Abstract
MicroRNA (miR)-101 copy loss is an early event in the development of human lung cancer, and it occurs in 29% of all lung cancer incidences. In addition, miR-101 expression in non-small cell lung cancer (NSCLC) is known to be downregulated. The aim of the present study was to explore the roles and mechanisms of the long non-coding (lnc)-RNA pro-transition associated RNA (PTAR) on NSCLC cell proliferation, migration and invasion in association with miR-101. Reverse transcription-quantitative PCR analysis was performed to detect the expression of lncRNA PTAR in 30 paired human NSCLC tissues and the corresponding para-tumor tissues. PTAR was amplified and cloned into the expression vector pCDNA3.1. Then, PTAR-overexpression plasmids or small interfering (si)-RNA-PTAR was transfected into A549 cells for 48 h, after which cell proliferation and the cell cycle distribution were evaluated. In addition, Transwell chamber and cell scratch-wound assays were conducted to analyze A549 cell migration and invasion. A luciferase activity assay was evaluated to determine the interaction between PTAR and miR-101. Furthermore, our results demonstrated that in human NSCLC tissues and cell lines, lncRNA PTAR expression was upregulated compared with normal lung tissues and cell lines, respectively. Additionally, PTAR transfection was observed to promote A549 cell proliferation, migration and invasion; opposing effects were observed with siRNA-PTAR transfection. The luciferase activity assay revealed that PTAR could act as a sponge to bind miR-101. Thus, miR-101 plays a role in NSCLC tumorigenesis and progression. In conclusion, lncRNA PTAR was proposed to promote NSCLC cell growth through sponging and inactivating miR-101, which may be a possible mechanism underlying miR-101 copy loss in human NSCLC.
Keywords: miR-101, lncRNA PTAR, competing endogenous RNA, NSCLC, cell migration and invasion
Introduction
According to the global cancer statistics in 2018, lung cancer is the most frequently diagnosed and the major cause of cancer-associated mortality in all cancer cases (1). Non-small cell lung cancer (NSCLC), with a 5-year survival rate of <19% at tumor-node-metastasis stages III or IV, accounts for ~80% of total lung cancer incidents (2).
Numerous microRNAs (miRNAs/miRs) exhibit aberrant expression in cancer, and function as oncogenes or tumor suppressors (3). miR-101 has two copies in the human genome, 1p31.3 (miR-101-1) and 9p24.1 (miR-101-2), and its expression is usually downregulated in various types of cancers, including bladder, liver, lung, gastric and prostate cancers (4–9). In human lung cancer, Thu et al (10) reported that ~29% lung cancer cases exhibit miR-101 copy loss, and that 90% of detected lung cancer cell lines possess the genomic loss of miR-101. These results indicate that the absence of miR-101 may serve an essential role in lung tumorigenesis and cancer development; therefore, similar approaches that suppress miR-101 function may also promote lung cancer progression.
Long non-coding RNAs (lncRNAs) are a type of non-coding RNA of 200 nucleotides in length that are mainly generated from gene introns, intergenic regions, promoter regions of coding mRNA, antisense strands of mRNAs and pseudogenes (11). The mature form of lncRNA has a similar structure to mRNA, including 5′ capping and 3′polyadenylation, but lncRNAs are not generally translated (12,13). Previous studies have demonstrated that the sponging of miRNAs is one of the critical regulatory functions of lncRNAs that act as competitive RNAs (14–16). Additionally, it has been reported that the regulation of miRNAs mediated by lncRNAs is involved in tumorigenesis and progression. For instance, the lncRNA nuclear paraspeckle assembly transcript 1 acts as a miR-193a-3p sponge to modulate the miR-193a-3p/KRAS pathway in colorectal cancer (17); lncRNA prostate cancer associated transcript-1 can promote NSCLC progression through competitively binding miR-149-5p and regulating the miR-149-5p/LRIG2 axis (18). In addition, lncATB serves as a molecular sponge to suppress interactions between miR-200b and Kindlin-2 in esophageal squamous cell carcinoma (19). LncRNAs have been suggested to have potential application in the diagnosis, prognosis and could serve as therapeutic targets of NSCLC (20); however, the roles and mechanisms of lncRNAs in cancer require further investigation.
Pro-transition associated RNA (PTAR; AP000695.4) is a novel lncRNA (21). The aim of the present study was to explore the roles and mechanisms of lncRNA PTAR in human NSCLC. The results indicated that lncRNA PTAR is upregulated in human NSCLC tissues than in the corresponding para-tumor tissues, and that it can accelerate tumor cell proliferation, migration and invasion in vitro. The effects of lncRNA PTAR and miR-101 were examined by a luciferase activity assay in 293T cells, which indicated that lncRNA PTAR serves as a tumor promoter by regulating miR-101.
