Abstract
Folding funnel is the essential concept of the free energy landscape for ordered proteins. How does this concept apply to intrinsically disordered proteins (IDPs)? Here, we address this fundamental question through the explicit characterization of the free energy landscapes of the representative α-helical (HP-35) and β-sheet (WW domain) proteins and of an IDP (pKID) that folds upon binding to its partner (KIX). We demonstrate that HP-35 and WW domain indeed exhibit the steep folding funnel: the landscape slope for these proteins is ca. −50 kcal/mol, meaning that the free energy decreases by ~5 kcal/mol upon the formation of 10% native contacts. On the other hand, the landscape of pKID is funneled but considerably shallower (slope of −24 kcal/mol), which explains why pKID is disordered in free environments. Upon binding to KIX, the landscape of pKID now becomes significantly steep (slope of −54 kcal/mol), which enables otherwise disordered pKID to fold. We also show that it is the pKID–KIX intermolecular interactions originating from hydrophobic residues that mainly confer the steep folding funnel. The present work not only provides the quantitative characterization of the protein folding free energy landscape, but also establishes the usefulness of the folding funnel concept to IDPs.
Subject terms: Computational chemistry, Biophysical chemistry, Computational biophysics
Introduction
Free energy landscape is the cornerstone in the study of protein folding. Its most fundamental aspect is that it is globally funneled such that the folding is energetically biased1–3. Indeed, this notion resolves the well-known paradox of Levinthal4, and accounts for why proteins fold in milliseconds to seconds instead of requiring astronomical timescales5,6. In recent years, the funneled landscape paradigm has been utilized also for understanding biomolecular binding as well as aggregation7–9. However, the usage of biomolecular free energy landscape has remained rather conceptual, which is in contrast to the quantitative role played by the potential energy surface in analyzing chemical reactions of small molecules. Herein, we develop a novel construction method for the protein free energy landscape to fill this gap. Pioneering works in this direction have been carried out through the density-of-state analysis of coarse-grained models10 and through the computation of enthalpy instead of free energy11. The method developed here can be distinguished from these previous works in that it is based on fully atomistic models for proteins and the direct evaluation of the free energy that defines the landscape12,13. We will apply this method to representative α-helical (HP-3514) and β-sheet (WW domain15) proteins to quantitatively argue the strength of the energetic bias toward the folded state.
Protein folding, on the other hand, does not always occur autonomously. In fact, the folding of numerous intrinsically disordered proteins, which is central to their functions, takes place only through the binding with their partners16–18. Can we understand the intrinsically disordered nature of a protein and rationalize its folding upon binding on the basis of the free energy landscape? This is the question we would like to address through the application of our construction method of the landscape. For this purpose, we investigate the pKID region of CREB protein, which is largely disordered when isolated, in the absence and presence of its binding partner, the KIX domain of CREB binding protein19. This is a well-studied paradigm that exhibits coupled folding and binding20. We aim to demonstrate that our explicit characterization of the landscape quantitatively captures common and distinctive features of ordered versus disordered proteins and that the folding funnel, which is steep enough for a disordered protein to fold, emerges as a result of the interaction with its binding partner.
Uncovering the molecular details of such an interaction underlying the folding upon binding of intrinsically disordered proteins is of fundamental importance in molecular biology and is of practical value in protein engineering. Site-directed mutagenesis is a powerful technique to probe effects on protein–protein interaction arising from specific amino acids in the sequence21,22. Related computational methods have also been developed such as computational alanine scanning of protein–protein interfaces23. These mutation-based approaches, however, necessarily invoke perturbations to the underlying protein structures, which sometimes exert disruptive effects in an unexpected and intricate manner24. Recently, we have developed a computational approach, termed the site-directed thermodynamic analysis method, that exactly decomposes protein thermodynamic functions into contributions from constituent amino acid residues13,25–27. Remarkably, this can be done without introducing any mutations, and our method is able to provide in situ characterization of protein–protein interaction at a detailed molecular level. By applying it to analyze the change in the pKID landscape induced by the binding with KIX, we will elucidate the detailed nature of the interaction relevant to the pKID–KIX coupled folding and binding.
