. oleracea speciation. Bna.FLC.C03b is considered to be a pseudogene, as we and others29 never observed expression. The predicted exon structure and protein of BnaFLC.C03b differs from the other expressed copies (Fig. 1B,C), and its promoter misses conserved motifs which seem to be important for expression. We hypothesise that BnaFLC.A01 occurred after the allopolyploidization, probably from the closest homolog Bna.FLC.A02, as no close homolog exists in any of the progenitors. The copies on A10 and C09 appear to be most conserved with A. thaliana, we therefore propose that the ancestor of this group is less recent and came from the second polyploidization step of the Brassica ancestor at the transition from the tetraploid to the hexaploid stage. Indeed, Bna.FLC.A10 and Bna.FLC.C09a are both located within the least fractionated parts of the subgenomes64, the youngest part of each subgenome. Given the observation that the copies which were most expressed in each subgenome are not homeologous to each other, subfunctionalisation is likely to have happened during the progenitors’ evolution. This is supported by the high conservation of motif composition in the promoter regions of orthologous copies, indicating that evolution of regulatory elements predates the interspecific hybridization.
Figure 8.
Proposed evolutionary path for Bna.FLC. The model was build based on synteny analysis and phylogenetic analysis. Pseudogenes were identified using protein sequence prediction and gene expression analysis. Gene copies are represented as colored rectangles. White color indicates a putative pseudogene. Arrows indicate ancestry. (A–C) Proposed evolutionary paths for the diploid Brassica species B. rapa, B. nigra and B. oleracea, respectively. (D) Copy configuration for the allotetraploid B. napus.
Different cold responsiveness points to subfunctionalisation of different Bna.FLC copies
B. napus is grown in diverse climates and therefore has developed different growth types. Due to the low genetic diversity within the growth type gene pools68, suitable markers for growth type would help to speed up introgression breeding and allow for better and more effective adaptation. We therefore aimed to study the gene expression patterns of the most prominent candidate gene for vernalisation. A functional vernalisation system prevents pre-winter flowering in winter rapeseed, but is generally missing in spring rapeseed. FLC copies have been proposed as candidate genes for flowering time in previous QTL studies and GWAS, both in B. napus and in the progenitor species B. oleracea and B. rapa. In the progenitor species, Bra.FLC.A10 and Bol.FLC.C02 have been found to underlie QTL for flowering time25,32,33,36. In B. napus, Bna.FLC.A10 has been proposed by different authors to be the most important vernalisation gene copy28,39,41,49. Others also found associations on C02 and A03 in different material29,43,46. Recently, it was shown that promoter variation for Bna.FLC.A10 was linked to strong selection signatures between winter and spring material49, but their data also show high variation in Bna.FLC.A10 expression especially in winter accessions. B. napus, however, has 9 annotated gene copies and therefore underwent gene amplification rather than gene loss. We were therefore interested in the gene expression patterns of all other copies in B. napus. Studies on gene expression of other Bna.FLC copies have been carried out by other authors. Tadege et al.28 was the first to qualitatively study Bna.FLC expression. They found that At.FLC delayed flowering in spring rapeseed, and that some copies of Bna.FLC also delayed flowering in A. thaliana, with Bna.FLC.A10 having the strongest effect. They also showed a strong difference between total Bna.FLC level between two spring and two winter accessions by Northern blotting, all aligning well with our findings. Moreover, they found Bna.FLC.A10 being more expressed in stems than in leaves, a finding we could not confirm. Hou et al.39 studied a large BC5F2 population derived from a cross between Tapidor (a winter type) and Ningyou7 (a semi-winter type). They could show that Bna.FLC.A10 downregulation was much quicker in semi-winter than in winter type, but original Bna.FLC.A10 expression was high in both accessions, indicating that the mechanism separating semi-winter and winter might be different from the mechanism separating winter and spring, as we have found the Bna.FLC:A10 expression is constantly low in spring types even before vernalization (Figs 4, 6). The authors found a 621 bp MITE insertion upstream of Bna.FLC.A10 being associated to this lower responsiveness. They did not find allelic variance between