Materials and methods
Tissues and cell lines
A total of 30 paired human NSCLC tissues and the corresponding para-tumor specimens were collected from patients (18 males and 12 females; average age, 48.4 years) in the Qingdao Municipal Hospital between May 2017 and January 2018. The human lung adenocarcinoma cell line A549, human lung squamous carcinoma cell line 95D and human normal lung epithelial cell line DEAS-2B were obtained from Land Biology Co., Ltd. 293T was maintained in our lab. Tissue specimens and cell lines were frozen in liquid nitrogen until subsequent use. All patients provided written consent prior to the study; the present study was approved by the Institutional Ethics Committee of the Qingdao Municipal Hospital.
Cell culture and transfection
All cells were cultured in Dulbecco's Modified Eagle's medium (Hyclone; GE Healthcare) supplemented with 10% fetal bovine serum (Hyclone; GE Healthcare) and 100 U/ml penicillin/streptomycin (Hyclone; GE Healthcare). Phosphate-buffered saline (PBS; Hyclone; GE Healthcare) solution was used for washing cells before dissociation and diluting cells for counting. Cells were maintained in a saturated and humidified 37°C incubator containing 5% CO2. Before transfection, A549 cells were sub-cultured in multiple-well plates (Corning Incorporated) at ~80% confluence. RNAs (30 nM) or plasmids (1.5 µg per well) were transfected into cells using Lipofectamine 2000® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's instructions. A549 cells were subjected to following experiments 48 h after transfection. lncRNA PTAR was amplified from the cDNA library from 293T cells and cloned into a pCDNA3.1 vector (Invitrogen; Thermo Fisher Scientific, Inc.). The sequences of the primers to amplify lncRNA PTAR were the following: Forward 5′-ACAGATGTAAACCAACCAGA-3′ and reverse 5′-ATGCTACTGGAGACTTTAGG-3′. The PCR was performed using the 2X SYBR Green qPCR SuperMix (Takara Bio, Inc.). The thermocycling condition were the following: Incubation at 50°C for 2 min, followed by an incubation at 95°C for 2 min, followed by 40 cycles of 95°C for 15 sec, 60°C for 32 sec.
PTAR small interfering (si)-RNA was synthesized by Sigma-Aldrich (Merck KGaA). miR-101 mimics and miR-101 inhibitor were obtained commercially from Guangzhou RiboBio Co., Ltd. Meanwhile, siRNA negative control, miRNA mimics and miRNA inhibitor were used as transfection control. The sequences of oligonucleotides used were as follows: miR-101 mimics, 5′-UACAGUACUGUGAUAACUGAA-3′; miRNA mimics control, 5′-UUUGUACUACACAAAAGUACUG-3′; miR-101 inhibitor, 5′-UUCAGUUAUCACAGUACUGUA-3′; miRNA inhibitor control, 5′-UCACAACCUCCUAGAAAGAGUAGA-3′.
Reverse transcription-quantitative PCR (RT-qPCR) analysis
The total RNA of lung tissues and cell lines were extracted using TRIzol reagent (Thermo Fisher Scientific, Inc.). RQ1 RNase-free DNase (Promega Corporation) was mixed with total RNAs for genomic DNA digestion, then extracted with 16.6% (v/v) chloroform and 50% (v/v) precipitated with isopropanol. The purity and concentration of total RNA was measured using a BioPhotometer plus spectrophotometer (Eppendorf). Total RNA (1 µg) was reverse transcribed into cDNA using the ImProm-II™ Reverse Transcription System kit (Promega Corporation) according to the manufacturer's protocol. Then, qPCR was performed using SYBR® GREEN qPCR Super Mix (Invitrogen; Thermo Fisher Scientific, Inc.) and the 7500 Sequence Detection System (Applied Biosystems; Thermo Fisher Scientific, Inc.). Briefly, a total volume of 20 µl reaction system, including 5 µl cDNA (1:20), 0.5 µl forward primer, 0.5 µl reverse primer, 10 µl 2X SYBR GREEN qPCR Super Mix and 4 µl ddH2O were mixed. The PCR thermocycling conditions were as follows: Initial denaturation at 95°C for 2 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 32 sec. GAPDH was regarded as the internal control for PTAR expression normalization. Relative expression of PTAR was evaluated using the 2−ΔΔCq method (22). The primers used were as follows: PTAR forward, 5′-ACAGATGTAAACCAACCAGA-3′ and reverse, 5′-ATGCTACTGGAGACTTTAGG-3′; and GAPDH forward, 5′-GGGAAACTGTGGCGTGAT-3′ and reverse, 5′-GAGTGGGTGTCGCTGTTGA-3′. Experiments for each group were repeated in triplicate.