Results
Constructing the folding free energy landscape
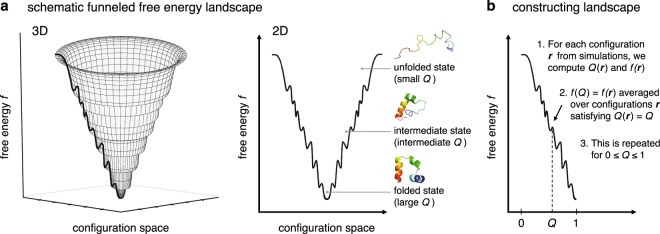
A typical diagram of the funneled free energy landscape is depicted in Fig. 1a, which schematically represents how the free energy decreases as the folding proceeds. To prepare for constructing such a diagram based on a fully microscopic approach, let us start from the precise definition of the landscape: it is the graph of the free energy expressed as a function of the positions (collectively abbreviated as r) of the atoms constituting a molecule of interest. Here, a molecule of interest is a protein, and all the rest of the system – surrounding water molecules and ions – is considered as solvent. The “free energy” f is then given by the gas-phase energy Eu and the solvation free energy , 12,13. (The connection of f to the thermodynamic free energy will be presented below.) Since is defined over the high dimensional configuration space even for small proteins, one necessarily needs to resort to the dimensionality reduction to visualize and practically utilize the landscape. This can be done by introducing an order parameter (or reaction coordinate) Q, defined such that it takes small and large values respectively for the unfolded and folded states. The reduced landscape is then defined by which is the average of over a set of configurations satisfying .
Figure 1.
(a) Schematic representation in 3D (left panel) and 2D (right panel) of the funneled free energy landscape. (b) Steps to construct the landscape from all-atom simulations.
Our method for the explicit construction of the landscape exactly follows what we just described (Fig. 1b). First, molecular dynamics simulations are performed that cover the protein’s unfolded and folded states. For each configuration r taken from the simulations, one computes and . The fraction of native amino acid contacts is chosen here as Q28. in can easily be calculated from the force field parameters, and for we employ the molecular integral-equation theory (see Supplementary Methods). Based on and for the simulated configurations, one can compute by averaging over those configurations having a specific , and this is repeated for . (This is illustrated in Supplementary Fig. S1 for HP-35.) The resulting -versus-Q plot corresponds to the reduced free landscape with which we shall argue the landscape characteristics. It also provides the outline for constructing the 3D representation (see Fig. 1a,b), which will also be used in the following for the visualization purpose.
Comments on the folding free energy landscape
Some comments might be in order here concerning the folding free energy landscape that we study in the present work. In the original work by Bryngelson et al.1, the concept of the folding funnel was introduced for the “energy landscape”. While an explicit expression was not given in that work, it was stated that the energy landscape is defined by “an effective free energy that is a function of the configuration of the protein to describe the protein–solvent system” and that “this description implicitly averages over the solvent coordinates”1. The explicit definition and derivation of the effective energy that defines the energy landscape can be found, e.g., in the article by Lazaridis and Karplus12: it is given by a sum of the gas-phase potential energy and the solvation free energy, that is, introduced above. We call the “free energy” since it includes the solvation free energy, and correspondingly, the energy landscape is referred to as the free energy landscape in the present work. The use of such a term for can be justified also by the fact that it is related to the probability distribution of observing a specific configuration r via with an inverse temperature . Finally, we notice that is defined for a single individual configuration r, and as such, it carries no configurational entropy.
It is important to recognize that the free energy landscape defined by , as well as the reduced one introduced by , are distinct from the free energy profile which is associated with the probability distribution of the order parameter Q, . In fact, these two free energies are related via in which is the configurational entropy1,29. and exhibit utterly different characteristics: While is globally funneled, i.e., there is an overall negative slope, toward the folded state, for a typical two-state folder shows the unfolded- and folded-state minima separated by a transition-state barrier. This is illustrated in Supplementary Fig. S2 displaying the - and -versus-Q curves for HP-35 and WW domain. Also, different computational approaches are necessary for and . Indeed, whereas the sampling of equilibrium configurations is sufficient for constructing the free energy profile , it is insufficient for obtaining the free energy landscape : one also needs to quantify the solvation free energies of the individually sampled configurations30.
