Abstract
The etiology of idiopathic membranous nephropathy (IMN) is considered to be closely associated with immunoregulation and genetic factors. Circular RNAs (circRNAs) have been found to regulate gene expression in various organisms, and to play an important role in multiple physiological and pathological processes, which may be involved in the pathogenesis of IMN. The purpose of the present study was to investigate the potential relationship between circRNAs in peripheral blood and disease. The diagnoses of IMN were confirmed using electron microscopy and immunofluorescence. Total RNA was isolated and microarray analysis was used to detect the expression levels of circRNAs in the peripheral blood of patients with IMN and in normal subjects. Selected genes from the microarray were selected and verified by reverse transcription-quantitative (RT-q)PCR. Bioinformatics tools were applied for further functional evaluation, and the potential disease predictability of circRNAs was determined using receiver-operating characteristic (ROC) curves. The results showed that a total of 955 differentially expressed circRNAs were found in blood samples, 645 of which were upregulated and 310 which were downregulated. In total, five candidate circRNAs were validated using RT-qPCR analysis. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analyses identified numerous types of target genes and their corresponding microRNAs (miRNAs). The miRNAs identified were involved in biological processes and enriched in multiple important pathways, including the mitogen-activated protein kinase, transforming growth factor-β and Ras signaling pathways. The levels of circ_101319 were significantly higher (P<0.001) and exhibited promising diagnostic value in patients with IMN (area under ROC =0.89). The co-expression network constructed for circ_101319 indicated that it may be associated with membranous nephropathy-related pathways by mediating miRNAs. In conclusion, the present study revealed the expression and functional profile of differentially expressed circRNAs in the peripheral blood of patients with IMN, and provided new perspectives to predict and elucidate the development of IMN.
Keywords: IMN, circRNA, gene expression, microarray, bioinformatics analysis
Introduction
Idiopathic membranous nephropathy (IMN), regarded as an organ-specific autoimmune disease, is one of the major causes of nephritic syndrome in adults, and is an important factor in the recurrence of nephritic syndrome in patients after renal transplantation (1). The massive deposition of immune complexes appearing in the epithelial side of the glomerular capillary loop is a characteristic pathological change, with immunoglobulin (Ig) G4 being the most prominent IgG subclass (2). Immune deposits are limited to the epithelial side of the glomerular basement membrane, generally without inherent glomerular cell proliferation and an inflammatory reaction (3). The clinical symptoms are manifested as nephritic syndrome, asymptomatic or non-nephritic proteinuria, and occasionally microscopic hematuria, hypertension, and renal damage (4,5). As the pathogenesis of the disease is not fully understood, and because few sensitive biomarkers have been found that reflect disease activity, an effective treatment for IMN is lacking in modern medicine. Expensive and potentially toxic drug treatment greatly limits the therapeutic regimen of the disease, and leads to serious life-threatening consequences for patients (6).
A large body of research has shown that epigenetics plays an important role in the pathophysiology of membranous nephropathy (MN) (7–9). Noncoding (nc)RNA is involved in different physiological and pathological processes, including embryonic development, cell differentiation, angiogenesis, tumor formation and DNA damage repair, and are important for regulating a variety of biological functions at both the transcriptional and posttranscriptional levels (10). As a type of ncRNA, circular RNAs (circRNAs) are abundant in bodily fluids and tissues (11). It has recently been found that circRNAs regulate gene expression in various ways, and that they may be a functional molecule in the occurrence and development of IMN (12).
circRNAs are endogenous ncRNAs that are formed as covalently closed loop structures without 5′caps or 3′tails by special variable splicing. circRNAs exist widely in human cells; they are found primarily in the cytoplasm of eukaryotic cells and also in the nucleus (13). circRNAs affect the regulation of gene expression at the transcriptional and posttranscriptional level, and specific expression profiles have been detected in different tissues and developmental phases (14). circRNAs have numerous biological functions, including the regulation of RNA binding proteins, microRNA (miRNA/miR) binding, protein translation and the regulation of gene transcription, which play important roles in gene expression throughout life (15–17). By contrast to the traditional linear RNAs with 5′ and 3′ends, the molecular structure of circRNA is a closed ring, which is characterized by a higher level of stability, including more stable expression, and resistance to exonucleases and degradation (18). Combined with their high specificity and conserved features among different species, circRNAs may become a novel candidate biomarker for disease diagnosis, treatment and prognosis. As the molecular mechanism of IMN pathogenesis has not been elucidated, to the best of our knowledge, there is no existing scientific research on circRNAs and IMN, and little is known regarding their relationship (19,20).
In the present study, >900 differently expressed circRNAs were identified by microarray analysis in the peripheral blood of patients with IMN compared with the normal population. Following bioinformatics screening of the results, five circRNAs were selected and the expression of these candidate molecules was verified by increasing the sample size. Among the five circRNAs, circ_101319 was found to be significantly upregulated by reverse transcription-quantitative RT-q(PCR). A receiver-operating characteristic (ROC) curve was used to investigate the potential diagnostic value for patients with IMN. A comprehensive network of its potential interaction was constructed using Cytoscape. The results identified the aberrant expression of circRNAs in IMN and suggested that circ_101319 had relevance in IMN.
Materials and methods
Clinical specimens and collection
Peripheral blood samples (2 ml) were collected from 30 patients at the People's Hospital of Guangxi Zhuang Autonomous Region between January 2016 and December 2017. Combined with the clinical index and the manifestation of the patients, the diagnosis of IMN was confirmed by renal biopsy and immunofluorescence. Inclusion criteria for patients with IMN were as follows: Proteinuria, with some patients having mild microscopic hematuria or renal insufficiency; diffuse thickening of the glomerular capillary wall and deposition of immune complexes in the subepithelial spaces, as observed by light microscopy; immunofluorescence results showing IgG4 deposited along the capillary loops, which could be combined with deposits of complement component 3 (C3); electron-dense deposits on the epithelial side or glomerular basement membrane, and foot process fusion by electron microscopy; and when the blood samples were collected, all the patients with IMN included in the disease group had not received glucocorticoid therapy or other immunosuppressive agents. The exclusion criteria for the IMN group were as follows: Patients with autoimmune diseases, including systemic lupus erythematosus, rheumatoid arthritis, multiple sclerosis, idiopathic thrombocytopenia or type 1 diabetes; patients with acute or chronic infectious diseases, including hepatitis virus, AIDS, inflammatory enteritis, parasitic infection and other acute or chronic infectious diseases; patients with cancer, a kidney transplant or heavy metal poisoning; and patients diagnosed with IMN who had previously received treated.
In total, 30 specimens were obtained from individuals who underwent annual checkups and were confirmed to be healthy between January 2016 and December 2017 at the medical examination center of the People's Hospital of Guangxi Zhuang Autonomous Region. These 30 specimens formed a control (NC) group. The following criteria were adopted to select the samples in the NC group: The age and gender were consistent with the patient group; the subjects had no other comorbidities; it was known that clinical test indexes, including renal function, were normal; and there were no clinical manifestations related to other diseases.
Blood samples were stored at −80°C immediately until further analysis. The protocol for this research was reviewed and approved by the Ethics Committee of the People's Hospital of Guangxi Zhuang Autonomous Region. All participants of the present study signed an informed consent form.
Total RNA extraction and quality control
Total RNA in the peripheral blood was extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The purity and concentration of the RNA were determined using a NanoDrop ND-1000 (Nanodrop Technologies; Thermo Fisher Scientific, Inc.), and the integrity of the RNA was determined using denaturing agarose gel electrophoresis. The RNA was stored at −80°C for the following experiments.
Sample preparation and circRNA microarray analysis
Each microarray contained 6 samples (3 patients with IMN and 3 controls), which were also included in the 30 samples used for PCR validation. Sample labeling and array hybridization were performed according to the manufacturer's protocol (Arraystar, Inc.). Briefly, total RNA was digested with RNase R (Epicentre; Illumina, Inc.) to remove linear RNAs. Then, the enriched circRNAs were amplified and transcribed into fluorescent complementary (c)RNA using a random priming method (Arraystar Super RNA Labeling kit; Arraystar, Inc.). The labeled cRNAs were hybridized onto the Arraystar Human circRNA Array V2 (8×15K; Arraystar, Inc.). After the slides were washed, the arrays were scanned using the Agilent Scanner G2505C (Agilent Technologies, Inc.). Scanned images of the raw data were imported into Agilent Feature Extraction software (version 11.0.1.1; Agilent Technologies, Inc.). Quantitative normalization of the raw data and subsequent data processing were performed using the R software package (version 3.1.2; http://cran.r-project.org/). The statistical significance of the differences were estimated using unpaired t-tests. CircRNAs having fold change (FC)>2.0 and P<0.01 were selected as significantly differentially expressed and were displayed by Volcano Plot filtering. miRNA targets of circRNAs and circRNA/miRNA interactions were predicted by the Arraystar Human circRNA Array V2 computer program based on TargetScan 7.1 (21) and miRanda (http://mirdb.org) (22).