Cell proliferation assay
A549 cell proliferation assay was evaluated using Cell Counting Kit (CCK)-8 reagent (Beyotime Institute of Biotechnology). A total of 1×104 cells were seeded into each well of a 96-well plate supplemented with 100 µl DMEM (Invitrogen; Thermo Fisher Scientific, Inc.) containing 10% FBS (Hyclone; GE Healthcare). Following 12 h, PTAR expression plasmids and si-PTAR were respectively transfected into cells. Cell proliferation was detected in the following 4 days. Briefly, 10 µl CCK-8 solution was applied to each well, and the plate was incubated at 37°C for 4 h. Then, after 0, 1, 2, 3 and 4 days, the cells were subjected to determine the absorbance at 450 nm by using a Multiscan MK3 microplate reader (Thermo Fisher Scientific, Inc.).
Cell cycle assay
A cell cycle assay was performed using a Cell Cycle Detection kit (Nanjing KeyGen Biotech Co., Ltd.). At 48 h after transfection, 1×106 cells were digested and collected. The cells were washed with pre-cooled PBS twice and fixed with 70% ethanol at 4°C overnight. Then, the ethanol was removed and cells were washed with PBS and suspended in 0.5 ml PBS containing 50 µg/ml propidium iodide, 100 µg/ml RNase A and 0.2% Triton X-100, and incubated in the dark at 4°C for 30 min. Finally, the cells were subjected to cell cycle analysis with a BD caliber flow cytometer using ModFit software version 3.2 (BD Biosciences).
Cell migration and invasion assay
Cell migration and invasion were analyzed using Transwell chambers with 8 µm pore size membrane inserts (BD Falcon™; BD Biosciences). At 48 h after transfection, A549 cells were subjected to cell migration detection. Briefly, 1×105 cells were seeded into the upper chamber supplemented with 100 µl serum-free medium, while 600 µl complete medium was added to the lower chamber as the inducer. Following 48 h of incubation with 5% CO2 at 37°C, non-migrated cells in the upper chamber were removed using a cotton swab, while migrated cells in the lower chamber were fixed with 4% paraformaldehyde for 20 min at room temperature, then washed with PBS, and stained with 0.1% crystal violet for 10 min at room temperature. Finally, the cells were washed with PBS and counted using an inverted microscope (Olympus Corporation; magnification, ×100) with the image processing program, ImageJ software version 1.8 (National Institute of Health). Images of six random fields of view were captured for each group to analyze. For cell invasion analysis, the procedures were the same as those performed for the cell migration assay, except the membranes were coated with 40 µl Matrigel and incubated at 37°C for 2 h. Experiments for each group were repeated in triplicate.
Cell scratch-wound assay
On the reverse side of 6-well plates 10 horizontal straight lines were drawn with a marker pen. A total 50 µl fibronectin (10 µg/ml) was added to each well of 6-well plates, and the plates were incubated at 4°C overnight. Then, 1×106 A549 cells were seeded into each well supplemented with 2 ml complete medium. At 12 h after incubation with 5% CO2 at 37°C, PTAR expression plasmids and si-PTAR were transfected into cells, respectively. At 4 h after transfection, cells were treated with mitomycin (20 µM; Roche Diagnostics) for 1 h, then a 10 µl pipette tip was used to create scratch wounds that were perpendicular to the marker lines. The cells were washed with PBS twice to remove the cell debris. Scratch-wound healing was then observed and images of eight random views were captured at 0, 24 and 48 h. The width of the wound was determined using Image Pro-Plus 6.0 software (Media Cybernetics, Inc.). The ratio of wound healing was represented as [width (at 0 h)-width (at 24 or 48 h)]/width (at 0 h). Experiments for each group were repeated in triplicate.
