Abstract
The chemical ecology and chemical defenses of sponges have been investigated for decades; consequently, sponges are among the best understood marine organisms in terms of their chemical ecology, from the level of molecules to ecosystems. Thousands of natural products have been isolated and characterized from sponges, and although relatively few of these compounds have been studied for their ecological functions, some are known to serve as chemical defenses against predators, microorganisms, fouling organisms, and other competitors. Sponges are hosts to an exceptional diversity of microorganisms, with almost 40 microbial phyla found in these associations to date. Microbial community composition and abundance are highly variable across host taxa, with a continuum from diverse assemblages of many microbial taxa to those that are dominated by a single microbial group. Microbial communities expand the nutritional repertoire of their hosts by providing access to inorganic and dissolved sources of nutrients. Not only does this continuum of microorganism–sponge associations lead to divergent nutritional characteristics in sponges, these associated microorganisms and symbionts have long been suspected, and are now known, to biosynthesize some of the natural products found in sponges. Modern “omics” tools provide ways to study these sponge–microbe associations that would have been difficult even a decade ago. Metabolomics facilitate comparisons of sponge compounds produced within and among taxa, and metagenomics and metatranscriptomics provide tools to understand the biology of host–microbe associations and the biosynthesis of ecologically relevant natural products. These combinations of ecological, microbiological, metabolomic and genomics tools, and techniques provide unprecedented opportunities to advance sponge biology and chemical ecology across many marine ecosystems.
Introduction
Marine sponges are an ancient group, with ∼9000 species currently recognized. Sponges have been widely successful in their expansion across diverse ecological niches (poles to tropics; soft and hard bottom habitats; presence in both shallow and deep seas and into freshwater habitats) (Van Soest et al. 2012; Webster and Thomas 2016). As a group, their success in these systems is linked to their ability to efficiently remove small particulate matter (phytoplankton, detritus, bacteria, and viruses) from the water via heterotrophic filter feeding (Maldonado et al. 2012; McMurray et al. 2016). Sponges also host a tremendous diversity of microbial symbionts, with at least 39 microbial (Bacterial and Archaeal) phyla detected in recent large-scale microbiome surveys, with a dominance of the bacterial groups γ-, α-, and β-proteobacteria, Actinobacteria, Cyanobacteria, Chloroflexi, and Bacteroidetes, as well as the presence of sponge-specific lineages like Poribacteria (Moya et al. 2008; Thomas et al. 2016; Pita et al. 2018; Podell et al. 2019). Their success in oligotrophic systems may therefore be linked to an expanded metabolic capacity from these symbiotic associations (Taylor et al. 2007; Thacker and Freeman 2012; Freeman et al. 2013; McMurray et al. 2018).
Marine sponges are a rich source of natural products, which have been reported in sponges from diverse ecosystems ranging from tropical to polar seas. Several decades of intensive searching have uncovered over 10,000 sponge-derived organic natural products (http://pubs.rsc.org/marinlit/; accessed January 1, 2019). Most research on sponge natural products has emphasized their biomedical relevance, such as anticancer activity (Gerwick and Moore 2012; Calcabrini et al. 2017). Sponge natural products encompass a range of structure types including alkaloids, terpenes (including sterols), polyketides, fatty acids, and peptides (Calcabrini et al. 2017). Because of their rich diversity of natural products and the notable bioactivities demonstrated for these compounds, sponges are among the best-studied marine invertebrates for their natural products chemistry (Blunt et al. 2018).
The numerous natural products that were being discovered in marine sponges in the 1970s and 1980s led to questions about why so many diverse compounds were produced in sponges, and collaborations between chemists and marine biologists beginning in the 1980s began to address these questions (reviewed by Pawlik et al. 2013). The field of chemical ecology integrates chemistry and biology to examine the ecological roles of natural products and chemically mediated interactions among organisms and their environment. Examples of ecological functions of natural products include chemical defenses against predators and pathogens, settlement cues for larvae, allelopathic effects that prevent fouling by epiphytes and overgrowth by competitors, protection against UV radiation, and pheromones for mate searching behavior. Marine chemical ecology relies on chemical isolation, structural elucidation, and ecological experimentation, often conducted in the field or at least with naturally co-occurring consumers, competitors, or pathogens, to determine the compounds responsible for mediating observed ecological interactions. Natural products derived from marine sponges have been shown to play a role in many of these behaviors and interactions (Paul et al. 2007; Baker et al. 2010; Pawlik 2011; Pawlik et al. 2013). Discovering the compounds responsible for ecological interactions can contribute to other research fields including animal behavior, neurobiology, sensory ecology, reproduction, symbiosis, larval biology, disease ecology, microbiology, and aquaculture.
While only a small fraction of the 10,000+ natural products known from marine sponges have been studied for their ecological functions, sponges are still the best studied marine invertebrate group in terms of their chemical ecology. Many reviews of marine chemical ecology have been written (McClintock and Baker 2001; Hay 2009; Paul et al. 2011; Pawlik et al. 2013; Puglisi et al. 2014, 2019; Puglisi and Becerro 2018), and it is not our intent to comprehensively review the chemical ecology of marine sponges. Instead, we provide highlights of recent research and will focus on some enduring questions in sponge chemical ecology that can be particularly aided by integrating “omics” methods into the chemical ecology toolkit. We examine the roles of sponge microbiomes and how associated microorganisms and symbionts diversify the nutritional capacities of sponges and are known to biosynthesize some of the natural products found in sponges. This knowledge about the source of different sponge natural products can provide insights into their evolution and ecology. Metabolomics methods can yield insights into the diversity of natural compounds present in sponges, including minor or unstable compounds that would be difficult to isolate and characterize, and we provide examples from our ongoing research. This new knowledge of sponge metabolomes provides greater insight into diversity, natural occurrence, and abundance of sponge compounds.
Sponge microbiology
Pioneering work in sponge microbiology recognized that microbial community composition was highly variable across sponge species, with some “bacteriosponges” hosting abundant symbiont communities (Reiswig 1974). These bacteriosponges (now termed high microbial abundance [HMA] sponges) generally have denser tissues and lower water pumping rates than sponges that host a lower diversity and abundance of microbial symbionts (low microbial abundance [LMA] sponges) (Vacelet and Donadey 1977; Hentschel et al. 2006). This dichotomy was originally established using electron microscopy to enumerate microbial abundance and has been widely used to group sponges into structural and functional groups over the past decade (Weisz et al. 2007, 2008; Gloeckner et al. 2014; Moitinho-Silva et al. 2017b). While microbiome surveys using higher resolution techniques like next generation 16S rDNA amplicon sequencing generally find higher microbial diversity in HMA sponges, it is also becoming increasingly evident that host sponge identity is the dominant driver of variation in microbial community composition (Easson and Thacker 2014; Thomas et al. 2016; Moitinho-Silva et al. 2017a). In fact, host species identity accounts for ∼60–75% of the variation in microbial community composition across individual sponges at regional scales (Caribbean Panama [Easson and Thacker 2014] and Vietnam [Turon et al. 2018]). Global assessments of sponge microbiome composition and diversity as part of the Global Sponge Microbiome Project also support these regional trends (Thomas et al. 2016; Moitinho-Silva et al. 2017a; Pita et al. 2018). This, along with strong phylogenetic relationships in microbiome diversity, implies strong evolutionary selection for divergent microbiomes across different sponge lineages (Easson and Thacker 2014; Thomas et al. 2016). Thus, the HMA/LMA dichotomy may represent the endpoints of a complex and nuanced continuum that we are only beginning to elucidate (Easson and Thacker 2014; Pita et al. 2018).