Free energy landscapes for ordered versus disordered proteins
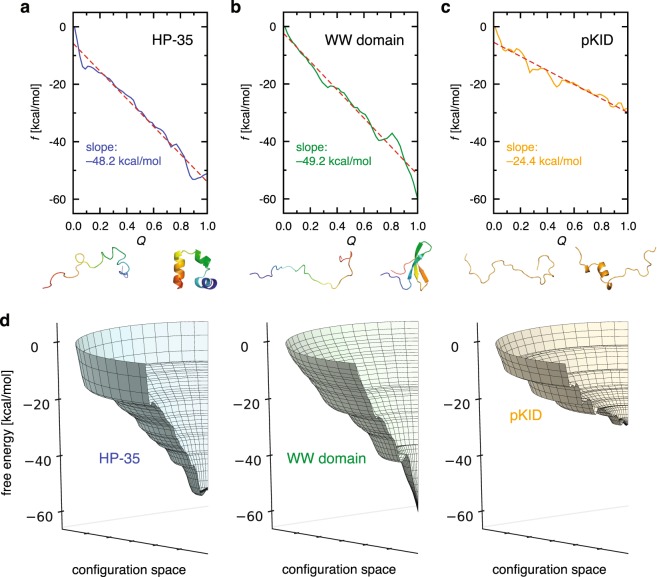
To extract common and distinctive characteristics of ordered and disordered proteins, we show and compare in Fig. 2 the free energy landscapes for HP-35, WW domain, and pKID. These landscapes were constructed based on their respective all-atom molecular dynamics simulations: for HP-35, we used the ~400 μs folding-unfolding simulation trajectory31, and the Q and f values along the trajectory, necessary for constructing the landscape, are displayed in Supplementary Fig. S3; for WW domain, we used 6 independent simulation trajectories of 100 μs32,33, and the Q and f values therefrom are shown in Supplementary Fig. S4; and for pKID, we conducted ~10 μs molecular dynamics simulations, and the simulation results for the systems involving pKID are presented in Supplementary Figs S5 to S7. The simulations for HP-35 and WW domain were performed at close to their respective in silico melting temperatures, whereas those for pKID at 300 K. The force fields used were FF99SB*-ILDN34–36 for HP-35, FF99SB-ILDN34,35 for WW domain, and CHARMM22*37–39 for pKID; and the TIP3P water model40 was adopted for simulating all the systems. HP-35 and WW domain are respectively representative α-helical and β-sheet proteins, and pKID is a well-studied intrinsically disordered protein. We have chosen these particular systems also because their sequence lengths are comparable (HP-35 and WW domain, 35 residues; pKID, 34 residues): this suppresses sequence-length dependent effects that may obscure our analysis.
Figure 2.
(a–d) Free energy landscapes for HP-35 (a), WW domain (b), pKID (c), and the comparison in 3-D representation (d). The free energy refers to the difference from the respective , and the dashed line in (a–c) denotes a linear fit.
The overall slope of the landscape characterizes the global funneledness (the strength of the energetic bias) toward the folded state. The slope of the landscape for HP-35 estimated in Fig. 2a, −48.2 ± 1.7 kcal/mol, means that, e.g., 10% of the native contacts is formed with the free energy gain (decrease) by 4.8 kcal/mol. (The error estimation was done based on the block analysis as described in Supplementary Methods. We also computed the standard errors for the landscape curves, and the results are shown in Supplementary Fig. S8). Interestingly, the slope of the β-sheet WW domain (−49.2 ± 0.5 kcal/mol; see Fig. 2b) is found to be comparable to that of the α-helical HP-35. Such a degree of funneledness may be a typical one that is necessary to fold proteins of this sequence length (35 residues) against the unfavorable force arising from the configurational entropy. The landscape of disordered pKID, on the other hand, shows intriguing characters. Like HP-35 and WW domain, the overall landscape for pKID is somewhat funneled. However, the slope of the landscape for pKID is −24.4 ± 3.6 kcal/mol (see Fig. 2c) which is significantly smaller than that for HP-35 and WW domain. Such common and distinctive characteristics clearly show up in the 3D representation of the respective landscapes (Fig. 2d). Since the sequence lengths of HP-35, WW domain, and pKID are about the same, the magnitude of the unfavorable entropic force is expected to be comparable. The net driving force for folding is determined by a balance of the energetic bias, given by the slope of the landscape, and the opposing force arising from the configurational entropy, and the intrinsically disordered nature of pKID can be accounted for by the insufficient energetic bias to overcome the unfolding force for this sequence length. Thus, pKID is disordered not because the landscape is not funneled, but because the landscape is not steep enough to allow its folding.
Binding-induced change in the landscape for pKID
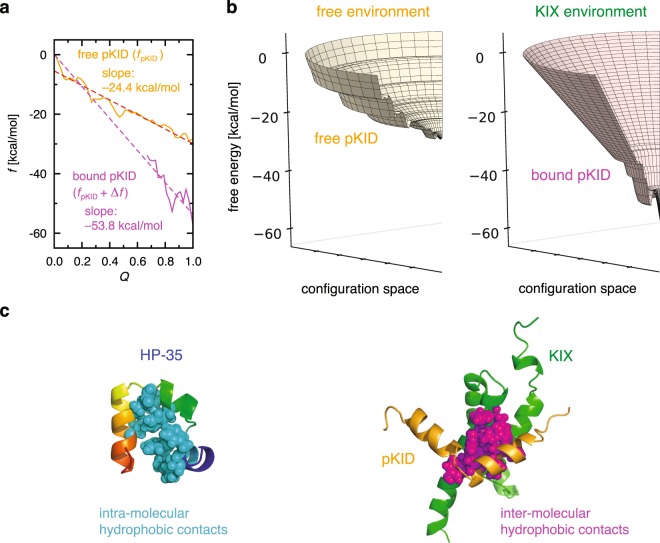
pKID is also known as a paradigmatic disordered protein exhibiting the folding upon binding with its partner (KIX)19,20. To characterize this fascinating phenomenon in landscape terms, we investigate the change in the landscape of pKID induced by the binding. This can be done through a comparison of the landscape for the free pKID (free environment) and the one for the bound pKID in the pKID–KIX complex (KIX environment). The latter landscape can be constructed based on molecular dynamics simulations for the pKID–KIX complex. Here, the free energy needs to be extended to , which is a sum of the free energy for pKID, denoted as , and the solvent-averaged binding potential, , incorporating the binding effect: is the direct pKID–KIX interaction potential, and is the solvent-induced potential defined by 41,42. The landscapes for the free and bound pKID are displayed in Fig. 3a,b. We find that the landscape for pKID gets significantly steeper upon binding, and its slope (−53.8 ± 12.9 kcal/mol) becomes comparable to that of HP-35 (−48.2 kcal/mol). Thus, the free energy landscape for pKID becomes steep in the KIX environment, and this provides the landscape explanation on why the binding with KIX is prerequisite for the folding of pKID.