Construction of circRNA/miRNA interaction networks, Gene Ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis
To further elucidate associations between circRNAs and miRNA, circRNA/miRNA interactions were predicted using Arraystar's miRNA target prediction software (Arraystar, Inc.) based on TargetScan 7.1 and mirdb 5.0. All differentially expressed circRNAs were annotated in detail with the circRNA/miRNA interaction information. GO analysis was (23,24) performed to explore the functional roles of target genes in terms of ‘biological processes’ (BPs), ‘cellular components’ and ‘molecular functions’ (MFs). Biological pathways were defined using KEGG (25,26) and identified using Database for Annotation, Visualization and Integrated Discovery (27,28)
Validation of candidate circRNAs using RT-qPCR
RT-qPCR analysis was performed to verify the results of the microarray analysis for five differentially expressed circRNAs. For RT reactions, 2 µg RNA, 1 µl 0.5 µg/µl random primers (N9) and 1.6 µl dNTP Mix (2.5 mM; HyTest, Ltd and H2O to a volume of 14.5 µl was incubated at 65°C for 5 min and then placed on ice for 2 min. After this, 4 µl 5X First-Strand Buffer, 1 µl 0.1 M DTT, 0.3 µl RNase Inhibitor (Epicentre; Illumina, Inc.) and 0.2 µl SuperScript III (Invitrogen; Thermo Fisher Scientific, Inc.) were added and incubated at 37°C for 1 min. A qPCR Master Mix (Arraystar, Inc.) and the ViiA 7 Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) were used for qPCR, following the manufacturer's instructions. qPCR was performed as follows: 95°C for 10 min followed by 40 cycles of 95°C for 10 sec and 60°C for 60 sec. Divergent primers, instead of the more commonly used convergent primers, were designed for the five selected circRNAs. β-actin was used as the internal control for circRNA normalization and quantification. The relative expression of each circRNA was analyzed by the 2−ΔΔCq method (29). All primers were synthesized by Invitrogen; Thermo Fisher Scientific, Inc. The results are expressed as the mean ± SEM of three independent experiments. Primer sequences are shown in Table SI.
Functional prediction for the circ_101319-miRNA-mRNA co-expression network
To explore the potential biological function of the candidate circRNA, a circ_101319-targeted miRNA-mRNA network was conducted using Cytoscape (version 3.6.0) (30), according to the analysis of TargetScan and miRanda. The target miRNAs for circ_101319 were identified and subsequently ranked based on their miRNA support vector regression (mirSVR) scores; the source code for the mirSVR scoring system was provided by Aksomics, Inc. The size of each node represents the different types and numbers of putative targets functionally connected to each circRNA. GO annotations and KEGG pathways were used to analyze the predicted gene functions in the network.
Transmission electron microscopy (TEM) and immunofluorescence
Kidney tissues from patients were obtained using renal puncture. A part of each tissue sample was sliced to 3–5 µm using freezing microtome (−20°C) and then cut into frozen sections. Immunofluorescence staining of IgA, IgG, IgM, C3, complement component 1q (C1q) was performed on the sections. The following fluorescently-labeled rabbit anti-human antibodies (1:40; Gene Tech Biotechnology Co., Ltd.) were used: IgG (cat. no. GF020229), IgA (cat. no. GF020429), IgM (cat. no. GF02032), C3 (cat. no. GF020129), C1q (cat. no. GF025429). Antibodies were added separately to each specimen and incubated in at 37°C for 40 min. Samples were washed three times with PBS for 3 min each time. The sections were observed under a fluorescence microscope (magnification, ×200), images were captured and the results were recorded. The remaining parts of the renal specimens were fixed using glutaraldehyde (2.5%) at 4°C for 2 h and embedded in epoxy resin. Samples were dehydrated using an ethanol series (50, 70, 80, 90 and 95%) and then incubated in propylene oxide (100%) for 45 min at 20°C. Ultra-thin sections (50–70 nm) were prepared after polymerization. The prepared sections were stained with uranyl acetate (3%) for 30 min at 25°C and lead citrate (3%) for 8 min at 25°C. Pathological changes in the renal tissue were observed and photographed using a transmission electron microscope.
Statistical analysis
The normalized data from the microarray processing were analyzed using the R software package (version 3.1.2, http://cran.r-project.org/) and P-value were calculated using unpaired t-tests. All other statistical data were visualized and analyzed using GraphPad Prism 5.0 (GraphPad Software, Inc.). Microarray and RT-qPCR data are presented as the means ± SEM, and are representative of at least three experiments. Differences between groups were evaluated using a two-tailed Student's t-test. P<0.05 was considered to indicate a statistically significant difference. Analysis of ROC curves was applied, and the area under the curve was calculated with 95% confidence intervals (CIs), which were used to assess the prediction value of the circRNAs in disease.
Results
Clinical characteristics of the patients
Between January 2016 and December 2017, >100 patients with renal diseases were screened. The results of renal biopsies and immunofluorescence analysis in these patients were observed. Combined with their clinical manifestation, 30 patients were diagnosed with IMN and were selected for the present study. The baseline characteristics are shown in Table I. The average age of the patients who underwent renal biopsy was 53±11 years, and 63% of the patients were male. Among the laboratory indexes listed, the level of proteinuria was 7.6±3.0 g/24 h, and urine and serum albumin was 23±3.8 g/l, which could reflect the nephrotic syndrome. Endogenous creatinine clearance was 81±24 ml/min per 1.73 m2, indicating that the kidney function of patients was well preserved. In addition, the mean level of serum cholesterol was 10±3.4 mmol/l. All the electron microscopy images and fluorescence analysis from the patients in disease group were consistent with IMN.
Table I.
Baseline characteristics of patients with idiopathic membranous nephropathy.
| Characteristic | Total population |
|---|---|
| N (% male) | 19 (63%) |
| Age at time of biopsy, years | 53±11 |
| Time between biopsy and urine analysis, months | 1 |
| Laboratory | |
| Serum creatinine (µmol/l) | 93±14 |
| Serum albumin (g/l) | 23±3.8 |
| Serum cholesterol (mmol/l) | 10±3.4 |
| Endogenous creatinine clearance (ml/min per 1.73 m2) | 81±24 |
| Urine samples | |
| Proteinuria (g/24 h urine) | 7.6±3.0 |
Data are presented as the mean ± SD.
Identification of differential expressed circRNA profiles in the peripheral blood of patients with IMN using microarray analysis
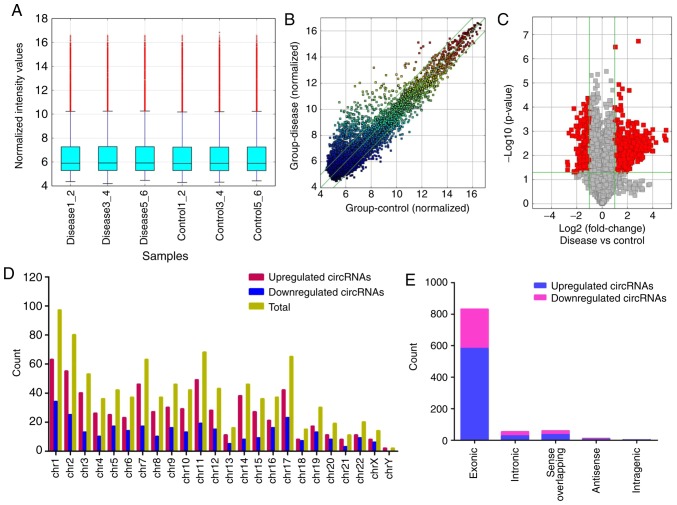
High-throughput microarray analysis of human peripheral blood was used to assess circRNA expression signatures in IMN. A total of 13,617 circRNA targets were identified using the microarray probes in three pairs of clinical specimens. In total, three microarrays were analysed for each group, with each microarray containing pooled RNA from two people. Box plots were used to visualize the distribution of intensities from all normalized data, and it was found that the distribution of circRNAs displayed using log2 ratios was almost equal in the IMN and NC groups (Fig. 1A). FC filtering revealed the differentially expressed circRNAs (Fig. 1B). In addition, the differentially expressed circRNAs with statistically significant differences between the two groups were identified by volcano plot filtering (Fig. 1C). The results of the microarray analysis showed that 955 circRNAs were differentially regulated (FC≥2.0; P<0.05; false discovery rate <0.05), of which 645 circRNAs were upregulated and 310 circRNAs were downregulated. Among these circRNAs, upregulated circRNAs were more prevalent than downregulated circRNAs and were more closely related to disease. The distribution of circRNAs on human chromosomes is described in Fig. 1D. The majority of differentially regulated circRNAs are transcribed from the exonic regions of protein coding genes (Fig. 1E). Certain circRNAs are also transcribed from intronic regions, and some are sense overlapping. Few circRNAs are transcribed from other sources (Fig. 1E). The top 10 upregulated and downregulated circRNAs identified by microarray analysis are listed in Table II. Hierarchical clustering analysis showed that the expression of circRNAs in peripheral blood was significantly different between the NC group and patients with IMN (Fig. 2).