Microbial symbionts confer a nutritional advantage to their host by allowing for the exploitation of novel sources of nutrients. Autotrophic symbionts may provide over 50% of their host’s carbon budget and may also allow for the assimilation of exogenous nitrogen sources like nitrate and ammonia (NO3− and NH3) or the recycling of NH3 from the host (Wilkinson 1983; Taylor et al. 2007; Southwell et al. 2008; Fan et al. 2012; Freeman et al. 2013). Symbionts may also fix nitrogen, carry out diverse nitrogen transformations, and facilitate phosphorus sequestration, potentially providing a selective advantage in oligotrophic systems like coral reefs (Hoffmann et al. 2009; Zhang et al. 2014; Pita et al. 2018). In addition, microbial symbionts may mediate the assimilation of dissolved organic matter (DOM) that ultimately contributes to the retention of carbon and nitrogen in reef environments through the “sponge loop” (de Goeij et al. 2013; McMurray et al. 2018; Pita et al. 2018). The breadth of metabolic innovations provided by microbial symbionts is likely to be considerable and is being expanded with metagenomic and meta-transcriptomic studies that report evidence of methylotrophy, vitamin synthesis, and even feeding on molecules from the extracellular matrix of the host (Fiore et al. 2015; Webster and Thomas 2016; Pita et al. 2018). How overall microbial abundance (HMA or LMA status) influences sponge holobiont (both sponge and microbial metabolic pathways) metabolism has been studied using the stable isotope ratios for C and N (δ13C and δ15N, respectively) to trace the sources of C and N assimilated and transformed by different sponge species (Weisz et al. 2007). A divergence in holobiont metabolism (δ13C and δ15N values based on bulk sponge tissue including both sponge and microbial cells) between HMA and LMA sponges was observed. Most, but not all, HMA sponges had depleted δ15N relative to LMA species, suggesting that nitrogen in HMA sponges is derived from microbial transformations, while LMA species rely more on external sources of nitrogen that they assimilate via heterotrophic filter feeding (Weisz et al. 2007).
Variation in microbial abundance or community composition may have important ecological and evolutionary consequences, especially if the presence of specific symbiont groups allows their host to exploit novel and unique sources of nutrients (Morganti et al. 2017). Recent evidence supports strong divergence in these interactions across host species (Erwin and Thacker 2007; Freeman and Thacker 2011; Freeman et al. 2013), with even closely related species being on distinct evolutionary trajectories with their symbiont communities (Freeman et al. 2015). In addition, ∼75% of the variation in δ13C and δ15N values of bulk sponge tissue from 19 Caribbean sponge species was driven by host sponge identity; overall microbial abundance (HMA or LMA) accounted for only 21% of this variation (Freeman et al. 2014). These data are beginning to highlight the fact that environmental conditions in some ocean basins may have favored ecological diversification in sponges at the species level, with unique microbial symbiont communities allowing coexisting sponge species to exploit novel sources (or combinations of sources) on these reefs. The evolutionary drivers of this diversification and the role that microbial symbionts have played in the successful expansion of sponges across coral reefs are still largely unknown, but this topic is increasingly discussed in the literature (Pawlik et al. 2018).
Chemical defenses are another important adaptation allowing some sponges to survive on coral reefs by deterring predators, competitors, and fouling organisms, and preventing disease (Pawlik et al. 1995; Pawlik 2011; Webster and Thomas 2016). Although microbial symbionts have been shown to provide their hosts with supplemental nutrients, an enduring question in sponge chemical ecology is whether the sponge or the microbial symbiont is responsible for the production of defensive compounds. In addition, it is unclear whether sponges use symbiont-derived nutrition to produce these metabolites. If the production of chemical defenses utilizes energy that would otherwise be used for host growth (Pawlik 2011), then symbiont production of these compounds might be energetically favorable. High-resolution methods that couple the use of enriched stable isotope tracer compounds (enriched in one or more of the heavy atoms 13C, 15N, or 2H) that target specific microbial metabolic pathways may allow future studies to investigate the fate of atoms assimilated or transformed by microbial symbionts. These tracers have proven useful in following the transfer of isotopically (13C- and 15N) labeled precursors from symbionts to host sponge cells (Freeman et al. 2013, 2015). Mass spectrometry techniques, such as imaging mass spectrometry, could trace the accumulation of isotopically enriched precursors in organelles or cells within the sponge holobiont (Buchberger et al. 2018). Tracing the metabolism and fates of these precursors via high resolution mass spectrometry of sponge metabolomes may also help to elucidate the source and the biosynthetic pathways involved in producing these metabolites.