Figure 3.
(a,b) Free energy landscape for the free pKID (colored orange) and the bound pKID (magenta) (a), and the comparison in 3-D representation (b). The free energy refers to the difference from the respective , and the dashed line in (a) denotes a linear fit. (c) Amino acid residues forming intra-molecular hydrophobic contacts in HP-35 (PDB entry 1YRF) and inter-molecular hydrophobic contacts in pKID–KIX complex (PDB entry 2LXT) are represented by spheres.
Site-directed analysis of the pKID–KIX interactions
It is thus the direct and solvent-mediated pKID–KIX interactions (both incorporated in ) that confer the folding funnel on otherwise disordered pKID. Using the simulated pKID–KIX complex configurations, we computed the average to be −25.4 kcal/mol. To further elucidate the molecular details of such interactions, we shall resort to the site-directed thermodynamic analysis method13,25–27. This method allows us to decompose into contributions from individual constituent amino acid residues (see Supplementary Methods). To facilitate the understanding of our results, we will separately deal with neutral- and charged-residue contributions. In fact, we find that neutral residues provide more significant contributions () than charged residues ().
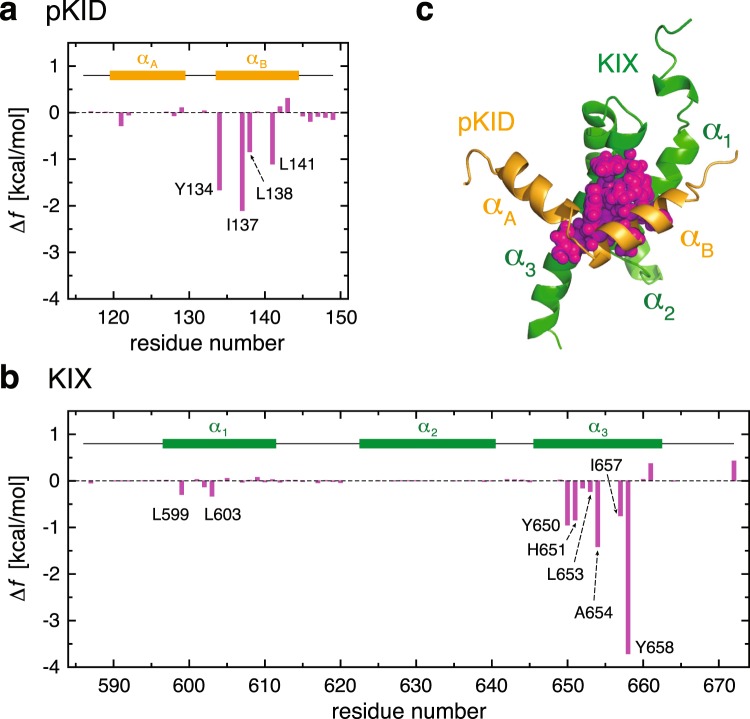
Site-resolved contributions to from neutral residues are shown in Fig. 4a,b, and the locations of the major contributing residues are displayed in Fig. 4c. We observe that major contributions arise from hydrophobic residues in the pKID αB helix and those in the KIX α3 helix. In particular, Tyr-134 and Ile-137 provide the two largest contributions to originating from pKID. This is in accord with the site-directed mutagenesis study, in which these two residues were found to be the most destabilizing residues in pKID when mutated to Ala43. Concerning the neutral residues in KIX, Tyr-658 and Ala-654 are the two most significant contributors to . The critical role of these residues in the pKID–KIX binding was discussed in the previous NMR study, and in particular, it was demonstrated that mutating Tyr-658 to Ala completely abolishes the complex formation19. Thus, our site-directed analysis method is able to identify those critical amino acid residues, and remarkably, this is achieved without introducing any mutations.
Figure 4.
(a,b) Contributions to from neutral residues of pKID (a) and KIX (b). (c) Amino acid residues forming inter-molecular hydrophobic contacts in pKID–KIX complex are represented by spheres.