Figure 1.
Distribution and differential expression of circRNA profiles between the IMN and NC groups. (A) In total, three microarrays were analysed for each group, with each microarray containing pooled RNA from two people in all possible combinations. In total, 6 samples from the IMN and NC groups were normalized and are presented using a box plot. (B) Scatter plots were used to assess the variation in circRNA expression between the two groups. The values plotted on the x- and y-axes are the averaged normalized signal values of the samples (log2 scale). The central green line indicates no difference between the two groups. The green lines above and below represent a 2-fold change. The circRNAs distributed above the top line and below the bottom line have ≥2 fold change between the two groups. (C) Volcano plots were constructed to visualize the differential expression of circRNAs using fold change values and P-values. The vertical lines correspond to a 2-fold (log2 scale) change between the IMN and NC groups while the horizontal line represents a P-value of 0.05 (-log10 scale). The red squares represent the statistically significant circRNAs. (D) Distribution of circRNAs throughout the genome. (E) Different circRNA categories are indicated. Most differentially expressed circRNAs are located in exonic regions. Differentially expressed circRNAs also originate from introns and from sense overlapping regions. IMN, idiopathic membranous nephropathy; NC, normal control; circRNA, circular RNA; Chr, chromosome.
Table II.
Annotation of the circRNA/miRNA interactions for the top 10 upregulated and downregulated circRNAs in the microarray analysis.
| A, Upregulated circRNAs | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| CircRNA ID | Fold change | CircRNA type | Chromosome location | Gene symbol | MRE1 | MRE2 | MRE3 | MRE4 | MRE5 |
| hsa_circ_102028 | 33.3140688 | Exonic | chr17(+):30267304-30274704 | SUZ12 | miR-19a-3p | miR-301a-3p | miR-516b-5p | miR-148b-5p | miR-335-3p |
| hsa_circ_104052 | 33.1883263 | Exonic | chr6(+):4891946-4892613 | CDYL | miR-892a | miR-378a-5p | miR-887-5p | miR-22-5p | miR-328-3p |
| hsa_circ_102027 | 31.6109823 | Exonic | chr17(+):30267304-30267505 | SUZ12 | miR-19a-3p | miR-301a-3p | miR-516b-5p | miR-148b-5p | miR-10b-3p |
| hsa_circ_104220 | 30.3375868 | Exonic | chr6(+):150092297-150094305 | PCMT1 | miR-338-3p | miR-19a-5p | miR-450a-1-3p | miR-769-5p | miR-153-3p |
| hsa_circ_102995 | 19.1417523 | Exonic | chr20(+):6011930-6012726 | CRLS1 | miR-424-5p | miR-16-5p | miR-195-5p | miR-15b-5p | miR-323a-3p |
| hsa_circ_101319 | 16.16546 | Exonic | chr14(−):23378691-23380612 | RBM23 | miR-138-5p | miR-135a-3p | miR-338-3p | miR-135b-5p | miR-135a-5p |
| hsa_circ_029937 | 15.9942885 | Exonic | chr13(−):33095509-33111164 | N4BP2L2 | miR-26b-3p | miR-135a-5p | miR-7107-3p | miR-135b-5p | miR-6780a-3p |
| hsa_circ_102611 | 15.889754 | Exonic | chr2(−):9083315-9098771 | MBOAT2 | miR-130b-3p | miR-130a-3p | miR-513a-3p | miR-136-5p | miR-301a-3p |
| hsa_circ_068655 | 15.4343859 | Exonic | chr3(−):196129822-196134264 | UBXN7 | miR-3140-3p | miR-4539 | miR-3660 | miR-4260 | miR-3118 |
| hsa_circ_102439 | 14.4829329 | Exonic | chr19(−):10286215-10288043 | DNMT1 | miR-876-3p | miR-450b-5p | miR-1468-5p | miR-1468-3p | miR-627-3p |
| B, Downregulated circRNAs | |||||||||
| CircRNA ID | Fold change | CircRNA type | Chromosome location | Gene symbol | MRE1 | MRE2 | MRE3 | MRE4 | MRE5 |
| hsa_circ_400019 | 6.7133031 | Intronic | chr11(+):8706439-8707219 | RPL27A | miR-1298-3p | miR-612 | miR-204-3p | miR-20b-3p | miR-135a-3p |
| hsa_circ_404819 | 6.6407844 | Intronic | chr10(−):126323958-126326678 | FAM53B | miR-7113-3p | miR-6862-5p | miR-1225-3p | miR-1909-3p | miR-4253 |
| hsa_circ_011474 | 6.5229429 | Exonic | chr1(+):33760537-33766320 | ZNF362 | miR-637 | miR-6756-5p | miR-6734-3p | miR-6799-5p | miR-328-5p |
| hsa_circ_001131 | 6.173524 | Intronic | chr2(−):98276327-98276412 | ACTR1B | miR-671-3p | miR-877-3p | miR-412-3p | miR-361-3p | miR-614 |
| hsa_circ_405788 | 5.3867752 | Exonic | chr19(−):44128265-44131942 | CADM4 | miR-4632-3p | miR-941 | miR-152-5p | hsa-miR-1278 | miR-6895-3p |
| hsa_circ_001359 | 5.1849713 | Intronic | chr1(−):169663839-169664181 | SELL | miR-130b-5p | miR-182-5p | miR-888-3p | miR-451a | miR-877-3p |
| hsa_circ_102645 | 4.6819455 | Exonic | chr2(−):26453059-26457223 | HADHA | miR-26a-1-3p | miR-26a-2-3p | miR-361-3p | miR-214-3p | miR-1323 |
| hsa_circ_084106 | 4.6683381 | Exonic | chr8(+):42202470-42210086 | POLB | miR-145-5p | miR-3134 | miR-5195-3p | miR-548j-3p | miR-6736-3p |
| hsa_circ_405240 | 4.5170015 | Exonic | chr14(−):60193636-60194386 | RTN1 | miR-423-3p | miR-4685-5p | miR-762 | miR-3196 | miR-6837-5p |
| hsa_circ_405614 | 4.1808481 | Exonic | chr17(−):60059558-60072726 | MED13 | miR-6782-5p | miR-3936 | miR-4760-3p | miR-325 | miR-337-3p |
Note: Fold change equals the absolute ratio (no log scale) of the normalized intensities between the patients with IMN and the controls. Hsa, Homo sapiens; circ/circRNA, circular RNA; MRE: microRNA response element.
Figure 2.