the winter and the semi-winter type and concluded that the difference between winter and semi-winter was entirely due to the expression difference. Raman 2016 studied Bna.FLC expression unspecifically for the pairs FLC1 (Bna.FLC.A10/C09ab), FLC2 (Bna.FLC.A02/C02) and FLC3 (Bna.FLC.A03a/C03a) in Australian and Japanese material46 and found that FLC2 expression was correlating to vernalization response, while FLC1 and FLC3 did not. However, we could not find a study showing expression patterns for all Bna.FLC copies in both winter and spring accessions to allow conclusions on possible subfunctionalisation. Therefore, we designed specific primers for all Bna.FLC copies and use them to test gene expression both in winter and spring accessions. We found strongly varying expression patterns in respect to expression level, response to cold and specificity to growth type, indicating a high degree of subfunctionalisation. The differential expression of Bna.FLC.A10 between winter and spring as found by others49 has been confirmed. At the same time, we also found Bna.FLC.A03b differentially expressed between winter and spring material. Bna.FLC.C02, however, was found to be differentially expressed in emerging leaves, but not in developed leaves, and population screening showed either a small (RNAseq data) or no difference (RT-qPCR). Moreover, the copy was shown to be deleted both in winter and spring accessions53, which in most cases also correlated with the expression pattern (data not shown), but had obviously no effect on life cycle characteristics. Both copies on C09 and partly Bna.FLC.C03a were found not to respond to cold, and Bna.FLC.C03b seems to be pseudogene as reported by others29. Bna.FLC.A02, Bna.FLC.A03a and partly Bna.FLC.C03a are expressed and react to cold, but do not show differences in expression between winter and spring. This patterns indicates that Bna.FLC.A10 is indeed the major source of variation in vernalisation requirement, while Bna.FLC.A03b and Bna.FLC.C02 might still be supportive.
Accessions with low Bna.FLC.A10 expression have a broken reading frame
When screening a population of 101 accessions for Bna.FLC.A10, we could show that winter accessions express Bna.FLC.A10 significantly stronger than spring accessions. However, our data along with others39,49 show that there are winter accessions with a very low Bna.FLC.A10 level, which still need vernalisation to flower, and spring accessions with high Bna.FLC.A10 levels which can flower without. Reassessment of deep-sequencing data on this copy allowed us to detect a sequence variant at the junction of the 5′UTR and exon 1 which was almost fully explanatory for Bna.FLC.A10 level. The variant overlapped with the binding site of the regulatory antisense RNA and also with a highly conserved 26 bp promoter motif shortly before the 5′UTR. We are therefore now able to describe the nature of the promoter variation which has been proposed by others before39,49. This also aligns with other findings, for example, the shorter 3′ end of COOLAIR in both C09 copies which do not react to cold. A similar mechanism has been found in A. thaliana, where variation in intron 1 caused a different splicing behavior of COOLAIR, increasing FLC expression69. While we now are able to explain the expression patterns in Bna.FLC.A10, accessions with unusual expression patterns indicate that Bna.FLC.A10 might be important, but not necessary to define the life cycle characteristics of winter and spring rapeseed. In winter accessions with low Bna.FLC.A10, we see upregulation of some downstream targets like Bna.SOC1 and Bna.FT. In spring accessions with high Bna.FLC.A10, we also see a decrease in downstream targets. This could indicate that those exceptions might also have a respective phenotype, but this is not the case, as both early and late flowering accessions are among those exceptional accessions. Therefore, some other genes seem to compensate the effect, but obviously not a different Bna.FLC copy. In spring accessions, Bna.TEM1.C02 is possibly serving this task, as it is known to be an early suppressor of FT in A. thaliana70. Apart from those exceptions, the analysis of the RNAseq data has shown that Bna.VRN2.Ann is also downregulated in spring accessions. Other vernalisation genes are not or not strongly differentially expressed at this time point. At the same time, copies of important downstream targets like Bna.FT (A02, C02) and Bna.SOC1 (A03, A05, C04) are significantly downregulated in winter accessions, indicating that the vernalisation system indeed is active, although Bna.FT expression levels remain low in spring accessions, probably due to non-inductive photoperiod.