Western blotting
Cells were lysed with pre-cooled RIPA lysis buffer supplemented with a cocktail of proteases inhibitors (both from Sigma-Aldrich; Merck KGaA). Total protein concentration was detected using a bicinchoninic acid protein assay kit (Nanjing KeyGen Biotech Co., Ltd.). Total proteins (10 µg) were mixed with 5X SDS-PAGE loading buffer and heated at 100°C for 5 min, then, proteins were separated by 4–12% SDS-PAGE. Proteins were transferred onto 0.45-µm-thick PVDF membranes (EMD Millipore) at 4°C for 2 h. After blocking with 5% non-fat milk for 1 h at room temperature, the PVDF membranes were incubated with primary antibodies at 4°C overnight and horseradish peroxidase (HRP)-conjugated secondary antibodies at 37°C for 1 h. Protein bands were visualized using Immobilon Western Chemilum HRP substrate (EMD Millipore). The primary antibodies used were anti-GAPDH (1:10,000; KangChen Bio-tech Co., Ltd.), anti-Cyclin D1 (1:500; Abcam), anti-ZEB1 (1:500; Abcam), anti-Vimentin (1:1,000; Abcam) and anti-E-cadherin (1:2,000; Abcam). The secondary antibody used was HRP-conjugated Goat Anti-Rabbit IgG (1:20,000; SouthernBiotech).
miRNA-lncRNA interactions and luciferase activity assay
The miRNA-lncRNA interactions were predicted using a public database (http://www.medsysbio.org/EMTRegulome). Luciferase activity assay was conducted using in 293T cells with a Dual-Luciferase Reporter Assay System (Promega Corporation) according to the instructions. Wild and mutant type lnc-RNA PTAR were cloned into psi-CHECK2 vector (Promega Corporation). Wild-type (wt)-PTAR-psi-CHECK2 and mutant (Mut)-PTAR-psi-CHECK2 were respectively co-transfected with miR-101 mimics or miR-101 inhibitor. At 48 h after transfection, cells were washed with PBS and lysed with 100 µl passive lysis buffer, included in the kit (Promega Corporation). Firefly luciferase activity was normalized to Renilla luciferase activity. Luciferase activity was measured using a GloMax 20/20 (Promega Corporation) detector following the manufacturer's instructions. Experiments for each group were repeated in triplicate.
Statistical analysis
Data analysis and the production of figures were conducted using Origin 8.5 software (OriginLab). All statistical data were presented as the mean ± standard deviation. Paired or independent samples t-tests were used to evaluate statistically significant differences between two groups. Difference among three groups were calculated by one-way analysis of variance followed by a Turkey's post-hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
PTAR expression is upregulated in human NSCLC tissues and cell lines
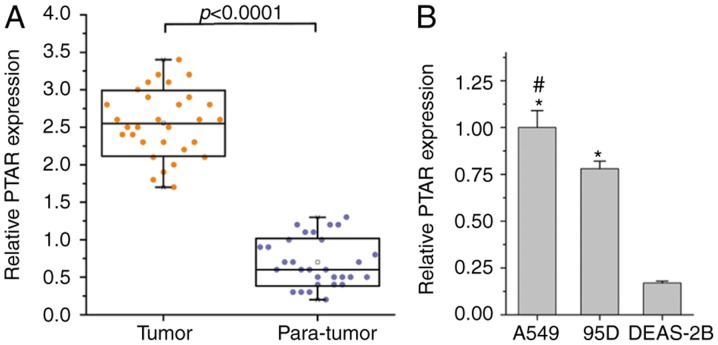
First, the present study determined the expression of lncRNA PTAR in NSCLC tissues and cell lines, and the results revealed that the expression of PTAR in NSCLC tumor tissue samples was significantly increased when compared with homologous para-tumor samples (Fig. 1A). In addition, the levels of PTAR expression were significantly increased in the A549 (lung adenocarcinoma) and 95D (squamous cell lung carcinoma) cell lines than in the human normal lung epithelial cells (DEAS-2B; Fig. 1B). Overall, PTAR expression was determined to be upregulated in human NSCLC tissues and cell lines.
Figure 1.
Long non-coding RNA PTAR expression in human NSCLC tissues and cell lines. (A) Levels of PTAR in 30 paired human NSCLC tumor and homologous para-tumor normal tissues specimens by RT-qPCR analysis. (B) PTAR expression in lung adenocarcinoma (A549), lung squamous cell carcinoma (95D), and lung normal epithelial (DEAS-2B) cell lines by RT-qPCR analysis. *P<0.05 vs. DEAS-2B cells, #P<0.05 vs. 95D cells. PTAR, pro-transition associated RNA; NSCLC, non-small cell lung cancer; RT-qPCR, reverse transcription-quantitative PCR.