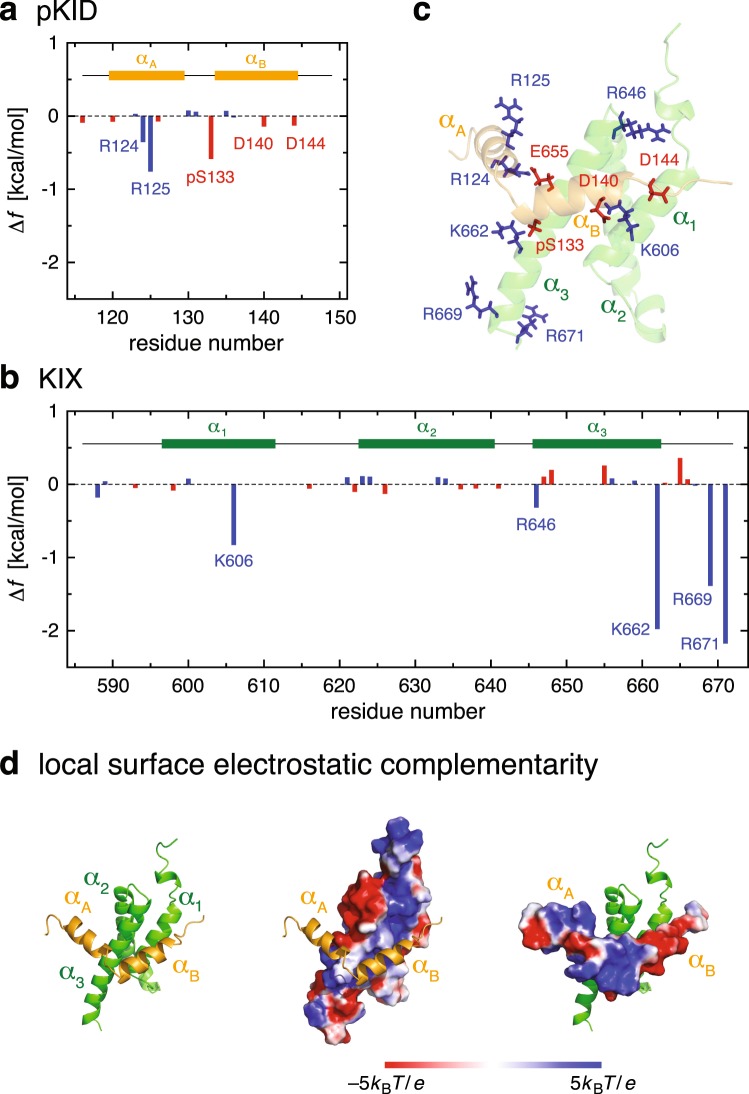
Site-resolved contributions to arising from charged residues are displayed in Fig. 5a,b (see also Fig. 5c for their locations). One observes large negative contributions from Lys-662, Arg-669 and Arg-671 of KIX. To understand these results, we have analyzed representative inter-protein contacts involving charged residues, and the results are summarized in Supplementary Table S1. As listed there, the phosphoserine residue (pSer-133) of pKID forms a hydrogen bond to Lys-662 of KIX with a large population (~90%) and it is also hydrogen bonded to the C-terminal basic residues (Arg-669 and Arg-671) of KIX with substantial probabilities (~50% and ~70%, respectively). Thus, the favorable negative contributions to from these residues reflect the presence of those stabilizing hydrogen-bond interactions between pKID and KIX.
Figure 5.
(a,b) Contributions to from charged residues of pKID (a) and KIX (b). (c) Amino acid residues forming inter-molecular hydrogen bonds/salt bridges are indicate by stick representation. (d) Surfaces of KIX (middle panel) and pKID (right panel) are color coded by the electrostatic potential.
We also find weak but non-negligible favorable contributions to originating from Arg-124, Arg-125, Asp-140 and Asp-144 in pKID and from Lys-606 and Arg-646 in KIX (Fig. 5a,b). As can be inferred from Supplementary Table S1, these contributions are associated with the inter-protein contacts between oppositely charged residues. Motivated by this observation, we examined the surface electrostatic potential of pKID and KIX. Interestingly, we find alternating local electrostatic complementarity at the binding faces between the pKID αA helix and the KIX α3 helix and between the pKID αB helix and the other side of the KIX α3 helix (Fig. 5d): the binding side of αA has positive electrostatic potential, which contacts with α3 having negative electrostatic potential; and the sign of electrostatic potential is reversed between αB and the other side of α3. Since pKID must be docked with a proper position and orientation at the KIX surface in order to maximize such an interaction reflecting the local surface electrostatic complementarity, this weak interaction must be responsible for the binding specificity. Its relevance in the pKID–KIX binding is also corroborated by noticing that those amino acid residues listed above, as well as Glu-648 and Glu-655 in KIX generating negative surface potential for the binding with the pKID αA helix, are well conserved in CREB and CBP family proteins19.