Conclusions
Our data support earlier findings showing that Bna.FLC.A10 is the most important copy of FLC regulating vernalisation in winter-type B. napus. Moreover, we were able to show that this expression pattern is linked to a sequence variant at the COOLAIR binding site. However, Bna.FLC.A10 expression level is not fully predictive of growth type, and more research needs to be done on other factors involved in vernalisation. Accessions showing exceptional patterns of Bna.FLC.A10 might be useful for such an approach, as they represent functional mutants in the respective background. Here, those mutants have shown that other Bna.FLC copies do not compensate lack or overexpression of Bna.FLC.A10. The question remains why Bna.FLC copy number increased so strongly if additional homologs are not needed for vernalisation. The answer may lie in the assumption of different regulatory roles besides vernalisation. Our finding that some copies are obviously no longer cold-responsive suggests this to be the case, and is reflected by the large variation in putative regulatory regions between the copies. Together these findings indicate beginning gene copy subfunctionalisation, implying ongoing functional diversification in this recent allopolyploid.
Supplementary information
Acknowledgements
The authors like to thank the Justus Liebig University for granting SVS with financial support for this project. The authors also thank Iulian Gabor for technical support in the lab and Franziska Hintermeier for sampling the Manitoba time series. We also thank Huey Tyng Lee for assembling the Bna.FLC.A10 sequences.
Author contributions
S.V.S., L.Q. and R.J.S. conceived the project. S.V.S. conducted the bioinformatics analysis of the RNAseq data, the phylogenetic analysis and the analysis of the regulatory regions. D.Q.M. and E.T. conducted the RT-qPCR. All authors have contributed in writing the manuscript and have read and approved the final version.
Data availability
All primer sequences are available from Table S3.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Sarah V. Schiessl and Daniela Quezada-Martinez.
Contributor Information
Sarah V. Schiessl, Email: sarah-veronica.schiessl@agrar.uni-giessen.de
Lunwen Qian, Email: qianlunwen@163.com.
Supplementary information
is available for this paper at 10.1038/s41598-019-51212-x.
References
- 1.Blümel M, Dally N, Jung C. Flowering time regulation in crops-what did we learn from Arabidopsis? Curr Opin Biotechnol. 2015;32:121–129. doi: 10.1016/j.copbio.2014.11.023. [DOI] [PubMed] [Google Scholar]
- 2.Song J, Irwin J, Dean C. Remembering the prolonged cold of winter. Curr Biol. 2013;23:R807–R811. doi: 10.1016/j.cub.2013.07.027. [DOI] [PubMed] [Google Scholar]
- 3.Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES. The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC) PNAS. 2000;97:3753–3758. doi: 10.1073/pnas.060023597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Srikanth A, Schmid M. Regulation of flowering time: all roads lead to Rome. Cell Mol Life Sci. 2011;68:2013–2037. doi: 10.1007/s00018-011-0673-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng W, et al. FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. PNAS. 2011;108:6680–6685. doi: 10.1073/pnas.1103175108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Searle I, et al. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 2006;20:898–912. doi: 10.1101/gad.373506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mateos JL, et al. Combinatorial activities of SHORT VEGETATIVE PHASE and FLOWERING LOCUS C define distinct modes of flowering regulation in Arabidopsis. Genome Biol. 2015;16:31. doi: 10.1186/s13059-015-0597-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abe M, et al. FD, a bZIP Protein Mediating Signals from the Floral Pathway Integrator FT at the Shoot Apex. Science. 2005;309:1052–1056. doi: 10.1126/science.1113095. [DOI] [PubMed] [Google Scholar]