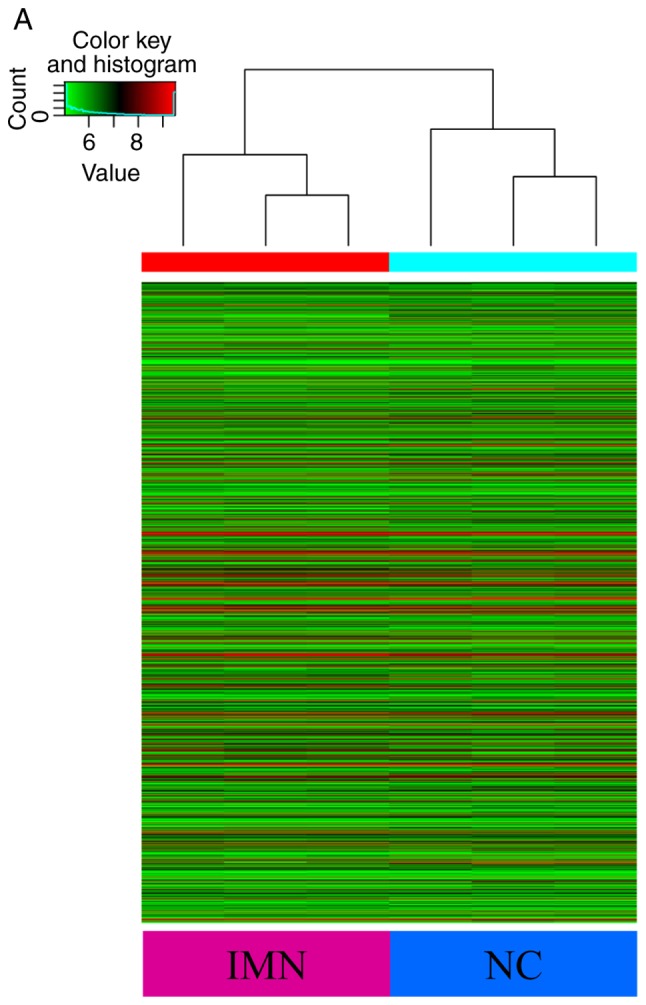
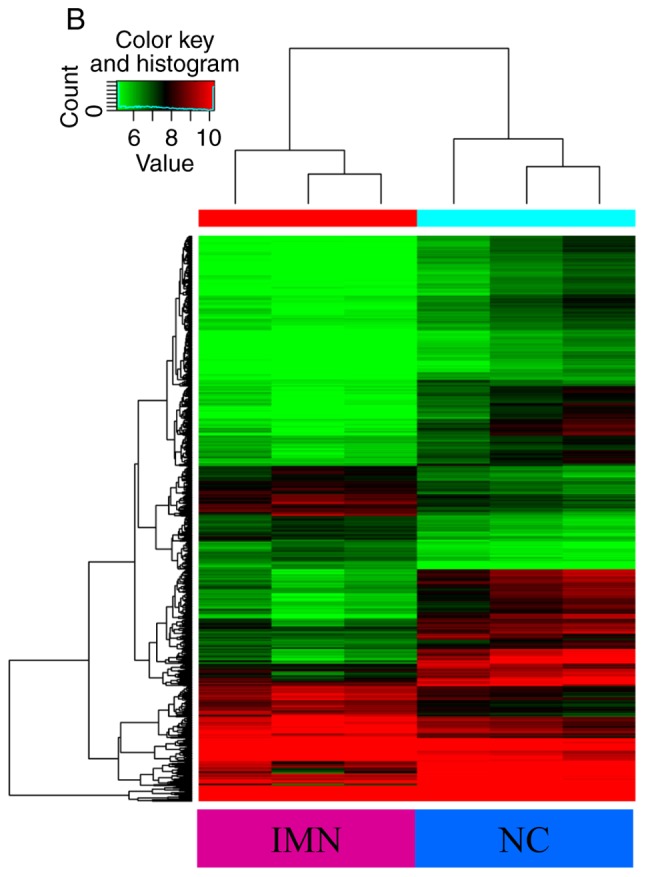
Heat map and hierarchical clustering showing the expression values of all circRNAs and dysregulated circRNAs. Each column represents a sample and each row represents a circRNA. The circRNAs that are upregulated are shown in red while those that are downregulated are shown in green. In total, three different samples from the IMN and NC groups are shown. (A) Heatmap displaying all detected circRNAs. (B) Hierarchical cluster analysis of the differently expressed circRNAs with statistical significance (IMN group vs. NC group, fold change >2.0, P<0.05). IMN, idiopathic membranous nephropathy group; NC, normal control.
Construction of the circRNA/miRNA interaction network
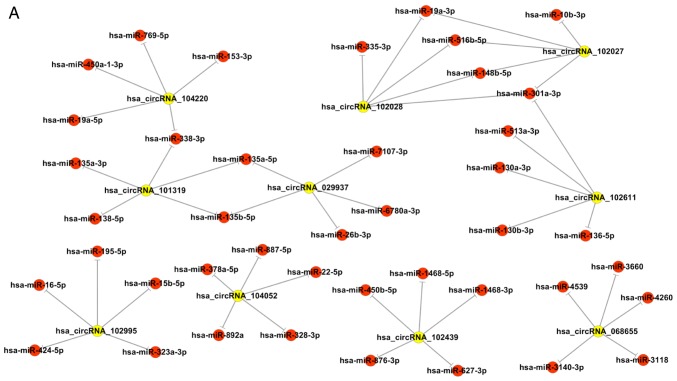
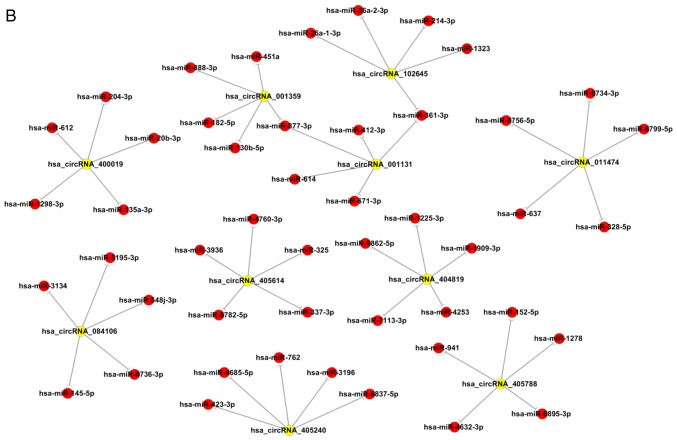
To further determine the biological function and potential regulators of circRNAs, software made by Arraystar to predict the miRNA targets of differentially expressed circRNAs based on bioinformatics analysis by TargetScan and miRanda databases was used. According to the analysis of complementary miRNA matching sequences and the microarray results, circRNA-miRNA interaction networks were constructed using Cytoscape. The interaction maps, including the top 10 upregulated and downregulated circRNAs and their corresponding miRNAs, are shown in Fig. 3. The five microRNA response elements with good mirSVR scores (a lower score indicates a more stable combination of miRNA and mRNA) for the top 10 dysregulated circRNAs are shown in Table II. Additionally, the host genes and genomic locations of these circRNAs are also provided for further functional prediction.
Figure 3.
circRNA/miR network analysis. Extended network based (A) on the top 10 upregulated circRNAs and (B) on the top 10 downregulated circRNAs. circRNA, circular RNA; miR, microRNA; hsa, Homo sapiens.