Sponge-derived natural products chemistry
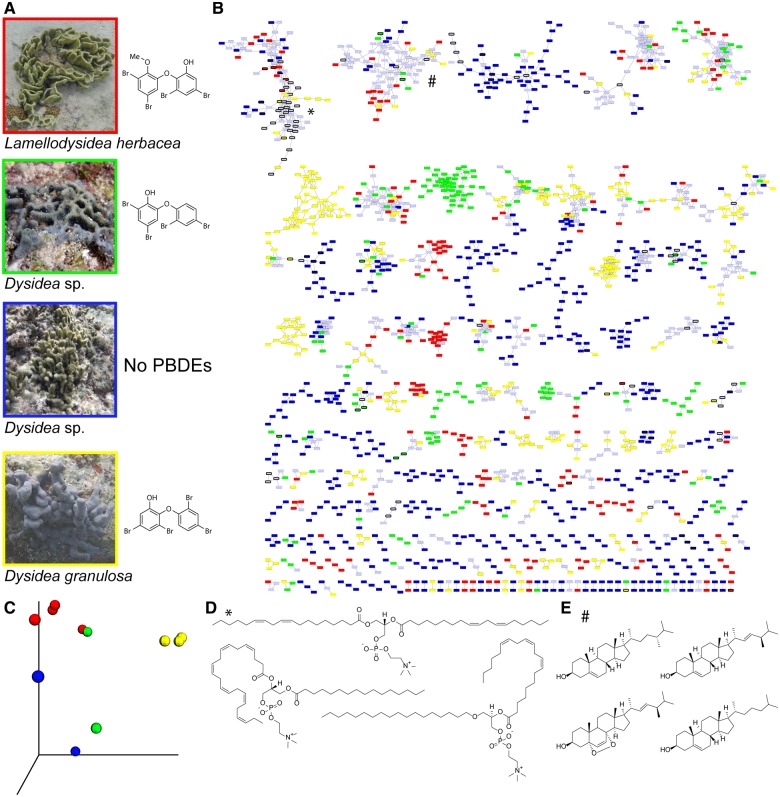
Sponges are a rich source of bioactive natural products and have been studied extensively for decades. Recent trends, however, suggest that the excitement for sponge natural products seems to be dwindling, suggesting that the abundant molecules from easier to access marine sponges have been tapped and that novel natural product discovery from sponges is becoming increasingly onerous (Blunt et al. 2018). The search for sponge-derived natural products is moving in multiple new directions. Mass spectrometry-based metabolomics reveals a much higher diversity of natural products in sponges than has been previously recognized. Tandem mass spectrometry-based molecular networking of extracts of Theonella swinhoei and enriched cell fractions of the sponge’s symbiotic filamentous bacteria “Candidatus Entotheonella” suggested the presence of unrecognized chemical diversity and previously unknown types of metabolites in the sponge (Wilson et al. 2014). Coupling mass spectrometry-guided fractionation of sponge extracts with the characterization of natural product chemical structures based on molecular fragmentation expanded the diversity of polybrominated diphenyl ethers (PBDEs) known from the Dysideidae family of sponges (Agarwal et al. 2015), a class of natural products that has been intensively studied for more than four decades from multiple geographical locations (Sharma and Vig 1972; Carte and Faulkner 1981; Calcul et al. 2009; Agarwal et al. 2015; Liu et al. 2016). Liquid chromatography mass spectrometry (LC–MS) datasets for marine sponge extracts are starting to be made accessible on metabolome mining tools, such as the Global Natural Products Social (GNPS) Molecular Networking platform, which enables a community-wide effort to curate natural products, dereplicate previously known natural products, and discover novel natural products based on chemical similarity to known natural product structures (Wang et al. 2016). Molecular networking was also used to advance the diversity of smenamides, hybrid polyketide–peptide natural products of cyanobacterial origin first described from the sponge Smenospongia aurea (Teta et al. 2013; Via et al. 2018). Advances in mass spectrometry instrumentation have the potential to transform sponge-derived natural product discovery. Using sub-gram amounts of sponge material, it is now facile to collect high resolution metabolomic datasets with high scan rate mass spectrometers that afford an in-depth representation of the chemical diversity harbored by marine sponges. Coupled with fingerprinting marker genes, such as 18S rDNA, metabolomics reveals that sponge species can be differentiated based on their metabolomic signatures (Fig. 1). As demonstrated for four Dysideidae sponges, molecular networking and principal component analyses reveal the clustering of sponge metabolomes according to species differences (Fig. 1C). Comparison of the sponge metabolomes against known natural product repositories reveals that a very small fraction of the sponge chemistry has been described. For the well-studied Dysideidae sponges, only very few molecules, such as fatty acids and sterols, can be dereplicated based on their mass spectrometric fragmentation signatures (Fig. 1D, E). A majority of the Dysideidae chemistry remains undescribed, and this is likely the case for most other sponge taxa.
Fig. 1.
Metabolomics for Dysideidae sponges. (A) Four representative Dysideidae sponges collected by the authors in Guam (years 2014–2015) with the dominant PBDEs present in each sponge. (B) A molecular network of metabolites detected in the extracts of Dysideidae sponges. Nodes unique to a Dysideidae species are color-coded as in panel A. Nodes shared between multiple species are colored gray. A molecular network was created using the online workflow at GNPS. The data were filtered by removing all MS/MS peaks within ±17 Da of the precursor m/z. The data were then clustered with MS-Cluster with a parent mass tolerance of 0.1 Da and a MS/MS fragment ion tolerance of 0.1 Da to create consensus spectra. Further, consensus spectra that contained less than 1 spectra were discarded. A network was then created where edges were filtered to have a cosine score above 0.7 and more than 4 matched peaks. Further edges between two nodes were kept in the network if and only if each of the nodes appeared in each other’s respective top 10 most similar nodes. The spectra in the network were then searched against GNPS’ spectral libraries. The library spectra were filtered in the same manner as the input data. All matches kept between network spectra and library spectra were required to have a score above 0.7 and at least 4 matched peaks. Multiple biological replicates were used for each sponge species. (C) Principal component analyses of the sponge metabolomes. Biological replicates of Lamellodysidea herbacea (in red) and of Dysidea granulosa (yellow) cluster together and can be neatly differentiated. A greater divergence is observed for the other two Dysidea sp. sponges which can be resolved by increasing the number of replicates in this analysis. Dereplicated (D) phospholipids (denoted by * in panel B) and (E) sterols (denoted by # in panel B) in the Dysideidae molecular network. Chemical identities of a large majority of Dysideidae metabolites remain unknown.
Microbial biosynthesis of sponge-derived natural products
As previously discussed, marine sponges harbor complex microbiomes that are being cataloged on a global scale (Thomas et al. 2016; Moitinho-Silva et al. 2017a). Many sponge-derived natural products are now known to be biosynthesized by sponge-associated microorganisms (Wilson et al. 2014; Ueoka et al. 2015; Agarwal et al. 2017; Mori et al. 2018; Morita and Schmidt 2018), with prominent examples discussed below. The biosynthetic capacity of these oftentimes uncultivable members of the sponge microbiome can be revealed by metagenomic and single-cell DNA sequencing. For instance, using metagenomics, the genes responsible for the construction of PBDEs in Dysideidae sponges were localized to the obligate cyanobacterial symbiont Hormoscilla spongeliae (previously named Oscillatoria spongeliae) residing within the sponge and experimentally characterized in an heterologous cyanobacterial host (Agarwal et al. 2017). The discovery of the hs_bmp gene cluster responsible for the production of PBDEs in H. spongeliae was facilitated by the prior discovery of the homologous bmp gene cluster in marine proteobacteria that encodes biosynthetic enzymes for the elaboration of numerous brominated phenols and pyrroles (Agarwal et al. 2014, 2017; El Gamal et al. 2016; Busch et al. 2019). Hybridization of fluorescent nucleotide probes revealed that cyanobacterial symbionts are also responsible for the biosynthesis of chlorinated peptidic natural products such as dysidinin in Dysideidae sponges (Flatt et al. 2005). Prior to these methods, physical separation of the cyanobacterial filaments from the sponge tissue was required to determine if Dysideidae cyanobacterial symbionts were responsible for the biosynthesis of PBDEs and chlorinated peptides (Unson et al. 1994; Flowers et al. 1998); cell separation methods also showed that sponge cells contained terpenes such as spirodysin (Flowers et al. 1998). These cyanobacteria–sponge associations are host-specific, with each sponge species hosting a distinct H. spongeliae strain (Thacker and Starnes 2003; Ridley et al. 2005), but it is unclear what the molecular drivers of this specificity are. The production of PBDEs or chlorinated peptidic natural products may provide the sponge with chemical defense against predation (Pennings et al. 1994), but they are not essential to maintain the cyanobacteria–sponge association as Dysideidae sponges that lack these types of natural products still maintain large population of H. spongeliae (Agarwal et al. 2017). Hormoscilla spongeliae likely plays a critical role in carbon fixation, as shading this species has led to a large reduction in sponge mass (Thacker 2005). Symbiotic cyanobacteria associated with other marine invertebrates are similarly prolific sources of natural products, with Prochloron cyanobacteria associated with tunicates producing diverse cyanobactins (Schmidt et al. 2012). The characteristic fluorescence of the photosynthetic pigment chlorophyll a allows for simple imaging of cyanobacterial symbionts within diverse holobionts, and these techniques can be paired with imaging mass spectrometry to determine the spatial co-localization of cyanobacteria with natural products in the sponge tissue (Simmons et al. 2008; Buchberger et al. 2018). As complete genomes of obligate cyanobacterial symbionts become available, it will be instructive to compare them against closely related free-living cyanobacterial genomes to determine the molecular determinants of cyanobacterial mutualism with various marine invertebrates.