- 9.Simpson GG. The autonomous pathway: epigenetic and post-transcriptional gene regulation in the control of Arabidopsis flowering time. Curr Opin Plant Biol. 2004;7:570–574. doi: 10.1016/j.pbi.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Johansson M, Staiger D. SRR1 is essential to repress flowering in non-inductive conditions in Arabidopsis thaliana. Ex Bot J. 2014;65:5811–5822. doi: 10.1093/jxb/eru317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He Y. Control of the transition to flowering by chromatin modifications. Mol Plant. 2009;2:554–564. doi: 10.1093/mp/ssp005. [DOI] [PubMed] [Google Scholar]
- 12.Turck F, Coupland G. When vernalization makes sense. Science. 2011;331:36–37. doi: 10.1126/science.1200786. [DOI] [PubMed] [Google Scholar]
- 13.Crevillén, P. et al. Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state. Nature; 10.1038/nature13722 (2014). [DOI] [PMC free article] [PubMed]
- 14.Choi K, et al. The FRIGIDA complex activates transcription of FLC, a strong flowering repressor in Arabidopsis, by recruiting chromatin modification factors. The Plant Cell. 2011;23:289–303. doi: 10.1105/tpc.110.075911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331:76–79. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- 16.Wood CC, et al. The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. PNAS. 2006;103:14631–14636. doi: 10.1073/pnas.0606385103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh S, Park S, van Nocker S. Genic and global functions for Paf1C in chromatin modification and gene expression in Arabidopsis. PLoS genet. 2008;4:e1000077. doi: 10.1371/journal.pgen.1000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Exner V, et al. The Chromodomain of LIKE HETEROCHROMATIN PROTEIN 1 Is Essential for H3K27me3 Binding and Function during Arabidopsis Development. PLoS ONE. 2009;4:e5335. doi: 10.1371/journal.pone.0005335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mylne JS, et al. LHP1, the Arabidopsis homologue of HETEROCHROMATIN PROTEIN1, is required for epigenetic silencing of FLC. PNAS. 2006;103:5012–5017. doi: 10.1073/pnas.0507427103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mozgova I, Köhler C, Hennig L. Keeping the gate closed: functions of the polycomb repressive complex PRC2 in development. PlantJ. 2015;83:121–132. doi: 10.1111/tpj.12828. [DOI] [PubMed] [Google Scholar]
- 21.Dittmar EL, Oakley CG, Agren J, Schemske DW. Flowering time QTL in natural populations of Arabidopsis thaliana and implications for their adaptive value. Mol Ecol. 2014;23:4291–4303. doi: 10.1111/mec.12857. [DOI] [PubMed] [Google Scholar]
- 22.Gazzani S, Gendall AR, Lister C, Dean C. Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol. 2003;132:1107–1114. doi: 10.1104/pp.103.021212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mendez-Vigo B, Pico FX, Ramiro M, Martinez-Zapater JM, Alonso-Blanco C. Altitudinal and Climatic Adaptation Is Mediated by Flowering Traits and FRI, FLC, and PHYC Genes in Arabidopsis. Plant Physiol. 2011;157:1942–1955. doi: 10.1104/pp.111.183426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Camargo LEA, Osborn TC. Mapping loci controlling flowering time in Brassica oleracea. Theor Appl Genet. 1996;92:610–616. doi: 10.1007/s001220050170. [DOI] [PubMed] [Google Scholar]
- 25.Okazaki K, et al. Mapping and characterization of FLC homologs and QTL analysis of flowering time in Brassica oleracea. Theor Appl Genet. 2007;114:595–608. doi: 10.1007/s00122-006-0460-6. [DOI] [PubMed] [Google Scholar]
- 26.Osborn, T. C. et al. Comparison of Flowering Time Genes in Brassica rapa, B. napus and Arabidopsis thaliana. Genetics 1123–1129 (1997). [DOI] [PMC free article] [PubMed]
- 27.Schiessl S, Samans B, Hüttel B, Reinhardt R, Snowdon RJ. Capturing sequence variation among flowering-time regulatory gene homologs in the allopolyploid crop species Brassica napus. FrontPlant Sci. 2014;5:404. doi: 10.3389/fpls.2014.00404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tadege, M. et al. Control of flowering time by FLC orthologues in Brassica napus. Plant J28 (2001). [DOI] [PubMed]