GO and KEGG pathway analysis for the biological function of genes
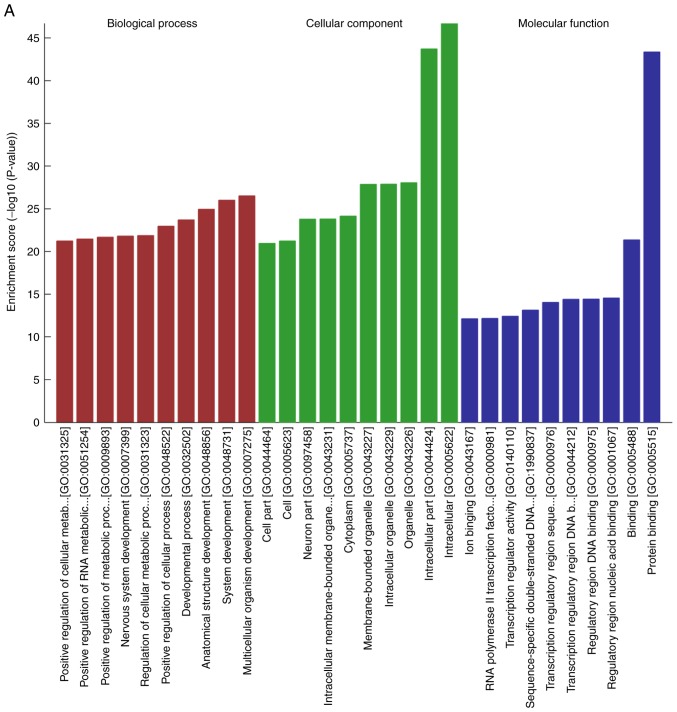
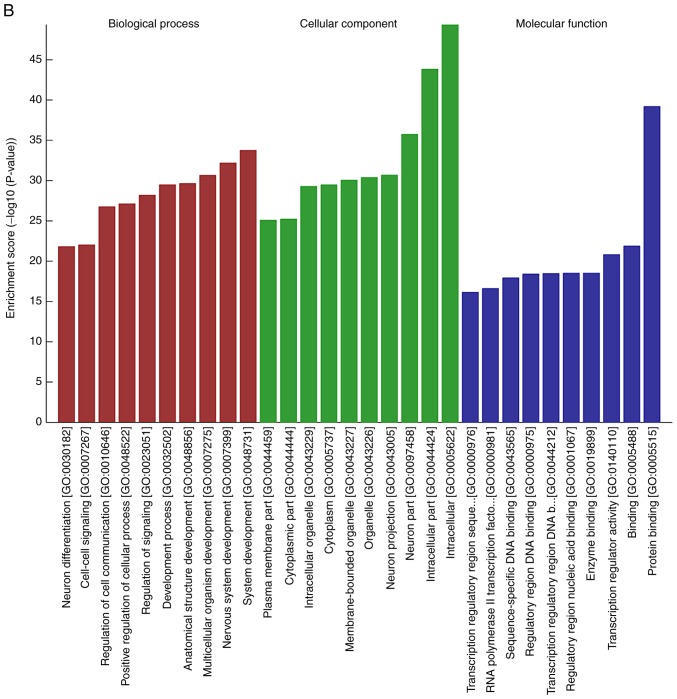
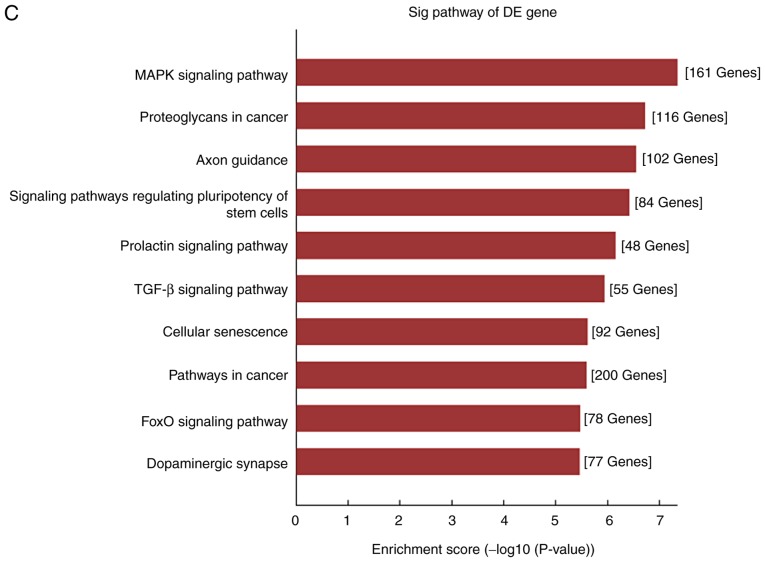
To explore the potential regulatory roles of circRNAs and related molecules in IMN, GO analysis of the predicted targets from the dysregulated circRNAs was performed. It was found that the BPs of the genes corresponding to the top 10 upregulated circRNAs were involved in the ‘positive regulation of the cellular metabolic process’ and ‘positive regulation of cellular process’, ‘positive regulation of the RNA metabolic process’ and ‘system development’. Their MFs were enriched in ‘regulatory region nucleic acid binding’, ‘transcription regulator activity’, ‘RNA polymerase II transcription factor activity’ and ‘protein binding’. Analysis of the downregulated circRNAs showed that the BPs included ‘system development’ and ‘positive regulation of cellular process’, whereas the MFs were consistent with the upregulated genes, with the exception of ‘enzyme binding’ (Fig. 4A and B). Following GO annotation, KEGG pathway analysis was used to identify the intermolecular signaling networks, which was informative for elucidating and interpreting the potential relationships between circRNAs and IMN pathogenesis. It was found that the mitogen-activated protein kinase (MAPK) signaling pathway, the transforming growth factor (TGF)-β signaling pathway and cellular senescence were ranked highly in the upregulated circRNA group, whereas the MAPK, Ras and Rap1 signaling pathways were ranked highly, with high enrichment scores, in the downregulated group (Fig. 4C and D).
Figure 4.
GO and KEGG analysis of the genes targeted by DE circRNAs. (A) GO annotations for the top 10 upregulated circRNAs and (B) for top 10 downregulated circRNAs. GO and KEGG analysis of the genes targeted by DE circRNAs. KEGG pathway enrichment analysis of (C) upregulated and (D) downregulated mRNAs with top 10 enrichment scores. circRNA, circular RNA; DE, differentially expressed; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Validation of the different expression levels of using RT-qPCR
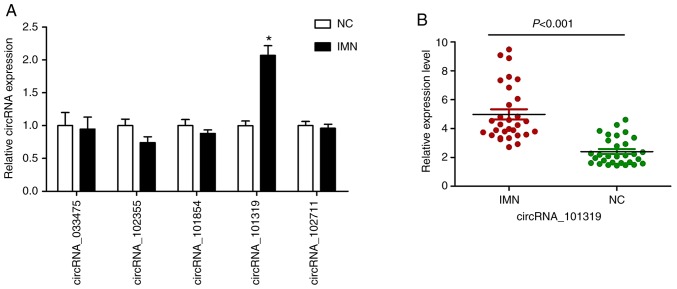
After the bioinformatics analysis on the circRNA/miRNA network and determination of the potential relationships between these molecules and the mechanisms underlying IMN, five candidate circRNAs were selected for further validation using RT-qPCR. At this stage, the sample size was increased by collecting the peripheral blood from 30 subjects in each group. The results showed no marked difference between the IMN and NC groups in terms of the expression of circ_033475, circ_102355, circ_101854 or circ_102711, whereas the expression of circ_101319 was significantly higher in the IMN group (Fig. 5A). This trend was consistent among the microarray and RT-qPCR analysis. The expression of circ_101319 in the IMN and NC groups is shown in Fig. 5B. These results suggested that there was a larger degree of variation in the expression of circ_101319 in the IMN group compared with the NC group.
Figure 5.
RT-qPCR validation of five differentially expressed circRNAs. (A) Expression levels of the following circRNAs were verified by RT-qPCR: circ_033475, circ_102355, circ_101854, circ_101319, and circ_102711. (B) Expression levels of circ_101319 in the IMN and NC groups, showing the concrete distribution of circ_101319 expression in the two groups. Expression was normalized to the expression of β-actin. NC group=30, IMN group=30. RT-qPCR was performed at least three times. Data are presented as the mean ± SEM. *P<0.05 vs. NC. RT-qPCR, reverse transcription-quantitative PCR; circRNA/circ, circular RNA; IMN, idiopathic membranous nephropathy group; NC, normal control.
Diagnostic value of circ_101319 in peripheral blood for IMN
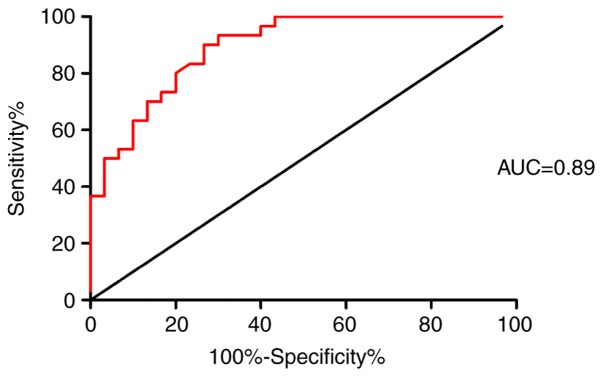
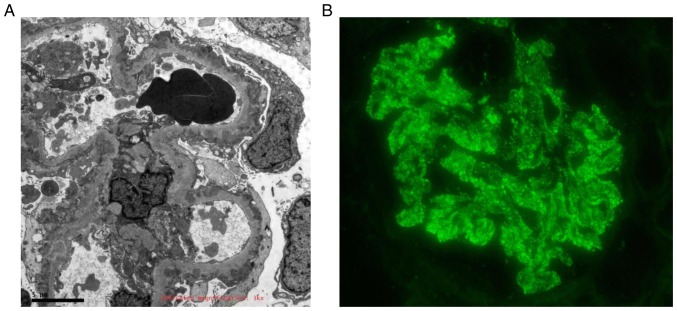
It was found that the expression of circRNAs in peripheral blood was different between the IMN and the NC groups via microarray analysis, and RT-qPCR analysis supported the upregulation of circ_101319 in the IMN group. Due to the characteristics of circRNAs, including resistance to exonucleases and degradation, and stable expression, circRNAs may have an important role to play in molecular diagnostics. ROC curves were used to explore the association between circ_101319 expression and IMN. When circ_101319 expression was applied for the purposes of this analysis, the area under the ROC curve (AUROC) reached 0.89 (95% CI; 0.815–0.971), suggesting that circ_101319 may be used as a predictor for IMN. Moreover, the results showed that the sensitivity and specificity of circ_101319 were 93.33 and 70.00%, respectively (Fig. 6), suggesting that the upregulated expression of circ_101319 was associated with the diagnosis of IMN, which is supported by electron microscopy and immunofluorescence images from renal biopsies (Fig. 7). The glomerular basement membrane (GBM) was thickened and the electron-dense deposits were observed on the epithelial side of the GBM by TEM. Immunofluorescence results showed that IgG4 was granular positive and distributed along the glomerular capillary loops. Negative staining results for IgM and IgA was observed and there was no deposition of complement components C4 or C1q in patients with IMN.