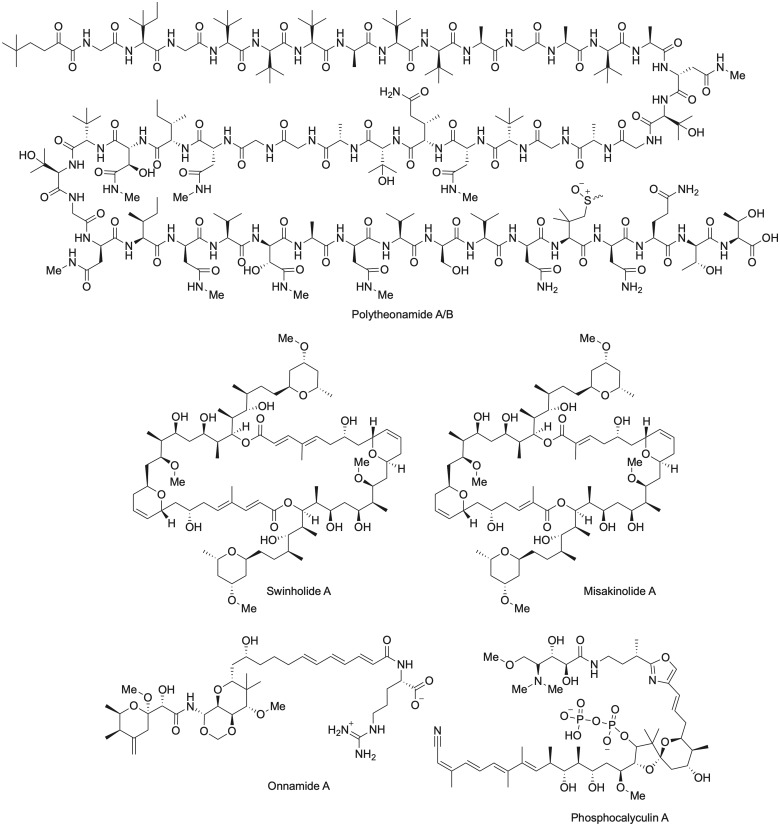
Another example of bacterial production of sponge-derived natural product biosynthesis is the production of polyketides and ribosomal and non-ribosomal peptide natural products by the filamentous “Candidatus Entotheonella sp.” within the sponge T. swinhoei (Fig. 2). These filaments were originally thought to be δ-proteobacteria (Bewley et al. 1996; Schmidt et al. 1998, 2000), but cell sorting, multiple displacement amplification of genomic DNA from individual cells, and DNA sequencing supported their placement into the independent candidate phylum “Tectomicrobia”; their phylogenetic status has been further refined as more member genomes are assembled from sponge microbiomes (Wilson et al. 2014; Lackner et al. 2017; Mori et al. 2018). Prominent examples of natural products for which the biosynthetic genes have been determined include the extensively modified, ribosomally synthesized peptidic polytheonamides (Freeman et al. 2012, 2016), and the polyketide onnamides (Piel et al. 2004), among several other natural products (Wilson et al. 2014). The biosynthetic genes for misakinolide, which structurally resembles swinholide A, were localized to another “Entotheonella” bacterium found in a distinct chemotype of T. swinhoei (Ueoka et al. 2015). Swinholide A has been isolated from T. swinhoei (Carmely and Kashman 1985; Bewley et al. 1996) as well as marine cyanobacteria (Andrianasolo et al. 2005), and the gene cluster involved in swinholide A biosynthesis was identified in a terrestrial cyanobacterium (Humisto et al. 2018). Furthermore, gene clusters involved in the biosynthesis of other related macrolides were also discovered in cyanobacteria (Ueoka et al. 2015). Taken together, these results are suggestive of the transfer of the swinholide biosynthetic gene cluster between free-living cyanobacteria and heterotrophic microbes within the microbiome of T. swinhoei. The ecological benefit of these compounds to T. swinhoei is largely unknown, but “Ca. Entotheonella sp.” in the marine sponge Discodermia calyx synthesizes the hybrid NRPS–PKS natural product phosphocalyculin A that is dephosphorylated upon damage to the sponge tissue, implying, perhaps, a role of this molecule to respond to host tissue damage or as a chemical defense (Wakimoto et al. 2014; Uria et al. 2018).
Fig. 2.
Chemical structures of key Theonella swinhoei natural products discussed in text.
Metagenomic and single cell genomics approaches delineated above are complemented by efforts to directly culture sponge-associated bacteria (Esteves et al. 2016; Indraningrat et al. 2016; Versluis et al. 2017). For example, a bacterial isolate from the sponge Arenosclera brasiliensis produces bromotyrosine alkaloids, which are commonly found in sponges (Nicacio et al. 2017). It is anticipated that a combination of these methods will continue to resolve members of the microbiome that are responsible for the production of natural products.
Sponges also host diverse eukaryotes that are biosynthetically active in producing natural products. For example, sponges harbor a wide diversity and high abundance of meroterpene natural products (Menna et al. 2013; Smith 2017). Fungi cultivated from sponge sources biosynthesize meroterpenes (Zhou et al. 2011; Zhang et al. 2018). However, a one-to-one correspondence between meroterpenes detected in the sponge metabolome, and those biosynthesized by sponge-derived fungi cultivated in the laboratory is still unrealized. While a recent report validates the prokaryotic potential to synthesize meroterpene natural products similar to those detected in sponges, no gene loci encoding meroterpene production have been characterized from sponge prokaryotic metagenomes (Moosmann et al. 2017). Other eukaryotic symbionts, such as dinoflagellates, may also contribute to the natural product biosynthetic milieu in sponge hosts. Given their genetic complexity, a metatranscriptomic approach may help to facilitate the identification of natural product biosynthetic genes in sponge eukaryotic symbionts and in the sponge cells themselves.