Standard binding free energy
Finally, we argue the relation between the free energy f defining the landscape and the thermodynamic free energy. The free energy is defined for individual protein configurations r, and hence, it carries no configurational entropy. The thermodynamic free energy, on the other hand, is given by with 12,13. With the probability distribution, , of observing a specific configuration r, and recalling the definition of the configurational entropy, , one understands that F consists of an ensemble average of f and the configurational entropy, . For the binding thermodynamics, one additional term, called the external entropy (to be denoted as ), needs to be incorporated44,45. The standard binding free energy is then given by 45. Here, ΔX for or is given by .
Using the simulated structures for the free pKID, free KIX, and pKID–KIX complex, we computed the terms that contribute to (see Supplementary Methods). The results of our computations, along with error estimations, are summarized in Supplementary Table S2. The resulting standard binding free energy, , is in reasonable agreement with experiment (−8.1 kcal/mol)46. The large standard error of mainly comes from that of the configurational entropy term, (see Supplementary Table S2). In this regard, we notice that the magnitude of standard error is quite small (<1%) for of the three individual systems, but this is significantly enlarged when the difference () is taken because of the large cancellation of the individual contributions.
Discussion
What could be the molecular origin of the different behavior between HP-35, an α-helical protein which autonomously folds, and pKID, which requires a partner for its folding into an α-helical structure? In this connection, we recall that a helical structure is in general not stable by itself, and additional stabilizing interactions must be present for its maintenance47. In fact, all the three α helices in HP-35 are tightly in contact with the hydrophobic core (left panel in Fig. 3c). On the other hand, intrinsically disordered proteins generally contain a low population of bulky hydrophobic residues48,49, and as such, pKID does not form intra-molecular hydrophobic contacts in the free environment. The presence/absence of the hydrophobic core in stabilizing the helical structure explains why the landscape for the free pKID is much shallower than that of HP-35. Upon the pKID–KIX binding, hydrophobic contacts can now be formed inter-molecularly (right panel in Fig. 3c), which contributes to stabilizing the helical structure of the pKID in KIX environment. The emergence of such additional intermolecular interactions upon binding renders the free energy landscape of the bound pKID to be steep enough to allow the folding of pKID.
Elucidating the molecular details of such interactions involving intrinsically disordered proteins is crucial to understand and eventually modify their function in gene regulation and signal transduction. While site-directed mutation is a common technique for identifying hot spots in protein–protein interactions, its application sometimes causes undesired significant alternations in protein structures. Here, we apply the site-directed thermodynamic analysis method – a computational approach that does not call for introducing any mutations – to provide in situ characterization of the pKID–KIX interactions. We find that interactions between hydrophobic residues that belong to the pKID αB helix and the KIX α3 helix play a dominant role in the pKID–KIX complex formation. In particular, Tyr-134 and Ile-137 are found to be the most significant amino acid residues in pKID, and Ala-654 and Tyr-658 are the corresponding residues in KIX, which is in accord with the experimental observations19,43. We also show that positively charged residues in the pKID αA helix and negatively charged residues in the KIX α3 helix provide weak but specific interactions between pKID and KIX.
Site-directed thermodynamic analysis thus reveals the presence of the strong interaction between the pKID αB helix and the KIX α3 helix, which mainly arises from hydrophobic contacts, and of the weak but specific interaction between the pKID αA helix and the other side of the KIX α3 helix, which is essentially of electrostatic origin. The presence of the two interactions that differ in strength will be responsible for the pKID–KIX binding process. In fact, it has been observed from the previous experimental studies that the binding of pKID to KIX involves an intermediate state where the transient complex is formed with the pKID αB helix anchored to the KIX hydrophobic residues20,43. Computer simulation studies also observe the initial encounter complex formed by the docking of the pKID αB helix to KIX, followed by the binding of the pKID αA helix50,51. Our results on the pKID–KIX interactions explain such a sequence of events observed in the pKID–KIX binding process.
Conclusions
Explicit characterization of the folding free energy landscape from fully microscopic approaches will significantly contribute to advancing our molecular-level understanding of protein folding phenomena. The present work develops a novel method for the explicit characterization based on atomistic simulations and the direct calculation of the free energy that defines the landscape. This method is applied to extract common and distinctive characteristics of the landscapes of ordered and intrinsically disordered proteins and to derive the landscape explanation on the folding upon binding. The method developed here is applicable to any atomistic simulations, and will be effective in expanding the scope of the funneled landscape perspective to a variety of processes that involve disordered proteins. We also apply the site-directed thermodynamic analysis method to provide detailed and in situ characterization of the interactions relevant to the coupled folding and binding. This analysis method identifies critical amino acid residues in protein–protein interactions without resorting to any mutations, and will also be valuable for identifying and characterizing hot spots in the protein–ligand interaction and the protein–DNA binding.