PTAR promotes NSCLC cell proliferation
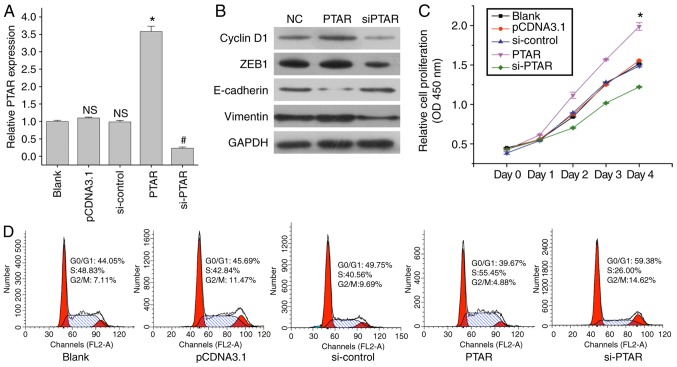
To further investigate the effects of PTAR in NSCLC, the present study transfected PTAR expression plasmids and si-PTAR into A549 cells. The transfection of a PTAR-overexpression plasmid and si-PTAR led to significant increases and decreases in PTAR expression, compared with the respective control (Fig. 2A). Additionally, the role of PTAR expression in A549 cell proliferation and protein expression was assessed. The results indicated that overexpression of PTAR promoted Cyclin D1 expression and tumor cell proliferation; opposing findings were reported in response to transfection with si-PTAR (Fig. 2B and C). Cell cycle analysis demonstrated that when compared with the blank and control groups, the ectopic expression of PTAR markedly increased the number of cells in S phase of the cell cycle; si-PTAR transfection arrested tumor cells at the G0/G1 and G2/M phases (Fig. 2D). All of these findings indicated that PTAR can promote NSCLC tumor cell proliferation.
Figure 2.
Effects of long non-coding RNA PTAR on A549 cell growth. (A) Expression of PTAR on A549 cells transfected with PTAR-overexpression plasmids and si-PTAR as determined by reverse transcription-quantitative PCR analysis. *P<0.05 vs. pCDNA3.1 group. #P<0.05 vs. si-control group. (B) Western blot analysis of Cyclin D1, ZEB1, E-cadherin and Vimentin protein expression in A549 cells transfected with PTAR-overexpression plasmids or si-PTAR. (C) Cell Counting Kit-8 analysis for the proliferation of A549 cells transfected with PTAR-overexpression plasmids or si-PTAR. *P<0.05 vs. pCDNA3.1 group. (D) Cell cycle analysis in A549 cells transfected with PTAR expression plasmids and PTAR siRNA. PTAR, pro-transition associated RNA; si, small interfering RNA; NC, negative control; N.S. indicates no significance compared with blank group. ZEB1, zinc finger E-box binding homeobox 1.
PTAR promotes NSCLC cell migration and invasion
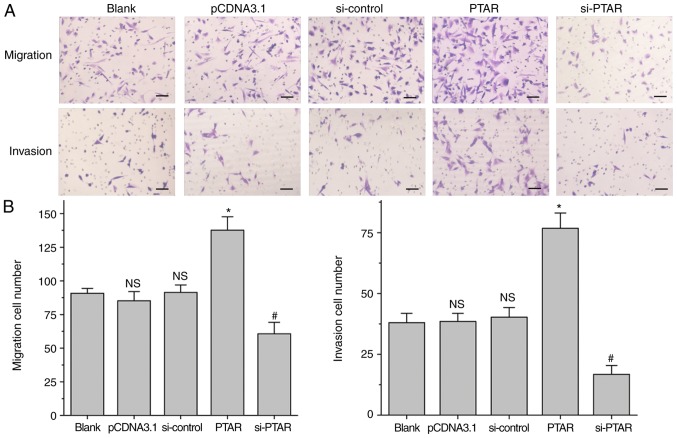
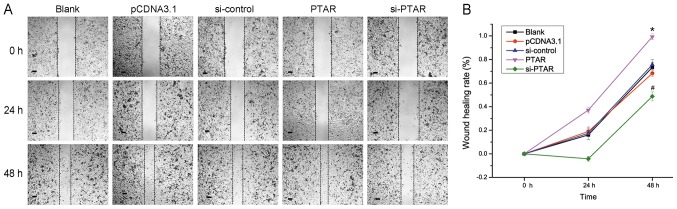
Transwell chamber based-cell migration and invasion assays were performed with transfected A549 cells. As shown in Fig. 3, PTAR overexpression significantly increased tumor cell migration and invasion compared with the blank or control groups. Furthermore, knockdown of PTAR significantly decreased A549 cell migration and invasion compared with the corresponding controls. In addition, PTAR-overexpression plasmid or si-PTAR transfection affected the expression of migration and invasion related proteins, such as ZEB1, E-cadherin and Vimentin. Overexpression of PTAR could promote the expression levels of Cyclin D1, ZEB1 and Vimentin, inhibiting the levels of E-cadherin in A549 cells. By contrast, knockdown of PTAR by si-PTAR suppressed the expression level of Cyclin D1, ZEB1 and Vimentin expression, and increased E-cadherin levels (Fig. 2B). A cell scratch-wound assay was also performed to continuously analyze tumor cell migration after transfection for 48 h. Similarly, the results indicated that PTAR significantly promoted scratch-wound healing rate at 48 h after transfection compared with the respective control (Fig. 4). Our results indicated that PTAR can effectively promote NSCLC cell migration and invasion.