Future directions
A combination of expanding metagenome sequencing capacity with the computational tools to mine for natural product biosynthetic gene clusters can potentially revolutionize natural product discovery from sponges (Medema and Fischbach 2015; Medema et al. 2015; Blin et al. 2017) and further our understanding of the functions of these compounds. Particularly for HMA sponges, metagenomes reveal that some microbial genomes are a tremendous source of silent or cryptic natural product gene clusters, the small molecule products of which are either not produced in the sponge holobiont or are produced in quantities that are undetectable by contemporary analytical methods. When cultivation of symbiotic bacteria is not possible, synthetic biology workflows, perhaps using synthetic DNA tailored for expression in specific heterologous hosts, will allow for the elaboration of small molecules encoded in gene loci detected in sponge metagenomes (Kim et al. 2015; Morita and Schmidt 2018). Marine metagenomes are already starting to yield interesting biocatalysts (Smith et al. 2017; Neubauer et al. 2018). We expect to observe a transformative expansion in this field as synthetic biologists and natural product chemists team up with geneticists and chemical ecologists to explore in greater molecular detail the biosynthetic potential of the sponge holobiont. Efforts to culture bacteria and fungi from sponges for discovery of new natural products and for the description of natural product biosynthetic gene clusters when successful can circumvent the complexity of the holobiont metagenomes and metatranscriptomes.
Finally, sponges appear to be less impacted by the predicted effects of ocean warming and ocean acidification that affect benthic organisms like corals, potentially leading to a structural and functional shift to “sponge reefs” in the near future (Bell et al. 2013; Rovellini et al. 2018). Changes in temperature, light, nutrients, and other environmental factors will impact sponges and their microbial associations in diverse ways. For example, cyanobacteria and other photosynthetic organisms will likely be affected by various environmental changes differently than heterotrophic microorganisms, with differing impacts on the nutritional ecology of sponges and the production of natural products. Changing environmental conditions may have profound effects on sponge ecology, with expected changes in their chemical defensive capabilities as well. A better understanding of the roles that symbionts and natural products chemistry have played in the past and current ecological success of sponges is therefore critical if we are to predict how these organisms will respond to continued environmental change in the future. With high abundance and diversity and a cosmopolitan distribution, sponges and their symbionts may act as sentinel species within their ecosystems.
Acknowledgments
We would like to acknowledge Dr Jason Biggs and the staff of the University of Guam Marine Laboratory for facilitating collection of sponge specimens. We thank the Guam Department of Agriculture Division of Aquatic and Wildlife Resources for permits to collect marine organisms. This is contribution number #1109 from the Smithsonian Marine Station.
Funding
This work was supported by the National Institutes of Health [R00-ES026620 to V.A.; R01-CA172310 to V.J.P.]; the National Science Foundation [award #1756114 to C.J.F.]; the Alfred P. Sloan Foundation [V.A.]; and the Gordon and Betty Moore Foundation through Grant GBMF5464 [V.J.P.].
References
- Agarwal V, Blanton JM, Podell S, Taton A, Schorn MA, Busch J, Lin Z, Schmidt EW, Jensen PR, Paul VJ, et al. 2017. Metagenomic discovery of polybrominated diphenyl ether biosynthesis by marine sponges. Nat Chem Biol 13:537–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal V, El Gamal AA, Yamanaka K, Poth D, Kersten RD, Schorn M, Allen EE, Moore BS.. 2014. Biosynthesis of polybrominated aromatic organic compounds by marine bacteria. Nat Chem Biol 10:640–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal V, Li J, Rahman I, Borgen M, Aluwihare LI, Biggs JS, Paul VJ, Moore BS.. 2015. Complexity of naturally produced polybrominated diphenyl ethers revealed via mass spectrometry. Environ Sci Technol 49:1339–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrianasolo EH, Gross H, Goeger D, Musafija-Girt M, McPhail K, Leal RM, Mooberry SL, Gerwick WH.. 2005. Isolation of swinholide A and related glycosylated derivatives from two field collections of marine cyanobacteria. Org Lett 7:1375–8. [DOI] [PubMed] [Google Scholar]
- Baker BJ, Amsler CD, McClintock JB.. 2010. Overview of the chemical ecology of benthic marine invertebrates along the Western Antarctic Peninsula. Int Comp Biol 50:967–80. [DOI] [PubMed] [Google Scholar]
- Bell JJ, Davy SK, Jones T, Taylor MW, Webster NS.. 2013. Could some coral reefs become sponge reefs as our climate changes? Glob Change Biol 19:2613–24. [DOI] [PubMed] [Google Scholar]
- Bewley CA, Holland ND, Faulkner DJ.. 1996. Two classes of metabolites from Theonella swinhoei are localized in distinct populations of bacterial symbionts. Experientia 52:716–22. [DOI] [PubMed] [Google Scholar]
- Blin K, Weber T, Kim HU, Lee SY, Takano E, Breitling R, Shelest E, Wolf T, Chevrette MG, Suarez Duran HG, et al. 2017. Antismash 4.0—improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res 45:W36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blunt JW, Carroll AR, Copp BR, Davis RA, Keyzers RA, Prinsep MR.. 2018. Marine natural products. Nat Prod Rep 35:8–53. [DOI] [PubMed] [Google Scholar]
- Buchberger AR, DeLaney K, Johnson J, Li L.. 2018. Mass spectrometry imaging: a review of emerging advancements and future insights. Anal Chem 90:240–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch J, Agarwal V, Schorn M, Machado H, Moore BS, Rouse GW, Gram L, Jensen PR.. 2019. Diversity and distribution of the bmp gene cluster and its products in the genus Pseudoalteromonas. Environ Microbiol 2019 (doi: 10.1111/1462-2920.14532). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcabrini C, Catanzaro E, Bishayee A, Turrini E, Fimognari C.. 2017. Marine sponge natural products with anticancer potential: an updated review. Mar Drugs 15:310.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcul L, Chow R, Oliver AG, Tenney K, White KN, Wood AW, Fiorilla C, Crews P.. 2009. NMR strategy for unraveling structures of bioactive sponge-derived oxy-polyhalogenated diphenyl ethers. J Nat Prod 72:443–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmely S, Kashman Y.. 1985. Structure of swinholide A, a new macrolide from the marine sponge Theonella swinhoei. Tetrahedron Lett 26:511–4. [Google Scholar]