Supplementary information
Acknowledgements
This work was supported by the Samsung Science and Technology Foundation under Project Number SSTF-BA1401-52. We are grateful to the D.E. Shaw Research for the simulation trajectories of HP-35 and WW domain.
Author contributions
S.-H.C. and S.H. conducted the research and wrote the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-50825-6.
References
- 1.Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG. Funnels, pathways, and the energy landscape of protein folding: A synthesis. Proteins. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 2.Wolynes PG, Onuchic JN, Thirumalai D. Navigating the folding routes. Science. 1995;267:1619–1620. doi: 10.1126/science.7886447. [DOI] [PubMed] [Google Scholar]
- 3.Dill KA, Chan HS. From Levinthal to pathways to funnels. Nat. Struct. Biol. 1997;4:10–19. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 4.Levinthal C. How to fold graciously. Mössbauer Spectroscopy in Biological Systems Proceedings. 1969;67:22–24. [Google Scholar]
- 5.Zwanzig R, Szabo A, Bagchi B. Levinthal’s paradox. Proc. Natl. Acad. Sci. USA. 1992;89:20–22. doi: 10.1073/pnas.89.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karplus M. Behind the folding funnel diagram. Nat. Chem. Biol. 2011;7:401–404. doi: 10.1038/nchembio.565. [DOI] [PubMed] [Google Scholar]
- 7.Hartl FU, Hayer-Hartl M. Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol. 2009;16:574–581. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- 8.Zheng W, Schafer NP, Davtyan A, Papoian GA, Wolynes PG. Predictive energy landscapes for protein–protein association. Proc. Natl. Acad. Sci. USA. 2012;109:19244–19249. doi: 10.1073/pnas.1216215109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adamcik J, Mezzenga R. Amyloid polymorphism in the protein folding and aggregation energy landscape. Angew. Chem. Int. Ed. 2018;57:8370–8382. doi: 10.1002/anie.201713416. [DOI] [PubMed] [Google Scholar]
- 10.Wang J, et al. Topography of funneled landscapes determines the thermodynamics and kinetics of protein folding. Proc. Natl. Acad. Sci. USA. 2012;109:15763–15768. doi: 10.1073/pnas.1212842109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piana S, Lindorff-Larsen K, Shaw DE. Atomic-level description of ubiquitin folding. Proc. Natl. Acad. Sci. USA. 2013;110:5915–5920. doi: 10.1073/pnas.1218321110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lazaridis T, Karplus M. Thermodynamics of protein folding: A microscopic view. Biophys. Chem. 2003;100:367–395. doi: 10.1016/S0301-4622(02)00293-4. [DOI] [PubMed] [Google Scholar]
- 13.Chong S-H, Ham S. Distinct role of hydration water in protein misfolding and aggregation revealed by fluctuating thermodynamics analysis. Acc. Chem. Res. 2015;48:956–965. doi: 10.1021/acs.accounts.5b00032. [DOI] [PubMed] [Google Scholar]
- 14.McKnight CJ, Matsudaira PT, Kim PS. NMR structure of the 35-residue villin headpiece subdomain. Nat. Struct. Biol. 1997;4:180–184. doi: 10.1038/nsb0397-180. [DOI] [PubMed] [Google Scholar]
- 15.Jäger M, et al. Structure–function–folding relationship in a WW domain. Proc. Natl. Acad. Sci. USA. 2006;103:10648–10653. doi: 10.1073/pnas.0600511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tompa P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 2005;579:3346–3354. doi: 10.1016/j.febslet.2005.03.072. [DOI] [PubMed] [Google Scholar]
- 17.Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uversky VN. A decade and a half of protein intrinsic disorder: Biology still waits for physics. Protein Sci. 2013;22:693–724. doi: 10.1002/pro.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radhakrishnan I, et al. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: A model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/S0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 20.Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447:1021–1025. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- 21.Fersht AR. Structure and Mechanism in Protein Science. New York: W. H. Freeman and Company; 1999. [Google Scholar]
- 22.Morrison KL, Weiss GA. Combinatorial alanine-scanning. Curr. Opin. Chem. Biol. 2001;5:302–307. doi: 10.1016/S1367-5931(00)00206-4. [DOI] [PubMed] [Google Scholar]
- 23.Massova I, Kollman PA. Computational alanine scanning to probe protein–protein interactions: A novel approach to evaluate binding free energies. J. Am. Chem. Soc. 1999;121:8133–8143. doi: 10.1021/ja990935j. [DOI] [Google Scholar]
- 24.Xiao S, et al. Rational modification of protein stability by targeting surface sites leads to complicated results. J. Am. Chem. Soc. 2013;110:11337–11342. doi: 10.1073/pnas.1222245110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chong S-H, Ham S. Atomic decomposition of the protein solvation free energy and its application to amyloid-beta protein in water. J. Chem. Phys. 2011;135:034506. doi: 10.1063/1.3610550. [DOI] [PubMed] [Google Scholar]