Figure 3.
Transwell assay to detect the effects of long non-coding RNA PTAR on A549 cell migration and invasion. (A and B) The number of migrated and invaded cells were counted following transfection with PTAR-overexpression plasmid or si-PTAR into A549 cells using Transwell chambers. *P<0.05 vs. pCDNA3.1 group. #P<0.05 vs. si-control group. Scale bar, 100 µm. N.S. indicates no significance compared with blank group. PTAR, pro-transition associated RNA; si, small interfering RNA; NC, negative control.
Figure 4.
Scratch-wound healing assay to evaluate cell migration at 48 h after transfection. (A) Images of scratch-wound healing respectively at 0, 24 and 48 h following transfection of PTAR-overexpression plasmid or si-PTAR. (B) Ratio of wound healing was represented as [width (at 0 h)-width (at 24 or 48 h)]/width (at 0 h). Scale bar, 100 µm. *P<0.05 vs. pCDNA3.1. #P<0.05 vs. si-control group. PTAR, pro-transition associated RNA; si, small interfering RNA.
PTAR can act as a sponge to bind to miR-101
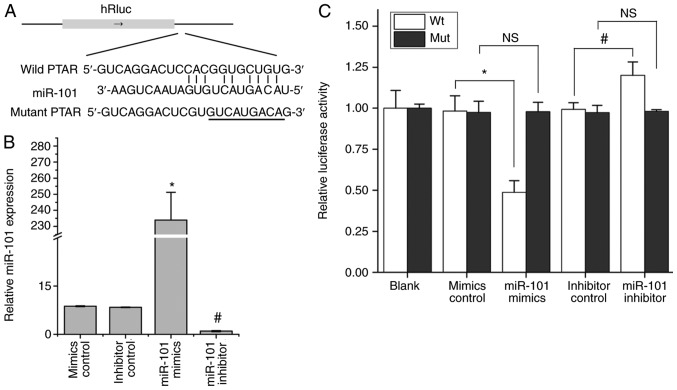
Subsequent analysis revealed there was a predicted combination of PTAR and miR-101. To confirm this, a luciferase activity assay was performed in 293T cells. Wt PTAR and Mut PTAR, containing a mutant fragment within the miR-101 predicted binding region, were constructed and cloned into the psi-CHECK2 luciferase reporter vector (Fig. 5A). Transfection with miR-101 mimics and inhibitor could effectively promote and inhibit the expression of miR-101 in 293T cells, respectively (Fig. 5B). Then, miR-101 mimics and miR-101 inhibitor were co-transfected with Wt or Mut PTAR psi-CHECK2 vectors. The results demonstrated that in the wt-PTAR-psi-CHECK2 transfection groups, miR-101 ectopic expression significantly decreased the luciferase activity, while miR-101 inhibitor transfection increased luciferase activity compared with the corresponding controls. However, following transfection with mut-PTAR-psi-CHECK2, no significant difference was observed in the miR-101 mimics or the miR-101 inhibitor co-transfected groups (Fig. 5C).
Figure 5.
Luciferase reporter assay to confirm the interaction between PTAR and miR-101. (A) Wt-PTAR and mut-PTAR, containing a mutant fragment within the miR-101 predicted binding region, were respectively inserted into psi-CHECK2 reporter vectors. (B) Reverse transcription-quantitative PCR was performed to detect the expression of miR-101 following transfection with miR-101 mimics or inhibitor into 293T cells. (C) Wt-PTAR-psi-CHECK2 and Mut-PTAR-psi-CHECK2 were respectively co-transfected with miR-101 mimics and miR-101 inhibitor into 293T cells. After 48 h, the luciferase activity of cells was determined. *P<0.05 vs. mimics control; #P<0.05 vs. inhibitor control Wt, wild type; N.S. indicates no significance; PTAR, pro-transition associated RNA; miR, microRNA; Mut, mutant; Wt, wild type.