- Carte B, Faulkner DJ.. 1981. Polybrominated diphenyl ethers from Dysidea herbacea, Dysidea chlorea and Phyllospongia foliascens. Tetrahedron 37:2335–9. [Google Scholar]
- de Goeij JM, van Oevelen D, Vermeij MJA, Osinga R, Middelburg JJ, de Goeij AFPM, Admiraal W.. 2013. Surviving in a marine desert: the sponge loop retains resources within coral reefs. Science 342:108–10. [DOI] [PubMed] [Google Scholar]
- Easson CG, Thacker RW.. 2014. Phylogenetic signal in the community structure of host-specific microbiomes of tropical marine sponges. Front Microbiol 5:532.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Gamal A, Agarwal V, Diethelm S, Rahman I, Schorn MA, Sneed JM, Louie GV, Whalen KE, Mincer TJ, Noel JP, et al. 2016. Biosynthesis of coral settlement cue tetrabromopyrrole in marine bacteria by a uniquely adapted brominase–thioesterase enzyme pair. Proc Natl Acad Sci U S A 113:3797–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin PM, Thacker RW.. 2007. Incidence and identity of photosynthetic symbionts in Caribbean coral reef sponge assemblages. J Mar Biol Assoc UK 87:1683–92. [Google Scholar]
- Esteves AIS, Amer N, Nguyen M, Thomas T.. 2016. Sample processing impacts the viability and cultivability of the sponge microbiome. Front Microbiol 7:499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Reynolds D, Liu M, Stark M, Kjelleberg S, Webster NS, Thomas T.. 2012. Functional equivalence and evolutionary convergence in complex communities of microbial sponge symbionts. Proc Natl Acad Sci U S A 109:E1878–87.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore CL, Labrie M, Jarett JK, Lesser MP.. 2015. Transcriptional activity of the giant barrel sponge, Xestospongia muta holobiont: molecular evidence for metabolic interchange. Front Microbiol 6:364.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt P, Gautschi J, Thacker R, Musafija-Girt M, Crews P, Gerwick W.. 2005. Identification of the cellular site of polychlorinated peptide biosynthesis in the marine sponge Dysidea (Lamellodysidea) herbacea and symbiotic cyanobacterium Oscillatoria spongeliae by CARD-FISH analysis. Mar Biol 147:761–74. [Google Scholar]
- Flowers AE, Garson MJ, Webb RI, Dumdei EJ, Charan RD.. 1998. Cellular origin of chlorinated diketopiperazines in the dictyoceratid sponge Dysidea herbacea (Keller). Cell Tissue Res 292:597–607. [DOI] [PubMed] [Google Scholar]
- Freeman CJ, Baker DM, Easson CG, Thacker RW.. 2015. Shifts in sponge–microbe mutualisms across an experimental irradiance gradient. Mar Ecol Prog Ser 526:41–53. [Google Scholar]
- Freeman CJ, Easson CG, Baker DM.. 2014. Metabolic diversity and niche structure in sponges from the Miskito Cays, Honduras. PeerJ 2:e695.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman CJ, Thacker RW.. 2011. Complex interactions between marine sponges and their symbiotic microbial communities. Limnol Oceanogr 56:1577–86. [Google Scholar]
- Freeman CJ, Thacker RW, Baker DM, Fogel ML.. 2013. Quality or quantity: is nutrient transfer driven more by symbiont identity and productivity than by symbiont abundance? ISME J 7:1116.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman MF, Gurgui C, Helf MJ, Morinaka BI, Uria AR, Oldham NJ, Sahl H-G, Matsunaga S, Piel J.. 2012. Metagenome mining reveals polytheonamides as posttranslationally modified ribosomal peptides. Science 338:387–90. [DOI] [PubMed] [Google Scholar]
- Freeman MF, Vagstad AL, Piel J.. 2016. Polytheonamide biosynthesis showcasing the metabolic potential of sponge-associated uncultivated ‘Entotheonella’ bacteria. Curr Opin Chem Biol 31:8–14. [DOI] [PubMed] [Google Scholar]
- Gerwick WH, Moore BS.. 2012. Lessons from the past and charting the future of marine natural products drug discovery and chemical biology. Chem Biol 19:85–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloeckner V, Wehrl M, Moitinho-Silva L, Gernert C, Schupp P, Pawlik JR, Lindquist NL, Erpenbeck D, Wörheide G, Hentschel U.. 2014. The HMA–LMA dichotomy revisited: an electron microscopical survey of 56 sponge species. Biol Bull 227:78–88. [DOI] [PubMed] [Google Scholar]
- Hay ME. 2009. Marine chemical ecology: chemical signals and cues structure marine populations, communities, and ecosystems. Ann Rev Mar Sci 1:193–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel U, Usher KM, Taylor MW.. 2006. Marine sponges as microbial fermenters. FEMS Microbiol Ecol 55:167–77. [DOI] [PubMed] [Google Scholar]
- Hoffmann F, Radax R, Woebken D, Holtappels M, Lavik G, Rapp HT, Schläppy M-L, Schleper C, Kuypers MM.. 2009. Complex nitrogen cycling in the sponge Geodia barretti. Environ Microbiol 11:2228–43. [DOI] [PubMed] [Google Scholar]
- Humisto A, Jokela J, Liu L, Wahlsten M, Wang H, Permi P, Machado JP, Antunes A, Fewer DP, Sivonen K.. 2018. The swinholide biosynthesis gene cluster from a terrestrial cyanobacterium, Nostoc sp. strain UHCC 0450. Appl Environ Microbiol 84:e02321–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indraningrat AA, Smidt H, Sipkema D.. 2016. Bioprospecting sponge-associated microbes for antimicrobial compounds. Mar Drugs 14:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Moore BS, Yoon YJ.. 2015. Reinvigorating natural product combinatorial biosynthesis with synthetic biology. Nat Chem Biol 11:649–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner G, Peters EE, Helfrich EJN, Piel J.. 2017. Insights into the lifestyle of uncultured bacterial natural product factories associated with marine sponges. Proc Natl Acad Sci U S A 114:E347.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Lohith K, Rosario M, Pulliam TH, O’Connor RD, Bell LJ, Bewley CA.. 2016. Polybrominated diphenyl ethers: structure determination and trends in antibacterial activity. J Nat Prod 79:1872–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado M, Ribes M, van Duyl FC.. 2012. Chapter 3—nutrient fluxes through sponges: biology, budgets, and ecological implications In: Becerro MA, Uriz MJ, Maldonado M, Turon X, editors. Advances in Sponge Science: Phylogeny, Systematics, Ecology. Advances in marine biology. Vol. 62. Amsterdam: Academic Press; p. 113–82. [DOI] [PubMed] [Google Scholar]
- McClintock JB, Baker BJ, (eds.). 2001. Marine chemical ecology. Boca Raton: CRC Press; 610 pp. [Google Scholar]
- McMurray SE, Johnson ZI, Hunt DE, Pawlik JR, Finelli CM.. 2016. Selective feeding by the giant barrel sponge enhances foraging efficiency. Limnol Oceanogr 61:1271–86. [Google Scholar]
- McMurray SE, Stubler AD, Erwin PM, Finelli CM, Pawlik JR.. 2018. A test of the sponge-loop hypothesis for emergent Caribbean reef sponges. Mar Ecol Prog Ser 588:1–14. [Google Scholar]