Figure 6.
ROC curve analysis shows an association between circ_101319 expression and idiopathic membranous nephropathy diagnosis. A large AUROC for circ_101319 indicated its potential as a biomarker (AUROC=0.89). The sensitivity was 93.33% and the specificity was 70.00%. AUROC, area under ROC curve; ROC, receiver-operating characteristic.
Figure 7.
Electron microscope and immunofluorescence images from the renal biopsies of patients with idiopathic membranous nephropathy. (A) Electron microscopy image showing that the GBM was thickened and the electron-dense deposits were on the epithelial side of GBM. Magnification, ×3,000. (B) Immunofluorescence results showing that IgG4 was granular positive and distributed along the glomerular capillary loops. Magnification, ×200. Staining for IgM and IgA was negative and there was no deposition of complement component C4 or C1q. GBM, glomerular basement membrane; Ig, immunoglobulin.
Prediction and annotation of circ_101319 using a circRNA-miRNA-mRNA network
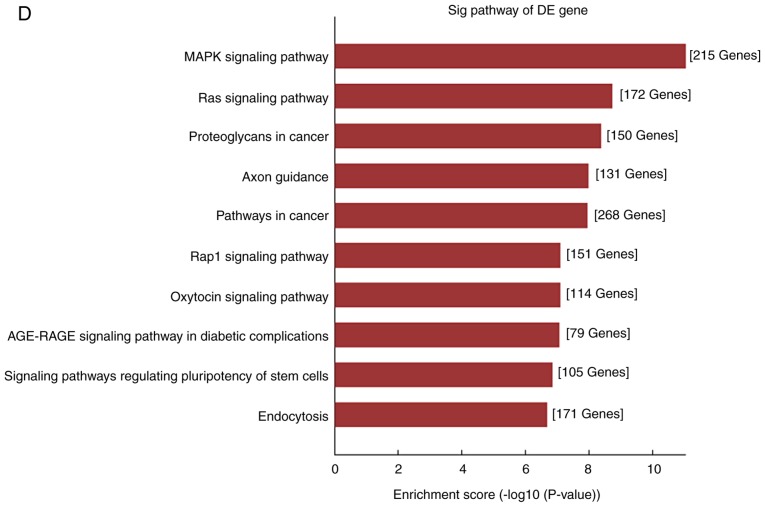
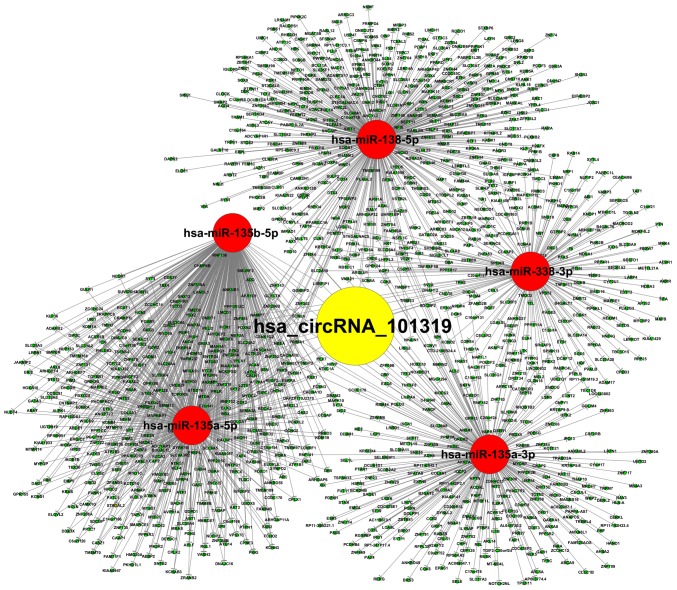
After clinical observation, the target miRNAs for circ_101319 were identified and subsequently ranked based on their mirSVR scores. The five highest ranking candidate miRNA binding targets were used for further analysis of underlying molecular mechanisms through specific base pairing. Combined with previous research concerning circRNA-mediated regulation, it was hypothesized that circ_101319 may also regulate a circRNA-miRNA-mRNA network through a competitive endogenous RNA mechanism (31–33). The potential interactions were predicted using TargetScan and miRanda. Based on these analysis tools, a map of five miRNAs and their predicted target mRNAs was constructed to demonstrate the circRNA-miRNA-mRNA network of circ_101319, which included miR-135a, −5p and −3p, miR-138-5p and miR-338-3p (Fig. 8). The co-expression network of circ_101319 indicated that the miR-135 family exhibited the largest interaction network. To gain further insight into the potential functions of circ_101319 in renal disease, GO annotation and KEGG pathway analysis were applied to the results. It was found that circ_101319 was involved in ‘cellular senescence’, ‘renal cell carcinoma’ and ‘renin secretion’ (Fig. 9).
Figure 8.
circRNA-miRNA-mRNA interaction network of circ_101319 constructed based on the prediction of sequence matching. The association between the top five targeted miRNAs of circ_101319 with the best miRNA support vector regression scores and their corresponding mRNAs. circ/circRNA, circular RNA/miR; miRNA, microRNA.
Figure 9.
Kyoto Encyclopedia of Genes and Genomes pathway analysis based on the circular RNA_101319-microRNA-mRNA network. The top 10 enriched pathways of the DE genes and enrichment scores are presented. DE, differentially expressed.
Discussion
IMN is one of the main causes of adult nephrotic syndrome. As the pathogenesis of the disease has not been elucidated, effective and safe therapies for IMN are lacking in modern medicine. The use of expensive and potentially toxic drugs has limited the treatment of this disease (34).
ncRNAs are not translated into proteins, and include ribosomal RNAs, transfer RNAs, miRNAs, long noncoding (lnc)RNAs and circRNAs (35). The aforementioned molecules are transcribed from the genome but are not translated into proteins, which is regarded as a common characteristic. These ncRNAs exercise biological functions at the RNA level, including in the biosynthesis of ribosomes, and the regulation of gene transcription and protein translation (36). Research has shown that ncRNAs are closely related with podocyte injury and the development of renal disease (37,38). With its molecular characteristics, it is possible that ncRNAs can be used as novel biomarkers for the diagnosis and screening of diseases (39–41). For example, the expression of miR-378-3p was found to be increased significantly in renal biopsies from patients with MN and was shown to cause glomerular dysfunction in animal experiments in vivo (42). Expression of the lncRNA X-inactive specific transcript in urine is associated with the pathogenesis of MN, and may be used for diagnosis (43). The expression of multiple ncRNAs is altered in tissues and fluids from patients and animals with nephropathy, and may be associated with glomerular dysfunction. Therefore, further studies are required to elucidate the mechanism and improve treatments.