Discussion
miR-101 expression is commonly downregulated in human cancers (4–9). The absence of miR-101 genomic copies occurs in ~40–80% of NSCLC cases, which is more frequent than that of SCLC, and the levels of miR-101 in NSCLC tend to be reduced (10). This difference in the expression of miR-101 induces the overexpression of downstream proteins in cancer (6,23,24). miR-101 suppression by the genomic loss of miR-101 has been reported to trigger the upregulation of enhancer of zeste homolog 2 (a histone methyltransferase) in a 3′-untranslated region (UTR)-regulatory manner, and promote lung cancer cell proliferation and invasion (6,23,24). Yan et al (25) reported that the restoration of miR-101 in lung cancer can suppress tumorigenesis via the regulation of DNA methyltransferase 3α 3′UTR. Notably, other modulatory approaches that inactivate miR-101 also exhibit pro-tumorigenic properties. Wan et al (26) have reported that protein patched homolog 1 3′UTR serves as an RNA sponge to inhibit the suppressive effects of miR-101 on the target gene SLC3916, and promotes NSCLC migration and invasion. Wang et al (27) demonstrated that lncRNA MALAT1 acts as a sponge to bind to miR-101, and inhibits the binding of miR-101 with the target gene, myeloid cell leukemia 1, to increase lung tumor cell drug-resistance. Thus, the suppression of miR-101 in lung cancer may serve a role in tumorigenesis.
PTAR, a novel ceRNA, has been reported to accelerate epithelial-to-mesenchymal transition (EMT) and metastasis by competitively binding miR-101 in ovarian cancer (21). However, the roles of PTAR and its underlying mechanisms in other types of cancer remain unclear. Accumulating evidence has indicated that miR-101 acts as a tumor suppressor factor in human lung cancer (28–30). Therefore, it is necessary to investigate the potential functions of PTAR via miR-101 in human lung cancer.
In present study, the levels of lncRNA PTAR were upregulated in NSCLC tissues, as well as in lung adenocarcinoma and lung squamous cell carcinoma cell lines. Knockdown of PTAR by siRNA significantly suppressed the proliferation, migration and invasion of A549 cells, and the opposite phenotype was reported with the ectopic expression of PTAR. It was hypothesized that these pro-tumorigenic effects may be associated with miR-101. The present study further confirmed the interaction between PTAR with miR-101 by a luciferase reporter assay, and concluded that the promoting effects on lung tumorigenesis may occur through the sponging of miR-101 via PTAR; however, further investigation is required to determine the mechanism underlying miR-101 copy loss in human NSCLC.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- NSCLC
non-small cell lung cancer
- lncRNA
long non-coding RNA
- PTAR
Pro-transition associated RNA
- miRNAs
microRNAs
Funding
The present study was supported by National Nature Science Foundation of China (grant no. 81573451).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
WY, ZS, LD and RY drafted and revised the manuscript. LD and RY collected the samples and conducted analysis. WY, ZS, LiY, YH and LuY designed the study, analyzed the data and drafted the paper. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Institutional Ethics Committee of the Qingdao Municipal Hospital. All patients provided written consent prior to the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Tsim S, O'Dowd CA, Milroy R, Davidson S. Staging of non-small cell lung cancer (NSCLC): A review. Respir Med. 2010;104:1767–1774. doi: 10.1016/j.rmed.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 3.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Friedman JM, Liang G, Liu CC, Wolff EM, Tsai YC, Ye W, Zhou X, Jones PA. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009;69:2623–2629. doi: 10.1158/0008-5472.CAN-08-3114. [DOI] [PubMed] [Google Scholar]
- 5.Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y, Zhuang SM. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69:1135–1142. doi: 10.1158/0008-5472.CAN-08-2886. [DOI] [PubMed] [Google Scholar]
- 6.Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, Laxman B, Cao X, Jing X, Ramnarayanan K, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang HJ, Ruan HJ, He XJ, Ma YY, Jiang XT, Xia YJ, Ye ZY, Tao HQ. MicroRNA-101 is down-regulated in gastric cancer and involved in cell migration and invasion. Eur J Cancer. 2010;46:2295–2303. doi: 10.1016/j.ejca.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 8.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 9.Zhang JG, Guo JF, Liu DL, Liu Q, Wang JJ. MicroRNA-101 exerts tumor-suppressive functions in non-small cell lung cancer through directly targeting enhancer of zeste homolog 2. J Thorac Oncol. 2011;6:671–678. doi: 10.1097/JTO.0b013e318208eb35. [DOI] [PubMed] [Google Scholar]