- 29.Zou X, et al. Comparative Analysis of FLC Homologues in Brassicaceae Provides Insight into Their Role in the Evolution of Oilseed Rape. PLoS ONE. 2012;7:e45751. doi: 10.1371/journal.pone.0045751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irwin JA, et al. Functional alleles of the flowering time regulator FRIGIDA in the Brassica oleracea genome. BMC plant biol. 2012;12:21. doi: 10.1186/1471-2229-12-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang N, et al. Flowering time variation in oilseed rape (Brassica napus L.) is associated with allelic variation in the FRIGIDA homologue BnaA.FRI.a. Ex Bot J. 2011;62:5641–5658. doi: 10.1093/jxb/err249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu J, et al. A naturally occurring InDel variation in BraA.FLC.b (BrFLC2) associated with flowering time variation in Brassica rapa. BMC plant biol. 2012;12:151. doi: 10.1186/1471-2229-12-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan Y-X, et al. A naturally occurring splicing site mutation in the Brassica rapa FLC1 gene is associated with variation in flowering time. Ex Bot J. 2009;60:1299–1308. doi: 10.1093/jxb/erp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao J, et al. BrFLC2 (FLOWERING LOCUS C) as a candidate gene for a vernalization response QTL in Brassica rapa. Ex Bot J. 2010;61:1817–1825. doi: 10.1093/jxb/erq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao D, et al. The Brassica rapa FLC homologue FLC2 is a key regulator of flowering time, identified through transcriptional co-expression networks. Ex Bot J. 2013;64:4503–4516. doi: 10.1093/jxb/ert264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Irwin JA, et al. Nucleotide polymorphism affecting FLC expression underpins heading date variation in horticultural brassicas. Plant J. 2016;87:597–605. doi: 10.1111/tpj.13221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Razi H, Howell EC, Newbury HJ, Kearsey MJ. Does sequence polymorphism of FLC paralogues underlie flowering time QTL in Brassica oleracea? Theor Appl Genet. 2008;116:179–192. doi: 10.1007/s00122-007-0657-3. [DOI] [PubMed] [Google Scholar]
- 38.Ridge S, Brown PH, Hecht V, Driessen RG, Weller JL. The role of BoFLC2 in cauliflower (Brassica oleracea var. botrytis L.) reproductive development. Ex Bot J. 2015;66:125–135. doi: 10.1093/jxb/eru408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hou J, et al. A Tourist-like MITE insertion in the upstream region of the BnFLC.A10 gene is associated with vernalization requirement in rapeseed (Brassica napus L.) BMC Plant Biol. 2012;12:238. doi: 10.1186/1471-2229-12-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pires JC, et al. Flowering time divergence and genomic rearrangements in resynthesized Brassica polyploids (Brassicaceae) Biol J Linn Soc. 2004;82:675–688. doi: 10.1111/j.1095-8312.2004.00350.x. [DOI] [Google Scholar]
- 41.Schiessl, Hüttel, B., Kuehn, D., Reinhardt, R. & Snowdon, R. J. Post-polyploidisation morphotype diversification associates with gene copy number variation. Sci Rep 41845, 10.1038/srep41845 (2017). [DOI] [PMC free article] [PubMed]
- 42.Chalhoub B, et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 2014;345:950–953. doi: 10.1126/science.1253435. [DOI] [PubMed] [Google Scholar]
- 43.Fletcher, R. S., Mullen, J. L., Heiliger, A. & McKay, J. K. QTL analysis of root morphology, flowering time, and yield reveals trade-offs in response to drought in Brassica napus. Ex Bot J; 10.1093/jxb/eru423 (2014). [DOI] [PMC free article] [PubMed]
- 44.Nelson MN, et al. Quantitative trait loci for thermal time to flowering and photoperiod responsiveness discovered in summer annual-type Brassica napus L. PLoS ONE. 2014;9:e102611. doi: 10.1371/journal.pone.0102611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quijada PA, Udall JA, Lambert B, Osborn TC. Quantitative trait analysis of seed yield and other complex traits in hybrid spring rapeseed (Brassica napus L.): 1. Identification of genomic regions from winter germplasm. Theor Appl Genet. 2006;113:549–561. doi: 10.1007/s00122-006-0323-1. [DOI] [PubMed] [Google Scholar]
- 46.Raman H, et al. Genome-wide association analyses reveal complex genetic architecture underlying natural variation for flowering time in canola. Plant Cell Environ. 2016;39:1228–1239. doi: 10.1111/pce.12644. [DOI] [PubMed] [Google Scholar]