As an endogenous cyclic ncRNA produced by nonclassical splicing, circRNAs were considered to be created by splicing errors, or to be secondary or by-products of posttranscriptional regulation since their discovery in the 1970s. With the rapid development of high-throughput RNA sequencing and continuous improvements to bioinformatics analysis, more and more circRNAs are being identified. circRNAs have been shown to possess important biological functions and have been associated with the development of various diseases (44–46). A previous study showed that circHLA-C is significantly elevated in the renal tissue of patients with lupus nephritis, and that it has a positive correlation with the urine protein, serum creatinine and renal activity indexes. Bioinformatics analysis and expression profiles show that circHLA-C has a binding site for miR-150 (47). Therefore, circRNAs may be involved in the pathogenesis of kidney disease. However, to the best of our knowledge, no research has been conducted into the interaction between circRNA and IMN.
In the present study, a diagnosis and case screening of IMN through renal biopsies and immunofluorescence analysis, combined with clinical manifestations, were first performed and peripheral blood samples were collected from patients. The expression of circRNAs in 3 patients from the IMN and the NC groups was determined via microarray analysis. To further understand the potential biological functions of the circRNAs, the predicted miRNA targets of the dysregulated circRNAs and target mRNAs were analyzed. GO and KEGG analyses were used to enrich the target genes and identify circRNA/miRNA interactions for the circRNAs with differential expression. GO annotation analysis demonstrated the BPs of the predicted mRNAs, with a primary focus on the ‘positive regulation of the cellular metabolic process’, ‘positive regulation of the RNA metabolic process’ and ‘system development’. The MFs of the mRNAs were concentrated on ‘regulatory region DNA binding’ and ‘regulatory region nucleic acid binding’, ‘transcription regulator activity’, ‘RNA polymerase II transcription factor activity’ and ‘protein binding’. After analyzing the predicted target groups of the top 10 upregulated and downregulated circRNAs, the MAPK, Ras and TGF-β signaling pathways, and cellular senescence were revealed to have high enrichment scores in the KEGG pathway analysis. Several TGF-β-related factors were found to be upregulated in the podocytes of patients with IMN. TGF-β was able to activate ERK and p38 MAPK in podocytes (48). Activation of p38 MAPK is required to induce apoptosis by TGF-β in podocytes (49). Considering the research previously carried out on MN pathogenesis and its underlying mechanisms, the aforementioned signaling pathways may be closely related to the occurrence and development of IMN.
According to the results of the microarray and bioinformatics analyses carried out in the present study combined with previous research (50,51), the following five candidate circRNAs were selected: circ_033475, circ_102355, circ_101854, circ_101319 and circ_102711. The gene regions from which these circRNAs are transcribed are microtubule affinity regulated kinase 3, PI3K catalytic subunit type 3, nuclear factor of activated T-cell (NFAT5), RNA binding motif protein 23 (RBM23) and protein phosphatase, Mg2+/Mn2+ dependent 1B (PPM1B). Regarded as an important target for therapeutic agents in MN, including cyclosporine and tacrolimus, NFAT is considered to play a key role in the development, maturation and function of the immune system (50). The PI3K pathway has been reported to be involved in podocyte proliferation and is synergistically activated to mediate the anti-apoptotic machinery with MAPK/ERK signaling (51). PPM1B is predicted to have connections with multiple pathways, including MAPK and cytokine signaling in the immune system, using KEGG and Reactome databases (52). The expression of the circRNAs was verified by RT-qPCR in 30 clinical samples from the IMN and NC groups. It was found that the expression of circRNA_101319, which originates from the RBM23 exon region, was significantly upregulated in the peripheral blood of patients with IMN.
The potential diagnostic value of circ_101319 in IMN was evaluated by using 30 clinical specimens. The results of the ROC curve based on the disease diagnosis showed that circ_101319 had a large AUROC with a good sensitivity and intensity, which indicated it may have a suitable prediction accuracy. It was observed that the expression of circ_101319 was consistent with the pathological indexes of patients with IMN. Combined with literature investigation and KEGG pathway analysis, it was found that a number of the predicted targets of circRNAs had important biological functions and relevance to renal disease. Therefore, it is proposed that circ_101319 may be a novel biomarker for IMN screening and treatment. Further studies investigating the mechanism of IMN and with larger sample sizes are required to support these findings.
A previous report demonstrated that circRNAs play an important role in fine-tuning miRNA-mediated regulation of gene expression by sequestering miRNAs. Their interactions with disease-associated miRNAs indicated that circular RNAs are important in disease development (53). The circ_101319 circRNA-miRNA-mRNA network identified target genes that are involved in various important signaling pathways, which may be related to podocyte injury and IMN mechanism. There are binding sites with high scores between circ_101319 and multiple molecules of the miR-135 family, including miR-135a and miR-135b. A previous study suggested that miR-135a and miR-135b can promote podocyte injury by activating the Wnt/β-catenin signaling pathway, a pathway of which is important in the production or repair of injury in kidney and renal disease progression (54). A previous study found that the activation of β-catenin can damage the integrity of podocytes and cause proteinuria in healthy rats (55). Targeted inhibition of the Wnt/β-catenin signaling pathway improved podocyte injury and proteinuria in model animals (56). Further research is required to elucidate the mechanism.
In summary, the expression profiles of circRNAs in peripheral blood were investigated in patients with IMN and healthy controls. The present study found that the expression of circ_101319 is significantly upregulated in patients with IMN, which may be related with the pathogenesis of the disease. Combined with ROC curve analysis and the clinicopathological images and parameters, it was revealed that circRNAs may be suitable for the diagnosis of IMN. The potential targets of circRNA were determined to be involved in potential disease mechanism using bioinformatics analysis. The present study provided novel ideas for the development of drug targets and is useful for further understanding the disease mechanisms. Future research should focus on the potential regulatory functions of circRNAs in the pathogenesis of IMN and further mechanistic studies will provide novel insights for targeted gene therapy.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- circRNAs
circular RNAs
- IMN
idiopathic membranous nephropathy
- NC
normal control
- circRNA_101319
circ_101319
- RT-qPCR
reverse transcription-quantitative PCR
- miRNAs
microRNAs
- ROC
receiver-operating characteristic
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- AUROC
area under the ROC curve
- mirSVR
miRNA support vector regression
- MAPK
mitogen-activated protein kinase
- TGF-β
transforming growth factor-β
- NFAT
nuclear factor of activated T-cell
Funding
The present study was supported by the Guangxi Zhuang Autonomous Region Science and Technology Key Project of China (grant no. AB16380185).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XJ, YC and XC participated in the design of the study. KY, XJ and ZY aided in collecting clinical samples. XJ, BD, KY, YH and ZY were responsible for completing experimental work. DY and XJ performed the statistical analysis. BD, XC and ZY edited the manuscript. XJ was responsible for revising the final version. All authors have read and approved the content of the manuscript.