- 10.Thu KL, Chari R, Lockwood WW, Lam S, Lam WL. miR-101 DNA copy loss is a prominent subtype specific event in lung cancer. J Thorac Oncol. 2011;6:1594–1598. doi: 10.1097/JTO.0b013e3182217d81. [DOI] [PubMed] [Google Scholar]
- 11.Ulitsky I, Bartel DP. lincRNAs: Genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154:240–251. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bánfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE, Jr, Kundaje A, Gunawardena HP, Yu Y, Xie L, et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012;22:1646–1657. doi: 10.1101/gr.134767.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 15.Cazalla D, Yario T, Steitz JA. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science. 2010;328:1563–1566. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Z, Du S, Yin K, Ai S, Yu M, Liu Y, Shen Y, Liu M, Jiao R, Chen X, Guan W. Knockdown long noncoding RNA nuclear paraspeckle assembly transcript 1 suppresses colorectal cancer through modulating miR-193a-3p/KRAS. Cancer Med. 2019;8:261–275. doi: 10.1002/cam4.1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Li Y, Wang B, Ma Y, Chen P. LncRNA-PCAT-1 promotes non-small cell lung cancer progression by regulating miR-149-5p/LRIG2 axis. J Cell Biochem. 2018 Dec 19; doi: 10.1002/jcb.28046. (Epub ahead of print) Doi: 10.1002/jcb.28046. [DOI] [PubMed] [Google Scholar]
- 19.Li Z, Wu X, Gu L, Shen Q, Luo W, Deng C, Zhou Q, Chen X, Li Y, Lim Z, et al. Long non-coding RNA ATB promotes malignancy of esophageal squamous cell carcinoma by regulating miR-200b/Kindlin-2 axis. Cell Death Dis. 2017;8:e2888. doi: 10.1038/cddis.2017.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu T, Wang Y, Chen D, Liu J, Jiao W. Potential clinical application of lncRNAs in non-small cell lung cancer. Onco Targets Ther. 2018;11:8045–8052. doi: 10.2147/OTT.S178431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang H, Yu T, Han Y, Jiang H, Wang C, You T, Zhao X, Shan H, Yang R, Yang L, et al. LncRNA PTAR promotes EMT and invasion-metastasis in serous ovarian cancer by competitively binding miR-101-3p to regulate ZEB1 expression. Mol Cancer. 2018;17:119. doi: 10.1186/s12943-018-0870-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Cho HM, Jeon HS, Lee SY, Jeong KJ, Park SY, Lee HY, Lee JU, Kim JH, Kwon SJ, Choi E, et al. microRNA-101 inhibits lung cancer invasion through the regulation of enhancer of zeste homolog 2. Exp Ther Med. 2011;2:963–967. doi: 10.3892/etm.2011.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hou Y, Li L, Ju Y, Lu Y, Chang L, Xiang X. MiR-101-3p regulates the viability of lung squamous carcinoma cells via targeting EZH2. J Cell Biochem. 2017;118:3142–3149. doi: 10.1002/jcb.25836. [DOI] [PubMed] [Google Scholar]
- 25.Yan F, Shen N, Pang J, Xie D, Deng B, Molina JR, Yang P, Liu S. Restoration of miR-101 suppresses lung tumorigenesis through inhibition of DNMT3a-dependent DNA methylation. Cell Death Dis. 2014;5:e1413. doi: 10.1038/cddis.2014.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wan X, Kong Z, Chu K, Yi C, Hu J, Qin R, Zhao C, Fu F, Wu H, Li Y, Huang Y. Co-expression analysis revealed PTCH1-3′UTR promoted cell migration and invasion by activating miR-101-3p/SLC39A6 axis in non-small cell lung cancer: Implicating the novel function of PTCH1. Oncotarget. 2017;9:4798–4813. doi: 10.18632/oncotarget.23219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H, Wang L, Zhang G, Lu C, Chu H, Yang R, Zhao G. MALAT1/miR-101-3p/MCL1 axis mediates cisplatin resistance in lung cancer. Oncotarget. 2017;9:7501–7512. doi: 10.18632/oncotarget.23483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lv P, Zhang P, Li X, Chen Y. Micro ribonucleic acid (RNA)-101 inhibits cell proliferation and invasion of lung cancer by regulating cyclooxygenase-2. Thorac Cancer. 2015;6:778–784. doi: 10.1111/1759-7714.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Yao J, Sun H, He K, Tong D, Song T, Huang C. MicroRNA-101 suppresses progression of lung cancer through the PTEN/AKT signaling pathway by targeting DNA methyltransferase 3A. Oncol Lett. 2017;13:329–338. doi: 10.3892/ol.2016.5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han L, Chen W, Xia Y, Song Y, Zhao Z, Cheng H, Jiang T. MiR-101 inhibits the proliferation and metastasis of lung cancer by targeting zinc finger E-box binding homeobox 1. Am J Transl Res. 2018;10:1172–1183. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.