- 47.Raman H, et al. Genetic and physical mapping of flowering time loci in canola (Brassica napus L.) Theor Appl Genet. 2013;126:119–132. doi: 10.1007/s00122-012-1966-8. [DOI] [PubMed] [Google Scholar]
- 48.Udall JA, Quijada PA, Lambert B, Osborn TC. Quantitative trait analysis of seed yield and other complex traits in hybrid spring rapeseed (Brassica napus L.): 2. Identification of alleles from unadapted germplasm. Theor Appl Genet. 2006;113:597–609. doi: 10.1007/s00122-006-0324-0. [DOI] [PubMed] [Google Scholar]
- 49.Wu D, et al. Whole-genome resequencing of a world-wide collection of rapeseed accessions reveals genetic basis of their ecotype divergence. Mol Plant. 2018 doi: 10.1016/j.molp.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 50.Wang X, et al. The genome of the mesopolyploid crop species Brassica rapa. Nat Genet. 2011;43:1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- 51.Parkin, I. A. P. et al. Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome Biol (2014). [DOI] [PMC free article] [PubMed]
- 52.Khan A, et al. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Research. 2018;46:D260–D266. doi: 10.1093/nar/gkx1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schiessl HB, Kuehn D, Reinhardt R, Snowdon RJ. Targeted deep sequencing of flowering regulators in Brassica napus reveals extensive copy number variation. Sci data. 2017;4:170013. doi: 10.1038/sdata.2017.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 55.Havlickova L, et al. Validation of an updated Associative Transcriptomics platform for the polyploid crop species Brassica napus by dissection of the genetic architecture of erucic acid and tocopherol isoform variation in seeds. PlantJ. 2018;93:181–192. doi: 10.1111/tpj.13767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bus A, Korber N, Snowdon RJ, Stich B. Patterns of molecular variation in a species-wide germplasm set of Brassica napus. Theor Appl Genet. 2011;123:1413–1423. doi: 10.1007/s00122-011-1676-7. [DOI] [PubMed] [Google Scholar]
- 57.Körber N, et al. Seedling development in a Brassica napus diversity set and its relationship to agronomic performance. Theor and Appl Genet. 2012;125:1275–1287. doi: 10.1007/s00122-012-1912-9. [DOI] [PubMed] [Google Scholar]
- 58.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Meth. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Quinlan AR. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr prot bioinf. 2014;47:11. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wagner, G. P., Kin, K. & Lynch, V. J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. 131, 281–285, 10.1007/s12064-012-0162-3 (2012). [DOI] [PubMed]
- 63.Schranz, M. E. et al. Characterization and Effects of the Replicated Flowering Time Gene FLC in Brassica rapa. Genetics (2002). [DOI] [PMC free article] [PubMed]
- 64.Cheng F, Wu J, Fang L, Wang X. Syntenic gene analysis between Brassica rapa and other Brassicaceae species. Front plant science. 2012;3:198. doi: 10.3389/fpls.2012.00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Becker A. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol Phylogen Evol. 2003;29:464–489. doi: 10.1016/S1055-7903(03)00207-0. [DOI] [PubMed] [Google Scholar]
- 66.Dreni L, Kater MM. MADS reloaded: evolution of the AGAMOUS subfamily genes. New Phytol. 2014;201:717–732. doi: 10.1111/nph.12555. [DOI] [PubMed] [Google Scholar]
- 67.Parkin IAP, et al. Towards unambiguous transcript mapping in the allotetraploid Brassica napus. Genome. 2010;53(11):929–38. doi: 10.1139/G10-053.. [DOI] [PubMed] [Google Scholar]
- 68.Snowdon RJ, Iniguez Luy FL. Potential to improve oilseed rape and canola breeding in the genomics era. Plant Breeding. 2012;131:351–360. doi: 10.1111/j.1439-0523.2012.01976.x. [DOI] [Google Scholar]
- 69.Li P, Tao Z, Dean C. Phenotypic evolution through variation in splicing of the noncoding RNA COOLAIR. Genes Dev. 2015;29:696–701. doi: 10.1101/gad.258814.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Matias-Hernandez L, Aguilar-Jaramillo AE, Marin-Gonzalez E, Suarez-Lopez P, Pelaz S. RAV genes: regulation of floral induction and beyond. 2014;114:1459–1470. doi: 10.1093/aob/mcu069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All primer sequences are available from Table S3.