Ethics approval and consent to participate
The protocol for this research was approved by the Ethics Committee of the People's Hospital of Guangxi Zhuang Autonomous Region and conducted in accordance with the Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ronco P, Debiec H. Pathophysiological advances in membranous nephropathy: Time for a shift in patient's care. Lancet. 2015;385:1983–1992. doi: 10.1016/S0140-6736(15)60731-0. [DOI] [PubMed] [Google Scholar]
- 2.Ronco P, Debiec H. Pathogenesis of membranous nephropathy: Recent advances and future challenges. Nat Rev Nephrol. 2012;8:203–213. doi: 10.1038/nrneph.2012.35. [DOI] [PubMed] [Google Scholar]
- 3.Herrmann SM, Sethi S, Fervenza FC. Membranous nephropathy: The start of a paradigm shift. Curr Opin Nephrol Hypertens. 2012;21:203–210. doi: 10.1097/MNH.0b013e32835026ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beck LH, Jr, Salant DJ. Membranous nephropathy: Recent travels and new roads ahead. Kidney Int. 2010;77:765–770. doi: 10.1038/ki.2010.34. [DOI] [PubMed] [Google Scholar]
- 5.Hofstra JM, Fervenza FC, Wetzels JF. Treatment of idiopathic membranous nephropathy. Nat Rev Nephrol. 2013;9:443–458. doi: 10.1038/nrneph.2013.125. [DOI] [PubMed] [Google Scholar]
- 6.Fervenza FC, Sethi S, Specks U. Idiopathic membranous nephropathy: Diagnosis and treatment. Clin J Am Soc Nephrol. 2008;3:905–919. doi: 10.2215/CJN.04321007. [DOI] [PubMed] [Google Scholar]
- 7.Fujino T, Hasebe N. Alteration of histone H3K4 methylation in glomerular podocytes associated with proteinuria in patients with membranous nephropathy. BMC Nephrol. 2016;17:179. doi: 10.1186/s12882-016-0390-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sha WG, Shen L, Zhou L, Xu DY, Lu GY. Down-regulation of miR-186 contributes to podocytes apoptosis in membranous nephropathy. Biomed Pharmacother. 2015;75:179–184. doi: 10.1016/j.biopha.2015.07.021. [DOI] [PubMed] [Google Scholar]
- 9.Cowley SM, Iritani BM, Mendrysa SM, Xu T, Cheng PF, Yada J, Liggitt HD, Eisenman RN. The mSin3A chromatin-modifying complex is essential for embryogenesis and T-cell development. Mol Cell Biol. 2005;25:6990–7004. doi: 10.1128/MCB.25.16.6990-7004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kota SK, Kota SB. Noncoding RNA and epigenetic gene regulation in renal diseases. Drug Discov Today. 2017;22:1112–1122. doi: 10.1016/j.drudis.2017.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang S, Yang B, Chen BJ, Bliim N, Ueberham U, Arendt T, Janitz M. The emerging role of circular RNAs in transcriptome regulation. Genomics. 2017;109:401–407. doi: 10.1016/j.ygeno.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 14.Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, Parast MM, Murry CE, Laurent LC, Salzman J. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015;16:126. doi: 10.1186/s13059-015-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 16.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 17.Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y, Li C, Tan C, Liu X. Circular RNAs: A new frontier in the study of human diseases. J Med Genet. 2016;53:359–365. doi: 10.1136/jmedgenet-2016-103758. [DOI] [PubMed] [Google Scholar]
- 19.Shen T, Han M, Wei G, Ni T. An intriguing RNA species-perspectives of circularized RNA. Protein Cell. 2015;6:871–880. doi: 10.1007/s13238-015-0202-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, Dinneny JR, Brown PO, Salzman J. Circular RNA is expressed across the eukaryotic tree of life. PLoS One. 2014;9:e90859. doi: 10.1371/journal.pone.0090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. ELife. 2015:4. doi: 10.7554/eLife.05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong N, Wang X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43((Database Issue)):D146–D152. doi: 10.1093/nar/gku1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The Gene Ontology Consortium, corp-author. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47:D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019;47:D590–D595. doi: 10.1093/nar/gky962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 28.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han D, Li J, Wang H, Su X, Hou J, Gu Y, Qian C, Lin Y, Liu X, Huang M, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–1164. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 32.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu J, Yang M, Zhou B, Luo J, Zhang Z, Zhang W, Yan Z. CircRNA-104718 acts as competing endogenous RNA and promotes hepatocellular carcinoma progression through microRNA-218-5p/TXNDC5 signaling pathway. Clin Sci (Lond) 2019;133:1487–1503. doi: 10.1042/CS20190394. [DOI] [PubMed] [Google Scholar]
- 34.Glassock RJ. The pathogenesis of idiopathic membranous nephropathy: A 50-year odyssey. Am J Kidney Dis. 2010;56:157–167. doi: 10.1053/j.ajkd.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 35.Eddy SR. Non-coding RNA genes and the modern RNA world. Nat Rev Genet. 2001;2:919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- 36.Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annu Rev Biophys. 2013;42:217–239. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu M, Wang R, Li X, Fan M, Lin J, Zhen J, Chen L, Lv Z. LncRNA MALAT1 is dysregulated in diabetic nephropathy and involved in high glucose-induced podocyte injury via its interplay with β-catenin. J Cell Mol Med. 2017;21:2732–2747. doi: 10.1111/jcmm.13189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elsenreich A, Langer S, Herlan L, Kreutz R. Regulation of podoplanin expression by microRNA-29b associates with its antiapoptotic effect in angiotensin II-induced injury of human podocytes. J Hypertens. 2016;34:323–331. doi: 10.1097/HJH.0000000000000799. [DOI] [PubMed] [Google Scholar]
- 39.Noureddine L, Hajarnis S, Patel V. MicroRNAs and polycystic kidney disease. Drug Discov Today Dis Models. 2013;10:e137–e1743. doi: 10.1016/j.ddmod.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Q, Chung AC, Huang XR, Dong Y, Yu X, Lan HY. Identification of novel long noncoding RNAs associated with TGF-β/Smad3-mediated renal inflammation and fibrosis by RNA sequencing. Am J Pathol. 2014;184:409–417. doi: 10.1016/j.ajpath.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 41.Kato M, Wang M, Chen Z, Bhatt K, Oh HJ, Lanting L, Deshpande S, Jia Y, Lai JY, O'Connor CL, et al. An endoplasmic reticulum stress-regulated lncRNA hosting a microRNA megacluster induces early features of diabetic nephropathy. Nat Commun. 2016;7:12864. doi: 10.1038/ncomms12864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Müller-Deile J, Dannenberg J, Schroder P, Lin MN, Miner JH, Chen R, Bräsen JH, Thum T, Nyström J, Staggs LB, et al. Podocytes regulate the glomerular basement membrane protein nephronectin by means of miR-378a-3p in glomerular diseases. Kidney Int. 2017;92:836–849. doi: 10.1016/j.kint.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang YS, Hsieh HY, Shih HM, Sytwu HK, Wu CC. Urinary Xist is a potential biomarker for membranous nephropathy. Biochem Biophys Res Commun. 2014;452:415–521. doi: 10.1016/j.bbrc.2014.08.077. [DOI] [PubMed] [Google Scholar]
- 44.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 45.You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18:603–610. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6:e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luan J, Jiao C, Kong W, Fu J, Qu W, Chen Y, Zhu X, Zeng Y, Guo G, Qi H, et al. CircHLA-C plays an important role in lupus nephritis by sponging miR-150. Mol Ther Nucleic Acids. 2018;10:245–253. doi: 10.1016/j.omtn.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng R, Deng Y, Chen Y, Fan J, Zhang M, Zhong Y, Zhu R, Wang L. Astragaloside IV attenuates complement membranous attack complex induced podocyte injury through the MAPK pathway. Phytother Res. 2012;26:892–898. doi: 10.1002/ptr.3656. [DOI] [PubMed] [Google Scholar]
- 49.Shankland SJ, Pippin J, Pichler RH, Gordon KL, Friedman S, Gold LI, Johnson RJ, Couser WG. Differential expression of transforming growth factor-beta isoforms and receptors in experimental membranous nephropathy. Kidney Int. 1996;50:116–124. doi: 10.1038/ki.1996.294. [DOI] [PubMed] [Google Scholar]
- 50.Kiani A, Rao A, Aramburu J. Manipulating immune responses with immunosuppressive agents that target NFAT. Immunity. 2000;12:359–372. doi: 10.1016/S1074-7613(00)80188-0. [DOI] [PubMed] [Google Scholar]
- 51.Davies CC, Mason J, Wakelam MJ, Young LS, Eliopoulos AG. Inhibition of phosphatidylinositol 3-kinase- and ERK MAPK-regulated protein synthesis reveals the pro-apoptotic properties of CD40 ligation in carcinoma cells. J Biol Chem. 2004;279:1010–1019. doi: 10.1074/jbc.M303820200. [DOI] [PubMed] [Google Scholar]
- 52.Geer LY, Marchler-Bauer A, Geer RC, Han L, He J, He S, Liu C, Shi W, Bryant SH. The NCBI BioSystems database. Nucleic Acids Res. 2010;38((Database Issue)):D492–D496. doi: 10.1093/nar/gkp858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ghosal S, Das S, Sen R, Basak P, Chakrabarti J. Circ2Traits: A comprehensive database for circular RNA potentially associated with disease and traits. Front Genet. 2013;4:283. doi: 10.3389/fgene.2013.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang X, Wang X, Nie F, Liu T, Yu X, Wang H, Li Q, Peng R, Mao Z, Zhou Q, Li G. miR-135 family members mediate podocyte injury through the activation of Wnt/β-catenin signaling. Int J Mol Med. 2015;36:669–677. doi: 10.3892/ijmm.2015.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou L, Liu Y. Wnt/β-catenin signalling and podocyte dysfunction in proteinuric kidney disease. Nat Rev Nephrol. 2015;11:535–545. doi: 10.1038/nrneph.2015.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He W, Kang YS, Dai C, Liu Y. Blockade of Wnt/β-catenin signaling by paricalcitol ameliorates proteinuria and kidney injury. J Am Soc Nephrol. 2011;22:90–103. doi: 10.1681/ASN.2009121236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.