- 26.Chong S-H, Ham S. Interaction with the surrounding water plays a key role in determining the aggregation propensity of proteins. Angew. Chem. Int. Ed. 2014;53:3961–3964. doi: 10.1002/anie.201309317. [DOI] [PubMed] [Google Scholar]
- 27.Chong S-H, Ham S. Site-directed analysis on protein hydrophobicity. J. Compute. Chem. 2014;35:1364–1370. doi: 10.1002/jcc.23631. [DOI] [PubMed] [Google Scholar]
- 28.Best RB, Hummer G, Eaton WA. Native contacts determine protein folding mechanisms in atomistic simulations. Proc. Natl. Acad. Sci. USA. 2013;110:17874–17879. doi: 10.1073/pnas.1311599110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chong S-H, Ham S. Dissecting protein configurational entropy into conformational and vibrational contributions. J. Phys. Chem. B. 2015;119:12623–12631. doi: 10.1021/acs.jpcb.5b07060. [DOI] [PubMed] [Google Scholar]
- 30.Ferreiro DU, Komives EA, Wolynes PG. Frustration in biomolecules. Q. Rev. Biophys. 2014;47:285–363. doi: 10.1017/S0033583514000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piana S, Lindorff-Larsen K, Shaw DE. Protein folding kinetics and thermodynamics from atomistic simulation. Proc. Natl. Acad. Sci. USA. 2012;109:17845–17850. doi: 10.1073/pnas.1201811109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaw DE, et al. Atomic-level characterization of the structural dynamics of proteins. Science. 2010;330:341–346. doi: 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- 33.Piana S, et al. Computational design and experimental testing of the fastest-folding β-sheet protein. J. Mol. Biol. 2011;405:43–48. doi: 10.1016/j.jmb.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 34.Hornak V, et al. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindorff-Larsen K, et al. Improved side-chain torsion potentials for the Amber ff99SB protein force fields. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Best RB, Hummer G. Optimized molecular dynamics force fields applied to the helix-coil transition of polypeptides. J. Chem. Phys. B. 2009;113:9004–9015. doi: 10.1021/jp901540t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacKerell AD, Jr., et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 38.MacKerell AD, Jr., Feig M, Brooks CL., III Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 39.Piana S, Lindorff-Larsen K, Shaw DE. How robust are protein folding simulations with respect to force field parameterization? Biophys. J. 2011;100:L47–L49. doi: 10.1016/j.bpj.2011.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 41.Ben-Naim A. Hydrophobic Interactions. New York: Plenum; 1980. [Google Scholar]
- 42.Chong S-H, Ham S. Impact of chemical heterogeneity on protein self-assembly in water. Proc. Natl. Acad. Sci. USA. 2012;109:7636–7641. doi: 10.1073/pnas.1120646109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dahal L, Kwan TOC, Shammas SL, Clarke J. pKID binds to KIX via an unstructured transition state with nonnative interactions. Biophys. J. 2017;113:2713–2722. doi: 10.1016/j.bpj.2017.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gilson MK, Given JA, Bush BL, McCammon JA. The statistical-thermodynamic basis for computation of binding affinities: A critical review. Biophys. J. 1997;72:1047–1069. doi: 10.1016/S0006-3495(97)78756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chong S-H, Ham S. New computational approach for external entropy in protein–protein binding. J. Chem. Theory Comput. 2016;12:2509–2516. doi: 10.1021/acs.jctc.6b00174. [DOI] [PubMed] [Google Scholar]
- 46.Goto NK, Zor T, Martinez-Yamout M, Dyson HJ, Wright PE. Cooperativity in transcription factor binding to the coactivator CREB-binding protein (CBP) J. Biol. Chem. 2002;277:43168–43174. doi: 10.1074/jbc.M207660200. [DOI] [PubMed] [Google Scholar]
- 47.Dill K. Dominant forces in protein folding. Biochemistry. 1990;29:7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 48.Uversky VN. Intrinsically disordered proteins from A to Z. Int. J. Biochem. Cell Biol. 2011;43:1090–1103. doi: 10.1016/j.biocel.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 49.Tompa P. Structure and Function of Intrinsically Disordered Proteins. Boca Raton: CRC Press; 2010. [Google Scholar]
- 50.Turjanski AG, Gutkind JS, Best RB, Hummer G. Binding-induced folding of a natively unstructured transcription factor. PLoS Comput. Biol. 2008;4:e1000060. doi: 10.1371/journal.pcbi.1000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ganguly D, Chen J. Topology-based modeling of intrinsically disordered proteins: Balancing intrinsic folding and intermolecular interactions. Proteins. 2011;79:1251–1266. doi: 10.1002/prot.22960. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.