- Medema MH, Fischbach MA.. 2015. Computational approaches to natural product discovery. Nat Chem Biol 11:639–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema MH, Kottmann R, Yilmaz P, Cummings M, Biggins JB, Blin K, de Bruijn I, Chooi YH, Claesen J, Coates RC, et al. 2015. Minimum information about a biosynthetic gene cluster. Nat Chem Biol 11:625–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menna M, Imperatore C, Aniello F, Aiello A.. 2013. Meroterpenes from marine invertebrates: structures, occurrence, and ecological implications. Mar Drugs 11:1602.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moitinho-Silva L, Nielsen S, Thomas T, Bell JJ, Vicente J, Björk JR, Montoya JM, Olson JB, Reveillaud J, Steindler L, et al. 2017a. The sponge microbiome project. GigaScience 6:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moitinho-Silva L, Steinert G, Nielsen S, Hardoim CCP, Wu Y-C, McCormack GP, López-Legentil S, Marchant R, Webster N, Thomas T, et al. 2017b. Predicting the HMA–LMA status in marine sponges by machine learning. Front Microbiol 8:752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moosmann P, Ueoka R, Grauso L, Mangoni A, Morinaka BI, Gugger M, Piel J.. 2017. Cyanobacterial ent-sterol-like natural products from a deviated ubiquinone pathway. Angew Chem Int Ed 56:4987–90. [DOI] [PubMed] [Google Scholar]
- Morganti T, Coma R, Yahel G, Ribes M.. 2017. Trophic niche separation that facilitates co-existence of high and low microbial abundance sponges is revealed by in situ study of carbon and nitrogen fluxes. Limnol Oceanogr 62:1963–83. [Google Scholar]
- Mori T, Cahn JKB, Wilson MC, Meoded RA, Wiebach V, Martinez AFC, Helfrich EJN, Albersmeier A, Wibberg D, Dätwyler S, et al. 2018. Single-bacterial genomics validates rich and varied specialized metabolism of uncultivated Entotheonella sponge symbionts. Proc Natl Acad Sci USA 115:1718–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Schmidt EW.. 2018. Parallel lives of symbionts and hosts: chemical mutualism in marine animals. Nat Prod Rep 35:357–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya A, Peretó J, Gil R, Latorre A.. 2008. Learning how to live together: genomic insights into prokaryote–animal symbioses. Nat Rev Genet 9:218–29. [DOI] [PubMed] [Google Scholar]
- Neubauer PR, Widmann C, Wibberg D, Schröder L, Frese M, Kottke T, Kalinowski J, Niemann HH, Sewald N.. 2018. A flavin-dependent halogenase from metagenomic analysis prefers bromination over chlorination. PLoS ONE 13:e0196797.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicacio KJ, Ióca LP, Fróes AM, Leomil L, Appolinario LR, Thompson CC, Thompson FL, Ferreira AG, Williams DE, Andersen RJ, et al. 2017. Cultures of the marine bacterium Pseudovibrio denitrificans AB134 produce bromotyrosine-derived alkaloids previously only isolated from marine sponges. J Nat Prod 80:235–40. [DOI] [PubMed] [Google Scholar]
- Paul VJ, Arthur KE, Ritson-Williams R, Ross C, Sharp K.. 2007. Chemical defenses: from compounds to communities. Biol Bull 213:226–51. [DOI] [PubMed] [Google Scholar]
- Paul VJ, Ritson-Williams R, Sharp K.. 2011. Marine chemical ecology in benthic environments. Nat Prod Rep 28:345–87. [DOI] [PubMed] [Google Scholar]
- Pawlik JR. 2011. The chemical ecology of sponges on Caribbean reefs: natural products shape natural systems. Bioscience 61:888–98. [Google Scholar]
- Pawlik JR, Amsler CD, Ritson-Williams R, McClintock JB, Baker BJ, Paul VJ.. 2013. Marine chemical ecology: a science born of scuba In: Lang M, Marinelli RL, Roberts SJ, Taylor PR, editors. Research and discoveries: the revolution of science through scuba. Smithsonian Contributions to the Marine Sciences, Vol. 39, 258 pp. Washington (DC: ): Smithsonian Institution Scholarly Press; p. 53–69. [Google Scholar]
- Pawlik JR, Chanas B, Toonen RJ, Fenical W.. 1995. Defenses of Caribbean sponges against predatory reef fish. I. Chemical deterrency. Mar Ecol Prog Ser 127:183–94. [Google Scholar]
- Pawlik JR, Loh T-L, McMurray SE.. 2018. A review of bottom-up vs. top-down control of sponges on Caribbean fore-reefs: what’s old, what’s new, and future directions. PeerJ 6:e4343.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel J, Hui D, Wen G, Butzke D, Platzer M, Fusetani N, Matsunaga S.. 2004. Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. Proc Natl Acad Sci U S A 101:16222–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pita L, Rix L, Slaby BM, Franke A, Hentschel U.. 2018. The sponge holobiont in a changing ocean: from microbes to ecosystems. Microbiome 6:46.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podell S, Blanton JM, Neu A, Agarwal V, Biggs JS, Moore BS, Allen EE.. 2019. Pangenomic comparison of globally distributed Poribacteria associated with sponge hosts and marine particles. ISME J 13:468–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi MP, Becerro MA, (eds.). 2018. Chemical ecology: the ecological impacts of marine natural products. Boca Raton: CRC Press; 400 pp. [Google Scholar]
- Puglisi MP, Sneed JM, Ritson-Williams R, Young R.. 2019. Marine chemical ecology in benthic environments. Nat Prod Rep published online (doi: 10.1039/C8NP00061A). [DOI] [PubMed] [Google Scholar]
- Puglisi MP, Sneed JM, Sharp KH, Ritson-Williams R, Paul VJ.. 2014. Marine chemical ecology in benthic environments. Nat Prod Rep 31:1510–53. [DOI] [PubMed] [Google Scholar]
- Reiswig HM. 1974. Water transport, respiration and energetics of three tropical marine sponges. J Exp Mar Biol Ecol 14:231–49. [Google Scholar]
- Ridley CP, Bergquist PR, Harper MK, Faulkner DJ, Hooper JN, Haygood MG.. 2005. Speciation and biosynthetic variation in four dictyoceratid sponges and their cyanobacterial symbiont, Oscillatoria spongeliae. Chem Biol 12:397–406. [DOI] [PubMed] [Google Scholar]
- Rovellini A, Bell JJ, Webster NS, Bennett HM.. 2018. Sponges to be winners under near-future climate scenarios. Bioscience 68:955–68. [Google Scholar]
- Schmidt EW, Bewley CA, Faulkner DJ.. 1998. Theopalauamide, a bicyclic glycopeptide from filamentous bacterial symbionts of the lithistid sponge Theonella swinhoei from Palau and Mozambique. J Org Chem 63:1254–8. [Google Scholar]
- Schmidt EW, Donia MS, McIntosh JA, Fricke WF, Ravel J.. 2012. Origin and variation of tunicate secondary metabolites. J Nat Prod 75:295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EW, Obraztsova AY, Davidson SK, Faulkner DJ, Haygood MG.. 2000. Identification of the antifungal peptide-containing symbiont of the marine sponge Theonella swinhoei as a novel δ-proteobacterium, “Candidatus Entotheonella palauensis.” Mar Biol 136:969–77. [Google Scholar]
- Sharma GM, Vig B.. 1972. Studies on the antimicrobial substances of sponges. VI. Structures of two antibacterial substances isolated from the marine sponge Dysidea herbacea. Tetrahedron Lett 13:1715–8. [Google Scholar]
- Simmons TL, Coates RC, Clark BR, Engene N, Gonzalez D, Esquenazi E, Dorrestein PC, Gerwick WH.. 2008. Biosynthetic origin of natural products isolated from marine microorganism-invertebrate assemblages. Proc Natl Acad Sci U S A 105:4587–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DRM, Uria AR, Helfrich EJN, Milbredt D, van Pée K-H, Piel J, Goss R.. 2017. An unusual flavin-dependent halogenase from the metagenome of the marine sponge Theonella swinhoei WA. ACS Chem Biol 12:1281–7. [DOI] [PubMed] [Google Scholar]
- Smith TE. 2017. Biogenetic relationships of bioactive sponge merotriterpenoids. Mar Drugs 15:285.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwell MW, Weisz JB, Martens CS, Lindquist N.. 2008. In situ fluxes of dissolved inorganic nitrogen from the sponge community on Conch Reef, Key Largo, Florida. Limnol Oceanogr 53:986–96. [Google Scholar]
- Taylor MW, Radax R, Steger D, Wagner M.. 2007. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol Mol Biol Rev 71:295.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teta R, Irollo E, Della Sala G, Pirozzi G, Mangoni A, Costantino V.. 2013. Smenamides A and B, chlorinated peptide/polyketide hybrids containing a dolapyrrolidinone unit from the Caribbean sponge Smenospongia aurea. Evaluation of their role as leads in antitumor drug research. Mar Drugs 11:4451–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker RW. 2005. Impacts of shading on sponge–cyanobacteria symbioses: a comparison between host-specific and generalist associations. Int Comp Biol 45:369–76. [DOI] [PubMed] [Google Scholar]
- Thacker RW, Freeman CJ.. 2012. Chapter two—sponge–microbe symbioses: recent advances and new directions In: Becerro MA, Uriz MJ, Maldonado M, Turon X, editors. Advances in Sponge Science: Phylogeny, Systematics, Ecology. Advances in marine biology. Vol. 62. Amsterdam: Elsevier/Academic Press. 57–111. [Google Scholar]
- Thacker RW, Starnes S.. 2003. Host specificity of the symbiotic cyanobacterium Oscillatoria spongeliae in marine sponges, Dysidea spp. Mar Biol 142:643–8. [Google Scholar]
- Thomas T, Moitinho-Silva L, Lurgi M, Björk JR, Easson C, Astudillo-García C, Olson JB, Erwin PM, López-Legentil S, Luter H, et al. 2016. Diversity, structure and convergent evolution of the global sponge microbiome. Nat Commun 7:11870.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turon M, Cáliz J, Garate L, Casamayor EO, Uriz MJ.. 2018. Showcasing the role of seawater in bacteria recruitment and microbiome stability in sponges. Sci Rep 8:15201.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueoka R, Uria AR, Reiter S, Mori T, Karbaum P, Peters EE, Helfrich EJN, Morinaka BI, Gugger M, Takeyama H, et al. 2015. Metabolic and evolutionary origin of actin-binding polyketides from diverse organisms. Nat Chem Biol 11:705–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unson MD, Holland ND, Faulkner DJ.. 1994. A brominated secondary metabolite synthesized by the cyanobacterial symbiont of a marine sponge and accumulation of the crystalline metabolite in the sponge tissue. Mar Biol 119:1–11. [Google Scholar]
- Uria AR, Piel J, Wakimoto T.. 2018. Chapter nine—biosynthetic insights of calyculin- and misakinolide-type compounds in “Candidatus Entotheonella sp.” In: Moore BS, editor. Methods in enzymology. Vol. 604. Cambridge, MA: Academic Press; p. 287–330. [DOI] [PubMed] [Google Scholar]
- Vacelet J, Donadey C.. 1977. Electron microscope study of the association between some sponges and bacteria. J Exp Mar Biol Ecol 30:301–14. [Google Scholar]
- Van Soest RWM, Boury-Esnault N, Vacelet J, Dohrmann M, Erpenbeck D, De Voogd NJ, Santodomingo N, Vanhoorne B, Kelly M, Hooper J.. 2012. Global diversity of sponges (Porifera). PLoS ONE 7:e35105.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versluis D, McPherson K, van Passel MWJ, Smidt H, Sipkema D.. 2017. Recovery of previously uncultured bacterial genera from three Mediterranean sponges. Mar Biotechnol 19:454–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via CW, Glukhov E, Costa S, Zimba PV, Moeller PDR, Gerwick WH, Bertin MJ.. 2018. The metabolome of a cyanobacterial bloom visualized by MS/MS-based molecular networking reveals new neurotoxic smenamide analogs (C, D, and E). Front Chem 6:316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakimoto T, Egami Y, Nakashima Y, Wakimoto Y, Mori T, Awakawa T, Ito T, Kenmoku H, Asakawa Y, Piel J, et al. 2014. Calyculin biogenesis from a pyrophosphate protoxin produced by a sponge symbiont. Nat Chem Biol 10:648–55. [DOI] [PubMed] [Google Scholar]
- Wang MX, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, Nguyen DD, Watrous J, Kapono CA, Luzzatto-Knaan T, et al. 2016. Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat Biotechnol 34:828–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster NS, Thomas T.. 2016. The sponge hologenome. mBio 7:e00135-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisz JB, Hentschel U, Lindquist N, Martens CS.. 2007. Linking abundance and diversity of sponge-associated microbial communities to metabolic differences in host sponges. Mar Biol 152:475–83. [Google Scholar]
- Weisz JB, Lindquist N, Martens CS.. 2008. Do associated microbial abundances impact marine demosponge pumping rates and tissue densities?. Oecologia 155:367–76. [DOI] [PubMed] [Google Scholar]
- Wilkinson CR. 1983. Net primary productivity in coral reef sponges. Science 219:410–2. [DOI] [PubMed] [Google Scholar]
- Wilson MC, Mori T, Rückert C, Uria AR, Helf MJ, Takada K, Gernert C, Steffens UAE, Heycke N, Schmitt S, et al. 2014. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 506:58–62. [DOI] [PubMed] [Google Scholar]
- Zhang F, Vicente J, Hill RT.. 2014. Temporal changes in the diazotrophic bacterial communities associated with Caribbean sponges Ircinia stroblina and Mycale laxissima. Front Microbiol 5:561.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Yuan B, Liu D, Gao S, Proksch P, Lin W.. 2018. Brasilianoids a–f, new meroterpenoids from the sponge-associated fungus Penicillium brasilianum. Front Chem 6:314.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Mándi A, Debbab A, Wray V, Schulz B, Müller WEG, Lin W, Proksch P, Kurtán T, Aly AH.. 2011. New austalides from the sponge-associated fungus Aspergillus sp. Eur J Org Chem 2011:6009–19. [Google Scholar]